Abstract
Candida auris is an emerging multidrug resistant yeast that causes nosocomial fungaemia and deep-seated infections. Notably, the emergence of this yeast is alarming as it exhibits resistance to azoles, amphotericin B and caspofungin, which may lead to clinical failure in patients. The multigene phylogeny and amplified fragment length polymorphism typing methods report the C. auris population as clonal. Here, using whole genome sequencing analysis, we decipher for the first time that C. auris strains from four Indian hospitals were highly related, suggesting clonal transmission. Further, all C. auris isolates originated from cases of fungaemia and were resistant to fluconazole (MIC >64 mg/L).
Keywords: Candida auris, India, multidrug resistant, outbreak, whole genome sequencing
Introduction
Candida auris is a multidrug-resistant yeast first described in 2009 as a species closely resembling Candida haemulonii [1], [2], [3]. The emergence of C. auris as an agent of nosocomial fungaemia and deep-tissue infections is alarming as this yeast is notorious for multidrug resistance [4], [5], [6]. Antifungal susceptibility patterns in various studies showed resistance to azoles, amphotericin B and caspofungin that may lead to clinical failure in patients [3], [7]. Many laboratories worldwide rely on phenotypic commercial automated systems for routine identification of yeasts, which may lead to misidentification of C. auris as C. haemulonii, so underestimating the spread of this yeast [7]. Recently, C. auris genetic and proteomic diversity analyses using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), amplified fragment length polymorphism and multi-gene phylogeny in Indian, South African, Japanese, Korean and Brazilian isolates demonstrated geographically specific clustering [8], [9]. In this study we confirm for the first time using whole genome sequencing that C. auris strains exhibit high clonality in different hospitals in India.
Materials and methods
Candida auris isolates
Five C. auris strains from candidaemia patients in four hospitals in India were sequenced for whole genome analysis [3], [8]. Their origin and clinical details are presented in Table 1. The isolates were identified by MALDI-TOF and sequencing of internal transcribed spacer and D1/D2 regions.
Table 1.
Clinical details and in vitro antifungal susceptibility profile of Candida auris (n = 5) isolates
| Isolate | Specimen/ year of isolation | Hospital | Age /Sex | Diagnosis | Risk factors | Therapy | Outcome | Drugsa (MIC, mg/L) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITC | VRC | ISA | POS | AMB | CFG | MFG | AFG | FLU | FC | ||||||||
| VPCI 669/P/12 | Blood/ 2012 | Hospital 1, New Delhi | 74/M | Jejunal perforation, peritonitis, septicaemia, MODS, DM | CVC, Broad-spectrum antibiotics, Surgery within 30 days, Intensive care, Antifungals within 30 days, Indwelling urinary catheter | CFG (loading dose of 70 mg, then 50 mg daily) for 2 weeks | Died 14 days after presentation | 0.125 | 0.125 | 0.125 | 0.06 | 0.5 | 0.25 | 0.125 | 0.5 | 64 | 0.125 |
| VPCI 692/P/12 | Blood/ 2012 | Hospital 1, New Delhi | 3 days/F | PT, TEF, ICH sepsis | Neutropenia, Broad-spectrum antibiotics, Intensive care, Indwelling urinary catheter | CFG (loading dose of 70 mg, then 50 mg daily) for 5 days | Died 8 days after admission | 0.125 | 0.25 | 0.06 | 0.125 | 1 | 0.25 | 0.125 | 0.125 | 64 | 0.125 |
| VPCI 479/P/13 | Blood/ 2013 | Hospital 2, Kochi | 48/F | RHD | CVC, Broad-spectrum antibiotics, Parenteral nutrition, Intensive care, Antifungals within 30 days, Indwelling urinary catheter | FLU (400 mg once daily) for 7 days | Died 1 month after admission | 0.125 | 0.5 | 0.125 | 0.06 | 0.25 | 0.5 | 0.06 | 0.125 | >64 | 0.5 |
| VPCI 510/P/14 | Blood/ 2014 | Hospital 3, New Delhi | 37/M | Chronic liver disease with decompensation | Broad-spectrum antibiotics, Antifungals within 30 days. | FLU( 400 mg once daily) for 7 days | Not followed | 0.5 | 16 | 0.25 | 0.25 | 4 | 0.5 | 0.125 | 0.5 | 64 | 0.25 |
| VPCI 550/P/14 | Blood/ 2014 | Hospital 4, New Delhi | 62/M | Hepato biliary carcinoma | Broad-spectrum antibiotics, Antifungal within 30 days, Urinary catheter, Intensive care, Chemotherapy | FLU | Died | 0.25 | 2 | 0.25 | ≤0.015 | 1 | 8 | >8 | >8 | >64 | >64 |
MODS, multi organ dysfunction syndrome; DM, diabetes mellitus; PT, pre-term; TEF, tracheo-oesophageal fistula; ICH, intracranial haemorrhage; RHD, rheumatic heart disease; CVC, central venous catheter.
