In the parasitic plant Phtheirospermum japonicum, the auxin biosynthesis gene YUCCA3 plays a pivotal role in the formation of haustoria, which invade host tissues and facilitate nutrient acquisition.
Abstract
Parasitic plants in the Orobanchaceae cause serious agricultural problems worldwide. Parasitic plants develop a multicellular infectious organ called a haustorium after recognition of host-released signals. To understand the molecular events associated with host signal perception and haustorium development, we identified differentially regulated genes expressed during early haustorium development in the facultative parasite Phtheirospermum japonicum using a de novo assembled transcriptome and a customized microarray. Among the genes that were upregulated during early haustorium development, we identified YUC3, which encodes a functional YUCCA (YUC) flavin monooxygenase involved in auxin biosynthesis. YUC3 was specifically expressed in the epidermal cells around the host contact site at an early time point in haustorium formation. The spatio-temporal expression patterns of YUC3 coincided with those of the auxin response marker DR5, suggesting generation of auxin response maxima at the haustorium apex. Roots transformed with YUC3 knockdown constructs formed haustoria less frequently than nontransgenic roots. Moreover, ectopic expression of YUC3 at the root epidermal cells induced the formation of haustorium-like structures in transgenic P. japonicum roots. Our results suggest that expression of the auxin biosynthesis gene YUC3 at the epidermal cells near the contact site plays a pivotal role in haustorium formation in the root parasitic plant P. japonicum.
INTRODUCTION
Most plants in the Orobanchaceae have adopted a parasitic lifestyle. Facultative parasites maintain their capability for autotrophic lifestyle, but often parasitize when a host is nearby. In contrast, obligate parasites are not able to complete their lifecycle without a host, at least under natural conditions. Several obligate parasites in the Orobanchaceae, such as witchweeds (Striga) and broomrapes (Orobanche and Phelipanche), are harmful agricultural pests that infect important crops and severely affect host growth and yields by depriving their hosts of water and nutrients. Significant economic losses place the parasitic plant Striga hermonthica as one of the most serious threats to food security (Pennisi, 2010). Fields infested by Striga species affect the agricultural income of 25 countries, causing billions of USD in damage yearly (reviewed in Spallek et al., 2013).
A common characteristic of parasitic plants is the formation of haustoria, i.e., multicellular organs that attach to and penetrate the host tissues. During the early stage of parasitization, the haustorium serves as the organ of penetration, establishing physical connections between parasite and host tissues. During later developmental stages, vascular cells develop in the haustorium, which absorb water and carbohydrates from the host (reviewed in Yoshida and Shirasu, 2012). Host-derived chemicals are able to induce the formation of haustoria in vitro; these chemicals are called haustorium-inducing factors (HIFs) (Lynn and Chang, 1990). Among these HIFs, 2,6-dimethoxy-p-benzoquinone (DMBQ) was originally identified from host root extracts and was shown to activate haustorium formation in parasite roots (Chang and Lynn, 1986; Smith et al., 1990). Although haustoria can be observed in different parts of the root, the area between the distal elongation zone and the meristematic root tip is the most sensitive to HIFs (Cui et al., 2016; Baird and Riopel, 1984). Typically, this area becomes swollen and forms a haustorium ∼24 h after DMBQ treatment (Albrecht et al., 1999; Ishida et al., 2011).
The morphological changes that occur during haustorium development have been described histologically in several parasitic plant species (Baird and Riopel, 1984; Ishida et al., 2011; Riopel and Musselman, 1979; Jamison and Yoder, 2001; Cui et al., 2016). In the facultative parasite Agalinis purpurea, cortex cells slightly expand radially 6 h after exposure to HIF. By 10 h, epidermal cells start dividing and rapidly differentiate into haustorial hairs. Between 24 and 36 h, the root continues to swell due to cell division and enlargement to form the haustorial structure. Similar morphological events were observed in other facultative parasitic plants in the Orobanchaceae, such as Triphysaria versicolor (Jamison and Yoder, 2001) and Phtheirospermum japonicum (Cui et al., 2016; Ishida et al., 2011), suggesting a conserved mechanism for haustorium induction in this plant family.
The molecular mechanism triggering the initiation of haustorium development is poorly understood. Redox signaling was suggested to play an essential role in this process. In addition to DMBQ, a range of quinones and flavonoids that have certain redox potentials are able to induce haustorium formation in vitro in T. versicolor (Matvienko et al., 2001b; Albrecht et al., 1999). A single electron-transferring quinone-reductase enzyme in T. versicolor (QR1) was found to play an important role in haustorium induction probably, via catalyzing the reduction of DMBQ-related quinones (Bandaranayake et al., 2010). In addition, Pirin, a nuclear protein associated with transcription factor activity, was reported to play regulatory roles in haustorium formation in T. versicolor (Bandaranayake et al., 2012). Accumulating information about the genes expressed in the parasitic Orobanchaceae, including ESTs of S. hermonthica (Yoshida et al., 2010), Phelipanche aegyptiaca, and T. versicolor (http://ppgp.huck.psu.edu/) (Yang et al., 2014; Torres et al., 2005), has contributed to our knowledge of the molecular basis of plant parasitism. Recent significant advances in this field include the identification of core parasitism genes through the sequencing of different tissues of three parasitic species in Orobanchaceae (Yang et al., 2014). However, our understanding of early haustorium development is still limited.
Early events in haustorium development involve hormonal accumulation and regulation. Haustorial tissues of the parasite Santalum album accumulate newly synthesized endogenous hormones, associating high auxin-to-cytokinin ratios with haustorium development (Zhang et al., 2012). Indeed, disruption of the auxin concentration gradients, either through the application of auxin transport/auxin activity inhibitors or by providing an excess amount of exogenous auxin, results in a reduction of infection frequency in the nonchlorophillic holoparasite P. aegyptiaca (Bar-Nun et al., 2008). Similar studies in the facultative parasitic plant T. versicolor have shown that disturbing auxin flow either by application of an auxin efflux inhibitor (2,3,5-triiodobenzoic acid) or auxin activity inhibitors (p-chlorophenoxyisobutyric acid) reduces the number of haustorium (Tomilov et al., 2005). Furthermore, T. versicolor roots transformed with ethylene- or auxin-inducible promoter-reporter constructs showed a positive response after DMBQ treatment, suggesting these hormones accumulate near the root tip (Tomilov et al., 2005). Auxin distribution in planta determines the formation patterns of organs such as leaves, flowers, and lateral roots (Reinhardt et al., 2000; Benková et al., 2003). Auxin gradients in tissues are largely modulated by the intercellular auxin transporters PIN proteins and AUX/LAX proteins (reviewed in Adamowski and Friml, 2015; Swarup and Péret, 2012). In addition, de novo biosynthesis of auxin at specific tissues significantly contributes to plant development (Zhao, 2010). The main pathway of auxin biosynthesis involves two steps. The TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS family of amino transferases first converts tryptophan to indole-3-pyruvic acid (Kasahara, 2016). Subsequently, the YUCCA (YUC) family of flavin monooxygenases catalyzes the formation of indole-3-acetic acid (IAA) (Mashiguchi et al., 2011; Zhao, 2012). The rate-limiting step for IAA production is regulated by YUC (Zhao et al., 2001). There are 11 YUC gene family members in the Arabidopsis thaliana genome and 14 in rice (Oryza sativa) (Yamamoto et al., 2007; Fujino et al., 2008). Disruption of a single YUC gene in Arabidopsis did not cause an apparent phenotype, whereas triple or quadruple yuc mutants show severe defects in various developmental processes, suggesting that spatially and temporally regulated auxin biosynthesis by multiple YUC genes is essential for plant development (Cheng et al., 2006).
In this study, we examined the facultative parasitic plant P. japonicum to identify differentially regulated genes during the initial stages of haustorium development. Since a transformation system was first established for P. japonicum (Ishida et al., 2011), this plant has served as an excellent model for parasitic plants. Our sequencing and microarray analyses identified 327 differentially expressed genes after DMBQ treatment, providing an overview of expression profile changes during early haustorium development. Among the differentially expressed genes, we focused on a highly upregulated gene, YUC3, encoding a member of the YUCCA flavin monooxygenase family that regulates auxin biosynthesis. Our functional studies indicate that YUC3 is a key regulator of haustorium formation in P. japonicum.
RESULTS
High-Throughput Sequencing and de Novo Assembly of the P. japonicum Root Transcriptome
To prepare for large-scale transcriptome analysis of the facultative parasite P. japonicum, we first tested its host specificity. P. japonicum is an annual herbaceous plant that grows to be 10 to 30 cm tall, with a generation time of 2 to 3 months (Figure 1A). P. japonicum is a generalist that can parasitize a number of species including rice, Arabidopsis, maize (Zea mays), and cowpea (Vigna unguiculata), but not Lotus japonicus (Figure 1; Supplemental Figure 1). We chose rice and Arabidopsis for further experiments because the complete genome sequences and molecular genetic tools are available for both species. We used rice for the de novo transcriptome analysis because P. japonicum haustoria from infected rice are larger than those from infected Arabidopsis, which increases the ease of sampling. Parasites with or without host were incubated in a rhizotron chamber for 4 to 6 weeks (Supplemental Figure 2). Parasites form lateral haustoria with host plants during various developmental stages. The newly formed P. japonicum roots frequently had immature haustoria (Figures 1F and 1G), in which the connection to the host vascular system was not yet established (Figure 1G). A haustorium in older roots was firmly attached to the host tissues (Figures 1H and 1I), forming vascular continuity with the host (Figure 1I). To sequence samples from distinct parasitic stages, a pool of haustoria tissues at different stages was carefully collected. When a haustorium was tightly attached to the host root, the attached host tissues were also sampled. As a control, parasite roots without host plants were similarly sampled.
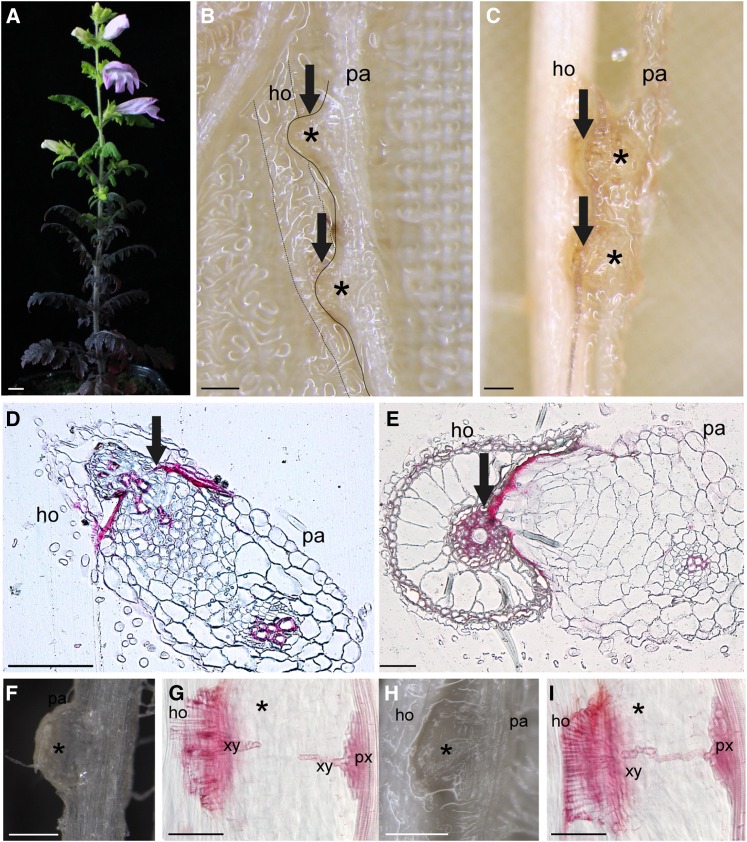
Figure 1.
