Abstract
The genetics of Ewing sarcoma (ES) are characterized by a canonical fusion involving EWSR1 gene and a member of the ETS family of transcription factors, such as FLI1 and ERG. In fact, ERG gene rearrangements represent the second most common molecular alteration, with EWSR1-ERG being identified in 5–10% of cases, while only a handful of reports document a FUS-ERG fusion. In this study, we focus on ES with ERG gene abnormalities, specifically to investigate the prevalence and clinicopathologic features of FUS-ERG fusions in a large cohort of small blue round cell tumors (SBRCTs) and compare to the eight reported FUS-positive ES. Among the 85 SBRCTs tested, seven (8.2%) cases harbored FUS gene rearrangements; six fused to ERG and one with FEV. During this investigation we came across a number of ERG-rearranged ES lacking both EWSR1 and FUS abnormalities by FISH. In one case, RNA sequencing identified an EWSR1-ERG transcript despite the negative EWSR1 rearrangements by FISH. Additional 3-color FISH fusion assay demonstrated the fusion of EWSR1 and ERG signals in all four cases negative for break-apart EWSR1 FISH. These results emphasize a potential pitfall of relying on EWSR1 FISH assay alone for diagnosis of ES. In cases with classic morphology and/or strong CD99 and ERG immunoreactivity, additional molecular testing should be applied, such as ERG FISH or RT-PCR/next generation sequencing, for a more definitive diagnosis. Although our study group is small, there were no differences noted between the clinical, morphologic features and immunoprofile of the different subsets of ERG-rearranged SBRCTs.
INTRODUCTION
The genetic hallmark of Ewing sarcoma (ES) is the recurrent t(11;22)(q24;q12) translocation resulting in the fusion of EWSR1 and FLI1 genes, identified in 90% of cases (Delattre et al., 1992, 1994). The second most common molecular abnormality is the t(21;22)(q22;q12), which accounts for 5% of the cases, resulting in an EWSR1-ERG fusion (Ginsberg et al., 1999). Only a small subset of ES shows EWSR1 fusions with other members of the ETS family of transcription factors, such as ETV1 (7p22) (Jeon et al., 1995), E1A-F (17q21) (Kaneko et al., 1996) and FEV (2q35-36) (Peter et al., 1997).
In addition to these well-defined and relatively common genetic abnormalities, alternative fusions have been reported, with either EWSR1 being fused to non-ETS partners or FUS substituting for EWSR1 in its fusions with ETS transcription factors. In both categories, the number of cases reported has been relatively small (case reports or small series) and insufficient to draw definitive conclusions as to whether any of these genetic subgroups belong to the EWSR1-ETS-positive conventional ES or as separate pathologic entities. In the first group, four different non-ETS partners have been described, including two members of the zinc-finger family of proteins, PATZ1 (Mastrangelo et al., 2000) and SP3 (Wang et al., 2007), NFATC2, a member of the nuclear factors of activated T cells transcription complex (Szuhai et al., 2009) and SMARCA5, a chromatin-remodeling gene (Sumegi et al., 2011). EWSR1 and FUS are members of the FET family of RNA binding proteins and presumed to have overlapping function as they can interchange in a number of translocation-associated sarcomas (Antonescu, 2014). However, only five cases of SBRCT with t(16;21)(q11.2;q22) resulting in FUS-ERG (Shing et al., 2003; Berg et al., 2009) and two cases harboring FUS-FEV fusions (Ng et al., 2007; Pierron et al., 2012) have been described to date.
In this study, we sought to investigate the prevalence of the FUS-ERG gene fusion in a large cohort of pathologically and molecularly well characterized SBRCTs, lacking other known gene rearrangements. Since most of the reported FUS-rearranged SBRCT have a limited pathologic description, we also carried out a detailed histologic and immunohistochemical analysis of these lesions, and compared to the reported cases.
MATERIALS AND METHODS
The files of the Pathology Department of MSKCC and personal consultation files (CRA) were reviewed for SBRCTs, in which material for molecular analysis was available. Criteria of selection for the study were tumors that were negative for EWSR1, CIC, and BCOR-CCNB3 gene abnormalities by FISH. The clinical data were obtained from the medical records. Hematoxylin and eosin stained sections and immunohistochemical stains were re-evaluated in all cases. Tumors were reviewed for growth pattern, cell size, cytomorphology (round, oval, spindle, rhabdoid), nuclear features including contour, pleomorphism, chromatin pattern, presence of nucleoli, “starry sky” appearance, mitotic activity, and necrosis. The stromal component was also assessed for abundance and type (myxoid, collagenous). The study was approved by the Institutional Review Board 02-060.
Fluorescence In Situ Hybridization Analysis
FISH for break-apart assay was applied on formalin-fixed and paraffin-embedded 4-micron sections in all cases. FISH was performed by applying custom probes using bacterial artificial chromosomes (BACs), covering and flanking the FUS, EWSR1, ERG, and FEV genes. BAC clones were chosen according to the UCSC genome browser (http://genome.ucsc.edu), see Supporting Information Table 1. The BAC clones were obtained from the BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer's instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with an incomplete set of signals were omitted from the score.