ITC, itraconazole; VRC, voriconazole; ISA, isavuconazole; POSA, posaconazole; AMB, amphotericin B; CFG, caspofungin; MFG, micafungin; AFG, anidulafungin; FLU, fluconazole; FC, flucytosine.
IlluminaMiSeq sequence determination
Genomic DNA was extracted using a column-based method with a QIAamp DNA minikit (Qiagen, Hilden, Germany) and processed using the Nextera XT DNA protocol (Illumina, Inc., San Diego, CA, USA) to generate sequencing-ready libraries. The genome was sequenced using an Illumina MiSeq platform with MiSeq v3 protocol (Paired-End, 300 × 2). Equal volumes of normalized libraries were combined, diluted and heat denatured and the resulting FASTQ files were imported into CLC Genomics Workbench for analysis. The genomes were assembled by the combination of Velvet v1.2.0 [10], SSPACE 2.0 and Gap-Filler v1.10 [11], [12]. Repetitive sequences were masked using RepeatMasker v4.0.5 (http://www.repeatmasker.org), followed by ab initio gene prediction using GeneMark-ES 2.0 [13]. Next, rRNA and tRNA were predicted with RNAmmer and tRNAscan-SE v1.21 respectively [14], [15]. Single-nucleotide polymorphism was detected using Unified Genotyper from the Genome Analysis Toolkit package [16]. The functional annotation was performed using the Clusters of Orthologous Groups of proteins (COGs) database [17]. To determine that C. auris belongs to the CUG Candida clade, a webserver Bagheera [18] was used to identify CUG codon usage in C. auris genomes.
Average nucleotide identity and phylogenetic analysis of C. auris
Average nucleotide identity (ANI) was calculated using the JSPECIES package [19] using MUMMER (ANIm). Five Candida auris genomes (contigs) were compared with other publically available Candida species genomes (n = 8) detailed in Table 2 by aligning the sequences using progressive MAUVE with the default settings [20]. The strip Subset ‘Locally Collinear Blocks’ script was used to extract core blocks, creating core alignments longer than 500 bp including all 13 genomes. A core genome alignment in XMFA format was obtained which was converted into FASTA format using a PerlScript (https://github.com/lskatz/lskScripts). A phylogenetic network was constructed using SplitsTree based on concatenated alignment of the core genes [21].
Table 2.
Results of average nucleotide identity analysis giving percentage similarity between Candida auris (n = 5) and other Candida species (n = 8)
| Strains | VPCI 479/P/13 | VPCI 669/P/12 | VPCI 692/P/12 | VPCI 510/P/14 | VPCI 550/P/14 | Candida dubliniensis (CD36) | Candida albicans (WO-1 and SC 5314) | Candida guillermondii (ATCC6260) | Candida lusitaniae (ATCC42720) | Candida tropicalis (MYA-3404) | Saccharomyces cerevisiae (S288C) | Candida parapsilosis (317) | Candida glabrata (CBS 138) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VPCI 479/P/13 | 100 | 99.84 | 99.85 | 99.83 | 99.85 | 84.31 | 83.89 | 85.77 | 86.34 | 83.41 | 83.95 | 84.18 | 84.14 |
| VPCI 669/P/12 | 99.85 | 100 | 99.84 | 99.81 | 99.83 | 85.24 | 84.72 | 86.35 | 86.38 | 84.43 | 84.21 | 85.54 | 84.83 |
| VPCI 692/P/12 | 99.84 | 99.83 | 100 | 99.81 | 99.81 | 84.23 | 83.45 | 85.83 | 86.36 | 83.51 | 84.83 | 84.38 | 84.44 |