P. japonicum Infecting Host Roots.
(A) Three-month-old P. japonicum. Bar = 1 cm.
(B) and (C) Interaction of P. japonicum root with Arabidopsis root (B) and rice root (C). Bars = 200 µm.
(D) and (E) Double-stained cross section of P. japonicum haustoria. Technovit 7200-embedded tissues were cross-sectioned at 5 weeks after host interaction and stained with safranin-O and Fast Green. P. japonicum penetrating Arabidopsis (D) and rice (E). Bar = 50 µm.
(F) Example of a haustorium at the immature stage at 3 dpi.
(G) Safranin-O-stained haustorium at 3 dpi. Note the vasculature connection has not yet been established.
(H) Haustorium at a later stage when the parasite is already tightly connected to the host rice root at 7 dpi.
(I) Safranin-O-stained haustorium at 7 dpi in rice interaction. The xylem bridge has been established. Bars = 250 µm in (F) to (I). pa, parasite; ho, host roots; xy, xylem bridge; px, plate xylem. Asterisks indicate haustoria. Arrows indicate the interface between a host and a parasite.
To perform de novo assembly of the P. japonicum transcriptome, two distinct sequencing platforms were adopted: the HiSeq 2000 (Illumina) and the GS-FLX pyrosequencer with FLX titanium reagents (454 Life Sciences). Total RNAs were extracted from autotrophic and parasitic stages of P. japonicum and respective Illumina libraries were generated. For 454 sequencing, a normalized library was constructed with mRNAs isolated from each sample and mixed in equal proportions. Approximately 24 million paired-end reads with a total length of ∼4.5 Gb were generated by the HiSeq 2000, whereas 888,638 reads with an average length of 333.8 bp and a total length of 296 Mb were generated by the 454 sequencer (Supplemental Table 1). The reads from parasitic tissues were mapped against the available full-length rice transcripts to remove reads derived from host tissues. The unmapped reads and the reads from the autotrophic stage libraries were then assembled. The assembly yielded 42 Mb distributed in 58,137 unigenes (nonredundant sequences; Supplemental Data Set 1). Among these, 198 unigenes were removed as likely rice contamination based on BLASTn analysis with e-value 1e−20 and 100% identity with rice sequences (Supplemental Data Set 1 and Supplemental Table 2). The final 57,939 unigenes had an average size of 812 bp and N50 contig length (the median contig length, the sum of the contig length larger than that which covers 50% of the entire assembly) of 1090 bp. Among unigenes, 47,817 (82.5%) included a putative coding region of more than 50 amino acids, as detected by ESTScan (version 2.0), and 17,044 (35.6% of protein coding sequences) had a predicted start codon (Supplemental Table 3).
Next, we assessed the coverage rate of the P. japonicum transcriptome by examining the proportion of conserved orthologous genes in the assembled data set. We used ultraconserved orthologs, which include 357 highly conserved single-copy genes detected in representative eukaryotes, Arabidopsis, humans, mice, yeast, fruit flies, and Caenorhabditis elegans, and APVO (Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, and Oryza sativa) sequences, which consist of 959 single-copy genes present in all four plant genomes (Duarte et al., 2010) for this assessment. We detected 100% of ultraconserved orthologs (with a threshold blast e-value 1e-10) and 95% of APVO sequences in the P. japonicum transcriptome assembly, indicating the high coverage rates of P. japonicum transcripts.
Local alignment using the BLASTx algorithm (cutoff e-value of 1e-10) showed that 34,897 (60.23%) and 37,259 (64.30%) of P. japonicum unigenes have homologous sequences in the Arabidopsis genome (TAIR10) and the nonredundant (nr) databases, respectively (Supplemental Table 3 and Supplemental Data Set 1). The remaining 20,546 are orphan sequences that have no BLASTx hits in the nr database. Among these, 2785 had at least 2 times higher expression values (measured as reads per kilobase of transcript per million reads [RPKM]) in the parasitic stages than in nonparasitizing roots, and 2481 sequences were specifically expressed in parasitic stages, indicating that these orphans found in the P. japonicum transcriptome are expressed genes, many of which may function in parasitic stages (Supplemental Data Set 1).
Identification and Characterization of P. japonicum Genes Induced by DMBQ
To identify genes involved in haustorium formation, we designed a customized microarray based on the P. japonicum transcriptome assembly. P. japonicum roots were treated with DMBQ, and gene transcriptional levels were analyzed by microarray hybridization. P. japonicum haustoria showed similar developmental sequences in response to DMBQ or host root exudate treatment (Figures 2A and 2B). The morphological changes in the roots were not apparent until 6 h after treatments, and initiation of haustorial hair proliferation was recognized at 12 h after treatments. Haustorial hair proliferation and root swelling became more apparent at 24 h after treatments (Figures 2A and 2B; Supplemental Movie 1). We chose eight time points across 48 h (0, 0.5, 1, 3, 6, 12, 24, and 48 h) for the microarray analysis to focus on the transcriptional changes that occur prior to morphological changes. We detected 327 differentially expressed genes that showed 2-fold higher or lower expression compared with 0 h samples (Supplemental Data Set 2). To validate the microarray results, we performed RT-qPCR analyses of several selected genes. For an internal control to normalize the expression values, the P. japonicum gene encoding the RNA binding polypyrimidine tract binding protein (PTB) was selected because of its homology to Arabidopsis housekeeping gene At3g01150 (Czechowski et al., 2005) and its stable expression levels across all eight time points in the microarray (Supplemental Data Set 3). Expression profiles of selected genes generated with RT-qPCR showed similar patterns to those obtained from microarray analysis (Supplemental Figure 3, left panels), verifying the reproducibility of the microarray data.
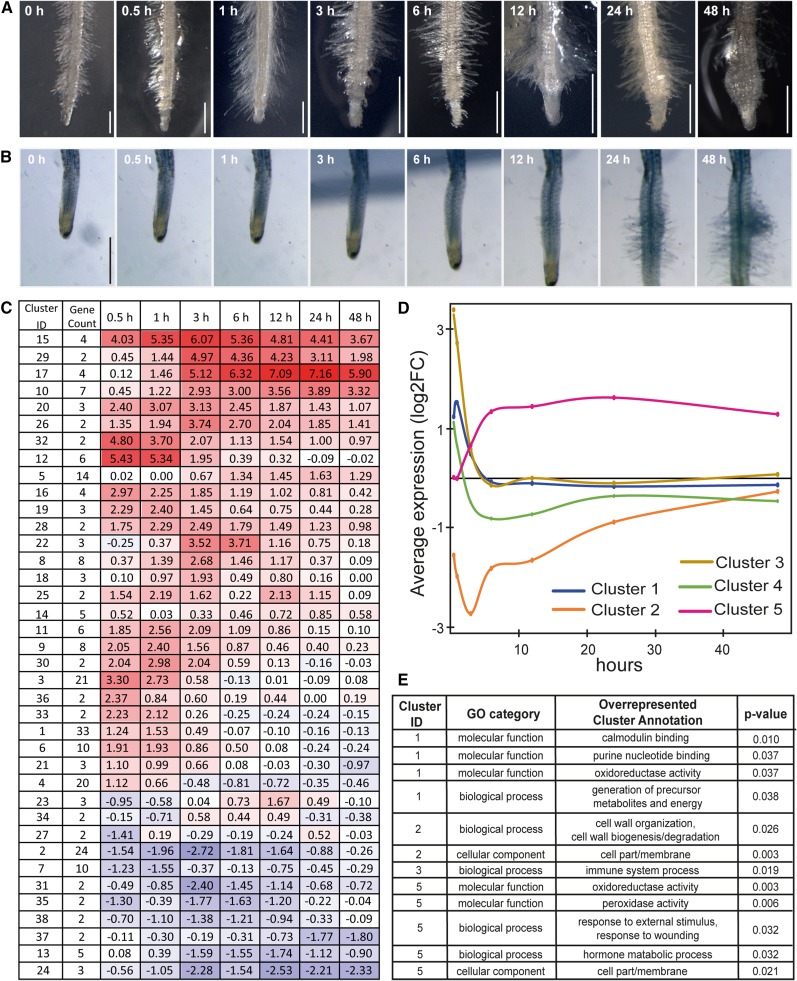
Figure 2.
Changes in Root Morphology and Gene Expression in Response to DMBQ Treatment.
(A) Morphological changes in P. japonicum roots exposed to 10 µM DMBQ.
(B) Time-lapse photographs of the haustorium after treatment with rice root exudate. The full-length movie is shown in Supplemental Movie 1. Bars = 500 µm.
(C) The average transcript abundance for all DE genes at each time point across three biological replications. The first column contains the unique cluster ID (Cluster ID) and the second column contains the number of unigenes belonging to each cluster (Gene Count). Colored boxes highlight the averaged expression values shown as log2 FC (fold change between DMBQ-treated/control [0 h]) across three biological replicates throughout the time course. Colors are based on a scale with red indicating upregulation and blue indicating downregulation. Rows were placed manually according to the cluster similarity. The unigenes in each cluster are listed in Supplemental Data Set 2.
(D) A graph of the expression profiles of the five largest clusters.
(E) Enriched gene ontology terms according to Fisher’s exact test (Benjamini and Hochberg-corrected FDR < 0.05).
Because chemical HIFs and host root exudate can induce the expression of different sets of genes as reported previously (Bandaranayake et al., 2010), we investigated the expression profiles of the DMBQ-induced P. japonicum genes after application of rice root exudate (Supplemental Figure 3, right panels). As shown in Supplemental Figure 4 and Supplemental Movie 1, rice root exudate initiates haustoria formation in P. japonicum roots. The tested gene set showed similar gene expression patterns between DMBQ-treated and host root exudate-treated roots (Supplemental Figure 3), confirming that the gene expression profiles in the DMBQ-treated samples most likely reflects gene expression during early haustorium development.
Functional Classification of P. japonicum Genes with Altered Expression Patterns in Response to the Haustoria-Inducing Factor DMBQ
To obtain an overview of the expression profiles of the P. japonicum transcriptome during haustorium formation, we grouped DMBQ-regulated genes with similar expression profiles (Olex and Fetrow, 2011). Among the 327 differentially expressed genes, 238 were divided into 38 clusters with two or more members (Supplemental Data Set 2 and Supplemental Figures 5 to 7). For each cluster, the average gene expression values (fold changes against the control sample) was calculated and displayed as a heat map (Figure 2C). The expression of 21 clusters comprising 157 unigenes peaked at very early time points (up to 3 h; Supplemental Figure 5), while the expression of 14 clusters with 73 genes peaked after 3 h (Supplemental Figure 6). Three clusters contained eight genes that were negatively modulated along the time course (Supplemental Figure 7).
Notably, the expression of genes in Cluster 15 peaked at 3 h after DMBQ treatment and showed one of the highest (log2FC >6) increases in expression among all clusters (Figure 2C; Supplemental Figure 5). This cluster includes homologs of Tv-Pirin and Tv-QR2 (Quinone reductase2), which are expressed in response to DMBQ in T. versicolor (Bandaranayake et al., 2012; Matvienko et al., 2001b), confirming the conservation of gene expression profiles among different Orobanchaceae species. Interestingly, the closest homolog of Tv-QR1, which functions in haustorium formation in T. versicolor, was not induced by DMBQ in P. japonicum (Supplemental Figure 8).
