Abstract
Bioluminescence is a ubiquitous imaging modality for visualizing biological processes in vivo. This technique employs visible light and interfaces readily with most cell and tissue types, making it a versatile technology for preclinical studies. Here we review basic bioluminescence imaging principles, along with applications of the technology that are relevant to the medicinal chemistry community. These include noninvasive cell tracking experiments, analyses of protein function, and methods to visualize small molecule metabolites. In each section, we also discuss how bioluminescent tools have revealed insights into experimental therapies and aided drug discovery. Last, we highlight the development of new bioluminescent tools that will enable more sensitive and multi-component imaging experiments and, thus, expand our broader understanding of living systems.
Introduction
A complete understanding of living systems requires methods to probe cells and discrete biomolecules within their native environs. Protein trafficking, metabolite production, and even whole cell movements are influenced by diverse spatial and environmental cues that cannot be easily recapitulated outside of a living organism. In recent years, a set of imaging technologies has emerged that enable biological features to be visualized—noninvasively—in whole cells and organisms.[1–10] These methods are providing unprecedented insight into cell and organismal biology, and revealing previously unknown mechanisms of disease formation.
Among the most popular noninvasive imaging tools are the bioluminescent proteins (luciferases).[1–10] Luciferases, unlike fluorescent probes, do not require incident radiation to produce light. Rather, these enzymes generate photons via the catalytic oxidation of small molecule substrates (luciferins). Luciferases can be expressed in numerous cell and tissue types, and when luciferin is present, light is produced.[1,11–13] There are virtually no endogenous light-emitting processes in cells and tissues, so the background signal for bioluminescence imaging in vivo is negligible. Such high signal-to-noise values are attractive for sensitive imaging applications within complex environments. Indeed, bioluminescence has been used to report on cell movements, gene expression patterns, and even the activities of individual biomolecules in whole tissues and living organisms.[11,14–16] In animals, bioluminescent light can usually be detected at depths of a few centimetres, although the exact limit is dependent on the brightness of the signal and the sensitivity of the detector. Based on these considerations, the technology is widely compatible with common rodent models and other small animals. The versatility and user-friendly features of bioluminescence have facilitated numerous biological discoveries.[3,17,18]
This review provides an overview of bioluminescence technology and how it can be used to monitor diverse biological features in complex environments. We first introduce luciferase–luciferin pairs commonly used for imaging and showcase their utility in visualizing cells and gene expression patterns in vivo. We then highlight methods to examine discrete biomolecules using engineered bioluminescent probes. These tools can illuminate the functions of proteins and other metabolites in cells and animals, providing a more detailed depiction of biological systems. Last, we describe ongoing work to expand the capabilities of bioluminescence technology using synthetic chemistry and molecular biology. Such advances promise to offer more refined views of cellular and biomolecular functions and potentially reveal new avenues for drug discovery.
Bioluminescent chemistries and colours
Several luciferase–luciferin combinations have been identified in nature, but only a few have been optimized for use in bioimaging applications (Table 1).[19] The bioluminescent pairs differ in size and shape, but all share a common mechanism for light generation: the luciferase catalyzes the oxidation of the complementary luciferin. During the enzymatic reaction, an electronically excited intermediate is produced; relaxation of this molecule to the ground state releases a photon of light. The colour (wavelength) of light is primarily dictated by the structure of the small molecule emitter, but also impacted by residues within the enzyme. Fluc and other insect luciferases release yellow photons (~550–600 nm) upon oxidation of a benzothiazole substrate (D-luciferin) with O2 and ATP.[20,21] While these luciferases utilize the same substrate, their emission spectra vary slightly owing to differences in enzyme architecture.[12,20,22,23] Luciferases from marine organisms, including Renilla reniformis (Rluc) and Gaussia princeps (Gluc), also catalyze the oxidation of a common imidazopyrazinone substrate, coelenterazine, and release primarily blue-green light (460–480 nm).[24] Rluc and Gluc, unlike Fluc, require no other cofactors (except O2), making them suitable for use in extracellular environments and other spaces lacking ATP.[25,26] Bioluminescent bacteria also release blue-green light, but require a combination of long-chain aldehydes, heterodimeric luciferases and flavin cofactors for the light-emitting reaction. Interestingly, the luciferase subunits and all enzymes required for bacterial luciferin production are coded within a single operon (lux).[27,28] Importing this entire gene sequence into non-luminescent bacteria—or even some eukaryotic cells—is sufficient to make them “glow” continuously.[29–31] By contrast, the luciferin substrates for Fluc, Gluc, and Rluc must be supplied exogenously in a given imaging experiment. The biosynthetic origins of coelenterazine and D-luciferin remain unknown, even though decades have passed since their chemical structures were first identified.[12,20,32–34] The elucidation of these biosynthetic pathways will be aided by efforts to isolate and characterize bioluminescent machinery from other organisms.[12,20,21,24,35–38]
Table 1. Luciferases and luciferins commonly used in bioluminescence imaging.
All luciferases catalyze the oxidation of small molecule substrates (luciferins) to release visible light. Popular luciferase–luciferin pairs, along with their characteristic features, are outlined below. Adapted from ref. 17, 19. *Emission wavelength at 25 °C

| |||||
|---|---|---|---|---|---|
luciferase
|
luciferin |
peak emission (nm) * | approximate size (kD) | comments | references |
| firefly luciferase (Fluc) |
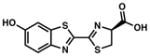 D-luciferin |
562 | 61 | Largest percentage of >600 nm light emitted among classes of luciferases. Primarily used intracellularly due to requirement for ATP. | 33, 27 |
| click beetle green (CB green) | 537 | 61 | |||
| click beetle red (CB red) | 613 | 61 | |||
| Renilla reniformis (Rluc/Rluc8) |
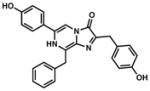 coelenterazine (CTZ) |
482/487 | 36 | All can be used extra-cellularly. Mutant versions (e.g., Rluc8/Rluc8.6/Gluc4) offer brighter and/or more sustained emission. | 25, 26, 32, 111, 137, 164 |
| Gaussia princeps (Gluc) | 482 | 20 | |||
| Aequorea victoria (Aequorin) | 470 | 22 | Aequorin is “pre-loaded” with CTZ, and light is emitted upon Ca2+ binding. | ||
| Lux AB |
long chain aldehydes + FMN cofactor |
480 | A: 42 B: 47 |
Mostly limited to use in bacteria ; lux operon (luxCDABE) encodes for all components necessary for light emission. | 27, 28, 31 |
| Vargula hilgendorfii (Vluc) |
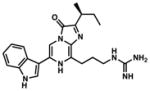 vargulin |
462 | 62 | Recently characterized bioluminescent system; can be used in tandem with other luciferase-luciferin pairs. | 38 |
Among the bioluminescent pairs, the luciferase and luciferin from the firefly (Fluc and D-luciferin, respectively) are the most widely used for imaging in vivo.[21] D-Luciferin is relatively stable and can penetrate most cell and tissue types following administration via standard i.p. injection.[39] Coelenterazine by contrast, is less bioavailable and typically requires intravenous administration to reach its targets in vivo. This luciferin is also cleared rapidly from animals and prone to air oxidation, resulting in non-specific background signal. Furthermore, coelenterazine luminescence with Rluc and Gluc is blue-green in colour; wavelengths of this sort are readily absorbed by haemoglobin and other chromophores in vivo, preventing their detection by imaging cameras.[16] Red light (>600 nm) more readily passes through blood and overlying tissues, and these wavelengths are the ones captured by detectors in a typical bioluminescence experiment.[11,16] While Rluc and Gluc emit mostly blue-green light, their emission spectra are sufficiently broad to contain wavelengths >600 nm, and are thus useful for routine bioluminescence imaging. Fluc and click beetle luciferases emit a larger percentage of red light, making them more suitable for in vivo applications.[40]
Traditional applications of bioluminescence imaging
Bioluminescence was first harnessed for in vivo imaging in 1995, when the lux operon was introduced into a non-luminescent strain of Salmonella typhimurium.[31] Upon inoculation into mice, the “glowing” bacteria could be readily identified and even localized (noninvasively) to discrete tissues. Moreover, changes in the infection profile were easily visualized in response to antibiotic treatment. This classic study showcased the remarkable sensitivity and broad dynamic range of bioluminescence for imaging in live animals, along with its potential for facilitating drug discovery.