RNA Sequencing and Fusion Sequencing Data Analysis
In one case lacking EWSR1, FUS, CIC, and BCOR-CCNB3 adequate RNA was available for RNA sequencing using paired-end sequencing for fusion gene discovery. Total RNA was prepared according to the Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (10 μg) followed by fragmentation at 94°C for 2.5 min in fragmentation buffer (Illumina). Prior to the adapter ligation step, an additional size-selection step (capturing 350–400 bp) was introduced to reduce inclusion of artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library (Quail et al., 2008). Enrichment of the adapter-ligated library was achieved by PCR for 15 cycles. After purification, the library was sized and quantified using DNA1000 Kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer's instructions. Paired-end RNA-sequencing at read lengths of 50 to 51 bp was performed with the HiSeq 2500 (Illumina).
Data analysis of RNA Sequencing Results was performed with the FusionSeq algorithm. Reads were independently aligned with STAR alignment software against the human genome reference sequence (hg19) and a splice junction library, simultaneously (Dobin et al., 2013). After conversion of mapped reads into Mapped Read Format (Habegger et al., 2011), analysis was performed using FusionSeq (Sboner et al., 2010). Briefly, in a first step, paired end reads mapping to different genes were used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcripts. After generation of a confident list of fusion candidates, they were ranked to prioritize experimental validation (Sboner et al., 2010).
RESULTS
FUS-Rearranged Small Blue Round Cell Tumors
We screened a total number of 85 SBRCTs that were negative for gene abnormalities in EWSR1, CIC, and BCOR by FISH and found seven (8.2%) cases that showed FUS gene rearrangements. These cases were then screened for ERG gene abnormalities by FISH and found to be positive in six of the seven cases (Fig. 1). The remaining ERG-negative FUS-rearranged SBRCT was then screened for FEV gene abnormalities and was found to be positive (Fig. 1).
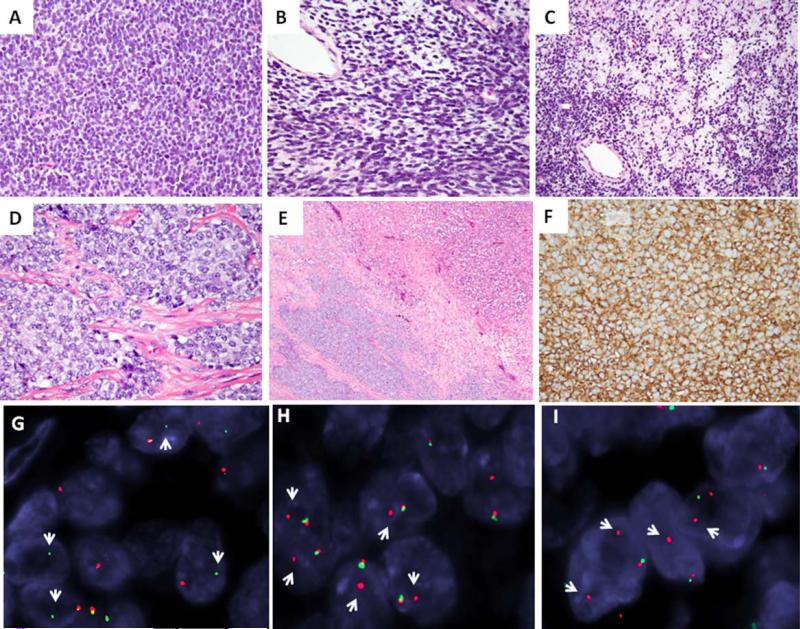
Figure 1.
The morphologic spectrum and FISH gene rearrangements in Ewing sarcoma with FUS gene abnormalities. FUS-ERG positive tumor characterized by solid growth of primitive, small blue cells with diffuse hyperchromasia and scant cytoplasm (A, ES1), focal areas of spindling and myxoid changes (B, C, ES1); Another FUS-ERG tumor with vesicular chromatin and vacuolated cytoplasm, separated in nests and compartments by fibrous bands and geographic areas of necrosis (D, E, ES2); All cases showed crisp and diffuse membranous staining pattern for CD99 (F, ES3); FISH break-apart assay showing unbalanced rearrangements for FUS (G, loss of red centromeric signal; ES2), ERG (H, loss of green telomeric signal; ES2), and FEV (I; loss of green telomeric signal; ES4) (red, centromeric, green, telomeric; arrows break-apart signals). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Clinicopathologic features of the seven FUS-rearranged SBRCT patients are summarized in Table 1. All patients except one were females, with a mean age 30.8 years (range 13–46, median 31). The anatomic site was distributed between bone and soft tissue of the extremities, with four and three cases, respectively. The skeletal tumors occurred in the chest wall, ilium, and distal femur, while the soft tissue lesions were deep seated in the arm and thigh.
TABLE 1.
