Abstract
SMARCB1 inactivation occurs in a variety of tumors, being caused by various genetic mechanisms. Since SMARCB1 and EWSR1 genes are located close to each other on chromosome 22, larger SMARCB1 deletions may encompass the EWSR1 locus. Herein, we report four cases with SMARCB1-deletions showing concurrent EWSR1 gene abnormalities by FISH, which lead initially to misinterpretations as EWSR1-rearranged tumors. Our study group included various morphologies: a poorly differentiated chordoma, an extrarenal rhabdoid tumor, a myoepithelial carcinoma, and a proximal-type epithelioid sarcoma. All cases showed loss of SMARCB1 (INI1) by immunohistochemistry (IHC) and displayed characteristic histologic features for the diagnoses. The SMARCB1 FISH revealed homozygous or heterozygous deletions in three and one case, respectively. The co-hybridized EWSR1 probes demonstrated either unbalanced split signals or heterozygous deletion in two cases each. The former suggested bona fide rearrangement, while the latter resembled an unbalanced translocation. However, all the FISH patterns were quite complex and distinct from the simple and uniform split signals seen in typical EWSR1 rearrangements. We conclude that in the context of 22q11-12 regional alterations present in SMARCB1-deleted tumors, simultaneous EWSR1 involvement may be misinterpreted as equivalent to EWSR1 rearrangement. A detailed clinicopathologic correlation and supplementing the EWSR1 FISH assay with complementary methodology is mandatory for correct diagnosis.
INTRODUCTION
SMARCB1 (also known as INI1, BAF47, or hSNF5) is a core subunit of the switch/sucrose nonfermenting (SWI/SNF) complexes, which consume ATP to modulate chromatin remodeling (Wilson and Roberts, 2011; Masliah-Planchon et al., 2015). The association between SWI/SNF complexes and neoplasia was first recognized nearly two decades ago, with the discovery of SMARCB1 truncating mutations and deletions in malignant rhabdoid tumors at different locations (brain, kidney, soft tissue) (Versteege et al., 1998; Biegel et al., 2002). Subsequently, SMARCB1 loss has been implicated in the pathogenesis of most epithelioid sarcomas and renal medullary carcinomas (Hollmann and Hornick, 2011; Agaimy, 2014). Loss of SMARCB1 protein expression was further described in a variety of epithelial and epithelioid mesenchymal tumors, such as myoepithelial carcinoma, extraskeletal myxoid chondrosarcoma, epithelioid malignant nerve sheath tumor, chordoma and a subset of GI and sinonasal carcinoma (Hollmann and Hornick, 2011; Agaimy, 2014). Remarkably, by whole genome sequencing, 19.6% of all human tumors harbor mutations in various SWI/SNF subunits, a similar rate to TP53 mutation (Kadoch et al., 2013).
Biallelic disruption of the SMARCB1 gene is the molecular hallmark of SMARCB1-deficient tumors and stems from a variety of mechanisms, including inactivating mutations, intragenic or whole-gene deletions, copy number-neutral loss of heterozygosity (LOH), and, infrequently, translocations (Jackson et al., 2009; Calderaro et al., 2015; Jamshidi et al., 2016). The incidence of homozygous or heterozygous SMARCB1 deletions varies depending on the tumor type and anatomic location of SMARCB1-deficient tumor. Furthermore, the involved region in 22q11-12 shows great diversity and complexity (Biegel et al., 2002; Jackson et al., 2009; Sullivan et al., 2013; Le Loarer et al., 2014). Among malignant rhabdoid tumors, brain tumors often exhibit large deletions or LOH of the whole chromosome arm 22q, while soft tissue counterparts usually harbor regional deletions across 22q11.22-11.23, and the renal tumors have either complex copy number deviation or copy number-neutral LOH (Jackson et al., 2009). Aside from 22q genomic abnormalities, malignant rhabdoid tumors are characterized by relatively simple karyotypes, containing only few other copy number variations or mutations (Rousseau-Merck et al., 1999; Jackson et al., 2009; Lee et al., 2012). In contrast, epithelioid sarcomas show a complex cytogenetic profile, with frequent aneuploidies, numerous structural aberrations, a high mutation burden and copy number changes (Lualdi et al., 2004; Jamshidi et al., 2016).
Since the SMARCB1 and EWSR1 genes are located only 5.5 Mb from each other in chromosome bands 22q11.23 and 22q12.2, respectively, secondary EWSR1 regional deletions may occur in SMARCB1-deleted tumors. In our previous study, we detected heterozygous telomeric EWSR1 deletions in 25% (9/36) of SMARCB1-deleted epithelioid sarcomas by fluorescence in situ hybridization (FISH) (Le Loarer et al., 2014). Herein, we describe four SMARCB1-deleted tumors associated with concurrent EWSR1 regional abnormalities that had been interpreted as primary gene rearrangements by outside cytogenetic reports, and therefore mis-diagnosed as EWSR1-rearranged tumors, to better illustrate the importance of clinicopathologic correlation and comprehensive interpretation of FISH findings.
MATERIALS AND METHODS
Case Collection
We identified three cases from the consultation files of the senior author (CRA) with a presumed EWSR1 gene rearrangement, which were sent for further classification or in an attempt to identify the EWSR1 fusion partner. An additional case with similar EWSR1 alteration was retrieved from the archive of MSKCC Surgical Pathology. The slides and correspondent immunohistochemical stains were reviewed to validate the diagnosis according to the current WHO criteria (Fletcher et al., 2013). In addition, potential fusion partners of EWSR1 were also investigated by FISH according to the initial diagnosis rendered, such as PBX1, ZNF444, POU5F1, PBX3 and KLF17 for myoepithelial tumors, (Huang et al., 2015) FLI1 and ERG for Ewing sarcoma, (Fletcher et al., 2013) and NR4A3 for extraskeletal myxoid chondrosarcoma (Agaram et al., 2014). The study was approved by the Institutional Review Board.