Since then, bioluminescence has evolved into a mainstream technique for visualizing not only bacteria, but also viruses and other pathogens, eukaryotic cells, and even gene expression patterns in live organisms.[41,42] Variants of Fluc, Rluc, and Gluc that offer brighter and more sustained light emission have been described (e.g., Rluc8, Rluc8.6), [26,43,44] and these can be readily introduced into numerous cell and tissue types using standard gene transfer techniques. For cells and tissues that are refractory to genetic manipulation, luciferase-labelled transgenic mice can serve as convenient sources of bioluminescent material.[1] Finally, the common luciferin substrates can be purchased from commercial vendors, and standard bioluminescence detectors are available in most research settings. The relative simplicity of bioluminescence imaging, combined with these user-friendly features, has enabled rapid discoveries in a broad spectrum of fields.[3,45,46]
Cell tracking
Luciferase-expressing cells can be imaged repeatedly in live animals, with the intensity of the signal correlating with the number of cells in a given area.[1] Thus, bioluminescence imaging is well suited for tracking cells over time in vivo, and many experiments—spanning several disciplines—have capitalized on this feature. For example, Contag and co-workers utilized bioluminescence to monitor haematopoiesis following bone marrow transplantation.[47] Such transplants are routinely used to treat leukaemia and other blood cancers. In a typical procedure, a patient’s diseased cells are first ablated (via radiation) and then replaced by blood cells from a healthy donor. The donor cells responsible for blood regeneration are haematopoietic stem cells (HSCs) found in bone marrow. HSCs initially engraft in the recipient before dividing and differentiating to reconstitute the haematopoietic system. The success of bone marrow transplantation has been mixed due to host immune clearance and other mechanisms. Thus, methods to visualize HSCs at early time points post-transplantation could offer insights into haematopoiesis and methods to improve the therapy. Toward this end, the authors harvested bone marrow from luciferase-expressing transgenic mice and delivered it into irradiated recipient mice. The engraftment and proliferation of the cells was visualized upon D-luciferin administration. The images revealed that HSCs take up residence at multiple sites post-transplantation during haematopoiesis, with no single site necessary for full haematopoiesis (Fig 1A). In addition, it was noted that transplantation of a single HSC could result in successful immune cell outgrowth.
Fig. 1. Tracking cells and gene expression with bioluminescence imaging.
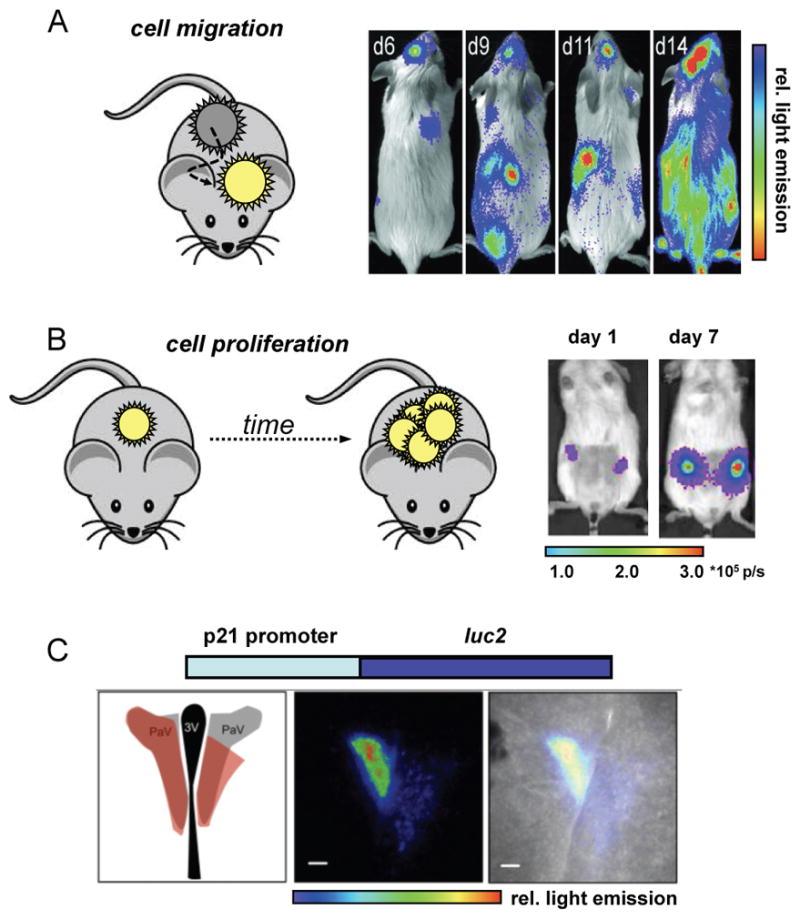
(A) Example of monitoring cell migration. Luciferase-labelled haematopoietic stem cells (HSCs) were transplanted into irradiated mice. Bioluminescence imaging with D-luciferin revealed multiple foci (sites of engraftment) at early time points. HSC differentiation and proliferation resulted in full regeneration of the blood system in these mice (as evident by the increase in spread and intensity of bioluminescent signal). Reprinted with permission from ref. 47. Copyright (2004) National Academy of Sciences, U.S.A. (B) Example of visualizing cell proliferation. Primary human cancer stem cells were transfected with luciferase genes and implanted into mouse mammary fat pads. Bioluminescence imaging revealed cell proliferation several weeks before palpable tumours emerged. Reprinted with permission from ref. 48. Copyright (2010) National Academy of Sciences, U. S. A. (C) To visualize gene expression, a luciferase gene (e.g., luc2) can be fused to the promoter for a gene of interest (e.g., p21). Cellular transcription of the target gene results in luciferase production. Luciferase was inserted downstream of an endogenous p21 promoter in transgenic mice, and regions of the mouse brain were probed for p21 activity using bioluminescence. From left to right: schematic of the mouse brain highlighting the paraventricular nucleus (PVN), the site of expected bioluminescence (red); bioluminescent image of brain tissue; overlay image showing localization of bioluminescent signal in the PVN. This research was originally published in ref. 61. Copyright (2013) the American Society for Biochemistry and Molecular Biology.
In a more recent example, bioluminescence technology was used to rapidly assay the proliferation of cancer stem cells in vivo and examine their roles in metastases.[48] The authors isolated cancer stem cells from patient breast tumour biopsies and transduced them with genes encoding Fluc or Rluc. The luciferase-expressing cells were implanted orthotopically in immunocompromised mice and monitored over time. The imaging studies revealed that only certain subsets of cells (i.e., “cancer stem cells”) were capable of proliferating in vivo and thus perpetuating tumour growth (Fig. 1B). These same subsets of cells were also located at secondary tumour sites, establishing one of the first connections between cancer stem cells and metastatic outgrowth. Impressively, as few as 10 Fluc-labelled cells could be visualized in vivo, suggesting that bioluminescent tools are amenable to monitoring patient tumour growth and therapeutic responses in surrogate hosts. Similar cell tracking experiments have been performed using immune cells, [49,50] stem cells, [51–55] and even pathogens.[31,45,56,57]
Visualizing gene expression
In addition to examining whole cell movements and behaviours, bioluminescence technology has been widely employed to monitor gene expression patterns in vivo.[26] Such studies can offer more detailed insights into biological mechanisms than cell tracking alone. In a typical experiment, luciferase expression is driven by the promoter sequence for a gene of interest. The bioluminescent enzyme is thus produced only when the target gene is being actively transcribed. In most cases, the amount of luciferase generated mirrors that of the target gene product, and can be readily visualized upon luciferin administration. Gene expression patterns relevant to inflammation, cancer, and Alzheimer’s disease have all been imaged (and often quantified) in this regard.1,58–60 In a recent example, Piwnica-Worms and colleagues utilized engineered mice to examine the dynamic expression of p21 in vivo. p21 is a cyclin-dependent protein kinase (CDK) inhibitor involved in cell cycle regulation.61 Mice expressing Fluc under the control of an endogenous p21 promoter were treated with D-luciferin and imaged. Interestingly, fluctuations in bioluminescent light were observed in discrete regions of the brain responsible for nutrient sensing (Fig. 1C). These results implicate p21 expression in the regulation of metabolism.
Recent advancements in probing biology with bioluminescence imaging
Monitoring protein abundance and function
A complete mechanistic understanding of living systems requires methods to probe not only cells and gene transcription, but also individual biomolecules. Proteins comprise one of the major classes of cellular biomolecules and fulfill an array of functions. Abnormal protein activities can disrupt major signalling networks, alter biosynthetic pathways, and compromise membrane integrity, all of which can potentiate disease. Given their central roles in cell structure and function, proteins are popular drug targets; new candidates for therapeutic targeting require an increased understanding of protein function in tissues and organisms. Bioluminescence imaging is already having an impact in this area.[3] Over the past decade, engineered luciferases and luciferins have been crafted to report on numerous facets of protein biology, including their localization and stability, interactions with other proteins, and enzymatic functions. Several examples are provided below.