| VPCI 510/P/14 | 99.84 | 99.82 | 99.83 | 100 | 99.84 | 84.16 | 83.73 | 85.83 | 86.3 | 83.47 | 84.35 | 84.04 | 84.1 |
| VPCI 550/P/14 | 99.85 | 99.83 | 99.81 | 99.84 | 100 | 84.26 | 83.50 | 85.58 | 86.42 | 84.43 | 84.85 | 85.23 | 84.27 |
| C. dubliniensis (CD36) | 84.07 | 85.11 | 84.37 | 84.07 | 84.26 | 100 | 87.84 | 85.53 | 85.16 | 85.33 | 84.54 | 84.96 | 84.45 |
| C. albicans (WO-1 and SC 5314) | 83.61 | 86.77 | 83.89 | 83.89 | 83.50 | 88.29 | 100 | 83.89 | 81.98 | 85.74 | 83.21 | 84.76 | 88.37 |
| C. guillermondii (ATCC6260) | 85.8 | 86.31 | 85.93 | 85.9 | 85.58 | 85.33 | 86.05 | 100 | 86.2 | 85.87 | 84.36 | 85.78 | 86.01 |
| C. lusitaniae (ATCC42720) | 85.95 | 86.07 | 86.07 | 86 | 86.42 | 84.68 | 84.96 | 86.1 | 100 | 84.47 | 84.87 | 84.76 | 85.58 |
| C. tropicalis (MYA-3404) | 83.52 | 83.51 | 83.54 | 83.35 | 84.43 | 85.18 | 85.39 | 84.31 | 84.14 | 100 | 83.85 | 84.62 | 83.1 |
| S. cerevisiae (S288C) | 84.86 | 85.13 | 85.08 | 84.95 | 84.85 | 84.3 | 84.85 | 83.82 | 85.09 | 84.18 | 100 | 84.5 | 86.62 |
| C. parapsilosis (317) | 84.07 | 85.01 | 84.03 | 84.07 | 85.23 | 84.86 | 84.91 | 85.45 | 84.95 | 84.91 | 84.05 | 100 | 84.95 |
| C. glabrata (CBS138) | 84.25 | 84.97 | 84.43 | 84.24 | 84.27 | 84.87 | 84.81 | 86.43 | 85.8 | 84.44 | 86.8 | 85.62 | 100 |
Antifungal susceptibility testing and antifungal resistance genes analysis
Antifungal susceptibility testing of C. auris (n = 5) was carried out using the CLSI broth microdilution method, following the M27-A3 guidelines [22]. In addition, a control group of 20 Candida albicans isolates originating from invasive and superficial candidiasis and stocked in the culture collection of Vallabhbhai Patel Chest Institute (VPCI) were subjected to antifungal susceptibility testing. Newly revised epidemiological cut-off MIC values for C. albicans for fluconazole (>0.5 mg/L), itraconazole (>0.12 mg/L), voriconazole (>0.03 mg/L), posaconazole (>0.06 mg/L), amphotericin B (>2 mg/L), flucytosine (>0.5 mg/L), anidulafungin (>0.12 mg/L), caspofungin (>0.12 mg/L) and micafungin (>0.03 mg/L) were used for comparison of C. auris MIC data [23]. The contigs from all the five C. auris strains were aligned and screened for the presence of ERG (ERG3 and ERG11) and FKS (FKS1, FKS2 and FKS3) genes and compared with the C. albicans and Candida glabrata ERG and FKS genes. Also, a solitary echinocandin-resistant C. auris isolate (VPCI 550/P/15, Table 1) was subjected to amplification and sequencing of the FKS1 and FKS2 genes to analyse the mutations similar to those reported in C. glabrata [7].
Results
The internal transcribed spacer and D1/D2 regions of the ribosomal DNA sequences of C. auris isolates showed 99% homology with the type C. auris isolates in GenBank. In all candidaemia cases the most common risk factor was the concomitant use of broad-spectrum antibiotics, intensive care unit stay and presence of indwelling urinary catheter. Other risk factors included parenteral nutrition and use of antifungals in three patients (Table 1). Four patients who developed breakthrough fungaemia had a fatal outcome.
