Abstract
The regulation of steroidogenic acute regulatory protein (StAR) gene transcription by cAMP-dependent mechanisms occurs in the absence of a consensus cAMP response element (CRE, TGACGTGA). This regulation is coordinated by multiple transcription factors that bind to sequence-specific elements located approximately 150 bp upstream of the transcription start site. Among the proteins that bind within this region, the basic leucine zipper (bZIP) family of transcription factors, i.e. CRE binding protein (CREB)/CRE modulator (CREM)/activating transcription factor (ATF), activator protein 1 (AP-1; Fos/Jun), and CCAAT enhancer binding protein β (C/EBPβ), interact with an overlapping region (−81/−72 bp) in the StAR promoter, mediate stimulus-transcription coupling of cAMP signaling and play integral roles in regulating StAR gene expression. These bZIP proteins are structurally similar and bind to DNA sequences as dimers; however, they exhibit discrete transcriptional activities, interact with several transcription factors and other properties that contribute in their regulatory functions. The 5′-flanking −81/−72 bp region of the StAR gene appears to function as a key element within a complex cAMP response unit by binding to different bZIP members, and the StAR promoter displays variable states of cAMP responsivity contingent upon the occupancy of these cis-elements with these transcription factors. The expression and activities of CREB/CREM/ATF, Fos/Jun and C/EBPβ have been demonstrated to be mediated by a plethora of extracellular signals, and the phosphorylation of these proteins at several Ser and Thr residues allows recruitment of the transcriptional coactivator CREB binding protein (CBP) or its functional homolog p300 to the StAR promoter. This review will focus on the current level of understanding of the roles of selective bZIP family proteins within the complex series of processes involved in regulating StAR gene transcription.
Keywords: CREB/CREM, Fos/Jun, C/EBPβ, cAMP signaling, phosphorylation, and StAR gene expression
1. Introduction
An indispensable event in the regulation of steroid hormone biosynthesis is the delivery of cholesterol from the outer to the inner mitochondrial membrane, a process that is mediated by the steroidogenic acute regulatory protein (StAR) (Clark et al., 1994; Lin et al., 1995; Waterman, 1995; Stocco and Clark, 1996; Christenson and Strauss, 2000; Miller, 2007). Expression of the StAR protein in the adrenals and gonads is regulated by cAMP signaling, and is tightly correlated with the acute steroidogenic response of these tissues to tropic hormone stimulation (Clark et al., 1995; Miller, 1995; Stocco, 2000; Manna and Stocco, 2005). Compelling evidence for the critical role StAR serves in regulating steroidogenesis came from patients suffering from lipoid congenital adrenal hyperplasia (lipoid CAH). These patients were determined to possess mutations in the StAR gene that leads to markedly impaired adrenal and gonadal steroid biosynthesis (Lin et al., 1995; Miller, 1997; Bose et al., 2000). The targeted disruption of the StAR gene in mice generates an essentially identical phenotype to that of lipoid CAH in humans (Caron et al., 1997, 1998; Hasegawa et al., 2000). Regulation of cAMP dependent StAR expression, and thus steroid synthesis, involves transcriptional induction; however, the StAR gene promoter lacks a consensus cAMP response element (CRE, 5′-TGACGTGA-3′) and resembles the promoters of several steroid hydroxylase genes that are regulated by cAMP signaling, suggesting the involvement of alternate regulatory factors in cAMP responsiveness (Waterman, 1994). Several lines of evidence indicate that the StAR gene is regulated by several trans-acting factors that bind to a region in the proximal StAR promoter which is highly homologous among different species (Wooton-Kee and Clark, 2000; Manna et al., 2003b, 2004, 2008; Hiroi et al., 2004a; Silverman et al., 2006; Yivgi-Ohana et al., 2008). Studies have identified a conserved motif within this region (−81/−72; 5′-TGACTGATGA-3′) that recognizes the CRE (CREB/CRE modulator (CREM)/activating transcription factor 1 (ATF-1)), activator protein 1 (AP-1, Fos and Jun), and CCAAT/enhancer binding protein β (C/EBPβ) families of proteins. This motif appears to function within a cAMP response unit in the StAR promoter, and serves a central role in the transcriptional regulation of the StAR gene (Manna et al., 2003b, 2004; Hiroi et al., 2004a; Clem et al., 2005; Silverman et al., 2006; Manna and Stocco, 2007, 2008; Yivgi-Ohana et al., 2008).
CREB/CREM/ATF-1, Fos and Jun, and C/EBPβ, are basic leucine zipper (bZIP) proteins that bind to specific double-stranded DNA sequences as homodimers and/or heterodimers, and result in varying effects on transcription (Montminy et al., 1986; Meyer and Habener, 1993; Montminy, 1997; De Cesare and Sassone-Corsi, 2000; Shaulian and Karin, 2001; Ramji and Foka, 2002; Hess et al., 2004; Deppmann et al., 2006). These factors constitute one of the most important classes of enhancer-type transcription factors and as a group are seen to play a role in a large array of physiological processes including development, differentiation, reproduction, tumor progression, metabolism, circadian rhythms, learning, memory and stress (Sassone-Corsi, 1995; Nantel et al., 1996; Sanyal et al., 2002; Manna et al., 2004; Sassone-Corsi, 2005; Hogeveen and Sassone-Corsi, 2006; Kehat et al., 2006). The bZIP proteins are structurally similar and typically possess a DNA binding domain which consists of a basic region involved in recognition and sequence-specific binding to DNA and a leucine zipper, which contains a coiled-coil structure with heptad repeats of leucines (appearing at an interval of six amino acids), responsible for dimerization (Fig. 1). The expression and activities of these proteins are mediated by a myriad of signaling pathways including cAMP-dependent protein kinase A (PKA), PKC and other kinases, and have been demonstrated to have diverse effects on DNA binding and gene transcription (Chrivia et al., 1993; Shaulian and Karin, 2001; Ramji and Foka, 2002; Johannessen et al., 2004; Tang et al., 2005).
Fig. 1.
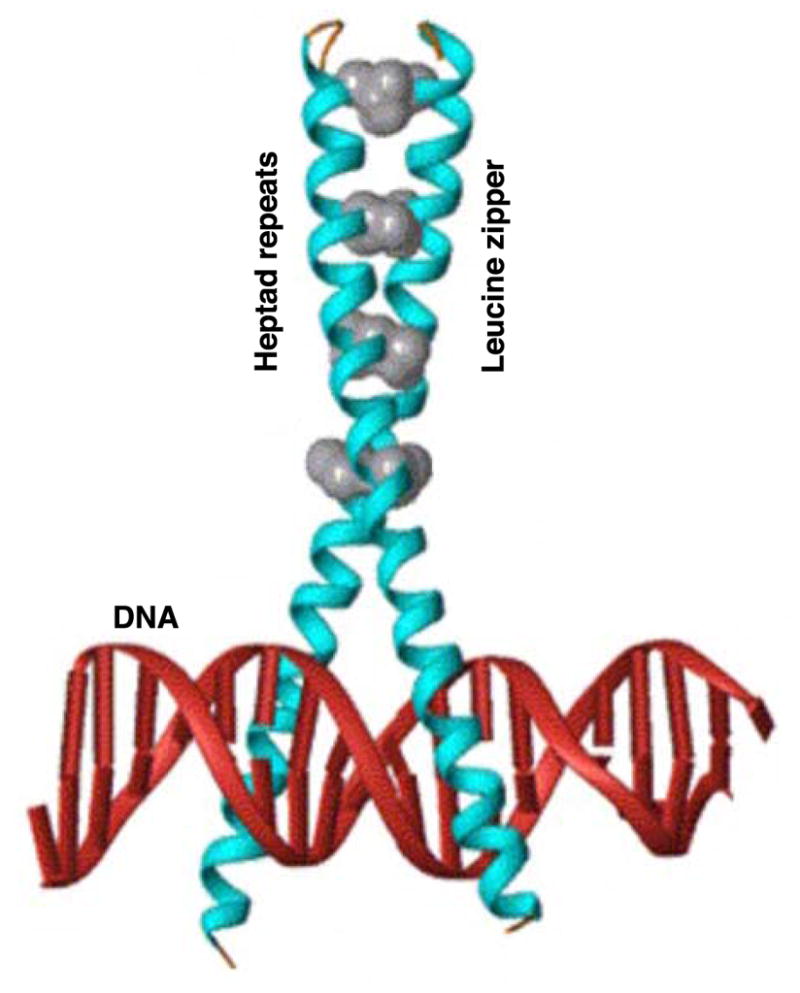
Schematic model of the bZIP motif bound to double stranded DNA. The DNA is in red, while bZIP α-helices are in blue and the leucines of their heptad repeats is in grey (diagram revised and reprinted with permission, Vinson et al., 2006).
The transcriptional coactivators, CREB binding protein (CBP) and closely related p300 (CBP/p300), structurally and functionally link specific transcriptional factors bound to elements of the promoter with the general transcriptional machinery (Cardinaux et al., 2000; Vo and Goodman, 2001). This facilitates RNA polymerase II recruitment and results in the activation of transcription of a number of genes including StAR (Goodman and Smolik, 2000; Hiroi et al., 2004a; Clem et al., 2005; Manna and Stocco, 2007). CBP/p300 harbor multiple functional domains, possess histone acetyltransferase (HAT) activity, and interact with a variety of transcription factors including CREB (Kwok et al., 1994; Manna and Stocco, 2007), Fos/Jun [Arias et al., 1994; Bannister and Kouzarides, 1995; Manna and Stocco, 2007), C/EBP (Kovacs et al., 2003; Silverman et al., 2006), and several nuclear receptors (Kamei et al., 1996; Horwitz et al., 1996; Fronsdal et al., 1998). Furthermore, these coactivators have been shown to perform an important role in the integration of diverse signaling pathways that lead to changes in gene expression (Kamei et al., 1996; Fronsdal et al., 1998). This review summarizes the transcriptional properties and potential functions of CREB/CREM/ATF-1, Fos/Jun and C/EBPβ, factors that are known to be involved in regulating StAR gene expression.