Direct attachment of luciferase to proteins of interest can be used to monitor the location and abundance of the biomolecules. Indeed, luciferase fusions have been used to visualize a variety of proteins, including the signalling biomolecules HIF-1α and β-catenin, [62,63] along with proteins destined for proteasomal degradation.[64] Luciferase fusions alone, though, cannot provide read-outs on discrete protein activities, including their interactions with other proteins. Protein–protein interactions drive major signalling cascades in human cells, and disruption of these contacts often contributes to disease.[65] Luciferase probes can provide noninvasive readouts on protein associations and facilitate screens of therapeutic agents to modulate these networks. In one well-known approach, luciferase and a spectrally matched fluorescent probe are respectively fused to the two proteins of interest.[66] When the proteins are far apart, only bioluminescent light emission is observed in the presence of the appropriate luciferin. When the targets are in close proximity, the bioluminescent photons drive the excitation of the fluorescent chromophore, resulting in light emission at a longer wavelength. This phenomenon, termed bioluminescence resonance energy transfer (BRET) has been used to examine kinase–kinase, receptor–peptide, and other protein–protein interactions, in cells and live animals.[7,66,67] BRET studies require spectrally matched probes: the luciferase emission wavelengths must overlap the fluorescent probe’s excitation spectrum. Many Rluc-yellow fluorescent protein and Rluc-quantum dot combinations meet this criterion and have been utilized for visualizing protein interactions and other biological processes.[64,67,68] For work in vivo, the identification of more red-shifted BRET pairs remains an important goal.[69–73] The Gambhir and Piwnica-Worms labs recently developed BRET systems comprising luciferase–fluorescent protein fusions that emit 550–650 nm light, making them more attractive for deep tissue imaging in vivo.[44,69,74] Further improvements in BRET technology are expected with the identification of more luciferase–fluorescent protein duos in nature, [75] along with the development of improved luciferase–nanomaterial conjugates.[76,77]
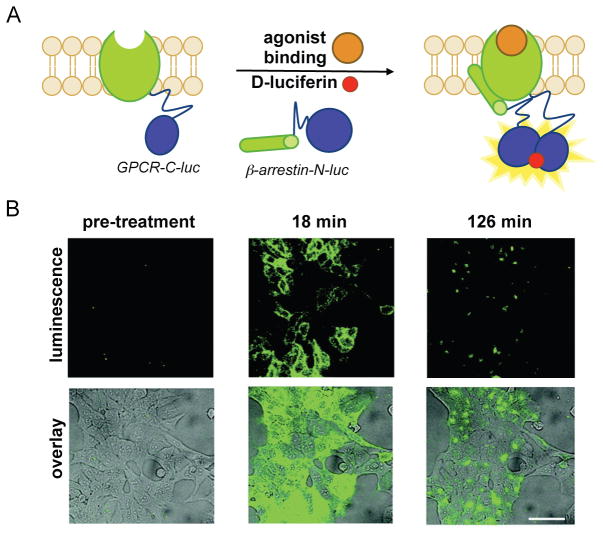
Protein–protein interactions can also be visualized using luciferase complementation.[70–73] In these assays, “split” fragments of a luciferase are attached to proteins of interest. Functional enzyme is produced only when the interacting proteins come into contact and drive the assembly of the complementary pieces.[72,73] “Split” versions of Fluc, Rluc, and Gluc have all been described, and used to monitor protein dimerization events, including the rapamycin-induced interaction of FRB and FKBP.[70,72,73,78] In a recent example, Ozawa and co-workers visualized the binding of β-arrestin to β-adrenergic receptor kinase (BARK), a membrane GPCR. When BARK is activated upon agonist binding, β-arrestin subsequently binds the GPCR to turn off the response and reset the system; the rate of β-arrestin association correlates with the potency of the ligand. The authors visualized BARK–β-arrestin binding using split Fluc probes.[79] Upon administration of small molecule agonists, conformational changes in BARK–C-luc promoted the binding of β-arrestin–N-luc (Fig. 2). This dimerization event enabled complementation between the two termini of Fluc, and ultimately light production. Using this split luciferase system, the authors quantified the relative potency of a panel of drugs.
Fig. 2. Visualizing protein–protein interactions with Fluc complementation.
(A) β-Arrestin is an antagonist of GPCRs, de-sensitizing the receptors upon external stimulation. When β-arrestin binds BARK (a known GPCR) in the presence of certain therapeutics, the GPCR is less responsive and, as a result, cells are less sensitive to hormones and other stimuli. BARK–β-arrestin dimerization can be visualized using “split” versions of Fluc. In the presence of GPCR agonists, β-arrestin binds the GPCR, enabling Fluc fragment complementation and bioluminescent signal production. (B) HEK cells expressing GPCR–C-luc and β-arrestin–N-luc were stimulated with a known agonist, isoproterenol and imaged over time. Bioluminescence signal is overlaid on bright field images. Reprinted with permission from ref. 79. Copyright (2012) American Chemical Society.
Measuring enzymatic activities in complex environments
Proteins drive cellular processes not only via their associations with other proteins, but also via enzymatic reactions. Several classes of enzymes have been identified to date, and many—including kinases and proteases—play integral roles in cell signalling. Aberrant enzyme activities are also associated with numerous pathologies and are commonly measured in clinical isolates to diagnose disease.[80,81] Engineered luciferases and luciferins have been developed to report on enzymatic activities in real time. In addition to providing a dynamic readout on protein function in complex environments, these imaging tools can facilitate screens for therapeutics designed to either inhibit or enhance enzymatic activity.[82]
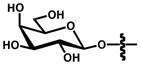
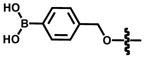
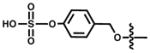
Among the most well recognized probes for measuring enzyme function are the “caged” luciferins. These molecules comprise luciferin cores outfitted with steric appendages (i.e., “cages”) or other groups that perturb its use in the bioluminescent reaction.[83] Most “ cages” are appended to the 6′ end of D-luciferin. A small, electron-donating group (e.g., OH or NH2) at this position is required for light production;[33] installing electron-withdrawing or bulky groups inhibits bioluminescence.[84,85] If the “cage” is labile to defined enzymatic activity, though, luciferin is released and available for the light emitting reaction. In these cases, light emission provides a direct readout on enzyme function. Phosphatase, sulfatase, and oxidase activity have all been visualized in this regard, and nearly a dozen caged luciferins are now commercially available to measure enzyme function in live cells or lysates (Table 2.)[83,86–93] Bertozzi and co-workers recently introduced a novel cage to report on the activity of certain sulfatases (row 5, Table 2).[94] The aryl sulphate caging group was found to be readily cleaved by sulfatases expressed in Mycobacterium tuberculosis, but not those found in human serum. These data suggest that the caged probe may have utility in the clinical diagnosis of pathogens.
Table 2.
Caged luciferins report on enzyme activities. “Caged” versions of D-luciferin have been used to image a variety of enzymatic activities and physiological states
In addition to engineered luciferins, “designer” luciferases can provide direct readouts on enzyme function.95–97 The majority of these probes comprise cyclic versions of luciferase that are locked into inactive conformations with various linkers. Upon cleavage of the tether (“activation”), a functional luciferase is produced. Wood and colleagues generated a series of such “activatable” luciferases comprising protease-specific linkers. Cleavage of the linkers in the presence of defined protease activity liberates Fluc to provide light emission, and ultimately a readout on protease function. One such “activatable” luciferase was recently used to monitor caspase-7 activity in both live cells and in vivo tumour models (Fig. 3).[98]
Fig. 3. Activatable luciferases report on enzymatic activities.
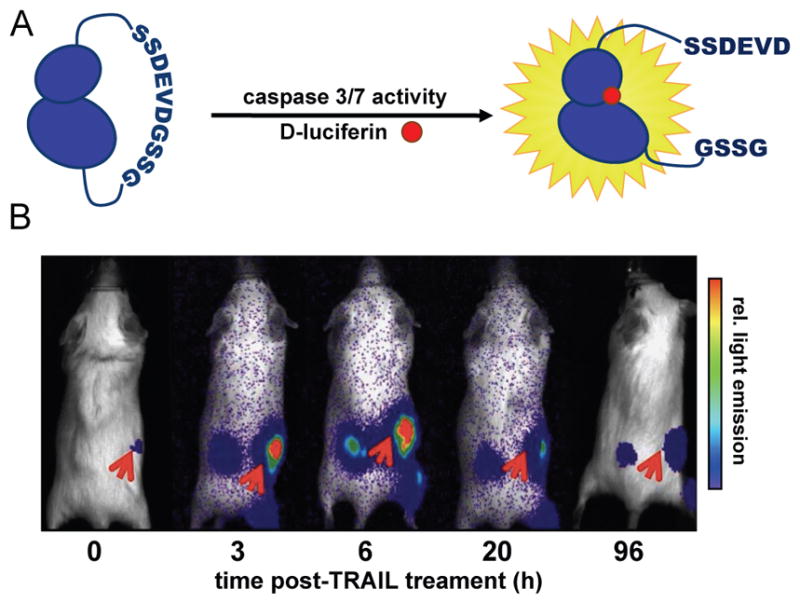
(A) A circular Fluc probe (caspase 3/7 GloSensor), comprising a specific caspase recognition sequence (DEVD) was prepared. In the absence of caspase activity, functional Fluc is not formed and bioluminescence is minimal. In the presence of caspase activity, the tether is cleaved, providing functional Fluc. (B) D54-MG glioma cells (2 × 106) stably expressing the caspase 3/7 bioluminescent reporter were implanted into NOD/SCID mice. When tumours emerged, the animals were treated with TRAIL, an activator of caspase activity. Bioluminescence imaging with D-luciferin revealed an increase in signal over time (red arrows), correlating with caspase induction. Reprinted in accordance with open-access license from ref. 98.