To investigate functional shifts in the gene population, Gene Ontology (GO) terms were assigned for each gene based on the best BLAST-hit annotations. The overrepresented GO terms were identified in the top five most abundant clusters, which cover 102 (31%) differentially expressed genes (Figures 2D and 2E). Cluster 1 contained 33 early-responsive genes with a peak of expression at 1 h, and the GO terms calmodulin binding and oxidoreductase activity were overrepresented (Figure 2E). Similarly, Clusters 3 and 4 also contained early-responsive genes whose expression peaks at 30 min after DMBQ treatment, followed by a rapid decrease in expression levels (Figure 2D). Although no particular GO term was overrepresented for Cluster 4, the term immune system process was significantly enriched for Cluster 3 (Figure 2E), including genes annotated as WRKY transcription factor family genes; members of this family are often associated with responses to abiotic and biotic stresses (Eulgem et al., 2000). Thus, DMBQ treatment may have provoked stress responses in parasitic plants. In contrast, Cluster 2 contained 24 unigenes whose expression was rapidly repressed after DMBQ treatment. The GO terms cell wall organization and membrane were overrepresented in this cluster (Figure 2E), probably because root elongation was halted upon DMBQ treatment (Riopel and Timko, 1995).
Cluster 5 consisted of late-responsive genes, with high expression levels observed at 6 h and sustained beyond 6 h (Figure 2D). The overrepresented GO terms in this cluster were oxidoreductase activity, peroxidase activity, response to external stimulus, response to wounding, and hormone metabolic pathway (Figure 2E). In particular, for hormone metabolic pathway, we found genes encoding CYTOKININ OXIDASE3, which catalyzes the degradation of cytokinin (Bartrina et al., 2011), as well as YUCCA and GH3 (Supplemental Data Set 2), which are involved in auxin metabolism (Kasahara, 2016). In addition, a member of the AUX/IAA transcription factor family was also grouped in Cluster 5 (Figure 2D; Supplemental Data Set 2). Finally, Cluster 17, which included an auxin responsive gene encoding gibberellin 2-β-dioxygenase 2 (GA2ox2) (Yamaguchi, 2008; Frigerio et al., 2006) showed a similar gene expression pattern to that of Cluster 5 (Supplemental Figure 6 and Supplemental Data Set 2). Thus, our expression analysis suggests that auxin may regulate multiple aspects of root cell reprogramming required for haustorium development.
A YUC Gene Is Upregulated during Haustorium Formation
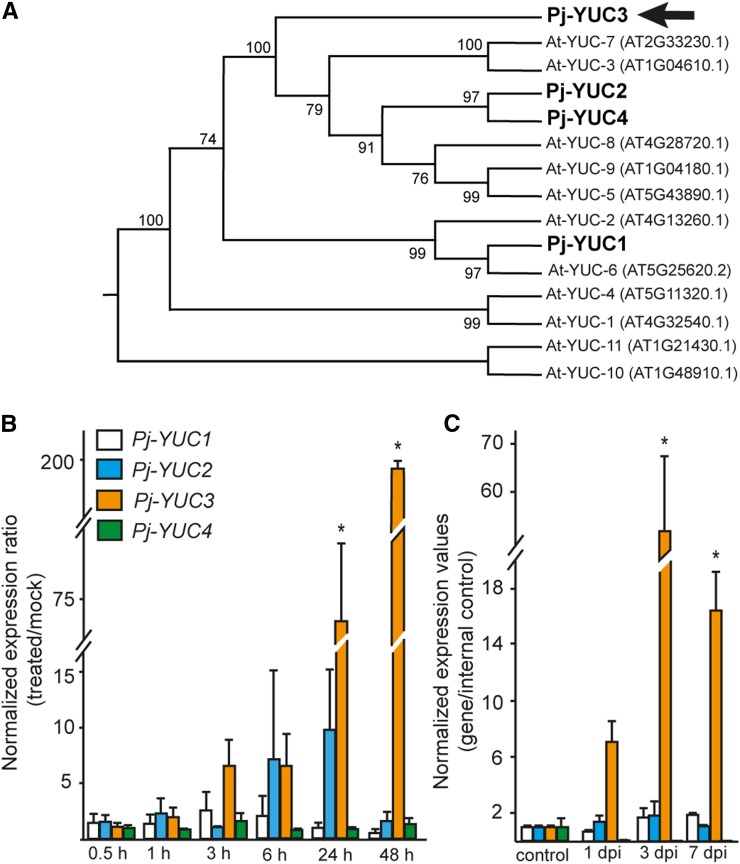
Given that a P. japonicum homolog of Arabidopsis YUC genes, which encode YUCCA enzymes that catalyze a rate-limiting step in auxin biosynthesis (Mashiguchi et al., 2011), was highly expressed during haustorium formation, we decided to further characterize this gene. First, we searched in the P. japonicum transcriptome for homologs of Arabidopsis YUC genes. Four YUC homologs, designated as P. japonicum YUC1, YUC2, YUC3, and YUC4, were identified in the P. japonicum transcriptome assembly, and their full-length sequences were determined (Supplemental Figure 9). Phylogenetic analysis revealed that P. japonicum YUC2, YUC3, and YUC4 are clustered together with Arabidopsis YUC genes that are expressed exclusively in roots (At-YUC5, 3, 7, 9, 8) (Figure 3A). Among these, Pj-YUC3 was identified as DMBQ-inducible in the microarray analysis and was included as a member of Cluster 5 (CUST_21965_PI426107926 in Supplemental Data Set 2 and Supplemental Figure 6). Consistent with the microarray analysis, the RT-qPCR data show that only YUC3 was highly upregulated at 24 h after host root exudate treatment and at 48 h posttreatment, with expression levels as high as 200-fold those of the mock treatment (Figure 3B). Similarly, the expression of YUC3 was upregulated in haustorial tissues at 1, 3, and 7 d postinfection (dpi). In contrast, three other Pj-YUC genes had expression patterns similar to those of the mock-treated samples (Figure 3B) and maintained a constant level of expression after host interaction (Figure 3C).
Figure 3.
YUC3 Is Upregulated during Haustorium Development.
(A) A maximum likelihood phylogenetic tree of Arabidopsis (At) and P. japonicum (Pj) YUC proteins. The bootstrap values are shown in percentage on the internal nodes.
(B) and (C) Expression levels of YUC genes measured by RT-qPCR. The values correspond to fold change compared with nontreated roots.
(B) YUC expression in 2-week-old P. japonicum root treated with host root exudate for 0.5, 1, 3, 24, and 48 h.
(C) YUC expression in host interaction. Haustorial region was excised after contact with rice roots for 1, 3, and 7 d postinfection (dpi). Values are shown in fold changes compared with the root without host interaction. All data show the average of three biological replicates with at least two technical replicates each. Error bars denote se. Asterisks indicate significant differences compared with mock treatment (B) and control samples (C) at α ≤ 0.05 by Student’s t test with equal variances.
YUC3 Encodes a Functional Auxin Biosynthesis Enzyme
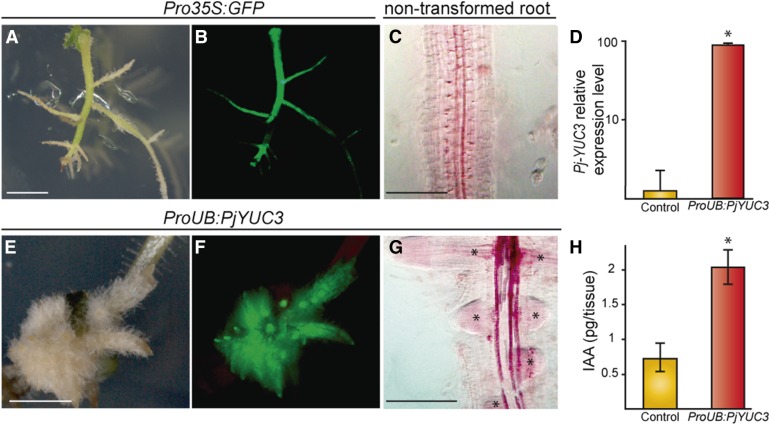
To determine whether Pj-YUC3 encodes a functional enzyme in the auxin biosynthetic pathway, transgenic P. japonicum roots overexpressing this gene under the control of the constitutively expressed ubiquitin promoter were generated with an Agrobacterium rhizogenes-based transformation system (Ishida et al., 2011). For comparison, we generated transgenic roots expressing GFP driven by the CaMV 35S promoter (Pro35S:GFP) (Figures 4A to 4B). RT-qPCR analysis verified that YUC3-transformed hairy roots had ∼70 times higher expression of YUC3 than nontransformed roots (Figure 4D). YUC3-overexpressing roots (ProUB:PjYUC3) had clear morphological phenotypes, including massive proliferation of lateral roots, short root length, and an increased number of root hairs (Figures 4A to 4C and 4E to 4G), as are often seen in auxin-overproducing plants (Boerjan et al., 1995). To confirm that endogenous auxin accumulated in YUC3-overexpressing roots, we measured the free IAA levels in the ProUB:PjYUC3 and empty vector-transformed roots. The free IAA level was ∼3 times higher in YUC3 overexpressing roots than the empty-vector transgenic roots (Figure 4H), indicating that P. japonicum YUC3 encodes a functional auxin biosynthesis enzyme.
Figure 4.
Overexpression of YUC3 in Transgenic P. japonicum Roots Shows an Auxin-Overproduction Phenotype.
(A) and (B) Control transgenic roots expressing CaMV 35S promoter upstream of the GFP gene (Pro35S:GFP) observed using bright-field (A) or fluorescence (B) microscopy.
(C) Nontransformed roots stained with safranin-O and cleared by chloral hydrate.
(D) Expression levels of YUC3 in nontransformed roots (control) and ProUB:PjYUC3 transformed root analyzed by RT-qPCR. Data are presented as the means with the se of two technical replicates of two biological replicates containing 5 to 10 roots in each experiment.
(E) to (G) Phenotype of transgenic roots overexpressing YUC3 observed under bright field (E) or fluorescence (F) microscopy. Transgenic root was stained with safranin-O and cleared with chloral hydrate (G). Asterisks denote lateral roots.
