The plant-specific F-box protein MEIOTIC F-BOX plays critical roles in male meiotic DNA double-strand break repair and is required for male meiotic progression in rice.
Abstract
F-box proteins constitute a large superfamily in plants and play important roles in controlling many biological processes, but the roles of F-box proteins in male meiosis in plants remain unclear. Here, we identify the rice (Oryza sativa) F-box gene MEIOTIC F-BOX (MOF), which is essential for male meiotic progression. MOF belongs to the FBX subfamily and is predominantly active during leptotene to pachytene of prophase I. mof meiocytes display disrupted telomere bouquet formation, impaired pairing and synapsis of homologous chromosomes, and arrested meiocytes at late prophase I, followed by apoptosis. Although normal, programmed double-stranded DNA breaks (DSBs) form in mof mutants, foci of the phosphorylated histone variant γH2AX, a marker for DSBs, persist in the mutant, indicating that many of the DSBs remained unrepaired. The recruitment of Completion of meiosis I (COM1) and Radiation sensitive51C (RAD51C) to DSBs is severely compromised in mutant meiocytes, indicating that MOF is crucial for DSB end-processing and repair. Further analyses showed that MOF could physically interact with the rice SKP1-like Protein1 (OSK1), indicating that MOF functions as a component of the SCF E3 ligase to regulate meiotic progression in rice. Thus, this study reveals the essential role of an F-box protein in plant meiosis and provides helpful information for elucidating the roles of the ubiquitin proteasome system in plant meiotic progression.
INTRODUCTION
Efficient chromosome segregation in meiosis depends on a highly coordinated series of events that occur during prophase I, involving homologous chromosome pairing, synapsis, and recombination. The cytological substages of meiotic prophase I (leptotene, zygotene, pachytene, diplotene, and diakinesis) are characterized by chromosome condensation, alignment, and chiasma formation (the cytological manifestation of genetic crossovers) (Zickler and Kleckner, 1998). Meiotic recombination is initiated by programmed DNA double-strand breaks (DSBs) catalyzed by the evolutionarily conserved type-II topoisomerase-like enzyme sporulation protein11 (SPO11) (Bergerat et al., 1997; Keeney et al., 1997). The DSB ends are resected by the meiotic recombination 11-radiation sensitive50 (RAD50)-Nijmegen breakage syndrome 1/X-ray-sensitive2 complex, together with completion of meiosis I (COM1)/sporulation in the absence of SPO11 (SAE2) to produce a long 3′-OH single-stranded DNA molecule. The nascent single-stranded DNA is bound by replication protein A (RPA) (Sakaguchi et al., 2009; Li et al., 2013; Aklilu et al., 2014). RAD51 and disrupted meiotic cDNA (DMC1) are then recruited to promote single-strand invasion of homologous regions of DNA. DMC1/RAD51 mediated stable strand-invasion produces a D-loop recombination intermediate that can then be stabilized by the ZMM proteins (Osman et al., 2011). The ZMM proteins cooperate to form double Holliday junctions, which are then either resolved as crossovers or non-crossovers (Bishop and Zickler, 2004; Osman et al., 2011).
DSBs are highly cytotoxic and lead to mutagenesis and genome disintegration if left unrepaired or repaired aberrantly. DSB formation, processing, and repair require tight regulation to avoid genome instability. Mutants of meiotic proteins involved in DSB processing and repair generally display severe chromosomal fragmentation and meiotic defects, including deficient meiotic recombination and synaptonemal complex assembly, which ultimately result in compromised fertility (Luo et al., 2014; Mercier et al., 2015). To coordinate multiple events through the meiotic program, many organisms have evolved checkpoint mechanisms to ensure faithful repair of DSBs. When an error occurs, checkpoint signaling pathways are activated to halt cell cycle progression and initiate repair responses (MacQueen and Hochwagen, 2011). In particular, the pachytene checkpoint, also known as the meiotic recombination checkpoint, prevents exit from pachytene when meiotic recombination and chromosome synapsis are incomplete (Roeder, 1997). Activation of the pachytene checkpoint leads to an arrest of the meiotic program that results in apoptosis. The pachytene checkpoint widely exists in yeast and animals, including Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, mice, and other mammals (Edelmann et al., 1996; McKee and Kleckner, 1997b; Pittman et al., 1998; Gartner et al., 2000; Abdu et al., 2002). However, it is not clear whether a similar pachytene checkpoint exists in plants because unlike yeast and animals that cause meiotic cell arrest or cell death, most plant meiotic mutants defective in SPO11 initiated recombination can complete meiosis but produce abnormal microspores (Li et al., 2009; Wijnker and Schnittger, 2013).
Ubiquitination is a posttranslational modification process by which ubiquitin, a highly conserved small peptide, becomes covalently linked to its target proteins. With a suite of sequential enzymes (ubiquitin-activating E1 enzymes, ubiquitin-conjugating E2 enzymes, and ubiquitin-protein E3 ligases), the ubiquitin-tagged protein will be degraded by the proteasome appropriately and efficiently. Alternatively, the ubiquitination modifies the location, structure, and function of the target protein or serves to recruit other proteins. Target proteins can be ubiquitinated through one of the seven lysine residues in the ubiquitin, either monoubiquitinated or polyubiquitinated. Generally, the canonical polyubiquitin chain via the Lys-48 linkage targets proteins for proteolysis (Pickart, 2001), whereas the monoubiquitylation or the Lys-63-linked Ub chain is often involved in localization or signaling events (Chen and Sun, 2009). E3 ubiquitin ligases are key enzymes in the ubiquitination process, as they recognize different substrates for ubiquitination (Hershko and Ciechanover, 1998). There are two major classes of E3 ligases, namely, HECT (Homologous to E6-associated protein C terminus) and RING (Really Interesting New Gene) (Metzger et al., 2012). The RING class of E3s can be further divided into single subunit type E3s, such as CONSTITUTIVE PHOTOMORPHOGENESIS1 and U-box family, and multisubunit type E3s, like SCF and APC complexes (Moon et al., 2004). The best characterized RING-type E3s that occur are multisubunit assemblies containing Cullin-family scaffold proteins (Cullin-RING Ligases-CRLs). A typical example is the SCF, the CRL1-type E3s formed by SKP1 (S-phase kinase-associated protein 1), Cullin-1, and a highly variable F-box protein. The N-terminal F-box motif of the F-box protein is responsible for interaction with SKP1, while its C-terminal protein binding domain binds targeted substrates to confer substrate specificity (Zheng et al., 2002; Ho et al., 2006).
The ubiquitination of meiotic proteins plays important regulatory roles in meiotic entry/exit, homologous recombination, and DSB repair (Bose et al., 2014). Interference with the ubiquitin-proteasome pathway leads to meiotic cell cycle arrest or germ cell apoptosis in Schizosaccharomyces pombe, S. cerevisiae, Drosophila, C. elegans, mice, rat, and other organisms (Bose et al., 2014). For example, disruption of the polyubiquitin Ubb in mice leads to infertility with a germ cell arrest at the prophase stage of meiosis I (Ryu et al., 2008). In C. elegans, silencing of the genes encoding PBS-4 and PAS-5, two subunits of the catalytic core of the proteasome, results in impaired entry into meiosis (Macdonald et al., 2008). Furthermore, defects in E3 ubiquitin ligases can cause aberrant meiocyte development. For instance, mouse mutants of a putative ubiquitin ligase MEI4, a mouse ortholog of Human Enhancer of Invasion 10, show defective assembly of chromosomes at the metaphase plate during meiosis I, leading to arrest and apoptosis (Ward et al., 2007). In addition, male mice deficient in another E3 ligase, Cul4A, are also infertile. Spermatocytes in Cul4A−/− mice are arrested at late prophase I and have persistent DSBs, likely due to failed homologous recombination (Kopanja et al., 2011). Compared with the extensive studies in yeast and animals, much less is known about the role of ubiquitination during plant meiosis. Arabidopsis SKP1-like1 (ASK1), a member of SCF complex, is essential for homologous chromosome pairing, synapsis, and nuclear reorganization during meiosis (Yang et al., 2006; Zhao et al., 2006), suggesting that the ubiquitination pathway is also active in regulating meiotic progression in plants. However, whether other members of the plant ubiquitination machinery are involved in the meiotic process remains unknown.
In this study, we report the identification and functional analysis of the F-box gene MEIOTIC F-BOX (MOF) in rice (Oryza sativa). Our study reveals that MOF is crucial for both meiotic DSB repair and prophase I progression. The male meiocytes of mof mutants are defective in early meiotic events, including telomere bouquet formation, homologous chromosome pairing, synapsis, and DSB repair. The mof mutant is completely male sterile due to the degradation of male meiocytes at late prophase I. Our results provide further evidence to support the crucial role of the ubiquitination pathway in meiotic progression in plants.
RESULTS
Identification of mof Mutants
To identify genes that are essential for rice anther development, we performed a forward genetics screen using 60Co γ-rays as the mutagen, which resulted in a rice mutant library in the japonica subspecies 9522 background (O. sativa ssp japonica) (Chen et al., 2006). From this library, we isolated one mutant, mof, by its complete male sterility. The mof plant was indistinguishable from the wild-type plant during the vegetative stage and developed normal panicles and floral organs (Figures 1A to 1C), except for smaller and pale-yellow anthers that failed to produce viable pollen grains (Figures 1D to 1G). When mof was pollinated with wild-type pollen, all F1 progeny displayed a normal phenotype, and its F2 progeny showed an approximate 3:1 ratio for phenotype segregation (fertility:sterility = 87:24; χ2 = 0.68, P > 0.05), indicating that mof specifically affects male but not female reproduction and is a monofactorial, recessive mutant.
Figure 1.
Phenotypic Analysis of mof.
(A) Wild-type and mof plants after bolting.
(B) Wild-type and mof inflorescences at the heading stage.
(C) Wild-type and mof spikelets before anthesis.
(D) Wild-type and mof spikelets after removing the lemma and palea.
(E) A wild-type anther (left) and a mof mutant pale-yellow, smaller anther (right).
(F) and (G) I2-KI staining of the pollen grains within the anther of the wild type (F) and mof (G).
