Abstract
Panallergens comprise various protein families of plant as well as animal origin and are responsible for wide IgE cross‐reactivity between related and unrelated allergenic sources. Such cross‐reactivities include reactions between various pollen sources, pollen and plant‐derived foods as well as invertebrate‐derived inhalants and foodstuff. Here, we provide an overview on the most clinically relevant panallergens from plants (profilins, polcalcins, non‐specific lipid transfer proteins, pathogenesis‐related protein family 10 members) and on the prominent animal‐derived panallergen family, tropomyosins. In addition, we explore the role of panallergens in the sensitization process and progress of the allergic disease. Emphasis is given on epidemiological aspects of panallergen sensitization and clinical manifestations. Finally, the issues related to diagnosis and therapy of patients sensitized to panallergens are outlined, and the use of panallergens as predictors for cross‐reactive allergy and as biomarkers for disease severity is discussed.
Keywords: allergy diagnosis, allergy treatment, IgE cross‐reactivity, non‐specific lipid transfer proteins, pathogenesis‐related protein family 10, polcalcins, profilins, tropomyosin
Allergens are harmless common environmental substances capable of inducing IgE‐mediated (type‐I) hypersensitivities in atopic individuals 1. Allergens have been identified in a variety of sources including over 80 plant species and over 100 animal species 2. In most cases, an allergenic source contains more than one allergen deriving from different protein families, which in turn exhibit differential sensitization rates 3. Many allergens have been implicated in cross‐reactivities, where a patient sensitized to a specific source is also reacting to other related and/or non‐related sources. The foundation of such cross‐recognitions is the presence of conserved linear or conformational IgE epitopes among members of the same protein family 4. Panallergens are proteins that take part in key processes of organisms and are therefore ubiquitously distributed with highly conserved sequences and structures 2. Thus, panallergens are implicated in a multitude of cross‐reactions, even between phylogenetically distant and unrelated organisms 2. Such cross‐reactivities include reactions between various pollen sources 5, 6, pollen and plant foods 7 as well as invertebrate inhalants and foods 8, 9. So far, panallergens have been classified into a few protein families with different levels of distribution and cross‐reactivity. In this review, we provide an overview on the most clinically relevant panallergens from plants (profilins, polcalcins, non‐specific lipid transfer proteins, pathogenesis‐related protein family 10 members) as well as a prominent animal panallergen family (tropomyosins). Until now, 44 profilins, 15 polcalcins, 45 nsLTPs, 24 PR‐10s and 29 tropomyosins have been identified and officially acknowledged by the WHO/IUIS allergen nomenclature subcommittee (www.allergen.org) (Tables 1 and 2).
Table 1.
List of plant panallergens profilins, polcalcins, nsLTPs and PR‐10s acknowledged by the IUIS nomenclature subcommittee
| Species | Protein family | ||||
|---|---|---|---|---|---|
| Scientific name | Common name | Profilin | Polcalcin | nsLTP | PR‐10 |
| Acacia farnesiana | Needle bush | Aca f 2 | |||
| Actinidia chinensis | Gold Kiwi fruit | Act c 10 | Act c 8 | ||
| Actinidia deliciosa | Kiwi fruit | Act d 9 | Act d 10 | Act d 8 | |
| Alnus glutinosa | Alder | Aln g 4 | Aln g 1 | ||
| Amaranthus retroflexus | Redroot pigweed | Ama r 2 | |||
| Ambrosia artemisifolia | Short ragweed | Amb a 8 |
Amb a 9 Amb a 10 |
Amb a 6 | |
| Ananas comosus | Pineapple | Ana c 1 | |||
| Apium graveolens | Celery | Api g 4 |
Api g 2 Api g 6 |
Api g 1 | |
| Arachis hypogaea | Peanut | Ara h 5 |
Ara h 9 Ara h 16 Ara h 17 |
Ara h 8 | |
| Artemisia vulgaris | Mugwort | Art v 4 | Art v 5 | Art v 3 | |
| Asparagus officinalis | Asparagus | Aspa o 1 | |||
| Beta vulgaris | Sugar beet | Beta v 2 | |||
| Betula verrucosa (pendula) | European white birch | Bet v 2 |
Bet v 3 Bet v 4 |
Bet v 1 | |
| Brassica oleracea | Cabbage | Bra o 3 | |||
| Brassica rapa | Turnip | Bra r 5 | |||
| Cannabis sativa | Indian hemp | Can s 3 | |||
| Capsicum annuum | Bell pepper | Cap a 2 | |||
| Carpinus betulus | Hornbeam | Car b 1 | |||
| Castanea sativa | Chestnut | Cas s 8 | Cas s 1 | ||
| Chenopodium album | Lambsquarters | Che a 2 | Che a 3 | ||
| Citrus limon | Lemon | Cit l 3 | |||
| Citrus retuculata | Tangerine | Cit r 3 | |||
| Citrus sinensis | Sweet orange | Cit s 2 | Cit s 3 | ||
| Corylus avellana | Hazel | Cor a 2 | Cor a 8 | Cor a 1a | |
| Crocus sativus | Saffron crocus | Cro s 2 | |||
| Cucumis melo | Muskmelon | Cuc m 2 | |||
| Cynodon dactylon | Bermuda grass | Cyn d 12 | Cyn d 7 | ||
| Daucus carota | Carrot | Dau c 4 | Dau c 1 | ||
| Fagus sylvatica | European beech | Fag s 1 | |||
| Fragaria ananassa | Strawberry | Fra a 4 | Fra a 3 | Fra a 1 | |
| Glycine max | Soybean | Gly m 3 | Gly m 4 | ||
| Helianthus annuus | Sunflower | Hel a 2 | Hel a 3 | ||