(H) Endogenous IAA levels in root tip tissues (2 mm form the root tips) overexpressing YUC3 (ProUB:PjYUC3) or empty vector (control). An asterisk indicates significant difference at α ≤ 0.05 by Student’s t test with equal variances (P value < 0.002). White bars = 2 mm, and black bars = 200 µm
YUC3 Expression Localizes to the Haustorium Apex and Haustorial Hairs
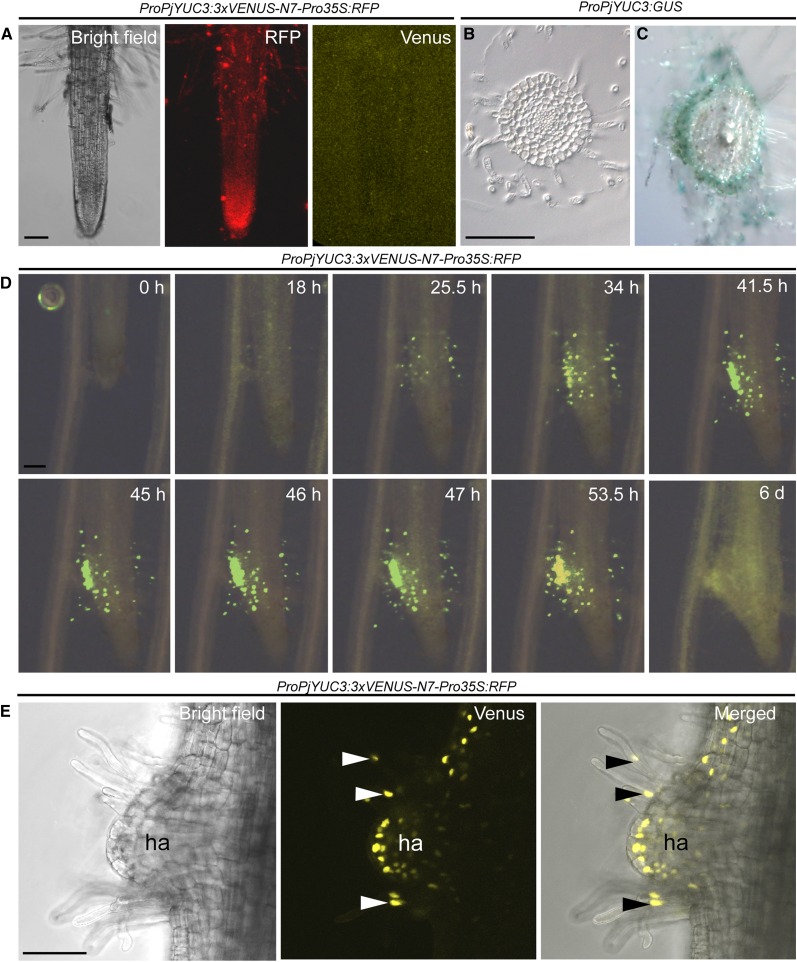
To characterize the roles of YUC3 in haustorium development, the spatio-temporal expression profile of YUC3 was investigated. The promoter region of YUC3 was cloned upstream of the reporter gene GUS and to nucleus-localized VENUS-N7, a modified version of YFP with a nuclear localization sequence, and introduced into P. japonicum hairy roots using A. rhizogenes. To facilitate the screening of transgenic roots, we modified the vectors by inserting the mRFP driven by the 35S promoter as a visible marker. The RFP-positive roots were selected under fluorescence microscopy and stained for GUS activity or visualized by VENUS-filtered fluorescence. Without any haustorium-inducing treatment, neither VENUS fluorescence (Figure 5A) nor GUS staining (Figure 5B) was detected in the transgenic roots. In contrast, when the transgenic roots were treated with the host root exudate or were infecting host roots, GUS reporter gene expression was visible (Figure 5C). The most pronounced GUS expression was observed within the epidermal and outer cortical layers of the haustorium initiation region, as well as in haustorial hairs (Figure 5C). Consistently, using time-lapse fluorescence photographs of Pj-YUC3 promoter-VENUS-N7 transformed roots coincubated with Arabidopsis roots, the earliest YUC3 promoter activity was detected in the haustorial hairs at 18 hpi (Figure 5D; Supplemental Movies 2 to 4). Initially, the expression activity was distributed evenly around the roots (Supplemental Movie 4). At 25.5 hpi, the fluorescence was detected mainly at the interface between the host and parasitic roots, reaching its peak at 46 hpi. The expression gradually declined from 47 hpi until being only weakly observed at 6 dpi (Figure 5D; Supplemental Movie 4). Confocal microscopy showed ProPjYUC3:3xVENUS-N7 expression in the epidermal cells and outer cortical cells, as well as in the haustorium hairs after 48 h of DMBQ treatment (Figure 5E).
Figure 5.
Tissue Localization of YUC3 Expression during Haustorium Development.
(A) Transgenic roots carrying a YUC3 promoter-driven VENUS fluorescent protein gene fused with a nuclear targeting signal. CaMV 35S promoter-driven RFP was inserted as a visual marker (ProPjYUC3:3xVENUS-N7-Pro35S:RFP). Roots without haustorium-inducing treatment were observed under bright-field, RFP, and VENUS fluorescence detecting filter sets.
(B) and (C) Transverse sections of a root transformed with the YUC3 promoter driving the GUS reporter gene (ProPjYUC3:GUS). Nontreated root (B) and 24 h after rice root-exudate treatment (C).
(D) Time-lapse series of ProPjYUC3:3xVENUS-N7-Pro35S:RFP root showing the interaction of a P. japonicum transgenic root with Arabidopsis. Representative photographs are shown from time-lapse images taken every 30 min for 6 d. Bright-field and YFP filter photos were merged. The complete movies are shown in Supplemental Movies 2 to 4.
(E) Confocal microscopy photos of ProPjYUC3:3xVENUS-N7-Pro35S:RFP roots under bright-field (right), YFP filter (middle), and merged pictures (left) after 48 h of DMBQ treatment. Arrowheads point to YUC3 expression at haustorial hairs. Bars = 100 µm.
Haustorium Initiation Site Shows Auxin Response
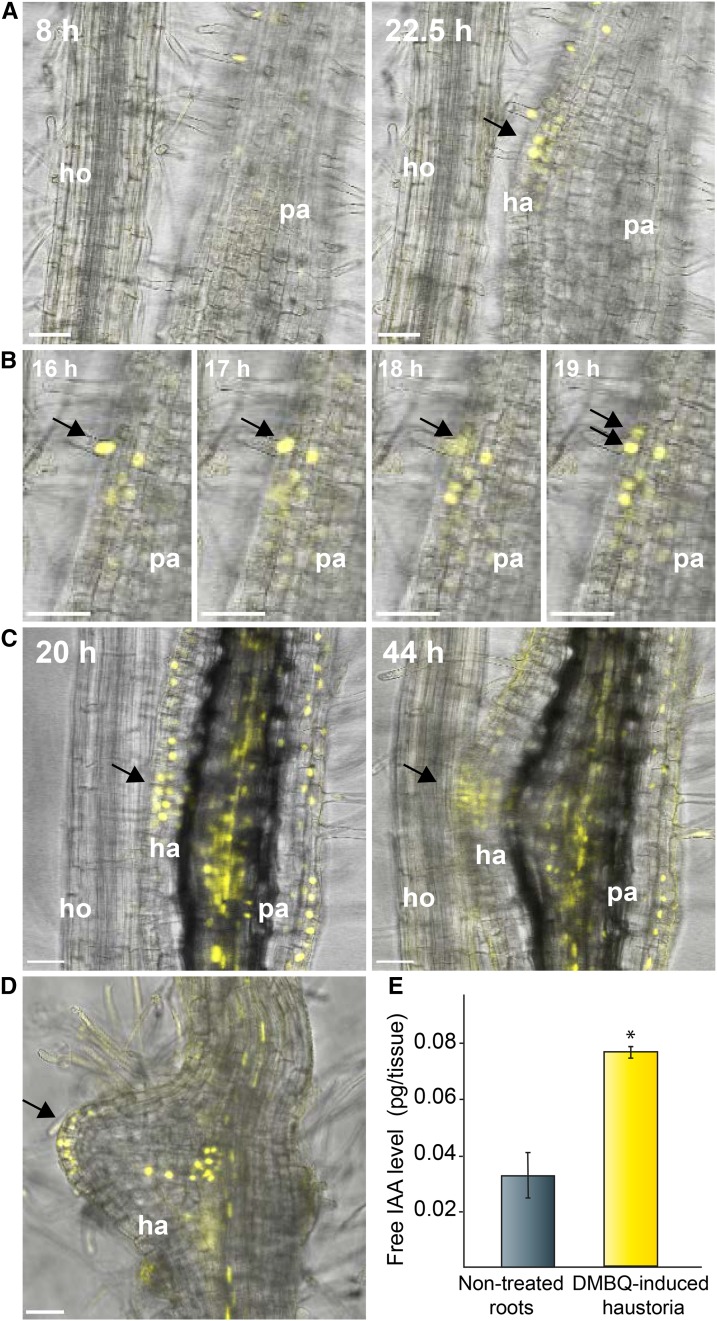
The expression pattern of YUC3 indicates that auxin is biosynthesized de novo at the haustorium apex. Therefore, we investigated whether an auxin response is observed at this site using the pDR5rev:3xVenus-N7 auxin-responsive reporter (Heisler et al., 2005) during haustorium initiation. In P. japonicum transgenic hairy roots carrying pDR5rev:3xVenus-N7, VENUS signals were detected at the root apical meristem and along the vasculature without host interaction (Supplemental Figure 10). This expression pattern is similar to the observations in other plant species’ roots, such as Arabidopsis and tomato (Solanum lycopersicum; Sabatini et al., 1999; Ottenschläger et al., 2003; Chaabouni et al., 2009). When Arabidopsis roots were placed near pDR5rev:3xVenus-N7 transgenic P. japonicum roots, the DR5-driven florescence signal was first observed at the root hair cells slightly above the haustorium initiation site. At later time points, the fluorescence signal was observed at the epidermal and outer cortical cells at the apex of a growing haustorium, similar to YUC3 promoter expression (Figure 6A; Supplemental Movie 5). At a higher magnification, cell division of epidermal cells including hair cells was observed (Figure 6B). These dividing cells showed DR5 fluorescence signals, suggesting that the high auxin response coincides with cell division. As haustorium development progressed, an auxin response maximum was observed in a haustorium apex that started to invade host root tissue (Figure 6C; Supplemental Movie 6). Similarly, the DMBQ-induced haustorium showed auxin responses at the haustorium apex (Figure 6D).
Figure 6.
Time-Lapse Observation of the Expression Pattern of DR5 Auxin-Responsive Promoter during Early Haustorium Development.
(A) to (D) Confocal images of P. japonicum roots transformed with pDR5rev:3xVenus-N7. Merged images of bright-field and Venus fluorescence (yellow) are shown.
(A) Arrow indicates newly established auxin maximum at haustorium initiation site facing the host root.
(B) Arrows track nucleus of a dividing root hair cell.
(C) and (D) Arrows indicate DR5 expression at haustorium apex. Maximum intensity projection images were used for Venus fluorescence. Images were derived from the complete time-lapse videos (Supplemental Movies 5 and 6). The numbers at the top left indicate time post host application or time post DMBQ application in each experiment. Bar = 50 µm. pa, parasite; ha, haustorium; ho, host root.
(E) IAA levels measured in DMBQ-induced haustoria region (0.5 to 1 mm from root tips) compared with the corresponding region in nontreated roots. Data are mean values of three biological replicates with 100 excised tissues for each experiment. An asterisk indicates significantly different at α ≤ 0.05 by Student’s t test with equal variances (P value < 0.0008). Error bars denote standard errors of the mean among three biological replicates.
Endogenous Auxin Levels Increase after DMBQ Treatment at the Haustorium-Forming Sites
Since the auxin-responsive promoter analysis revealed a high auxin response in the haustorium, we investigated the endogenous auxin levels in this organ. P. japonicum roots were treated with or without DMBQ for 24 h and the endogenous free IAA levels of tissues within 1 mm from the root tip, where most new haustoria are frequently formed, were measured. Free IAA contents were more than 2-fold higher in DMBQ-treated roots than control roots (Figure 6E). This suggests that IAA levels are elevated at the root segment containing the haustorium forming area after DMBQ treatment, which is consistent with the high levels of DR5 auxin response marker expression at the haustorium.