Clinicopathologic Features of FUS Rearranged SBRCT From Our Study and Literature Review
| Present study/Reference | Age/gender | Site | CD99 | FUS-fusiontypeby FISH | Follow-up (months) |
|---|---|---|---|---|---|
| ES1 | 31/F | Thigh | Diffuse membranous | FUS-ERG | NED (16) |
| ES2 | 15/F | Femur | Diffuse membranous | FUS-ERG | NED (12) |
| ES3 | 46/F | Chest wall | Diffuse membranous | FUS-ERG | NED (5) |
| ES4 | 46/F | Arm | Diffuse membranous | FUS-FEV | AWD (24) Lung mets |
| ES5 | 13/F | Ilium | Diffuse membranous | FUS-ERG | – |
| ES6 | 45/M | Chest wall | Diffuse membranous | FUS-ERG | AWD (12) Lung mets |
| ES7 | 19/F | Thigh | Diffuse membranous | FUS-ERG | – |
| ES8 (Shing et al., 2003) | 9/M | Chest wall | Membranous | FUS-ERG | DOD (24) |
| ES9 (Shing et al., 2003) | 7/F | Chest wall | Patchy membranous | FUS-ERG | – |
| ES10 (Shing et al., 2003) | 15/F | Chest wall | Membranous | FUS-ERG | – |
| ES11 (Shing et al., 2003) | 21/M | Femur | Membranous | FUS-ERG | – |
| ES12 (Berg et al., 2009) | 3/F | Kidney | Strong membranous | FUS-ERG | NED (16) |
| ES13 (Pierron et al., 2012) | – | – | – | FUS-FEV | – |
| ES14 (Ng et al., 2007) | 33/M | Clavicle | Strong membranous | FUS-FEV | NED (10) |
| ES15 (Brohl et al., 2014) | 15/M | Femur | – | FUS-NFATC2 | – |
M, male; F, female; DOD, dead of disease at the time of publication; NED, no evidence of disease.
Histologically, all FUS-rearranged SBRCTs showed a solid growth of monomorphic, mainly small-sized, round to oval neoplastic cells. Five of the six FUS-ERG positive tumors showed hyperchromatic nuclei (Figs. 1A–1C), while one case showed vesicular nuclei with small, inconspicuous nucleoli (Fig. 1D). Only one of the cases showed focal areas of spindling and focal myxoid changes (Figs. 1B and 1C). One case showed thick collagenous bands dividing the tumor in wide compartments (Figs. 1D and 1E). There was no significant nuclear pleomorphism in any of the cases, with only mild variation in size and shape, and mostly smooth nuclear borders. The tumor cells had indistinct cell borders, with mostly scant cytoplasm, ranging from clear, vacuolated to light eosinophilic. The mitotic count varied from 8 to 32/10HPFs, with a mean of 17.5 ± 10.2. Four cases showed geographic necrosis (Fig. 1E). The only FUS-FEV positive tumor showed a more abundant clear cytoplasm and had a diffusely myxoid background.
All seven FUS-rearranged SBRCT showed diffuse membranous staining for CD99 (Fig.1F). Focal positivity for CAM5.2 was seen in one case, while S100 protein was noted in two cases, one case (ES7) showing diffuse and strong reactivity (also co-expressing SOX10), while the other only focal reactivity (ES3).
Meta-Analysis of the Eight FUS-Rearranged SBRCTs Reported in the Literature
The clinicopathologic features of these eight ES patients harboring FUS-related fusions from the literature (Shing et al., 2003; Ng et al., 2007; Berg et al., 2009; Brohl et al., 2014) are summarized in Table 1. Clinicopathologic and immunohistochemical information for one of the cases was not available (Pierron et al., 2012). Of the remaining patients there were four males and three females, with a mean age of 14.7 years (range: 3–33 years; median 15). All except one were located in the bone, with three involving chest wall, two in the femur and one in the clavicle. The remaining case occurred in the kidney.
Histologically, the growth pattern reported varied among nodular/nested, solid/sheet-like, and infiltrative. None of the reported cases demonstrated a spindled appearance. Only one case was described displaying mild nuclear pleomorphism, while remaining showed a monomorphic cytologic appearance. Half of the cases showed necrosis, extensive in one and focal in two. Immunohistochemically, all except one displayed a diffuse membranous pattern of CD99 staining.
Of the two reported FUS-FEV-rearranged SBRCT, only one case had available clinical information. This SBRCT occurred in the clavicle of a 33-year-old male and showed diffuse strong membranous positivity for CD99 (Ng et al., 2007). Although no clinical information was available for the second reported case (Pierron et al., 2012), it was described as clinically and morphologically similar to ES.