| Hevea brasiliensis | Para rubber tree (latex) | Hev b 8 | Hev b 12 | ||
| Hordeum vulgare | Barley | Hor v 12 | |||
| Juglans regia | English walnut | Jug r 3 | |||
| Juniperus oxycedrus | Prickly juniper | Jun o 4 | |||
| Kochia scoparia | Burning bush | Koc s 2 | |||
| Lactuca sativa | Cultivated lettuce | Lac s 1 | |||
| Lens culinaris | Lentil | Len c 3 | |||
| Litchi chinensis | Litchi | Lit c 1 | |||
| Malus domestica | Apple | Mal d 4 | Mal d 3 | Mal d 1 | |
| Mercurialis annua | Annual mercury | Mer a 1 | |||
| Morus nigra | Mulberry | Mor n 3 | |||
| Musa acuminata | Banana | Mus a 1 | Mus a 3 | ||
| Olea europea | Olive | Ole e 2 |
Ole e 3 Ole e 8 |
Ole e 7 | |
| Oryza sativa | Rice | Ory s 12 | |||
| Ostrya carpinifilia | European Hophornbeam | Ost c 1 | |||
| Parietaria judaica | Pellitory‐of‐the‐Wall | Par j 3 | Par j 4 | ||
| Phaseolus vulgaris | Green bean | Pha v 3 | |||
| Phleum pratense | Timothy | Phl p 12 | Phl p 7 | ||
| Phoenix dactylifera | Date palm | Pho d 2 | |||
| Platanus acerifolia | London plane tree | Pla l 2 | Pla a 3 | ||
| Platanus orientalis | Oriental plane | Pla or 3 | |||
| Prosopis juliflora | Mesquite | Pro j 2 | |||
| Prunus armeniaca | Apricot | Pru ar 3 | Pru ar 1 | ||
| Prunus avium | Sweet cherry | Pru av 4 | Pru av 3 | Pru av 1 | |
| Prunus domestica | European plum | Pru d 3 | |||
| Prunus dulcis | Almond | Pru du 4 | Pru du 3 | ||
| Prunus persica | Peach | Pru p 4 | Pru p 3 | Pru p 1 | |
| Punica granatum | Pomegranate | Pun g 1 | |||
| Pyrus communis | Pear | Pyr c 4 | Pyr c 3 | Pyr c 1 | |
| Quercus alba | White oak | Que a 1 | |||
| Rubus idaeus | Red raspberry | Rub i 3 | Rub i 1 | ||
| Salsola kali | Russian thistle | Sal k 4 | |||
| Sinapis alba | Yellow mustard | Sin a 4 | Sin a 3 | ||
| Solanum lycopersicum | Tomato | Sola l 1 |
Sola l 3 Sola l 6 Sola l 7 |
Sola l 4 | |
| Syringa vulgaris | Lilac | Syr v 3 | |||
| Triticum aestivum | Wheat | Tri a 12 | Tri a 14 | ||
| Vigna radiata | Mung bean | Vig r 1 | |||
| Vitis vinifera | Grape | Vit v 1 | |||
| Zea mays | Maize | Zea m 12 | Zea m 14 | ||
Hazel trees contain Cor a 1 as a food allergen in the nut and as an inhalant allergen in the pollen.
Table 2.
List of tropomyosins acknowledged by the IUIS nomenclature subcommittee
| Species | Protein family | ||
|---|---|---|---|
| Scientific name | Common name | Tropomyosin | |
| Arachnids | Blomia tropicalis | Mite | Blo t 10 |
| Chortoglyphus arcuatus | Storage mite | Cho a 10 | |
| Dermatophagoides farinae | American house dust mite | Der f 10 | |
| Dermatophagoides pteronyssinus | European house dust mite | Der p 10 | |
| Lepidoglyphus destructor | Storage mite | Lep d 10 | |
| Tyrophagus putrescentiae | Storage mite | Tyr p 10 | |
| Insects | Aedes aegypti | Yellow fever mosquito | Aed a 10 |
| Blattella germanica | German cockroach | Bla g 7 | |
| Chironomus kiiensis | Midge | Chi k 10 | |
| Lepisma saccharina | Silverfish | Lep s 1 | |
| Periplaneta americana | American cockroach | Per a 7 | |
| Mollusks | Helix aspersa | Brown garden snail | Hel as 1 |
| Todarodes pacificus | Squid | Tod p 1 | |
| Parasites | Anisakis simplex | Nematode | Ani s 3 |
| Ascaris lumbricoides | Common roundworm | Asc l 3 | |
| Seafood | Charybdis feriatus | Crab | Cha f 1 |
| Crangon crangon | North Sea shrimp | Cra c 1 | |
| Homarus americanus | American lobster | Hom a 1 | |
| Litopenaeus vannamei | White shrimp | Lit v 1 | |
| Macrobrachium rosenbergii | Giant freshwater prawn | Mac r 1 | |
| Melicertus latisulcatus | King prawn | Mel l 1 | |
| Metapenaeus ensis | Shrimp | Met e 1 | |
| Oreochromis mossambicus | Mozambique tilapia | Ore m 4 | |
| Pandalus borealis | Northern shrimp | Pan b 1 | |
| Panulirus stimpsoni | Spiny lobster | Pan s 1 | |
| Penaeus aztecus | Shrimp | Pen a 1 | |
| Penaeus indicus | Shrimp | Pen i 1 | |
| Penaeus monodon | Black tiger shrimp | Pen m 1 | |
| Portunus pelagicus | Blue swimmer crab | Por p 1 | |
This review also undertakes the task of clarifying the role of panallergens in the sensitization process and the progress of the allergic disease. Whilst controversially discussed, panallergen sensitizations have been shown to provide further complexity to the allergic profile of patients 10. If developed, panallergen sensitization further complicates the allergic disease notably due to its most defining feature, cross‐reactivity. Panallergen cross‐reactivities can result in a wider range of IgE responses to distinct allergen sources, hence creating false positives in diagnosis and further complicating treatment 11. Although just a minority of individuals become sensitized and go onto develop allergy towards panallergens, this phenomenon appears to be highly influenced by many variables including allergen source exposure, geographic differences 12, the type of allergen source and age demography 13. Such variations are reflected in the disparity of results obtained within the literature and highlight the need for understanding panallergens and their sensitization profiles in the development of future diagnostics and treatments of allergy.