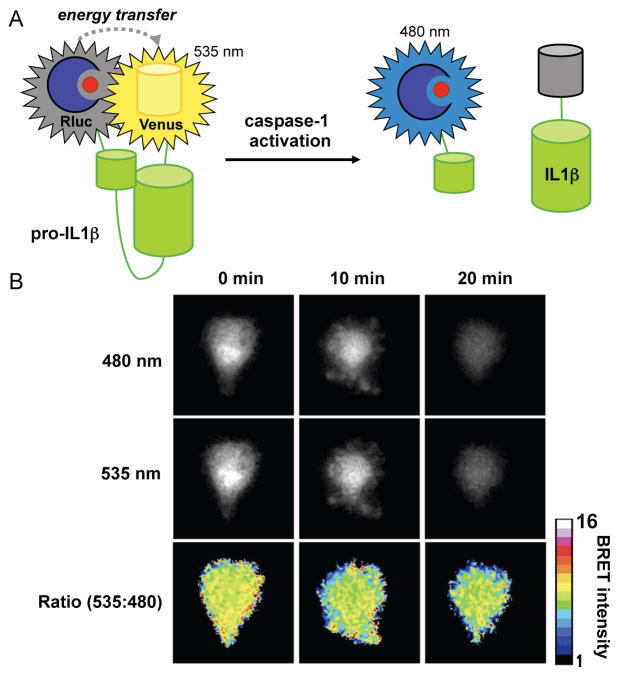
BRET sensors represent another major class of bioluminescent reporters for enzyme activity. These sensors comprise luciferases that are physically linked to a BRET-matched fluorescent protein.[66,99] If the adjoining link is severed by enzymatic activity, the luciferase and fluorescent protein diffuse apart, and the ensuing reduction in BRET signal correlates with enzyme function. A BRET sensor was recently developed to measure caspase activity in primary macrophages.[68] Caspase-1 is known to activate several inflammatory signalling molecules via proteolytic cleavage. The chemokine IL-1β is one such substrate that plays a pivotal role in cellular apoptosis. Active IL-1β is generated via caspase cleavage of a proprotein (pro-IL-1β). The pro-IL-1β cleavage is rapid and not easily monitored via Western blot or other traditional cell biology assays. To capture this activity, Pelegrin and co-workers designed a BRET sensor comprising Rluc8 and a yellow fluorescent protein (Venus) linked by pro-IL-1β. When expressed in cells devoid of high levels of caspase activity, the sensor remains intact and yellow light is produced upon coelenterazine treatment (Fig. 4). (The blue photons emitted by Rluc8 are absorbed by Venus and emitted as yellow light.) Upon caspase-1 activation and cleavage of the sensor, Rluc and Venus separate and mostly blue bioluminescent emission is observed (Fig 4A). The ratio of blue to yellow light in each case can provide a measure of caspase activity and IL-1β activation and, more broadly, the inflammatory response (Fig. 4B).
Fig. 4. Visualizing enzyme function with BRET sensors.
(A) IL-1β is produced as a non-active pro-protein in macrophages. When cleaved by the cysteine protease caspase-1, IL-1β stimulates a host of pro-inflammatory responses. The release of active IL-1β can be visualized using a BRET sensor. This probe comprises Rluc, Venus fluorescent protein, and an intervening IL-1β pro-protein sequence. Prior to caspase-1 activation, the sensor remains intact, with the luciferase and fluorescent protein in close proximity (BRET is observed). Upon caspase-mediated cleavage of the pro-protein, Rluc and Venus separate, resulting in reduced BRET signal. In the former case, 535 nm light (Venus emission) is observed in the presence of coelenterazine. In the latter case, mostly bioluminescent light (480 nm) is observed. (B) The Gluc-IL-1β-Venus sensor was transfected into primary macrophages. Upon caspase activation, images were acquired at both 535 nm and 480 nm. Decreased levels of 535:480 nm light were observed over time, correlating with of caspase activity and the presence of active IL-1β. Reprinted with permission from ref. 68. Copyright 2012, The American Association of Immunologists, Inc.
Probing metabolites with bioluminescent sensors
While proteins comprise the bulk of cellular matter, other biopolymers including glycans and lipids, in addition to metabolites and ions also play pivotal roles in cell biology. Methods to visualize the abundance and activities of these species are thus required for a complete understanding of living systems. Imaging such non-primary gene products has been historically challenging.[100] Such molecules cannot be fused genetically to luciferase or other optical reporters for direct tracking. Rather, indirect methods for visualization must often be employed.[101] Bioluminescent probes can be engineered to report on a diverse array of biomolecules, and examples from the recent literature are highlighted below.
Nucleic acids, like proteins, are often associated with certain physiological states and can report on the presence of pathogens. For example, methods to rapidly detect DNA from “foreign” microorganisms would simplify and hasten clinical diagnoses, where positive pathogen identification can take days. Toward this end, Ikebukuro and colleagues utilized bioluminescence to detect bacterial DNA in biologically relevant sample sizes.[102] Their platform involved zinc finger domains conjugated to Fluc. Zinc fingers recognize specific stretches of nucleic acids and can be readily evolved to bind virtually any target. Bioluminescence imaging of the DNA samples provided a visual readout of the desired sequence with femtomolar sensitivity without the need for gel analysis. Eventual automation this type of procedure is promising for clinical translation. Bioluminescent tools have also been applied to image other major classes of biopolymers, including glycans and lipids.[103–105]
Certain small molecule metabolites can also be visualized with bioluminescence technology. For example, all luciferases require molecular oxygen for light production, and bioluminescent emission can be used to approximate O2 levels in solid tumours.106–108 Bioluminescence can also report on metabolites that are required for luciferase activities. For instance, Fluc is routinely employed to report on ATP levels in cell extracts and nucleotide identity in sequencing analyses.[109–111] The photoprotein aequorin requires exogenous Ca2+ for light production, and can be employed for visualizing calcium flux in nerve cells.[112] For other metabolites and analytes of interest, luciferase reporter genes are often used. Levels of glycolysis intermediates, hormones, and metals have been measured in this regard.[109,113–118] Small molecule metabolites have also been imaged using BRET constructs[119–121] and engineered luciferins.[83] In a recent example of the latter, cellular peroxide levels were measured with a phenyl boronate probe (entry 4, Table 2). The boronate group is labile to peroxide, one of many reactive oxygen species generated in activated cells.[93,122] More recently, the Chang lab modified this caged probe to report on two analytes relevant to inflammation.[123] This luciferin was ultimately used to image both ROS production and protease activity in various cancer cell lines. We anticipate further multi-analyte imaging studies as new methods for luciferin caging are developed.
Drug screening and drug development
Since Fluc light emission requires ATP and new protein production, bioluminescence is a simple and direct assay for cell viability and proliferation.[40,48,50,55,73] Indeed, global reductions in light emission have been used as readouts of cell death in many examples introduced in this article.[124–128] Bioluminescence is also an attractive choice for monitoring cell viability and potential therapies in heterogeneous models. The McMillin group recently demonstrated the utility of bioluminescence for drug screening with mixtures of stroma and tumour cells—a more realistic model of human tumours.[129,130] The authors cultured various Fluc-expressing tumour cells with stromal cells in the presence and absence of common chemotherapies. Cytotoxicities were correlated with reductions in light output. While many of the cell lines demonstrated increased resistance to certain drug treatments in the presence of stromal cells, the effect was not common to all cell lines or drugs. These results demonstrate that bioluminescence imaging can provide a facile and high throughput screening method for models that better recapitulate the heterogeneity of human cancers and other diseases. That said, non-specific interactions between pharmacophores and firefly luciferase have historically confounded the discovery of small molecule therapeutics.[131–133] The development of new luciferases and luciferins could potentially circumvent these issues, and work is ongoing in this area.[134]
Beyond direct measures of cell death, bioluminescence has aided drug discovery efforts in additional areas. For example, this imaging technology can be used for the direct tracking of therapeutics in vivo. The ability to visualize both protein and small molecule biologics provides insights into delivery and targeting. In a recent example, Gluc was appended to an anti-CEA antibody and used to detect tumours expressing the CEA antigen in vivo.[135] Gluc is among the brightest luciferases characterized to date and is functional in extracellular environments, making it an attractive choice for diagnostic imaging.[136,137] Similarly, small molecule delivery can be probed using luciferins as surrogate drugs. In one example, Wender and co-workers used an octaarginine tailed luciferin to optimize the design of cationic peptide tags for intracellular drug delivery.[138] In these studies, bioluminescent emission correlated with successful drug localization. The authors ultimately utilized the arginine-rich tail to enhance the delivery of Taxol and other chemotherapeutics.