A proportion of ERG-rearranged ES lack FISH abnormalities in EWSR1 and FUS genes
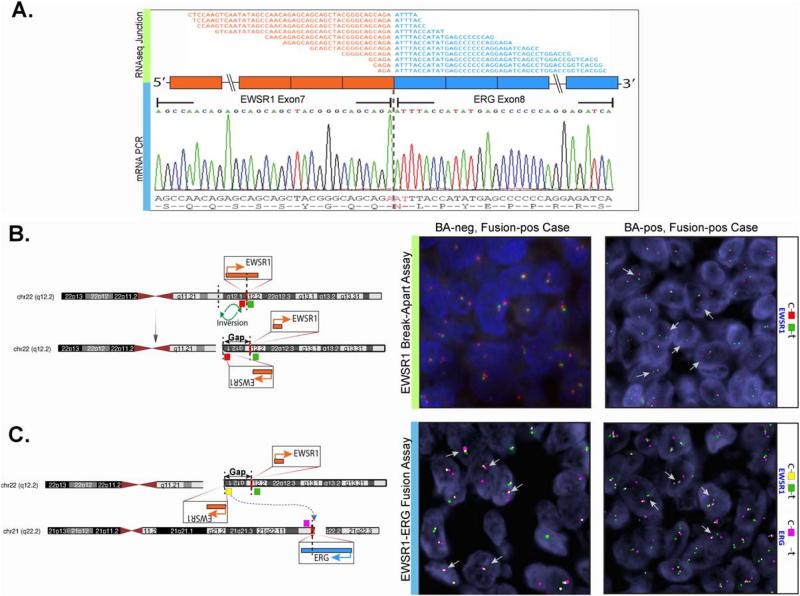
Among the 15 ERG-rearranged ES identified in our database, 4 (27%) lacked gene abnormalities in either EWSR1 or FUS by FISH. The remaining cases showed EWSR1 in four cases and FUS gene rearrangements in seven cases. The total number of EWSR1-ERG positive ES is under-represented in this cohort, since most EWSR1-rearranged ES with typical clinical and pathologic findings are not routinely screened to confirm its specific fusion partner (i.e., FLI1 or ERG). Furthermore, the aim of this study was not designed to establish an accurate incidence of EWSR1-ERG fusion positive ES, but rather investigate the frequency and clinicopathologic characteristics of FUS-ERG positive cases. Despite this bias, these results point out an important pitfall in screening ES only with EWSR1 FISH. In some of these cases ERG FISH testing was performed despite negative FISH results for both EWSR1 and FUS, due to typical morphologic features and/or strong and diffuse reactivity for ERG immunoreactivity. Furthermore, in one case RNA sequencing was performed due to negative FISH results for all known fusions, including EWSR1, FUS, CIC, and BCOR, in an attempt to discover novel fusion genes. Only after the EWSR1-ERG fusion transcript was detected by FusionSeq and subsequently validated by RT-PCR, FISH was performed to confirm the ERG gene rearrangement (Fig. 2A).
Figure 2.
Complex or masked gene rearrangements in Ewing sarcomas with EWSR1-ERG fusions. A. RNA sequencing (upper panel, fusion junction reads) and RT-PCR (lower panel) confirmation of EWSR1-ERG fusion (exon 7 of EWSR1 fused to exon 8 of ERG) in ES17. B. Diagrammatic representation of the multistep, complex mechanism of unbalanced t(21;22) translocation, with one such example involving two 22q12 breaks, including the EWSR1 break, followed by inversion; depending on the size of the inverted segment (gap, green arrows), the EWSR1 FISH break-apart might not (central panel, ES17) or may (right panel, ES23) be positive (red, centromeric; green, telomeric). C. The 5’EWSR1 fragment insertion on derivative 21, schematic depiction (left) and 3-color FISH fusion assay confirming the association of 5’EWSR1 orange centromeric signal with the red ERG signal (central and right panels, ES17 and ES23)(green, EWSR1 telomeric). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
FISH Fusion Assays Confirm the EWSR1-ERG Fusion in Cases with Negative EWSR1 FISH Break-Apart Results
As EWSR1 and ERG have opposite directions of transcription, the EWSR1-ERG fusion typically does not result through a simple balanced translocation, as seen with EWSR1-FLI1 or FUS-ERG fusion. As shown in Figure 2, two breaks are required in the 22q12 locus resulting in EWSR1 break and inversion before being inserted in 21q22.3. Depending on the distance between the two breaks in 22q12, the EWSR1 break-apart FISH assay (red, centromeric; green, telomeric) may or may not detect the EWSR1 gene rearrangements (Fig. 2B). As the resolution of FISH is suboptimal for breaks < 2 Mb apart, some of the smaller breaks/insertions in ES with EWSR1-ERG fusion are not detected by this method, the so-called “cryptic” EWSR1-ERG fusions. Thus, in order to confirm this result, we carried out a three-color FISH fusion assay: orange, EWSR1 centromeric, green, EWSR1 telomeric, and red, ERG centromeric BACs (Fig. 2C). Indeed, the remaining three cases with EWSR1 negative FISH break-apart but positive for ERG rearrangement showed a fused red-orange signal, in keeping with a 5′ centromeric EWSR1-ERG fusion (Fig. 2C). Thus, a diagnosis of EWSR1-ERG positive ES was confirmed in all eight cases; clinical findings being summarized in Table 2. There were six females and two males, with a mean age at diagnosis of 15 years (range: 1–23; median 19). The tumors were more commonly located in the bone (ilium, spine, mandible), four cases, followed by soft tissue (retroperitoneum, intra-abdominal), two cases and one case each in the brain and scalp.
TABLE 2.
Clinicopathologic Features of the EWSR1-ERG Positive Ewing Sarcoma
| Patient | Age/gender | Site | ERG IHC | CD99 IHC | FUS FISH | FISH EWS | FISH ERG | Follow-up (months) |
|---|---|---|---|---|---|---|---|---|
| ES16 | 1/F | T10 | Diffuse | Diffuse membranous | ND | Positivea | Positive | NED (27) |
| ES17 | 16/F | Ilium | Diffuse | Diffuse membranous | Negative | Negativeb | Positive | NED (6) |
| ES18 | 4/F | Retro-peritoneum | Diffuse | Diffuse membranous | ND | Positive | Positive | AWD (2) |
| ES19 | 22/M | Mandible | Diffuse | Diffuse membranous | Negative | Negativec | Positive | – |
| ES20 | 23/M | Brain | Diffuse | Diffuse membranous | Negative | Negative | Positive | LFU (6) |
| ES21 | 19/F | Chest wall | ND | Diffuse membranous | Negative | Negative | Positive | AWD (16) |
| ES22 | 20/F | Scalp | Diffuse | Diffuse membranous | ND | Positive | Positive | NED (9) |
| ES23 | 22/F | Intra-abdominal | Diffuse | Diffuse membranous | Negative | Positive | Positive | NED (4) |
FISH detected only 4% of cells with standard pattern and an additional 10% with complex rearrangement (loss of 3’EWSR1 or duplication of rear-ranged EWSR1).