The YUC3 Contributes to Haustorium Formation
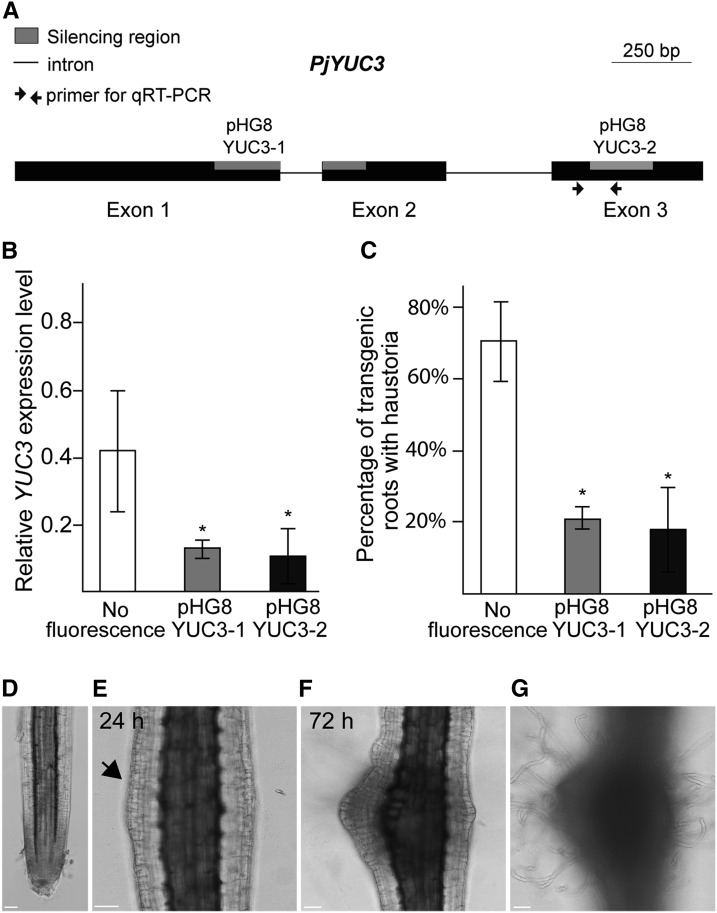
To investigate the function of YUC3 in haustorium formation, we knocked down its expression using the RNA interference (RNAi) method. Two target sites for RNAi were selected from the YUC3 cDNA sequence and were inserted into the pHG8YFP vector (Bandaranayake et al., 2010), generating pHG8YUC3-1 and pHG8YUC3-2 (Figure 7A). Efficient and specific silencing of YUC3 was verified by RT-qPCR using specific primer sets for Pj-YUC3 (Supplemental Data Set 4) and its homologs P. japonicum YUC1, YUC2, and YUC4 (Figure 7B; Supplemental Figure 11). The knockdown root lines did not show any obvious developmental defects (Supplemental Figures 12A to 12C) or changes in primary root length (Supplemental Figure 12D) and number of lateral roots formed (Supplemental Figure 12E) without chemical treatments or during host absence. However, in the presence of host Arabidopsis roots, the transgenic roots harboring RNAi constructs displayed a significant reduction in host infection (21.5 and 17.8% for pHG8YUC3-1 and pHG8YUC3-2, respectively) compared with the control hairy roots (70.6%; Figure 7C). This indicates that YUC3 positively contributes to host interaction. We further tested the frequency of haustorium induction upon host rice root exudate treatment in the pHG8YUC3-1 transgenic roots. The frequency of haustorium formation was significantly reduced in the RNAi construct-expressing roots compared with control roots (Supplemental Figure 13), suggesting that YUC3 plays an important role in haustorium formation.
Figure 7.
YUC3 Expression Levels Positively Correlate with Haustoria Formation.
(A) Schematic view of the YUC3 gene. Exons are shown as black boxes, introns are shown as simple lines, primer pairs used for RT-qPCR analyses are indicated with arrows, and the gray boxes correspond to target regions for silencing constructs. Bar = 250 bp.
(B) Transcriptional levels of YUC3 in host-induced transgenic roots. Values represent the relative expression levels normalized by the internal control Pj-PTB. The controls are cotyledon-emerged hairy roots with no florescence.
(C) Percentage of transgenic roots that formed at least one haustorium when placed next to Arabidopsis roots. Values are means (±se) of three to five biological replicates. Each replicate had 5 to 15 independently transformed roots. Asterisks indicate significant difference at α ≤ 0.05 by Student’s t test with equal variances. Error bars denote standard errors of the mean among three biological replicates.
(D) to (G) Transgenic hairy roots with the AtPGP4>>PjYUC3 construct. Confocal bright-field images of the root before DEX treatment (D), 24 h after 10 µM DEX treatment, with an arrow indicating cell division in the epidermis (E), and 72 h after DEX treatment (F) are shown. Surface focal plane is shown in (G) to focus on hair cells. Bar = 50 µm.
Expression of YUC3 at the Epidermal Cells Induces Haustorium-Like Structure Formation in P. japonicum Roots
To investigate if the expression of YUC3 at the epidermal cells alone is sufficient to induce haustorium formation without host signals, we expressed YUC3 specifically in epidermal cells, using the dexamethasone (DEX)-inducible CRE-lox system (Brocard et al., 1998). In this system, the CRE recombinase fused with the hormone binding domain of glucocorticoid receptor (GR) was placed under the control of the epidermal cell-specific promoter At-PGP4 (Terasaka et al., 2005; Supplemental Figure 14A), and the Pj-YUC3 coding region sequence was driven by the constitutively active At-RPS5a promoter with interruption by 4× terminator sequences sandwiched between two LoxP sequences (Weijers et al., 2001; Supplemental Figures 14B and 14C, AtPGP4>>PjYUC3). Using this construct, YUC3 can be induced in epidermal cells upon DEX treatment due to the release of CRE recombinase in epidermal cells followed by the removal of LoxP-sandwiched terminator sequences inserted between the promoter and the coding regions. The construct was transformed into P. japonicum roots, and morphological changes were observed after DEX treatment. Without DEX treatment, no morphological changes in transgenic P. japonicum root were observed (Figure 7D). At 24 h after DEX treatment, part of the transgenic root began to swell (Figure 7E), and cell division was observed in the epidermis. This site continued to swell and proliferate and eventually formed a haustorium-like structure (Figure 7F). Furthermore, this structure was often associated with dense and curly hairs, which are similar to haustorial hairs (Figure 7G). Taken together, our results demonstrate that the expression of YUC3 is sufficient to induce a haustorium-like structure in P. japonicum roots.
DISCUSSION
Parasitic plants in the Orobanchaceae are serious threats to agriculture worldwide. Despite their economic importance, the molecular mechanisms regulating plant parasitism are poorly understood. Here, our combination of de novo transcriptome and customized microarray analysis of the facultative hemiparasite P. japonicum provides a comprehensive view of the molecular events during the early stages of haustorium development. Using this information, we showed that the parasite gene Pj-YUC3, encoding a functional auxin biosynthesis enzyme, plays an unequivocal role in haustorium development.
Comprehensive Survey of Genes Involved in the Early Stages of Haustorium Development in P. japonicum
The currently available EST sequences in parasitic plants are still limited to several species; ESTs for three members of the Orobanchaceae family, i.e., S. hermonthica, P. aegyptiaca, and T. versicolor (Yoshida et al., 2010; Yang et al., 2014), the root parasite in Santalales, S. album (sandalwood), and the stem parasite Cuscuta pentagona (Kim et al., 2014; Ranjan et al., 2014) have been reported. The P. japonicum sequences analyzed in this study provide a valuable addition to parasitic plant gene databases and are suitable for conducting comparative analyses with other parasitic species. Our assembly yielded 57,939 contigs, 64.3% of which have a BLASTx hit with the nr database (Supplemental Table 3). This value is higher than those in other published de novo dicot transcriptome data sets; e.g., 44.7% in the chickpea de novo assembly based solely on 454 reads, 43.3% in tea (Camellia sinensis) unigenes (Shi et al., 2011), and 59.65% in Litchi chinensis (Li et al., 2013). Thus, our de novo assembled transcriptome provides reasonable coverage of P. japonicum genes with an enrichment of transcripts expressed at different stages of haustorium development, thereby representing an excellent resource for mining essential genes during haustorium development.
Our microarray analysis further identified the kinetics of transcriptional changes during early haustorial development processes. We selected a detailed time course, in particular the initial events during haustorium development, with the aim to identify molecular events that correlate with haustorium development. Interestingly, the earliest expression modulation occurs with genes related to calmodulin binding and oxidoreductase activity (Figure 2C), suggesting that calcium and reactive oxygen species (ROS)-related signaling may be triggered in the early stage of development. Indeed, the involvement of ROS in haustorium initiation signaling has been proposed (Kim et al., 1998; Ngo et al., 2013; Matvienko et al., 2001a, 2001b; Honaas et al., 2013; Bandaranayake et al., 2010; Yang et al., 2015). Consistent with other published studies, our analysis identified homologous genes, which were previously identified as DMBQ-responsive genes in T. versicolor, such as QR2 and Pirin, that are upregulated after DMBQ treatment in P. japonicum, suggesting that gene expression profiles are conserved among Orobanchaceae species during haustorium formation. However, a homolog of Tv-QR1 known to be crucial for haustorium induction signaling in T. versicolor was not upregulated in P. japonicum. This may indicate the diversity of haustorium induction signaling between species. The involvement of calcium metabolism-related transcripts was also suggested by comparative transcriptome analysis between three Orobanchaceae species (Yang et al., 2015). Thus, our data and previous reports suggest that calcium and ROS signaling are likely to be involved in the initial steps of haustorium formation.
The other early responsive genes in Cluster 3 include genes associated with immune system process. In particular, homologs of WRKY genes, encoding transcription factors that regulate defense responses, were grouped in this cluster. The WRKY gene family is known to regulate defense responses either positively or negatively (Zheng et al., 2006; Grunewald et al., 2008; Pandey et al., 2010; Xu et al., 2006). The upregulation of WRKY genes in response to DMBQ suggests that the initial recognition of the host provokes a defense-like response in parasitic plants. The upregulation of plant genes responsive to pathogen attack during the infection process has also been described in T. versicolor (Honaas et al., 2013) and C. pentagona (Ranjan et al., 2014). Thus, parasitic plants in general may recognize the host signal as a biotic stress or potential pathogen at the beginning of the host interaction.
Cluster 5 contains genes categorized in GO terms related to ROS generation, wound response, and hormone metabolism. The increase in gene expression in this cluster occurred just prior to visible morphological changes in the roots; thus, these genes may have regulatory functions in haustorium formation. Indeed, it was postulated that ROS generation acts as a signal that triggers haustorium formation in a parasitic plant (Kim et al., 1998; Bandaranayake et al., 2010). The wound response is associated with tissue regeneration processes, including reprogramming of differentiated cells (Ikeuchi et al., 2016). The enrichment of the GO term response to wounding suggests that haustorium formation may occur through reprogramming of root tissues, despite the lack of injury. Proliferation of haustorial hairs and activation of cell division in haustorial tissues indicate that the root cells undergo redifferentiation processes during haustorium formation. The upregulation of the metabolism of plant hormones suggests that auxin plays important roles in haustorium formation.
YUC3 Functions Specifically in Haustorium Formation
An interesting finding from our transcriptome analysis is that the auxin biosynthesis gene YUC3 is transcriptionally upregulated during haustorium development. In Arabidopsis, each of the 11 YUC gene family members has specific and overlapping functions (Zhao, 2008). The coordinated functions of Arabidopsis YUC3, YUC5, YUC7, YUC8, and YUC9 are responsible for root development, and the quintuple mutants have severely disturbed root growth (Chen et al., 2014). In the P. japonicum transcriptome, we identified four YUC sequences; P. japonicum YUC2 and YUC4 are in the same node with root-specific At-YUC genes in the phylogenetic tree. Although Pj-YUC3 is close to the root-specific clade, this gene was placed just outside of this group (Figure 3A). Transgenic P. japonicum roots with silenced YUC3 did not show any defects in root growth (Supplemental Figure 12). However, knocking down YUC3 led to a reduction in haustorium formation (Figure 7C), implying that YUC3 may function specifically during parasitism. A search of the available EST sequences of other parasitic species in the Orobanchaceae family (Yang et al., 2015), including S. hermonthica, P. aegyptiaca, and T. versicolor, revealed Pj-YUC3 homologs (e-value <1e−100 in tBLASTn) in haustorium-induced libraries but not in prehaustorial or shoot libraries, indicating the conserved function of Pj-YUC3 homologs in this plant family.