(H) to (Q) Transverse section analysis of the anthers. The images are cross sections of a single locule. The wild-type anther is shown in (H), (J), (L), (N), and (P) and the mof mutant anther in (I), (K), (M), (O), and (Q). Stage 7 (meiosis prophase I) ([H] and [I]); stage 8a (dyad stage) ([J] and [K]); stage 8b (tetrad stage) ([L] and [M]); stage 9 (early microspore stage) ([N] and [O]); and stage 12 ([P] and [Q]).
DMC, degenerated meiocyte cell; DMs, degenerated microspores; Dy, dyad; E, epidermis; En, endothecium; M, middle layer; MC, meiocyte cell; Mp, mature pollen; Ms, microspores; T, tapetal layer; Tds, tetrads; gl, glume; le, lemma; pa, palea; st, stamen. Bars = 20 cm in (A), 2 cm in (B), 2 mm in (C) to (E), 10 μm in (F) and (G), and 15 μm in (H) to (Q).
Defective Anther and Meiocyte Development in mof
To characterize developmental defects in the mof mutant, anther transverse sections were examined. The early stages of anther development preceding meiosis were indistinguishable between the wild type and mof, suggesting that the mutation does not affect the differentiation and early mitotic division of germ cells. However, aberrant male meiocytes that were irregular in size and shape were found in mof shortly after the initiation of meiosis (Figures 1H and 1I). In wild-type anthers, male meiocytes produced dyads and tetrads (Figures 1J and 1L) from which haploid microspores were released after the completion of meiosis (Figure 1N). However, in the mof mutant male meiocytes completely degraded after meiosis (Figures 1K, 1M, and 1O). Unlike wild-type anthers, which were full of pollen grains, anther locules in mof were empty at the mature stage (Figures 1P and 1Q).
To obtain more detailed observations of mof anther development, we performed transmission electron microscopy (TEM). Consistent with the observations of light microscopy, mof male meiocytes were aberrant (Supplemental Figure 1). Meiocytes in wild-type anthers were vacuolated, whereas in mof they were overvacuolated (Supplemental Figures 1A, 1B, 1I, and 1J). These vacuoles appeared ruptured during early meiosis and were similar to those exhibiting programmed cell death in the wild-type tapetum (Wu and Cheun, 2000). At later stages, these aberrant meiocytes collapsed to debris and totally degraded (Supplemental Figures 1K to 1P).
The Male Sterility of mof Is Caused by Genome Fragmentation during Meiosis
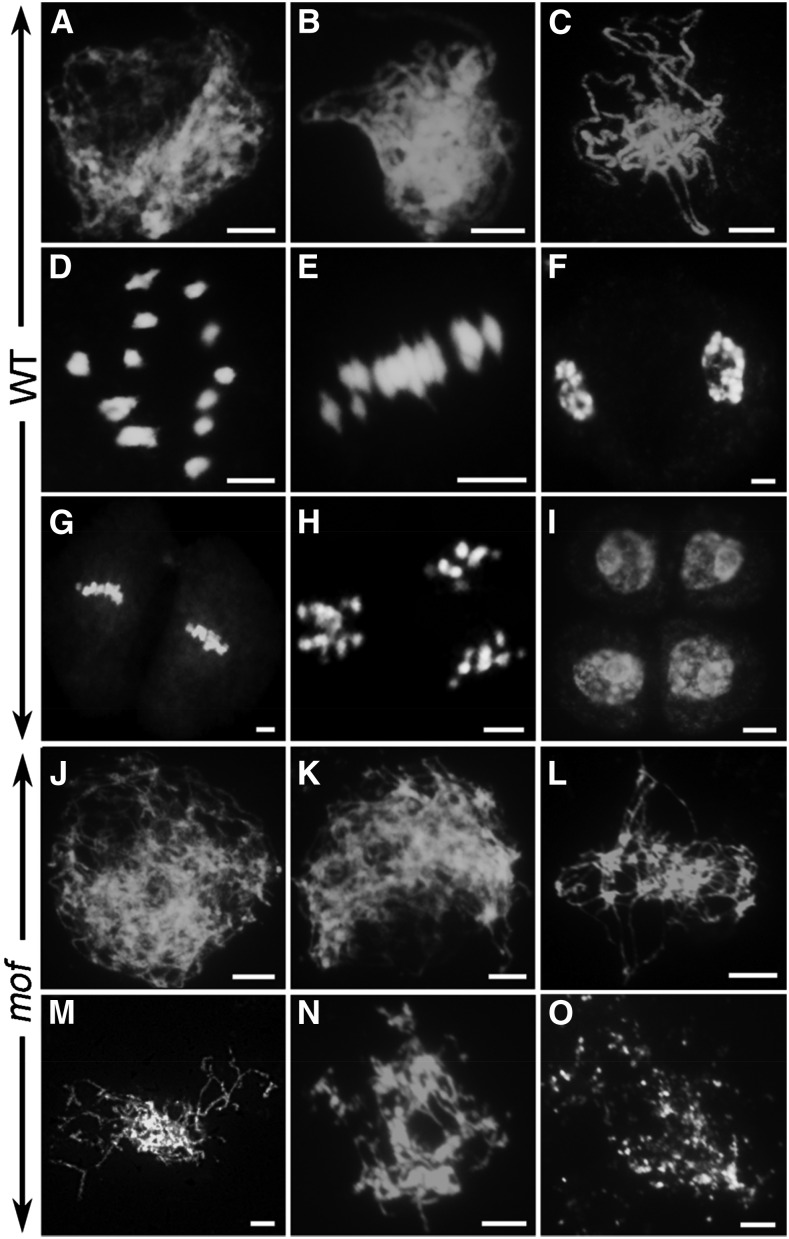
The degradation of pollen mother cells suggested that the male sterility might be a result of meiotic defects in the mof mutant. To further investigate the cause of male sterility in mof, we examined and compared the meiotic chromosome behavior of meiocytes in the wild type and the mof mutant by 4′,6-diamidino-2-phenylindole (DAPI) staining. In the wild type, individual chromosomes condensed into visible strands as thin threads at leptotene (Figure 2A) that began pairing and undergoing synapsis at zygotene with the assembly of the central element of the synaptonemal complex (SC) (Figure 2B). At pachytene, the homologous chromosome pairs were fully synapsed (Figure 2C). At diplotene, the SCs were disassembled and homologous chromosomes separated except for the physical associations of chiasmata, which are the cytological sites of crossovers. The homologous chromosomes further condensed as 12 bivalents at diakinesis (Figure 2D). The homologous chromosomes then segregated to opposing poles and began the process of meiosis II without interphase before finally producing tetrads (Figures 2E to 2I).
Figure 2.
Meiotic Chromosome Behaviors of Male Meiocytes in the Wild Type and mof.
Chromosome behavior of male meiocytes of wild type ([A] to [I]) and mof ([J] to [O]) at various stages. Leptotene ([A] and [J]); zygotene ([B] and [K]); pachytene (C); diakinesis (D); metaphase I (E); anaphase I (F); metaphase II (G); anaphase II (H); telophase II (I); stages after zygotene ([L] to [O]). Bars = 5 μm.
In mof, chromosomes appeared normal at leptotene in male meiocytes, indicating that the cohesion of sister chromatids was not affected in the mutant (Figure 2J); however, subsequent stages were severely impaired. Homologous chromosome pairing was not observed at zygotene and later stages (Figures 2K and 2L). At late prophase I, chromosomes aggregated into a chromosome mass (Figure 2L). Subsequently, these aberrant chromosomes became fragmented and degraded (Figures 2M to 2O). The later stages of meiosis I and II were not observed in the mof mutant. Next, we examined the distribution of meiocytes at various stages in different flower lengths. In 4.5- and 5.0-mm-long flowers, tetrads and microspores were observed in the wild type, whereas no meiocytes were detected in mof. Meiosis appeared to initiate normally in mutant meiocytes compared with the wild type in 3.0-mm flowers (Supplemental Figure 2).
Cell disintegration and chromosome degradation observed in the mof indicated that mutant meiocytes might undergo apoptosis. To test this possibility, we performed the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in wild-type and mof meiocytes. TUNEL signals were not detected in the wild-type male meiocytes at any stages of meiosis (Supplemental Figure 3). In contrast, strong TUNEL signals were observed in the mof male meiocytes (Supplemental Figure 3). The TUNEL analysis, consistent with the semi-thin-section observation, TEM, and DAPI analysis, confirmed that apoptotic processes occur in mof male meiosis.
MOF Is Essential for Telomere Bouquet Formation and Homologous Chromosome Pairing
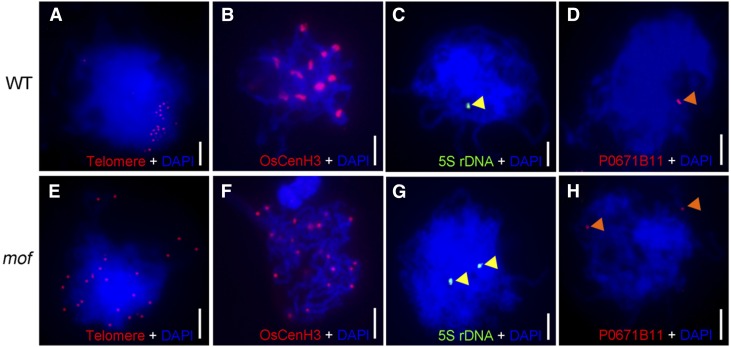
The clustering of telomeres into a “bouquet” is one of the mechanisms thought to facilitate the subsequent meiotic prophase I processes, such as homologous pairing and synapsis, by bringing chromosomes into close proximity, which increases searching efficiency by collecting chromosome ends into a limited region and shortening the distance between homologs (Tsai and McKee, 2011). To investigate telomere bouquet formation in the wild type and mof, we conducted fluorescent in situ hybridization (FISH) analysis using Cy3-probe labeled telomere-specific sequences as described (Mizuno et al., 2006). In wild-type rice male meiocytes, telomeres attached to the nuclear envelope and clustered together at early prophase I, showing a typical bouquet configuration (Figure 3A). By contrast, telomeres in mof meiocytes did not cluster to a certain region but were scattered throughout the nuclear volume (Figure 3E), indicating that bouquet formation is defective in the mof mutant.
