Abstract
Using 3D telomere quantitative fluorescence in situ hybridization, we determined the 3D telomere organization of 74 neuroblastoma tissue samples. Hierarchical cluster analysis of the measured telomere parameters identified three subgroups from our patient cohort. These subgroups have unique telomere profiles based on telomere length and nuclear architecture. Subgroups with higher levels of telomere dysfunction were comprised of tumors with greater numbers of telomeres, telomeric aggregates, and short telomeres (P < .0001). Tumors with greater telomere dysfunction were associated with unfavorable tumor characteristics (greater age at diagnosis, unfavorable histology, higher stage of disease, MYCN amplification, and higher MYCN expression) and poor prognostic risk (P < .001). Subgroups with greater telomere dysfunction also had higher intratumor heterogeneity. MYCN overexpression in two neuroblastoma cell lines with constitutively low MYCN expression induced changes in their telomere profile that were consistent with increased telomere dysfunction; this illustrates a functional relationship between MYCN and 3D telomere organization. This study demonstrates the ability to classify neuroblastomas based on the level of telomere dysfunction, which is a novel approach for this cancer.
Introduction
Neuroblastoma is the most common extracranial malignancy in children. This neoplasm is characterized by both clinical and molecular heterogeneity, and the prognostic risk calculation is multifactorial. The most important parameters predictive of an unfavorable outcome include MYCN amplification [1], [2], MYCN protein overexpression [3], [4], age greater than 18 months at diagnosis [5], [6], loss of chromosome arm 1p [7], [8] and 11q [9], and gain of chromosome arm 17q [10].
Neuroblastomas have been subgrouped according to their pattern of chromosomal instability: whereas some tumors exhibit numerical and few or no structural aberrations, others are dominated by structural rearrangements, including intrachromosomal rearrangements [11], [12]. Chromosome instability has been linked to telomere length aberrations in many cancers, including neuroblastoma [12], [13]. However, there have been conflicting findings on whether an increase, decrease, or unchanging telomere length is associated with a better outcome [13], [14], [15]. These inconclusive results may be due to the methods employed to measure telomere length, wherein only the average telomere length for each cell was determined.
Nuclear architecture is key to cellular function [16], and changes in nuclear architecture contribute to the pathogenesis and progression of cancer [17]. Our laboratory developed a method of analyzing the interphase nuclear organization of telomeres as a novel and more in-depth approach to study telomere length and telomere dysfunction in disease [18]. Our method uses 3D quantitative fluorescence in situ hybridization (Q-FISH) to label all telomeres in interphase nuclei while preserving the nuclear architecture of the sample. Our software measures multiple telomere parameters for each cell including the number of telomeres, the length of each telomere, the number of telomeric aggregates, the nuclear volume, and the spatial organization of the telomeres [18], [19]. These measurements create a telomere profile unique to an individual sample. Increases in the number of telomeres, telomeric aggregates, and short telomeres are frequently associated with tumor compared to nontumor cells, aggressive forms of disease, and poor patient outcomes [19], [20], [21], [22].
In this study, we examined for the first time the 3D telomere organization in 74 archived neuroblastoma tissue samples. Using hierarchical cluster analysis of the measured telomere parameters, we identified three tumor subgroups representing unique levels of telomere dysfunction. We found that tumors with greater telomere dysfunction were associated with unfavorable tumor characteristics including MYCN amplification and higher MYCN expression. To test the hypothesis that high levels of MYCN present in a subgroup of patients (11/74) lead to increased telomere dysfunction, we overexpressed MYCN in two neuroblastoma cell lines with constitutively low MYCN expression. This induced changes in the telomere parameters similar to those seen in high MYCN–expressing neuroblastoma tissue samples, demonstrating a functional relationship between MYCN expression and 3D telomere organization.
Materials and Methods
Patient Samples
A total of 74 primary neuroblastoma tissue samples, 5 μm in thickness, were obtained from the Health Sciences Centre (Winnipeg, Manitoba, Canada) (n = 16) and Children's Oncology Group (COG) (n = 58). All of the tumor samples were derived from untreated patients. Hematoxylin and eosin–stained sections were used to identify tumor areas. The experimenters were blinded to the tumor characteristics and outcome data until after the experiments and measurements were complete. After decoding this information, it was discovered that only 31 out of the 74 patients had clinical follow-up, and therefore survival analyses for the whole cohort was not feasible.
Patients were classified according to the International Neuroblastoma Staging System (INSS) [23] and divided into clinical-genetic risk groups using the COG risk scoring system [7]. The MYCN amplification status was determined by FISH for the COG samples. The MYCN amplification status of the Manitoban samples was provided to us by the Health Sciences Centre (Winnipeg, Manitoba, Canada). MYCN protein expression was determined for all samples by tissue immunofluorescence. Tumor characteristics of the study cohort are shown in Table 1.
Table 1.
Neuroblastoma Cohort Characteristics
| Cases (n) | |
|---|---|
| Stage | |
| 1 | 13 |
| 2 | 11 |
| 3 | 8 |
| 4 | 30 |
| 4S | 11 |
| Unknown | 1 |
| Histology | |
| Favorable | 28 |
| Unfavorable | 35 |
| Unknown | 11 |
| Age at diagnosis | |
| <18 months | 41 |
| >18 months | 33 |
| MYCN amplification | |
| No | 63 |
| Yes | 11 |
| MYCN expression | |
| Low | 43 |
| Medium | 20 |
| High | 11 |
| COG risk score | |
| Low | 36 |
| Intermediate | 8 |
| High | 28 |
| Unknown | 2 |
This study was approved by Pathology Access Committee for Tissue (12-0048), Health Science Centre Research Impact committee (2012:187), CancerCare Manitoba Research Resource Impact Committee (92-2012), and Research Ethics Board (H2012:391).