Local Expression of YUC3 Coincides with High Auxin Responses at the Haustorium Apex
Our promoter-reporter analysis revealed that the expression of YUC3 precedes haustorium formation. YUC3 promoter-driven reporter florescence was observed at the root hairs and epidermal cells at the haustorium initiation sites. Overexpression of YUC3 resulted in more than 2-fold higher auxin accumulation in P. japonicum roots compared with control roots, indicating that YUC3 encodes a functional auxin biosynthesis enzyme. Thus, it is likely that cells where YUC3 is expressed perform de novo auxin biosynthesis. Coincidently, the auxin-responsive DR5 promoter construct showed a similar localization pattern. Previous studies have shown that the auxin-responsive IAA2 promoter was activated in DMBQ-treated roots using a GUS reporter protein, but the resolution was not sufficiently high to specify which cells were responding to auxin (Tomilov et al., 2005). Our DR5-promoter analysis localized auxin responses in specific cells (i.e., the epidermis and the outer cortex of the haustorium apex as well as the haustorial hairs near the host) in an initiating haustorium, forming auxin response maxima.
Localized auxin responses in particular cells may contribute to the dedifferentiation and differentiation of the cells. Indeed, proliferation of haustorial hairs at the haustorium surface is one of the earliest events in haustorium formation (Cui et al., 2016). Along with the expression of YUC3 and DR5, the haustorial hair cells begin cell division, and one daughter cell becomes an epidermal cell while the other becomes a haustorial hair cell (Supplemental Movie 5). These data suggest that auxin responses correlate with epidermis cell differentiation at the first stage of haustorium formation. Measurement of IAA confirmed that IAA levels are elevated at the root segment containing the haustorium-forming sites after DMBQ treatment. These data indicate that DR5 expression is likely to reflect the accumulation of newly synthesized auxin at the haustorial apex near the host. It is also the possible that local de novo production of this hormone could result in the local redistribution of auxin transporters because auxin itself can serve as a signal for relocalization of auxin transporters such as PIN proteins (Sauer et al., 2006; Xu, 2006).
Local auxin maxima in organ initiation site are commonly observed during plant development (Benková et al., 2003; Heisler et al., 2005) and are thought to be sufficient to induce developmental programs in lateral organs (Reinhardt et al., 2003; Dubrovsky et al., 2008). We reconstructed the YUC3 expression pattern in the epidermis using the CRE/lox system aiming to mimic DMBQ treatment. Induction of YUC3 expression at the epidermis cells by DEX treatment led to epidermis cell proliferation and dense hair cell differentiation similar to haustorial hair formation, resulting in the formation of haustorium-like morphology. This observation implies that local YUC3 expression at epidermis cells, to a certain extent, can initiate haustorium development similar to that observed after DMBQ treatment. Taken together, our results demonstrate that localized expression of YUC3 in epidermis cells is necessary and sufficient for priming haustorium formation in P. japonicum. Further studies on the genetic components upstream and downstream of YUC3 will increase our understanding of the mechanism underlying plant parasitism.
METHODS
Plant Materials and Growth Conditions
Phtheirospermum japonicum (Okayama strain) seeds were surface-sterilized and germinated as previously described (Ishida et al., 2011). The outer coats of rice seeds (Oryza sativa cultivar Nipponbare) were mechanically removed, and the seeds were immersed with agitation in 70% (v/v) ethanol for 3 min, washed with 50% (v/v) commercial bleach solution (including 6% [v/v] sodium hypochlorite; Kao Chemicals) for 30 min, and extensively rinsed with sterile water. The rice seeds were sown in a Petri dish (50 seeds per dish) containing a moistened filter paper (10 to 15 mL water per dish). Arabidopsis thaliana Col-0 seeds were surface sterilized with 10% commercial bleach solution for 5 min and rinsed with sterile water. Sterilized seeds were sown on half-strength MS medium containing 1.0% sucrose and 0.8% agar, and kept at 4°C for 2 d in the dark. Seven- to ten-day-old plants grown vertically at 22°C under long-day conditions (16 h day/8 h night) were used for haustorium induction. One-week-old rice and P. japonicum seeds were separately grown, and 10 rice seedlings and 6 to 10 P. japonicum seedlings were carefully placed in a rhizotron system as previously described (Yoshida and Shirasu, 2009). The rhizotrons were placed vertically at 25°C under a photoperiod of 16 h light/8 h dark with a light intensity of ∼701 mol·m−2 s−1 illuminated by the daylight-white fluorescence lump (FL40SEX-N-FL; NEC), unless otherwise specified.
Total RNA Extraction
RNA was extracted using a CTAB/LiCl based-protocol as previously reported (Yoshida et al., 2010). Genomic DNA was removed from the RNA samples by treatment with DNase I, Amplification Grade (Life Technologies; Cat. No. 18068-015), followed by a cleanup step with an RNeasy Mini Spin Column from an RNeasy Plant Mini Kit (Qiagen; Cat. No. 74904). RNA was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies) using an RNA 600 Nano Kit (Cat. No. 5067-1511). RNA qualities for sequencing and microarray experiments were set at a minimum RNA Integrity Number of 8.0.
RNA Sequencing and de Novo Transcriptome Assembly
P. japonicum and rice seeds were germinated and grown separately for 1 week, and P. japonicum seedlings were transferred to rhizotron chambers without host plants (autotrophic stages) (Supplemental Figure 2, left photograph) or in contact with rice (parasitic stages) (Supplemental Figure 2, right photograph) for 4 to 6 weeks. For the parasitic-stage library, haustoria parts were carefully excised from P. japonicum roots under a stereoscope and immediately frozen in liquid nitrogen. When haustoria could not be disconnected from rice roots (Figures 1H and 1I), both tissues were collected. RNAs from P. japonicum roots grown without rice (nonparasitizing) were also collected for the autotrophic-stage library.
Twenty micrograms\ of total RNA was used for constructing the Illumina HiSeq 2000 libraries. Paired-end 90-bp sequences with 200-bp insert lengths were analyzed with the Illumina HiSeq 2000 platform according to the manufacturer’s instructions. The Roche 454 library was constructed using normalized mRNA from tissues with and without host interactions. Approximately 4 µg of mRNA was purified from total RNAs using the Illustra mRNA purification kit (GE Healthcare; Cat. No. 27-9258-01). mRNA normalization was performed using duplex-specific nuclease, and single read sequences were obtained with GS FLX titanium chemistry. Library construction and sequencing were performed by Hokkaido Systems Science Company for the 454 sequencer and Riken Genesis for the HiSeq 2000 sequencer.
For the de novo transcriptome assembly, the adapter oligonucleotide sequences from raw Roche 454 reads were trimmed off using CLC Genome Workbench Software 4.9. The trimmed 454 reads and the paired-end Illumina-HiSeq reads from the parasitic stage library were mapped against the annotated cDNA sequences of O. sativa (version 6) using Mosaik, allowing a maximum of four mismatches in a 45-bp overlap (http://bioinformatics.bc.edu/marthlab/Mosaik). The unmapped reads were used for the assembly. The rice sequence-filtered reads and the Illumina reads from the autotrophic stage were assembled using a combination of CLC Genome Workbench Software 4.9 and CAP3 software (Huang and Madan, 1999) to ensure nonredundant sequence assembly. Contigs longer than 300 bp were used for subsequent analyses. BLAST annotation and GO analysis of the translated sequences were analyzed by blast2GO (Conesa et al., 2005). The contigs were then assigned to PlantTribes 2.0 orthogroups (Wall et al., 2008). The expression values were computed through mapping of reads against the assembled contigs using CLC Genome Workbench 4.9 and the RPKM estimation function.
DMBQ and Rice Root Exudate Treatment
To induce in vitro haustorium formation, P. japonicum roots were treated with the haustorium-inducer DMBQ (Sigma-Aldrich; Cat. No. 565032). P. japonicum seeds were sown on modified MS medium (1× MS supplemented with 1% [w/v] sucrose, 0.01% [w/v] myo-inositol, 0.06% [w/v] MES, and 0.8% [w/v] agar) in 10 × 14 × 1.5-cm square plates and kept in a vertical position in 25°C under long-day conditions. The bottom halves of the plates were covered with aluminum foil to avoid exposure of the roots to light. After 2 weeks, 7 mL of 10 µM DMBQ solution was dropped onto the P. japonicum roots and the plates were incubated horizontally. For microarray analysis, the DMBQ- or mock-treated (0.1% [v/v] DMSO) roots were sampled at 0, 0.5, 1, 3, 6, 12, 24, and 48 h after treatments. Each experiment was repeated three times with 12 to 15 plants per time point.
For the rice root exudate treatment, 50 rice seedlings were soaked in 12 to 15 mL water for 1 week, after which 5 mL of the water was solidified with agar (0.8% [w/v]). In addition, the roots of 1- to 2-week-old rice seedlings were cut into ∼1- to 3-mm pieces with a razor blade, and 180 mg of chopped rice roots was added to 20 mL of warm agar (0.8% [w/v]) containing 5 mL rice exudate. The solidified agar was carefully placed onto 2-week-old P. japonicum roots. For the control treatment, only the agar (0.8% [w/v]) blocks were placed on the parasite roots. The plates were incubated horizontally at 25°C with the bottom halves of the plates covered with aluminum foil.
Custom Microarray Design, Labeling, and Hybridization
For custom microarray slides, putative coding regions of the assembled contigs were detected by ESTScan2 (Iseli et al., 1999), and 47,817 unigenes with a putative coding region of at least 50 amino acids were selected. Approximately 80% of these unigenes (38,120 unigenes) showed at least one hit in the nr database in a BLASTx search (e-value threshold 1e-5), suggesting that most of the unigenes are protein-coding genes. The gene-specific 60-mer probes were designed using the Web-based application e-Array Tools from Agilent Technologies (http://www.chem.agilent.com, design number 253261110001/253416810002) and fixed on Agilent 8 × 60K microarray slides. As a control to check for uneven hybridization, 10 groups of probes with each group containing 50 randomly selected sequences in the P. japonicum transcriptome (a total of 500 probes) were designed and fixed in various locations on the array. Cyanine-3 (Cy3)-labeled cRNA was prepared from 0.25 µg RNA using the Low Input Quick Amp Labeling Kit one-color (Agilent Technologies) according to the manufacturer's instructions. Dye incorporation and cRNA yield were checked with a NanoDrop spectrophotometer. Microarray hybridization, washing, and scanning were performed according to the instructions provided by Agilent. Raw data were extracted using Feature Extraction Software (v 10.7.3), and spots without signals or signals under background level were removed from the analysis. Feature extraction files were imported to GeneSpring GX (v. 11.0) using baseline transformation and the normalization option set to 75% of the median shift.
Microarray raw data were filtered to select the differentially expressed genes using the method described previously (Lewis et al., 2013). The base-line transformed values were compared with the control (0 h) sample, and genes showing 2-fold up- or downregulation in all three biological replicates were selected. In addition, the data from a replicate were compared with mock treatment at each time point, and those showing less than 2-fold differences from mock-treated samples were eliminated. The expression values were calculated in fold changes compared with control (0 h) samples. The Euclidean distance (ED) and Pearson’s correlation coefficient (PCC) of expression profiles between biological replicates were calculated and filtered on thresholds of PCC 0.7 and ED 3.94. The threshold values were chosen for ∼0.07 P value for PCC and median value for ED. The unigenes that fulfilled these criteria in two of three combinations were selected. We selected 327 consistently differently expressed (DE) genes.
Clustering analysis was performed according to a previous report using SC2ATmd software (Olex and Fetrow, 2011). First, figure of metric analysis was applied to obtain parameters for the accurate clustering algorithm using ED metric for a given input. Next, the filtered DE genes were submitted to consensus clustering with the option of hierarchical clustering (k-means=16, 16 repetitions) with a consensus threshold of 90%. GO terms were assigned for each gene based on the best BLAST-hit annotations, and the enrichment test was performed using the R package GOstat (Beissbarth and Speed, 2004). False discovery rate (FDR) correction was done using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995) with statistical significance of FDR < 0.05.