Building better bioluminescent tools
Despite the broad utility and user-friendly features of existing bioluminescent tools, challenges remain in expanding the scope of the technology. Some of the obstacles arise from the probes themselves. Many luciferases are only quasi-stable in mammalian cells.[59] This instability is useful for visualizing dynamic biological processes, where rapid turnover of the reporter is required. However, it is less desirable for long-term tracking studies or when large photon outputs and sustained emissions are desired. Many luciferases are also sensitive to pH, reactive-oxygen species and other environmental factors, although variants with improved, broad-range cellular compatibilities have been identified.[43,44,139–144] In a recent example, Tannous and co-workers evolved a variant of Gluc (Gluc4) that is more stable in standard mammalian cell assays and provides more sustained light emission.[145] These features were found to improve drug screening results by decreasing the number of false positives associated with rapid loss of emission with native Gluc. Luciferins, too, can be rapidly degraded in cellular environments or cleared readily from living systems. Ongoing work to identify more chemically robust substrates and formulations will benefit experiments requiring prolonged luciferin circulation.[138,148] Osmotic pumps and alternative delivery mechanisms will also be useful in this regard.[61,147]
The photon output from all luciferase enzymes is inherently weak, and only a fraction of the emitted light typically reaches the detector; the majority of bioluminescent photons are absorbed or scattered by endogenous chromophores in blood and tissues.[44,69] Improvements in bioluminescence sensitivity and depth are possible by engineering luciferases to produce more tissue penetrant (i.e., red-shifted) light. Branchini and coworkers generated one such luciferase by conjugating small molecule fluorophores (AlexaFluor 650 and 680) onto the surface of Fluc.[149] Following administration of D-luciferin, near infrared light is produced via BRET transfer from the bioluminescent reaction to the fluorophores on the enzyme surface. This chemically modified luciferase was used to image factor Xa in human blood samples, an assay that is problematic for traditional luciferase probes owing to photon absorption by haeme. While the protein conjugate is applicable to a variety of sensing assays ex vivo, it is not amenable to long-term cell tracking in vivo. For these studies, luciferase mutants with red-shifted emission spectra are desirable. A handful of such mutants have been described, although the gains in sensitivity due to altered emission wavelengths are often offset by lower enzymatic turnover.[11,22,23,150–152] One notable exception is an Rluc variant recently reported by the Gambhir group (Rluc8.6-535). This luciferase not only provides a nearly six-fold improvement in the fraction of red light emitted, but also exhibits greater overall photon output than native Rluc.[153] Additional improvements in luciferase function are expected as more structural information becomes available.[154]
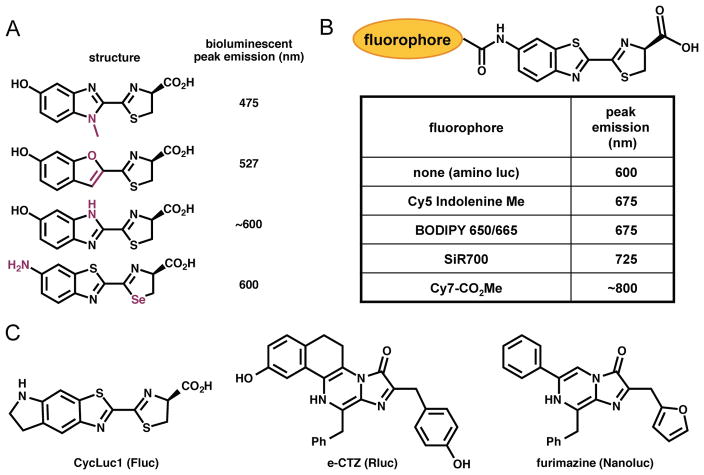
The majority of methods to alter bioluminescent spectra and function have focused on modifications to the luciferase scaffold.[43,145,152,155] An alternative strategy for generating new or brighter “colours” of bioluminescent light involves modulating the small molecule luciferins. Thus, modifying the luciferin core itself can result in altered emission spectra.[85,156–158] In a recent example, Conley, et al. synthesized a luciferin analogue comprising a selenium atom in place of sulfur (Fig. 5A). The altered electronic density in this heterocycle resulted in significant red-shifting of the emitted light.[158] Luciferin–fluorophore conjugates that exhibit altered wavelengths of emission have also been prepared (Fig. 5B).[159] Analogous to the fluorophore–luciferase conjugate, these molecules exploit BRET: the electronically excited luciferin core is capable of energy transfer to a covalently bound fluorophore. Urano and colleagues synthesized a variety of these conjugates and found that both luciferin–Cy7 and luciferin–SiR700 provided a significant enhancement in >600 nm photons. Not surprisingly, though, these bulky luciferins were poorly utilized by native luciferase, resulting in reduced overall photon production. Identifying mutant luciferases that can efficiently utilize these probes, along with more spectrally tuned luciferin–fluorophore conjugates (for maximal BRET efficiency) are important next steps.
Fig. 5. Luciferin architectures for improved bioluminescence imaging.
(A) Examples of heterocyclic analogues of D-luciferin that exhibit altered bioluminescent emission spectra. (B) Luciferin–fluorophore conjugates also exhibit red-shifted emissions. (C) Chemical structures of three luciferins that exhibit robust emission with various luciferases.
Synthetic modifications to the luciferin core cannot only provide altered bioluminescence spectra, but also improve the sensitivity of the imaging technique. In recent work, Miller and co-workers synthesized a cyclic aminoluciferin variant that is efficiently utilized by Fluc.[160] The rigidified structure limits the non-radiative relaxation of the excited state molecule, thus improving the quantum yield of bioluminescence. We further demonstrated that this molecule has improved cell and tissue permeance in a variety of cultured cell and mouse models, resulting in more sensitive imaging.[146] Improved signal-to-noise ratios with Rluc and Gluc are also possible using synthetic variants of coelenterazine.[161,162] Both rigidified analogues and shelf-stable coelenterazine variants (i.e., molecules less prone to auto-oxidation) have been found to exhibit better signal-to-noise ratios in Rluc imaging studies (Fig 5B).[163,164] Unfortunately, these and most other coelenterazine analogues are not efficiently utilized by Gluc.
Several groups are attempting to address the need for improved coelenterazines and other luciferins that are efficiently processed by luciferases. Wood and co-workers recently designed a coelenterazine derivative (furimazine, Fig. 5C) that is more stable than the native small molecule with lower rates of autoluminescence.[36] To capitalize on these features, the authors engineered a new enzyme specific to this designer luciferin. As a starting point, they turned to the luciferase from the bioluminescent deep-sea shrimp Oplophorus gracilirostris. This enzyme (106 kDa) comprises two large regulatory units and two smaller catalytic cores. A single catalytic unit (19 kDa) is capable of light-emission with coelenterazine, but in the absence of the regulatory piece, the enzyme is unstable. The authors used rational mutagenesis to generate more robust variants of the catalytic unit. To impart selectivity for furimazine over coelenterazine, they used additional rounds of mutagenesis and screened clones for stable light emission and overall light output. They eventually identified a variant, termed Nanoluc, that was over 100-fold brighter than Rluc and currently ranks among the smallest luciferases for biological imaging. Miller and co-workers used a similar approach to identify mutant versions of firefly luciferase that more efficiently catalyze light production with aminoluciferin variants.[165]
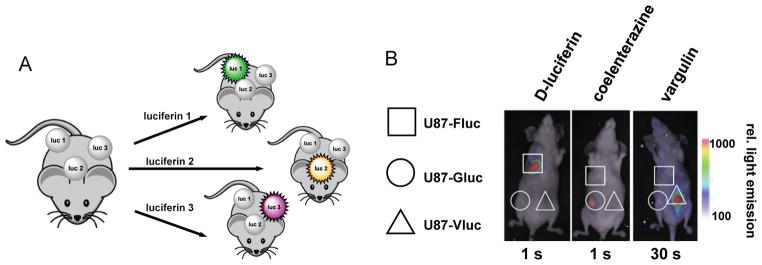
The development of brighter and spectrally altered tools will continue to expand the capabilities of bioluminescence imaging in vivo. The ultimate goal would be to generate a diverse collection luciferase–luciferin pairs that could be used simultaneously for imaging in vivo. Such tools would enable multi-component cell tracking experiments, and monitoring of signal transduction in real time. So far, the majority of tools designed to expand the set of luminescent probes have focused on mutants with altered emission spectra (analogous to the fluorescent protein palette). Separating the bioluminescent pairs by colour alone, though, is a lofty challenge, especially in vivo where the “colour” of light observed by the detector is skewed by the depth of the emitter in tissue. This means that separating the blue-green-emitting Rluc from the yellow-emitting Fluc, by colour, is difficult.[16,166] For this reason, multi-spectral bioluminescence imaging has largely been limited to studies in vitro.[78,151,167] While luciferases are not easy to spectrally resolve, they can be “biochemically” distinguished based on their specificities for distinct substrates. Sequential application of the substrates enables the desired targets to be visualized in a single organism. This approach has already been applied to imaging mesenchymal stem cells and their interactions with breast cancer in vivo where the stem cells were tagged with an Fluc reporter, while breast cancer cells were monitored with Rluc.[168] Most recently, the Vargula (Vluc) luciferase–luciferin pair was used in tandem with Rluc and Fluc, enabling three distinct tumour cell populations to be imaged in mice (Fig. 6).[169]
Fig. 6. Orthogonal luciferase–luciferin pairs enable multi-component imaging in vivo.
(A) Three unique cell types can be visualized using distinct luciferase–luciferin pairs. Sequential administration of the luciferins enables the target cells to be visualized. (B) Fluc- (square), Gluc- (circle) and Vluc- (triangle) expressing glioma cells were implanted in distinct regions in a mouse model. The cells were selectively illuminated with D-luciferin, coelenterazine, and vargulin respectively, with one day separating each injection. Reprinted from ref. 169 by permission from Macmillan Publishers, Lt: Molecular Therapy – Nucleic Acids.
Moving beyond the naturally occurring systems, we expect that the design and implementation of orthogonal luciferase probes will be aided by new, improved and rapid syntheses of luciferin analogues.[19,157,170] We recently introduced a highly divergent luciferin synthesis that enables facile access to a variety of potential substrates.[157] Coupled with efficient means for the generation of luciferase mutants, we anticipate the development of more orthogonal bioluminescence systems.[10,169,171–173]
With the continued development of new luciferases and luciferins, along with other luminescent platforms, [174,175] researchers will soon have access to an expanded set of bioimaging tools. These probes will broaden the scope of noninvasive imaging, providing insights into macroscopic, multi-cellular behaviours ranging from immune function to tumour heterogeneity. We also expect creative new applications of bioluminescence in visualizing cell–cell contacts and other microscopic behaviours. Collectively, these studies will continue to refine our understanding of biological systems and reveal new opportunities for therapeutic development.