Cell Lines
The established neuroblastoma cell lines SHEP and GIMEN were a gift from Manfred Schwab (The German Cancer Research Center, Heidelberg, Germany). The cells were cultured in RPMI 1640 with 10% fetal bovine serum with 1% l-glutamine, 1% sodium pyruvate, and 1% penicillin-streptomycin (Life Technologies Inc., Burlington, Ontario, Canada) at 37°C in a humidified atmosphere and 5% CO2.
MYCN Transfection of SHEP and GIMEN Neuroblastoma Cell Lines
SHEP and GIMEN cell lines were transfected with MYCN pUHD 10-3, a gift from Manfred Schwab (The German Cancer Research Center, Heidelberg, Germany), which contains the entire MYCN coding sequence under the control of an hCMV minimal promoter. Before transfection, 6 × 105 SHEP and 2 × 105 GIMEN cells were seeded per well and grown to near confluence. Twenty-four hours postseeding, cells were cotransfected with TransIT-X2 (MIR6004; Mirus Bio LLC, Madison, WI) and pmaxGFP (Lonza, Allendale, NJ) in a ratio of reagent:vector DNA of 2:1, as per the company's protocol. Cells were cultured in the absence of serum for the first 12 hours. The transfection efficiency was 40% at 15 hours when the cells were sterile sorted by GFP expression. The sorted transfected cells were put back into culture and harvested at 72 hours for 3D nuclei fixation. Mock-transfected cells were used as a control.
3D Nuclei Fixation
Cells were harvested from the cultures of SHEP and GIMEN and underwent 3D fixation to prepare nuclei for Q-FISH experiments. 3D nuclear fixation was performed according to the protocol published by Solovei et al. [24].
Immunofluorescence
MYCN immunofluorescence, imaging, and analysis were performed on the neuroblastoma tissue samples and SHEP and GIMEN cell lines as previously described [25]. The MYCN antibody, a gift from Manfred Schwab, is described in Wenzel et al. (1991) and was used at a titer of 1/2000. The slides were imaged using an AxioImager Z2 microscope (Carl Zeiss, Toronto, Ontario, Canada), an AxioCam HR charge-coupled device (Carl Zeiss) with a 63×/1.4 oil objective lens (Carl Zeiss), and DAPI and Cy3 filters (Carl Zeiss). Cy3 exposure times were kept constant between cell lines and between tissue samples. Thirty cells per experimental replicate were analyzed for the cell lines, and 100 cells were analyzed in each tumor sample. We used the following ranges to classify MYCN expression: low as <2000 relative fluorescence units (RFU), medium as 2000 to 4999 RFU, and high as ≥5000 RFU.
3D Telomere Q-FISH
Nuclei from the neuroblastoma cell lines and 5-μm tissue sections underwent 3D telomere Q-FISH with a telomere probe (DAKO, Glostrup, Denmark). The hybridization and image acquisition were performed according to previously published protocols [26], [27]. The slides were imaged using an AxioImager Z2 microscope (Carl Zeiss), an AxioCam HR charge-coupled device (Carl Zeiss) with a 63×/1.4 oil objective lens (Carl Zeiss), and DAPI and Cy3 filters for detection of nuclear DNA staining and telomere probe signals, respectively (Carl Zeiss). Cy3 exposure times were kept constant between cell lines and between tumor samples, and tricolor beads were used to standardize the bulb strength. Eighty z-stacks for cell lines and 40 z-stacks for tissue at 200 nm each, with Δx = Δy = 106 nm and Δz = 200 nm, were acquired and then deconvolved using the constrained iterative algorithm [28]. Thirty cells per experimental replicate were analyzed for the cell lines, and 100 cells were analyzed in each tumor sample.
The 3D interphase telomere signals were quantitatively analyzed with TeloView software [18] to measure the following parameters in each cell: the number of signals, which corresponds to the number of telomeres [18]; the intensity of each signal, which correlates with telomere length [29]; the number of telomeric aggregates, which are telomeres in close proximity that cannot be resolved as separate entities at an optical resolution of 200 nm [18]; the nuclear volume [18]; and the a/c ratio, which is a measurement of the spherical nature of the telomere distribution [18]. From these measurements, the following parameters were calculated: the percentage of cells per patient with aggregates, the number of telomeres per nuclear volume, and the mean telomere intensity [20], [30], [31]. With these measurements, we generated a 3D telomere profile for each cell and then combined the data from multiple cells to create an overall profile for each tumor sample and each cell line replicate.
Statistical Analysis
Statistical analysis was conducted using SAS 9.3 (SAS Institute Inc., Cary, NC) software. Hierarchical clustering analysis was used to subgroup the neuroblastoma tissue samples based on the following telomere parameters: number of telomeres per cell, number of telomeric aggregates per cell, percentage of cells per patient with aggregates, number of telomeres per nuclear volume, and a/c ratio. To visualize the results of the clustering analysis, canonical discriminant analysis was performed to generate a plot of can1 versus can2. Can1 is a calculated value for each tumor sample based on a linear combination of the clustering variables that has the highest correlation with the clusters. Can2 is the second linear combination of the clustering variables uncorrelated with can1 but also showing correlation with the clusters.
The nested factorial analysis of variance with Tukey multiple-comparisons test was used to compare the telomere parameters between the subgroups. The χ2 test was employed to compare tumor characteristics among the subgroups. Telomere signal intensities were binned at intervals of 1000 and divided into 3 groups formed by the 25th and 75th percentiles. In tumor samples, telomeres were defined as short with an RFU less than 6000, medium length with an RFU between 6000 and 11,999, and long with an RFU greater than or equal to 12,000. In cell lines, telomeres were defined as short with an RFU less than 7000, medium length with an RFU between 7000 and 16,999, and long with an RFU greater than or equal to 17,000. The χ2 test was used to compare the distribution of short, medium, and long telomeres. The unpaired t test was used to compare the telomere parameters of the cell lines after MYCN and mock transfection. P < .05 was considered significant.