EWSR1-ERG fusion was confirmed by RNAseq, and subsequently validated by ERG FISH.
ERG FISH was done due to ERG IHC positivity in the absence of EWSR1 rearrangement by FISH; NED: No evidence of disease; LFU: Lost to follow up; AWD: Alive with disease.
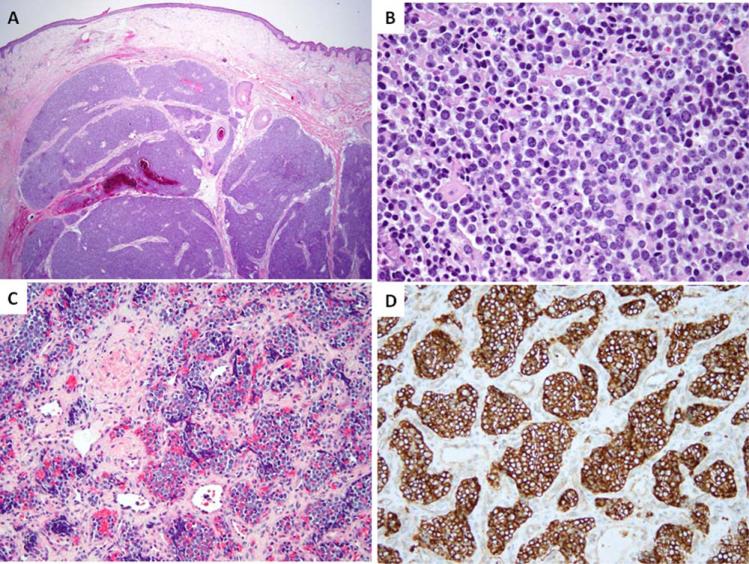
Histologically, the EWSR1-ERG positive ES were composed of uniform small round to oval neoplastic cells with scant light eosinophilic to vacuolated cytoplasm and indistinct cell borders. Four cases displayed solid growth, three had a nodular architecture (Figs. 3A and 3B), and one a nested appearance (Fig. 3C). Most cases showed a fine chromatin pattern (Fig. 3B), while two had vesicular nuclei with smooth nuclear contours. Only one tumor showed a coarser chromatin pattern with irregular nuclear membranes and a mild nuclear pleomorphism. The mitotic count varied from 1 to 33/10 HPFs, with a mean of 11 ± 10.8. Most of the cases showed geographic or multifocal necrosis (5/8 cases), which in two was focal. The stromal component was typically scant, except for one brain tumor, showing extensive hyalinization without any prior treatment. None of the cases had myxoid stroma. All cases showed diffuse membranous staining for CD99 (Fig. 3D). FLI1 was positive in one case.
Figure 3.
Morphologic spectrum of EWSR1-ERG Ewing sarcomas. Nodular growth of ES involving deep dermis and subcutis of the scalp (A, ES22); solid growth showing ill-defined cell borders and round nuclei with fine chromatin (B, ES17); a distinctive nested growth in an infant with ES from the vertebral body, showing diffuse CD99 reactivity (C, D; ES16). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
The histologic and molecular spectrum of the Ewing sarcoma (ES) family of tumors is still evolving since the initial description by Dr. James Ewing in 1921 (Ewing, 1921). Classic ES is characterized by a monotonous cytomorphology, with round nuclei, smooth nuclear contours, and fine to vesicular chromatin. Intra-tumoral architectural and cytologic heterogeneity is not a feature of classic ES. Atypical Ewing sarcoma was first described as a morphologic variant of classic ES in the 1970s, showing deviations from its typical monomorphic proliferation of primitive round cells (Llombart-Bosch et al., 1978), such as the presence of complex epithelial cell differentiation, that is, adamantinoma-like ES (Bridge et al., 1999), or large cell variants (Folpe et al., 2005; Llombart-Bosch et al., 2009). With the increased application of next generation sequencing and molecular screening, novel genetic alterations were described in tumors resembling ES. Thus CIC-DUX4 gene fusions and BCOR-CCNB3 inversions were found in different clinical subsets of SBRCT sharing some but also displaying distinct morphologic and clinical characteristics from classic ES (Kawamura-Saito et al., 2006; Italiano et al., 2012; Pierron et al., 2012).