Immunohistochemistry
IHC staining, including INI1 (BAF47), was performed on whole sections of formalin-fixed, paraffin-embedded tissue blocks. The primary antibody used was a mouse monoclonal anti-BAF47 antibody (23, 1:30; BD bioscience, San Jose, CA). Antigen retrieval, antibody incubation, and chromogen counterstaining were performed in a BenchMark ULTRA automated immunostainer (Ventana, Tucson, AZ).
Fluorescence in situ hybridization
FISH for SMARCB1 deletion and EWSR1 break-apart assay was applied on formalin-fixed and paraffin-embedded 4 μm-sections in all cases. FISH was performed by applying custom probes using bacterial artificial chromosomes (BACs), covering or flanking the SMARCB1 or EWSR1 genes (Fig. 1A and Supporting Information Table 1). The BAC clones were obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA)(http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer's instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Huang et al., 2015). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a deleted or break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
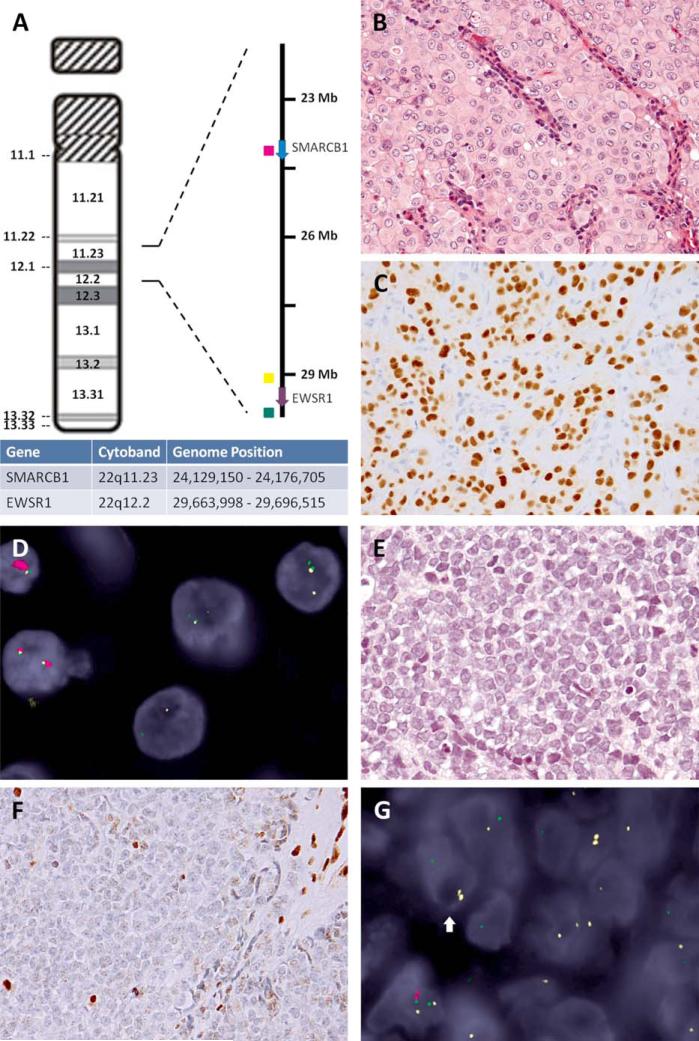
Figure 1.
Secondary break-apart EWSR1 patterns in SMARCB1-deleted tumors. A: SMARCB1 and EWSR1 genomic positions on chromosome 22 ideogram, with correspondent fluorochrome-labeled DNA probes used in the 3-color FISH assays (red: intragenic SMARCB1, yellow: centromeric EWSR1, green: telomeric EWSR1). B: Poorly differentiated chordoma showing compact sheets of epithelioid cells with moderate nuclear pleomorphism (Case 1, 400x) and (C) strong T-brachyury nuclear immunoreactivity (400x). D: The 3-color FISH assay showed homozygous SMARCB1 deletion (loss of both red signals) and EWSR1 break-apart pattern (yellow and green split signals with lower intensities). E: Extrarenal rhabdoid tumor resembling Ewing sarcoma, characterized by solid sheets of small round cells with prominent nucleoli (Case 2, 400x) and (F) loss of SMARCB1 expression (internal positive control, inflammatory and endothelial cells; 400x). G: The 3-color FISH revealed a homozygous SMARCB1 deletion (loss of both red signals) and coexistent EWSR1 regional abnormalities (the arrow): one intact yellow signal (telomeric EWSR1 deletion) and one split pair of smaller-sized yellow and green signals (EWSR1 regional break and deletion). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
RESULTS
A SMARCB1-Deficient Clival Tumor with EWSR1 Regional Abnormalities
Case no. 1 was a 3-year-old girl presenting with a large clivus tumor, who subsequently developed distant metastases (Table 1). The histology revealed interconnecting solid nests of large polygonal cells, separated by thin fibrovascular septae infiltrated by lymphocytes and plasma cells. The tumor cells showed distinctive cell borders, abundant eosinophilic cytoplasm, focal nuclear atypia and coarse chromatin with prominent nucleoli (Fig. 1B). Variable rhabdoid cells were noted, but no cytoplasmic vacuoles, physaliferous cells, or myxoid matrix was present. The mitotic activity was <1/10 HPFs. Immunohistochemically, the tumor cells expressed cytokeratin and S100 protein, with complete loss of SMARCB1 immunoexpression. T-brachyury was reported as negative on the initial evaluation. Under the suspicion of a myoepithelial carcinoma, EWSR1 FISH was performed and initially reported as positive for rearrangement. We received the case for further molecular evaluation, in an attempt to determine the EWSR1 fusion partner. However, all known EWSR1 gene partners in the setting of myoepithelial tumor were negative by FISH, including: PBX1, ZNF444, POU5F1, PBX3 and KLF17. The FISH assay displayed homozygous SMARCB1 deletion associated with a heterogeneous and complex EWSR1 split pattern (Fig. 1D). The latter showed EWSR1 split signals of smaller size and lower intensity, compared to the normal copies, suggesting a more complex EWSR1 regional deletion rather than a simple, balanced rearrangement. The T-brachyury immunostain performed in our lab was positive and, corroborated with the homozygous SMARCB1 FISH deletions, was consistent with a diagnosis of poorly differentiated chordoma, rather than an EWSR1-rearranged myoepithelial carcinoma (Fig. 1C) (Hoch et al., 2006).
TABLE 1.
Clinicopathologic and Genetic Summary of SMARCB1-Deleted Tumors with EWSR1 FISH Alterations
| Case | Age (yrs)/Sex | Location | Initial diagnosis | Final diagnosis | SMARCB1 deletion | EWSR1 break apart FISH result | Other pertinent results |
|---|---|---|---|---|---|---|---|
| 1 | 3/F | Clivus | Myoepithelial carcinoma | Poorly differentiated chordoma | Homozygous | Break-apart with deletion | T-brachyury IHC positive |
| 2 | 5/M | Abdominal wall | Ewing sarcoma | Malignant rhabdoid tumor | Homozygous | Break-apart with deletion | FLI1, ERG FISH negative |
| 3 | 16/F | Dura mater | Myoepithelial carcinoma | Myoepithelial carcinoma | Homozygous | Heterozygous deletion and break-apart | POU5F1, PBX1/3, KLF17 FISH negative |
| 4 | 21/M | Groin | Extraskeletal myxoid chondrosarcoma | Epithelioid sarcoma | Heterozygous | Heterozygous deletion | NR4A5 FISH negative |
FISH, fluorescence in situ hybridization; IHC, immunohistochemistry..
A Pediatric Round Cell Sarcoma with INI1 Loss and EWSR1 Genes Abnormalities
Case no. 2 was a round cell sarcoma of the abdominal wall in a 5-year-old boy. Microscopically, the tumor was composed of compact sheets of small round cells, with scant cytoplasm, and eccentric nuclei with open chromatin and prominent nucleoli (Fig. 1E). Necrosis, brisk mitotic activity (>50/10 HPF), and abundant karyorrhexis was noted. The tumor was negative for CD99, cytokeratin, S100, desmin, myogenin, CD45, TdT and synaptophysin, and showed complete INI1 loss (Fig. 1F). The outside FISH analysis showed a positive EWSR1 rearrangement, and a diagnosis of an atypical Ewing sarcoma with loss of SMARCB1 expression was entertained. The case was sent for further molecular characterization and diagnosis confirmation. Our FISH results failed to show abnormalities in FLI1 and ERG genes. The combined SMARCB1 and EWSR1 FISH assay demonstrated a homozygous SMARCB1 deletion, accompanied by loss of the telomeric EWSR1 signal on one allele and a split EWSR1 signal on the other allele (Fig. 1G). The split-apart EWSR1 signals were smaller in size, as observed in Case no. 1. The overall histologic and molecular findings fit the diagnosis of an extrarenal malignant rhabdoid tumor (Hollmann and Hornick, 2011; Fletcher et al., 2013).
A Meningeal Myoepithelial Neoplasm with SMARCB1 Deletions and Heterogeneous EWSR1 Alterations
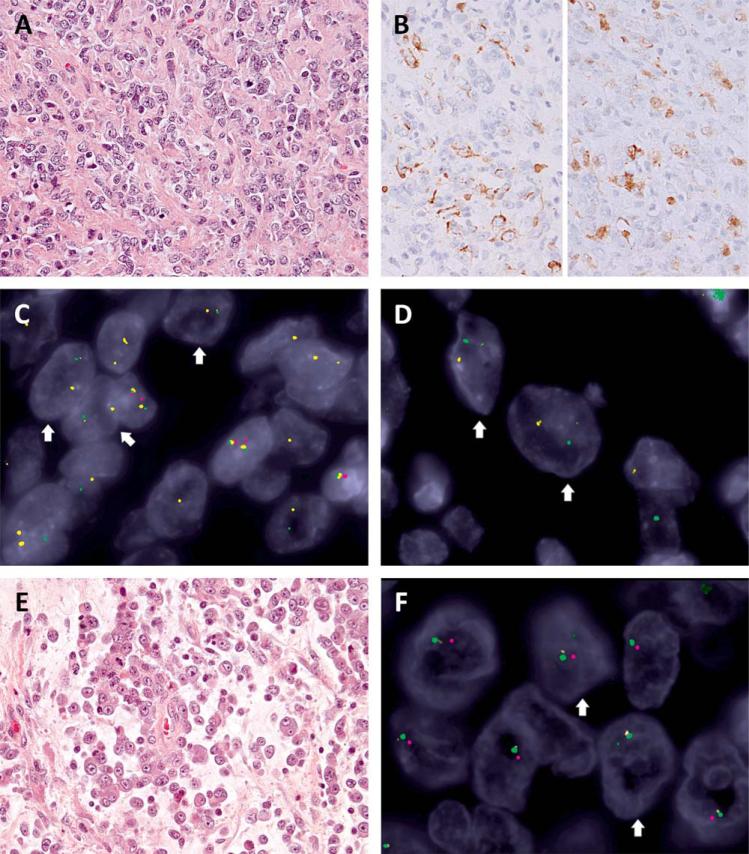
Case no. 3 was a large dural-based tumor occurring in a 16-year-old girl. Morphologically, the tumor consisted of cords and small nests embedded in a hyalinized stroma (Fig. 2A), but solid sheets with central necrosis were also seen. The tumor cells ranged from round, ovoid, to spindle, without rhabdoid features. Frequent mitotic figures (>10/10 HPFs), moderate nuclear pleomorphism and distinct nucleoli were noted. The tumor showed focal immunoreactivity for EMA, cytokeratin and S100 (Fig. 2B), but negativity for estrogen receptor and T-brachyury. SMARCB1 expression was lost. The initial FISH examination reported EWSR1 rearrangement and a presumed diagnosis of myoepithelial carcinoma was rendered. Our combined SMARCB1 and EWSR1 assay revealed homozygous SMARCB1 deletion with heterogeneous alterations of the EWSR1 locus (Fig 2C, 2D). Most tumor cells exhibited deletion of the telomeric EWSR1 region (loss of green signals) on one allele, while the other harbored a 22q11 deletion extending to the centromeric part of EWSR1 region (residual tiny yellow signals)(Fig. 2C). This FISH pattern was misleading for an unbalanced EWSR1 translocation with telomeric deletion (chromosome 22 monosomy with break-apart signals), as the other allele abnormalities with small yellow signals were easy to miss. Overall findings were in keeping with a myoepithelial carcinoma with homozygous SMARCB1 deletion and secondary EWSR1 genetic abnormalities. This case illustrates the complexity and heterogeneity of 22q11-12 regional alterations in SMARCB1-deficient myoepithelial tumors.
Figure 2.
Heterozygous EWSR1 deletion in SMARCB1-deleted tumors. A: Myoepithelial carcinoma composed of relatively monotonous epithelioid to round cells arranged in cords in a collagenous stroma (Case 3, 400x) showing (B) focal reactivity for cytokeratin (left, 400x) and S100 (right, 400x). The 3-color FISH revealed homozygous SMARCB1 deletions (lack of both red signals) with heterogeneous EWSR1 abnormalities, including (C) heterozygous deletions (arrows: one allele retained the paired yellow-and-green signals but with diminished yellow intensity, indicating partial deletion of centromeric part, while the other allele showed loss of green signal, consistent with deletion of telomeric part) and (D) break-apart (arrows: yellow-and-green split signals of smaller size, in keeping with concurrent deletions of centromeric and telomeric parts outside the EWSR1 gene locus). E: Proximal-type epithelioid sarcoma with myxoid changes and rhabdoid morphology (Case 4, 400x). F: The 3-color FISH showed heterozygous SMARCB1 (one retained red signal) and EWSR1 regional deletions (arrows: the involved allele containing a tiny residual green signal in keeping with a larger deletion across centromeric and telomeric part). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
A High-Grade Myxoid Sarcoma Showing Epithelioid/Rhabdoid Cell Features and Heterozygous SMARCB1 and EWSR1 Deletions
Case no. 4 was a left groin tumor in a 21-year-old male patient. The tumor showed solid areas with necrosis alternating with myxoid regions where the tumor cells floated singly or in cords (Fig. 2E). The neoplastic cells showed an epithelioid to rhabdoid morphology, with large vesicular nuclei and macronucleoli. Intracytoplasmic hyaline inclusions, occasional binucleation, and brisk mitotic activity (>20/10 HPFs) were noted. By IHC, the tumor was positive for EMA, but negative for cytokeratin, CD34, desmin, and S100. The tumor showed complete loss of SMARCB1. The original EWSR1 FISH reported one residual copy of normal EWSR1 signals and a diagnosis of extraskeletal myxoid chondrosarcoma was suspected. Our NR4A3 FISH testing was negative for rearrangement. The combined SMARCB1 and EWSR1 FISH assay revealed heterozygous SMARCB1 deletion extending to the telomeric-EWSR1 region (small residual green signals) (Fig. 2F). These heterozygous deletions resulted in a misinterpretation as an unbalanced EWSR1 translocation with centromeric loss, even though the 5’-end of EWSR1 gene is always represented in the EWSR1 chimeric fusion transcript. Corroborating all the genetic findings of SMARCB1 deletion and absence of NR4A3 abnormalities the diagnosis was reclassified as a proximal-type epithelioid sarcoma (Hollmann and Hornick, 2011; Fletcher et al., 2013).
DISCUSSION
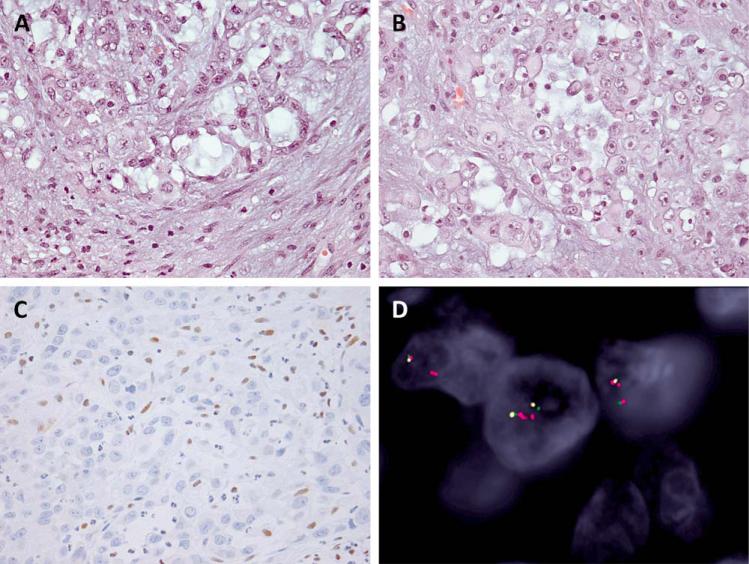
FISH analysis on paraffin-embedded tissue sections has emerged as a reliable and widely applied clinical assay for detection of gene rearrangements, deletions, and amplifications (Speicher and Carter, 2005; Bayani and Squire, 2007). In the setting of SMARCB1-deficient tumors, FISH can potentially be used to investigate SMARCB1 gene deletions as the size range of the genomic regions (100 kb–18 Mb) involved are often within the resolution of FISH probes (50 kb–2 Mb) (Jackson et al., 2009; Lee et al., 2012; Le Loarer et al., 2014). Although FISH analysis is not suitable to identify other SMARCB1 molecular alterations, such as mutations, small deletions, or LOH, it remains a reliable test in SMARCB1-deficient tumors where large homozygous or heterozygous 22q11-12 deletions predominate. For example, homozygous SMARCB1 deletions are present in the overwhelming majority of both proximal and distal epithelioid sarcomas (83–90%) (Sullivan et al., 2013; Le Loarer et al., 2014). In malignant rhabdoid tumors, the genetic abnormalities are equally divided between homozygous SMARCB1 deletions and SMARCB1 missense mutations with heterozygous deletion or LOH (Jackson et al., 2009). Despite limited case numbers, SMARCB1-deficient chordoma and sinonasal rhabdoid/basaloid carcinoma appear to have a comparable high rate of homozygous or heterozygous SMARCB1 deletion (Mobley et al., 2010; Bishop et al., 2014). At least a small subset of extraskeletal myxoid chondrosarcoma and myoepithelial carcinoma with loss of SMARCB1 expression show concurrent SMARCB1 deletion, although none of the cases reported to date show co-existing NR4A3 or EWSR1 rearrangements, suggesting a mutually exclusive mechanism (Kohashi et al., 2008; Le Loarer et al., 2014). The absence of SMARCB1 deletions by FISH in the settings of SMARCB1-deficient tumors harboring a high rate of SMARCB1 deletions should raise concerns about the correct diagnosis. To illustrate this point, we have recently encountered a 28-year-old female African American presenting with disseminated disease involving lymph nodes, lung, liver, and kidney. The lymph node biopsy revealed malignant epithelioid and rhabdoid tumor cells with a nested arrangement in a fibromyxoid background (Fig. 3). The tumor was positive for cytokeratin and vimentin and had complete loss of SMARCB1 expression; thus an epithelioid sarcoma diagnosis was considered. By FISH examination no SMARCB1 deletions were identified, which was unusual for an epithelioid sarcoma and alerted us to obtain additional clinical information, which revealed that the patient's sister had sickle cell trait. Thus a diagnosis of a renal medullary carcinoma was rendered, in which the preferred mechanism of SMARCB1 inactivation is more likely through a SMARCB1 translocation or LOH (Swartz et al., 2002; Liu et al., 2013; Calderaro et al., 2015).
Figure 3.
Metastatic renal medullary carcinoma. A,B: The lymph node biopsy showed epithelioid cells with light eosinophilic cytoplasm and scattered rhabdoid cells in a variably fibromyxoid stroma (200x, 400x). C: The SMARCB1 IHC was lost. D: The three-color FISH found two to four copies of intact SMARCB1 gene. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
However, FISH assays entail certain limitations and pitfalls (Gozzetti and Le Beau, 2000) and their applicability is largely restricted by the narrow selection of commercially available labeled DNA probes. As no FISH probes are available for SMARCB1, a number of labs and studies use as a surrogate the anti-BCR telomeric probe, which targets a 645-kb long region (23,986–24,631 kb genomic position, spanning the SMARCB1 gene, 24,129–24,176 kb) (Papp et al., 2013). However, this commercial probe covers a bigger target region (fourfold larger than our specific custom probe, 148 kb), including the centromeric end of the SMARCB1 gene. Thus the smaller size or centromeric-centered SMARCB1 deletions might result in partial or minimal loss of signal intensity, which can be easily missed by the low FISH resolution. Using this approach, Papp et al. reported a significantly lower percentage (15/31, 48%) of SMARCB1 deletions in epithelioid sarcoma using the Vysis LSI BCR SpectrumGreen probes (BCR/ABL Dual Color Translocation Probe Set, Abbott Molecular, Illinois, USA) (Papp et al., 2013). An identical setback occurred in our Case no. 2 in which the anti-BCR FISH probe failed to identify a SMARCB1 deletion, and thus a diagnosis of rhabdoid tumor was excluded. In contrast, our custom SMARCB1-specific strategy detected homozygous deletion in this case, which also explained the secondary nearby EWSR1 gene abnormalities. Furthermore, another limitation of FISH assay is that it examines only the targeted region without providing information on a more global scale of the whole chromosome structure. In this regard, we have recently identified that 27% of Ewing sarcoma with EWSR1-ERG fusions show a false negative result by FISH using the EWSR1 break-apart probes, due to a complex pattern of inversions and rearrangements (Chen et al., 2016).
In this study, we describe four cases of SMARCB1-deleted tumors harboring concurrent EWSR1 regional alterations, mimicking either balanced or unbalanced EWSR1 rearrangements. These four cases spanned a variety of diagnoses, including a poorly differentiated chordoma, myoepithelial carcinoma, extrarenal malignant rhabdoid tumor, and an epithelioid sarcoma. In two cases, the SMARCB1 deletion involved the EWSR1 gene, and the FISH result showed a split signal, although the size of the signals was smaller than the ones seen in a typical balanced rearrangement. In the other two cases, FISH showed a heterozygous deletion of EWSR1 FISH probes. Although our previous study showed one-fourth of SMARCB1-deleted epithelioid sarcoma contained synchronous deletion of telomeric EWSR1 region, (Le Loarer et al., 2014) the prevalence of concurrent break-apart or heterozygous EWSR1 abnormalities reminiscent of EWSR1 rearrangement in SMARCB1-deleted tumors remains unclear. The single nucleotide polymorphism-based oligonucleotide array showed heterozygous 22q12.2 deletion in 8.5% (3/35) of malignant rhabdoid tumors (Jackson et al., 2009). Spectral karyotyping depicted complex chromosomal aberrations encompassing a t(12;22)(q12;q12) without EWSR1 break-apart FISH signal in one of the six epithelioid sarcoma (Lualdi et al., 2004). Conversely, most of EWSR1-rearranged soft tissue sarcomas, such as Ewing sarcoma, clear cell sarcoma, desmoplastic small round cell tumor, and myxoid liposarcoma, are not associated with SMARCB1 protein loss (Kohashi et al., 2008). In two studies, none of the SMARCB1-deficient myoepithelial carcinomas or extraskeletal myxoid chondrosarcomas had concurrent EWSR1 rearrangement and SMARCB1 deletion (Kohashi et al., 2008; Le Loarer et al., 2014).
Chordomas arising in children and adolescents account for less than 5% of all chordoma cases, have a predilection for the skull base and behave more aggressively, especially the subset exhibiting poorly differentiated features (Hoch et al., 2006; Ridenour et al., 2010). They often have loss of SMARCB1 protein expression as a result of SMARCB1 deletions (Mobley et al., 2010; Yadav et al., 2014). The co-expression of cytokeratin and S100 in Case no. 1 raised the differential diagnosis between myoepithelial carcinoma and chordoma. The initial negative result of T-brachyury immunostaining and FISH positivity for EWSR1 rearrangement favored a myoepithelial carcinoma. However, our FISH findings of SMARCB1 deletions encompassing the EWSR1 locus, as well as the lack of rearrangements in all EWSR1-gene partners described in myoepithelial tumors, questioned the outside EWSR1 gene break-apart result. Furthermore the repeat T-brachyury showed nuclear reactivity confirming a diagnosis of chordoma. Indeed, chromosomal 22q translocation or mitotic recombination with variable regional deletion, outside the hot spot EWSR1 breakpoints, represents one of the main mechanisms for SMARCB1 inactivation (Rousseau-Merck et al., 1999; Lualdi et al., 2004).
Rare round cell sarcomas harboring SMARCB1 genetic alterations and consequent loss of expression have been previously described, having a predilection for young children and a favorable outcome (Kreiger et al., 2009). Their morphology includes undifferentiated round cells arranged in sheet-like pattern, with variably prominent nucleoli but lacking a classic rhabdoid phenotype. Jahromi et al. (2012) recently reported that 10/106 Ewing sarcomas had complete SMARCB1 protein loss, most of them occurring in the soft tissue and displaying atypical features and even rhabdoid cells. EWSR1 rearrangements were only demonstrated in 2/4 such cases using break-apart FISH strategy, but not confirmed by other methods. It is possible that some of the so-called “EWSR1-rearranged SMARCB1-deficient Ewing sarcomas” are equivalent to our Case no. 2, representing rhabdoid tumors with secondary EWSR1 abnormalities due to SMARCB1 deletions. Additional FISH or RT-PCR testing to assess FLI1 or ERG gene rearrangements would settle this dilemma as confirming the diagnosis of Ewing sarcoma.
Loss of SMARCB1 protein expression has been described in 10–40% of myoepithelial carcinoma, with pediatric cases showing a higher frequency (Hollmann and Hornick, 2011). Our previous study showed homozygous SMARCB1 deletions in three of five myoepithelial carcinomas with SMARCB1 immunonegativity, but none of them harbored EWSR1 gene rearrangement (Le Loarer et al., 2014). Even though Kohashi et al. (2008) reported SMARCB1 deletions in 2/4 SMARCB1-deficient extraskeletal myxoid chondrosarcoma, none of them harbored EWSR1-NR4A3 or TAF2A-NR4A3 fusion transcript by RT-PCR. Furthermore, in our prior study, none of seven SMARCB1-deficient extraskeletal myxoid chondrosarcoma with NR4A3 gene rearrangements showed SMARCB1 abnormalities by FISH (Le Loarer et al., 2014). In this setting, we highly recommend evaluation of NR4A3 gene rearrangement by FISH or RT-PCR to confirm the diagnosis of extraskeletal myxoid chondrosarcoma, as EWSR1 gene abnormalities might be secondary to large SMARCB1 deletions. Except for the above-mentioned Ewing sarcoma study (Jahromi et al., 2012), there are three additional case reports describing SMARCB1-deficient tumors with co-existing EWSR1 rearrangements, including two myoepithelial carcinomas and one desmoplastic small round cell tumor (Machado et al., 2015; Soon and Petersson, 2015; Thway et al., 2015). The first myoepithelial carcinoma case was reported to have an unbalanced EWSR1 translocation (one normal EWSR1 pair signal and an isolated green telomeric signal), (Thway et al., 2015) similar to our Case no. 4, which suggests the loss of derivative chromosome 22 that contained the critical 5’ end of EWSR1 gene. The second myoepithelial carcinoma case showed rhabdoid cells and myxoid background, and despite the EWSR1 rearrangement reported by FISH there was no identified fusion partner (Machado et al., 2015). The third case was a cytokeratin and desmin-negative desmoplastic small round cell tumor which by RT-PCR showed an EWSR1-WT1 transcript, but by FISH had one pair of EWSR1 break-apart signals, indicating monosomy 22 or allelic deletion (Soon and Petersson, 2015).
In summary, this study illustrates a recurring pitfall in interpreting the EWSR1 FISH results in the setting of SMARCB1 deficient and SMARCB1-deleted tumors. We describe four index cases, spanning four different diagnoses, which were misinterpreted mainly due to the positive FISH results for EWSR1 gene rearrangement. Despite the fact that all cases demonstrated SMARCB1 loss of expression, the possibility of a primary SMARCB1 genetic inactivation event was dismissed once the EWSR1 abnormality was found. Our results emphasize that a subset of SMARCB1-deleted tumors contain secondary EWSR1 regional alterations by FISH, including break-apart and heterozygous deletions; both of these EWSR1 FISH abnormalities patterns having the potential for misdiagnosis. The key feature in recognizing this abnormal EWSR1 pattern includes the presence of smaller size signals of lower intensity by FISH compared to the normal cells. Our results corroborated with a careful literature review further emphasizes that most EWSR1 rearranged tumors do not show concurrent SMARCB1 gene inactivations, and most likely represent independent mechanisms of pathogenesis. Our recommendation for a case showing loss of SMARCB1 expression by IHC and found concurrently to display rearrangements of the EWSR1 gene region by FISH, is that additional genetic investigation should be applied, including complementary molecular methods to exclude a primary SMARCB1 gene abnormality. Additional FISH testing for confirming rearrangements in the potential EWSR1 gene partners or applying complementary methods (RT-PCR, NGS) should be recommended in challenging cases, and close clinicopathologic correlation with the FISH results should be carried out to avoid these pitfalls.
Supplementary Material
Acknowledgments
Supported by: Cycle for Survival (CRA); Grant number: P50CA140146-01.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Agaimy A. The expanding family of SMARCB1(INI1)-deficient neoplasia: Implications of phenotypic, biological, and molecular heterogeneity. Adv Anat Pathol. 2014;21:394–410. doi: 10.1097/PAP.0000000000000038. [DOI] [PubMed] [Google Scholar]
- Agaram NP, Zhang L, Sung YS, Singer S, Antonescu CR. Extraskeletal myxoid chondrosarcoma with non-EWSR1-NR4A3 variant fusions correlate with rhabdoid phenotype and high-grade morphology. Hum Pathol. 2014;45:1084–1091. doi: 10.1016/j.humpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayani J, Squire JA. Application and interpretation of FISH in biomarker studies. Cancer Lett. 2007;249:97–109. doi: 10.1016/j.canlet.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–3467. [PubMed] [Google Scholar]
- Bishop JA, Antonescu CR, Westra WH. SMARCB1 (INI-1)-deficient carcinomas of the sinonasal tract. Am J Surg Pathol. 2014;38:1282–1289. doi: 10.1097/PAS.0000000000000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderaro J, Masliah-Planchon J, Richer W, Maillot L, Maille P, Mansuy L, Bastien C, de la Taille A, Boussion H, Charpy C, Jourdain A, Blechet C, Pierron G, Gentien D, Choudat L, Tournigand C, Delattre O, Allory Y, Bourdeaut F. Balanced translocations disrupting SMARCB1 are hallmark recurrent genetic alterations in renal medullary carcinomas. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.09.027. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Chen S, Deniz K, Sung YS, Zhang L, Dry S, Antonescu CR. Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer. 2016;55:340–349. doi: 10.1002/gcc.22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CDM, Bridge JA, Hogendoorn P, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. 4th ed. IARC; Lyon, France: 2013. [Google Scholar]
- Gozzetti A, Le Beau MM. Fluorescence in situ hybridization: Uses and limitations. Semin Hematol. 2000;37:320–333. doi: 10.1016/s0037-1963(00)90013-1. [DOI] [PubMed] [Google Scholar]
- Hoch BL, Nielsen GP, Liebsch NJ, Rosenberg AE. Base of skull chordomas in children and adolescents: A clinicopatho-logic study of 73 cases. Am J Surg Pathol. 2006;30:811–818. doi: 10.1097/01.pas.0000209828.39477.ab. [DOI] [PubMed] [Google Scholar]
- Hollmann TJ, Hornick JL. INI1-deficient tumors: Diagnostic features and molecular genetics. Am J Surg Pathol. 2011;35:e47–e63. doi: 10.1097/PAS.0b013e31822b325b. [DOI] [PubMed] [Google Scholar]
- Huang SC, Chen HW, Zhang L, Sung YS, Agaram NP, Davis M, Edelman M, Fletcher CD, Antonescu CR. Novel FUSKLF17 and EWSR1-KLF17 fusions in myoepithelial tumors. Genes Chromosomes Cancer. 2015;54:267–275. doi: 10.1002/gcc.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EM, Sievert AJ, Gai X, Hakonarson H, Judkins AR, Tooke L, Perin JC, Xie H, Shaikh TH, Biegel JA. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15:1923–1930. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi MS, Putnam AR, Druzgal C, Wright J, Spraker-Perlman H, Kinsey M, Zhou H, Boucher KM, Randall RL, Jones KB, Lucas D, Rosenberg A, Thomas D, Lessnick SL Schiffman JD. Molecular inversion probe analysis detects novel copy number alterations in Ewing sarcoma. Cancer Genet. 2012;205:391–404. doi: 10.1016/j.cancergen.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi F, Bashashati A, Shumansky K, Dickson B, Gokgoz N, Wunder JS, Andrulis IL, Lazar AJ, Shah SP, Huntsman DG, Nielsen TO. The genomic landscape of epithelioid sarcoma cell lines and tumours. J Pathol. 2016;238:63–73. doi: 10.1002/path.4636. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohashi K, Oda Y, Yamamoto H, Tamiya S, Oshiro Y, Izumi T, Taguchi T, Tsuneyoshi M. SMARCB1/INI1 protein expression in round cell soft tissue sarcomas associated with chromosomal translocations involving EWS: A special reference to SMARCB1/INI1 negative variant extraskeletal myxoid chondrosarcoma. Am J Surg Pathol. 2008;32:1168–1174. doi: 10.1097/PAS.0b013e318161781a. [DOI] [PubMed] [Google Scholar]
- Kreiger PA, Judkins AR, Russo PA, Biegel JA, Lestini BJ, Assanasen C, Pawel BR. Loss of INI1 expression defines a unique subset of pediatric undifferentiated soft tissue sarcomas. Mod Pathol. 2009;22:142–150. doi: 10.1038/modpathol.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loarer F, Zhang L, Fletcher CD, Ribeiro A, Singer S, Italiano A, Neuville A, Houlier A, Chibon F, Coindre JM, Antonescu CR. Consistent SMARCB1 homozygous deletions in epithelioid sarcoma and in a subset of myoepithelial carcinomas can be reliably detected by FISH in archival material. Genes Chromosomes Cancer. 2014;53:475–486. doi: 10.1002/gcc.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, Biegel JA, Getz G, Roberts CW. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Galli S, Srinivasan R, Linehan WM, Tsokos M, Merino MJ. Renal medullary carcinoma: Molecular, immunohisto-chemistry, and morphologic correlation. Am J Surg Pathol. 2013;37:368–374. doi: 10.1097/PAS.0b013e3182770406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lualdi E, Modena P, Debiec-Rychter M, Pedeutour F, Teixeira MR, Facchinetti F, Dagrada GP, Pilotti S, Sozzi G. Molecular cytogenetic characterization of proximal-type epithelioid sarcoma. Genes Chromosomes Cancer. 2004;41:283–290. doi: 10.1002/gcc.20086. [DOI] [PubMed] [Google Scholar]
- Machado I, Lopez-Soto MV, Rubio L, Navarro L, Llombart-Bosch A. Soft tissue myoepithelial carcinoma with rhabdoid-like features and EWSR1 rearrangement: Fine needle aspiration cytology with histologic correlation. Diagn Cytopathol. 2015;43:421–426. doi: 10.1002/dc.23259. [DOI] [PubMed] [Google Scholar]
- Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- Mobley BC, McKenney JK, Bangs CD, Callahan K, Yeom KW, Schneppenheim R, Hayden MG, Cherry AM, Gokden M, Edwards MS, Fisher PG, Vogel H. Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathol. 2010;120:745–753. doi: 10.1007/s00401-010-0767-x. [DOI] [PubMed] [Google Scholar]
- Papp G, Changchien YC, Peterfia B, Pecsenka L, Krausz T, Stricker TP, Khoor A, Donner L, Sapi Z. SMARCB1 protein and mRNA loss is not caused by promoter and histone hypermethylation in epithelioid sarcoma. Mod Pathol. 2013;26:393–403. doi: 10.1038/modpathol.2012.190. [DOI] [PubMed] [Google Scholar]
- Ridenour RV, III, Ahrens WA, Folpe AL, Miller DV. Clinical and histopathologic features of chordomas in children and young adults. Pediatr Dev Pathol. 2010;13:9–17. doi: 10.2350/09-01-0584.1. [DOI] [PubMed] [Google Scholar]
- Rousseau-Merck MF, Versteege I, Legrand I, Couturier J, Mairal A, Delattre O, Aurias A. hSNF5/INI1 inactivation is mainly associated with homozygous deletions and mitotic recombinations in rhabdoid tumors. Cancer Res. 1999;59:3152–3156. [PubMed] [Google Scholar]
- Soon GS, Petersson F. Beware of immunohistochemistry–report of a cytokeratin-, desmin- and INI-1-negative pelvic desmoplastic small round cell tumor in a 51 year old woman. Int J Clin Exp Pathol. 2015;8:973–982. [PMC free article] [PubMed] [Google Scholar]
- Speicher MR, Carter NP. The new cytogenetics: Blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- Sullivan LM, Folpe AL, Pawel BR, Judkins AR, Biegel JA. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod Pathol. 2013;26:385–392. doi: 10.1038/modpathol.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz MA, Karth J, Schneider DT, Rodriguez R, Beckwith JB, Perlman EJ. Renal medullary carcinoma: Clinical, pathologic, immunohistochemical, and genetic analysis with pathogenetic implications. Urology. 2002;60:1083–1089. doi: 10.1016/s0090-4295(02)02154-4. [DOI] [PubMed] [Google Scholar]
- Thway K, Bown N, Miah A, Turner R, Fisher C. Rhabdoid variant of myoepithelial carcinoma, with EWSR1 rearrangement: Expanding the spectrum of EWSR1-rearranged myoepithelial tumors. Head Neck Pathol. 2015;9:273–279. doi: 10.1007/s12105-014-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Yadav R, Sharma MC, Malgulwar PB, Pathak P, Sigamani E, Suri V, Sarkar C, Kumar A, Singh M, Sharma BS, Garg A, Bakhshi S, Faruq M. Prognostic value of MIB-1, p53, epidermal growth factor receptor, and INI1 in childhood chordomas. Neuro Oncol. 2014;16:372–381. doi: 10.1093/neuonc/not228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.