Results
3D Telomere Organization of Neuroblastoma Tissue Samples
We investigated the 3D telomere organization in 74 neuroblastoma tissue samples using 3D telomere Q-FISH while blinded to tumor characteristics. After image acquisition and deconvolution [28], 100 nuclear segments from each tumor were analyzed with TeloView software [18] to determine the telomere profile associated with that sample (see Materials and Methods). Hierarchical cluster analysis was used to stratify the study cohort into subgroups based on the telomere parameters.
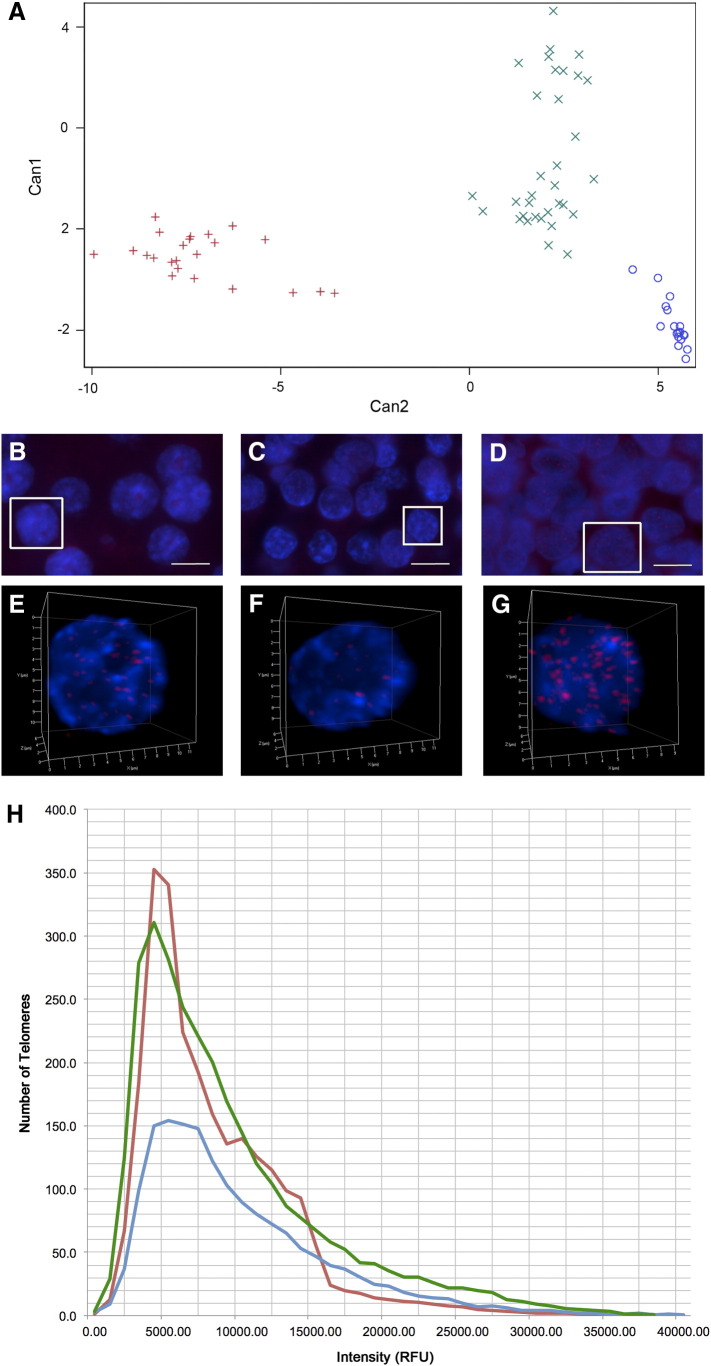
Cluster analysis revealed three unique subgroups, shown graphically in Figure 1A. The number of neuroblastomas in each subgroup was as follows: 23 in subgroup I (red), 18 in subgroup II (blue), and 33 in subgroup III (green). A tissue overview as well as a nucleus illustrating the telomere hybridization from a representative sample in each of the subgroups is shown in Figure 1, B–G. The scatterplot in Figure 1H illustrates the distribution of telomere lengths in each subgroup, with a plot of the number of telomeres found at each relative fluorescence intensity interval measured.
Figure 1.
3D telomere organization of neuroblastoma subgroups. Hierarchical cluster analysis based on the measured telomere parameters of the 74 neuroblastoma tissue samples in our study cohort revealed three unique subgroups. Subgroup I (red), subgroup II (blue), and subgroup III (green) are illustrated in a graphical representation (A). 3D telomere Q-FISH of the tumors labels all telomeres (red) in the nuclei, counterstained with DAPI (blue). Images of multiple cells in 2D and 3D renderings of individual representative nuclei (outlined in white in the overview) are illustrated for a neuroblastoma in subgroup I (B & E), subgroup II (C & F), and subgroup III (D & G). Scale bars represent 10 μm. The distribution of telomere lengths in each subgroup is illustrated in a scatterplot (H) of the number of telomeres found at each relative fluorescence intensity interval measured.
χ2 analysis revealed a significantly different distribution of telomere lengths between the subgroups (P < .0001). As shown in Table 2, there was also a significant difference in the percentage of short, medium, and long telomeres per cell (P < .001 for all comparisons). Subgroups I and III are comprised of neuroblastomas with the greatest percentage of short telomeres per cell (38.7% and 35.9%). Subgroup II contains tumors with the highest percentage of medium-length telomeres (42.92%). Tumors with the highest percentage of long telomeres per cell are found in subgroup III (31.9%). These findings illustrate that subgroup III, which has a high percentage of both short and long telomeres, has the most telomere length heterogeneity.