Profilins
Profilins are 12–16 kDa, actin(–monomer)‐binding proteins, expressed in all eukaryotic cells and certain viruses, with the exception of some protists 14, 15. Profilins promote polymerization of actin filaments and monomers and are thus involved in the generation of the cytoskeleton and movement 15. A variety of 50 additional identified profilin ligands, like phosphoinositides or proteins containing proline rich domains (also used for purification), suggest an important role in many more complex molecular processes as well as signal transduction 16, 17. They belong to the α‐β class of proteins with a highly conserved structure consisting of a central antiparallel β‐sheet core surrounded by α‐helices 18. The first allergenic profilin, Bet v 2 from birch pollen, was identified in 1991 19, and since then, many allergenic members have been identified in pollen, plant foods and latex (www.allergen.org). The involvement of profilins in such essential cellular processes explains their ubiquitous expression and high levels of conservation. This, in turn, can justify the strong serologic cross‐reactivity of the molecules 20. The worldwide prevalence of profilin sensitization, which appears to be initiated by sensitization to pollen, lies between 5% and 42%, whilst the sensitization profile is influenced by geographic factors and can vary greatly between different countries 20, 21. For some profilins, the sensitization rates can reach proportions of major allergens >50% (e.g. Pho d 2 from date palm) 22. As profilins are labile against heat and gastric digestion, reactions to food profilins manifested as pollen‐food syndromes are not uncommon 2.
Polcalcins
Polcalcins are a pollen‐restricted portion of widespread Ca2+‐binding proteins comprising a major group of allergenic molecules of this kind. The mostly α‐helical proteins contain the characteristic EF‐hand motifs (helix‐loop‐helix structure), which are responsible for the binding of calcium. Polcalcins have a molecular mass of 9–28 kDa, depending on the number of EF‐hand motifs they hold (2, 3 or 4 EF‐hand motifs), and show a monomeric or dimeric structure. They can assume two different conformations; a closed, Ca2+‐free apo‐form and an open, Ca2+‐bound holo form which is more stable and results in stronger interactions with IgE antibodies 23. Although the exact physiologic function of polcalcins remains elusive, their localization in the pollen and their ability to regulate intracellular Ca2+ levels suggest a contribution in pollen tube outgrowth 24. Allergenic polcalcins derive from tree, grass and weed pollen; are highly cross‐reactive; and induce sensitization rates between 5% and 10% among pollen‐allergic patients 2. It seems that the clinical relevance of polcalcin sensitization is highly dependent on geographic factors and therefore exposure. Phl p 7 from timothy grass pollen is, among all allergenic polcalcins, the most cross‐reactive molecule and could hence be used as a marker to identify multiple pollen sensitizations 25.
Non‐specific lipid transfer proteins (nsLTPs)
Originally named after their ability to transfer lipid molecules between membranes in vitro, which seems unlikely in vivo, lipid transfer proteins (LTPs) are widely distributed small and basic proteins in higher plants 26. LTPs can be either lipid specific or can non‐specifically accommodate several classes of lipids (nsLTPs); all allergenic LTP members have been found within the nsLTP cluster. According to their size, nsLTPs can be further subdivided into the 9 kDa nsLTP1 and the 7 kDa nsLTP2 subfamilies 27, 28. Their physiologic function is still unknown; however, accumulating evidence shows that they play a role in cytology, growth and development. Furthermore, the implication of nsLTPs in the general defence of plants classifies them as members of the pathogenesis‐related protein family (PR‐14 family) 26, 29. nsLTPs share a common conserved tertiary structure consisting of four α‐helices stabilized by 4 disulphide bonds (eight‐cysteine motif), forming a hydrophobic tunnel‐like cavity, where interactions with lipids occur 30, 31. Allergenic nsLTPs show a wide distribution in pollen of trees and weeds, in foods (fruits, nuts, seeds and vegetables) as well as in latex 2. Due to their high thermal and proteolytic stability, nsLTPs are potent class I food allergens. Nevertheless, it is still unclear whether pollen or food nsLTPs are the primary sensitizers 32. nsLTPs are clinically highly relevant and cross‐reactive allergens. In Mediterranean areas, nsLTP representatives like Pru p 3 from peach constitute major allergens, whilst in Central and Northern areas, sensitization to Pru p 3 is limited 33.
Pathogenesis‐related protein family 10 (PR‐10)
PR‐10 proteins function in the general defence mechanisms of plants, and their expression is induced by stress and pathogens. They constitute mostly intracellular, intrinsic and highly similar globular proteins with a length of around 160 amino acids. They share a molecular mass of around 17 kDa and are encoded by a diverse multigene family 34. Their 3D fold consists of 3 α‐helices embedded in an antiparallel β‐sheet consisting of 7 β‐strands forming an amphiphilic intrinsic solvent‐accessible y‐shaped cavity. Crystallographic analyses of Bet v 1 in combination with a variety of different ligands revealed the availability of a promiscuous binding site in the pocket, which could help to unravel the biologic function of PR‐10s 35, 36. Recently, the physiologic ligand quercetin‐3‐O‐sophoroside in birch pollen for Bet v 1 has been identified 37. A subclass of PR‐10 proteins forms a group of pollen and food allergens. The botanically related Fagales species of birch, alder, hornbeam, hop‐hornbeam, hazelnut, beech, chestnut and oak are the main elicitors of early seasonal rhinitis in the temperate climate zone of the Northern Hemisphere. Whilst Bet v 1 is known to act as the main sensitizer, a potential (co)‐sensitization of other Fagales members is hypothesized 38. A high percentage of Fagales pollen‐allergic patients develop oral reactions against a variety of fresh fruits, nuts and vegetables. Such clinical reactions, known as pollen‐food syndrome, are triggered by IgE antibodies, which are produced against aeroallergens and cross‐react with food allergens 7.
Tropomyosin
Tropomyosins are α‐helical and, in their native form, dimeric coiled‐coil fibrous structural proteins consisting of two subunits (α, β), each with a molecular weight ranging from 34 to 38 kDa. They form co‐polymers with actin and regulate actin–myosin interactions in skeletal muscles and diversify the functions of actin filaments in different intracellular compartments 39. Evolutionarily, tropomyosins were introduced in metazoa and fungi to expand the functional capacity of actin filaments without enlarging the number of actin isoforms and are therefore not present in plants. In vertebrates, four highly conserved tropomyosin genes are expressed. The increasing number of tropomyosins and splicing variants parallels the increasing complexity of the animal kingdom 15. Tropomyosins are considered important food allergens, whereas tropomyosins from vertebrates are typically non‐allergenic, with the exception of the fresh water fish tilapia 9. Crustacean shellfish form one group of ‘the big‐8’ major food allergens (www.fda.gov). The highly heat‐stable tropomyosins comprise the major allergens in crustaceans and mollusks, making them significant food allergens in exposed populations 40. In addition, these patients show frequent reactions towards (house dust) mites and insects, especially in warmer climates where the growth of house dust mites is favoured. Several studies have already established cross‐reactivity between seafood, mollusks, insects and some parasites 41, 42. However, the route of sensitization remains a matter of debate. There is a controversy as to whether primary sensitization is happening via the gastrointestinal or the respiratory tract. Nonetheless, there are clear indications that inhalation of aeroallergens or cooking vapours is an important sensitising route in the context of tropomyosin hypersensitivity 41, 43. A schematic diagram describing potential exposure routes and sensitization to tropomyosins is presented in Fig. 1.