RT-qPCR Analysis
For RT-qPCR analysis, we first identified constitutively expressed genes to be used as normalization controls. We selected homologs of Arabidopsis housekeeping genes (Czechowski et al., 2005) using a local BLAST search (e-value <1e-20) as well as the constitutively expressed genes (in the range of ±1.2 fold change) (Supplemental Data Set 3). We identified four P. japonicum candidates, and primer sets for each gene were tested for amplification efficiency and specificity for P. japonicum by testing with the genomic DNAs of Lotus japonicus and rice. A primer pair designed for PTB was used for the internal control in subsequent experiments. The first-strand cDNA was synthesized with a ReverTra Ace-α- kit (Toyobo; code FSK-101), and qPCR was performed with a Thunderbird SYBR qPCR Mix kit (Toyobo; code A4251K). The relative expression was calculated with the standard curve method. All primers used in this study are listed in Supplemental Data Set 4. All experiments were repeated at least three times with at least two technical replications each. For statistical analysis, the t test was performed across the replicates.
Cloning of YUC Genes in the P. japonicum Transcriptome
To identify YUC gene homologs in P. japonicum, the amino acid sequences of Arabidopsis YUC genes (AT4G32540, AT1G04180, AT1G04610, AT1G48910, AT2G33230, AT4G13260, AT4G28720, AT5G25620, and AT5G43890) were aligned against the P. japonicum transcriptome using the tBLASTn algorithm. Sequences showing an alignment score below an e-value of 1e-35 and amino acid identity over 35% were considered to be homologs of YUC genes. The RACE method was used to determine the full-length cDNA sequences of Pj-YUC homologs (GeneRacer core kit; Invitrogen). The RACE library was constructed from a pool of RNA extracted from P. japonicum roots at the parasitic and autotrophic stages. To clone a full-length cDNA, primers were designed at both ends of the cDNA, and PCR amplifications were performed using the first-strand cDNA library from parasite-infected roots as a template. All PCR amplification reactions were performed using Advantage 2 PCR Enzyme (Clontech; Cat. No. 639137).
Phylogenetic Analysis
Predicted amino acid sequences were aligned using the option Muscle in the CLC genomic workbench (ver. 4.8) program with default arguments. Based on these alignments (Supplemental Files 1 and 2), phylogenetic trees were generated using the UPGMA method, and the reliability of the trees was tested by bootstrap with 1000 resamplings. The topology of the trees was also confirmed by the maximum-likelihood method with the MEGA program (Tamura et al., 2011).
Plasmid Construction
For the Pj-YUC3 promoter construct, oligonucleotides flanking the putative promoter region and the second exon of YUC3 were designed and used for PCR amplification using the P. japonicum genome as a template. The 4924-bp genomic fragment was cloned, and the sequence was confirmed using an ABI3770 sequencer. The promoter region (3137 bp) was amplified by specific primers (Supplemental Data Set 4). For the GUS and VENUS reporter construct, the CaMV 35S promoter and mRFP sequences were PCR amplified from the pEZS-CL vector (Yoshimoto et al., 2004) and inserted within the HindIII site into the linearized R4L1pGWB532 vector (Nakamura et al., 2009) or into the SacI site of pDR5rev:3XVenus-N7 (Heisler et al., 2005), respectively. pDR5rev:3XVenus-N7 was kindly provided by Elliot Meyerowitz. The YUC3 fragment with attB4 and attB1 recombination sites was cloned into the pDONRG-P4P1R vector (Oshima et al., 2011) using the Gateway BP reaction and subsequently transferred to the RFP-inserted R4L1pGWB532 vector through the LR reaction catalyzed by Gateway LR Clonase (Invitrogen). For the VENUS reporter construct, the auxin-specific DR5 promoter from pDR5rev:3XVenus-N7 was replaced with the YUC3 promoter through the multiple fragments cloning strategy using an In-fusion Cloning Kit (Takara).
For overexpression and silencing constructs, the plasmid pUB-GW-GFP (Maekawa et al., 2008) (http://www.legumebase.brc.miyazaki-u.ac.jp) and the silencing vector pHG8YFP (Bandaranayake et al., 2010) were used, respectively. The YUC3 coding sequence was amplified with the primers listed in Supplemental Data Set 4 and inserted into the pENTR/D-TOPO vector using a TOPO cloning kit (Invitrogen). The resulting clones were verified by sequencing, and the targeted sequence was inserted into the destination vectors through a recombination reaction catalyzed by LR Clonase II. The empty vector was used for the silencing control. Transgenic hairy roots were selected based on the presence of GFP or YFP signals.
For the DEX-inducible CRE-lox system construct, Golden Gate cloning technology was used (Engler et al., 2014). The Arabidopsis PGP4 and RPS5a promoter region and the Pj-YUC3 sequence from the start codon to the 3′ untranslated region were divided into two parts and amplified by PCR from Arabidopsis genomic DNA and P. japonicum genomic DNA, respectively. Each fragment was inserted into vector pAGM1311. These sequences were combined into vector pICH41295 for the promoter module (AtPGP4-pro and AtRPS5a-pro) and pICH41308 for the CDS module (PjYUC3-CDS). The At-RPS5a promoter sequence was recloned into pAGM1251 using At-RPS5a-pro as a template for the promoter (f) module [At-RPS5a-pro(f)]. The ligand binding domain of human glucocorticoid receptor was PCR-amplified from vector p35SGRG (Iwase et al., 2013) and inserted into pAGM1311. This vector was combined with the synthesized sequence of CRE recombinase and cloned into pICH41308, yielding the CDS module (CRE-GR-CDS). The HSP18.2 terminator sequence was amplified from Arabidopsis genomic DNA and inserted into vector pICH41276 (At-HSP-ter). The 35S terminator sequences were amplified from pICH41414 (35S-ter) with the primer including the LoxP sequence and tandemly assembled into pAGM1311, yielding LoxP-2xTer-LoxP. To increase the spacer region, two HSP18.2 terminator sequences were inserted between the 35S terminator sequences and cloned into vector pAGM1276 for the NT1 module, yielding LoxP-4xTer-LoxP-NT1. AtPGP4-pro and CRE-GR-CDS and 35S-ter were combined into pICH47732, yielding AtPGP4:CRE-GR. AtRPS5a-pro(f) and LoxP-4xTer-NT1 and PjYUC3-CDS and AtHSP-ter were combined into pICH47781, yielding AtRPS5a-LoxP:PjYUC3. These gene cassettes were further combined into pAGM4723, yielding AtPGP4>>PjYUC3, which was and used for transformation.
Transformation of P. japonicum
Transformation of P. japonicum was performed as previously described by Ishida et al. (2011) with minor modifications. Three-day-old P. japonicum seedlings were immersed in a bacterial solution (OD600 = 1.0) and submitted to ultrasonication using a bath sonicator (Ultrasonic Automatic Washer; AS ONE) for 15 to 25 s. The sonicated seedlings were vacuum infiltrated for 5 min. The seedlings were transferred to cocultivation medium (Gamborg B5 agar media, 1% [w/v] sucrose, and 450 µM, acetosyringone) and kept in the dark at 22°C for 1 to 2 days. After the cocultivation period, the seedlings were transferred to B5 agar medium containing cefotaxime (300 µg/mL). After 3 to 4 weeks, the transformed roots were analyzed. All plasmids, except pDR5rev:3xVenus-N7, contained a visible marker in which a constitutive promoter was fused upstream of a gene encoding fluorescent protein. To identify transgenic roots, the marker protein fluorescence was detected using a Leica M165 FC stereoscope.
Analysis of Transgenic Roots
Hairy roots emerging after 3 to 4 weeks were used for haustorium-inducing assays. Fluorescent roots were selected and placed on either host root exudate containing medium or 0.7% agarose together with 10-d-old Arabidopsis seedlings. The number of fluorescent roots that formed at least one haustorium was counted after 24 or 48 h. To verify silencing and overexpression in the transformed roots, RNA was extracted using an Arcturus PicoPure RNA isolation kit (Applied Biosystems Life Technologies) and RT-qPCR was performed. For statistical analysis, at least 30 independent fluorescent roots were analyzed across three to five biological replicates, unless otherwise described. For the DEX-induced CRE-lox system, the hairy roots were treated with 10 µM DEX-containing agar.
Time-Lapse Photography and Microscopy
Arabidopsis Col-0 was grown vertically for 10 d on full-strength MS medium supplemented with 1% (w/v) sucrose. Transgenic P. japonicum roots were placed carefully near Arabidopsis roots in a Petri dish containing agar 0.7% (w/v). Seedlings and roots were covered with 0.7% (w/v) agarose to avoid dehydration. Time-lapse photographs were automatically taken with a Leica M165 FC stereoscope or by confocal Leica TCS-SP5 II microscopy at 30-min intervals. The double staining of the haustorium thin sections and safranin-staining of whole haustorium were performed as previously described (Yoshida and Shirasu, 2009). GUS staining was performed as described (Vitha et al., 1995) and observed under an Olympus BX-50 microscope. Confocal photographs were taken with a Leica TCS-SP5 II confocal microscope.
Measurement of Endogenous IAA
For auxin measurements in YUC3-overexpressing roots, root tip tissues (2 mm from the root tips) were carefully excised from pUB:PjYUC3- and empty vector-transformed roots. For IAA measurements, a haustorium region (∼1 to 2 mm from the root tip) was excised after treatment with DMBQ for 24 h. The root tip region was excluded because high levels of auxin accumulation in root tips may mask the detection of increases in auxin levels in haustoria. All measurements were performed in three biological replicates, and each biological experiment contained 100 excised tissues. Free IAA measurement was performed as previously reported (Okumura et al., 2013) with slight modifications. HPLC was performed with a gradient of 3 to 15% of acetonitrile/0.05% acetic acid over 20 min. Parameters for LC-ESI-MS/MS analysis were modified as follows: fragmentor, 120 V; MS/MS transition for [phenyl-13C6]IAA, 182/136 (m/z). Data were analyzed using Student’s t test, assuming equal variances among three biological replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession number SAMN03271814. MIAME-compliant (minimum information about a microarray experiment) raw microarray data were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65723.
Supplemental Data
Supplemental Figure 1. Host specificity of the parasitic plant P. japonicum.
Supplemental Figure 2. P. japonicum grown in a rhizotron with or without rice host plants.
Supplemental Figure 3. RT-qPCR validation of the expression profiles of selected genes.
Supplemental Figure 4. P. japonicum roots at 48 h after rice root exudate treatment.
Supplemental Figure 5. Expression profiles of early-responsive genes.
Supplemental Figure 6. Expression profiles of late-responsive genes.
Supplemental Figure 7. Expression profiles of downregulated genes.
Supplemental Figure 8. QR2 rather than QR1 is differentially regulated during parasitism.
Supplemental Figure 9. Alignment of Pj-YUC and At-YUC amino acid sequences.
Supplemental Figure 10. Expression pattern of DR5 promoter in P. japonicum roots.
Supplemental Figure 11. Transcript levels of YUCCA homologs in transgenic roots.
Supplemental Figure 12. Morphology of transformed hairy roots.
Supplemental Figure 13. RNAi lines of P. japonicum roots targeted to YUC3 show reduced haustorium formation induced by rice root exudates.
Supplemental Figure 14. Expression patterns of At-PGP4 promoter and At-RPS5a promoter in P. japonicum.
Supplemental Table 1. Total number of reads obtained in each RNA-seq library.
Supplemental Table 2. Investigation of rice contamination in P. japonicum unigenes
Supplemental Table 3. Summary of de novo assembly of P. japonicum transcriptome.
Supplemental Data Set 1. Annotation and expression values of the P. japonicum transcriptome at the autotrophic and parasitic stages.