Table 2.
Statistical Analysis of Telomere Parameters by Neuroblastoma Subgroup
| Subgroup I | Subgroup II | Subgroup III | P Value between Subgroups⁎ | |
|---|---|---|---|---|
| Average number of telomeres per nuclear segment (SD†) | 24.6‡ (3.4) | 16.1 (2.3) | 29.5 (6.2) | <.0001 |
| Average number of telomeric aggregates per nuclear segment (SD) | 1.1 (0.1) | 1.9 (0.1) | 3.4 (1.0) | <.0001 |
| Average percentage of cells per patient with telomeric aggregates (SD, %) | 42.4 (6.9) | 99.5 (0.7) | 90.3 (5.2) | <.0001 |
| Average nuclear volume (SD, μm3) | 351.30 (71.20) | 335.78 (76.76) | 318.03 (85.55) | NS§ |
| Average number of telomeres per nuclear volume (SD) | 0.072 (0.013) | 0.060 (0.017) | 0.092 (0.029) | <.0001 |
| Average a/c ratio per cell (SD) | 2.57 (0.64) | 2.18 (0.56) | 3.81 (0.87) | <.0001 |
| Average mean telomere intensity per patient (SD, RFU) | 8833.0 (341.7) | 10,322.6 (649.1) | 10,118.3 (472.3) | <.0001 |
| Average percentage of short telomeres per cell (SD, %) | 38.7 (4.8) | 31.0 (4.9) | 35.9 (7.3) | <.001 |
| Average percentage of medium telomeres per cell (SD, %) | 40.0 (3.5) | 42.9 (2.7) | 32.2 (4.1) | <.0001 |
| Average percentage of long telomeres per cell (SD, %) | 21.4 (2.7) | 26.4 (4.3) | 31.9 (4.7) | <.0001 |
P value determined by one-way analysis of variance.
Standard deviation.
Data range indicated in Supplementary Table 6.
Not significant.
In addition to measuring individual telomere lengths, 3D telomere organization calculates telomere parameters that reflect tumor cell nuclear architecture. Telomeric aggregates are telomeres in close proximity that cannot be resolved as separate entities at a resolution of 200 nm. The subgroups displayed significantly different numbers of telomeric aggregates per cell and percentages of cells per patient with telomeric aggregates (P < .0001 for both comparisons). Subgroup III was comprised of neuroblastomas with the most telomeric aggregates (3.4), followed by subgroup II (1.9) and subgroup I (1.1). The highest percentage of cells with telomeric aggregates was found in tumors belonging to subgroup II (99.5%), followed by subgroup III (90.3%) and subgroup I (42.4%).
The greatest number of telomeres per cell was observed in the neuroblastomas found in subgroup III (29.5), followed by subgroup I (24.6), and subgroup II (16.1) (P < .0001). As the nuclear volume is similar for all subgroups, subgroup III, which has tumors with the most telomeres per cell, exhibits the most dense telomere distribution: the average number of telomeres per nuclear volume is 0.092 for subgroup III, 0.072 for subgroup I, and 0.060 for subgroup II. The a/c ratio, which measures the spherical nature of the telomere distribution, was also significantly different between the subgroups (P < .0001). Subgroup III was comprised of tumors with the largest a/c ratio (3.81), followed by subgroup I (2.57), and subgroup II (2.18).
In summary, the three subgroups contain neuroblastomas with unique telomere profiles based on both telomere length and nuclear organization. The lowest level of telomere dysfunction is seen in subgroup I, which contains tumors with the fewest telomeric aggregates per nucleus and lowest percentage of cells per patient with telomeric aggregates. Subgroup III illustrates the highest level of telomere dysfunction; it is comprised of tumors with the most telomeres per nucleus, the highest number of telomeric aggregates per nucleus, and a high percentage of short telomeres per cell and cells per patient with telomeric aggregates. A moderate level of telomere dysfunction is seen in subgroup II, which contains tumors with the least number of telomeres per cell but has more telomeric aggregates per cell than subgroup I and the highest percentage of cells per patient with telomeric aggregates. Telomere dysfunction has been linked to genomic instability, with higher levels of telomere dysfunction indicating greater genomic instability [32], [33]. Therefore, neuroblastomas with the highest level of genomic instability are found subgroup III, followed by those in subgroup II, and the tumors in subgroup I have the lowest level of genomic instability.
Neuroblastomas are known to exhibit intratumor heterogeneity [34], with increasing diversity of telomere length strongly associated with disease progression and mortality [35]. Our study supports these findings as we determined that subgroup III had the greatest standard deviation in all the telomere parameters, except percentage of cells per patient with telomeric aggregates and mean telomere intensity; therefore, subgroup III is comprised of the neuroblastomas with the greatest intratumor heterogeneity. As tumors with the highest level of telomere dysfunction are also found in subgroup III, our findings illustrate that greater intratumor heterogeneity correlates with higher levels of telomere dysfunction.
Tumor Characteristics in the Neuroblastoma Subgroups
We decoded the neuroblastoma tissue samples to determine the histopathological characteristics of the tumors in each of the three subgroups identified by hierarchical cluster analysis of the 3D telomere parameters. The histology, age at diagnosis, MYCN amplification status, and MYCN expression level of each sample are summarized in Table 3. χ2 analysis indicated that there was an association between the cluster to which a tumor belonged and every tumor characteristic (P < .001 for all comparisons).
Table 3.