Although EWSR1 and FUS belong to the same family of RNA binding proteins and appear to be functionally interchangeable, only rare ES have been reported to date harboring FUS gene rearrangements. A novel t(16;21)(p11;q22) translocation was first reported in a small series of four ES cases (Shing et al., 2003), resulting in the fusion of FUS (fused in sarcoma) and ERG (erythroblast transformation specific related) genes. A number of different fusion transcripts have been reported, fusing one of the FUS exons 5–7 to ERG exon 6–9 (Shing et al., 2003; Berg et al., 2009). Subsequently, a t(2;16) translocation resulting in an in-frame fusion of FUS-FEV was reported (Ng et al., 2007), involving FUS exon 10 to FEV exon 2. A second putative case of FUS-FEV fusion was detected by transcriptome sequencing and confirmed by RT-PCR (Pierron et al., 2012), but the exact fusion transcript was not disclosed. More recently a novel FUS-NFATC2 fusion was detected by RNA sequencing in a large genomic study of Ewing sarcoma, in which FUS exon 6 was fused to exon 2 of NFATC2 (Brohl et al., 2014). This novel fusion occurred in a 15-year-old male with a femur lesion; its morphologic description included some atypical features, such as focal areas of spindling as well as foci of chondroid differentiation. However, no previous study to date has investigated the incidence of FUS gene abnormalities in a molecularly well-characterized cohort of ES. Furthermore, since most of the reported FUS-rearranged SBRCTs have a limited pathologic description, it is not known if the clinical and pathologic features of these cases overlap with the classic ES with EWSR1-FLI1 fusion.
Our findings confirm the rarity of FUS gene rearrangements in ES, with only seven (8.2%) cases harboring this genetic abnormality among 85 SBRCTs negative for all other known fusions. Our results further confirm that ERG is the most common ETS member partner participating in the fusion, a FUS-ERG fusion being detected in 6/7 cases. Only one SBRCT with FUS-FEV fusion was identified, occurring in a skeletal location of a young adult.
Compared to the eight reported cases, the patients from our series of seven FUS-rearranged ESs were older (median age at diagnosis of 31 years versus 15 years in the literature) and occurred preferentially in women compared to equal gender distribution. Furthermore the tumors in our study were evenly distributed between bone and soft tissue, while the reported cases occurred predominantly in the bone. No other significant differences were discerned in their morphologic appearance or immunoprofile. Combining our current series to the reported data, there are 11 FUS-ERG positive ESs, eight of them occurring in the bone, two in soft tissue and one in the kidney. Cumulative patient data revealed a median age of 15 years (mean 20; range 3–46), with an overall female predominance (73% of patients). Morphologically both the reported and our FUS-ERG fusion positive SBRCTs had a monomorphic round cell cytomorphology in keeping with a classic ES phenotype. Furthermore, tumors showed diffuse membranous CD99 reactivity in all except one case, which showed a patchy staining pattern. Moreover, compared to the EWSR1-ERG fusion positive ES there was no difference noted in the age at presentation, gender, anatomic location and morphologic appearance. Similarly, all EWSR1-ERG ES demonstrated diffuse CD99 membranous staining.
The presence of diffuse and strong CD99 membranous pattern of staining in a SBRCT with a typical morphology reminiscent of ES but with FISH negative for EWSR1 rearrangement, should raise suspicion for the possibility of FUS-ERG fusion. In contrast, other EWSR1-negative SBRCTs with alternative fusions, such as CIC-DUX4 or BCOR-CCNB3, more often exhibit patchy CD99 positivity (Fisher, 2014; Puls et al., 2014). In the case of CIC-DUX4 positive SBRCTs the morphologic appearance is less monomorphic, with prominent nucleoli, more abundant cytoplasm and often myxoid stroma (Italiano et al., 2012). Furthermore, one of our cases showed FLI1 immunopositivity; aberrant FLI1 expression has been previously described in rare ES cases with EWSR1-ERG fusion (Folpe et al., 2005).
In the process of our investigation we identified a group of ERG-rearranged ES, lacking FISH abnormalities in either EWSR1 or FUS genes. The RNA sequencing performed in one case confirmed the presence of an EWSR1-ERG fusion transcript, suggesting an unbalanced, complex rearrangement beyond the FISH resolution. These results are in keeping with the few prior reports demonstrating a complex and unbalanced exchange of chromosomal material between chromosomes 21 and 22 (Desmaze et al., 1997; Szuhai et al., 2006). It was previously reported by FISH and RT-PCR that the chromosome 22 fragment containing the 5′ portion of EWSR1 is inverted and inserted into chromosome 21, fusing to the 3′; portion of ERG; this finding being often not visible on routine cytogenetics (Kaneko et al., 1997). In their study, the two color fusion assay FISH showed that EWSR1 and ERG clones fuse signals on the der(21) chromosome, but no ERG signals on the chromosome 22 homologs. Since the t(21;22) in Ewing sarcomas are mostly unbalanced, the size of the inversion/insertion chromosomal segment transferred between chromosomes 21 and 22 is variable in size and often quite small, it is not surprising that EWSR1-ERG fusion is often masked, beyond the resolution of routine cytogenetics, and in some cases even beyond the FISH break-apart resolution according to our results. As these two genes have opposite orientations, this complex pattern of rearrangement and subsequent inversion is required for a functional transcript to occur. In contrast, EWSR1-FLI1 or FUS-ERG fusions typically result from simple balanced translocation events, as the gene partners have similar directions of transcription. In this study, the EWSR1-ERG fusion was confirmed by a fusion FISH assay in all four SBRCTs with ERG gene rearrangements and negative break-apart FISH for EWSR1 and FUS abnormalities. Due to its relatively low-cost, low failure rates and a 24-hour turn-around time, FISH testing has gained a wide applicability recently in the translocation work-up of sarcomas, replacing most other more laborious molecular techniques, such as conventional karyotyping and RT-PCR, especially in small biopsy material or when only archival material is available. Thus EWSR1 gene status as reflected by a single FISH assay has nowadays too often become the first and last resource in confirming the diagnosis of ES at the molecular level. Our results show that in the setting of EWSR1-ERG fusions, standard break-apart FISH for EWSR1 will yield false negative results in half of the cases as a consequence of the complex rearrangement pattern between these two genes with inverse orientations, requiring often a multistep mechanism, with multiple, unbalanced breaks, inversions and insertions events (Desmaze et al., 1997; Maire et al., 2008). Thus other molecular techniques, that is, RT-PCR or NGS designed to span known fusion breakpoints, might be required for a more definitive classification. Alternatively, FISH for ERG gene rearrangements and/or 3-color FISH fusion assay for EWSR1 and ERG can be used to prevent this pitfall. We recommend following one of these approaches when dealing with a SBRCT displaying the typical ES monomorphic cytomorphology and diffuse reactivity for both CD99 and ERG.