Figure 3.
MOF Is Required for Bouquet Formation and Homologous Pairing.
(A) and (E) Telomere bouquet formation analysis revealed by FISH using the telomere sequence as a probe (red) in wild-type (A) and mof (E) nuclei. Chromosomes (blue) are stained with DAPI.
(B) and (F) Immunolocalization of OsCenH3 (red) showing paired and unpaired homologous chromosomes in wild-type (B) and mof (F) nuclei.
(C) and (G) Homologous pairing analysis revealed by FISH using 5S rDNA probe (green; indicated by yellow triangle) in wild-type (C) and mof (G) nuclei.
(D) and (H) Homologous pairing analysis revealed by FISH using probes prepared from BAC P0671B11 (red; indicated by orange triangle) in wild-type (D) and mof (H) nuclei. Bars = 5 μm.
To understand whether homologous chromosome pairing was defective in the mutant, we performed immunolocalization and FISH analysis. The anti-OsCenH3 antibody recognizes functional centromeric regions on chromosomes in rice somatic and meiotic cells (Nonomura et al., 2006). Rice 5S rDNA is located on the short arm of chromosome 11 (Zhang et al., 2005), and P0671B11 locates near the telomere of Chromosome 1. Combination of the use of these cytological markers is useful for monitoring chromosome pairing and separation. In the wild type, 12 OsCenH3 signals were observed (Figure 3B), but >20 signals were detected in the mof mutant (Figure 3F). In the wild type, 5S rDNA and BAC P0671B11 produced only one signal (Figures 3C and 3D), indicative of synapsed chromosomes, whereas in mof mutants, the two probes produced two totally separated signals (Figures 3G and 3H), suggesting that homologous chromosome pairing is perturbed in mof.
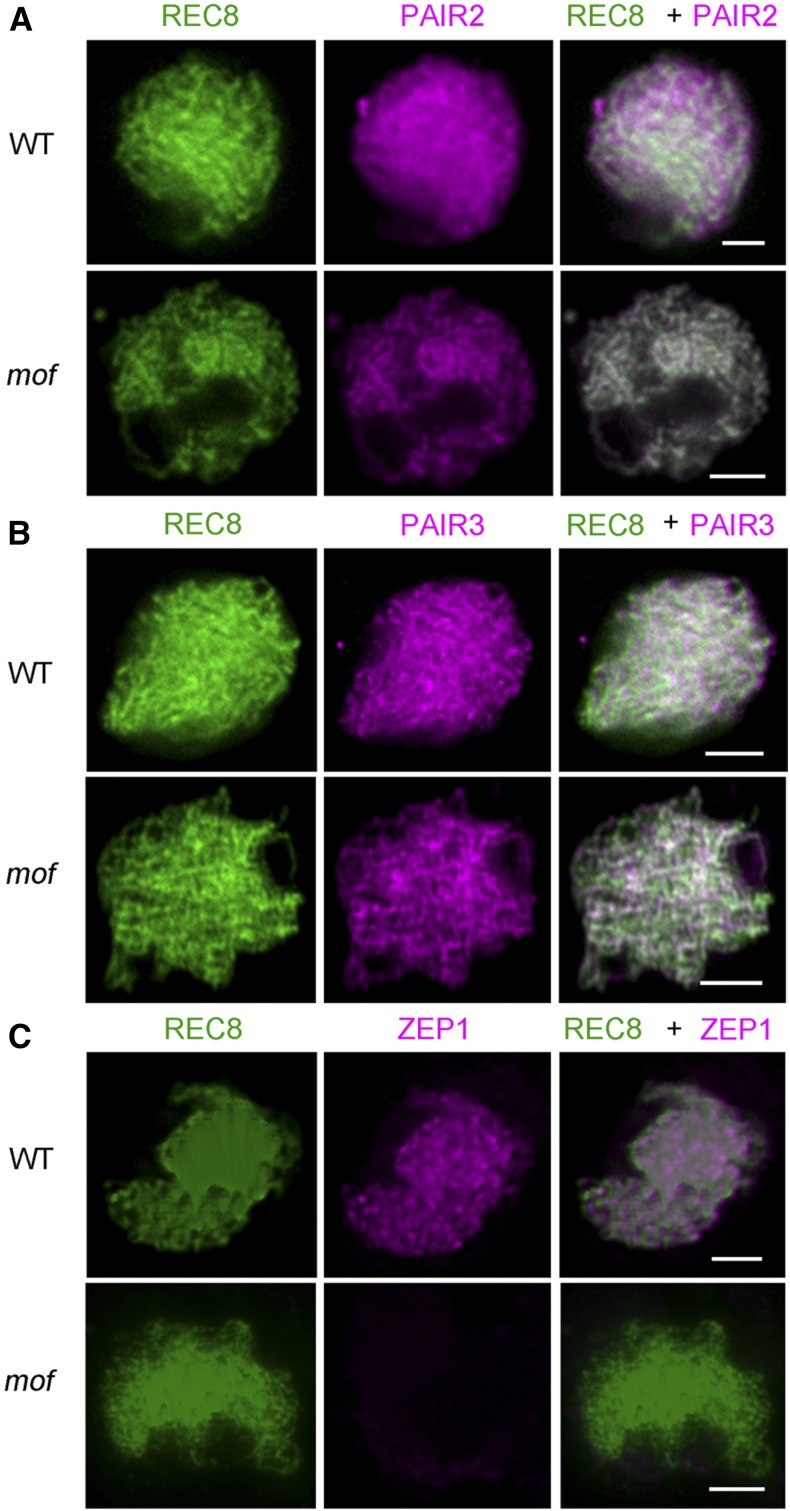
MOF Is Required for SC Formation
The synaptonemal complex stabilizes initial chromosomal axial associations and promotes crossover formation, until the SC is disassembled at diplotene. To further explore the role of MOF during meiosis, we performed immunolocalization on several rice SC-related proteins, including PAIR2, PAIR3, ZEP1, and REC8 in wild-type and mof male meiocytes. Rice REC8 is a cohesin critical for sister-chromatid cohesion, axial element formation, and homologous pairing (Shao et al., 2011). In mof, REC8 localization to chromosomes was identical to the wild type (Figure 4) and thus was used as a marker for further meiotic analysis. PAIR2, the rice ortholog of S. cerevisiae Hop1 and Arabidopsis ASY1, is essential for homologous chromosome pairing (Nonomura et al., 2006). PAIR3 localizes to the chromosome axes and is essential for homologous pairing, recombination, and SC assembly and is the functional homolog of Arabidopsis ASY3 (Wang et al., 2011; Ferdous et al., 2012). The transverse filament protein ZEP1, a homolog of Arabidopsis ZYP1, is the central element of SC in rice (Higgins et al., 2005; Wang et al., 2010). In mof meiocytes, PAIR2 and PAIR3 loaded normally onto the chromosome axes (Figures 4A and 4B). However, no signals of ZEP1 were observed in mof (Figure 4C), indicating that SC formation is severely disrupted in mof meiocytes.
Figure 4.
Immunolocalization of SC-Related Proteins in mof.
Immunolocalization of PAIR2 (magenta) (A), PAIR3 (magenta) (B), and ZEP1 (magenta) (C) at zygotene in both the wild type and mof. Bars = 5 μm.
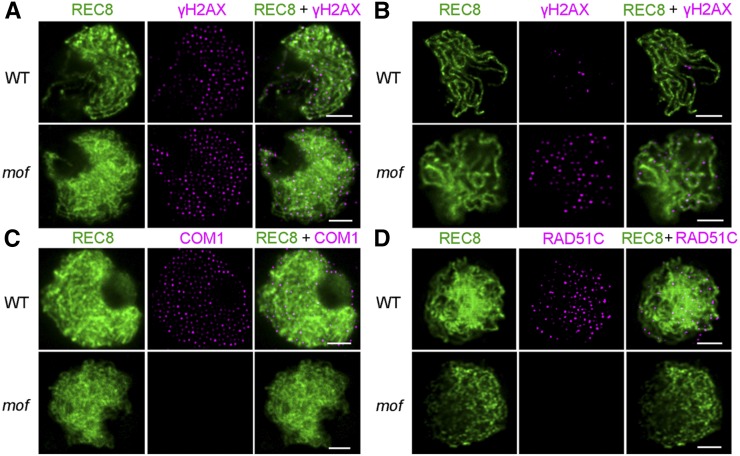
MOF Is Not Required for DSB Formation but Is Essential for DSB Repair
Meiotic recombination is initiated by programmed DNA DSBs, which are generated by the conserved topoisomerase II-like enzyme SPO11. The formation and repair of DSBs is essential for correct homologous chromosome pairing, synapsis, and recombination in most species (Inagaki et al., 2010). Unrepaired DSBs can lead to severe chromosomal aberrations that may ultimately result in apoptosis or aneuploidy (Pastink et al., 2001). To investigate whether defective homologous chromosome pairing and synapsis were correlated with aberrant DSB formation or processing, we applied antibodies to REC8, γH2AX, COM1, and RAD51C in wild-type and mof male meiocytes. The histone variant H2AX is rapidly phosphorylated to γH2AX at DSB sites, so γH2AX foci are considered to be a reliable marker for DSBs (Hunter et al., 2001). At zygotene, no significant difference in numbers of γH2AX foci was observed between the wild type (163 ± 30; n = 33) and mof (165 ± 29; n = 27), indicating that DSB formation is normal in the mutant (Figure 5A). In wild-type meiosis, all DSBs are repaired. If unrepaired, chromosome fragments will cause genome instability. In the wild type, the number of γH2AX foci decreased to 19 ± 9 (n = 17) at late pachytene, while the number of γH2AX foci was still high in mof (74 ± 19; n = 16; P = 8.2E-12) (Figure 5B). COM1 and RAD51C have been reported to play important roles in DSB end processing and repair by homologous recombination (Ji et al., 2012; Tang et al., 2014). In wild-type meiocytes, COM1 was first visible in meiotic interphase and peaked during zygotene (Figure 5C). The RAD51C signals were first detected as punctate foci at leptotene and reached a maximum at zygotene (Figure 5D). However, in mof zygotene cells, 85% (n = 55) and 58% (n = 36) were found with no signals of COM1 and RAD51C, respectively (Figures 5C and 5D), and the foci numbers of COM1 and RAD51C decreased in the remaining meiocytes, indicating that normally formed DSB ends are not processed and repaired in the mof mutant.