2. CRE signaling and StAR gene transcription
Transcriptional regulation, a key control mechanism in fundamental biological processes that results from stimulation of the adenylate cyclase signaling pathway, is mediated by a family of cAMP-responsive factors. This family consists of a large number of proteins encoded by the CREB, CREM, and ATF genes that bind the 8-bp palindromic sequence 5′-TGACGTCA-3′ or a minor variation thereof. This specific sequence, termed the cAMP response element (CRE), is commonly found within 100 nucleotides of the TATA box in the promoters of eukaryotic genes (Montminy et al., 1986; Meyer and Habener, 1993; Sassone-Corsi, 1995; Montminy, 1997; Roesler, 2000). Studies have demonstrated that the 5′-TGACG half of the palindrome is more highly conserved with respect to the 3′-TCA (Sassone-Corsi, 1995). CREB, a 43-kDa bZIP transcription factor that binds to DNA as a homodimer, not only mediates stimulus-transcription coupling in the cAMP pathway but also functions as a point of convergence among a variety of signaling molecules. CREB cDNAs cloned from human, rat and mouse reveal the presence of two CREB protein isoforms, CREB341 and CREB327, which arise from alternatively spliced mRNAs of the same gene (Fig. 2). Members of the CREB family (including CREM and ATF-1) are distinguished by their DNA-binding bZIP domains, interact with each other, and mediate the transcriptional response to cAMP signaling (Meyer and Habener, 1993; Della Fazia et al., 1997). Although CREB has been shown to serve as the principal agent in mediating the transcriptional response at CREs for a variety of biological functions, CREB knockout studies demonstrated that these functions can be compensated for by the involvement of the other CRE-binding proteins such as CREM and ATF-1 (Hummler et al., 1994). Such a mechanism appears to be in place in regulating steroidogenesis, since CREB family members are known to directly accelerate the transcription of the StAR gene (Manna et al., 2002, 2003a, 2006; Hiroi et al., 2004a; Clem et al., 2005; Yivgi-Ohana et al., 2008). We have identified and characterized three CRE-like elements within the −96/−67 bp region (−71/−42 from the TATA box) of the mouse StAR promoter that recognize CREB and other CRE-binding proteins (Manna et al., 2002, 2003a, 2004, Manna and Stocco, 2007). In examining the functional impact of these three sites, termed CRE1, CRE2, and CRE3, it has been shown that CREB-mediated StAR gene transcription is most heavily influenced by the CRE2 site when compared to the CRE1 and CRE3 sites and that CREB shows higher sequence-specific binding to the CRE2 site than the other CREs in in vitro binding assays (Manna et al., 2003a, 2004; Manna and Stocco, 2007). Recently, the functional importance of the CRE2 site in CREB/ATF binding has also been demonstrated in placental StAR transcription (Yivgi-Ohana et al., 2008). Notably, however, all three CRE elements are required in order for the StAR promoter to exhibit full CREB responsiveness. Consistent with these findings, the ability of CREB to activate promoters frequently occurs by having multiple CRE binding factors recognize tandem sets of CREs rather than relying on the binding of a single dimer to the DNA (Roesler et al., 1995; Roesler, 2000; Wilson et al., 2002).
Fig. 2.
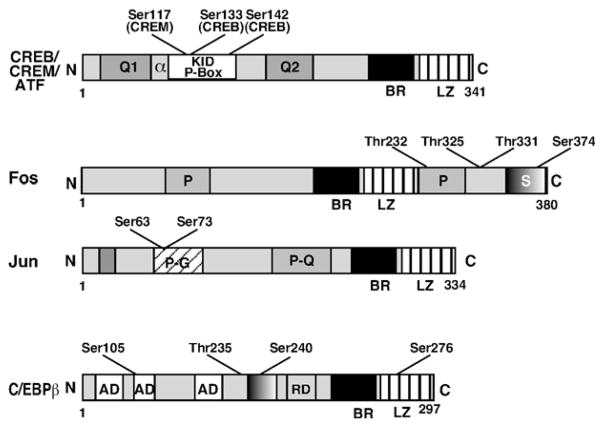
Schematic representations of the CREB/CREM/ATF-1, Fos and Jun, and C/EBPβ proteins. These families of proteins are characterized by a highly conserved bZIP DNA binding domain consisting of basic region (BR) and leucine zipper (LZ). Two glutamine rich regions Q1 and Q2 for CREB, CREM and ATF-1 are labeled along with the phosphorylated region/kinase inducible domain (P-Box/KID). Phosphorylation of Ser133 and Ser117, in CREB and CREM respectively, turns them into powerful activators through interaction with CBP. Ser142 in CREB attenuates its activity. The Fos and Jun proteins possess several domains including the bZIP domain, transactivation domains and docking sites for several kinases. The regions rich in prolines (P), prolines and glutamines (P–Q), prolines and glycines (P–G), and threonines (S) in Fos and Jun are labeled. Structure of the C/EBPβ protein indicating different regions (AD, activation domain; RD, repression domain) is provided. All of these bZIP proteins can be phosphorylated on several Ser and Thr residues by multiple kinases; however, only selected phosphorylation sites are illustrated in the diagrams. Numbers indicate the approximate positions of amino acids in the proteins; note that protein structures are not drawn to scales.
Whereas CREB gene products are generally observed to be positive trans-activators, CREM is found to either activate or repress CRE-mediated transcription depending upon whether the glutamine-rich exons C and G are spliced in or out, respectively (Foulkes and Sassone-Corsi, 1992; Meyer and Habener, 1992). CREB and CREM are cyclically expressed during spermatogenesis, where the function of CREM is converted from antagonist to activator (Foulkes et al., 1992, 1993). Moreover, the regulation of CREM in the testis is under the control of the hypothalamo-pituitary-gonadal axis, especially through FSH signaling. Alternative splicing of the CREM gene can give rise to several isoforms that act as either activators (τ, τ1 and τ2) or repressors (α, β and γ) of transcription (Walker and Habener, 1996; Sassone-Corsi, 1998). Of these isoforms, CREMτ is abundant in the adult testis, but exhibits low expression in prepubertal animals where the repressive isoforms are more readily detected (Zhou et al., 1996). It has been demonstrated that functional regions in the CREB protein are identical to CREMτ (Meyer and Habener, 1992; Ruppert et al., 1992; Lee and Masson, 1993), and the overexpression of either CREB or CREMτ has been shown to have qualitatively similar effects in activating cAMP-mediated StAR gene expression in mouse adrenal and gonadal cells (Manna et al., 2002, 2003a, 2006; Sugawara et al., 2006; Manna and Stocco, 2007). In contrast, CREMα and CREMβ are to repress transcription of the StAR gene (Manna et al., 2003a). As pointed out, both the CREM and CREB genes can express numerous isoforms generated though a multiplicity of alternative exon splicing sites; however, the CREM gene is unique from CREB and the other bZIP transcription factors in that it encodes two distinct DNA binding domains, which may be differentially utilized through splicing events (Ruppert et al., 1992; Meyer and Habener, 1993). The CREM proteins can recognize and bind to CREs as homodimers or as heterodimers with CREB/ATF, and these dimers show similar functional responses to those of CREB (Meyer and Habener, 1993; Sassone-Corsi, 1998). For example, oligonucleotide probes containing one or more CRE sites found in the StAR promoter can readily bind recombinant CREB protein and endogenous CREB in nuclear extracts (NE) from mouse Leydig, adrenal and placental trophoblast giant cells (Manna et al., 2002, 2003a, 2004; Sugawara et al., 2006; Manna and Stocco, 2007; Yivgi-Ohana et al., 2008). Closer study, however, identified CREM proteins as the predominant binding species, as the protein:DNA binding was essentially abolished with CREM1 antibody (Ab), but not with Abs specific for other CREB family members. In addition, an ATF-1 Ab supershifted the protein:DNA binding that occurred in connection with the StAR CRE element(s) (Manna et al., 2002; Yivgi-Ohana et al., 2008). Importantly, CREB and CREM have been demonstrated to associate with the proximal rather than the distal StAR promoter following treatment with a cAMP analog (Hiroi et al., 2004a; Clem et al., 2005; Manna and Stocco, 2007), although the specific balance of these genes in regulating the StAR gene is still a matter of contention. A comparison between the involvement of CREB and CREM in cAMP-mediated StAR gene expression identified CREM as the principal mediator in adrenal NCI-H295R cells (Sugawara et al., 2006). However, another study demonstrated that CREB and ATF-1, but not CREM, are predominantly bound to the StAR promoter in response to cAMP signaling (Clem et al., 2005). Taken together, these findings demonstrate the functional relevance of different CRE DNA binding proteins in the transcriptional regulation of the StAR gene.
CREM is known to play a pivotal role in reproduction as mice lacking the CREM gene, while known survive to adulthood, are sterile due to an arrest in spermatogenesis resulting from germ cell apoptosis instead of differentiation (Nantel et al., 1996). It is noteworthy, however, that the induction of CREM activators by phosphorylation results in a marked increase in the expression of the ICER (inducible cAMP early repressor) (Delmas et al., 1993; Walker et al., 1994; Sassone-Corsi, 1998). ICER arises via an alternative intronic promoter within the CREM gene. While ICER contains the DNA binding and dimerization domains, it lacks the typical trans-activation domains. Furthermore, the DNA binding domain of ICER can efficiently bind to CREs, heterodimerizes with CREB, CREMτ or related trans-activators, and thus functions as a powerful repressor of cAMP-induced transcription (Walker et al., 1998; Sassone-Corsi, 1998; Zwermann et al., 2007). It has been demonstrated that ICER represses the CREB/CREM-mediated activation of StAR gene transcription (Manna et al., 2002), and it is presumed that heterodimerization of ICER with CREB/CREM inhibits their normal capacity to induce StAR expression in response to cAMP signaling. ICER is capable of down regulating the CRE-mediated transcription of the CREB gene in Sertoli cells and suppresses both basal and cAMP-induced expression of the inhibin α-subunit gene in granulosa cells (Walker et al., 1998; Mukherjee et al., 1998; Zwermann et al., 2007). It is worth noting that the functional effects of CREM in the testis are also known to involve ACT (activator of CREM in testis), a testis specific factor consisting of four complete LIM domains and one N-terminal half-LIM motif (Fimia et al., 2000; Kotaja et al., 2004). ACT is co-localized with CREM in germ cells, coordinately regulated during spermatogenesis, interacts with CREB/CREM and acts as a trans-activator (Fimia et al., 2000, 2001); however, its role in StAR gene transcription has not been described.