Figure 1.
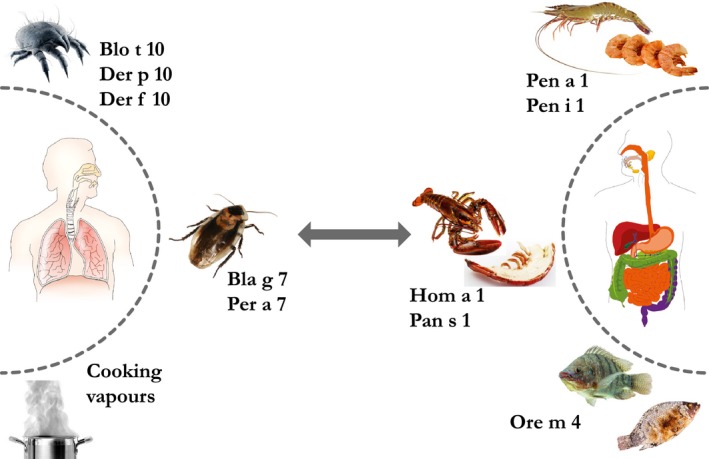
The importance of panallergens in multiple sensitizations. Potential allergic reactions to tropomyosins can occur as a result of primary sensitization via different routes or via IgE cross‐reactivities. The left hand side describes arthropod tropomyosins to which individuals are commonly exposed to via the inhalation route. Cooking vapours of crustaceans are also a possible route of exposure and sensitization. Ingestion of crustaceans and fish, as described in the right hand side, can also cause sensitization to tropomyosins (Table 2 catalogues the mentioned tropomyosins in detail). Images were acquired from ©Ammit; psdesign1/fotolia.com and ©daagron; roblan; suchatbky; tunedin123/123RF.com.
Sensitization patterns
Panallergen sensitizations tend to occur as part of a co‐ or poly‐sensitization profile, with panallergen monosensitizations rarely arising 44, 45. In particular, panallergen sensitizations are commonly associated with high or prolonged exposure to a given allergen source, often correlating with increased severity of disease 10, 12. The clinical relevance of panallergens is highly influenced by a number of factors including differences in geographic region 12, allergenic source, the host response against the allergen and exposure 13. Furthermore, differences between child and adult populations have exhibited great variability 12, 46. Hence, studying sensitization profiles of a given source can be a useful tool in understanding the clinical relevance of panallergens. Hatzler et al. exemplifies such a predictable pattern of sensitization for the allergen source Phleum pratense (timothy grass). Initial sensitizations towards major (Phl p 1 and Phl p 5) and minor (Phl p 4, 2, 6, 11) allergens gradually result in the onset of allergic disease, following such disease onset the occurrence of panallergen sensitizations, towards Phl p 12 and 7, increases for a minority of patients in correlation with disease severity. Hence, whilst not being an indicative marker for allergic disease onset, panallergen‐specific IgE responses have the potential as clinical biomarkers for increased severity of disease, although it must be emphasized that only a minority of patients become sensitized 10. Such rates of sensitization are influenced by the level of exposure to an allergen source. Feliu et al. demonstrated that children, even with a short disease history, were able to become panallergen sensitized to both date palm profilin (Pho d 2) and peach nsLTP (Pru p 3), with 12% and 13% incidence of IgE positivities, respectively 47. Moreover, high olive pollen rates in southern Spain have been reported to drive increases in sensitizations to the olive nsLTP Ole e 7 12, further showing high exposure rates are strongly correlated with increases in panallergen IgE‐positive patients.
Furthermore, such increased rates of panallergen sensitization have been shown to correlate with an increased severity of allergic symptoms (and in the absence of panallergen allergy). A study carried out by Alverado et al. investigating profilin‐related allergic reactions over both an intense and a mild grass pollen season showed that more severe profilin allergy occurred during the intense season again emphasising the relationship between higher allergen exposures and increased panallergen sensitization rates 44. This phenomenon has been further demonstrated in a large study of 891 allergic patients from Spain where sensitizations to grass pollen profilin correlated with the severity of the allergic disease 12. Conversely, recent studies have emerged describing sensitizations to multiple panallergen families and their effects in reducing the severity of allergic reactions. In particular, such co‐sensitizations have been investigated in peach allergy due to the multiple panallergens present within the fruit, namely Pru p 1 (PR‐10), Pru p 3 (nsLTP) and Pru p 4 (profilin). Patients sensitized to nsLTPs along with PR‐10/profilins present a lower severity of symptoms when compared to patients with sensitizations to nsLTP alone 3. Considering that many allergenic sources contain multiple panallergen families, exploring this avenue and the theories behind such interactions may be of key benefit for developing therapeutic strategies 3, 46. Further investigations into allergies of different plant species have revealed that certain pollen exhibits extraordinarily high rates of panallergen sensitization 48, 49. For Chenopodium album pollen (white goosefoot), sensitization rates of 55% and 46% to profilin and polcalcin, respectively, have been reported 48. A further study by Nouri et al., carried out in an Iranian cohort, showed that 81% of patients tested positive to the profilin panallergen Che a 2 49. Disparity of panallergen sensitizations is also exhibited between child and adult populations. Interestingly, Barber et al. also show sensitizations towards the peach nsLTP Pru p 3 to be more prevalent within children than in adult populations in areas of high pollen sensitization in Spain 12. Furthermore, in a Mediterranean study observing nsLTP sensitizations, it was shown that children below the age of six were more frequently sensitized by Pru p 3. However, for the adult population, sensitizations to the walnut nsLTP Jug r 3 reached comparable levels, suggesting an alternative source of sensitization within this age group 46. Such epidemiological differences must be considered when performing clinical investigations, and understanding such profiles of panallergen sensitizations may be of high clinical benefit in both diagnosis and treatment of allergy.