Figure 5.
MOF Is Not Required for DSB Formation but Is Essential for Meiotic Progression.
Immunolocalization of γH2AX (magenta) at zygotene (A) and pachytene (B), and COM1 (magenta) (C) and RAD51C (magenta) (D) at zygotene in both the wild type and mof. REC8 signals (green) were used to indicate the meiotic chromosome axes. Bars = 5 μm.
MOF Encodes an F-Box-Containing Protein
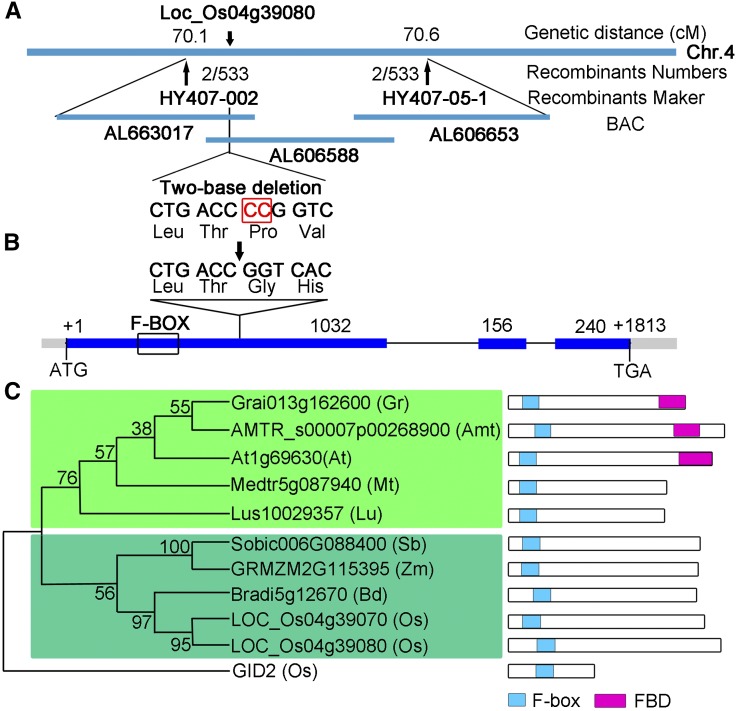
To identify the MOF gene, we initially used a map-based cloning approach to locate the MOF locus between two InDel molecular markers, OS407 and OS408, on chromosome 4. To finely map MOF, we used 533 mutants from an F2 mapping population that enabled us to narrow down the region to 160 kb between two InDel markers HY407-002 and HY407-05-1 (Figure 6A). We then used MutMap, a method based on whole-genome resequencing of pooled DNA (Abe et al., 2012), to identify the mutation in the mof gene. There was a two-nucleotide deletion in the first exon of the candidate gene (LOC_Os04g39080) (Figure 6B), causing a frameshift and premature translational termination (Supplemental Figure 4). To confirm that the mutation in LOC_Os04g39080 is responsible for the meiotic defects in the mof mutant, a binary plasmid containing an ∼5.4-kb wild-type genomic fragment subcloned from the BAC OSJNBa0060P14 was introduced into the mof homozygous plants. The wild-type genomic fragment was able to rescue the male sterile phenotype of mof (Supplemental Figures 5E and 5F), demonstrating that LOC_Os04g39080 is the MOF gene.
Figure 6.
Molecular Characterization of MOF.
(A) Fine mapping of MOF on chromosome 4. Names and positions of the markers are noted. cM, centimorgans.
(B) A schematic representation of three exons and two introns of Os04g39080. The +1 indicates the putative starting nucleotide of translation, and the stop codon (TGA) is +1813. Blue boxes indicate exons, and intervening lines indicate introns. Numbers indicate the exon length (bp). The deletion site in mof is shown (arrow).
(C) Phylogenetic analysis of MOF and its related homologs. A maximum likelihood analysis was performed using MEGA 4.0 using MOF-related sequences from Amborella trichopoda (Amt), Arabidopsis thaliana (At), Brachypodium distachyon (Bd), Gossypium raimondii (Gr), Linum usitatissimum (Lu), Medicago truncatula (Mt), Oryza sativa (Os), Sorghum bicolor (Sb), and Zea mays (Zm). The green shaded box indicates the dicotyledon cluster, and the turquoise shaded box indicates the monocotyledon cluster.
The predicted MOF protein is 475 amino acids in length and contains a putative F-box domain at the N terminus (amino acids 61 to 101) (Figure 6B; Supplemental Figure 4). In rice, 687 potential F-box proteins have been identified with several other functional domains in the C terminus, including leucine-rich repeats, Kelch repeats, tetratricopeptide repeat, WD-40, tubby, and a domain of unknown function (Jain et al., 2007). MOF belongs to the FBX subfamily of F-box proteins in rice and contains no other known functional domains (Jain et al., 2007). F-box proteins have been reported to be associated with a broad spectrum of biological events in plant development and stress signaling, including morphogenesis, hormone perception and signaling, cell cycle, circadian clock regulation, senescence, responses to abiotic stress, and plant-pathogen interactions (Stefanowicz et al., 2015).
To gain insights into the phylogenetic relationship between MOF and its close homologs, BLASTP searches were conducted with the full-length amino acid sequence of MOF. Homologs of MOF can only be found in angiosperm species. In rice, MOF and LOC_Os04g39070, one homolog of MOF, are tandemly located on chromosome 4, suggesting that they may be derived from gene duplication. An unrooted tree of MOF and its nine homologs showed that MOF was closely related to Bradi5g12670 from Brachypodium distachyon and two other genes from monocots, and distant from dicots, indicating that MOF is specific to angiosperms (Figure 6C).
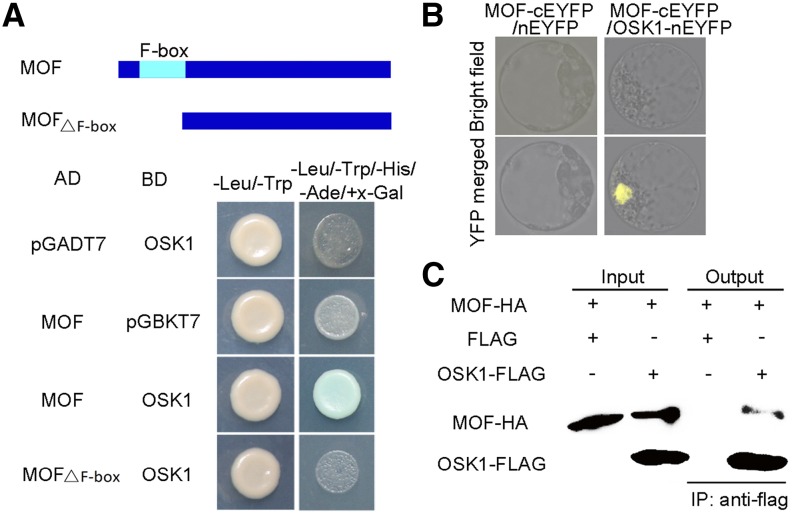
MOF Interacts with OSK1 as Part of an SCF Complex
MOF contains a conserved F-box motif in the N terminus (Figure 6B; Supplemental Figure 4). To determine if MOF functioned as a canonical F-box protein, we examined its interactions with known components of the rice SCF complex. OSKs are SKP1-like proteins in rice, which are thought to interact with Cullin and F-box proteins to function as an SCF E3 ubiquitin ligase. In rice, there are at least 32 annotated OSKs, all derived from a single ancestral gene (Kahloul et al., 2013). OSK genes display various expression patterns, and seven of them, including OSK1, OSK8, OSK19, OSK20, OSK21, OSK22, and OSK24, are highly expressed during early anther development (Supplemental Figure 6). A yeast two-hybrid assay revealed that among these OSKs, only OSK1 was able to interact with the full-length MOF (Figure 7A; Supplemental Figure 7), while no interaction was detected between OSK1 and a truncated MOF without the F-box domain (Figure 7A). Furthermore, we validated this interaction using bimolecular fluorescence complementation (BiFC) in rice protoplasts. In contrast to the negative control, YFP signals were observed in the nucleus of the cells expressing MOF-cYFP and OSK1-nYFP (Figure 7B). We also performed coimmunoprecipitation analysis to confirm the interaction between MOF and OSK1. HA-tagged MOF coimmunoprecipitated with FLAG-tagged OSK1 (Figure 7C). These results indicate that MOF interacts with OSK1 to function within an SCF complex in plant cells.
Figure 7.
Physical Interaction between MOF and OSK1.
(A) Yeast two-hybrid assay for interaction of MOF with OSK1. A schematic diagram of MOF and the truncations used is shown. The interactions were verified by the growth of yeast strains on the -Leu-Trp-His-Ade+X-Gal selection medium.
(B) BiFC assay for interaction between the MOF and OSK1 in rice protoplasts.
(C) Coimmunoprecipitation of MOF-HA and OSK1-FLAG based on anti-FLAG immunoprecipitation from transfected Nicotiana benthamiana leaves.