In addition to CREB and CREM, the CRE-binding protein ATF-1 also appears to be an important regulator of StAR expression. Although the nucleic acid sequences found in these factors have diverged considerably, ATF-1 is structurally and functionally similar to the activators of CREB and CREM. ATF-1 differs from CREB and CREM in that it lacks the glutamine-rich Q1 domain, but this does not prevent ATF-1 from functioning effectively as a transcriptional activator (Meyer and Habener, 1993; Della Fazia et al., 1997; Sassone-Corsi, 1998). As such, ATF-1 is thought to serve in both complementary and substitutionary capacities relative to the other CRE-binding proteins. In support of this mechanism, recombinant CREB bound to CRE-like element(s) in the StAR promoter can be recognized by ATF-1 and/or ATF-2 Abs in different steroidogenic cells (Manna et al., 2002, 2006; Yivgi-Ohana et al., 2008). DNA-affinity chromatography has demonstrated that ATF-1 can bind to the C/EBPβ/non-consensus AP-1/nuclear receptor half-site site of the StAR promoter (Clem et al., 2005), which is the same region described earlier that can bind recombinant CREB, and the binding was found to be supershifted by ATF-1 (Manna et al., 2002; Yivgi-Ohana et al., 2008). Given the large number of CRE-binding proteins involved in regulating the StAR promoter, it is interesting to note that two paralogs of ATF-1, termed CRE binding protein 1 (CRE-BP1, also known as ATF-2) and ATF-a, which have evolved by gene duplication, exhibit alternative exon splicing and bind to CREs, but do not mediate cAMP responsive trans-activations (Meyer and Habener, 1993).
3. Role of Fos/Jun in StAR gene expression
The AP-1 family of transcription factors, which includes Fos and Jun, share many properties, bind to a DNA sequence known as the AP-1/phorbol 12-O-tetradecanoate 13-acetate responsive element (AP-1/TRE; TGA(C/G)TCA), and have been reported to play central roles in regulating many biological functions including proliferation, differentiation, and transformation (Angel and Karin, 1991; Hai and Curran, 1991; Kerppola and Curran, 1991; Shaulian and Karin, 2001; Masquilier and Sassone-Corsi, 1992; Manna et al., 2004; Zenz et al., 2008). Fos members heterodimerize with Jun proteins and with select members of the CREB/ATF family, but do not form homodimers, whereas Jun members function as homodimers or heterodimers among themselves or with members of the Fos and CREB/ATF families (Hai and Curran, 1991; Angel and Karin, 1991; O’Shea et al., 1992; Hess et al., 2004). Examination of the mouse StAR promoter identified a highly conserved element (TGACTGA, −81/−75 bp) that was homologous to the AP-1/TRE sequence, and was also noted to overlap with the CRE2 sequence (Manna et al., 2002). This element, referred to as CRE2/AP-1, was demonstrated to recruit Fos and Jun, and through binding to the CRE2/AP-1 motif these proteins can have both positive and negative effects on StAR gene transcription (Manna et al., 2004; Manna and Stocco, 2007, 2008). Notably, however, the CRE2/AP-1 region (TGACTGA) in the mouse StAR promoter does not bind to AP-1 proteins in primary cultures of ovarian cells, unless it is mutated to TGACTcA (Silverman et al., 1999). Results of a number of studies point to the CRE2/AP-1 element as a key site for AP-1 family members to mediate these specific effects (Wooton-Kee and Clark, 2000; Manna et al., 2002, 2003a; Clem et al., 2005; Manna and Stocco, 2007, 2008). First, the effects of Fos and Jun on the StAR promoter are lost when mutations are introduced in the CRE2/AP-1 motif. Second, oligonucleotide probes encompassing the CRE2/AP-1 element are seen to bind proteins in MA-10 NE, and this protein:DNA complex is abolished by c-Fos and Fra-2 Abs. Third, chromatin immunoprecipitation assays targeting this region show that CREB as well as Fos/Jun are targeted to the proximal StAR promoter in response to cAMP signaling (Hiroi et al., 2004a; Clem et al., 2005; Manna and Stocco, 2007, 2008). Therefore, it appears that CREB and other bZIP members, such as AP-1 family members, serve in modulating StAR promoter activity, and that the CRE2/AP-1 site plays a predominant role in coordinating the functions of these transcription factors. In the ovary, the patterns of StAR regulation suggested developmental controlled modes of StAR transcription, i.e. during follicular phase cAMP/FSH signaling activates CREB-mediated StAR gene expression which is cAMP/CREB independent upon luteinization associated with CREB to FRA-2 replacement and the requirement for C/EBPβ and SF-1 that bind to the −117/−95 motif (Yivgi-Ohana et al., 2008). Nevertheless, the inhibition of protein binding to CRE2/AP-1 DNA by Abs specific for c-Fos, Fra-2 and CREM1 supports the notion that AP-1 and CREB/CREM proteins can form heterodimers (Hai and Curran, 1991; Chatton et al., 1994). It is now generally accepted that AP-1 complexes are not limited to dimers consisting solely of Fos and Jun, and these proteins are frequently known to dimerize with other bZIP proteins (Hai and Curran, 1991; Chinenov and Kerppola, 2001). Just as with the CRE-binding proteins, the increased diversity in AP-1 dimerization partners is thought to yield a greater or more dynamic range of regulation for TRE-containing genes. While this appears to be the case for the StAR promoter, it is likely more complex in that two additional putative AP-1 binding sites have been identified in the rat StAR promoter, where c-Fos was able to diminish basal, cAMP and c-Jun mediated rat StAR gene transcription in Y-1 adrenocortical cells (Shea-Eaton et al., 2002). Fos and Jun are known to have opposite effects on the transcription of a number of genes including phosphoenolpyruvate carboxykinase (PEPCK) (Gurney et al., 1992), gonadotropin-releasing hormone, and myogenic helix-loop-helix (Gurney et al., 1992; Bruder et al., 1996). Nevertheless, studies have shown that repression of the PEPCK gene by Fos does not require a consensus TRE but it does require an intact DNA binding region and dimerization domain, and that it binds to the C/EBP protein family (Gurney et al., 1992). In accordance with this, a similar overlapping region in the StAR promoter has also been shown to bind C/EBPβ (see below) (Silverman et al., 1999, 2006).
A functional comparison of Fos and Jun suggests that c-Jun is the most potent AP-1 family member to trans-activate the StAR gene. Recently we observed that the c-Jun proto-oncogene, but not other AP-1 factors, plays an essential role in PKC-mediated regulation of StAR transcription and steroidogenesis in mouse Leydig cells (Manna and Stocco, 2008). The loss of c-Jun in mice is embryonic lethal, with embryos dying between mid- and late-gestation due to several tissue defects (Hilberg et al., 1993; Johnson et al., 1993). Studies have shown that c-Jun containing tethered Fos dimers could efficiently bind to consensus TRE, is recognized by appropriate Abs with a mobility shift similar to that of a mixture of Fos and Jun, and have distinct promoter specificity and biological responses (Bakiri et al., 2002; Manna and Stocco, 2007). Likewise, the tethered c-Jun~c-Fos has been demonstrated to be the strongest activator of the human collagenase gene promoter, which possesses a canonical TRE, followed by c-Jun~c-Fra-1, c-Jun~c-Fra-2, and c-Jun~ATF2 (Jonat et al., 1992; Ito et al., 2001; Bakiri et al., 2002). In addition, c-Jun~c-Fos dimers have been shown to prefer TRE like motifs while Jun~ATF2 complexes prefer CRE like motifs (Bakiri et al., 2002). Since the CRE2/AP-1 region of the StAR promoter more closely resembles a consensus TRE, it is likely that c-Jun~c-Fos and/or c-Jun~Fos (Fra-2) dimers play a more dominant role in regulating the transcription of the StAR gene, as opposed to a direct effect of c-Jun alone (Manna et al., 2004, Manna and Stocco, 2008). Therefore, at the StAR promoter it appears that, much like in other genes, the effects of Fos and Jun can be mediated through binding sites closely related to the consensus AP-1/TRE (Hai and Curran, 1991; Rutberg et al., 1999).
4. C/EBPs and StAR gene transcription
The C/EBP family, a bZIP class of transcription factors, has been shown to play important roles in regulating genes that control differentiation and function of multiple cell types (Johnson and Williams, 1994; Lekstrom-Himes and Xanthopoulos, 1998; McKnight, 2001; Roesler, 2001; Wilson and Roesler, 2002). The C/EBP family is comparable to other bZIP proteins with respect to its modular organization. Members of the C/EBP family (C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ (Ig/EBP), C/EBPε and C/EBPζ (CHOP)) (Table 1) are highly similar in their dimerization and DNA binding domains, but show significant variation in both the structure and function of their trans-activation domains. C/EBPs bind with varying affinities to a consensus site consisting of a dyad symmetrical repeat (A/GTTGCGC/TAAC/T) (Osada et al., 1996; Grimm and Rosen, 2003). The C/EBP protein was initially characterized based upon its ability to bind viral enhancer elements and was later identified to bind to the CCAAT box element in different cellular promoters. C/EBPs mediate cAMP responsiveness by indirect mechanisms since the cAMP-inducible domains of C/EBPs, with the exception of C/EBPβ, lack a PKA phosphorylation site. The C/EBPβ isoform can be phosphorylated within its bZIP domain, which alters its DNA binding activity (Roesler, 2001; Wilson et al., 2002). C/EBPs are expressed differentially in a number of tissues including liver, adipose, lung, intestine, testis and ovary (McKnight, 2001; Wilson et al., 2002; Ramji and Foka, 2002; Grimm and Rosen, 2003). Interestingly, C/EBPα and C/EBPβ are expressed in steroidogenic cells, including Leydig and granulosa cells, where levels of nuclear C/EBPβ have been shown to be increased by LH and cAMP analogs (Nalbant et al., 1998; Piontkewitz et al., 1996; Silverman et al., 1999, 2006). In mice lacking either the C/EBPα or the C/EBPβ gene, ovarian development fails to occur in response to tropic hormone, leading to reduced or halted ovulation and an inability to form the corpus luteum (Piontkewitz et al., 1996; Sterneck et al., 1997). Three putative C/EBP binding sites have been identified within the proximal −151/−1 bp region in the human and rodent StAR promoters, and the roles of C/EBPα and C/EBPβ in cAMP-mediated regulation of StAR gene transcription have been demonstrated (Reinhart et al., 1999; Silverman et al., 1999; Christenson et al., 1999; Wooton-Kee and Clark, 2000; Tremblay et al., 2002; Silverman et al., 2006). Notably, one of the C/EBP recognition motifs, (−81/−72 bp, TGACTGATGA) that is known to bind C/EBPβ, overlaps with the CRE2/AP-1 site, and its disruption reduces both basal and FSH-mediated StAR gene expression in rat granulosa cells (Silverman et al., 1999). EMSA studies targeting the −81/−72 bp region illustrated that it bound C/EBPβ. The requirement of C/EBPβ in luteal cells has recently been demonstrated to be associated with StAR gene transcription in concurrence with cAMP independent signaling (Yivgi-Ohana et al., 2008). In addition to C/EBPβ, studies have also revealed that two truncated forms of C/EBPβ lacking the N-terminal activation domain (Table 1), known as LAP (liver-enriched activating protein) and LIP (liver-enriched inhibitory protein), were both up-regulated in response to FSH and immediately ahead of StAR expression (Silverman et al., 1999). In a recent study, C/EBPβ has been shown to associate with the proximal, but not with the distal, region of the StAR promoter and that treatment with prostaglandin E2 increased the levels of C/EBPβ bound to the StAR promoter in vivo (Hsu et al., 2008). Studies have demonstrated that both the CREB/ATF and C/EBP family members can efficiently compete for binding to the elements that show sequence similarity to either CRE or CCAAT motifs present within a promoter, and direct the transcription of several genes (Bakker and Parker, 1991; Liu et al., 1991; Wilson et al., 2002). In the case of the androgen-regulated C3 gene, CREB/ATF sites have been demonstrated to compete with C/EBP protein that binds to a CCAAT box motif, an observation in agreement with what has been shown to occur at the somatostatin promoter ATF site (Zhang et al., 1990). Thus C/EBP may act either as a transcriptional activator or may interfere with the activity of CREB and/or AP-1 family members bound to a similar recognition site (Fig. 3); however, the incidence of the latter in StAR gene transcription remains to be elucidated.