In the context of SBRCTs, ERG reactivity should be interpreted with caution and only in addition to other supporting evidence for ES diagnosis. ERG immunopositivity has in fact also been described in other SBRCTs, mainly CIC-DUX4 fusion positive, which often display WT1 and ERG immunoreactivity (Specht et al., 2014). Furthermore, identical FUS-ERG fusions have been identified in pediatric acute myelocytic leukemia, and ERG-immunopositive leukemic soft tissue infiltrates may suggest a diagnosis of ERG-rearranged SBRCT and should be interpreted in the context of other leukemia markers, such as CD43 and myeloperoxidase (Choi et al., 2006). Moreover, high grade undifferentiated angiosarcomas with an epithelioid to small blue round cell phenotype strongly express ERG immunoreactivity and may lack any vasoformative properties. Thus adding CD31 immunostaining is required for any ERG-positive neoplasm, since CD31 is a highly reliable endothelial marker in angiosarcoma and has more specificity compared to ERG in this context.
In conclusion, our study is the first molecular investigation to establish the prevalence of FUS gene rearrangements in a well-characterized cohort of SBRCT, which accounts for 8.2% of cases lacking all other known fusions. The most common FUS gene partner is ERG, with very infrequent cases showing FEV gene involvement. FUS-ERG positive SBRCTs show a monotonous morphologic phenotype and strong CD99 membranous immunoreactivity, which are similar to classic ES. We suggest that FISH for FUS gene rearrangements should be examined in EWSR1 negative SBRCTs with classic histology and membranous CD99 immunopositivity. Our study also highlights a significant pitfall in the setting of EWSR1-ERG fusion SBRCTs, where FISH assay may not be able to detect the EWSR1 gene rearrangements due to the complex pattern of t(21;22) translocation. These results also caution interpreting discrepant molecular results from the overall morphologic and immunohistochemical findings. As detection of EWSR1 break-apart by FISH is emerging as the gold standard for diagnosis of ES, these results raise awareness to the use of a single method in the molecular diagnosis of complex gene rearrangements.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Drs. Noah Federman, Department of Pediatric Oncology, and Fritz Eilber, Department of Surgery, UCLA, for providing clinical follow-up; and Milagros Soto, MSKCC, for editorial assistance.
Supported by: Cancer Center Support Grant of the National Institute of Health/National Cancer Institute, Grant numbers: P50 CA140146-01 and P30 CA008748 (to CRA); Kristen Ann Carr Foundation (to CRA).
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Antonescu C. Round cell sarcomas beyond Ewing: Emerging entities. Histopathology. 2014;64:26–37. doi: 10.1111/his.12281. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Kalsaas AH, Buechner J, Busund LT. Ewing sarcoma-peripheral neuroectodermal tumor of the kidney with a FUS-ERG fusion transcript. Cancer Genet Cytogenet. 2009;194:53–57. doi: 10.1016/j.cancergencyto.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Fidler ME, Neff JR, Degenhardt J, Wang M, Walker C, Dorfman HD, Baker KS, Seemayer TA. Adamantinoma-like Ewing's sarcoma: Genomic confirmation, phenotypic drift. Am J Surg Pathol. 1999;23:159–165. doi: 10.1097/00000478-199902000-00004. [DOI] [PubMed] [Google Scholar]
- Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, Liao H, Wen X, Gerard J, Kim JS, Lopez Guerrero JA, Machado I, Wai DH, Picci P, Triche T, Horvai AE, Miettinen M, Wei JS, Catchpool D, Llombart-Bosch A, Waldman T, Khan J. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Shin MG, Sawyer JR, Cho D, Kee SJ, Baek HJ, Kook H, Kim HJ, Shin JH, Suh SP, Hwang TJ, Ryang DW. Unusual type of TLS/FUS-ERG chimeric transcript in a pediatric acute myelocytic leukemia with 47,XX,110,t(16;21)(p11; q22). Cancer Genet Cytogenet. 2006;167:172–176. doi: 10.1016/j.cancergencyto.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, Ambros PF, Sheer D, Turc-Carel C, Triche TJ, et al. The Ewing family of tumors–a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- Desmaze C, Brizard F, Turc-Carel C, Melot T, Delattre O, Thomas G, Aurias A. Multiple chromosomal mechanisms generate an EWS/FLI1 or an EWS/ERG fusion gene in Ewing tumors. Cancer Genet Cytogenet. 1997;97:12–19. doi: 10.1016/s0165-4608(96)00326-3. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing J. Diffuse endothelioma of bone. Proc NY Path Soc. 1921;21:17–24. [Google Scholar]
- Fisher C. The diversity of soft tissue tumours with EWSR1 gene rearrangements: A review. Histopathology. 2014;64:134–150. doi: 10.1111/his.12269. [DOI] [PubMed] [Google Scholar]