Expression Pattern of MOF
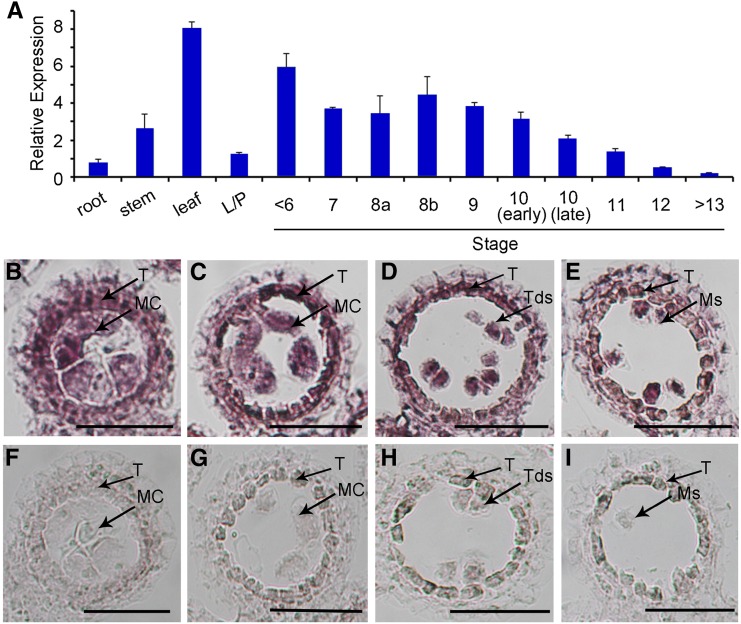
The spatio-temporal expression pattern of MOF was examined by RT-qPCR analyses. Based on the results of RT-qPCR, MOF transcripts were highly expressed in the anther during meiosis and then gradually decreased after meiosis. MOF was also highly expressed in the stem, leaf, and weakly expressed in roots and lemma/palea (Figure 8A). To further reveal the MOF expression pattern in the anther, we performed RNA in situ hybridization with wild-type floral sections. From stage 7 to stage 9, MOF transcripts were detected in both the tapetum and male meiocytes (Figures 8B to 8E). Only background levels of signal were detected with the sense probe (Figures 8F to 8I).
Figure 8.
Expression Pattern of MOF.
(A) Spatial and temporal expression analysis of MOF by RT-qPCR. RNAs were extracted from the root, stem, leaf, lemma/palea, and anthers of <stage 6, stage 7, stage 8a, stage 8b, stage 9, stage 10 (early), stage 10 (late), stage 11, stage 12, and stage 13/14. L/P, lemma/palea. Each reaction had three biological repeats. Error bars indicate sd. Actin served as a control.
(B) to (I) In situ analyses of MOF in wild-type anthers. Anthers at stage 7, stage 8a, stage 8b, and stage 9 with MOF antisense probe ([B] to [E]) and sense probe ([F] to [I]) showing MOF expression in tapetal cells and meiocytes. MC, meiocyte cell; Ms, microspores; T, tapetal layer. Bars = 50 μm.
MOF is widely expressed, implicating that it may function in other biological processes except meiosis. To confirm that the sterility of mof was caused by defective MOF function in meiosis, the coding sequence of MOF driven by a male meiocyte specific promoter was introduced into the mof mutant. MTR1 encodes a secretory fasciclin glycoprotein and is specifically expressed in male meiocytes (Tan et al., 2012). The fact that MTR1:MOF was capable of rescuing the male sterile phenotype of mof (Supplemental Figures 5I and 5J) demonstrated that MOF plays an indispensable role during male meiosis.
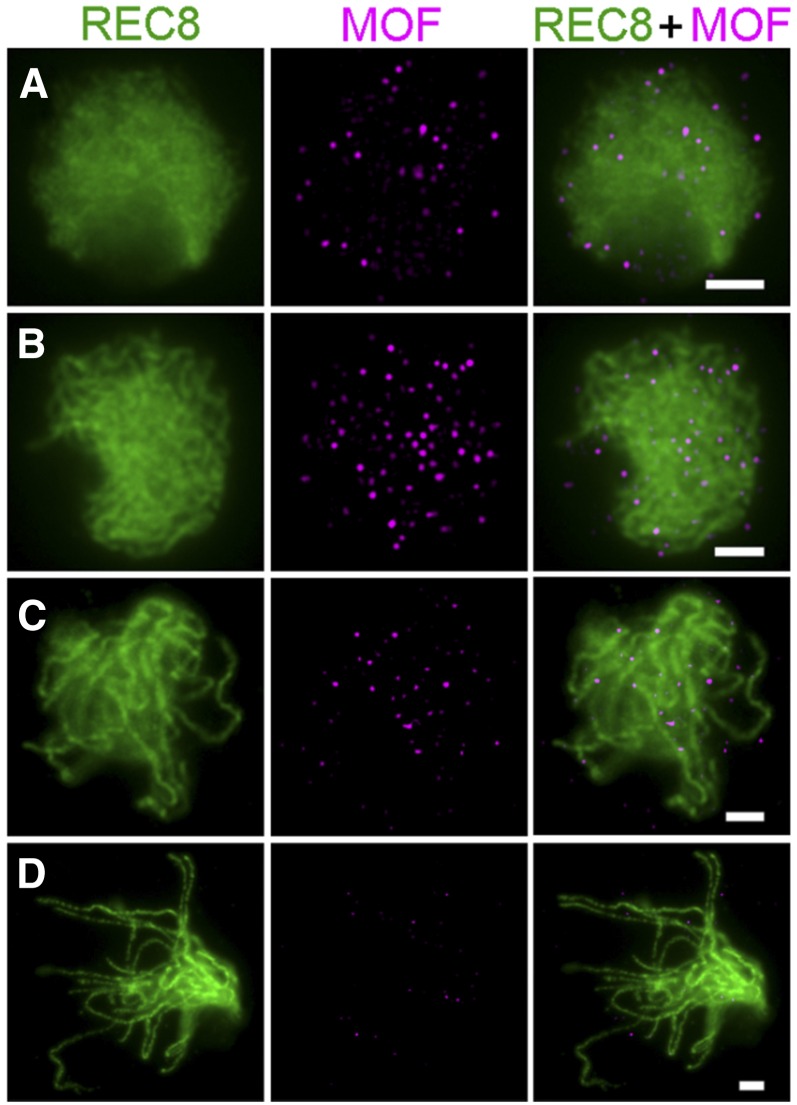
MOF Localizes to Meiotic Chromosomes from Leptotene to Pachytene
To investigate the spatial and temporal distribution of MOF during meiosis, dual immunolocalization with polyclonal antibodies against REC8 and GFP tag was conducted in male meiocytes of mof/proMOF:MOF:eGFP transgenic lines. proMOF:MOF:eGFP could fully rescue the fertility of the mof mutant (Supplemental Figures 5G and 5H), indicating that the GFP-tag did not interfere with the function of MOF. In the transgenic line, MOF/GFP foci were visible at leptotene (Figure 9A). The number of MOF/GFP foci rapidly increased from leptotene to zygotene and reached a peak during zygotene (Figure 9B). In early pachytene, MOF/GFP foci began to diminish and very few signals were detected after late pachytene (Figures 9C and 9D). No MOF/GFP signal was detected in the wild type, indicating that the antibody is specific.
Figure 9.
Dual Immunolocalization of REC8 and MOF in Meiocyte Cells of the Complemented Transgenic Line.
(A) Leptotene; (B) zygotene; (C) early pachytene; and (D) late pachytene. REC8, green; MOF, magenta. Bars = 5 μm.
DISCUSSION
MOF Is Essential for Early Meiotic Events in Rice
Meiosis is a complex and highly controlled process, which is not only regulated by transcriptional and translational controls, but also by posttranslational aspects such as targeted protein degradation and modification. The ubiquitin-proteasome pathway plays critical roles in several key events during meiosis such as meiotic entry/exit, DSB processing, repair, and genetic recombination in yeast and animals (Bose et al., 2014). However, very little is known about the role of ubiquitin-proteasome system in the plant meiosis program. In this study, we described the functional characterization of a rice F-box protein, MOF, which is involved in male meiotic progression in rice.
MOF encodes a putative F-box containing protein that belongs to the FBX subfamily. MOF can interact with the SKP1-like protein OSK1, suggesting that MOF is a component of SCF ubiquitin ligases and may function through the ubiquitin-mediated protein degradation or signaling pathway. Based on the gene structure, expression profile, and the ability to interact with a wide spectrum of F-box proteins, OSK1 has been proposed to be the functional equivalent of ASK1 in Arabidopsis (Kahloul et al., 2013). Arabidopsis ASK1 is the homolog of the yeast and human SKP1 and is involved in many aspects of plant growth and development. During meiosis, ASK1 is mainly expressed from leptotene to pachytene. The ask1 mutant is defective in meiotic chromosome condensation, homologous chromosome pairing, synapsis, recombination, and separation (Yang et al., 1999; Wang and Yang, 2006). It is reported that the wide range of defects in ask1 are likely caused by the failure of meiotic chromosomes released from the nucleolus or nuclear membrane at leptotene, suggesting that ASK1 plays essential role in the reorganization of the nucleus and the juxtaposition of homologous chromosomes at early prophase I (Yang et al., 2006; Zhao et al., 2006). However, how ASK1 regulates nuclear remodeling has not yet been determined.
Similar to ASK1, MOF also acts predominantly during early prophase I, from leptotene to pachytene. In contrast to ask1, the mof mutant is not able to produce tetrads and microspores. The mof mutation leads to several chromosome abnormities, including defective telomere bouquet formation, homologous chromosome pairing, and synapsis in male meiocytes. In particular, the mof male meiotic chromosomes are arrested at late prophase I and become severely fragmented and degenerated at a later stage. Our analysis indicated that these abnormal chromosomal behaviors could be the consequence of defects in DSB processing and repair. In higher plants, DSB formation and repair is a prerequisite for homologous chromosome pairing, SC formation, and recombination during meiosis (Borde and de Massy, 2013). Many aspects of the mof mutant phenotype resemble those described for the mutants of genes involved in DSB processing and repair, such as osmre11, oscom1, osrad51c, rpa1a, and xrcc3 in rice (Chang et al., 2009; Ji et al., 2012, 2013; Tang et al., 2014; Zhang et al., 2015). OsCOM1 is the homolog of COM1/SAE2 in S. cerevisiae, Ctp1 in fission yeast, and CtBP-interacting protein in mammals (McKee and Kleckner, 1997a; Prinz et al., 1997; Schaeper et al., 1998; Limbo et al., 2007). Loss-of-function mutants of COM1/SAE2 homologs show defects in DSB resection and subsequent meiotic processes. The COM1 and RAD51C foci were undetectable in most of the mof meiocytes, but the protein levels were not reduced (Supplemental Figure 8), indicating that the mof deficiency interferes with the recruitment of DSB processing and repair proteins. Consistent with this, the immunolocalization results showed a prolonged presence of γH2AX foci, supporting that DSBs were normally formed but not correctly processed and repaired in the mof male meiocytes. In addition, over half of the MOF foci colocalize with γH2AX, and one-third of MOF foci overlapped with those of COM1 and RAD51C signals (Supplemental Figure 9), indicating that MOF is dynamically localized around DSB sites. Collectively, these results suggest that MOF is an essential regulator of DSB processing, repair and meiotic progression in rice.