Panallergen allergy
For only a minority of patients sensitized to panallergens, allergy arises 50. It is in such cases that the cross‐reactivity of panallergens plays a role in worsening the allergic profile of patients via increasing the amount of potential allergenic reactions to allergens in unrelated sources 51. Tropomyosins 43, profilins, PR‐10s and nsLTPs are commonly found in food and plant sources (Tables 1 and 2) and are strongly associated to food allergy arising as a consequence of cross‐reactivity to inhaled allergens 51, 52. Pollen‐food allergy syndrome is occurring commonly for profilins, PR‐10s and nsLTPs 51, especially when exposure rates are increased 12. Associated symptoms range from oral allergy to anaphylaxis and high co‐occurrence with other atopic diseases including allergic rhinitis and asthma, further emphasising the clinical relevance of panallergens 51, 53, 54, 55. Identification of such cross‐reactivities may be vital in finding biomarkers for allergy. For example, the discovery of cross‐reactions between the plane pollen nsLTP Pla a 3 and the peach nsLTP Pru p 3 has elucidated the potential use of Pla a 3 as a biomarker in identifying plane pollen‐allergic patients at risk of developing plant food‐related nsLTP allergies, thereby improving diagnosis and treatment 56. Such strategies may be helpful in avoiding unnecessary dietary restrictions associated to food allergy 57.
It is of note that these different panallergen families present varying levels of severity with regard to the symptoms induced. Tropomyosin represents the only panallergen family with the potential to induce autoimmune responses 58; however, all of the families elicit their effect through the allergic response. Most severely, cases of anaphylaxis have been documented for tropomyosins, nsLTPs 59 and PR‐10s 51, 60. Polcalcins and profilins, whilst presenting slight correlations to anaphylactic reactions, have not been described as direct initiators 61, although wide associations with local reactions (i.e. oral allergy syndrome) have been documented 62. Most frequently, profilins act as minor respiratory allergens resulting in mild allergic symptoms. However, variations in epidemiological factors may present profilin as a potentially severe food allergen and a marker for severe allergy, in particular for grass pollen allergy 44, 47, 63. PR‐10s are associated with both mild and more severe reactions 60, 64, whilst nsLTPs are linked to more severe allergic reactions 65. Polcalcins, whilst exclusively found in pollen sources, have not been implicated in any pollen‐food cross‐reactivities but are more heavily involved in mild respiratory reactions due to inhalation 66.
Diagnosis and treatment
Panallergen sensitizations may be indicative of allergic disease severity and key in identifying a specific allergenic source, especially when sensitization rates are high 67. Currently, a main treatment of such panallergen allergy is avoidance of the allergy causing foods. This course of action may present detrimental side effects to the patient, with regular intake of fresh fruit having associations with increased health and reduced risk in the development of several chronic diseases 52. For the interests of patients’ quality of life and nutritional satisfaction, it is of benefit to understand the demographics of panallergen sensitization in a specific detail, especially in order to be able to exactly determine the specific allergenic cause. Moreover, with evidence that panallergen sensitizations are linked to increased severity of symptoms, it is crucial to efficiently identify those patients with a higher risk of a severe reaction.
In vitro molecular‐based diagnosis (MBD) uses allergenic proteins, either natural, purified or recombinantly produced, to quantify the amount of allergen‐specific IgE antibodies in the blood circulation 59. The use of microarray systems, such as the Immuno‐Sorbent Allergen Chip (ISAC) used for detecting and quantifying the reactivity of specific IgE antibodies towards more than 100 allergens and allergen components, is helpful in the identification and interpretation of allergic sensitization profiles 68. However, it must be noted that IgE positivity alone is not entirely indicative of allergy, and further information obtained by oral/respiratory provocation challenges or basophil activation tests is key in obtaining a true allergic diagnosis 47, 57, 69. Successful diagnosis is essential for deciphering treatment; allergen immunotherapy (AIT) currently represents one of the few ways to modulate allergic disease, and success is highly variable 70. Whilst there is potential in moderating panallergen sensitizations, in particular for food‐related allergy, current commercial availability of therapeutic extracts is limited for minor allergens/panallergens 57. Moreover, lack of standardization provides further difficulty in the use of commercial extracts 11, 71.
Exploration of the hierarchical patterns of cross‐reactive molecules may also provide further insight into panallergen sensitizations and potentially elucidate novel therapeutics based on the concept that some molecules are more cross‐reactive than others. A recent microarray study by Pfiffner et al. investigates the structural basis of cross‐reactivity using an iterative motif detection algorithm 72. Identifying such hierarchies raises the question as to whether treatments using AIT should constitute of more frequently cross‐reacting allergens or an allergen that less frequently cross‐reacts with other allergens, but targets a larger repertoire of allergens within the hierarchy. Further exploration into individual panallergen sensitization profiles (e.g. nsLTPs) 46 may provide key insight into the mechanisms of panallergens in allergy and reveal predicable patterns of cross‐reactivities, thus improving diagnosis. Furthermore, using panallergens as biomarkers for disease severity and as predictors of cross‐reactive allergens may pose the most clinically relevant use of panallergens in allergic disease 47, 69.
Summary and conclusions
In daily clinical practice, it is often observed that individuals allergic to fruits, nuts and vegetables have sensitivities to more than one food, not necessarily belonging to the same family. In addition, many of them are also allergic to pollens and/or seafood. These associations occur when the same IgE antibody is able to recognize similar epitopes in different panallergens and appear to be highly influenced by many variables. This phenomenon explains why some patients develop a severe allergic reaction upon ingesting allergenic foods they have never before encountered 73. Cross‐reactivity can also occur due to conformational diversity of antibodies 74 and through T‐cell epitopes 75, which in turn may lead to disease. Multiple reactivity to inhalant or allergenic food sources appeared to be caused by the sensitization to at least one representative molecule from a single panallergen group, emphasising the importance of panallergens in multiple sensitizations (Fig. 1).
In conclusion, panallergens comprise a variety of protein families of plant and animal origin and are responsible for wide IgE cross‐reactivity between different allergenic sources. Panallergen sensitizations are clinical ‘warning signs’ for elevated risk of multiple allergies as well as severe symptoms. Thus, improving the understanding of this immunologic cross‐reactivity is essential for the interpretation of their clinical relevance, the development of improved diagnostic tests and the generation of more efficacious immunotherapy agents. Future studies focusing on the identification and evaluation of cross‐reactive epitopes between different members of panallergenic families might provide the basis for understanding their role in sensitization and allergic reactions.