Histopathological Characteristics and Classification of Neuroblastoma Subgroups
| Subgroup I (n) | Subgroup II (n) | Subgroup III (n) | P Value* | |
|---|---|---|---|---|
| Histology | .0005 | |||
| Favorable | 13 | 9 | 6 | |
| Unfavorable | 4 | 7 | 24 | |
| Unknown | 6 | 2 | 3 | |
| Age at diagnosis | <.0001 | |||
| <18 months | 19 | 13 | 9 | |
| >18 months | 4 | 5 | 24 | |
| MYCN amplification | .0003 | |||
| No | 23 | 18 | 22 | |
| Yes | 0 | 0 | 11 | |
| MYCN expression | <.0001 | |||
| Low | 23 | 18 | 2 | |
| Medium | 0 | 0 | 20 | |
| High | 0 | 0 | 11 | |
| Stage | <.0001 | |||
| 1 | 9 | 3 | 1 | |
| 2 | 7 | 4 | 0 | |
| 3 | 2 | 1 | 5 | |
| 4 | 0 | 5 | 25 | |
| 4S | 5 | 5 | 1 | |
| Unknown | 0 | 0 | 1 | |
| COG risk score | <.0001 | |||
| Low | 21 | 13 | 2 | |
| Intermediate | 1 | 2 | 5 | |
| High | 0 | 3 | 25 | |
| Unknown | 1 | 0 | 1 |
P value determined by χ2 test.
The majority of neuroblastomas with unfavorable histology and an age of diagnosis of more than 18 months were found in subgroup III (24/35 and 24/33, respectively), whereas most tumors with favorable histology and an age of diagnosis of less than 18 months were found in subgroups I and II. All tumors with MYCN amplification and medium or high levels of MYCN expression were found in subgroup III. Subgroups I and II were comprised of tumors with low MYCN expression and no MYCN amplification; however, tumors in subgroup II had a higher average MYCN expression than those in subgroup I (1281.09 RFU vs 285.26 RFU, P < .0001). Our data clearly demonstrate that neuroblastomas with unfavorable histopathological characteristics were found in the subgroups associated with more telomere dysfunction.
3D Telomere Profiles Versus Current Neuroblastoma Classification Systems
After decoding the neuroblastoma cohort, we determined the INSS stage and COG risk score of the tumors in each of the subgroups to gauge their prognosis (summarized in Table 3). χ2 analysis indicated that there was an association between the cluster to which a tumor belonged and both INSS stage and COG risk (P < .0001 for both comparisons).
The majority of neuroblastomas with stage 1 disease were found in subgroup I (9/13), which also contained no tumors of stage 4 disease. Subgroup III was comprised of mainly stage 4 tumors (25/33). Similar numbers of tumors of all stages were found in subgroup II (3 stage 1, 4 stage 2, 1 stage 3, 5 stage 4, 5 stage 4S). Subgroups I and II were comprised almost solely of low–COG risk neuroblastomas (21/23 and 13/18, respectively); however, subgroup II contained three tumors of high risk. The majority of tumors in subgroup III were of high COG risk (25/33). Therefore, neuroblastomas of high stage and poor prognostic risk were found in the subgroups with more telomere dysfunction.
Changes in 3D Telomere Organization following MYCN Transfection in Neuroblastoma Cell Lines
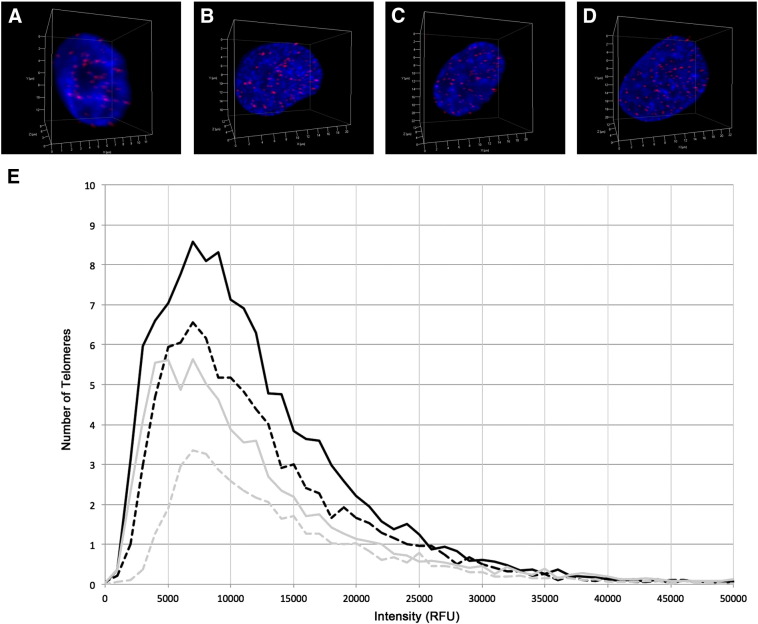
3D telomere profiles that indicated more telomere dysfunction were associated with tumor subgroups with higher MYCN expression. To determine whether there is a functional relationship between MYCN expression and 3D telomere organization, we transfected two neuroblastoma cell lines with constitutive low MYCN expression, SHEP and GIMEN, with a MYCN expression vector. 3D telomere Q-FISH was conducted on transfected cells and mock-transfected controls to analyze the nuclei for changes in their telomere profiles. Transfected cells were sorted to ensure that only cells with MYCN overexpression were analyzed (see Materials and Methods). Representative nuclei illustrating the telomere hybridization in the control and MYCN-transfected SHEP and GIMEN cell lines are shown in Figure 2, A–D. The scatterplot in Figure 2E illustrates the distribution of telomere lengths from each condition, with a plot of the number of telomeres found at each relative fluorescence intensity interval measured: SHEP (gray) and GIMEN (black) cell lines after MYCN (solid lines) and mock (dashed lines) transfection.
Figure 2.