- Folpe AL, Goldblum JR, Rubin BP, Shehata BM, Liu W, Dei Tos AP, Weiss SW. Morphologic and immunophenotypic diversity in Ewing family tumors: A study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29:1025–1033. [PubMed] [Google Scholar]
- Ginsberg JP, de Alava E, Ladanyi M, Wexler LH, Kovar H, Paulussen M, Zoubek A, Dockhorn-Dworniczak B, Juergens H, Wunder JS, Andrulis IL, Malik R, Sorensen PH, Womer RB, Barr FG. EWS-FLI1 and EWS-ERG gene fusions are associated with similar clinical phenotypes in Ewing's sarcoma. J Clin Oncol. 1999;17:1809–1814. doi: 10.1200/JCO.1999.17.6.1809. [DOI] [PubMed] [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: A modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre JM, Antonescu CR. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer. 1996;15:115–121. doi: 10.1002/(SICI)1098-2264(199602)15:2<115::AID-GCC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Kobayashi H, Handa M, Satake N, Maseki N. EWS-ERG fusion transcript produced by chromosomal insertion in a Ewing sarcoma. Genes Chromosomes Cancer. 1997;18:228–231. [PubMed] [Google Scholar]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, Takizawa T, Nakamura T. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- Llombart-Bosch A, Blache R, Peydro-Olaya A. Ultrastructural study of 28 cases of Ewing's sarcoma: Typical and atypical forms. Cancer. 1978;41:1362–1373. doi: 10.1002/1097-0142(197804)41:4<1362::aid-cncr2820410421>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Llombart-Bosch A, Machado I, Navarro S, Bertoni F, Bacchini P, Alberghini M, Karzeladze A, Savelov N, Petrov S, Alvarado-Cabrero I, Mihaila D, Terrier P, Lopez-Guerrero JA, Picci P. Histological heterogeneity of Ewing's sarcoma/PNET: An immunohistochemical analysis of 415 genetically confirmed cases with clinical support. Virchows Arch. 2009;455:397–411. doi: 10.1007/s00428-009-0842-7. [DOI] [PubMed] [Google Scholar]
- Maire G, Brown CW, Bayani J, Pereira C, Gravel DH, Bell JC, Zielenska M, Squire JA. Complex rearrangement of chromosomes 19, 21, and 22 in Ewing sarcoma involving a novel reciprocal inversion-insertion mechanism of EWS-ERG fusion gene formation: A case analysis and literature review. Cancer Genet Cytogenet. 2008;181:81–92. doi: 10.1016/j.cancergencyto.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, Radice P, Azzarelli A, Pilotti S, Croce CM, Pierotti MA, Sozzi G. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–3804. doi: 10.1038/sj.onc.1203762. [DOI] [PubMed] [Google Scholar]
- Ng TL, O'Sullivan MJ, Pallen CJ, Hayes M, Clarkson PW, Winstanley M, Sorensen PH, Nielsen TO, Horsman DE. Ewing sarcoma with novel translocation t(2;16) producing an in-frame fusion of FUS and FEV. J Mol Diagn. 2007;9:459–463. doi: 10.2353/jmoldx.2007.070009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new sub-type of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Puls F, Niblett A, Marland G, Gaston CL, Douis H, Mangham DC, Sumathi VP, Kindblom LG. BCOR-CCNB3 (Ewing-like) sarcoma: A clinicopathologic analysis of 10 cases, in comparison with conventional Ewing sarcoma. Am J Surg Pathol. 2014;38:1307–1318. doi: 10.1097/PAS.0000000000000223. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: A modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing DC, McMullan DJ, Roberts P, Smith K, Chin SF, Nicholson J, Tillman RM, Ramani P, Cullinane C, Coleman N. FUS/ERG gene fusions in Ewing's tumors. Cancer Res. 2003;63:4568–4576. [PubMed] [Google Scholar]
- Specht K, Sung YS, Zhang L, Richter GH, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 2014;53:622–633. doi: 10.1002/gcc.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumegi J, Nishio J, Nelson M, Frayer RW, Perry D, Bridge JA. A novel t(4;22)(q31;q12) produces an EWSR1-SMARCA5 fusion in extraskeletal Ewing sarcoma/primitive neuroectodermal tumor. Mod Pathol. 2011;24:333–342. doi: 10.1038/modpathol.2010.201. [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ijszenga M, Tanke HJ, Rosenberg C, Hogendoorn PC. Molecular cytogenetic characterization of four previously established and two newly established Ewing sarcoma cell lines. Cancer Genet Cytogenet. 2006;166:173–179. doi: 10.1016/j.cancergencyto.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ijszenga M, de Jong D, Karseladze A, Tanke HJ, Hogendoorn PC. The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res. 2009;15:2259–2268. doi: 10.1158/1078-0432.CCR-08-2184. [DOI] [PubMed] [Google Scholar]
- Wang L, Bhargava R, Zheng T, Wexler L, Collins MH, Roulston D, Ladanyi M. Undifferentiated small round cell sarcomas with rare EWS gene fusions: Identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagn. 2007;9:498–509. doi: 10.2353/jmoldx.2007.070053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.