Acknowledgments
Olivia E. McKenna is supported by the Austrian Science Funds (FWF) International PhD Program ‘Immunity in Cancer and Allergy’, ICA, Project W1213); Claudia Asam and Anargyros Roulias are supported by the FWF project P27589; Galber R. Araujo is post‐doctorate fellow from ‘Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant N. 153753/2015‐3’, and Luiz R. Goulart is supported by CNPq, ‘grant N. 401131/2014‐9’. The authors are grateful to the University of Salzburg's Priority Program ‘Allergy‐Cancer‐BioNano Research Centre’ for supporting their work.
McKenna OE, Asam C, Araujo GR, Roulias A, Goulart LR, Ferreira F. How relevant is panallergen sensitization in the development of allergies? Pediatr Allergy Immunol 2016: 27: 560–568.
References
- 1. Pfaar O, Bachert C, Bufe A, et al. Guideline on allergen‐specific immunotherapy in IgE‐mediated allergic diseases: S2k Guideline of the German Society for Allergology and Clinical Immunology (DGAKI), the Society for Pediatric Allergy and Environmental Medicine (GPA), the Medical Association of German Allergologists (AeDA), the Austrian Society for Allergy and Immunology (OGAI), the Swiss Society for Allergy and Immunology (SGAI), the German Society of Dermatology (DDG), the German Society of Oto‐ Rhino‐Laryngology, Head and Neck Surgery (DGHNO‐KHC), the German Society of Pediatrics and Adolescent Medicine (DGKJ), the Society for Pediatric Pneumology (GPP), the German Respiratory Society (DGP), the German Association of ENT Surgeons (BV‐HNO), the Professional Federation of Paediatricians and Youth Doctors (BVKJ), the Federal Association of Pulmonologists (BDP) and the German Dermatologists Association (BVDD). Allergo J Int 2014: 23: 282–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hauser M, Roulias A, Ferreira F, Egger M. Panallergens and their impact on the allergic patient. Allergy Asthma Clin Immunol 2010: 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pastorello EA, Farioli L, Stafylaraki C, et al. Anti‐rPru p 3 IgE levels are inversely related to the age at onset of peach‐induced severe symptoms reported by peach‐allergic adults. Int Arch Allergy Immunol 2013: 162: 45–9. [DOI] [PubMed] [Google Scholar]
- 4. Kazatsky AM, Wood RA. Classification of Food Allergens and Cross‐Reactivity. Curr Allergy Asthma Rep 2016: 16: 22. [DOI] [PubMed] [Google Scholar]
- 5. Asam C, Hofer H, Wolf M, Aglas L, Wallner M. Tree pollen allergens‐an update from a molecular perspective. Allergy 2015: 70: 1201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gadermaier G, Dedic A, Obermeyer G, Frank S, Himly M, Ferreira F. Biology of weed pollen allergens. Curr Allergy Asthma Rep 2004: 4: 391–400. [DOI] [PubMed] [Google Scholar]
- 7. Price A, Ramachandran S, Smith GP, Stevenson ML, Pomeranz MK, Cohen DE. Oral allergy syndrome (pollen‐food allergy syndrome). Dermatitis 2015: 26: 78–88. [DOI] [PubMed] [Google Scholar]
- 8. Pedrosa M, Boyano‐Martinez T, Garcia‐Ara C, Quirce S. Shellfish allergy: a comprehensive review. Clin Rev Allergy Immunol 2015: 49: 203–16. [DOI] [PubMed] [Google Scholar]
- 9. Reese G, Ayuso R, Lehrer SB. Tropomyosin: an invertebrate pan‐allergen. Int Arch Allergy Immunol 1999: 119: 247–58. [DOI] [PubMed] [Google Scholar]
- 10. Hatzler L, Panetta V, Lau S, et al. Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J Allergy Clin Immunol 2012: 130: 894–901. e5. [DOI] [PubMed] [Google Scholar]
- 11. Egger M, Hauser M, Himly M, Wopfner N, Wallner M, Ferreira F. Development of recombinant allergens for diagnosis and therapy. Front Biosci (Elite Ed) 2009: 1: 77–90. [DOI] [PubMed] [Google Scholar]
- 12. Barber D, de la Torre F, Feo F, et al. Understanding patient sensitization profiles in complex pollen areas: a molecular epidemiological study. Allergy 2008: 63: 1550–8. [DOI] [PubMed] [Google Scholar]
- 13. Ferreira F, Hawranek T, Gruber P, Wopfner N, Mari A. Allergic cross‐reactivity: from gene to the clinic. Allergy 2004: 59: 243–67. [DOI] [PubMed] [Google Scholar]
- 14. Machesky LM, Cole NB, Moss B, Pollard TD. Vaccinia virus expresses a novel profilin with a higher affinity for polyphosphoinositides than actin. Biochemistry 1994: 33: 10815–24. [DOI] [PubMed] [Google Scholar]
- 15. Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 2015: 128: 2009–19. [DOI] [PubMed] [Google Scholar]
- 16. Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 2004: 14: 461–9. [DOI] [PubMed] [Google Scholar]
- 17. Sun T, Li S, Ren H. Profilin as a regulator of the membrane‐actin cytoskeleton interface in plant cells. Front Plant Sci 2013: 4: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krishnan K, Holub O, Gratton E, Clayton AH, Cody S, Moens PD. Profilin interaction with phosphatidylinositol (4,5)‐bisphosphate destabilizes the membrane of giant unilamellar vesicles. Biophys J 2009: 96: 5112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valenta R, Duchene M, Pettenburger K, et al. Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 1991: 253: 557–60. [DOI] [PubMed] [Google Scholar]
- 20. Santos A, Van Ree R. Profilins: mimickers of allergy or relevant allergens? Int Arch Allergy Immunol 2011: 155: 191–204. [DOI] [PubMed] [Google Scholar]
- 21. Wopfner N, Gruber P, Wallner M, et al. Molecular and immunological characterization of novel weed pollen pan‐allergens. Allergy 2008: 63: 872–81. [DOI] [PubMed] [Google Scholar]
- 22. Asturias JA, Ibarrola I, Fernandez J, Arilla MC, Gonzalez‐Rioja R, Martinez A. Pho d 2, a major allergen from date palm pollen, is a profilin: cloning, sequencing, and immunoglobulin E cross‐reactivity with other profilins. Clin Exp Allergy 2005: 35: 374–81. [DOI] [PubMed] [Google Scholar]