The interruption of DSB formation may cause the DNA fragmentation and cell death phenotypes in mof mutants. There are five SPO11 homologs in rice and currently it is not clear which of them participate in meiotic DSB formation (An et al., 2011). The identification of the functional meiotic SPO11 will provide further clues to understand the role of MOF in DSB processing and repair.
What Is the Function of MOF in DSB Repair?
Our results showed that MOF is important for the recruitment of DSB repair proteins to the DSB sites. Although the exact role of MOF in this process is not clear, it is possible that MOF is involved in the posttranslational modification of chromatin and nonchromatin proteins to provide a platform for DSB repair protein docking.
Accumulating evidence demonstrates that nondegradative ubiquitin signaling predominates in the DNA damage response in animals. Histone ubiquitination at the chromatin surrounding DSBs is one of the initial steps in DNA damage response activation (Lukas et al., 2011). In vertebrates, phosphorylation of the histone variant H2AX (known as γH2AX) promotes the association of the E3 ligase RING-finger-containing nuclear factors 8 (RNF8) through the mediator of DNA damage checkpoint protein 1 (MDC1) to the DSB site. RNF8 recruits a second E3 ligase RNF168 and RNF8/RNF168 coordinately interacts with the E2 conjugate enzyme UBC13 to catalyze the ubiquitination of H2A and H2AX, which is a prerequisite for the stable accumulation of effectors at the DSB site (Huen et al., 2007; Wang and Elledge, 2007). Alternatively, MOF may be required for the removal of proteins that hinder the recruitment of DSB repair proteins through proteolysis pathway. It has been reported that the E3 ligase RNF4 can promote the turnover of MDC1 and RPA by ubiquitylation-mediated protein degradation. The proteasome component PSMD4 is recruited to the DSB sites in a DNA damage and RNF4 dependent manner. The RNF4 depletion causes the failure of RAD51C and BRCA2 loading on the resected single-stranded DNA (Galanty et al., 2012). These ubiquitylation events are indispensable for male meiocyte development. Loss of function of RNF8 does not affect the female ovary development but causes disruption of spermatogenesis within the testis (Li et al., 2010). Mice lacking RNF4 have defective spermatogenesis characterized by increased apoptosis and depletion of spermatocytes (Vyas et al., 2013).
None of the E3 ligase homologs have been reported in plants, while MOF may represent the functional counterpart of one of these or other yet unknown modifiers in rice and facilitate the formation of a DNA repair-stimulating microenvironment around the DSB sites via proteolysis or nonproteolysis pathway.
MOF Is Required for Meiotic Progression in Rice
In eukaryotic organisms, the integrity of genetic information is maintained through the operation of cell cycle checkpoints. Checkpoints ensure the proper order of events in the cell cycle by preventing the initiation of late events until earlier events have been successfully completed. During meiosis, errors frequently occur along with the intricate chromosomal events, so meiotic checkpoints have been evolved in most organisms to ensure normal chromosome synapsis and DNA damage repair. A prolonged checkpoint may result in the destruction of defective meiocytes, thus preventing the generation of defective gametes (Li et al., 2009). In particular, the pachytene checkpoint is triggered in meiocytes impaired in synapsis and recombination (Roeder and Bailis, 2000). It widely exists in many species, including S. cerevisiae, Drosophila, C. elegans, and mice, and leads to cell cycle arrest during the pachytene stage and cell death thereafter (Edelmann et al., 1996; McKee and Kleckner, 1997b; Pittman et al., 1998; Gartner et al., 2000; Roeder and Bailis, 2000; Abdu et al., 2002).
It has been generally thought that plants may not have a typical pachytene checkpoint because most plant meiotic mutants are able to complete the meiotic program and produce abnormal microspores (Caryl et al., 2003). However, recent studies on meiosis arrested at leptotene (mel2) and ameiotic1 (am1) mutants suggest the existence of a meiotic leptotene/zygotene checkpoint in plants (Nonomura et al., 2011). The MEL2 gene encodes an RNA recognition motif-containing protein. In the rice mel2 mutant, male meiocytes and tapetal cells were hypervacuolated and degenerated due to apoptosis. The mutation disturbed 80% of germ cells progressing into premeiotic S-phase and the remaining 20% into late zygotene (Nonomura et al., 2011). AM1 homologs encode a plant-specific protein with a coiled-coil domain. Mutations of OsAM1 in rice result in meiotic arrest at early prophase I, especially during leptotene/zygotene transition (Che et al., 2011). In contrast to most of the maize am1 alleles that are defective in meiosis entry, meiocytes in maize am1-praI enter meiosis but are arrested in leptotene/zygotene (Pawlowski et al., 2009). The cell cycle arrest in these mutants suggests a novel leptotene/zygotene checkpoint in plants.
In our study, we found that the meiotic progression in mof was arrested at a later stage than in am1 mutants. The aberrant chromosome behavior in mof is similar to the am1/osam1 mutants, including a lack of telomere bouquet formation, defective homologous chromosome pairing, and synapsis as well as failure to recruit DSB repair proteins. However, compared with the zygotene-like chromosomes in am1/osam1 mutants, most chromosomes in mof male meiocytes displayed the characteristics of pachytene. In addition, although SC formation was lost in both mof and am1/osam1 mutants, these events were arrested at different steps. In the osam1 mutant, no PAIR2 and ZEP1 signals were detected in the meiocytes, indicating the absence of axial element formation (Che et al., 2011). However, axial element formation was not affected in the mof mutant, as evidenced by the detection of normal PAIR2 and PAIR3 signals. These observations suggest that MOF may function at a later stage than MEL2 and OsAM1 and the meiotic arrest in the mof mutant resembles the arrest caused by the pachytene checkpoint in yeast and metazoans.
MOF Is Required Specifically for Male Meiotic Progression
Depletion of most meiotic genes in plants leads to defects in both male and female cells. However, the fact that the mof mutant is female fertile suggests that MOF serves a nonredundant function in male meiosis in rice. Other F-box genes in rice are likely to provide essential functions in the female meiotic programs. This possibility is supported by the existence of a large number of F-box proteins and many SKP1 homologs in rice (Jain et al., 2007). In addition, there is one MOF closely related homolog in rice. It may be essential for female meiosis and other developmental processes. MOF may also share redundant functions with its homolog during vegetative growth as MOF is also expressed in vegetative tissues. In animals, mutation of a protein involved in the ubiquitin-proteasome system pathway always has different impacts on male and female reproduction. The E3 ligase Cul4A is indispensable for male meiosis but not essential for female meiosis. The male knockout mice are infertile, while the female knockout mice are still able to generate pups although with a smaller litter size (Kopanja et al., 2011). The Drosophila polyubiquitin Ubi-p63E is critical for meiotic cell cycle progress in male germ cells. Loss of function of Ubi-p63E causes meiotic arrest sterility specifically in males (Lu et al., 2013). A similar phenomenon was also observed in plants. The Arabidopsis ask1 mutant is also male sterile but female fertile (Yang et al., 1999; Wang and Yang, 2006). These studies reveal that machinery of the ubiquitination system has diverged to acquire specialized functions to meet the unique requirements of protein turnover or modification during male and female meiotic programs.
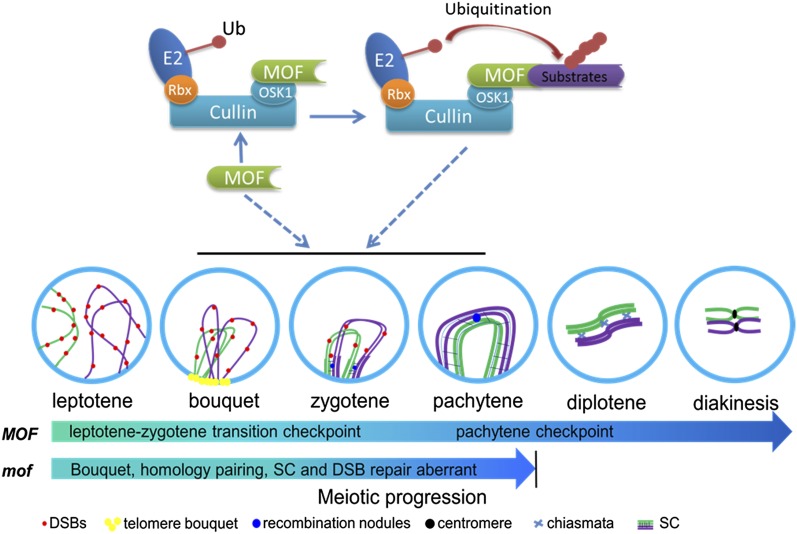
Understanding on the molecular control of meiosis in plants is of importance for crop breeding and fundamental biology (Zhang and Liang 2016). Based on our data, we propose a model for how MOF regulates rice male meiotic progression (Figure 10). At early prophase I, MOF interacts with OSK1 to form an SCF-complex E3 ligase complex that work together to promote early meiotic events, especially the DSB processing and repair. The mof deficiency leads to failure of homologous chromosome synapsis and recombination, which triggers meiotic arrest at late prophase I. A prolonged arrest induces apoptosis and meiocyte elimination. These findings reveal a novel regulatory role for an F-box protein in early meiotic events. Further analysis of MOF function will contribute to a better understanding of posttranslational regulation of rice germ cell development and meiotic progression.
Figure 10.
Proposed Working Model for MOF in the Regulation of Meiotic Progression.
At early prophase I, MOF interacts with OSK1 to form an SCF complex E3 ligase complex that works together to promote early meiotic events, especially DSB processing and repair. Loss of function of MOF results in failure of homologous chromosome synapsis and recombination, which triggers meiotic arrest at late prophase I, and a prolonged arrest induces apoptosis and meiocyte elimination.