3D telomere organization of neuroblastoma cell lines. 3D telomere Q-FISH labels all telomeres (red) in the SHEP neuroblastoma cell line mock (A) and MYCN transfection (B) and the GIMEN neuroblastoma cell line mock (C) and MYCN transfection (D). Nuclei are counterstained with DAPI (blue). The distribution of telomere lengths for each condition is illustrated in a scatterplot (E) of the number of telomeres found at each relative fluorescence intensity interval measured: SHEP (gray) and GIMEN (black) cell lines after MYCN (solid lines) and mock (dashed lines) transfection.
The average MYCN expression was significantly higher in the post- compared to mock-transfection nuclei for both the SHEP and GIMEN cell lines: 1480.78 versus 6962.54 RFU in the SHEP cell line (P < .0001) and 1577.19 versus 7839.49 RFU in the GIMEN cell line (P < .0001). As shown in Table 4, all telomere parameters were significantly different between the MYCN-transfected and control cells for both cell lines (P < .05 for all comparisons), except nuclear volume in both cell lines and percentage of long telomeres in the GIMEN cell line. Similar to the results seen in low– compared to higher–MYCN-expressing tumor samples, increased MYCN expression in the cell lines is associated with an increase in the number of telomeres, number of telomeric aggregates, and percent of cells with telomeric aggregates (P < .05 for all comparisons). For both cell lines, there is a significantly different distribution of telomere lengths between the control and MYCN-transfected cells (P < .0001), with a higher percentage of short telomeres correlated with increased MYCN expression (P < .05 for both comparisons). An increase in the average number of telomeres per nuclear volume and the a/c ratio indicates that there is also a different nuclear distribution of the telomeres after MYCN transfection (P < .05 for all comparisons).
Table 4.
Statistical Analysis of Telomere Parameters by Neuroblastoma Cell Line
| SHEP Control | SHEP MYCN-Transfection | GIMEN Control | GIMEN MYCN-Transfection | P Value between SHEP Conditions* | P Value between GIMEN Conditions* | |
|---|---|---|---|---|---|---|
| Average MYCN expression per cell (RFU) | 1480.78 | 6962.54 | 1577.19 | 7839.49 | <.0001 | <.0001 |
| Average number of telomeres per nucleus | 41.8 | 74.9 | 85.7 | 120.3 | <.0001 | <.01 |
| Average number of aggregates per nucleus | 4.9 | 9.6 | 10.6 | 15.1 | <.05 | <.01 |
| Average percentage of cells per replicate with aggregates (%) | 71.3 | 100.0 | 80.7 | 100.0 | <.001 | <.01 |
| Average nuclear volume (μm3) | 1053.31 | 1262.03 | 1934.85 | 2225.43 | NS† | NS† |
| Average number of telomeres per nuclear volume | 0.040 | 0.060 | 0.044 | 0.054 | <.05 | <.05 |
| Average a/c ratio per cell | 3.34 | 6.34 | 6.32 | 11.32 | <.01 | <.05 |
| Average percentage of short telomeres per cell (%) | 15.1 | 30.5 | 24.7 | 39.1 | <.001 | <.01 |
| Average percentage of medium telomeres per cell (%) | 55.7 | 47.1 | 52.2 | 38.5 | <.05 | <.05 |
| Average percentage of long telomeres per cell (%) | 29.3 | 22.4 | 23.1 | 22.4 | <.05 | NS |
P value determined by unpaired t test.
Not significant.
In summary, the increased expression of MYCN in both cell lines following MYCN transfection was associated with altered telomere profiles that indicated increased telomere dysfunction. These findings closely parallel those in the neuroblastoma tissue samples and illustrate a functional relationship between MYCN expression and 3D telomere organization.
Discussion
Using 3D telomere Q-FISH, we determined the telomere profiles of 74 neuroblastoma tissue samples. Statistical hierarchical cluster analysis identified three patient subgroups based on the measured 3D nuclear telomere parameters. These subgroups had unique levels of telomere dysfunction. We found that tumors with more telomere dysfunction were associated with unfavorable histopathologic characteristics and poor prognosis. Subgroup III, which had the highest level of telomere dysfunction, clearly identified a group of neuroblastomas that are of high risk as the majority are stage 4, have MYCN amplification and high MYCN expression, are greater than 18 months at diagnosis, and have unfavorable histology. Although tumors in subgroups I and II had similar characteristics, subgroup II had a higher level of telomere dysfunction and may identify neuroblastomas with poorer prognosis among this group; accordingly, subgroup II contained more tumors of higher stage and COG risk than subgroup I. Previous studies have also found that telomere profiles can differentiate tumors that are clinically similar. For example, 3D telomere organization identified recurrent compared to nonrecurrent Hodgkin's lymphoma patients at diagnosis [31]. And previously unknown subgroups of glioblastoma that corresponded with patient survival were discovered using 3D telomere organization [36].
Telomere dysfunction has previously been implicated in neuroblastoma pathogenesis. However, previous studies that determined mean telomere length have been inconclusive as to whether no change, an increase, or a decrease in telomere length is associated with poor patient prognosis [13], [14], [15]. Our method of 3D telomere analysis determines the length of each individual telomere in addition to mean telomere length. All subgroups had a significantly different distribution of telomere lengths. Interestingly, subgroups II and III did not have significantly different mean telomere intensities, but they had unique percentages of short, medium, and long telomeres (P ≤ .05 for all comparisons, see Supplementary Table 1, Table 2, Table 3, Table 4). This finding supports using our approach of measuring all individual telomere lengths as it can detect differences that may be overlooked by methods that solely determine mean telomere length.