Acknowledgments
We thank Will Porterfield and Rachel Steinhardt for critical reading of the manuscript, as well as all members of the Prescher lab for informative discussions.
References
- 1.Contag CH, Bachmann MH. Annu Rev Biomed Eng. 2002;4:235. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 2.Massoud TF, Gambhir SS. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 3.Duda J, Karimi M, Negrin RS, Contag CH. Methods Mol Med. 2007;134:17. doi: 10.1007/978-1-59745-223-6_2. [DOI] [PubMed] [Google Scholar]
- 4.Chudakov DM, Lukyanov KA. Biochemistry (Mosc) 2003;68:952. doi: 10.1023/a:1026048109654. [DOI] [PubMed] [Google Scholar]
- 5.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Nat Biotechnol. 2005;23:313. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 6.Sarantopoulos A, Beziere N, Ntziachristos V. Annu Biomed Eng. 40:346. doi: 10.1007/s10439-011-0501-4. [DOI] [PubMed] [Google Scholar]
- 7.Rao J, Dragulescu-Andrasi A, Yao H. Curr Opin Biotechnol. 2007;18:17. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Germain RN, Robey EA, Cahalan MD. Science. 336:1676. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnsson N, Johnsson K. ACS Chem Biol. 2007;2:31. doi: 10.1021/cb6003977. [DOI] [PubMed] [Google Scholar]
- 10.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 12.Viviani VR. Cell Mol Life Sci. 2002;59:1833. doi: 10.1007/PL00012509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greer LF, 3rd, Szalay AA. Luminescence. 2002;17:43. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 14.Choy G, O’Connor S, Diehn FE, Costouros N, Alexander HR, Choyke P, Libutti SK. BioTechniques. 2003;35:1022. doi: 10.2144/03355rr02. [DOI] [PubMed] [Google Scholar]
- 15.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, Cooper LJ, Gelovani J, Hwu P. Proc Natl Acad Sci USA. 2008;105:14342. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice BW, Cable MD, Nelson MB. J Biomed Opt. 2001;6:432. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 17.Luker KE, Luker GD. Antiviral Res. 86:93. doi: 10.1016/j.antiviral.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang NF, Okogbaa J, Babakhanyan A, Cooke JP. Theranostics. 2:346. doi: 10.7150/thno.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescher JA, Contag CH. Curr Opin Chem Biol. 2010;14:80. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Hastings JW. Gene. 1996;173:5. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- 21.Fraga H. Photochem Photobiol Sci. 2008;7:146. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 22.Branchini BR, Magyar RA, Murtiashaw MH, Anderson SM, Helgerson LC, Zimmer M. Biochemistry. 1999;38:13223. doi: 10.1021/bi991181o. [DOI] [PubMed] [Google Scholar]
- 23.Miloud T, Henrich C, Hammerling GJ. J Biomed Opt. 2007;12:54018. doi: 10.1117/1.2800386. [DOI] [PubMed] [Google Scholar]
- 24.Haddock SH, Moline MA, Case JF. Annu Rev Mar Sci. 2010;2:443. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- 25.Bhaumik S, Gambhir SS. Proc Natl Acad Sci USA. 2002;99:377. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Mol Ther. 2005;11:435. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Close D, Xu T, Smartt A, Rogers A, Crossley R, Price S, Ripp S, Sayler G. Sensors. 2012;12:732. doi: 10.3390/s120100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashiro T, Ruby EG. Mol Microbiol. 2012;84:795. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Hook M, Cirillo JD, Skare JT. Mol Microbiol. 2011;82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrissey R, Hill C, Begley M. Trends Food Sci Technol. 2013;32:4. [Google Scholar]
- 31.Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Mol Microbiol. 1995;18:593. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 32.Hori K, Charbonneau H, Hart RC, Cormier MJ. Proc Natl Acad Sci USA. 1977;74:4285. doi: 10.1073/pnas.74.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White E, McCapra F, Field G, McElroy W. J Am Chem Soc. 1961;83:2402. [Google Scholar]
- 34.Day JC, Tisi LC, Bailey MJ. Luminescence. 2004;19:8. doi: 10.1002/bio.749. [DOI] [PubMed] [Google Scholar]
- 35.Viviani VR, Bechara EJ, Ohmiya Y. Biochemistry. 1999;38:8271. doi: 10.1021/bi9900830. [DOI] [PubMed] [Google Scholar]
- 36.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. ACS Chem Biol. 2012;7:1848. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Branchini BR, Behney CE, Southworth TL, Rawat R, Deheyn DD. Photochem Photobiol. 2013 doi: 10.1111/php.12169. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 38.Otsuji T, Okuda-Ashitaka E, Kojima S, Akiyama H, Ito S, Ohmiya Y. Anal Biochem. 2004;329:230. doi: 10.1016/j.ab.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Berger F, Paulmurugan R, Bhaumik S, Gambhir SS. Eur J Nucl Med Mol Imaging. 2008;35:2275. doi: 10.1007/s00259-008-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer LF, Szalay AA. Luminescence. 2002;17:43. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 41.De Almeida PE, Van Rappard JRM, Wu JC. Am J Physiol: Heart Circ Physiol. 2011;301:H663. doi: 10.1152/ajpheart.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadikot RT, Blackwell TS. Methods Mol Biol. 2008;477:383. doi: 10.1007/978-1-60327-517-0_29. [DOI] [PubMed] [Google Scholar]
- 43.Maguire CA, van der Mijn JC, Degeling MH, Morse D, Tannous BA. Mol Imaging. 11:13. [PubMed] [Google Scholar]
- 44.Loening AM, Wu AM, Gambhir SS. Nat Methods. 2007;4:641. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 45.Badr CE, Tannous BA. Trends Biotechnol. 29:624. doi: 10.1016/j.tibtech.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Curr Opin Biotech. 2009;20:45. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao YA, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Proc Natl Acad Sci USA. 2004;101:221. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF. Proc Natl Acad Sci USA. 2010;107:18115. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy CT, Moloney G, Hall LJ, Quinlan A, Faivre E, Casey P, Shanahan F, Melgar S, Nally K. Clin Exp Immunol. 2010;162:188. doi: 10.1111/j.1365-2249.2010.04234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olson JA, Zeiser R, Beilhack A, Goldman JJ, Negrin RS. J Immunol. 2009;183:3219. doi: 10.4049/jimmunol.0804268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vocht ND, Lin D, Praet J, Hoornaert C, Reekmans K, Blon DL, Daans J, Pauwels P, Goossens H, Hens N, Berneman Z, Linden AVd, Ponsaerts P. Immunobiology. 2013;218:696. doi: 10.1016/j.imbio.2012.08.266. [DOI] [PubMed] [Google Scholar]
- 52.Reagan MR, Kaplan DL. Stem Cells. 2011;29:920. doi: 10.1002/stem.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutschka I, Chen IY, Kofidis T, von Degenfeld G, Sheikh AY, Hendry SL, Hoyt G, Pearl J, Blau HM, Gambhir SS, Robbins RC. J Heart Lung Transplant. 2007;26:273. doi: 10.1016/j.healun.2006.11.604. [DOI] [PubMed] [Google Scholar]
- 54.Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Science. 2010;329:1078. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Nature. 2008;456:502. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah K, Weissleder R. NeuroRx. 2005;2:215. doi: 10.1602/neurorx.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keyaerts M, Caveliers V, Lahoutte T. Trends Mol Med. 18:164. doi: 10.1016/j.molmed.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Watts JC, Giles K, Grillo SK, Lemus A, Dearmond SJ, Prusiner SB. Proc Natl Acad Sci. 2011;108:2528. doi: 10.1073/pnas.1019034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terashima M, Ehara S, Yang E, Kosuge H, Tsao PS, Quertermous T, Contag CH, Mcconnell MV. Mol Imaging Biol. 2011;13:1061. doi: 10.1007/s11307-010-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moss BL, Elhammali A, Fowlkes T, Gross S, Vinjamoori A, Contag CH, Piwnica-Worms D. J Biol Chem. 2012;287:31359. doi: 10.1074/jbc.M112.364018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tinkum KL, White LS, Marpegan L, Herzog E, Piwnica-Worms D, Piwnica-Worms H. J Biol Chem. 2013;39:27999. doi: 10.1074/jbc.M113.494328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, Serganova I. PLoS One. 2009;4:e5077. doi: 10.1371/journal.pone.0005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naik S, Piwnica-Worms D. Proc Natl Acad Sci USA. 2007;104:17465. doi: 10.1073/pnas.0704465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perroy J, Pontier S, Charest PG, Aubry M, Bouvier M. Nat Methods. 2004;1:203. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- 65.Ryan DP, Matthews JM. Curr Opin Struct Biol. 2005;15:441. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 66.De A. Curr Pharm Biotechnol. 2011;12:558. doi: 10.2174/138920111795163922. [DOI] [PubMed] [Google Scholar]