Phylogenetic analysis
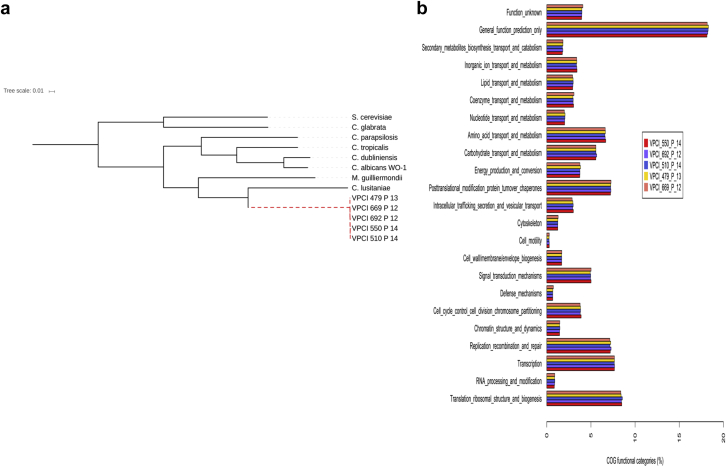
The assembled C. auris genome is diploid, comprising 12.3 Mb with a G+C content of 44.8%. A total of 6675 coding sequences among all the isolates were found with one 5.8S rRNA, 184 tRNA and 3262 repetitive elements. A phylogeny based on the concatenated sequences of 136 conserved core genes of the C. auris genomes (n = 5) and other Candida species (n = 8) confirms that the C. auris isolates are more similar to each other than to other Candida species analysed (Fig. 1a). Remarkably, the genome of all C. auris isolates analysed was highly related with only 0.2% variation observed among their genomes (Table 2). Among all sequenced C. auris isolates, single-nucleotide polymorphism differences ranging from 3048 to 4330 were observed. ANI calculated for the C. auris isolates is consistently above 99% (Table 2). In contrast the low ANI value (<95%) among C. auris and other Candida species suggested its divergence from the Candida genomes listed (Fig. 1a, Table 2). Among the eight Candida species aligned with C. auris, the highest ANI value (85.9%–86.4%) was observed for Candida lusitaniae. Also, the functional annotation data suggest 40% of the C. auris proteins to be orthologous to C. lusitaniae with most of them assigned as hypothetical or functionally uncharacterized.
Fig. 1.
(a) Phylogenetic tree based on 136 core/conserved genes of the Candida auris isolates and other Candida species. (b) Functional annotation of Candida auris genome based on Clusters of Orthologous Groups of proteins (COG) database classification.
Functional annotation of C. auris
Although a major proportion of the C. auris genome remains uncharacterized, a large number of proteins possessed transporter along with binding and catalytic activity involved in cellular and metabolic processes followed by proteins involved in signal transduction (Fig. 1b). Candida auris shares virulence traits common to C. albicans. The presence of orthologues including ion transporters, amino acid and metabolite transporters, oligopeptide transporters, secreted proteinases, lipases, phospholipases, adhesins, secreted aspartyl proteases, mannosyl transferases, MADS-box (for Minichromosome Maintenance1, Agamous, Deficiens and Serum Response Factor) and STE (Serine/threonine enzyme)-related proteins in C. auris genome were observed when compared with C. albicans as a reference genome (Fig. 1b). The above-mentioned gene classes may contribute to virulence acquisition in C. auris isolates. The codon usage analysis in C. auris appears to be more similar and overlapping with C. lusitaniae than with any other Candida species, indicating that C. auris is part of the CUG clade. This similarity has been supported by the phylogenetic and functional data.
Antifungal susceptibility patterns and analysis of antifungal resistance genes
The in vitro susceptibility data of C. auris (n = 5) is presented in Table 1. All isolates were resistant to fluconazole, one each was resistant to voriconazole and amphotericin B and the other isolate was resistant to voriconazole, flucytosine and echinocandins. The PCR targeting FKS1 and FKS2 genes of a solitary echinocandin-resistant C. auris isolate (VPCI 550/P/15) generated amplicons of 391 bp and 460 bp, respectively. Echinocandin-resistant C. auris did not reveal any mutation in the hot spot regions previously reported for caspofungin-resistant C. glabrata. In contrast to C. auris, all C. albicans isolates were found to be highly susceptible to all azoles including itraconazole (MIC50, 0.03 mg/L; MIC90, 0.03 mg/L); voriconazole (MIC50, 0.03 mg/L; MIC90, 0.03 mg/L); posaconazole (MIC50, 0.015 mg/L; MIC90, 0.015 mg/L) and amphotericin B (MIC50, 0.5 mg/L; MIC90, 0.5 mg/L). The alignment of the C. auris contigs with C. albicans and C. glabrata ERG and FKS genes revealed that ERG3, ERG11, FKS1, FKS2 and FKS3 genes were present as a single copy in the C. auris genome. ERG and FKS genes of C. auris exhibited 78%–85% similarity with those of C. albicans and C. glabrata.