- 23. Henzl MT, Sirianni AG, Wycoff WG, Tan A, Tanner JJ. Solution structures of polcalcin Phl p 7 in three ligation states: Apo‐, hemi‐Mg2 + ‐bound, and fully Ca2 + ‐bound. Proteins 2013: 81: 300–15. [DOI] [PubMed] [Google Scholar]
- 24. Wopfner N, Dissertori O, Ferreira F, Lackner P. Calcium‐binding proteins and their role in allergic diseases. Immunol Allergy Clin North Am 2007: 27: 29–44. [DOI] [PubMed] [Google Scholar]
- 25. Tinghino R, Twardosz A, Barletta B, et al. Molecular, structural, and immunologic relationships between different families of recombinant calcium‐binding pollen allergens. J Allergy Clin Immunol 2002: 109: 314–20. [DOI] [PubMed] [Google Scholar]
- 26. Liu F, Zhang X, Lu C, et al. Non‐specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J Exp Bot 2015: 66: 5663–81. [DOI] [PubMed] [Google Scholar]
- 27. Richard C, Leduc V, Battais F. Plant lipid transfer proteins (LTPS): biochemical aspect in panallergen–structural and functional features, and allergenicity. Eur Ann Allergy Clin Immunol 2007: 39: 76–84. [PubMed] [Google Scholar]
- 28. Salcedo G, Sanchez‐Monge R, Barber D, Diaz‐Perales A. Plant non‐specific lipid transfer proteins: an interface between plant defence and human allergy. Biochim Biophys Acta 2007: 1771: 781–91. [DOI] [PubMed] [Google Scholar]
- 29. Sinha M, Singh RP, Kushwaha GS, et al. Current overview of allergens of plant pathogenesis related protein families. ScientificWorldJournal 2014: 2014: 543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jose‐Estanyol M, Gomis‐Ruth FX, Puigdomenech P. The eight‐cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 2004: 42: 355–65. [DOI] [PubMed] [Google Scholar]
- 31. Hauser M, Egger M, Wallner M, Wopfner N, Schmidt G, Ferreira F. Molecular properties of plant food allergens: a current classification into protein families. Open Immunol J 2008: 1: 1–12. [Google Scholar]
- 32. Zuidmeer L, van Ree R. Lipid transfer protein allergy: primary food allergy or pollen/food syndrome in some cases. Curr Opin Allergy Clin Immunol 2007: 7: 269–73. [DOI] [PubMed] [Google Scholar]
- 33. Egger M, Hauser M, Mari A, Ferreira F, Gadermaier G. The role of lipid transfer proteins in allergic diseases. Curr Allergy Asthma Rep 2010: 10: 326–35. [DOI] [PubMed] [Google Scholar]
- 34. Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR‐10 proteins. FEBS J 2013: 280: 1169–99. [DOI] [PubMed] [Google Scholar]
- 35. Kofler S, Asam C, Eckhard U, Wallner M, Ferreira F, Brandstetter H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen bet v 1. J Mol Biol 2012: 422: 109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asam C, Batista AL, Moraes AH, et al. Bet v 1–a Trojan horse for small ligands boosting allergic sensitization? Clin Exp Allergy 2014: 44: 1083–93. [DOI] [PubMed] [Google Scholar]
- 37. Seutter von Loetzen C, Hoffmann T, Hartl MJ, et al. Secret of the major birch pollen allergen Bet v 1: identification of the physiological ligand. Biochem J 2014: 457: 379–90. [DOI] [PubMed] [Google Scholar]
- 38. Hauser M, Asam C, Himly M, et al. Bet v 1‐like pollen allergens of multiple Fagales species can sensitize atopic individuals. Clin Exp Allergy 2011: 41: 1804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 2005: 15: 333–41. [DOI] [PubMed] [Google Scholar]
- 40. Lopata AL, Lehrer SB. New insights into seafood allergy. Curr Opin Allergy Clin Immunol 2009: 9: 270–7. [DOI] [PubMed] [Google Scholar]
- 41. Wong L, Huang CH, Lee BW. Shellfish and house dust mite allergies: is the link tropomyosin? Allergy Asthma Immunol Res 2016: 8: 101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santos AB, Rocha GM, Oliver C, et al. Cross‐reactive IgE antibody responses to tropomyosins from Ascaris lumbricoides and cockroach. J Allergy Clin Immunol 2008: 121(1040–6): e1. [DOI] [PubMed] [Google Scholar]
- 43. Fernandes J, Reshef A, Patton L, Ayuso R, Reese G, Lehrer SB. Immunoglobulin E antibody reactivity to the major shrimp allergen, tropomyosin, in unexposed Orthodox Jews. Clin Exp Allergy 2003: 33: 956–61. [DOI] [PubMed] [Google Scholar]
- 44. Alvarado MI, Jimeno L, De La Torre F, et al. Profilin as a severe food allergen in allergic patients overexposed to grass pollen. Allergy 2014: 69: 1610–6. [DOI] [PubMed] [Google Scholar]
- 45. Ortolani C, Ispano M, Ansaloni R, Rotondo F, Incorvaia C, Pastorello EA. Diagnostic problems due to cross‐reactions in food allergy. Allergy 1998: 53: 58–61. [DOI] [PubMed] [Google Scholar]
- 46. Scala E, Till SJ, Asero R, et al. Lipid transfer protein sensitization: reactivity profiles and clinical risk assessment in an Italian cohort. Allergy 2015: 70: 933–43. [DOI] [PubMed] [Google Scholar]
- 47. Feliu A, Gonzalez‐de‐Olano D, Gonzalez E, et al. A multicenter study of sensitization profiles in an allergic pediatric population in an area with high allergen exposure. J Investig Allergol Clin Immunol 2013: 23: 337–44. [PubMed] [Google Scholar]
- 48. Barderas R, Villalba M, Pascual CY, Batanero E, Rodriguez R. Profilin (Che a 2) and polcalcin (Che a 3) are relevant allergens of Chenopodium album pollen: isolation, amino acid sequences, and immunologic properties. J Allergy Clin Immunol 2004: 113: 1192–8. [DOI] [PubMed] [Google Scholar]
- 49. Nouri HR, Varasteh A, Vahedi F, Chamani J, Afsharzadeh D, Sankian M. Constructing a hybrid molecule with low capacity of IgE binding from Chenopodium album pollen allergens. Immunol Lett 2012: 144: 67–77. [DOI] [PubMed] [Google Scholar]