Supplemental Data Set 2. List of 327 differentially expressed genes divided into 38 clusters in response to DMBQ.
Supplemental Data Set 3. List of genes with unaltered expression levels in microarray samples treated with 10 µM DMBQ.
Supplemental Data Set 4. List of primers.
Supplemental Movie 1. Time-lapse images of haustorium development.
Supplemental Movie 2. Time-lapse images of root harboring ProPjYUC3:3XVenus-N7-Pro35S:RFP vector during infection of Arabidopsis roots viewed under a bright-field microscopy.
Supplemental Movie 3. Time-lapse images of root transformed with the ProPjYUC3:3XVenus-N7-Pro35S:RFP vector during infection of Arabidopsis roots shown in Supplemental Movie 2 observed under RFP fluorescence conditions.
Supplemental Movie 4. Time-lapse images of root transformed with the ProPjYUC3:3XVenus-N7-Pro35S:RFP vector during infection of Arabidopsis roots shown in Supplemental Movie 2 observed under VENUS fluorescence conditions.
Supplemental Movie 5. Confocal time-lapse images of root transformed with the pDR5rev:3XVenus-N7 vector during infection of Arabidopsis roots.
Supplemental Movie 6. Confocal time-lapse images of root transformed with the pDR5rev:3XVenus-N7 vector during infection of Arabidopsis roots.
Supplemental File 1. Alignments of predicted amino acid sequences of Pj-YUC and At-YUC genes.
Supplemental File 2. Alignments of predicted amino acid sequences of Pj-QR and Tv-QR genes
Supplementary Material
Acknowledgments
We thank John Yoder (University of California, Davis), Elliot Meyerowitz (California Institute of Technology), and Akira Iwase (RIKEN) for kindly providing pHG8-YFP, pDR5rev:3xVENUS-N7, and p35SGRG, respectively. We also thank Mika Kawashima and Minami Matsui (RIKEN Center for Sustainable Resource Science) for helping with the hybridization of microarray slides. We also thank Anuphon Laohavisit, Thomas Spallek, and Yasunori Ichihashi for their careful reading of this manuscript. Our work is partially supported by MEXT KAKENHI grants (24228008 and 15H05959 to K.S. and 25114521, 25711019, and 25128716 to S.Y.) and the PhD fellowship programs (MEXT to J.K.I. and JSPS to T.W.).
AUTHOR CONTRIBUTIONS
J.K.I., T.W., S.Y., and K.S. conceived the project and design the experiments. J.K.I., S.Y., E.W., and C.W.D. performed transcriptome data generation and analysis. Y.T. and H.K. performed auxin measurements. J.K.I. and T.W. performed plasmid construction, plant transformation, and phenotypic analyses. E.W., H.K., S.N., and C.W.D. critically revised the manuscript. J.K.I., T.W., S.Y., and K.S. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht H., Yoder J.I., Phillips D.A. (1999). Flavonoids promote haustoria formation in the root parasite triphysaria versicolor. Plant Physiol. 119: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird W.M.V., Riopel J.L. (1984). Experimental studies of haustorium initiation and early development in Agalinis purpurea (L.) RAF. (Scrophulariaceae). Am. J. Bot. 71: 803–814. [Google Scholar]
- Bandaranayake P.C.G., Filappova T., Tomilov A., Tomilova N.B., Jamison-McClung D., Ngo Q., Inoue K., Yoder J.I. (2010). A single-electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria. Plant Cell 22: 1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaranayake P.C.G., Tomilov A., Tomilova N.B., Ngo Q.A., Wickett N., dePamphilis C.W., Yoder J.I. (2012). The TvPirin gene is necessary for haustorium development in the parasitic plant Triphysaria versicolor. Plant Physiol. 158: 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nun N., Sachs T., Mayer A.M. (2008). A role for IAA in the infection of Arabidopsis thaliana by Orobanche aegyptiaca. Ann. Bot. (Lond.) 101: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T., Schmülling T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P. (2004). GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Boerjan W., Cervera M.T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., Inzé D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard J., Feil R., Chambon P., Metzger D. (1998). A chimeric Cre recombinase inducible by synthetic,but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 26: 4086–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S., Jones B., Delalande C., Wang H., Li Z., Mila I., Frasse P., Latché A., Pech J.C., Bouzayen M. (2009). Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 60: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Lynn D.G. (1986). The haustorium and the chemistry of host recognition in parasitic angiosperms. J. Chem. Ecol. 12: 561–579. [DOI] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55: 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cui S., Wakatake T., Hashimoto K., Saucet S., Toyooka K., Yoshida S. Shirasu K. (2016). Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant, Phtheirospermum japonicum. Plant Physiol. 170: 1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J.M., Wall P.K., Edger P.P., Landherr L.L., Ma H., Pires J.C., Leebens-Mack J., dePamphilis C.W. (2010). Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Youles M., Gruetzner R., Ehnert T.M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S. (2014). A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Frigerio M., Alabadí D., Pérez-Gómez J., García-Cárcel L., Phillips A.L., Hedden P., Blázquez M.A. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142: 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K., Matsuda Y., Ozawa K., Nishimura T., Koshiba T., Fraaije M.W., Sekiguchi H. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics 279: 499–507. [DOI] [PubMed] [Google Scholar]
- Grunewald W., Karimi M., Wieczorek K., Van de Cappelle E., Wischnitzki E., Grundler F., Inzé D., Beeckman T., Gheysen G. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Honaas L.A., Wafula E.K., Yang Z., Der J.P., Wickett N.J., Altman N.S., Taylor C.G., Yoder J.I., Timko M.P., Westwood J.H., dePamphilis C.W. (2013). Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 13: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Iseli C., Jongeneel C.V., Bucher P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 99: 138–148. [PubMed] [Google Scholar]
- Ishida J.K., Yoshida S., Ito M., Namba S., Shirasu K. (2011). Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6: e25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A., Mitsuda N., Ikeuchi M., Ohnuma M., Koizuka C., Kawamoto K., Imamura J., Ezura H., Sugimoto K. (2013). Arabidopsis WIND1 induces callus formation in rapeseed, tomato, and tobacco. Plant Signal. Behav. 8: e27432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison D.S., Yoder J.I. (2001). Heritable variation in quinone-induced haustorium development in the parasitic plant Triphysaria. Plant Physiol. 125: 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H. (2016). Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80: 34–42. [DOI] [PubMed] [Google Scholar]
- Kim D., Kocz R., Boone L., Keyes W.J., Lynn D.G. (1998). On becoming a parasite: evaluating the role of wall oxidases in parasitic plant development. Chem. Biol. 5: 103–117. [DOI] [PubMed] [Google Scholar]
- Kim G., LeBlanc M.L., Wafula E.K., dePamphilis C.W., Westwood J.H. (2014). Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345: 808–811. [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Olex A.L., Lundy S.R., Turkett W.H., Fetrow J.S., Muday G.K. (2013). A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell 25: 3329–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wang Y., Huang X., Li J., Wang H., Li J. (2013). De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genomics 14: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn D.G., Chang M. (1990). Phenolic signals in cohabitation: implications for plant development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 497–526. [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M., Torres M.J., Yoder J.I. (2001a). Transcriptional responses in the hemiparasitic plant Triphysaria versicolor to host plant signals. Plant Physiol. 127: 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M., Wojtowicz A., Wrobel R., Jamison D., Goldwasser Y., Yoder J.I. (2001b). Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 25: 375–387. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Nakao A., Kawamukai M., Kimura T., Ishiguro S., Nakagawa T. (2009). Development of Gateway binary vectors, R4L1pGWBs, for promoter analysis in higher plants. Biosci. Biotechnol. Biochem. 73: 2556–2559. [DOI] [PubMed] [Google Scholar]
- Ngo Q.A., Albrecht H., Tsuchimatsu T., Grossniklaus U. (2013). The differentially regulated genes TvQR1 and TvPirin of the parasitic plant Triphysaria exhibit distinctive natural allelic diversity. BMC Plant Biol. 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K., Goh T., Toyokura K., Kasahara H., Takebayashi Y., Mimura T., Kamiya Y., Fukaki H. (2013). GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for arabidopsis lateral root initiation. Plant Cell Physiol. 54: 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olex A.L., Fetrow J.S. (2011). SCATmd: a tool for integration of the figure of merit with cluster analysis for gene expression data. Bioinformatics 27: 1330–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y., Mitsuda N., Nakata M., Nakagawa T., Nagaya S., Kato K., Ohme-Takagi M. (2011). Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol. 28: 201–210. [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Roccaro M., Schön M., Logemann E., Somssich I.E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 64: 912–923. [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2010). Armed and dangerous. Science 327: 804–805. [DOI] [PubMed] [Google Scholar]
- Ranjan A., Ichihashi Y., Farhi M., Zumstein K., Townsley B., David-Schwartz R., Sinha N.R. (2014). De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol. 166: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Riopel J.L., Musselman L.J. (1979). Experimental initiation of haustoria in Agalinis purpurea (Scrophulariaceae). Am. J. Bot. 66: 570–575. [Google Scholar]
- Riopel J.L., Timko M.P. (1995). Haustorial initiation and differentiation. In Parasitic Plant, M.C. Press and J.D. Graves, eds (London: Chapman & Hall; ), pp. 40–79. [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sauer M., Balla J., Luschnig C., Wisniewska J., Reinöhl V., Friml J., Benková E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20: 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-Y., Yang H., Wei C.-L., Yu O., Zhang Z.-Z., Jiang C.-J., Sun J., Li Y.-Y., Chen Q., Xia T., Wan X.-C. (2011). Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.E., Dudley M.W., Lynn D.G. (1990). Vegetative/parasitic transition: control and plasticity in striga development. Plant Physiol. 93: 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T., Mutuku M., Shirasu K. (2013). The genus Striga: a witch profile. Mol. Plant Pathol. 14: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Péret B. (2012). AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 3: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka K., Blakeslee J.J., Titapiwatanakun B., Peer W.A., Bandyopadhyay A., Makam S.N., Lee O.R., Richards E.L., Murphy A.S., Sato F., Yazaki K. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilov A.A., Tomilova N.B., Abdallah I., Yoder J.I. (2005). Localized hormone fluxes and early haustorium development in the hemiparasitic plant. Plant Physiol. 138: 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.J., Tomilov A.A., Tomilova N., Reagan R.L., Yoder J.I. (2005). Pscroph, a parasitic plant EST database enriched for parasite associated transcripts. BMC Plant Biol. 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Benes K., Phillips J.P., Gartland K.M. (1995). Histochemical GUS analysis. Methods Mol. Biol. 44: 185–193. [DOI] [PubMed] [Google Scholar]
- Wall P.K., Leebens-Mack J., Müller K.F., Field D., Altman N.S., dePamphilis C.W. (2008). PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res. 36: D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Franke-van Dijk M., Vencken R.J., Quint A., Hooykaas P., Offringa R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299. [DOI] [PubMed] [Google Scholar]
- Xu J. (2006). A Molecular Framework for Plant Regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143: 1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., et al. (2015). Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 32: 767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ishida J.K., Kamal N.M., Ali A.M., Namba S., Shirasu K. (2010). A full-length enriched cDNA library and expressed sequence tag analysis of the parasitic weed, Striga hermonthica. BMC Plant Biol. 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Shirasu K. (2009). Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 183: 180–189. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Shirasu K. (2012). Plants that attack plants: molecular elucidation of plant parasitism. Curr. Opin. Plant Biol. 15: 708–713. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Teixeira da Silva J.A., Duan J., Deng R., Xu X., Ma G. (2012). Endogenous hormone levels and anatomical characters of haustoria in Santalum album L. seedlings before and after attachment to the host. J. Plant Physiol. 169: 859–866. [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2008). The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 11: 16–22. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.