Table 1.
Transcription of the StAR gene involves three bZIP families of proteins. Formation of selective dimers among these family members is listed.
| BZIP factors | CREB/CREM/ATF | Fos/Jun | C/EBP |
|---|---|---|---|
| Families | CREB CREMα, β, γ, τ ATF-1, ATF-2 (CRE-BP1), ATF-3, ATFa |
c-Fos, Fos B, Fra-1, and Fra-2 c-Jun, Jun B, and Jun D |
C/EBPα C/EBPβ (NF-IL6), LIP, LAP C/EBPγ (Ig/EBP1) C/EBPδ C/EBPε (CRP-1) C/EBPζ (CHOP-10) |
| Dimers | CREB:CREB CREB:CREM CREB:ATF-1 CREM:CREM ATF-1:ATF-1 |
Fos:Jun Jun:Jun c-Jun:c-Fos c-Jun:ATF-1 c-Fos:ATF-4 |
C/EBP C/EBP:Ig/EBP1 Ig/EBP1 C/EBPβ:C/ATF C/EBPβ:CHOP-10 |
| Target elements | TGACGTCA | TGA(C/G)TCA | A/GTTGCGC/TAAC/T |
Fig. 3.
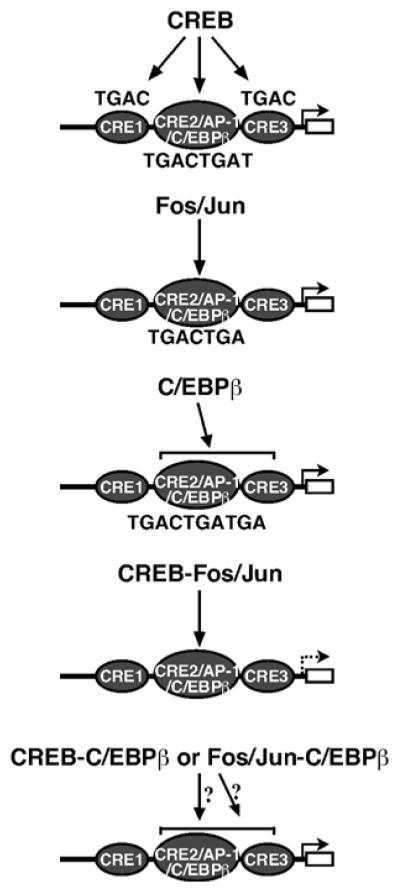
Proposed models of CREB, c-Fos/c-Jun, C/EBPβ, or CREB and c-Fos/c-Jun interactions on a single cis-element known to coordinate both the positive and negative regulation of StAR gene transcription. Three different families of proteins have been shown to bind to a motif, labeled as CRE2/AP-1/C/EBPβ (−81/−72), which displays overlapping similarity to multiple consensus binding sites. Crosstalk of CREB and c-Fos/c-Jun at the −81/−72 site results in trans-repression of the StAR gene due to competition of these factors with limiting amounts of intracellular CBP. The functional cooperation, interaction and/or inhibition of CREB-C/EBPβ or Fos/Jun-C/EBPβ at the same site, is not currently known.
5. Phosphorylation of CREB/CREM, Fos/Jun and C/EBP and its relevance to StAR gene expression
The transduction of signals via hormones and neurotransmitters is known to rapidly activate adenylate cyclase, which in turn generates intracellular cAMP. The cAMP signaling pathway has a profound impact in regulating a number of physiological processes by influencing basic patterns of gene expression through stimulus-coupled transcription. The response to cAMP at the genetic level is typically mediated through a palindromic conserved CRE sequence found in the promoters of many cAMP-responsive genes (Montminy et al., 1986; Meyer and Habener, 1993; Sassone-Corsi, 1995, 1998). As mentioned above, the StAR gene lacks a consensus CRE even though it is regulated by cAMP-dependent mechanisms. One of the first events regulating transcription factor activity is their translocation into the nucleus. The phosphorylation status of a transcription factor can positively or negatively modulate its sub-cellular localization as well as its DNA binding activity, both of which can dictate a transcription factor’s subsequent interactions with the transcriptional machinery as well as modulate potential enzymatic activity by influencing conformational changes. CREB/CREM/ATF is activated by PKA, PKC, and other kinases, which phosphorylate the transcription factors at specific residues within their N-terminal region, a domain common to these activators known as either the P-box (phosphorylation box) or KID (kinase inducible domain) (Fig. 2). A large variety of extracellular stimuli and environmental conditions have been shown to induce phosphorylation of CREB at Ser133 (Johannessen et al., 2004). Specifically, transcriptional increases of CREB and CREM are observed following the phosphorylation of CREB341/327 at Ser133/119 or CREM at Ser117, events that are indispensable for their interactions with CBP/p300 (Gonzalez and Montminy, 1989; Chrivia et al., 1993; Kwok et al., 1994; Parker et al., 1996; Fimia et al., 1999; Clem et al., 2005; Manna and Stocco, 2007). In Leydig and other steroidogenic cells, LH/hCG and cAMP analogs are capable of increasing the phosphorylation of CREB in a time-dependent manner, and this event is tightly correlated with the association of both phosphorylated CREB and CBP with the proximal StAR promoter (Hiroi et al., 2004a; Clem et al., 2005; Manna et al., 2006; Silverman et al., 2006; Manna and Stocco, 2007). More recently, the binding of CREB/CREM to the proximal StAR promoter following cAMP analog treatment, and the subsequent recruitment of CBP, has been shown to be associated with histone modifications (Hiroi et al., 2004a, 2004b; Clem et al., 2005; Manna and Stocco, 2007). Although the phosphorylation of Ser133 is crucial for the activation of CREB, this single modification alone is not sufficient to render the protein fully active, and typically a short region C-terminal to the PKA phosphorylation site (140-DLSSD) is required for transcriptional activation of CREB (Meyer and Habener, 1993; Sassone-Corsi, 1995, 1998; Bonni et al., 1995; Johannessen et al., 2004). Compelling evidence for the role of CREB phosphorylation by PKA is also seen in vivo where transgenic mice expressing a non-phosphorylatable mutant of CREB (termed CREB-M1, Ser133→Ala substitution), suffer from somatotroph hypoplasia and dwarfism (Struthers et al., 1991). Subsequently, expression of CREB-M1 in adrenal and gonadal cells has been shown to markedly diminish cAMP-induced StAR gene expression (Manna et al., 2002, 2003a; Sugawara et al., 2006), suggesting that the phosphorylation of CREB or a similar CRE-binding protein is required for the transcription of the StAR gene. Additionally, CREB-M1 has been shown to be ineffective in inducing somatostatin gene transcription and adversely affects survival in transfected F9 teratocarcinoma and rat granulosa cells, respectively (Gonzalez and Montminy, 1989; Scobey et al., 2001). Recent studies have demonstrated that activating PKC with phorbol 12-myristate 13-acetate is able to augment CREB phosphorylation at Ser133, which in turn increases in trans-activation of the StAR gene (Jo et al., 2005; Manna et al. 2006). On the other hand, conflicting reports on the phosphorylation of CREB at Ser142 are found in the literature, demonstrating both positive (increasing the interaction with the CBP) and negative (impairing the interaction with the CBP) effects on CREB activity (Sun et al., 1994; Wu and McMurray, 2001). Nonetheless, the activation of CREB has been demonstrated to be mediated by a large number of agents and/or signals (Johannessen et al., 2004), and the results suggest that in addition to phosphorylation of CREB at Ser133, one or more events is required for CREB to fully activate transcription.
The AP-1 transcription factors, Fos and Jun, are immediate-early response genes and function to couple extracellular signals with cellular responses by influencing the expression of target genes. It is important to note that members of the Fos and Jun proteins are expressed and regulated in a cell type specific manner, suggesting different cells have a complex mixture of AP-1 dimers with subtly discrete functions (Shaulian and Karin, 2001; Wagner, 2001; Hess et al., 2004; Zenz et al., 2008). Regulation of AP-1 activity can be dependent on the constituency of its dimers, which possess varying specificity, as well as changes in the transcription of genes encoding Fos and Jun. AP-1 effects can be further modulated by the interactions that can occur between AP-1 and other transcription factors and cofactors (Hess et al., 2004). Post-translational processing of Fos and Jun has been studied extensively and demonstrated that several Ser and Thr residues are substrates for phosphorylation by PKA, PKC and also by kinases associated with mitogenesis, differentiation, and neuronal excitation (Chen et al., 2001; Hess et al., 2004; Manna et al., 2006; Manna and Stocco, 2007, 2008). In particular, Thr232, Thr325, Thr331, and Ser374 are phosphorylated in Fos, whereas Jun is phosphorylated at Ser63 and Ser73. Treatment with a cAMP analog or a growth factor has been demonstrated to increase the phosphorylation of Ser63 of c-Jun and Thr325 of c-Fos, which are coordinately associated with StAR expression and steroidogenesis in mouse Leydig cells (Clem et al., 2005; Manna et al., 2006; Manna and Stocco, 2007, 2008). In addition, PKC is also found to phosphorylate c-Jun at Ser63 and results in increased transcription of StAR gene expression (Manna and Stocco, 2008). The consequences of c-Jun and c-Fos phosphorylation that occur in response to cAMP signaling are seen in the increased association between P-c-Jun/P-c-Fos and CBP, and the resulting recruitment of this cofactor to the StAR promoter (Clem et al., 2005; Manna and Stocco, 2007, 2008). Fos and Jun phosphorylation has also been shown to alter the interaction with other transcription factor(s) affecting DNA binding and dimerization (Hai and Curran, 1991; Masquilier and Sassone-Corsi, 1992; Hunter and Karin, 1992; Rutberg et al., 1999; Hess et al., 2004). Recent studies show that the interaction of c-Fos/c-Jun with CREB on the CRE2/AP-1 site resulted in an inhibition of the CREB responsiveness involved in StAR gene transcription (Manna and Stocco, 2007). Nevertheless, the phosphorylation of Fos and Jun has also been demonstrated to be associated with both the activation and inhibition of DNA binding and gene transcription, indicating a complex relationship between phosphorylation and function (Boyle et al., 1991; Baker et al., 1992; Abate et al., 1993).