- 50. Masilamani M, Commins S, Shreffler W. Determinants of food allergy. Immunol Allergy Clin North Am 2012: 32: 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Popescu F‐D. Cross‐reactivity between aeroallergens and food allergens. World J Methodol 2015: 5: 31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andersen MB, Hall S, Dragsted LO. Identification of european allergy patterns to the allergen families PR‐10, LTP, and profilin from Rosaceae fruits. Clin Rev Allergy Immunol 2011: 41: 4–19. [DOI] [PubMed] [Google Scholar]
- 53. Nowak‐Wegrzyn A, Sampson HA. Future therapies for food allergies. J Allergy Clin Immunol 2011: 127: 558–73; quiz 74‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoffmann‐Sommergruber K, Mills ENC. Food allergen protein families and their structural characteristics and application in component‐resolved diagnosis: new data from the EuroPrevall project. Anal Bioanal Chem 2009: 395: 25–35. [DOI] [PubMed] [Google Scholar]
- 55. Migueres M, Davila I, Frati F, et al. Types of sensitization to aeroallergens: definitions, prevalences and impact on the diagnosis and treatment of allergic respiratory disease. Clin Transl Allergy 2014: 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wangorsch A, Larsson H, Messmer M, et al. Molecular cloning of plane pollen allergen Pla a 3 and its utility as diagnostic marker for plane pollen associated peach allergy. Clin Exp Allergy 2016: 46: 764–74. [DOI] [PubMed] [Google Scholar]
- 57. Kattan JD, Sicherer SH. Optimizing the diagnosis of food allergy. Immunol Allergy Clin North Am 2015: 35: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu R, Holck AL, Yang E, Liu C, Xue W. Tropomyosin from tilapia (Oreochromis mossambicus) as an allergen. Clin Exp Allergy 2013: 43: 365–77. [DOI] [PubMed] [Google Scholar]
- 59. Heaps A, Carter S, Selwood C, et al. The utility of the ISAC allergen array in the investigation of idiopathic anaphylaxis. Clin Exp Immunol 2014: 177: 483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kleine‐Tebbe J, Vogel L, Crowell DN, Haustein UF, Vieths S. Severe oral allergy syndrome and anaphylactic reactions caused by a Bet v 1‐ related PR‐10 protein in soybean, SAM22. J Allergy Clin Immunol 2002: 110: 797–804. [DOI] [PubMed] [Google Scholar]
- 61. Florido Lopez JF, Quiralte Enriquez J, Arias de Saavedra Alias JM, Saenz de San Pedro B, Martin Casanez E. An allergen from Olea europaea pollen (Ole e 7) is associated with plant‐derived food anaphylaxis. Allergy 2002: 57 (Suppl. 71): 53–9. [DOI] [PubMed] [Google Scholar]
- 62. Ibáñez MD, Sastre J, San Ireneo MM, Laso MT, et al. Differents patterns of allergen recognition in children allergic to orange. J Allergy Clin Immunol 2004: 113: 175–7. [DOI] [PubMed] [Google Scholar]
- 63. Worm M, Jappe U, Kleine‐Tebbe J, et al. Food allergies resulting from immunological cross‐reactivity with inhalant allergens. Allergologie 2014: 37: 170–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wensing M, Akkerdaas JH, van Leeuwen WA, et al. IgE to Bet v 1 and profilin: cross‐reactivity patterns and clinical relevance. J Allergy Clin Immunol 2002: 110: 435–42. [DOI] [PubMed] [Google Scholar]
- 65. Pascal M, Sanchez‐Lopez J, Munoz‐Cano R, et al. Association of Pla a 1, Pla a 2 and Cor a 8 with Pru p 3 food allergy. Allergy 2012: 67: 376–76. [Google Scholar]
- 66. Mas S, Garrido‐Arandia M, Batanero E, et al. Characterization of profilin and polcalcin panallergens from ash pollen. J Investig Allergol Clin Immunol 2014: 24: 257–66. [PubMed] [Google Scholar]
- 67. Huertas AJ, Carreno A, Merida C, Pajaron‐Fernandez MJ, Ramirez‐Hernandez M, Carnes J. Profilin sensitisation in a Mediterranean population. Allergol Immunopathol (Madr) 2014: 42: 387–94. [DOI] [PubMed] [Google Scholar]
- 68. Pomponi D, Bernardi ML, Liso M, et al. Allergen micro‐bead array for IgE detection: a feasibility study using allergenic molecules tested on a flexible multiplex flow cytometric immunoassay. PLoS ONE 2012: 7: e35697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scala E, Alessandri C, Palazzo P, et al. IgE recognition patterns of profilin, PR‐10, and tropomyosin panallergens tested in 3,113 allergic patients by allergen microarray‐based technology. PLoS ONE 2011: 6: e24912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Douladiris N, Savvatianos S, Roumpedaki I, Skevaki C, Mitsias D, Papadopoulos NG. A molecular diagnostic algorithm to guide pollen immunotherapy in southern Europe: towards component‐resolved management of allergic diseases. Int Arch Allergy Immunol 2013: 162: 65–74. [DOI] [PubMed] [Google Scholar]
- 71. Popescu F‐D. Molecular biomarkers for grass pollen immunotherapy. World J Methodol 2014: 4: 26–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pfiffner P, Stadler BM, Rasi C, Scala E, Mari A. Cross‐reactions vs co‐sensitization evaluated by in silico motifs and in vitro IgE microarray testing. Allergy 2012: 67: 210–6. [DOI] [PubMed] [Google Scholar]
- 73. Bonds RS, Midoro‐Horiuti T, Goldblum R. A structural basis for food allergy: the role of cross‐reactivity. Curr Opin Allergy Clin Immunol 2008: 8: 82–6. [DOI] [PubMed] [Google Scholar]
- 74. James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science 2003: 299: 1362–7. [DOI] [PubMed] [Google Scholar]
- 75. Burastero SE. Pollen‐cross allergenicity mediated by panallergens: a clue to the patho‐genesis of multiple sensitizations. Inflamm Allergy Drug Targets 2006: 5: 203–9. [DOI] [PubMed] [Google Scholar]