- 67.Prinz A, Diskar M, Herberg FW. Chembiochem. 2006;7:1007. doi: 10.1002/cbic.200600048. [DOI] [PubMed] [Google Scholar]
- 68.Compan V, Baroja-Mazo A, Bragg L, Verkhratsky A, Perroy J, Pelegrin P. J Immunol. 2012;189:2131. doi: 10.4049/jimmunol.1201349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Proc Natl Acad Sci USA. 108:12060. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Massoud TF, Paulmurugan R, Gambhir SS. FASEB J. 2004;18:1105. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nyfeler B, Michnick SW, Hauri HP. Proc Natl Acad Sci USA. 2005;102:6350. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Massoud TF, Paulmurugan R, De A, Ray P, Gambhir SS. Curr Opin Biotechnol. 2007;18:31. doi: 10.1016/j.copbio.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villalobos V, Naik S, Piwnica-Worms D. Annu Rev Biomed Eng. 2007;9:321. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 74.Gammon ST, Villalobos VM, Roshal M, Samrakandi M, Piwnica-Worms D. Biotechnol Prog. 2009;25:559. doi: 10.1002/btpr.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Widder EA. Science. 2010;328:704. doi: 10.1126/science.1174269. [DOI] [PubMed] [Google Scholar]
- 76.Ma N, Marshall AF, Rao J. J Am Chem Soc. 132:6884. doi: 10.1021/ja101378g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Nat Biotechnol. 2006;24:339. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 78.Paulmurugan R, Massoud TF, Huang J, Gambhir SS. Cancer Res. 2004;64:2113. doi: 10.1158/0008-5472.can-03-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takakura H, Hattori M, Takeuchi M, Ozawa T. ACS Chem Biol. 2012;7:901. doi: 10.1021/cb200360z. [DOI] [PubMed] [Google Scholar]
- 80.Reichling J, Kaplan M. Dig Dis Sci. 1988;33:1601. doi: 10.1007/BF01535953. [DOI] [PubMed] [Google Scholar]
- 81.Straub V, De Waele L, Barresi R. In: Muscle Disease: Pathology and Genetics. Goebel CSaRWHH., editor. John Wiley & Sons; Oxford: 2013. [Google Scholar]
- 82.Razgulin A, Ma N, Rao J. Chem Soc Rev. 40:4186. doi: 10.1039/c1cs15035a. [DOI] [PubMed] [Google Scholar]
- 83.Li J, Chen L, Du L, Li M. Chem Soc Rev. 2012;42:662. doi: 10.1039/c2cs35249d. [DOI] [PubMed] [Google Scholar]
- 84.Branchini BR. Meth Enzymol. 2000;305:188. doi: 10.1016/s0076-6879(00)05488-4. [DOI] [PubMed] [Google Scholar]
- 85.Meroni E, Santaniello G, Rajabi M. ARKIVOC. 2009:265. [Google Scholar]
- 86.Kindermann M, Roschitzki-Voser H, Caglic D, Repnik U, Miniejew C, Mittl PR, Kosec G, Grutter MG, Turk B, Wendt KU. Chem Biol. 2010;17:999. doi: 10.1016/j.chembiol.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 87.O’Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV. J Biomol Screen. 2005;10:137. doi: 10.1177/1087057104271865. [DOI] [PubMed] [Google Scholar]
- 88.Wehrman TS, von Degenfeld G, Krutzik PO, Nolan GP, Blau HM. Nat Methods. 2006;3:295. doi: 10.1038/nmeth868. [DOI] [PubMed] [Google Scholar]
- 89.O’Brien MA, Moravec RA, Riss TL, Bulleit RF. Methods Mol Biol. 2008;414:163. doi: 10.1007/978-1-59745-339-4_13. [DOI] [PubMed] [Google Scholar]
- 90.Dragulescu-Andrasi A, Liang G, Rao J. Bioconjugate Chem. 2009;20:1660. doi: 10.1021/bc9002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leippe DM, Nguyen D, Zhou M, Good T, Kirkland TA, Scurria M, Bernad L, Ugo T, Vidugiriene J, Cali JJ, Klaubert DH, O’Brien MA. Biotechniques. 2011;51:105. doi: 10.2144/000113716. [DOI] [PubMed] [Google Scholar]
- 92.Zhou W, Valley MP, Shultz J, Hawkins EM, Bernad L, Good T, Good D, Riss TL, Klaubert DH, Wood KV. J Am Chem Soc. 2006;128:3122. doi: 10.1021/ja058519o. [DOI] [PubMed] [Google Scholar]
- 93.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. Proc Natl Acad Sci USA. 2010;107:21316. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rush JS, Beatty KE, Bertozzi CR. Chembiochem. 2010;11:2096–2099. doi: 10.1002/cbic.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stevens GB, Silver DA, Zgaga-Griesz A, Bessler WG, Vashist SK, Patel P, Achazi K, Strotmeier J, Worbs S, Dorner MB, Dorner BG, Pauly D, Rummel A, Urban GA, Krueger M. Analyst. 2013;138:6154. doi: 10.1039/c3an00525a. [DOI] [PubMed] [Google Scholar]
- 96.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. ACS Chem Biol. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 97.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Angew Chem Int Ed Engl. 2007;46:7595. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 98.Galban S, Jeon YH, Bowman BM, Stevenson J, Sebolt KA, Sharkey LM, Lafferty M, Hoff BA, Butler BL, Wigdal SS, Binkowski BF, Otto P, Zimmerman K, Vidugiris G, Encell LP, Fan F, Wood KV, Galban CJ, Ross BD, Rehemtulla A. PLoS One. 2013;8:e66248. doi: 10.1371/journal.pone.0066248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xia Z, Rao J. Curr Opin Biotech. 2009;20:37. doi: 10.1016/j.copbio.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 101.Sletten EM, Bertozzi CR. Angew Chem Int Ed Engl. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abe K, Kumagai T, Takahashi C, Kezuka A, Murakami Y, Osawa Y, Motoki H, Matsuo T, Horiuchi M, Sode K, Igimi S, Ikebukuro K. Anal Chem. 2012;84:8028. doi: 10.1021/ac3018845. [DOI] [PubMed] [Google Scholar]
- 103.Contessa JN, Bhojani MS, Freeze HH, Ross BD, Rehemtulla A, Lawrence TS. Clin Cancer Res. 16:3205. doi: 10.1158/1078-0432.CCR-09-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shekhawat SS, Ghosh I. Curr Opin Chem Biol. 15:789. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Henkin AH, Cohen AS, Dubikovskaya EA, Park HM, Nikitin GF, Auzias MG, Kazantzis M, Bertozzi CR, Stahl A. ACS Chem Biol. 7:1884. doi: 10.1021/cb300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cairns RA, Papandreou I, Sutphin PD, Denko NC. Proc Natl Acad Sci USA. 2007;104:9445. doi: 10.1073/pnas.0611662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moriyama EH, Niedre MJ, Jarvi MT, Mocanu JD, Moriyama Y, Subarsky P, Li B, Lilge LD, Wilson BC. Photochem Photobiol Sci. 2008;7:675. doi: 10.1039/b719231b. [DOI] [PubMed] [Google Scholar]
- 108.Saha D, Dunn H, Zhou H, Harada H, Hiraoka M, Mason RP, Zhao D. J Vis Exp. 2011;56:e3175. doi: 10.3791/3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kricka LJ. Anal Biochem. 1988;175:14. doi: 10.1016/0003-2697(88)90354-5. [DOI] [PubMed] [Google Scholar]
- 110.Umezawa Y, Ozawa T, Sato M. Anal Sci. 2002;18:503. doi: 10.2116/analsci.18.503. [DOI] [PubMed] [Google Scholar]
- 111.Ahmadian A, Ehn M, Hober S. Clin Chim Acta. 2006;363:83. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 112.Naumann EA, Kampff AR, Prober DA, Schier AF, Engert F. Nat Neurosci. 2010;4:513. doi: 10.1038/nn.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hattori M, Haga S, Takakura H, Ozaki M, Ozawa T. Proc Natl Acad Sci USA. 2013;110:9332. doi: 10.1073/pnas.1304056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hemeon I, Gutierrez JA, Ho MC, Schramm VL. Anal Chem. 2011;83:4996. doi: 10.1021/ac200816m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taneoka A, Sakaguchi-Mikami A, Yamazaki T, Tsugawa W, Sode K. Biosens Bioelectron. 2009;25:76. doi: 10.1016/j.bios.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 116.Walenta S, Schroeder T, Mueller-Klieser W. Biomol Eng. 2002;18:249. doi: 10.1016/s1389-0344(01)00107-1. [DOI] [PubMed] [Google Scholar]
- 117.Kim SB, Sato M, Tao H. Anal Chem. 2009;81:3760–3768. doi: 10.1021/ac802674w. [DOI] [PubMed] [Google Scholar]
- 118.Kabiersch G, Rajasärkkä J, Tuomela M, Hatakka A, Virta M, Steffen K. Anal Chem. 2013;85:5740. doi: 10.1021/ac4003062. [DOI] [PubMed] [Google Scholar]
- 119.Binkowski B, Fan F, Wood K. Curr Opin Biotechnol. 2009;20:14. doi: 10.1016/j.copbio.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 120.Paulmurugan R, Gambhir SS. Proc Natl Acad Sci USA. 2006;103:15883. doi: 10.1073/pnas.0607385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim SB, Umezawa Y, Kanno KA, Tao H. ACS Chem Biol. 2008;3:359. doi: 10.1021/cb800004s. [DOI] [PubMed] [Google Scholar]