The activation domain of C/EBP is also subjected to post-translational regulation. The C/EBP proteins exert pleiotropic effects based on tissue and stage-specific gene expression, and are able to interact with other transcription factors and coactivators (Johnson and Williams, 1994; Reinhart et al., 1999; Christenson et al., 1999; McKnight, 2001; Tremblay et al., 2002; Ramji and Foka, 2002; Silverman et al., 2006). Phosphorylation of Ser and Thr residues by a number of kinases has been shown to regulate the ability of C/EBP proteins to bind DNA as well as their capacity to induce gene transcription. Specifically, the phosphorylation of Thr235, Ser105 and Ser276 by MAPK, PKC and Ca2+-calmodulin dependent protein kinases, respectively, have been reported to influence the transcriptional activity of C/EBPβ (Zahnow, 2002; Ramji and Foka, 2002; Tang et al., 2005). Signaling through cAMP is also capable of enhancing the phosphorylation of C/EBPβ at Thr235, and this event increases the association of C/EBPβ with the StAR promoter (Manna and Stocco, unpublished observation). This suggests that phosphorylation of C/EBPβ increases its DNA binding activity and results in trans-activation of the StAR gene. It is noted, however, that the phosphorylation and de-phosphorylation of C/EBPβ by growth hormones can coordinately modulate both its transcriptional activity and DNA binding capacity (Piwien-Pilipuk et al., 2002). Conversely, the trans-activation potential of C/EBPβ is repressed in response to being phosphorylated at Ser240 by PKC (Trautwein et al., 1994). PKC also phosphorylates C/EBPα at Ser248, Ser277, and Ser299 and results in a suppression of its DNA binding activity (Mahoney et al., 1992; Ramji and Foka, 2002).
6. Dimerization of bZIP proteins and crosstalk in StAR gene regulation
As discussed above, CREB/CREM/ATF-1, Fos/Jun and C/EBP are three families of dimeric transcription factors and each generally binds to a DNA sequence that is unique to each family. Dimerization is mediated by a structure known as the ‘leucine zipper’, and electrostatic interactions between amino acids along the dimerization interface can permit or favor the formation of either homodimers or heterodimers (Meyer and Habener, 1993; Deppmann et al., 2004; Vinson et al., 2006). The leucine zipper is typically composed of four to five heptad repeats of amino acids that are required to form a productive, coiled-coil dimerization interface (Meyer and Habener, 1993; Poels and Vanden Broeck, 2004; Vinson et al., 2006; Kehat et al., 2006). Analyses of the human genome have identified 53 genes that contain bZIP motifs with the potential to form over 2700 dimers (Poels and Vanden Broeck, 2004; Vinson et al., 2006; Kehat et al., 2006). As such, many forms of CREB/ATF-1, Fos/Jun and C/EBP have the potential to dimerize with either themselves or each other, which can result in a continuum of transcription factors with varied activities (Hai and Curran, 1991; Masquilier and Sassone-Corsi, 1992; Meyer and Habener, 1993; Millhouse et al., 1998; Rutberg et al., 1999). Several examples of dimer combinations that are known to form among CREB/CREM/ATF-1, Fos/Jun and C/EBPs are provided in Table 1. The CRE/ATF (TGACGTGA) and AP-1/TRE (TGA(C/G)TCA) sequence motifs are two of the major classes of regulatory elements that contribute to transcriptional regulation by recruiting bZIP proteins in response to a variety of extracellular signals (Sassone-Corsi et al., 1990; Hai and Curran, 1991; Montminy, 1997). The difference between the CRE and AP-1 consensus sequences is only one nucleotide, and thus overlap and/or crosstalk can occur affecting the versatility of the transcriptional response to signal transduction (Hai and Curran, 1991; Masquilier and Sassone-Corsi, 1992; Millhouse et al., 1998; Rutberg et al., 1999). Indeed, CREB/CREM/ATF-1, Fos/Jun and C/EBPβ families of proteins bind to an overlapping motif in the StAR promoter, and confer the potential for multiple effects in regulating transcription of the StAR gene (Manna et al., 2002, 2003a,b, 2004; Hiroi et al., 2004a; Clem et al., 2005; Silverman et al., 2006; Sugawara et al., 2006; Manna and Stocco, 2007, 2008; Yivgi-Ohana et al., 2008). Several lines of evidence indicate that CRE DNA binding proteins heterodimerize with Fos/Jun and repress AP-1 transcriptional activity (Masquilier and Sassone-Corsi, 1992; Millhouse et al., 1998; Rutberg et al., 1999). On the other hand, heterodimers formed between C/EBPs and CREB/ATF can bind to CRE site(s) and activate transcription (Ross et al., 2001). We have recently demonstrated that CREB and c-Fos/c-Jun can form heterodimers that bind to the closely related CRE2/AP-1 sequence with an altered DNA binding affinity that results in the repression of StAR gene transcription (Manna and Stocco, 2007). It has been demonstrated that bZIP heterodimers have distinct binding activities and functional specificities than those of their parental homodimers, and that they can bind to related CRE and/or AP-1 site depending on the dimer composition (Hai and Curran, 1991; Masquilier and Sassone-Corsi, 1992; Millhouse et al., 1998; Ross et al., 2001; Ramji, 2002 #1400). Whereas CREB/ATF and C/EBP and/or Fos/Jun and C/EBP dimers influence transcription of many genes, the role of these events in the regulation of StAR gene expression requires further investigation (Fig. 3).
Previous studied have indicated that CREB/CREM, steroidogenic factor 1 (SF-1), GATA-4, C/EBPβ and Fos/Jun binding following cAMP analog treatment occurred in parallel to the association of CBP with the proximal StAR promoter (Hiroi et al., 2004a; Clem et al., 2005; Silverman et al., 2006; Manna and Stocco, 2007). Consequently, the simultaneous interactions of one or more of these factors are involved in trans-activation of the StAR gene. However, competition for CBP binding by CREB and c-Fos/c-Jun and the resulting repression of the StAR gene suggest the involvement of similar regulatory events in diverse physiological responses that require stimulus-transcription coupling (Manna and Stocco, 2007). Regulation of the transcriptional machinery involves the interaction of multi-protein complexes, including enhanceosomes and coactivators. In addition to phosphorylation, protein-protein interactions of non-bZIP factors with bZIP dimers have also been reported to regulate their trans-activation potential and DNA binding activity. This appears particularly important at the StAR promoter where studies have shown that the AP-1 family of proteins, especially c-Fos and c-Jun, can interact with SF-1 and GATA-4 to control StAR gene transcription (Shea-Eaton et al., 2002; Manna et al., 2004). C/EBPβ and GATA-4 are known to cooperate and interact both in vitro and in vivo, and this interaction results in a synergistic activation of the mouse StAR promoter (Silverman et al., 1999; Tremblay et al., 2002; Silverman et al., 2006). Furthermore, SF-1 can physically interact with C/EBPβ, Sp1 and CREB/CREMτ in regulating the expression of the StAR gene (Reinhart et al., 1999; Sugawara et al., 2000; Wooton-Kee and Clark, 2000; Manna et al., 2003a, 2004). Therefore, a multiplicity of transcription factors binding in concert to most, if not all, of the cis-elements of the StAR gene are responsible for fine-tuning the regulatory events associated with StAR gene transcription.
7. CBP/p300 and StAR gene transcription
CBP and its functional homolog p300, are transcriptional coactivators, which harbor multiple functional domains, possess HAT activity, and are known to participate in the activities of hundreds of different transcription factors (Kwok et al., 1994; Bannister and Kouzarides, 1995; Arias et al., 1994; Bannister et al., 1995; Vo and Goodman, 2001). The activation of second messenger pathways trigger the phosphorylation of sequence-specific transcription factors, which can then bind and recruit CBP/p300 (Parker et al., 1996; Fronsdal et al., 1998; Ray et al., 2002; Kovacs et al., 2003). A prime example of this, mentioned in part above, is the ability of CREB to interact with CBP when phosphorylated at Ser133 (Kwok et al., 1994; Chrivia et al., 1993; Parker et al., 1996; Goodman and Smolik, 2000). CBP/p300 do not bind to DNA but rather act as a bridge between the sequence-specific transcription factors and the general transcriptional machinery to promote transcriptional activation (Goodman and Smolik, 2000; Vo and Goodman, 2001). CBP/p300 possess PKA consensus sites and their phosphorylation by PKA has been proposed to be involved in regulating their functions. Studies have demonstrated that CBP/p300 play integral roles in transcriptional regulation of the StAR gene (Fig. 4). In steroidogenic cells, cAMP analog induced phosphorylation of CREB at Ser133, c-Jun at Ser63, and c-Fos at Thr325 was demonstrated to enhance the association and recruitment of CBP/p300 to the proximal StAR promoter (Hiroi et al., 2004a; Clem et al., 2005; Silverman et al., 2006; Manna and Stocco, 2007). Similarly, treatment with cAMP analog results in the phosphorylation of C/EBPβ at Thr 325 and increases the association of C/EBPβ to the proximal StAR promoter (Tremblay et al., 2002). Therefore, in addition to CREB/CREM, Fos/Jun and C/EBPβ, other factors that are bound to the proximal region of the StAR promoter and phosphorylated in response to cAMP signaling (for example, SF-1, GATA-4) may also enhance CBP/p300 recruitment to the StAR promoter. In accordance with this, CBP/p300 have been shown to increase the effects of C/EBPβ and GATA-4 on StAR gene expression (Silverman et al., 2006). It has been reported that DNA binding factors that utilize CBP/p300 as coactivators may function either cooperatively or antagonistically depending upon the identity and conformation of complexes bound to the composite element (Kamei et al., 1996; Blobel et al., 1998; Vo and Goodman, 2001; Silverman et al., 2006; Manna and Stocco, 2007). It was also shown that overexpression of CBP/p300 was able to potentiate the activity of CREB, Fos/Jun, C/EBPβ and GATA-4 in StAR gene transcription, an event that was attenuated by the adenovirus E1A oncoprotein (Silverman et al., 2006; Manna and Stocco, 2007). Thus, CBP/p300 appear to act as integrators with respect to their role in modulating CREB, Fos/Jun, C/EBPβ and GATA-4 mediated StAR gene transcription in the face of activation of diverse signaling pathways. In support of this, CREB and Fos/Jun were found to compete with each other for limiting amounts of intracellular CBP, resulting in the repression of the StAR gene. Furthermore, the interference in transcriptional crosstalk between nuclear receptors and AP-1 has been demonstrated to be associated with relatively low levels of CBP/p300 in different cells (Kamei et al., 1996; Fronsdal et al., 1998). The loss of a single CBP allele, as seen in the human Rubinstein-Taybi syndrome, results in severe developmental defects, illustrating that a small decrease in the concentration of CBP can be deleterious (Petrij et al., 1995). E1A has been shown to be a potent inhibitor of CBP/p300 HAT activity and binds to the same cystein/histidine-rich domain through which several other key regulatory proteins interact (Perissi et al., 1999; Ray et al., 2002; Vo and Goodman, 2001). It is likely that E1A inhibits CBP/p300 and subsequently represses StAR gene transcription either by preventing the HAT activity of CBP/p300, and/or by inhibiting the interaction of CBP/p300 with other transcription factors or with the basal transcriptional machinery.
Fig. 4.
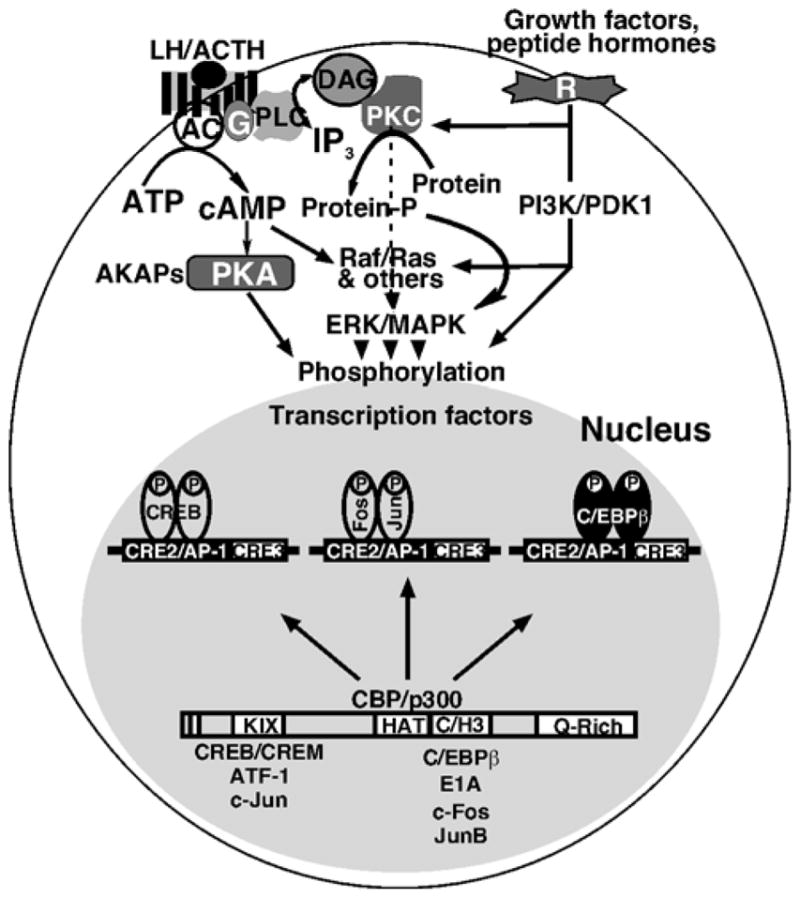
A model illustrating the involvement of CBP/p300 function as a cointegrator of different signaling. The interaction of LH/ACTH with specific receptors results in the activation of G proteins (G), which in turn activate membrane associated adenylyl cyclases (AC) that catalyzes cAMP formation from ATP. cAMP then activates PKA which results in the phosphorylation of transcription factors regulating StAR gene expression. Activation of the PKC pathway results in an increase in the transcription and translation of StAR. Growth factors and peptide hormones bind to specific membrane receptors and can stimulate the steroidogenic response via different signaling pathways. cAMP and/or different factors are capable of activating a cascade of protein kinases including Raf/Ras or related kinases leading to the ERK/MAPK pathway that phosphorylates different transcription factors. Trans-activation of the StAR gene requires simultaneous interaction of CREB/CREM, Fos/Jun and C/EBPβ to the CRE2/AP-1/C/EBPβ overlapping site in the StAR promoter. cAMP-dependent PKA, PKC and other kinases phosphorylate these bZIP proteins, and the subsequent activation of these factors lead to the recruitment of CBP/p300 to the StAR promoter. CBP/p300 act as bridging proteins between the transcription factors and the general transcription machinery, and thus enhance the transcriptional activation of the StAR gene.
8. Conclusions
The cAMP signal transduction pathway regulates a number of physiological processes by directing the activation and DNA-binding capacities of cAMP-responsive transcription factors. In eukaryotes, the dimerization of transcription factors serves as a fundamental mechanism for defining the specificity of DNA-binding and modulating the transcriptional activity of the genes to which these proteins bind. The vast majority of research has focused on the nuclear transcription factors from the bZIP family and underscores the pivotal roles these genes play, and the wide breadth of biological functions that these proteins have on development, differentiation, tumor progression, metabolism, circadian rhythm, learning, memory and stress. The synthesis of steroid hormones is effectively governed by regulating the expression of StAR, thus, it is not surprising that the StAR gene is under extensive regulation by bZIP transcription factors. Considerable progress has been made towards defining the specific bZIP proteins that regulate the StAR gene, elucidating the regions of DNA through which these proteins mediate their effects and on correlating the effects of these transcription factors with the regulatory events observed in StAR gene transcription. It is now evident that in the StAR promoter the CREB/CREM/ATF-1, Fos/Jun, and C/EBP (especially C/EBPβ) families of proteins can bind to a single sequence composed of overlapping recognition sites and that this complex interplay serves a key role in the cAMP-mediated regulation of StAR gene transcription. The involvement of coregulator(s) in the activities of several transcription factors, associated in StAR gene expression, is beginning to come into focus, although much remains unknown. Molecular events directing CREB/CREM, Fos/Jun, and C/EBPβ binding to a single cis-element in the StAR promoter and the concurrent cofactor recruitment provide insights into the convergence of multiple signaling pathways in the regulation of StAR expression. Notably, tissue- and cell-specific abundance of these transcription factors likely have an impact on the transcriptional machinery. Thus, additional studies revealing the biochemical and functional characteristics of these bZIP proteins will further enhance our understanding of the hormonal regulation of StAR gene transcription.
Acknowledgments
The authors wish to thank our many collaborators and the studies of several research groups whose contribution helped in preparing this review. This review was supported in part by NIH grant HD-17481 and with funds from the Robert A. Welch Foundation grant B1-0028.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate C, Baker SJ, Lees-Miller SP, Anderson CW, Marshak DR, Curran T. Dimerization and DNA binding alter phosphorylation of Fos and Jun. Proc Natl Acad Sci USA. 1993;90:6766–6770. doi: 10.1073/pnas.90.14.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- Arias J, Alberts AS, Brindle P, Claret FX, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Kerppola TK, Luk D, Vandenberg MT, Marshak DR, Curran T, Abate C. Jun is phosphorylated by several protein kinases at the same sites that are modified in serum-stimulated fibroblasts. Mol Cell Biol. 1992;12:4694–4705. doi: 10.1128/mcb.12.10.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker O, Parker MG. CAAT/enhancer binding protein is able to bind to ATF/CRE elements. Nucleic Acids Res. 1991;19:1213–1217. doi: 10.1093/nar/19.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Ginty DD, Dudek H, Greenberg ME. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- Bose HS, Sato S, Aisenberg J, Shalev SA, Matsuo N, Miller WL. Mutations in the steroidogenic acute regulatory protein (StAR) in six patients with congenital lipoid adrenal hyperplasia. J Clin Endocrinol Metab. 2000;85:3636–3639. doi: 10.1210/jcem.85.10.6896. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M, Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- Bruder JM, Spaulding AJ, Wierman ME. Phorbol ester inhibition of rat gonadotropin-releasing hormone promoter activity: role of Fos and Jun in the repression of transcription. Mol Endocrinol. 1996;10:35–44. doi: 10.1210/mend.10.1.8838143. [DOI] [PubMed] [Google Scholar]
- Cardinaux JR, Notis JC, Zhang Q, Vo N, Craig JC, Fass DM, Brennan RG, Goodman RH. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron KM, Soo SC, Parker KL. Targeted disruption of StAR provides novel insights into congenital adrenal hyperplasia. Endocr Res. 1998;24:827–834. doi: 10.3109/07435809809032693. [DOI] [PubMed] [Google Scholar]
- Caron KM, Soo SC, Wetsel WC, Stocco DM, Clark BJ, Parker KL. Targeted disruption of the mouse gene encoding steroidogenic acute regulatory protein provides insights into congenital lipoid adrenal hyperplasia. Proc Natl Acad Sci USA. 1997;94:11540–11545. doi: 10.1073/pnas.94.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B, Bocco JL, Goetz J, Gaire M, Lutz Y, Kedinger C. Jun and Fos heterodimerize with ATFa, a member of the ATF/CREB family and modulate its transcriptional activity. Oncogene. 1994;9:375–385. [PubMed] [Google Scholar]
- Chen D, Fong HW, Davis JS. Induction of c-fos and c-jun messenger ribonucleic acid expression by prostaglandin F2alpha is mediated by a protein kinase C-dependent extracellular signal-regulated kinase mitogen-activated protein kinase pathway in bovine luteal cells. Endocrinology. 2001;142:887–895. doi: 10.1210/endo.142.2.7938. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Christenson LK, Johnson PF, McAllister JM, Strauss JF., III CCAAT/enhancer-binding proteins regulate expression of the human steroidogenic acute regulatory protein (StAR) gene. J Biol Chem. 1999;274:26591–26598. doi: 10.1074/jbc.274.37.26591. [DOI] [PubMed] [Google Scholar]
- Christenson LK, Strauss JF., III Steroidogenic acute regulatory protein (StAR) and the intramitochondrial translocation of cholesterol. Biochim Biophys Acta. 2000;1529:175–187. doi: 10.1016/s1388-1981(00)00147-5. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Combs R, Hales KH, Hales DB, Stocco DM. Inhibition of transcription affects synthesis of steroidogenic acute regulatory protein and steroidogenesis in MA-10 mouse Leydig tumor cells. Endocrinology. 1997;138:4893–4901. doi: 10.1210/endo.138.11.5535. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Soo SC, Caron KM, Ikeda Y, Parker KL, Stocco DM. Hormonal and developmental regulation of the steroidogenic acute regulatory protein. Mol Endocrinol. 1995;9:1346–1355. doi: 10.1210/mend.9.10.8544843. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Clem BF, Hudson EA, Clark BJ. Cyclic adenosine 3′,5′-monophosphate (cAMP) enhances cAMP-responsive element binding (CREB) protein phosphorylation and phospho-CREB interaction with the mouse steroidogenic acute regulatory protein gene promoter. Endocrinology. 2005;146:1348–1356. doi: 10.1210/en.2004-0761. [DOI] [PubMed] [Google Scholar]
- De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- Della Fazia MA, Servillo G, Sassone-Corsi P. Cyclic AMP signalling and cellular proliferation: regulation of CREB and CREM. FEBS Lett. 1997;410:22–24. doi: 10.1016/s0014-5793(97)00445-6. [DOI] [PubMed] [Google Scholar]
- Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: down-stream targets and implications for haploid germ cell differentiation. Mol Endocrinol. 1993;7:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004;32:3435–3445. doi: 10.1093/nar/gkh653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Alvania RS, Taparowsky EJ. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol Cell Biol. 2000;20:8613–8622. doi: 10.1128/mcb.20.22.8613-8622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Morlon A, Macho B, De Cesare D, Sassone-Corsi P. Transcriptional cascades during spermatogenesis: pivotal role of CREM and ACT. Mol Cell Endocrinol. 2001;179:17–23. doi: 10.1016/s0303-7207(01)00463-4. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Mellstrom B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Sassone-Corsi P. More is better: activators and repressors from the same gene. Cell. 1992;68:411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Schlotter F, Pevet P, Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature. 1993;362:264–267. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- Fronsdal K, Engedal N, Slagsvold T, Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Grimm SL, Rosen JM. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:191–204. doi: 10.1023/a:1025900908026. [DOI] [PubMed] [Google Scholar]
- Gurney AL, Park EA, Giralt M, Liu J, Hanson RW. Opposing actions of Fos and Jun on transcription of the phosphoenolpyruvate carboxykinase (GTP) gene. Dominant negative regulation by Fos. J Biol Chem. 1992;267:18133–18139. [PubMed] [Google Scholar]
- Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T, Zhao L, Caron KM, Majdic G, Suzuki T, Shizawa S, Sasano H, Parker KL. Developmental roles of the steroidogenic acute regulatory protein (StAR) as revealed by StAR knockout mice. Mol Endocrinol. 2000;14:1462–1471. doi: 10.1210/mend.14.9.0515. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hilberg F, Aguzzi A, Howells N, Wagner EF. C-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Hiroi H, Christenson LK, Chang L, Sammel MD, Berger SL, Strauss JF., III Temporal and spatial changes in transcription factor binding and histone modifications at the steroidogenic acute regulatory protein (stAR) locus associated with stAR transcription. Mol Endocrinol. 2004a;18:791–806. doi: 10.1210/me.2003-0305. [DOI] [PubMed] [Google Scholar]
- Hiroi H, Christenson LK, Strauss JF., III Regulation of transcription of the steroidogenic acute regulatory protein (StAR) gene: temporal and spatial changes in transcription factor binding and histone modification. Mol Cell Endocrinol. 2004b;215:119–126. doi: 10.1016/j.mce.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Hogeveen KN, Sassone-Corsi P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum Fertil. 2006;9:73–79. doi: 10.1080/14647270500463400. [DOI] [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Lu CW, Huang BM, Wu MH, Tsai SJ. Cyclic adenosine 3′,5′-monophosphate response element-binding protein and CCAAT/enhancer-binding protein mediate prostaglandin E2-induced steroidogenic acute regulatory protein expression in endometriotic stromal cells. Am J Pathol. 2008;173:433–441. doi: 10.2353/ajpath.2008.080199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummler E, Cole TJ, Blendy JA, Ganss R, Aguzzi A, Schmid W, Beermann F, Schutz G. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI. SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- Jo Y, King SR, Khan SA, Stocco DM. Involvement of protein kinase C and cyclic adenosine 3′,5′-monophosphate-dependent kinase in steroidogenic acute regulatory protein expression and steroid biosynthesis in Leydig cells. Biol Reprod. 2005;73:244–255. doi: 10.1095/biolreprod.104.037721. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Johnson PF, Williams SC. CCAAT/Enhancer Binding (C/EBP) Proteins. In: Tronche F, Yaniv M, editors. Liver Gene Expression. R. G. Landes Co; Austin: 1994. pp. 231–258. [Google Scholar]
- Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- Jonat C, Stein B, Ponta H, Herrlich P, Rahmsdorf HJ. Positive and negative regulation of collagenase gene expression. Matrix Suppl. 1992;1:145–155. [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kehat I, Hasin T, Aronheim A. The role of basic leucine zipper protein-mediated transcription in physiological and pathological myocardial hypertrophy. Ann N Y Acad Sci. 2006;1080:97–109. doi: 10.1196/annals.1380.009. [DOI] [PubMed] [Google Scholar]
- Kerppola TK, Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kotaja N, De Cesare D, Macho B, Monaco L, Brancorsini S, Goossens E, Tournaye H, Gansmuller A, Sassone-Corsi P. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proc Natl Acad Sci USA. 2004;101:10620–10625. doi: 10.1073/pnas.0401947101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lee KA, Masson N. Transcriptional regulation by CREB and its relatives. Biochim Biophys Acta. 1993;1174:221–233. doi: 10.1016/0167-4781(93)90191-f. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Lin D, Sugawara T, Strauss JF, III, Clark BJ, Stocco DM, Saenger P, Rogol A, Miller WL. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science. 1995;267:1828–1831. doi: 10.1126/science.7892608. [DOI] [PubMed] [Google Scholar]
- Liu JS, Park EA, Gurney AL, Roesler WJ, Hanson RW. Cyclic AMP induction of phosphoenolpyruvate carboxykinase (GTP) gene transcription is mediated by multiple promoter elements. J Biol Chem. 1991;266:19095–19102. [PubMed] [Google Scholar]
- Mahoney CW, Shuman J, McKnight SL, Chen HC, Huang KP. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J Biol Chem. 1992;267:19396–19403. [PubMed] [Google Scholar]
- Manna PR, Chandrala SP, King SR, Jo Y, Counis R, Huhtaniemi IT, Stocco DM. Molecular mechanisms of insulin-like growth factor-I mediated regulation of the steroidogenic acute regulatory protein in mouse leydig cells. Mol Endocrinol. 2006;20:362–378. doi: 10.1210/me.2004-0526. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, Zeleznik AJ, Stocco DM. Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol Endocrinol. 2002;16:184–199. doi: 10.1210/mend.16.1.0759. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Jo Y, Stocco DM. Role of DAX-1 in PKA- and PKC-Mediated Regulation of the Steroidogenic Acute Regulatory Protein Expression in Mouse Leydig Tumor Cells: Mechanism of Action. Endocrinology. 2008 doi: 10.1210/en.2008-0368. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PR, Eubank DW, Lalli E, Sassone-Corsi P, Stocco DM. Transcriptional regulation of the mouse steroidogenic acute regulatory protein gene by the cAMP response-element binding protein and steroidogenic factor 1. J Mol Endocrinol. 2003a;30:381–397. doi: 10.1677/jme.0.0300381. [DOI] [PubMed] [Google Scholar]
- Manna PR, Eubank DW, Stocco DM. Assessment of the role of activator protein-1 on transcription of the mouse steroidogenic acute regulatory protein gene. Mol Endocrinol. 2004;18:558–573. doi: 10.1210/me.2003-0223. [DOI] [PubMed] [Google Scholar]
- Manna PR, Kero J, Tena-Sempere M, Pakarinen P, Stocco DM, Huhtaniemi IT. Assessment of mechanisms of thyroid hormone action in mouse Leydig cells: regulation of the steroidogenic acute regulatory protein, steroidogenesis, and luteinizing hormone receptor function. Endocrinology. 2001;142:319–331. doi: 10.1210/endo.142.1.7900. [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM. Regulation of the steroidogenic acute regulatory protein expression: functional and physiological consequences. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:93–108. doi: 10.2174/1568008053174714. [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM. Crosstalk of CREB and Fos/Jun on a single cis-element: transcriptional repression of the steroidogenic acute regulatory protein gene. J Mol Endocrinol. 2007;39:261–277. doi: 10.1677/JME-07-0065. [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM. The role of JUN in the regulation of PRKCC-mediated STAR expression and steroidogenesis in mouse Leydig cells. J Mol Endocrinol. 2008;41:329–341. doi: 10.1677/JME-08-0077. [DOI] [PubMed] [Google Scholar]
- Manna PR, Wang XJ, Stocco DM. Involvement of multiple transcription factors in the regulation of steroidogenic acute regulatory protein gene expression. Steroids. 2003b;68:1125–1134. doi: 10.1016/j.steroids.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Masquilier D, Sassone-Corsi P. Transcriptional cross-talk: nuclear factors CREM and CREB bind to AP-1 sites and inhibit activation by Jun. J Biol Chem. 1992;267:22460–22466. [PubMed] [Google Scholar]
- McKnight SL. McBindall--a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/s0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- Meyer TE, Habener JF. Cyclic AMP response element binding protein CREB and modulator protein CREM are products of distinct genes. Nucleic Acids Res. 1992;20:6106–6106. doi: 10.1093/nar/20.22.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TE, Habener JF. Cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid- binding proteins. Endocr Rev. 1993;14:269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- Miller WL. Mitochondrial specificity of the early steps in steroidogenesis. J Steroid Biochem Mol Biol. 1995;55:607–616. doi: 10.1016/0960-0760(95)00212-x. [DOI] [PubMed] [Google Scholar]
- Miller WL. Congenital lipoid adrenal hyperplasia: the human gene knockout for the steroidogenic acute regulatory protein. J Mol Endocrinol. 1997;19:227–240. doi: 10.1677/jme.0.0190227. [DOI] [PubMed] [Google Scholar]
- Miller WL. StAR search--what we know about how the steroidogenic acute regulatory protein mediates mitochondrial cholesterol import. Mol Endocrinol. 2007;21:589–601. doi: 10.1210/me.2006-0303. [DOI] [PubMed] [Google Scholar]
- Millhouse S, Kenny JJ, Quinn PG, Lee V, Wigdahl B. ATF/CREB elements in the herpes simplex virus type 1 latency-associated transcript promoter interact with members of the ATF/CREB and AP-1 transcription factor families. J Biomed Sci. 1998;5:451–464. doi: 10.1007/BF02255935. [DOI] [PubMed] [Google Scholar]
- Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Urban J, Sassone-Corsi P, Mayo KE. Gonadotropins regulate inducible cyclic adenosine 3′,5′-monophosphate early repressor in the rat ovary: implications for inhibin alpha subunit gene expression. Mol Endocrinol. 1998;12:785–800. doi: 10.1210/mend.12.6.0126. [DOI] [PubMed] [Google Scholar]
- Nalbant D, Williams SC, Stocco DM, Khan SA. Luteinizing hormone-dependent gene regulation in Leydig cells may be mediated by CCAAT/enhancer-binding protein-beta. Endocrinology. 1998;139:272–279. doi: 10.1210/endo.139.1.5663. [DOI] [PubMed] [Google Scholar]
- Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- O’Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Dasen JS, Kurokawa R, Wang Z, Korzus E, Rose DW, Glass CK, Rosenfeld MG. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc Natl Acad Sci USA. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, Breuning MH. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Piontkewitz Y, Enerback S, Hedin L. Expression of CCAAT enhancer binding protein-alpha (C/EBPα) in the rat ovary: implications for follicular development and ovulation. Dev Biol. 1996;179:288–296. doi: 10.1006/dbio.1996.0258. [DOI] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- Poels J, Vanden Broeck J. Insect basic leucine zipper proteins and their role in cyclic AMP-dependent regulation of gene expression. Int Rev Cytol. 2004;241:277–309. doi: 10.1016/S0074-7696(04)41005-5. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Sherman CT, Lu M, Brasier AR. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol Endocrinol. 2002;16:824–836. doi: 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- Reinhart AJ, Williams SC, Clark BJ, Stocco DM. SF-1 (steroidogenic factor-1) and C/EBP beta (CCAAT/enhancer binding protein-beta) cooperate to regulate the murine StAR (steroidogenic acute regulatory) promoter. Mol Endocrinol. 1999;13:729–741. doi: 10.1210/mend.13.5.0279. [DOI] [PubMed] [Google Scholar]
- Roesler WJ. What is a cAMP response unit? Mol Cell Endocrinol. 2000;162:1–7. doi: 10.1016/s0303-7207(00)00198-2. [DOI] [PubMed] [Google Scholar]
- Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. [DOI] [PubMed] [Google Scholar]
- Roesler WJ, Graham JG, Kolen R, Klemm DJ, McFie PJ. The cAMP response element binding protein synergizes with other transcription factors to mediate cAMP responsiveness. J Biol Chem. 1995;270:8225–8232. doi: 10.1074/jbc.270.14.8225. [DOI] [PubMed] [Google Scholar]
- Ross HL, Nonnemacher MR, Hogan TH, Quiterio SJ, Henderson A, McAllister JJ, Krebs FC, Wigdahl B. Interaction between CCAAT/enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte/macrophage lineage. J Virol. 2001;75:1842–1856. doi: 10.1128/JVI.75.4.1842-1856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert S, Cole TJ, Boshart M, Schmid E, Schutz G. Multiple mRNA isoforms of the transcription activator protein CREB: generation by alternative splicing and specific expression in primary spermatocytes. EMBO J. 1992;11:1503–1512. doi: 10.1002/j.1460-2075.1992.tb05195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg SE, Adams TL, Olive M, Alexander N, Vinson C, Yuspa SH. CRE DNA binding proteins bind to the AP-1 target sequence and suppress AP-1 transcriptional activity in mouse keratinocytes. Oncogene. 1999;18:1569–1579. doi: 10.1038/sj.onc.1202463. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Coupling gene expression to cAMP signalling: role of CREB and CREM. Int J Biochem Cell Biol. 1998;30:27–38. doi: 10.1016/s1357-2725(97)00093-9. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Transcription factors governing male fertility. Andrologia. 2005;37:228–229. doi: 10.1111/j.1439-0272.2005.00701.x. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P, Ransone LJ, Verma IM. Cross-talk in signal transduction: TPA-inducible factor jun/AP-1 activates cAMP-responsive enhancer elements. Oncogene. 1990;5:427–431. [PubMed] [Google Scholar]
- Scobey M, Bertera S, Somers J, Watkins S, Zeleznik A, Walker W. Delivery of a cyclic adenosine 3′,5′-monophosphate response element- binding protein (CREB) mutant to seminiferous tubules results in impaired spermatogenesis. Endocrinology. 2001;142:948–954. doi: 10.1210/endo.142.2.7948. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Shea-Eaton W, Sandhoff TW, Lopez D, Hales DB, McLean MP. Transcriptional repression of the rat steroidogenic acute regulatory (StAR) protein gene by the AP-1 family member c-Fos. Mol Cell Endocrinol. 2002;188:161–170. doi: 10.1016/s0303-7207(01)00715-8. [DOI] [PubMed] [Google Scholar]
- Silverman E, Eimerl S, Orly J. CCAAT enhancer-binding protein beta and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J Biol Chem. 1999;274:17987–17996. doi: 10.1074/jbc.274.25.17987. [DOI] [PubMed] [Google Scholar]
- Silverman E, Yivgi-Ohana N, Sher N, Bell M, Eimerl S, Orly J. Transcriptional activation of the steroidogenic acute regulatory protein (StAR) gene: GATA-4 and CCAAT/enhancer-binding protein beta confer synergistic responsiveness in hormone-treated rat granulosa and HEK293 cell models. Mol Cell Endocrinol. 2006;252:92–101. doi: 10.1016/j.mce.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPβ in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocco DM. The role of the StAR protein in steroidogenesis: challenges for the future. J Endocrinol. 2000;164:247–253. doi: 10.1677/joe.0.1640247. [DOI] [PubMed] [Google Scholar]
- Stocco DM, Clark BJ. Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev. 1996;17:221–144. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- Struthers RS, Vale WW, Arias C, Sawchenko PE, Montminy MR. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non- phosphorylatable CREB mutant. Nature. 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Saito M, Fujimoto S. Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression. Endocrinology. 2000;141:2895–2903. doi: 10.1210/endo.141.8.7602. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Sakuragi N, Minakami H. CREM confers cAMP responsiveness in human steroidogenic acute regulatory protein expression in NCI-H295R cells rather than SF-1/Ad4BP. J Endocrinol. 2006;191:327–337. doi: 10.1677/joe.1.06601. [DOI] [PubMed] [Google Scholar]
- Sun P, Enslen H, Myung PS, Maurer RA. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci USA. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. Protein kinase A and C site-specific phosphorylations of LAP (NF-IL6) modulate its binding affinity to DNA recognition elements. J Clin Invest. 1994;93:2554–2561. doi: 10.1172/JCI117266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Hamel F, Viger RS. Protein kinase A-dependent cooperation between GATA and CCAAT/enhancer-binding protein transcription factors regulates steroidogenic acute regulatory protein promoter activity. Endocrinology. 2002;143:3935–3945. doi: 10.1210/en.2002-220413. [DOI] [PubMed] [Google Scholar]
- Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wagner EF. AP-1--Introductory remarks. Oncogene. 2001;20:2334–235. doi: 10.1038/sj.onc.1204416. [DOI] [PubMed] [Google Scholar]
- Walker WH, Daniel PB, Habener JF. Inducible cAMP early repressor ICER down-regulation of CREB gene expression in Sertoli cells. Mol Cell Endocrinol. 1998;143:167–178. doi: 10.1016/s0303-7207(98)00082-3. [DOI] [PubMed] [Google Scholar]
- Walker WH, Habener JF. Role of transcription factors CREB and CREM in cAMP-regulated transcription during spermatogenesis. Trends Endocrinol Metab. 1996;7:133–138. doi: 10.1016/1043-2760(96)00035-5. [DOI] [PubMed] [Google Scholar]
- Walker WH, Sanborn BM, Habener JF. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc Natl Acad Sci USA. 1994;91:12423–12427. doi: 10.1073/pnas.91.26.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MR. Biochemical diversity of cAMP-dependent transcription of steroid hydroxylase genes in the adrenal cortex. J Biol Chem. 1994;269:27783–27786. [PubMed] [Google Scholar]
- Waterman MR. A rising StAR: an essential role in cholesterol transport. Science. 1995;267:1780–1781. doi: 10.1126/science.7892600. [DOI] [PubMed] [Google Scholar]
- Wilson HL, McFie PJ, Roesler WJ. Different transcription factor binding arrays modulate the cAMP responsivity of the phosphoenolpyruvate carboxykinase gene promoter. J Biol Chem. 2002;277:43895–43902. doi: 10.1074/jbc.M203169200. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Roesler WJ. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity? Mol Cell Endocrinol. 2002;188:15–20. doi: 10.1016/s0303-7207(01)00754-7. [DOI] [PubMed] [Google Scholar]
- Wooton-Kee CR, Clark BJ. Steroidogenic factor-1 influences protein-deoxyribonucleic acid interactions within the cyclic adenosine 3,5-monophosphate-responsive regions of the murine steroidogenic acute regulatory protein gene. Endocrinology. 2000;141:1345–1355. doi: 10.1210/endo.141.4.7412. [DOI] [PubMed] [Google Scholar]
- Wu X, McMurray CT. Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the CREB-binding protein. J Biol Chem. 2001;276:1735–1741. doi: 10.1074/jbc.M006727200. [DOI] [PubMed] [Google Scholar]
- Yivgi-Ohana N, Sher N, Melamed-Book N, Eimerl S, Koler M, Manna PR, Stocco DM, Orly J. Transcription of Steroidogenic Acute Regulatory protein (STAR) in the rodent ovary and placenta: alternative modes of cyclic adenosine 3′, 5′-monophosphate dependent and independent regulation. Endocrinology. 2008 doi: 10.1210/en.2008-0541. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnow CA. CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 2002;4:113–121. doi: 10.1186/bcr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Eferl R, Scheinecker C, Redlich K, Smolen J, Schonthaler HB, Kenner L, Tschachler E, Wagner EF. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res Ther. 2008;10:201–201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Parker MG, Bakker O. Tissue-specific differences in the binding of nuclear proteins to a CCAAT motif in the promoter of the androgen-regulated C3 gene. Mol Endocrinol. 1990;4:1219–1225. doi: 10.1210/mend-4-8-1219. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Sun Z, Means AR, Sassone-Corsi P, Bernstein KE. cAMP-response element modulator tau is a positive regulator of testis angiotensin converting enzyme transcription. Proc Natl Acad Sci USA. 1996;93:12262–12266. doi: 10.1073/pnas.93.22.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwermann O, Braun AC, Lalli E, Sassone-Corsi P, Beuschlein F, Reincke M. Regulation of human MC2-R gene expression by CREB, CREM, and ICER in the adrenocortical cell line Y1. Horm Metab Res. 2007;39:560–566. doi: 10.1055/s-2007-985141. [DOI] [PubMed] [Google Scholar]