Furthermore, the C. auris genome when analysed with C. albicans and Saccharomyces cerevisiae revealed that a significant portion of its genome encodes ABC and MFS transporter family along with drug transporters. A number of zinc cluster transcription factor orthologues such as TAC1 (29% similarity with C. albicans) and protein kinases, which may contribute to the acquisition of drug resistance were observed (Fig. 1b). The protein kinases such as protein kinase A, HOG1 have been reported to be activated on perceiving stress thereby regulating the stress signalling pathways to enhance tolerance of pathogenic fungi to various fungicides [24].
Mating locus analysis
Mining of the de novo assembly of C. auris sequence reads the presence of MATα locus, similar to Naumovozyma castellii CBS 4309, in all the C. auris strains investigated. However, the MATa gene was absent in all the isolates in the present study.
Discussion
The application of whole genome sequencing confirms for the first time that C. auris strains in Indian settings are highly clonal, suggesting the possibility of a common source of origin or a phenomenon of their recent differentiation. It is noteworthy to mention that ANI analysis showed C. auris strains exhibiting a highly divergent relationship with other clinically significant Candida species namely C. albicans, C. tropicalis, C. parapsilosis and C. glabrata. However, the genome of C. auris may be more closely related to C. haemulonii with whom C. auris is commonly misidentified using phenotypic methods but whose genome is not yet sequenced. Future studies analysing the whole genome sequencing of other related Candida species might provide more insight about their similarity with C. auris. The lack of information on the ecological niche of C. auris hampers the understanding of its divergence from other Candida species. Further, all the strains of C. auris analysed in the present study were recovered from five individual patients in four hospitals located 50–2000 km apart and at a different time period. However, irrespective of the variability in the above parameters all strains exhibited low genetic diversity. Also, all the C. auris strains exhibited MATα mating locus and its presence was also validated by gene-specific PCR. Further experiments analysing the mating locus in a large number of C. auris isolates will determine the true sexual status of this pathogen. Furthermore, the antifungal resistance profile of the isolates was variable but uniformly showed the presence of ABC and MFS transporters, which may explain the multidrug resistance. It is also possible that the undiscerning use of antifungals has resulted in its emergence as a successful multidrug-resistant pathogen. Overall our study provides the first comparative analysis of C. auris genomes using whole genome sequencing and highlights clonal expansion of C. auris isolates in India.
Conflict of interest
JFM received grants from Astellas, Basilea and Merck. He has been a consultant to Astellas, Basilea and Merck and received speaker's fees from Merck, United Medical and Gilead. All other authors have no potential conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee W.G., Shin J.H., Uh Y., Kang M.G., Kim S.H., Park K.H. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011;49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A., Sharma C., Duggal S., Agarwal K., Prakash A., Singh P.K. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 2013;19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magobo R.E., Corcoran C., Seetharam S., Govender N.P. Candida auris associated candidemia, South Africa. Emerg Infect Dis. 2014;20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhary A., Anil Kumar V., Sharma C., Prakash A., Agarwal K., Babu R. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014;33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 6.Emara M., Ahmad S., Khan Z., Joseph L., Al-Obaid I., Purohit P. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis. 2015;21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathuria S., Singh P.K., Sharma C., Prakash A., Masih A., Kumar A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI Broth Microdilution, and Etest method. J Clin Microbiol. 2015;53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakash A., Sharma C., Singh A., Singh P.K., Kumar A., Hagen F. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect. 2016;22:277.e1–277.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Girard V., Mailler S., Chetry M., Vidal C., Durand G., van Belkum A. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 10.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 12.Boetzer M., Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mühlhausen S., Kollmar M. Predicting the fungal CUG codon translation with Bagheera. BMC Genomics. 2014;15:411. doi: 10.1186/1471-2164-15-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darling A.E., Mau B., Perna N.T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huson D., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard-3rd ed. CLSI document M27-A3. [Google Scholar]
- 23.Pfaller M.A., Diekema D.J. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes B.M., Anderson M.A., Traven A., van der Weerden N.L., Bleackley M.R. Activation of stress signalling pathways enhances tolerance of fungi to chemical fungicides and antifungal proteins. Cell Mol Life Sci. 2014;71:2651–2666. doi: 10.1007/s00018-014-1573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]