In addition to measuring telomere length, our method also analyzes the 3D nuclear telomere distribution. The difference in these parameters indicated that each neuroblastoma subgroup had a unique nuclear architecture. The a/c ratio measures the spherical nature of the telomere distribution and correlates with the cell cycle [18], [19]. A higher ratio indicates more cells at the G2/M boundary; therefore, subgroup III, which has the largest a/c ratio, has the highest level of cellular proliferation. As telomeres shorten with each cell cycle, high rates of proliferation can lead to critically short telomeres [37]. Critically short telomeres trigger DNA damage responses and trigger end-to-end fusions of sister chromatids or of different chromosomes [38]; subsequently, telomere fusions generate dicentric chromosomes and breakage-fusion-bridge cycles that lead to chromosome rearrangements and ongoing genomic instability [39]. Telomeric aggregates can represent telomere fusions [39]. Subgroup III also has the highest number of telomeric aggregates.
3D telomere organization reflects genomic reflect genomic stability [32], [33]. As the neuroblastoma subgroups had different telomere profiles, they will also have unique levels of genomic instability: subgroups with higher levels of telomere dysfunction will have greater genomic instability. Previously, it has been found that MYCN amplified neuroblastomas have multiple structural chromosome aberrations; however, a subset of high-risk MYCN nonamplified tumors harbor more segmental chromosome aberrations including intrachromosomal rearrangements [12], suggesting that these tumors have the most genomic instability. It has also been proposed that these high-risk MYCN nonamplified tumors employ the alternative lengthening of telomeres mechanism [13], a feature of which is heterogeneous telomere lengths [40]. To compare our data with these findings, we further examined subgroup III, which contained tumors with the highest level of genomic instability and highest prognostic risk. We divided subgroup III into two groups: IIIa, composed of 22 MYCN nonamplified tumors, and IIIb, composed of 11 MYCN amplified tumors. Subgroup IIIb had tumors with more telomeres and telomeric aggregates per cell than those in IIIa (P < .0001, see Supplementary Table 5); however, the tumors in subgroup IIIa had more telomeres and telomeric aggregates per cell than both subgroups I and II. The neuroblastomas in subgroups IIIa and IIIb also had a significantly different distribution of telomere lengths (P < .0001); MYCN amplified neuroblastomas in IIIb had predominately short telomeres, whereas MYCN nonamplified tumors in IIIa had similar percentages of short, medium-length, and long telomeres. Therefore, our results are consistent with the previous findings of a subgroup of high-risk MYCN nonamplified neuroblastomas that exhibit a high level of genomic instability and use alternative lengthening of telomeres.
As tissue samples with MYCN amplification and higher MYCN expression were associated with more telomere dysfunction, we examined whether MYCN overexpression could reproduce these findings. MYCN transfection in two neuroblastoma cell lines with constitutive low MYCN expression generated changes in their telomere profiles that were consistent with increased telomere dysfunction. Therefore, we suggest that MYCN overexpression in neuroblastoma may cause telomere dysfunction. Previously, it has been shown that MYC deregulation alters 3D telomere organization with increases in telomere aberrations and genomic instability [41].
In conclusion, this study demonstrates that 3D telomere organization can be used to subgroup neuroblastomas. Classifying neuroblastomas based on the level of telomere dysfunction is a novel approach for this cancer. Similar to previous studies in acute myeloid leukemia, multiple myeloma, glioblastoma, and Hodgkin's lymphoma [20], [21], [22], [36], we found telomere profiles in neuroblastoma to correlate with tumor characteristics and patient prognosis. We propose that 3D telomere organization may be a novel prognostic marker in neuroblastoma, and further studies are needed to validate its potential.
Acknowledgements
The authors thank Mary Cheang for her assistance with statistical analysis. The authors also thank the Children's Oncology Group for development of the neuroblastoma tumor microarray. This work was supported by Canadian Institutes of Health Research funding received by S. M.
Footnotes
Financial support: This work was funded by a Canadian Institutes of Health Research operating grant to S. M. (MOP-123379) and a Canadian Institutes of Health Research studentship to A. K.
Disclosures: S. M. is a founder of and currently a director, officer (CSO), and shareholder of 3D Signatures Inc.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2016.07.001.
Appendix A. Supplementary data
Supplementary Tables.