- 122.Ren H, Xiao F, Zhan K, Kim YP, Xie H, Xia Z, Rao J. Angew Chem Int Ed. 2009;48:9658. doi: 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Godinat A, Park HM, Miller SC, Cheng K, Hanahan D, Sanman LE, Bogyo M, Yu A, Nikitin GF, Stahl A, Dubikovskaya EA. ACS Chem Biol. 2013;8:987. doi: 10.1021/cb3007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petty RD, Sutherland LA, Hunter EM, Cree IA. J Biolumin Chemilumin. 1995;10:29. doi: 10.1002/bio.1170100105. [DOI] [PubMed] [Google Scholar]
- 125.Kalra J, Anantha M, Warburton C, Waterhouse D, Yan H, Yang YJ, Strut D, Osooly M, Masin D, Bally MB. Cancer Biol Ther. 11:826. doi: 10.4161/cbt.11.9.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kasahara N, Kikuchi T, Doi J, Teratani T, Fujimoto Y, Uemoto S, Yasuda Y, Kobayashi E. Transplant Proc. 45:2486. doi: 10.1016/j.transproceed.2013.02.117. [DOI] [PubMed] [Google Scholar]
- 127.Kodack DP, Chung E, Yamashita H, Incio J, Duyverman AMMJ, Song Y, Farrar CT, Huang Y, Ager E, Kamoun W, Goel S, Snuderl M, Lussiez A, Hiddingh L, Mahmood S, Tannous BA, Eichler AF, Fukumura D, Engelman JA, Jain RK. Proc Natl Acad Sci USA. 2012;109:E3119. doi: 10.1073/pnas.1216078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kolibab K, Derrick SC, Jacobs WR, Morris SL. J Microbiol Methods. 2012;90:245. doi: 10.1016/j.mimet.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 129.McMillin DW, Delmore J, Negri JM, Vanneman M, Koyama S, Schlossman RL, Munshi NC, Laubach J, Richardson PG, Dranoff G, Anderson KC, Mitsiades CS. Blood. 2012;119:e131. doi: 10.1182/blood-2011-04-348490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, Mitsiades N, Schlossman RL, Munshi NC, Kung AL, Griffin JD, Richardson PG, Anderson KC, Mitsiades CS. Nat Med. 2010;16:483. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Auld DS, Thorne N, Nguyen DT, Inglese J. ACS Chem Biol. 2008;3:463. doi: 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thorne N, Auld DS, Inglese J. Curr Opin Chem Biol. 2010;14:315. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat Chem Biol. 2007;3:466. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 134.Ho P-i, Yue K, Pandey P, Breault L, Harbinski F, McBride AJ, Webb B, Narahari J, Karassina N, Wood KV, Hill A, Auld DS. ACS Chem Biol. 2013;8:1009. doi: 10.1021/cb3007264. [DOI] [PubMed] [Google Scholar]
- 135.Venisnik KM, Olafsen T, Gambhir SS, Wu AM. Mol Imaging Biol. 2007;9:267. doi: 10.1007/s11307-007-0101-8. [DOI] [PubMed] [Google Scholar]
- 136.Kelkar M, De A. Curr Opin Pharmacol. 2012;12:592. doi: 10.1016/j.coph.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 137.El-Sayed R, Eita M, Barrefelt A, Ye F, Jain H, Fares M, Lundin A, Crona M, Abu-Salah K, Muhammed M, Hassan M. Nano Lett. 2013;13:1393. doi: 10.1021/nl304123u. [DOI] [PubMed] [Google Scholar]
- 138.Dubikovskaya EA, Thorne SH, Pillow TH, Contag CH, Wender PA. Proc Natl Acad Sci USA. 2008;105:12128. doi: 10.1073/pnas.0805374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bovenberg MS, Degeling MH, Tannous BA. Anal Chem. 2013;84:1189. doi: 10.1021/ac202833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thompson JF, Hayes LS, Lloyd DB. Gene. 1991;103:171. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 141.Leclerc GM, Boockfor FR, Faught WJ, Frawley LS. Biotechniques. 2000;29:590. doi: 10.2144/00293rr02. [DOI] [PubMed] [Google Scholar]
- 142.Koksharov MI, Ugarova NN. Biochemistry Moscow. 2008;73:862. doi: 10.1134/s0006297908080038. [DOI] [PubMed] [Google Scholar]
- 143.Ugarova NN. Moscow Univ Chem Bull. 2010;65:139. [Google Scholar]
- 144.Czupryna J, Tsourkas A. PLoS One. 2011;6:e20073. doi: 10.1371/journal.pone.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Degeling MH, Bovenberg MS, Lewandrowski GK, de Gooijer MC, Vleggeert-Lankamp CL, Tannous M, Maguire CA, Tannous BA. Anal Chem. 2013;85:3006. doi: 10.1021/ac4003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Evans ME, et al. in revision. [Google Scholar]
- 147.Gross S, Abraham U, Prior JL, Herzog ED, Piwnica-Worms D. Molecular Imaging. 2007;6:121. [PubMed] [Google Scholar]
- 148.Kheirolomoom A, Kruse DE, Qin S, Watson KE, Lai CY, Young LJ, Cardiff RD, Ferrara KW. J Control Release. 2010;141:128. doi: 10.1016/j.jconrel.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Branchini BR, Ablamsky DM, Rosenberg JC. Bioconjugate Chem. 2010;21:2023. doi: 10.1021/bc100256d. [DOI] [PubMed] [Google Scholar]
- 150.Branchini BR, Ablamsky DM, Rosenman JM, Uzasci L, Southworth TL, Zimmer M. Biochemistry. 2007;46:13847. doi: 10.1021/bi7015052. [DOI] [PubMed] [Google Scholar]
- 151.Branchini BR, Southworth TL, Khattak NF, Michelini E, Roda A. Anal Biochem. 2005;345:140. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 152.Caysa H, Jacob R, Muther N, Branchini B, Messerle M, Soling A. Photochem Photobiol Sci. 2009;8:52. doi: 10.1039/b814566k. [DOI] [PubMed] [Google Scholar]
- 153.Loening AM, Dragulescu-Andrasi A, Gambhir SS. Nat Methods. 2010;7:5. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM. Biochemistry. 2012;51:6493–6495. doi: 10.1021/bi300934s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li X, Nakajima Y, Niwa K, Viviani VR, Ohmiya Y. Protein Sci. 2010;19:26. doi: 10.1002/pro.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Woodroofe CC, Meisenheimer PL, Klaubert DH, Kovic Y, Rosenberg JC, Behney CE, Southworth TL, Branchini BR. Biochemistry. 2012;51:9807. doi: 10.1021/bi301411d. [DOI] [PubMed] [Google Scholar]
- 157.McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA. J Am Chem Soc. 2010;134:7604. doi: 10.1021/ja301493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Conley NR, Dragulescu-Andrasi A, Rao J, Moerner WE. Angew Chem Int Ed. 2012;51:3350. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kojima R, Takakura H, Ozawa T, Tada Y, Nagano T, Urano Y. Angew Chem Int Ed. 2012;52:1175. doi: 10.1002/anie.201205151. [DOI] [PubMed] [Google Scholar]
- 160.Reddy GR, Thompson WC, Miller SC. J Am Chem Soc. 2010;132:13586. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morse D, Tannous BA. Mol Ther. 2012;20:692. doi: 10.1038/mt.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Levi J, De A, Cheng Z, Gambhir SS. J Am Chem Soc. 2007;129:11900. doi: 10.1021/ja073936h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shimomura O, Musicki B, Kishi Y. Biochem J. 1989;261:913. doi: 10.1042/bj2610913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Inouye S, Shimomura O. Biochem Biophys Res Commun. 1997;233:349. doi: 10.1006/bbrc.1997.6452. [DOI] [PubMed] [Google Scholar]
- 165.Harwood KR, Mofford DM, Reddy GR, Miller SC. Chem Biol. 18:1649. doi: 10.1016/j.chembiol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. PLoS One. 6:e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kwon HJ, Ohmiya Y, Yasuda K. Anal Biochem. 2012;430:45. doi: 10.1016/j.ab.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 168.Wang H, Cao F, De A, Cao Y, Contag C, Gambhir SS, Wu JC, Chen X. Stem Cells. 2009;27:1548. doi: 10.1002/stem.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maguire CA, Bovenberg MS, Crommentuijn MH, Niers JM, Kerami M, Teng J, Sena-Esteves M, Badr CE, Tannous BA. Mol Ther Nucleic Acids. 2013;2:e99. doi: 10.1038/mtna.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Woodroofe CC, Meisenheimer PL, Klaubert DH, Kovic Y, Rosenberg JC, Behney CE, Southworth TL, Branchini BR. Biochemistry. 51:9807. doi: 10.1021/bi301411d. [DOI] [PubMed] [Google Scholar]
- 171.Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. Biochemistry. 2008;47:10383. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]
- 172.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Nat Methods. 10:540. [PubMed] [Google Scholar]
- 173.Wendt MK, Molter J, Flask CA, Schiemann WP. J Vis Exp. 2011;56:e3245. doi: 10.3791/3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yuan H, Chong H, Wang B, Zhu C, Liu L, Yang Q, Lv F, Wang S. J Am Chem Soc. 2012;134:13184. doi: 10.1021/ja304986t. [DOI] [PubMed] [Google Scholar]
- 175.Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, Ratner L, Piwnica-Worms D. Nat Med. 2009;15:455. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]