METHODS
Plant Materials and Growth Conditions
All rice plants (Oryza sativa) were grown in the paddy field of Shanghai Jiao Tong University. The F2 mapping population and MutMap mutants were generated from a cross between mof (O. sativa ssp japonica) and Guang Lu Ai4 (indica). The Nicotiana benthamiana (tobacco) plants were grown in the green house under 16-h-white light/8-h-dark conditions.
Characterization of Mutant Phenotypes
The photography of plant materials and nuclei staining using DAPI, scanning electron microscopy, TEM, and TUNEL were performed according to a previous study (Li et al., 2006).
Antibody Production
The antibodies against OsCenH3, REC8, γH2AX, COM1, RAD51C, PAIR2, PAIR3, and ZEP1 were produced in mice and the latter six in rabbits, as previously described (Nonomura et al., 2006; Shao et al., 2011; Wang et al., 2011; Ji et al., 2012; Tang et al., 2014). All antibodies were prepared by ABclonal of China. The anti-GFP antibody was obtained from Abcam (Catalog No. ab290).
Meiotic Chromosome Preparation and FISH Analysis
Young panicles of mof mutants and the wild type were harvested and fixed with Carnoy’s solution (ethanol:glacial acetic, 3:1). Anthers at the proper stages were squashed in a 45% acetocarmine solution. Slides containing the chromosomes were frozen in liquid nitrogen and then quickly remove the cover slip with a razor blade. Chromosomes were counterstained with DAPI (Vector Laboratories). The FISH procedure was performed as described (Cheng, 2013). The sequence of the telomere FISH probe is (TTTAGGG)7 labeled with Cy3 as described (Mizuno et al., 2006). BAC clone P0671B11 was kindly provided by B. Han (National Center for Gene Research, Shanghai, Institutes for Biological Sciences, Chinese Academy of Sciences).
Immunolocalization Assays
Immunolocalization was conducted as described by a previous study (Wang et al., 2010). Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 20 min at room temperature. Anthers in the proper stages were squashed on a slide with PBS solution and soaked in liquid nitrogen and then the cover slip was quickly removed with a blade. The slides were dehydrated through an ethanol series (70, 90, and 100%) and then incubated in a humid chamber at 37°C for 4 h with different antibody combinations diluted 1:500 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent). After three rounds of washing in PBS, Texas red-conjugated goat anti-rabbit antibody and fluorescein isothiocyanate-conjugated sheep anti-mouse antibody (1:1000) were added to the slides. Finally, after incubation in the humid chamber at 37°C for 30 min, the slides were counterstained with DAPI in antifade solution (Vector Laboratories). The slides were photographed using a Nikon Eclipse Ni-E microscope and analyzed with the NIS-Elements AR software (Nikon).
Cloning of MOF and Complementation
For mapping of the MOF locus, bulked segregant analysis was used. InDel (insertion-deletion) markers were developed according to the sequence difference between the genome sequence of japonica Nipponpare and indica 9311 (Li et al., 2006). Genome sequencing using MutMap (Abe et al., 2012) was performed by Novel Bioinformatics Company (China). For functional complementation, an ∼5.4-kb genomic sequence of MOF (MOF-infusion-1/2; primers listed in Supplemental Table 1) was amplified from the BAC clone OSJNBa0060P14 (kindly provided by B. Han). The genomic fragment was subcloned into the binary vector pCAMBIA1301 (CAMBIA) by In-fusion (In-Fusion HD Cloning Kit; PT5162-1). The MTR1:MOF was constructed by fusing the MOF cDNA to the native MTR1 promoter and then was inserted to the binary vector pCAMBIA1301. MOF:MOF:eGFP was constructed by fusing an eGFP to the C-terminal of MOF cDNA, which was driven by the native MOF promoter and then inserted into the binary vector pCAMBIA1301. Calli induced from young panicles of the homozygous mof plants were used for transformation with Agrobacterium tumefaciens EHA105, as previously described (Li et al., 2006). The primers are listed in Supplemental Table 1.
Phylogenetic Analysis
The amino acid sequences were aligned using MUSCLE 3.6 with the default settings (Edgar, 2004) and then adjusted manually using GeneDoc software (version 2.6.002; http://genedoc.software.informer.com/). Using the MEGA software (version 4.0; http://www.megasoftware.net/index.html) and based on the full-length protein sequences, neighbor-joining trees were constructed with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
RT-qPCR Assays
Total RNA isolation, RT-qPCR procedures, and data calculation were performed as described (Zhang et al., 2010). Total RNA was isolated from rice tissues (root, shoot, leaf, lemma/palea, and anthers) at different stages with the Trizol Reagent kit (Invitrogen) according to the manufacturer’s protocol. The stages of rice anthers were classified according to a previous article (Zhang and Wilson, 2009). The first-strand cDNAs were synthesized using MLV reverse transcriptase (Fermentas). Quantitative real-time PCR analyses were performed on a CFX96 (Bio-Rad) using a SYBR green detection protocol according to the manufacturer’s instructions. All reactions were performed in three separate experiments.
In Situ Hybridization
RNA in situ hybridizations were performed as described by Zhang et al. (2010). Two MOF cDNA fragments generated by PCR were used for preparing antisense and sense probes under the T7 promoter with RNA polymerase using the DIG RNA labeling kit (Roche) (Supplemental Table 1).
Yeast Two-Hybrid Assay
The yeast two-hybrid assays were performed using the Matchmaker Yeast Two-Hybrid System (Clontech). All protocols were performed according to the manufacturer’s user manual. Plasmids pGADT7 and pGBKT7 were used as the bait and prey vectors. The yeast strain used was AH109.
BiFC Assay
The cDNA of MOF was cloned into the pSAT-cEYFP-c1 vector, and the cDNA of O. sativa SKP1-like (OSK1) was cloned into the pSAT-nEYFP-c1 vector. BiFC assays followed the described protocol (Zhang et al., 2011).
Coimmunoprecipitation
The cDNA of MOF was cloned into the pGREEN-HA vector, and the cDNA of O. sativa SKP1-like (OSK1) was cloned into the pGREEN-FLAG vector (pGREEN vector was kindly provided by H. Yu, Department of Biological Sciences, National University of Singapore). Coimmunoprecipitation experiments were performed as described (Liu et al., 2012). The protein from the N. benthamiana leaves coinfiltrated with the appropriate plasmids was extracted with native extraction buffer as described by Liu et al. (2012). A 15-μL volume of anti-FLAG antibody (ABclonal) was added to 1 mL of cell lysates. Then, binding was performed at 4°C for 4 h with gentle shaking and was followed by the addition of 20 μL of protein G agarose beads (Roche). After 2 h of incubation at 4°C, the precipitated samples were washed, separated by SDS-PAGE, and subjected to immunoblot analysis using anti-HA antibody (ABclonal) and anti-FLAG antibody (ABclonal).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MOF (Os04g0464966), OSK1 (Os11g0456300), OSK8 (Os11g0707700), OSK19 (Os07g0625600), OSK20 (Os09g0539500), OSK21 (Os07g0409500), OSK22 (Os07g0625500), OSK24 (Os12g0594600), and MTR1 (Os02g0491300). Accession numbers for the sequences used in the phylogenetic analysis are listed on the tree.
Supplemental Data
Supplemental Figure 1. TEM of Wild-Type and mof Anthers.
Supplemental Figure 2. Distribution of Prophase I Cells in Wild-Type and mof Anthers.
Supplemental Figure 3. DNA Fragmentation Assay Analysis of Wild-Type and mof Anthers.
Supplemental Figure 4. Nucleotide and Amino Acid Sequences of MOF.
Supplemental Figure 5. Complementation Analysis of mof.
Supplemental Figure 6. Expression Pattern of OSK Genes in Rice.
Supplemental Figure 7. Yeast Two-Hybrid Assay for Interaction of MOF with Selected OSK Proteins.
Supplemental Figure 8. Protein Levels of Meiosis-Related Components in Wild-Type and mof Plants.
Supplemental Figure 9. Dual Immunolocalization of MOF, γH2AX, COM1, and RAD51C in Male Meiocytes of the Complemented Transgenic Line.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Figure 6C.
Supplementary Material
Acknowledgments
We thank Bin Han (Rice Genome Resource Center) and Hao Yu for providing BAC clone, cDNA clone, and pGREEN vector. We thank Zhukuan Cheng for providing RAD51C polyclonal antibody. We thank Zhijin Luo, Mingjiao Chen, and Zibo Chen for mutant screening and generation of F2 populations for the mapping. This work was supported by funds from the National Key Basic Research Developments Program, Ministry of Science and Technology, China (2013CB126902); National Key Technologies Research and Development Program of China (2016YFD0100804); National Natural Science Foundation of China (31322040); National Transgenic Major Program Grants (2016ZX08009003-003-007); China Innovative Research Team, Ministry of Education, and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016); and the Science and Technology Commission of Shanghai Municipality (Grant 13JC1408200).
AUTHOR CONTRIBUTIONS
W.L. and D.Z. designed the experiments. Y.H. carried out most of the experiments. C.W. and J.Y. assisted in TUNEL, FISH, and immunolocalization analysis. J.Z. analyzed the high-throughput sequencing data by using MutMap. P.L. and J.D.H. assisted in analyzing the data. Y.H., J.D.H., and W.L. wrote the article.
Glossary
- DSB
double-strand break
- TEM
transmission electron microscopy
- DAPI
4′,6-diamidino-2-phenylindole
- SC
synaptonemal complex
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- FISH
fluorescent in situ hybridization
- BiFC
bimolecular fluorescence complementation
Footnotes
Articles can be viewed without a subscription.
References
- Abdu U., Brodsky M., Schüpbach T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12: 1645–1651. [DOI] [PubMed] [Google Scholar]
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Aklilu B.B., Soderquist R.S., Culligan K.M. (2014). Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 42: 3104–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X.J., Deng Z.Y., Wang T. (2011). OsSpo11-4, a rice homologue of the archaeal TopVIA protein, mediates double-strand DNA cleavage and interacts with OsTopVIB. PLoS One 6: e20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Zickler D. (2004). Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Borde V., de Massy B. (2013). Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23: 147–155. [DOI] [PubMed] [Google Scholar]
- Bose R., Manku G., Culty M., Wing S.S. (2014). Ubiquitin-proteasome system in spermatogenesis. Adv. Exp. Med. Biol. 759: 181–213. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Jones G.H., Franklin F.C.H. (2003). Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54: 25–38. [DOI] [PubMed] [Google Scholar]
- Chang Y., Gong L., Yuan W., Li X., Chen G., Li X., Zhang Q., Wu C. (2009). Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol. 151: 2162–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L., Tang D., Wang K., Wang M., Zhu K., Yu H., Gu M., Cheng Z. (2011). OsAM1 is required for leptotene-zygotene transition in rice. Cell Res. 21: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chu H., Yuan Z., Pan A.h., Liang W., Huang H., Shen M., Zhang D., Chen L. (2006). Isolation and genetic analysis for rice mutants treated with 60 Co γ-Ray. J. Xiamen Univ. 45: 82–85. [Google Scholar]
- Chen Z.J., Sun L.J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33: 275–286. [DOI] [PubMed] [Google Scholar]
- Cheng Z. (2013). Analyzing meiotic chromosomes in rice. Methods Mol. Biol. 990: 125–134. [DOI] [PubMed] [Google Scholar]
- Edelmann W., et al. (1996). Meiotic pachytene arrest in MLH1-deficient mice. Cell 85: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous M., Higgins J.D., Osman K., Lambing C., Roitinger E., Mechtler K., Armstrong S.J., Perry R., Pradillo M., Cuñado N., Franklin F.C. (2012). Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8: e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Jackson S.P. (2012). RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M.O. (2000). A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479. [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M.S., Tsai P.I., Chien C.T. (2006). F-box proteins: the key to protein degradation. J. Biomed. Sci. 13: 181–191. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Börner G.V., Lichten M., Kleckner N. (2001). Gamma-H2AX illuminates meiosis. Nat. Genet. 27: 236–238. [DOI] [PubMed] [Google Scholar]
- Inagaki A., Schoenmakers S., Baarends W.M. (2010). DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 5: 255–266. [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., Kapoor S., Tyagi A.K., Khurana J.P. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Tang D., Wang K., Wang M., Che L., Li M., Cheng Z. (2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72: 18–30. [DOI] [PubMed] [Google Scholar]
- Ji J., Tang D., Wang M., Li Y., Zhang L., Wang K., Li M., Cheng Z. (2013). MRE11 is required for homologous synapsis and DSB processing in rice meiosis. Chromosoma 122: 363–376. [DOI] [PubMed] [Google Scholar]
- Kahloul S., HajSalah El Beji I., Boulaflous A., Ferchichi A., Kong H., Mouzeyar S., Bouzidi M.F. (2013). Structural, expression and interaction analysis of rice SKP1-like genes. DNA Res. 20: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kopanja D., Roy N., Stoyanova T., Hess R.A., Bagchi S., Raychaudhuri P. (2011). Cul4A is essential for spermatogenesis and male fertility. Dev. Biol. 352: 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Halaby M.J., Hakem A., Cardoso R., El Ghamrasni S., Harding S., Chan N., Bristow R., Sanchez O., Durocher D., Hakem R. (2010). Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 207: 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chang Y., Xin X., Zhu C., Li X., Higgins J.D., Wu C. (2013). Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice. Plant Cell 25: 3885–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.C., Barringer B.C., Barbash D.A. (2009). The pachytene checkpoint and its relationship to evolutionary patterns of polyploidization and hybrid sterility. Heredity (Edinb) 102: 24–30. [DOI] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. (2007). Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhao Q., Xie Q. (2012). In vivo ubiquitination assay by agroinfiltration. Methods Mol. Biol. 876: 153–162. [DOI] [PubMed] [Google Scholar]
- Lu C., Kim J., Fuller M.T. (2013). The polyubiquitin gene Ubi-p63E is essential for male meiotic cell cycle progression and germ cell differentiation in Drosophila. Development 140: 3522–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. (2011). More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Luo Q., Li Y., Shen Y., Cheng Z. (2014). Ten years of gene discovery for meiotic event control in rice. J. Genet. Genomics 41: 125–137. [DOI] [PubMed] [Google Scholar]
- Macdonald L.D., Knox A., Hansen D. (2008). Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180: 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Hochwagen A. (2011). Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends Cell Biol. 21: 393–400. [DOI] [PubMed] [Google Scholar]
- McKee A.H., Kleckner N. (1997a). A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A.H., Kleckner N. (1997b). Mutations in Saccharomyces cerevisiae that block meiotic prophase chromosome metabolism and confer cell cycle arrest at pachytene identify two new meiosis-specific genes SAE1 and SAE3. Genetics 146: 817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66: 297–327. [DOI] [PubMed] [Google Scholar]
- Metzger M.B., Hristova V.A., Weissman A.M. (2012). HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H., Wu J., Kanamori H., Fujisawa M., Namiki N., Saji S., Katagiri S., Katayose Y., Sasaki T., Matsumoto T. (2006). Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J. 46: 206–217. [DOI] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Eiguchi M., Suzuki T., Kurata N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119: 217–225. [DOI] [PubMed] [Google Scholar]
- Nonomura K., Eiguchi M., Nakano M., Takashima K., Komeda N., Fukuchi S., Miyazaki S., Miyao A., Hirochika H., Kurata N. (2011). A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet. 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Higgins J.D., Sanchez-Moran E., Armstrong S.J., Franklin F.C. (2011). Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190: 523–544. [DOI] [PubMed] [Google Scholar]
- Pastink A., Eeken J.C., Lohman P.H. (2001). Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 480-481: 37–50. [DOI] [PubMed] [Google Scholar]
- Pawlowski W.P., Wang C.J., Golubovskaya I.N., Szymaniak J.M., Shi L., Hamant O., Zhu T., Harper L., Sheridan W.F., Cande W.Z. (2009). Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc. Natl. Acad. Sci. USA 106: 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503–533. [DOI] [PubMed] [Google Scholar]
- Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1: 697–705. [DOI] [PubMed] [Google Scholar]
- Prinz S., Amon A., Klein F. (1997). Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. (1997). Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder G.S., Bailis J.M. (2000). The pachytene checkpoint. Trends Genet. 16: 395–403. [DOI] [PubMed] [Google Scholar]
- Ryu K.Y., Sinnar S.A., Reinholdt L.G., Vaccari S., Hall S., Garcia M.A., Zaitseva T.S., Bouley D.M., Boekelheide K., Handel M.A., Conti M., Kopito R.R. (2008). The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol. Cell. Biol. 28: 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Ishibashi T., Uchiyama Y., Iwabata K. (2009). The multi-replication protein A (RPA) system--a new perspective. FEBS J. 276: 943–963. [DOI] [PubMed] [Google Scholar]
- Schaeper U., Subramanian T., Lim L., Boyd J.M., Chinnadurai G. (1998). Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273: 8549–8552. [DOI] [PubMed] [Google Scholar]
- Shao T., Tang D., Wang K., Wang M., Che L., Qin B., Yu H., Li M., Gu M., Cheng Z. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz K., Lannoo N., Van Damme E. (2015). Plant F-box proteins: judges between life and death. Crit. Rev. Plant Sci. 34: 523–552. [Google Scholar]
- Tan H., Liang W., Hu J., Zhang D. (2012). MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 22: 1127–1137. [DOI] [PubMed] [Google Scholar]
- Tang D., Miao C., Li Y., Wang H., Liu X., Yu H., Cheng Z. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.H., McKee B.D. (2011). Homologous pairing and the role of pairing centers in meiosis. J. Cell Sci. 124: 1955–1963. [DOI] [PubMed] [Google Scholar]
- Vyas R., et al. (2013). RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 20: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. (2007). Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA 104: 20759–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Qin B., Li M., Cheng Z. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang K., Tang D., Wei C., Li M., Shen Y., Chi Z., Gu M., Cheng Z. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang M. (2006). The ARABIDOPSIS SKP1-LIKE1 (ASK1) protein acts predominately from leptotene to pachytene and represses homologous recombination in male meiosis. Planta 223: 613–617. [DOI] [PubMed] [Google Scholar]
- Ward J.O., Reinholdt L.G., Motley W.W., Niswander L.M., Deacon D.C., Griffin L.B., Langlais K.K., Backus V.L., Schimenti K.J., O’Brien M.J., Eppig J.J., Schimenti J.C. (2007). Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., Schnittger A. (2013). Control of the meiotic cell division program in plants. Plant Reprod. 26: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.M., Cheun A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44: 267–281. [DOI] [PubMed] [Google Scholar]
- Yang M., Hu Y., Lodhi M., McCombie W.R., Ma H. (1999). The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Timofejeva L., Ma H., Makaroff C.A. (2006). The Arabidopsis SKP1 homolog ASK1 controls meiotic chromosome remodeling and release of chromatin from the nuclear membrane and nucleolus. J. Cell Sci. 119: 3754–3763. [DOI] [PubMed] [Google Scholar]
- Zhang D., Liang W. (2016). Pushing the Boundaries of Scientific Research: 120 Years of Addressing Global Issues. (Washington, DC: Science/AAAS; ). [Google Scholar]
- Zhang H., Liang W., Yang X., Luo X., Jiang N., Ma H., Zhang D. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang M., Tang D., Li Y., Xu M., Gu M., Cheng Z., Yu H. (2015). XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis. J. Exp. Bot. 66: 5713–5725. [DOI] [PubMed] [Google Scholar]
- Zhang D., Wilson Z. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 2342–2353. [Google Scholar]
- Zhang W., Yi C., Bao W., Liu B., Cui J., Yu H., Cao X., Gu M., Liu M., Cheng Z. (2005). The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 139: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Yang X., Quan L., Timofejeva L., Rigel N.W., Ma H., Makaroff C.A. (2006). ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol. Biol. 62: 99–110. [DOI] [PubMed] [Google Scholar]
- Zheng N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1998). The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.