References
- 1.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 2.Schwab M. Amplification of N-myc as a prognostic marker for patients with neuroblastoma. Semin Cancer Biol. 1993;4:13–18. [PubMed] [Google Scholar]
- 3.Hiyama E, Hiyama K, Yokoyama T, Ishii T. Immunohistochemical analysis of N-myc protein expression in neuroblastoma: correlation with prognosis of patients. J Pediatr Surg. 1991;26:838–843. doi: 10.1016/0022-3468(91)90151-i. [DOI] [PubMed] [Google Scholar]
- 4.Bordow SB, Norris MD, Haber PS, Marshall GM, Haber M. Prognostic significance of MYCN oncogene expression in childhood neuroblastoma. J Clin Oncol. 1998;16:3286–3294. doi: 10.1200/JCO.1998.16.10.3286. [DOI] [PubMed] [Google Scholar]
- 5.Breslow N, McCann B. Statistical estimation of prognosis for children with neuroblastoma. Cancer Res. 1971;31:2098–2103. [PubMed] [Google Scholar]
- 6.George RE, London WB, Cohn SL, Maris JM, Kretschmar C, Diller L. Hyperdiploidy plus nonamplified MYCN confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol. 2005;23:6466–6473. doi: 10.1200/JCO.2005.05.582. [DOI] [PubMed] [Google Scholar]
- 7.Maris JM. The biologic basis for neuroblastoma heterogeneity and risk stratification. Curr Opin Pediatr. 2005;17:7–13. doi: 10.1097/01.mop.0000150631.60571.89. [DOI] [PubMed] [Google Scholar]
- 8.Caron H. Allelic loss of chromosome 1 and additional chromosome 17 material are both unfavourable prognostic markers in neuroblastoma. Med Pediatr Oncol. 1995;24:215–221. doi: 10.1002/mpo.2950240402. [DOI] [PubMed] [Google Scholar]
- 9.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 10.Bown N, Cotterill S, Lastowska M, O'Neill S, Pearson AD, Plantaz D. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 11.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 12.Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer. 2012;131:1591–1600. doi: 10.1002/ijc.27432. [DOI] [PubMed] [Google Scholar]
- 13.Lundberg G, Sehic D, Lansberg JK, Ora I, Frigyesi A, Castel V. Alternative lengthening of telomeres—an enhanced chromosomal instability in aggressive non-MYCN amplified and telomere elongated neuroblastomas. Genes Chromosomes Cancer. 2011;50:250–262. doi: 10.1002/gcc.20850. [DOI] [PubMed] [Google Scholar]
- 14.Ohali A, Avigad S, Ash S, Goshen Y, Luria D, Feinmesser M. Telomere length is a prognostic factor in neuroblastoma. Cancer. 2006;107:1391–1399. doi: 10.1002/cncr.22132. [DOI] [PubMed] [Google Scholar]
- 15.Onitake Y, Hiyama E, Kamei N, Yamaoka H, Sueda T, Hiyama K. Telomere biology in neuroblastoma: telomere binding proteins and alternative strengthening of telomeres. J Pediatr Surg. 2009;44:2258–2266. doi: 10.1016/j.jpedsurg.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 16.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Pienta KJ, Partin AW, Coffey DS. Cancer as a disease of DNA organization and dynamic cell structure. Cancer Res. 1989;49:2525–2532. [PubMed] [Google Scholar]
- 18.Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- 19.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadji M, Adebayo Awe J, Rodrigues P, Kumar R, Houston DS, Klewes L. Profiling three-dimensional nuclear telomeric architecture of myelodysplastic syndromes and acute myeloid leukemia defines patient subgroups. Clin Cancer Res. 2012;18:3293–3304. doi: 10.1158/1078-0432.CCR-12-0087. [DOI] [PubMed] [Google Scholar]
- 21.Knecht H, Sawan B, Lichtensztejn D, Lemieux B, Wellinger RJ, Mai S. The 3D nuclear organization of telomeres marks the transition from Hodgkin to Reed-Sternberg cells. Leukemia. 2009;23:565–573. doi: 10.1038/leu.2008.314. [DOI] [PubMed] [Google Scholar]
- 22.Klewes L, Vallente R, Dupas E, Brand C, Grun D, Guffei A. Three-dimensional nuclear telomere organization in multiple myeloma. Transl Oncol. 2013;6:749–756. doi: 10.1593/tlo.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 24.Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- 25.Kuzyk A, Booth S, Righolt C, Mathur S, Gartner J, Mai S. MYCN overexpression is associated with unbalanced copy number gain, altered nuclear location, and overexpression of chromosome arm 17q genes in neuroblastoma tumors and cell lines. Genes Chromosomes Cancer. 2015;54:616–628. doi: 10.1002/gcc.22273. [DOI] [PubMed] [Google Scholar]
- 26.Knecht H, Kongruttanachok N, Sawan B, Brossard J, Prevost S, Turcotte E. Three-dimensional telomere signatures of Hodgkin- and Reed-Sternberg cells at diagnosis identify patients with poor response to conventional chemotherapy. Transl Oncol. 2012;5:269–277. doi: 10.1593/tlo.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzyk A, Mai S. Selected telomere length changes and aberrant three-dimensional nuclear telomere organization during fast-onset mouse plasmacytomas. Neoplasia (New York, NY) 2012;14:344–351. doi: 10.1593/neo.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204:99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 29.Poon SS, Martens UM, Ward RK, Lansdorp PM. Telomere length measurements using digital fluorescence microscopy. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Knecht H, Bruderlein S, Wegener S, Lichtensztejn D, Lichtensztejn Z, Lemieux B. 3D nuclear organization of telomeres in the Hodgkin cell lines U-HO1 and U-HO1-PTPN1: PTPN1 expression prevents the formation of very short telomeres including "t-stumps". BMC Cell Biol. 2010;11:99. doi: 10.1186/1471-2121-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knecht H, Bruderlein S, Mai S, Moller P, Sawan B. 3D structural and functional characterization of the transition from Hodgkin to Reed-Sternberg cells. Ann Anat. 2010;192:302–308. doi: 10.1016/j.aanat.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell cycle (Georgetown, Tex) 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. [DOI] [PubMed] [Google Scholar]
- 33.Gadji M, Vallente R, Klewes L, Righolt C, Wark L, Kongruttanachok N. Nuclear remodeling as a mechanism for genomic instability in cancer. Adv Cancer Res. 2011;112:77–126. doi: 10.1016/B978-0-12-387688-1.00004-1. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg G, Jin Y, Sehic D, Ora I, Versteeg R, Gisselsson D. Intratumour diversity of chromosome copy numbers in neuroblastoma mediated by on-going chromosome loss from a polyploid state. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pezzolo A, Pistorio A, Gambini C, Haupt R, Ferraro M, Erminio G. Intratumoral diversity of telomere length in individual neuroblastoma tumors. Oncotarget. 2015;6:7493–7503. doi: 10.18632/oncotarget.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gadji M, Fortin D, Tsanaclis AM, Garini Y, Katzir N, Wienburg Y. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia (New York, NY) 2010;12:183–191. doi: 10.1593/neo.91752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Lange T. How telomeres solve the end-protection problem. Science (New York, NY) 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murnane JP. Telomeres and chromosome instability. DNA Repair. 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 39.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 40.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci U S A. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables.