Abstract
The NIH Pharmacogenetics Research Network (PGRN) is a collaborative group of investigators with a wide range of research interests, but all attempting to correlate drug response with genetic variation. Several research groups concentrate on drugs used to treat specific medical disorders (asthma, depression, cardiovascular disease, addiction of nicotine, and cancer), whereas others are focused on specific groups of proteins that interact with drugs (membrane transporters and phase II drug-metabolizing enzymes). The diverse scientific information is stored and annotated in a publicly accessible knowledge base, the Pharmacogenetics and Pharmacogenomics Knowledge base (PharmGKB). This report highlights selected achievements and scientific approaches as well as hypotheses about future directions of each of the groups within the PGRN. Seven major topics are included: informatics (PharmGKB), cardiovascular, pulmonary, addiction, cancer, transport, and metabolism.
The explosive and simultaneous development of molecular pharmacology, biotechnology, and genomics is revolutionizing basic principles of drug therapy and development. Although the concept that an individual’s DNA sequence could be an integral determinant of drug therapy has not yet become a standard part of clinical medicine, progress in linking inheritance to drug discovery and therapeutics has been the topic of innumerable basic science reports, clinical reviews, and articles in the lay press. The goal of individualized drug therapy has already had a powerful impact on key funding agencies such as the National Institutes of Health (NIH) (e.g., the NIH Roadmap), regulatory agencies such as the Food and Drug Administration (FDA) (e.g., 2003 Draft Guidance for Industry Pharmacogenomic Data Submissions), and the pharmaceutical and biotechnology industries (e.g., package insert for irinotecan). Nonetheless, the critical element for moving the relationship of drug therapy and genetics closer to the clinical realm is solid scientific evidence and clear advice for the practicing clinician on how to modify dosages or therapies based on the results of pharmacogenetic tests.
The NIH Pharmacogenetics Research Network (PGRN) functions as a collaborative team of multidisciplinary research groups focused on a wide range of scientific questions, but all that attempting to correlate drug response phenotypes with genetic variation. Creating an infrastructure that includes a network of investigators with complementary areas of expertise generates an impetus to share resources, tools, and statistical approaches. The research activity of PGRN encompasses a variety of disorders, including asthma, depression, cardiovascular disease, addiction to nicotine, and cancer. Another major component of the PGRN focuses on specific groups of proteins, membrane transporters, and phase II drug-metabolizing enzymes, which have critical roles in drug absorption and elimination in patients with a wide variety of disease states. Thus, the unifying, fundamental focus of PGRN investigation is pharmacogenetics; aiming to identify the genetic sources of interindividual variability in response to drugs by using and exploiting diverse research strategies. Finally, coordinating this diverse scientific activity into a single, publicly accessible knowledge database, Pharmacogenomics and Pharmacogenetics Knowledge base (PharmGKB),1 remains an ongoing and vital function of the PGRN. Such a knowledge base is likely to attain consistent quality and significant depth with greater diversity than an individual research team.
In drug development and therapy, the role of genetics must be tied to both maximizing effective therapy and avoiding adverse effects; recognizing that variable responses are secondary to many overlapping factors that interact with an individual’s genetic make-up (e.g., age, co-morbidities, drug–drug interactions, environment, diet, etc). Although currently there is a paucity of genetics-guided recommendations, eventually mainstream medicine will include targeted drug therapies that will be prescribed based on genotypic information. In this case, the cost of and morbidity from drug toxicity or side effects will be minimized and efficacy maximized.
Early pharmacogenetic research focused on obtaining DNA from outliers in drug response (e.g., succinylcholine, isoniazid) to identify inherited variation in one or few enzymes that metabolized that particular drug. The sequencing of the entire human genome created the foundation for further studies to identify genetic factors that can be aligned with drug response or toxicity. In the postgenomic era, the ready availability of DNA sequencing via high-throughput analyses has propelled the field of pharmacogenetics beyond recognizing and defining abnormal phenotypes (e.g., unexpected toxicity associated with a “normal” dose of a drug) toward determining underlying molecular mechanisms responsible for pharmacodynamics (PDs) and pharmacokinetics (PKs) of drug therapy. For example, samples from anonymized volunteers are screened for common single-nucleotide polymorphisms (SNPs) and haplotypes (combinations of SNPs within a contiguous segment of DNA), and then these candidate variants are searched within populations expressing an abnormal phenotype. Genome-wide association studies are now possible and can link multiple SNPs and haplotypes to drug response with no a priori assumptions, thereby facilitating new discoveries. Eventually, from such complex data sets, objective data will be gained that will serve as the basis for designing drug therapy based on the specific molecular genetic profile of a patient. However, interpreting this massive amount of data to prospectively guide dosage and drug regimens will require another level of integrated investigation, access to large databases rich in well-defined phenotypic and genotypic data, and systems to maintain patient confidentiality.
PGRN investigators employ three primary strategies to identify genetic factors that associate with drug response. The first strategy involves phenotype–to-genotype studies in which SNPs in candidate genes are associated with variation in drug response including adverse drug reactions. Candidate genes are selected by constructing PK and PD pathway diagrams for the drug (see Figure 1, for statins). SNPs in any gene in the PK or PD pathways are candidates for association with drug response. The second strategy involves genotype-to-phenotype studies. In these controlled studies, individuals with particular genotypes are given drugs and a drug phenotype (e.g., QT interval) is measured to provide powerful in vivo proof that an SNP is associated with a drug response. The third strategy involves whole-genome analysis. Here, investigators perform whole-genome association studies and/or linkage analysis, identifying portions of the genome that contain genetic variants associated with a specific phenotype. Several groups within PGRN use cell lines from the International HapMap project that are rich in genotypic information, whereas others perform these studies on clinical samples from individuals. In some cases, these SNPs or haplotypes may be causative, i.e., they may be responsible for the mechanism of altered drug response. In other cases, the SNPs or haplotypes are in linkage disequilibrium with the actual causative SNP and further studies will be needed to identify the causative SNPs. One important caveat is the problem of multiple testing, resulting in false positives. Therefore, strategies to corroborate initial associations through replication in independent cohorts and/or through assessment of a functional role of the associated SNPs or haplotypes are critical.
Figure 1.
PD and PK pathways of HMGCo A reductase inhibitors (Statins). (a) Cholesterol and lipoprotein transport: genes involved in mediating statin effects on hepatic cholesterol metabolism and consequent effects on plasma lipoprotein transport. Statins inhibit endogenous cholesterol production by competitive inhibition of HMG-CoA reductase (HMGCR), the enzyme that catalyzes conversion of HMG-CoA to mevalonate, an early rate-limiting step in cholesterol synthesis. This pathway delineates genes involved in statin pharmacogenomics, including genes involved in mediating the PD effects of statins on plasma lipoprotein metabolism and those involved in the PKs effects of the drug transport and metabolism. Note the effects of inhibition of HMG-CoA reductase on major aspects of hepatic cholesterol metabolism and selected gene products that can modulate the effects of statins on metabolism and transport of plasma lipoproteins. (b) PKs of Statins: representation of the superset of all genes involved in the transport, metabolism, and clearance of statin class drugs. This figure depicts a generalized view of the PKs of statins, representing the superset of all genes with a reported influence on statin transport and metabolism. Statins are dosed orally and enter the systemic circulation through intestinal cells both passively and by active transport via the ABC and SLC gene family transporters. The major organs of metabolism and elimination include the liver and, to a lesser extent, the kidney. Metabolism is catalyzed by enzymes of the CYP and UGT gene family. The main pathway of elimination is ABC-transporter-mediated biliary excretion. The more hydrophilic compounds (e.g., pravastatin) require active transport into the liver, are less metabolized by the CYP family, and exhibit more pronounced active renal excretion, whereas the less hydrophilic compounds are transported by passive diffusion and are better substrates for both CYP enzymes and transporters involved in biliary excretion. These figures are available at www.pharmgkb.org.
The initial research groups within the PGRN were funded for 5 years beginning in 2000. After competitive renewal, three new projects were added in 2005. The intention of this document is to highlight the achievements and approaches of each of the research groups within the PGRN, including hypotheses about future directions of the various collaborating groups within PGRN. There are seven major topics including informatics (PharmGKB): cardiovascular, pulmonary, addiction, cancer, transport, and metabolism. In this overview, we present goals, findings, and future directions of each of the research groups in the PGRN organized under the seven major topics.
INFORMATICS
PharmGKB
Introduction
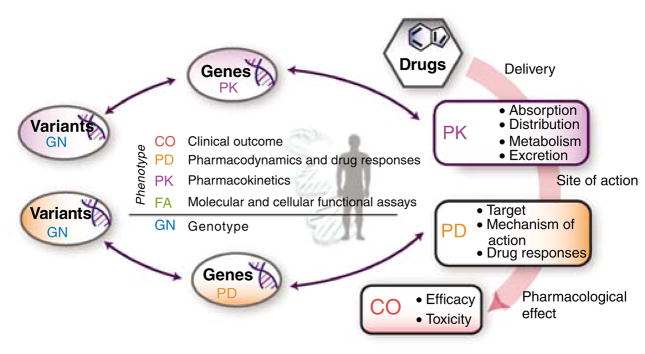
PharmGKB (http://www.pharmgkb.org/) is a publicly available internet research tool1 that curates information to establish knowledge about the relationships among drugs, genes, and disease (Figures 2, 3), as described in detail below. In spite of the wide range of projects included in the PGRN, the PharmGKB serves as a central bank of data that is readily accessible by investigators both within and beyond the PGRN network and as a system to correlate variant data with those in other databases (e.g., dbSNP, HapMap, and jSNP). As a result, PharmGKB provides the scientific and lay community with an integrated knowledge base.
Figure 2.
The home page of PharmGKB provides a straightforward schema for understanding pharmacogenomics. After drugs are delivered, PKs and PDs both involve sets of genes and lead to both efficacious and toxic effects. Variation in response can be related to the PK and PD genes by studying their variations in the human population. All data in the PharmGKB are indexed as being relevant to PK, PD, clinical outcomes (CO), genetic variation (GN), or functional assays at the molecular and cellular level (FN).
Figure 3.
Browsing function in PharmGKB. PharmGKB allows users to browse the major classes of data (genetic variation in pharmacogenes, curated literature, drugs associated with genotype, phenotype, pathway or other information, pathways, diseases, and phenotype data files). The number of data objects in each category is displayed, and there is a full-text search capability to allow more focused searching.
Goals
The PharmGKB strives to establish the definitive source of information about the interaction of genetic variability and drug response in five primary ways. First, primary genotyping data that are important for the PKs or PDs of drugs are stored and presented in an organized and clear format. Second, phenotype measures of drug response at the molecular, cellular, and organismal level are correlated with genotypic data. Third, curating the major findings of the published literature in establishing gene–drug interactions provides easy access to multiples areas of research. For example, more than 1,500 articles in pharmacogenetics can be accessed through PharmGKB. Fourth, providing information about drug response pathways (both PK and PD) allows visual integration of several different projects. Fifth, PharmGKB highlights very important pharmacogenes (VIP genes) that are critical for understanding pharmacogenomics, including information on variant genes, drugs, diseases, and pathways and phenotypes of drug response (Figure 1).
Findings and future directions
PharmGKB provides accurate information about genetic variation in more than 200 genes important for PK or PD (Figure 3). This site hosts more than 35,000 unique internet visitors per month and has more than 2,100 registered users (who gain access to individual level genotype and phenotype data). For example, PharmGKB contains more than 1.2 million individual SNPs measured in at least 13,000 subjects, corresponding to multiple loci in the human genome showing variation. This knowledge base provides visual and spreadsheet mapping of SNPs (Figure 4) and links to other resources, such as dbSNP and vendor genotyping platforms. Currently, the knowledge base is a repository for data on more than 200 genes and their variants and includes information on more than 300 diseases and 400 drugs. Collaborating with investigators in the PGRN, PharmGKB has established novel displays of drug response pathways and specific pages summarizing data about VIP genes that may facilitate research design and data analysis. The site can be reached directly (http://www.pharmgkb.org/search/annotatedGene/index.jsp) or via the main PharmGKB site. As new directions of research and discovery evolve, this site is updated.
Figure 4.
Example of the PharmGKB gene variant browser: nitric oxide synthetase 3 (NOS3). NOS3 is involved in the angiotensin and vascular endothelial growth factor (agents inhibiting the vascular endothelial growth factor signaling pathway have been developed as a new class of anticancer agents) pathways and the response to a number of drugs. The genomic DNA is the thick bar, with exons marked in brown. SNPs in PharmGKB are shown above the genomic DNA with a graphical indication of minor allele frequency. The location of SNPs in dbSNP and jSNP are shown below the genomic DNA. The table shows the chromosomal position, with links to the Golden Path genome browser, dbSNP, and with links to detailed information about the alleles, assays, frequencies, and individual-level data.
PharmGKB is developing as an enhanced system for annotating pharmacogenomic information, both contained within PharmGKB and present in other data resources, in order to allow users to conduct more powerful searches and discovery of relevant datasets. As part of this effort, PharmGKB is conducting studies to understand the primary applications and anticipated requirements of those who frequently access this site. PharmGKB is focusing on integrating, aggregating, and annotating important data sets for pharmacogenomics to catalyze this research, particularly for investigators who are new to the area of genetics of drug response.
PharmGKB can be accessed for in-depth analysis, detailed data sets, and additional references for each of the topics discussed below.
CARDIOVASCULAR
Pharmacogenomic Evaluation of Antihypertensive Responses
Introduction
Hypertension is a common, chronic disease, affecting an estimated 65 million Americans.2 Although five drug classes are available for the first-line treatment of hypertension,3 only about a third of hypertensive patients have their blood pressure (BP) controlled to <140/90 mm Hg.3 The underlying mechanisms for this poor rate of response to antihypertensives are unclear, but one contributory factor is that selecting an agent for an individual is empiric, and any given antihypertensive drug is effective in only about 50% of the population.4 Similar to the response to the wide range of drugs available to treat other complex polygenic diseases such as asthma and various arrhythmias, genetic mechanisms are likely to contribute to the variable response to antihypertensive agents.
Goals
The overriding goal of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEARs) is to identify the genetic determinants of response to two major antihypertensive drug classes, thiazide diuretics and β-adrenergic-receptor blockers. As with other complex, multifactorial diseases, the key tactic is correlating underlying genetic factors with carefully documented clinical phenotypes to design rational, individualized antihypertensive therapy and to base drug development on genetic principles.
Although recent studies have suggested that genetic variability influences the response to hydrochlorothiazide, a thiazide diuretic, many genes seem to contribute to the overall effect. For example, GNB3, WNK1¸ AGTR1, SCNN1G, and NOS35–8 each seem to explain only a small portion (<5%) of the response variability. Clearly, a more in-depth analysis of genes involved in variable response to diuretics is critical before such data can be rationally applied to predict drug response to hydrochlorothiazide.
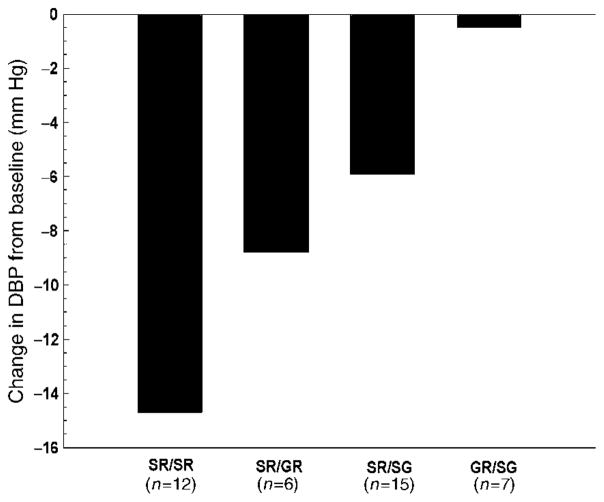
A similar challenge has been identified for β-blockers. The β1-adrenergic receptor (ADRB1) may explain up to 20% of the variable response to a β-blocker.9 For example, two of four diplotype groups responded to metoprolol, but two did not (Figure 5). Clearly, as with the thiazide diuretics, establishing an algorithm that predicts poor or excellent response based on genotype would dramatically promote establishing an antihypertensive regimen tailored to a specific patient. Not only is the genetic contribution to treatment of hypertension incomplete, data about the genetic basis for adverse metabolic responses to diuretics and β-blockers are nonexistent. Metabolic side effects often limit the clinical use of these drugs and contribute to noncompliance. Accurately predicting which hypertensive patients are predisposed to adverse events would also facilitate rational antihypertensive therapy.
Figure 5.
Diastolic blood pressure response to metoprolol in hypertensive patients is predicted by ADRB1 diplotype. Diplotype is defined by the SNPs at codons 49 and 389. SR = Ser49Arg389; SG = Ser49Gly389; GR = Gly49Arg389. Reproduced from Clinical Pharmacology and Therapeutics 37, 44–52 (2003).
Future directions
As PEAR is one of three newly funded centers in the PGRN, a description of approaches, rather than findings, follows. PEAR is a research group primarily focused on an 800-subject clinical study enrolling patients with mild to moderate hypertension, including approximately 45% African Americans. Key aspects of this effort are that all antihypertensive drugs will be discontinued, followed by re-defining the profile of each patient’s hypertension by collecting BP data both at home and at a clinic. Because a high-quality, reproducible phenotype in genetic association studies is critical, focusing on home and ambulatory BP over the clinic BP is especially important in these initial efforts to establish a reliable, less variable phenotype of uncomplicated, mild-to-moderate hypertension.10–12 Along with precisely defining the clinical phenotype, blood and urine samples will be collected for genetic and biomarker studies. Transformed lymphocyte cell lines will be created for other genetic in vitro investigations.
Patients will be randomized to receive either atenolol 50 mg daily or hydrochlorothiazide 12.5 mg daily, with one dose-doubling step, after which the BP profile will be repeated. In those patients whose BP remains greater than 120/70 mm Hg (expected to be >95% of the study population), the other antihypertensive agent will be added, with a similar dose-doubling step for BP >120/70 mm Hg, followed by an additional collection of the BP profile.
The hypothesis in PEAR is that genetic polymorphisms influence the antihypertensive and adverse metabolic responses to atenolol and hydrochlorothiazide. Candidate gene and whole-genome approaches will be combined in an attempt to define a predictive model for response to these two pharmacologically distinct agents. The candidate gene studies will include 70 genes, using a tag SNP approach to identify variability in the gene. The primary BP response phenotype will be defined according to home BP measurements, and the primary metabolic response phenotypes will be insulin sensitivity and change in plasma triglycerides. The BP response–genetic associations will include analyses of responses to monotherapy, add-on therapy, and the combination of the two drugs. This candidate gene approach will be supplemented through genome-wide associations.
Given that the response to β-blockers is strongly correlated with that of angiotensin-converting enzyme inhibitors and angiotensin-receptor blockers, and diuretic response is strongly correlated with calcium channel blocker response, these studies may also provide insights into the genes involved in responses to the other first-line anti-hypertensive drug classes.
Pharmacogenomics and Risk of Cardiovascular Disease
Introduction
Although statins are the most prescribed class of drugs worldwide, and therapy with these drugs is generally associated with a reduction in risk for cardiovascular events by 20–30%, clinical response can be highly variable and adverse drug responses are well described. In fact, as many as 30% of patients do not achieve the lipid-lowering goals set before onset of therapy. Six statins are currently marketed in the United States, including atorvastatin, simvastatin, rosuvastatin, pravastatin, fluvastatin, and lovastatin. These statins all target HMGCoA reductase, but differ in terms of their potencies and PK properties. Dose-limiting side effects include myopathies and liver function abnormalities. Rhabdomyolysis is a rare, but serious adverse effect of this class of drugs.
Goals
The overall objective of the project Pharmacogenomics and Risk of Cardiovascular Disease (PARC) is to identify genetic determinants of the wide range of inter-individual variability in phenotypic and clinical response to statins. A key feature is the use of multiple statin-treated population samples to test the reproducibility and generalizability of findings derived from both candidate gene and genome-wide searches for SNP associations with markers of statin efficacy as well as muscle toxicity.
Findings and future directions
In a group of 944 African-American and European-American subjects, treatment with simvastatin was associated with variable response in levels of lipid and lipoproteins. Of note, European Americans had a greater low-density lipoprotein reduction and a slightly greater increase in high-density lipoprotein. Older subjects, women, and nonsmokers had a greater decrease in low-density lipoprotein cholesterol.13
In this study population, the magnitude of statin-induced low-density lipoprotein cholesterol reduction was associated with a common haplotype in the HMGCoA reductase gene.14 This haplotype also has been reported to be associated with low-density lipoprotein response in a second independent study population, in which a different statin was administered (pravastatin);15 this population is now being further studied in PARC. Such replication of results is not frequently observed in pharmacogenetic studies. Moreover, the minor allele of this haplotype, in combination with a second haplotype in HMGCoA reductase, was found to contribute significantly to reduced statin efficacy in African Americans.13,14 The SNPs in both of these haplotypes are exclusively in non-coding regions, and evidence has been obtained that variation in transcriptional regulation of HMGCoA reductase may underlie this genetic effect on statin response (Medina M and Krauss RM, unpublished).
A genome-wide SNP association study, with replication of significant findings in a total of four study populations treated with various statins, is currently underway to provide a more comprehensive analysis of the contribution of genetic polymorphisms to variation in statin response. Future goals include testing for associations of the most informative SNPs with clinical cardiac end points in several of the largest statin trials. In addition, studies utilizing a genome-wide SNP panel along with candidate gene SNPs are aimed at identifying genetic susceptibility to statin-related myopathy. PARC presents a comprehensive approach for determining effects of specific genotypes on clinically meaningful variations in responsiveness to the class of drugs most widely used to prevent cardiovascular disease.
Pharmacogenomics of Arrhythmia Therapy
Introduction
Sudden cardiac death due to ventricular fibrillation accounts for about 20% of all deaths in US adults, about 400,000/year,16 and atrial fibrillation (AF), a major cause of stroke, affects 2,000,000–5,000,000 Americans.17 In some cases, drugs successfully treat arrhythmias, but the effects of antiarrhythmics are unpredictable in an individual patient; indeed, commonly used antiarrhythmic drugs can themselves elicit fatal rhythm disturbances in some patients.18 One well-described “proarrhythmia” syndrome includes marked prolongation of the QT interval on the surface electrocardiogram. This rhythm is particularly high risk because it can elicit the potentially fatal ventricular arrhythmia, torsades de pointes.19 Marked QT prolongation (and torsades de pointes) has been clearly associated with the well-described congenital long QT syndromes and also has been induced by a variety of antiarrhythmics. In addition, exposure to certain non-cardiovascular drugs (e.g., certain antipsychotics, erythromycin) may also elicit long QT syndrome. These and other causes of sudden cardiac death can be effectively managed by implanting cardioverter/defibrillator devices.20,21 In fact, non-pharmacologic therapies (e.g., implanting a cardioverter/defibrillator device, ablation procedures) have evolved, in part, secondary to the limited efficacy and proarrhythmic side effects of antiarrhythmics. As the concepts of pharmacogenetics evolve to fine-tune the process of individualized drug selection, better pharmacologic options may become available to patients at risk for sudden cardiac death.
Goals
The overall hypothesis of the Pharmacogenomics of Arrhythmia Therapy (PAT) Center of PGRN is that susceptibility to spontaneous cardiac arrhythmias, as well as those induced by exposure to certain drugs, may be associated with inherited polymorphisms in genes involved with this complex phenotype. Defining the genetic associations of antiarrhythmics may allow identification of the high-risk patient, as well as define the most effective and least toxic therapy.
Findings and future directions
Among PAT’s current studies of pharmacogenetics of arrhythmia generation and treatment are two distinct foci: control of the QT interval and studies of AF.
Reduction in cardiac sodium current either by drugs22 or as a result of loss of function in the sodium channel gene, SCN5A,23,24 predisposes to ventricular fibrillation. Recent studies identified multiple variants with altered promoter activity in SCN5A. Notably, a common set of linked polymorphisms was identified in Asian subjects, with a minor allele frequency of ~25%.25,26 The variant haplotype displays markedly reduced promoter activity in vitro, but also predicts QRS duration (an electrocardiogram index of sodium channel function) at baseline and during a challenge with sodium-channel-blocking drugs. Identifying interspecies conserved nucleotide sequences and noting variations in these conserved areas may identify additional polymorphisms in candidate genes that regulate SCN5A expression.
Screening another gene, KCNA5, which regulates human atrial potassium current IKur, has revealed new coding region polymorphisms.27,28 Notably, a polymorphism resulting in P532L (minor allele frequency 2% in African American subjects) in the C terminus generated a potassium current that was unexpectedly associated with resistance to drug therapy. Of note, structural studies identified a probable α-helix in P532L, absent in wild-type channels. Further, structural data support a model where an α-helix impairs access of the drug to a pore-binding site. Variants in KCNA5 are now being described in AF.
Evolving from these studies, the effort of PAT can be divided into three overall goals. First, candidate genes are screened for variants and then function is characterized in heterologous expression systems. Second, the genetic determinants of variability in the response of the QT interval to drug challenge are evaluated. To accomplish this, large numbers of subjects with well-phenotyped responses to QT-prolonging drugs are systematically accumulated. DNA samples are screened for variants in the relevant genes. In addition, new genes modulating the response to challenge with QT-prolonging drugs are being sought in a validated and reproducible assay system using the model organism Danio rerio (zebrafish). Third, genetic determinants of drug response in AF provide another arena to define genetic contribution to a complex clinical phenotype. Two databases are being developed; one including patients and kindreds with AF, and the other including subjects undergoing cardiac surgery, as 20% develop AF postoperatively. These data sets of clinical profile plus DNA samples can then be screened for variants with high-throughput methods. Applying high-throughput genomic analyses to DNA samples that can be correlated to highly characterized clinical data constitutes the major strategy that PAT is pursuing to both identify the genetic component of risk for the various arrhythmias as well as to contribute to rational drug therapy.
Pharmacogenomics of Antiplatelet Therapy Intervention
Introduction
Of those who die suddenly from coronary heart disease (CHD), 50% of men and 64% of women have no previous symptoms. This clinical observation, coupled with an increased appreciation of the interdependence of atherosclerosis, inflammation, and thrombosis, have led to the conclusion that platelet aggregation and thrombosis are major factors leading to vasculo-occlusive or atherothrombotic events. Antiplatelet agents such as aspirin and clopidogrel are effective for primary and secondary prevention of coronary events; decreasing incidence rates of myocardial infarction by approximately 20–25%. However, numerous studies indicate that there is substantial variability in response to antiplatelet agents with up to 30% of subjects considered nonresponders to aspirin,29,30 and 25% of subjects considered to nonresponders to clopidogrel.31–34 The ability to predict which individuals will respond to and which will be resistant to antiplatelet therapy would have a profound impact on the prevention and treatment of CHD and would benefit millions of Americans with CHD or those who are at risk for CHD.
Goals
The mechanism underlying interindividual variability in response to antiplatelet agents is not known. However, evidence supports an important genetic component. The goal of the pharmacogenomics of antiplatelet therapy intervention (PAP) group, one of the three new groups in the PGRN, is to identify specific gene variants that predict response to aspirin and clopidogrel therapy.
Future directions
Studies in pharmacogenomics of anti-platelet therapy intervention are being conducted in the Old Order Amish of Lancaster, Pennsylvania, a genetically homogeneous closed founder population ideal for genetic studies. The Amish Heredity and Phenotype Intervention (HAPI) Heart Study is part of the NHLBI-funded PROgram for GENetic Interaction Network and was designed to examine gene–environment interactions in defining risk for CHD. A total of 868 healthy Amish subjects from large families were recruited for HAPI.
Currently, members of the Amish HAPI Heart Study are being recalled for entry into the pharmacogenomics of antiplatelet therapy intervention study. The study design will evaluate response to clopidogrel alone and clopidogrel plus aspirin. Candidate gene and genome-wide association studies will be conducted to identify the genetic underpinnings of interindividual variation in response to these antiplatelet agents. Specifically, the studies will: (1) determine the frequency and heritability of clopidogrel response and the relationship between clopidogrel resistance and aspirin resistance; (2) exhaustively define sequence variation and haplotype structure of 100 candidate genes and include association analysis of SNPs/haplotypes with platelet function phenotypes; and (3) perform genome-wide association analysis using 500K Affymetrix SNP chips, already available in all HAPI Heart subjects.
The proposed study will provide important genetic, molecular, and mechanistic insights into aspirin and clopidogrel resistance. These insights will lead to diagnostic testing to identify patients who are clopidogrel- and/or aspirin-resistant so that more effective therapies can be prescribed for these individuals. Furthermore, understanding the molecular underpinnings of clopidogrel and aspirin resistance will provide mechanistic insights from which new medications can be designed.
PULMONARY
Pharmacogenetics of Asthma Treatment
Introduction
Although asthma is a common entity affecting an estimated 300 million individuals worldwide,35 a specific etiology has not been identified. Treatment modalities vary and must be adjusted depending on the response of an individual patient. That is, although the diagnosis of “asthma” implies a common constellation of physical symptoms and signs, the disease is a complex syndrome with a spectrum of severity in clinical findings. The response to treatment of asthma is characterized by marked inter- and intraindividual variability36–38 and frequent adverse drug reactions. Available data suggest that genetic factors may contribute as much as 60–80% to the variability in treatment response. Such a disease entity provides a model system, though complicated, to investigate the functional genomics of candidate gene and whole genome associations.
Goals
Pharmacogenetics of Asthma Treatment (PHAT) (http://www.pharmgat.org/) has focused on the hypothesis that genetic determinants of the response to drug therapy for asthma can be identified and studied in model systems, and eventually a prognostic genetic test determining responder vs nonresponder for each class of asthma medication can be clearly defined. Such a project moves from the “classic” monogenic Mendelian inheritance to the more complicated arena of variable, complex phenotypes that characterize asthma and many clinical diseases (e.g., diabetes, hypertension, depression). PHAT studies three classes of drugs: β-adrenergic agonists, inhaled corticosteroids, and leukotriene antagonists.
Findings and future directions
As with many pharmacogenetic investigations, PHAT uses multiple approaches to identify genetic determinants of response to antiasthmatic drugs. Each approach has particular advantages (see Table 1). PHAT initially focused on single candidate genes, or small groups of genes,39–42 including associations of CRHR1, TBX21, and AC9 with response to inhaled corticosteroids. Of significance, more recent efforts have expanded to apply a well-described, family-based screening algorithm to genome-wide association studies.43 The screening technique identifies markers with the highest conditional power for association without biasing any subsequent test statistic.44–46 PHAT investigators have adapted this approach to current studies and are currently evaluating 2,013 SNPs in 220 candidate genes for response to inhaled corticosteroids and β-agonists.
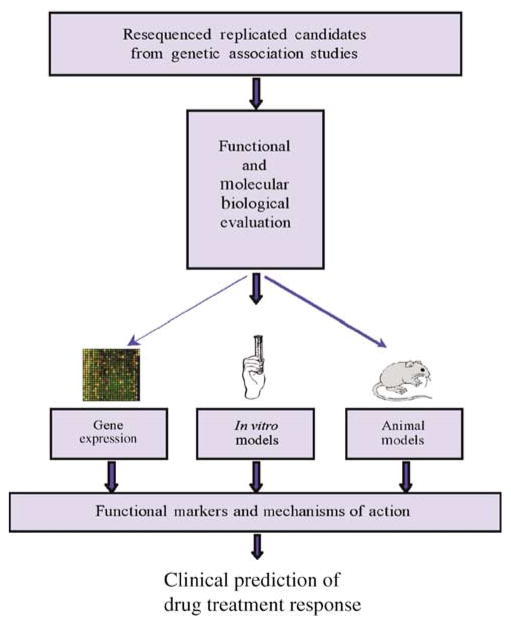
By combining biologic, pathway, and expression array analyses, variants are identified in candidate genes powered for association in a population of 464 European-American trios who were participants in the Childhood Asthma Management Program clinical trial.47 Genes that indicate association with pharmacogenetic phenotypes in Childhood Asthma Management Program are then sequentially explored in a second and third clinical trial population. Next, genes with confirmed replications are completely resequenced and functionally evaluated to characterize the etiologic basis underlying the association. Finally, upon completion of the association testing and functional work, the findings are utilized to develop a predictive model of treatment response, which will be tested on multiple population samples to assess its predictive capabilities (see Figure 6). This approach has been presented in more detail in a recent review.48
Figure 6.
Flow chart for the functional evaluation of genes with replicated associations. Replicated genes from clinical trial populations are explored for functional basis of genetic effects, using gene expression studies, cellular models, and animal models (reprinted from: Pharmacogenomics J. 6, 311–326 (2006).
This approach strives to address many of the potential problems inherent in genetic association studies, including multiple comparisons, lack of statistical power, population stratification, and failure to replicate.48 Moreover, this group of collaborative investigators is well positioned to investigate the molecular biology and functional genomics of any identified associations. To accomplish this, it is essential to explore variations of the screening algorithm (e.g., candidate gene vs candidate SNP approaches) to determine the optimal combination of sensitivity and specificity. Extensions of these methods can be readily applied to planned whole-genome association studies. This current modification to the approach to identifying the genetic contribution to response to pharmacotherapy for asthma marks a striking sophistication to defining the genetics of a complicated phenotype.
ADDICTION
Pharmacogenetics of Nicotine Addiction and Treatment
Introduction
Tobacco use is arguably the most important preventable cause of premature disability and death. Smokers tend to maintain exposure to similar amounts of nicotine, the psychoactive substance in tobacco, from day to day, so as to optimize nicotine-mediated reinforcements and to minimize nicotine withdrawal symptoms. Of note, rapid metabolizers of nicotine smoke more cigarettes, take in more tobacco smoke, and have altered responses to treatment medications, compared with slower metabolizers.49 Nicotine is metabolized to cotinine (COT) primarily via C-oxidation by the liver enzyme CYP2A6. COT is further metabolized to trans-3′-hydroxycotinine (3HC) by the same enzyme. Although the rate of metabolism of nicotine by CYP2A6 is highly correlated with CYP2A6 genotype, variability in nicotine metabolism remains large among smokers with the functional wild-type allele of CYP2A6. That is, most of the individual variability in CYP2A6 activity cannot be fully explained by the current available data about variant alleles of CYP2A6.50 Validating the nicotine metabolic ratio (3HC/COT) has introduced a noninvasive, accurate phenotypic marker of CYP2A6 activity to allow investigation of nicotine metabolism with smoking behavior.
Goals
The Pharmacogenetics of the Nicotine Addiction Treatment (PNAT) group, one of the three new PGRN groups, is investigating the genetic basis of addiction to tobacco and the variation in response to medications used to treat tobacco dependence. Genetic risk contributes to various aspects of smoking, including, for example, persistence in smoking in spite of multiple attempts to stop, as well as the number of cigarettes smoked per day (50% genetic heritability).
Findings and future directions
The impact of four known and common CYP2A6 alleles (*2, *4, *9, *12) have been correlated with in vivo metabolism of nicotine and COT, allowing genotype to predict an average level of nicotine clearance.51 However, in view of the current incomplete knowledge about the role of genotype on nicotine metabolism, novel variants (e.g., *14–*22), regulatory variants (e.g., *1B), and other completely uncharacterized variants are under study in well-characterized cohorts of nicotine delivery.
Designing individualized smoking cessation programs based on the likelihood of success is a long-term goal of the Pharmacogenetics of Nicotine Addiction and Treatment program. The 3HC/COT ratio, measured in the blood of smokers before nicotine transdermal treatment, was predictive of outcome. That is, smokers with a low ratio (slow metabolizers) had a twofold greater smoking cessation response to nicotine patch treatment than smokers with a high ratio (rapid metabolizers), suggesting that rapid metabolizers need higher doses of the nicotine patch.50 In contrast, when given nicotine spray, which is a titratable form, those with genetically slow CYP2A6 used fewer sprays (titrated) and had similar abstinence rates.49,50 Bupropion, a commonly used smoking cessation drug, is metabolized predominantly by CYP2B6. CYP2B6 genotype was significantly correlated with outcome at the end of treatment, which was similar at 6-month follow-up. Of interest, the CYP2B6*4 allele has been associated with an increased 3HC/COT ratio, implying its role in nicotine as well as in bupropion metabolism.
Thus, by focusing on genotyping a large number of candidate genes believed to be involved in nicotine addiction pathways in smokers who have participated in clinical trials of nicotine replacement or bupropion for smoking cessation, PNAT aims to comprehensively evaluate the interaction of genetic factors and pharmacologic interventions via pathway-based Bayesian hierarchical modeling. Ultimately, this modeling may predict individual outcomes so that pharmacologic interventions for smoking cessation treatment can be tailored individually.
CANCER
Pharmacogenetics of Anticancer Agents Research
Introduction
Although research over the last decade has led to new and improved therapies for a variety of different diseases, anticancer drug therapy continues to have unacceptable outcomes, including both poor response and severe toxicity. In addition to the critical need to discover new drugs, it is important to optimize existing drugs to minimize adverse drug reactions and maximize efficacy.
Goals
Pharmacogenetics of Anticancer Agents Research (PAAR) investigators primarily follow a phenotype-driven approach to identify those genetic polymorphisms that are most important for anticancer drug efficacy and adverse events. In particular, the group investigates genetic polymorphisms in both PK and PD pathways of several anticancer drugs. In addition, PAAR focuses on the pharmacogenetics of treatment of childhood acute lymphoblastic leukemia (ALL).
Findings and future directions
PK pathways
Many drugs that effectively treat adult and pediatric tumors are substrates for the enzyme CYP3A, and the efflux transporter P-glycoprotein (ABCB1, MDR1). Cytochrome P450 (CYP) refers to a complex group of enzymes that catalyze the metabolism of a large number of endogenous and exogenous compounds, as well as affect circulating steroid levels and the response to at least 50% of all oxidatively metabolized drugs. More than 50 different CYP genes operate in humans, and these are classified into different families based on sequence homology. The CYP3A subfamily is the most abundant CYP in human liver and small intestine. Furthermore, one member, CYP3A5, is expressed polymorphically, varying with ethnicity, and contributing significantly to the inter-individual and interracial differences in drug responses and clearance. CYP3A5 is also the main CYP3A enzyme expressed in extra-hepatic tissue, such as intestine, kidney, lung, and white blood cells.
Identifying that the genetic basis for polymorphic expression of CYP3A552 is caused by a remarkable intronic SNP that abolishes expression via creation of an alternative splice site dramatically influenced general approaches to pinpointing other molecular mechanisms that explain variability in drug metabolism. That is, linking an alteration in splicing to the expression of a gene (in this case, expression of a specific allele, CYP3A5*1) defined its contribution to total CYP3A as 50%. This discovery identified a specific molecular entity that predicted a patient’s ability to clear substrates of an enzyme and, therefore, also suggested a possible source of drug–drug interactions. In addition to the direct impact on pharmacology of drugs dependent on this metabolic pathway, CYP3A5’s prevalence in renal tissue has led to investigations of its role in the risk of hypertension.53,54
Although the tactic of linking specific variants in a gene to change in function of its product has been productive, in many cases, variability in the gene does not completely account for a phenotype. For example, cis variability in CYP3A or PXR (the primary transcription factor for CYP3A4) genes55,56 does not appear to totally account for CYP3A activity. Instead, the final expression of the gene must be more complex and probably depends on variation in the genotype of trans genes, such as ABCB1 (P-glycoprotein, MDR1). In fact, MDR1 may predict the basal CYP3A4 expression, and in that way, mediate various drug interactions for substances that are substrates of both CYP3A4 and MDR1/P-glycoprotein.57,58
Another PK pathway of major relevance to cancer pharmacotherapy involves glucuronidation, particularly the UGT1 gene, which is alternatively spliced to produce nine different glucouronosyltransferases.59 For example, toxicity after irinotecan, a standard option for relapsed/refractory advanced colorectal cancer, is inversely correlated with glucuronidation of its active metabolite, SN-38, by UGT1A1.60–62 Variability in glucuronidation of SN-38 is associated with a common polymorphism in the promoter of UGT1A1. Of significance, this polymorphism is also associated with the magnitude of myelosuppression coronary to irinotecan.63,64 This critical finding is directly applicable to the clinical realm and promptly led to the FDA incorporating this finding into the package insert of irinotecan. In addition, a 510(k) diagnostic test for UGT1A1 genotyping is now available.
PD pathways
Identifying polymorphisms that explain variability in PDs of anticancer agents is another major aspect of PAAR’s current investigations. For example, cisplatin heritability has been of particular interest. A linkage analysis study of the cytotoxicity of cisplatin using CEPH cell lines identified clear heritability (0.47), with the strongest linkage signal on chromosome 1.65 Further studies are ongoing to identify genes that may be associated with sensitivity and resistance to cisplatin.
A drug target of particular interest to PAAR is the epidermal growth factor receptor (EGFR) that includes an intracellular tyrosine kinase domain, is located on most epithelial cells and malignant tumors of epithelial cell origin, and has a critical role in regulating cell proliferation and differentiation. A class of chemotherapeutic agents has been developed to treat tumors that overexpress EGFR, which include ~30% of various primary cancer of the breast, colon and rectum, lung, prostate, pancreas, head and neck, and ovary. Overexpression of EGFR implies an adverse disease stage, poor prognosis, and higher risk for metastases. In part, the poor outcome is secondary to lack of response to chemotherapy or, after an initial response, development of drug resistance. Of particular challenge in developing rational drug therapy is the fact that the regulation of EGFR expression is complex and incompletely defined. In addition, evidence suggests that overexpression of the EGFR gene contributes not only to variable response to therapy, but also to an increased genetic risk for developing cancer.
Response to EGFR inhibitors (i.e., genitinib and eriotinib) is unpredictable and does not seem to correlate highly with the level of overexpression, and only a small portion of the variability can be attributed to somatic mutations.66,67 The PAAR Group has focused on germline polymorphisms in the promoter region and intron 1 of EGFR, and has identified a common functional SNP in an Sp1-binding site in the promoter.68,69 Of note, binding of Sp1 proteins in the promoter region is essential to EGFR gene transcription, and SNPs in this area have been associated with changes in Sp1 binding affinity that may affect gene expression. Of particular interest to PGRN, marked interethnic variability exists in some of these variants, which may be associated with interethnic variability in response to EGFR inhibitors. Clearly, the molecular mechanisms for variable response as well as resistance to anti-EGFR therapy is multifactorial (e.g., redundant tyrosine kinase receptors, activation of alternative activators, etc.), but the role of SNPs or haplotypes in the promoter region may be critical to individualized therapy with these agents.
ALL
One of the primary disease-oriented pharmacogenetic efforts of the PAAR group is childhood ALL, the most common pediatric malignancy, and a model tumor in that it is highly curable with medications alone.70 Until recently, treatment for ALL was initiated based on the presence of various genetic markers of leukaemia, but without considering genetic variability of the patient. As in other cancers, chemotherapy for ALL involves agents with narrow therapeutic-to-toxic dose ranges and outcome of ALL may be affected by modest changes in doses of effective agents. Clearly, if genetic variation to chemotherapy could be determined, treatment could be optimized to further improve cure while avoiding toxicity. In fact, the PAAR group has identified germline genetic polymorphisms that predict risk for hematologic relapse; the most common cause of failure of treatment in ALL.
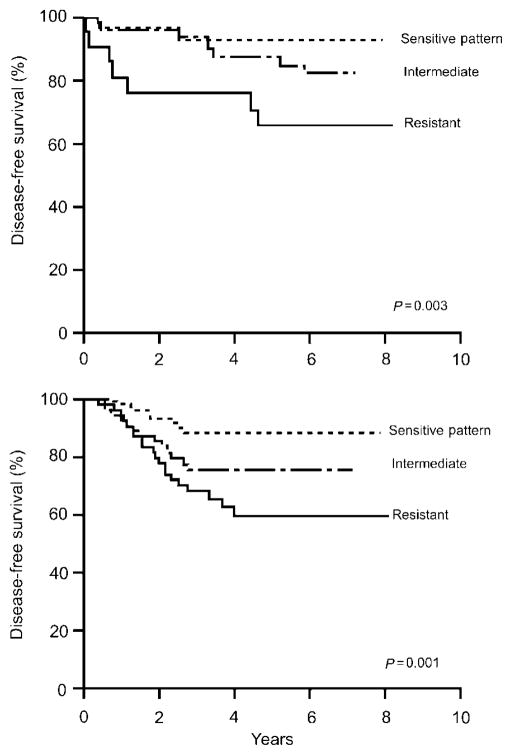
Based on pathways affecting the drugs used to cure the disease, the PAAR group has focused on polymorphisms in genes that code for proteins involved in the metabolism of antileukemic agents. Several specific enzymes (and their genes) have been investigated: glutathione transferase (GSTM1), thymidylate synthase (TYMS), vitamin D receptor (VDR), and thiopurine methyltransferase (TPMT). Polymorphisms that affect GSTM1 and TYMS correlate with the probability of relapse,71 whereas polymorphisms in VDR and TYMS correlate with the risk of one of the primary dose-limiting toxicities of therapy, glucocorticoid-induced osteonecrosis.72 In addition, polymorphisms in TPMT have been associated with drug resistance and may be particularly relevant to the concept of individualized drug dosages to avoid toxicity without compromising efficacy (Figure 7).73 A genome-wide approach has been used to show that germline-variability affects gene expression,74 that modest differences in therapy (e.g., high- vs low-dose methotrexate) cause substantially different effects on gene expression,75 and to identify novel genetic variation that contributes to drug resistance ex vivo, which also predicts long-term relapse risk in vivo in at least two independent clinical trials (Figure 8).76,77 Because fewer than 5% of genes associated with resistance were predicted based on a candidate gene pathway approach, these studies have demonstrated the critical importance of whole-genome approaches to pharmacogenetics.
Figure 7.
Impact of germline TPMT genotype on incidence of toxicity (upper). The lower graph indicated that when dose is individualized based on TMPT germline status, the cumulative incidence (CI) of relapse is not compromised.
Figure 8.
Whole-genome approach to identify genes that predict survival. Genes whose expression predicted in vitro drug sensitivity also predicted probability of disease-free survival in St Jude patients (upper) and independent group of patients treated on Dutch/CoAll studies (lower).
In cancer, one must consider both germline genetic variability and the acquired genetic variability of the target tumor tissue. PAAR group investigators have demonstrated that the acquired karyotype of the malignancy can create a discordance between germline pharmacogenetic genotypes and the genotype of the malignant cells, which can accentuate or diminish the tumor response relative to host genetic variability, depending on the allelic distribution of chromosomal aberrations in the tumor.78 Thus, outcome after treatment of ALL is a function of multiple factors: drug interactions, tumor sensitivity/genetics, and host factors (including germline variants). Of particular interest to the PGRN investigations, genetic factors in pediatric patients with ALL have been identified that in the future may allow therapy to be intensified in particular patients and relaxed in others in direct response to pharmacogenetic profiles.
Consortium on breast cancer pharmacogenomics
Introduction
More than 70% of all breast cancers are estrogen dependent. Studies over the last 120 years have demonstrated that strategies that target the interaction of estrogen with its cellular receptor (the estrogen receptor, ER) result in dramatic reductions in the morbidity, mortality, and incidence of this common disease. Several pharmacologic agents have been shown to be effective endocrine therapies for breast cancer. These include the selective ER modulator tamoxifen and agents that inhibit the aromatization of steroidal precursors into estradiol and estrone (aromatase inhibitors, AIs) in peripheral tissue in postmenopausal women. There are two classes of AIs: triazoles (letrozole, anastrozole) and steroids (exemestane). More recently, a third category of antiestrogens has been studied: the so-called selective ER downregulators, for which fulvestrant is the prototype.
These drugs represent a cornerstone in the treatment of breast cancer, and they also offer an opportunity to study the actions of estrogen in women without breast cancer. The wide variety of pharmacologic strategies to interfere with the estrogen/ER interaction is an ideal situation to study pharmacogenomics. Tamoxifen, the AIs, and fulvestrant are all active against estrogen-receptor-positive breast cancer, but only incompletely, with benefit rates ranging from 30% to 70%. Furthermore, toxicities are highly variable. For example, although hot flashes are a common complaint among treated women, they are not universal (approximately 50–70%). Musculoskeletal complaints occur in approximately 40% of women treated with an AI, and are the reason for drug discontinuation in approximately 10%. Until recently, most investigators focused their studies of the variability of activity on somatic changes within the tumor (such as the presence or absence of ER or associated molecules). Few, if any, studies have, investigated the effects of inherited genetic differences on activity and almost none have addressed toxicities. Translational research studies of inherited differences in genes that alter these drugs to active or inactive metabolites (which represent many different chemical compounds) and in genes that serve as the targets or modulators for these agents (such as the aromatases and/or the ER) are rich areas of opportunity to individualize therapy.
The Consortium on Breast Cancer Pharmacogenomics (COBRA) represents a unique collaboration between the Division of Clinical Pharmacology at Indiana University and five large breast oncology research groups at NCI-funded Comprehensive Cancer Centers: Indiana University, Johns Hopkins University, the Mayo Clinic, University of Michigan, and Baylor College of Medicine. The considerable, practical clinical experience in these centers and the remarkable willingness of patients with breast cancer to contribute to research has been harnessed to enroll a large number of patients willing to contribute genetic information and carefully-curated phenotypes to the PharmGKB. These include a series of serum biomarkers in response to anti-estrogen treatments, PK assessments of AI concentrations, measures of bone density, breast density, hot flashes, and validated diaries designed to record subjective symptoms that document psychiatric symptoms and quality of life changes.
Goals
The goals of COBRA are to facilitate studies that test for associations between variations in candidate genes and response to treatments for breast cancer. Given the strong role that anti-estrogens have historically played in the treatment of breast cancer, COBRA has chosen to focus on genes in the estrogen metabolism and response pathways and those that control the activation, distribution, and elimination of these drugs.
Findings and future directions
The early work of COBRA demonstrated new routes of activation for tamoxifen that are under genetic control and identified a new and active metabolite of tamoxifen, endoxifen. These data showed a strong association between CYP2D6 polymorphisms and plasma concentrations of N-desmethyl 4-hydroxy tamoxifen (“endoxifen”).78 In the laboratory, CYP2D6 was demonstrated to catalyze the metabolism of tamoxifen to endoxifen,79 and endoxifen was shown to be as potent an anti-estrogen as 4-hydroxy-tamoxifen, previously thought to be the active form of tamoxifen.80 Endoxifen and 4-hydroxy-tamoxifen appeared to have almost identical effects as measured by microarray expression analysis.81 Early clinical studies demonstrated that women who carry germline-inactiving SNPs of CYP2D6, such as CYP2D6 *4*4, produce little or no endoxifen after tamoxifen administration. Further, CYP2D6 inhibitors, such as the selective serotonin reuptake inhibitor (SSRIs) paroxetine and fluoxetine,78,82 which are frequently prescribed to breast cancer patients to treat depression and hot flashes, result in lower concentrations of this active metabolite. As a result, patients are now routinely advised to avoid CYP2D6 inhibitors while taking tamoxifen.
To investigate whether this PK observation is clinically important, COBRA has completed accrual to a prospective registry of women initiating tamoxifen therapy to study correlations between several phenotypes (hot flashes, quality of life, bone mineral density, circulating lipids, weight gain) and multiple genotypes (CYP2D6, sulfatases, ER, coactivators, and repressors) will be studied. Importantly, CYP2D6 status will be evaluated to identify any association with tamoxifen activity against breast cancer.
COBRA investigators have demonstrated that genotyping CYP2D6 from archived formalin-fixed, paraffin-embedded breast cancer tissue and from white blood cells from the same individuals produced identical results. These data were the basis of a retrospective collaboration with PGRN-funded investigators from the Mayo Clinic. That is, DNA from participants enrolled in an earlier prospective trial of tamoxifen was utilized in an investigation that suggested that poor metabolizers of tamoxifen (several variants of CYP2D6) have greater rates of disease recurrence in the adjuvant setting83 and that CYP2D6 inhibitors mimic this effect.84,85 The FDA is considering changing the label of tamoxifen to include the link between CYP2D6 genotype and activation/response to tamoxifen. Although these data are clinically significant for certain patients, investigators in COBRA are sensitive to the concern that this single retrospective study should not lead to the general practice of withholding tamoxifen in favor of other drugs, such as the AIs. This newer class of agent has a different mechanism of action, distinct activity and toxicity profiles, and cannot be used in premenopausal women. Thus, COBRA has initiated a large number of collaborations with the NCI cooperative groups and with the large international trial community to validate the utility of CYP2D6 genotyping for patients considering tamoxifen therapy using archived formalin-fixed, paraffin-embedded tissue.
The increasing importance of the AIs in the treatment and prevention of breast cancer is clear. Thus, in addition to the ongoing prospective and retrospective studies of tamoxifen, COBRA has initiated a prospective randomized clinical trial to compare the pharmacogenomics of agents from the two classes of AIs (letrozole and exemestane) in postmenopausal breast cancer patients in the adjuvant setting. DNA will be collected to perform pharmacogenetic studies that can then be correlated with a large number of curated phenotypes to potentially identify biomarkers of response. Moreover, COBRA is collaborating with other investigators to study the pharmacogenomics of selected candidate genes involved in metabolism and activity of anastrozole and fulvestrant.
Comprehensive research on expressed alleles in therapeutic evaluation
Introduction
Similar to most common diseases, the clinical response to a medication is under the control of a network of genes (i.e., polygenic trait), each contributing to the patient’s phenotype, in this case, drug response. Comprehensive Research on Expressed Alleles in Therapeutic Evaluation (CREATE) aims to identify common polymorphisms in various drug response pathway genes most relevant to association studies, concentrating on the multispecies conserved sequences. Then, by using computational biology approaches to evaluate the identified variants, the goal is to predict functional importance. The identified variants are correlated with common ethnic/racial groups.
Goals
A central challenge in pharmacogenetic studies is selecting the “right” genes to study to improve clinical decision-making in the setting of variability in response to drug therapy. Constructing drug response pathways has provided a framework to prioritize candidate gene investigations as well as a method to then increase the comprehensive impact of SNP/haplotype discovery on maximizing appropriate drug choice across diverse groups of patients. To accomplish its investigations, CREATE has interacted with several groups of the PGRN network: PAAR, Pharmacogenetics of Membrane Transporter (PMT), and COBRA.
Findings and future directions
The concept that multiple subpopulations exist within a cohort of patients with a diagnosis of breast or colon (and other) cancer is well recognized. However, focusing on pathways of pharmacologically relevant proteins to define drug response in such clinical settings is novel. Three variations of this emphasis on pathways rather than individual genes have led to innovative views of individualized therapy for cancer by the CREATE group. First, the CREATE group recently evaluated the irinotecan (topoisomerase I inhibitor used to treat colorectal, lung, cervical, and esophageal cancers) pathway in a spectrum of common tumors. Of note, irinotecan’s metabolism is complicated, with contributions from a wide variety of enzymes that define not only efficacy, but also toxicity. After studying 255 tumors with 11 predefined markers, two distinct patient groups were identified.86 The data demonstrate that patients with lymphoma, melanoma, and brain, colon, breast, as well as ovarian, and prostate cancer have the same pharmacologic profile with distinct homogeneity in sensitivity to irinotecan. This sets the stage for further investigation into anatomy-independent therapy of cancer.
A second view of pharmacologic profiles was established in a study of polymorphism discovery in 51 chemotherapy pathway genes of nine commonly used anticancer agents, sampling three different ethnic populations (African American, Asian Americans, and European Americans).87 Of note, 346 novel variants were identified in this focused resequencing project. From these data, a more comprehensive set of polymorphisms was generated that will be important to consider in defining therapeutic response and toxicity targeted as a function of specific ethnic groups.
The third focus on pathways evaluated DNA repair and microsatellite instability in one specific cancer, Dukes’ C colorectal.88 Instead of evaluating a specific DNA repair gene, the mRNA expression level of 20 DNA repair pathway genes were evaluated together. The variability identified again emphasizes that predicting outcome and response to chemotherapy results from the interplay of multiple genes. Thus, these and future investigations of pharmacologic pathways advance the possibility for selecting the “best” patients for a given drug or combination therapy, increasing efficiency, and decreasing toxicity for groups of patients as well as for individuals with variant genetics.
TRANSPORT
This section describes the Pharmacogenetics of Membrane Transporters (PMT) project and the Genetics of Response to Antidepressants (GRAD) project, a subproject within PMT.
Pharmacogenetics of Membrane Transporters
Introduction
Membrane transporters are of great pharmacological importance as they provide the target for many commonly used prescription medications and are a major determinant of the absorption, distribution, and elimination of a large number of clinically used drugs. In humans and other mammals, there are two major superfamilies of membrane transporters: (1) the solute carrier superfamily (SLC); and (2) the ATP-binding cassette superfamily (ABC). SLC transporters are primarily facilitated influx pumps, whereas ABC transporters are ATP-dependent efflux pumps.89,90 In many cases, SLC and ABC transporters work in concert to regulate systemic and intracellular drug levels.
Goals
The overall goal of the PMT project is to understand how genetic variation in membrane transporters contributes to variation in drug response. The first step in the genotype-to-phenotype strategy used by PMT investigators is to identify common and rare sequence variants in SLC and ABC membrane transporter genes in ethnically diverse populations. Nonsynonymous and promoter sequence variants are then phenotypically characterized in cellular systems. Finally, the biological relevance of functionally significant membrane transporter variants in determining drug response is investigated.
Findings and future directions
Deep resequencing of the coding and flanking intronic regions of almost 50 membrane transporter genes by the PMT project has led to the discovery of many new variants in ethnically diverse US populations. Statistical genetic analysis of membrane transporter sequence variant data indicates that for the majority of transporters there is a three- to fourfold enrichment of variants at synonymous sites than at nonsynonymous sites and that high-frequency variants are less likely to change an evolutionarily conserved amino acid in the transporter sequence.91,92 These findings suggest that there is selective pressure against significant changes in transporter sequence, and therefore function, and support an important role for ABC and SLC membrane transporters in human fitness.
PMT investigators have pioneered efforts to functionally characterize amino-acid variants of membrane transporters. The functional analysis of 88 protein-altering variants of transporters from the SLC22A, SLC28A, and SLC29A families has provided insight into the effect of the alteration of an amino acid on the function of a transport protein.93 Twenty two variants resulted in more than a 40% decrease in transport function, with 14 of these showing an almost complete loss of function (<20%). Both the degree of chemical change of the amino acid and evolutionary conservation predicted the impact of a protein-altering variation on transporter function. An allele frequency distribution that is skewed toward lower frequencies was found for variants that decreased function, as well as for variants at evolutionarily conserved sites that retain function, providing strong support of selective pressure.93 An important consideration for using these functional data in clinical association studies is that substrate-specific effects on transporter function were noted for several variants.93
Resequencing and cellular phenotyping efforts by PMT investigators are continuing, with a current emphasis on understanding, the extent and functional significance of genetic variation in non-coding regions, especially promoter regions of membrane transporter genes. Computational methods are also being developed to aid in the prediction of the functional consequences of amino acid-altering variants. The important question of whether genetic polymorphisms in membrane transporters influence drug response or toxicity is an emerging focus of PMT. Clinical questions are being addressed both in genotype-to-phenotype studies in healthy subjects and in phenotype-to-genotype studies in relevant patient populations. An increased understanding of the role of membrane transporters in drug response pathways will emerge from these continued efforts by PMT investigators and their PGRN colleagues.
Genetic Responses to Antidepression
Introduction
Medications belonging to the selective sera-tonic reuptake inhibitor (SSRI) class of antidepressants are highly effective in the treatment of depression, although 30–40% of individuals with depression fail to respond to the first SSRI agent that is prescribed. Effectiveness is assessed after a trial of 6–8 weeks of pharmacotherapy, during which interval the individual is at risk for suicide and other negative outcomes associated with major depression. Currently, there are no reliable demographic or genetic predictors of response to these medications.
Goals
The goals of the GRAD study during the first period of funding of the PGRN were to examine potential associations between both therapeutic response to and side effects of SSRI antidepressants and genetic variation in monoamine membrane transporters, as well as other candidates directly involved in serotonin production, transmission, or response. A prospective study population of 1,025 adults initiating pharmacotherapy with fluoxetine or paroxetine for treatment of unipolar depression was recruited from Kaiser Permanente psychiatric and primary care clinics in Northern California. Standardized interviews confirmed DSM-IV major depression and assessed baseline depression severity, side effects, and potential covariates. Follow-up was conducted after 4 and 8 weeks to observe treatment response. The sample was ethnically diverse; African Americans and Hispanics comprised 15% each of the sample, Asians and “Others” comprised 5% each, and non-Hispanic Whites were 60% of the sample. DNA samples from the patients were genotyped for functional polymorphisms in candidate genes involved in serotonin neurotransmission, focusing particularly on variants in the serotonin transporter (SLC6A4), which is the target of SSRIs. Of particular interest was a common insertion/deletion polymorphism in the promoter region of the serotonin transporter (5-HTTLPR) that had previously shown mixed results in studies of antidepressant response. The two common alleles of 5-HHTLPR are 14 (or s (short)), and 16 (or l (long)), with the l allele associated with increased transcription.94,95
Findings and future directions
There were no differences in therapeutic response by medication or ethnic group. In linear regression analyses of change in symptoms of depression from baseline to 8 weeks following initiation of therapy, the serotonin transporter-linked promoter region polymorphism (5-HTTLPR) was not significantly associated with treatment response (P = 0.06). However, in multivariable logistic regression analysis that compared those who responded initially but relapsed before 8 weeks (relapse or placebo response – 9% of sample) with those who had a sustained or improving response at 8 weeks (sustained response – 72%), both the 16/16 (l/l) homozygote and 14/16 (s/l) heterozygote genotypes were associated with a twofold odds of sustained response to treatment (95% confidence intervals excluded 1.0) compared with the 14/14 (s/s) genotype. Among African Americans, the s/l and l/l genotypes were associated with over five times the likelihood of sustained response compared with the s/s genotype. No association was found between 5-HTTLPR genotype and sustained response vs nonresponse to medication (i.e., no significant reduction in symptoms of depression). There were no other significant associations between therapeutic response to SSRIs and variants in other candidate genes. These results suggest that a common functional variant in the serotonin membrane transporter may be associated with greater likelihood of a sustained therapeutic response to fluoxetine and paroxetine, two common SSRI medications.
Future directions for the GRAD study include new efforts to identify candidate genes that affect therapeutic response and side effects in the GRAD sample. Further, all GRAD participants will be contacted and a standard interview conducted to determine the effects of antidepressant treatment over the intervening 4–6 years following acute therapy. In this way, GRAD may be able to identify potential genetic determinants of the longer-term neurotrophic response to antidepressants reported in recent studies.
METABOLISM
Pharmacogenetics of Phase II Drug Metabolizing Enzymes
Introduction
Metabolism is often a critical component of the final clinical effect of a drug. That is, metabolism often converts drugs to more water-soluble compounds that are more easily excreted. In addition, in some cases, a drug must be metabolized to become therapeutically active. Two categories of metabolic pathways are commonly described: phase I (oxidation, reduction, hydrolysis) and phase II (conjugation reactions, such as glucuronidation, sulfation, and methylation). Many of the well-described pharmacogenetic traits have involved drug metabolism (e.g., N-acetylation and the NAT2 gene). Pharmacogenetics of Phase II Drug Metabolizing Enzymes (PPII) has focused on Phase II enzymes. Early studies resulted in the molecular cloning and characterization of genes encoding a series of methyl-transferase and sulfotransferase enzymes. The genetic polymorphisms in the prototypic pharmacogenomic and clinically relevant thiopurine S-methyltransferase gene (TPMT)96 were correlated with low and high activity of the enzyme and have been shown to directly impact the clinical use of thiopurine drugs (e.g., 6-mercaptopurine and azathioprine) and to vary among ethnic groups. Of major significance is the fact that clinically applicable pharmacogenetic tests for the TPMT polymorphism were developed and are being widely applied in medical practice.
Goals
The PPII has applied a genotype-to-phenotype strategy that involves resequencing of genes encoding proteins that catalyze phase II drug metabolism to fully identify variant alleles. A major focus for PPII has been characterizing both the functional implications and mechanisms responsible for the effects of nonsynonymous coding SNPs. These SNPs alter the amino-acid sequence of the encoded protein and are a common cause of pharmacologically relevant functional variation.
Findings and future directions
PPII has now identified and functionally characterized many polymorphisms, including coding SNPs in scores of genes and has studied their functional implications. Most often, altered function of genetically variant allozymes is due to an alteration in protein quantity.97–100 A striking example of this phenomenon is provided by the allozyme encoded by the most common variant allele for TPMT (TPMT*3A). Decreased levels of this allozyme have been shown to result from a variety of mechanisms including aberrant folding and accelerated degradation.101–103 The accelerated degradation of the protein product of the most common variant allele has been shown to be associated, at least in part, with molecular chaperones such as hsp90 and hsp70 that play a role in protein folding and targeting misfolded proteins for degradation.102 TPMT*3A has also been shown to aggregate and form “aggresomes”.103
Recently, PPII has used a yeast genetic system to identify a series of genes involved in trafficking and targeting TPMT*3A for degradation and aggregation. These yeast proteins can be used to identify mammalian homologs to better characterize pathways involved in the degradation and aggregation of TPMT*3A and, perhaps, other genetic polymorphisms of pharmacogenetic importance.
Finally, two major translational pharmacogenomic studies comprise a major PPII effort that has evolved from the gene sequencing effort. One is focused on anastrozole, an Aromatase Inhibitor that is used to treat breast cancer. These studies are being performed in collaboration with the MD Anderson Cancer Center, Houston, Texas and with the COBRA PGRN Center at Indiana University, Indiana. The other translational study involves escitalopram, a selective serotonin inhibitor, and is based on a collaboration between the Department of Psychiatry at the Mayo Clinic and the University of Arkansas, Little Rock, Arkansas.
This series of studies demonstrates the complementary nature of basic and translational pharmacogenomic science, and that the application of discovery science can lead to mechanistic hypothesis-based studies that serve to increase our understanding of biologic mechanisms responsible for clinically relevant pharmacogenomic effects of proteins such as TPMT.
SUMMARY
The PGRN comprises a group of investigators with various approaches to ultimately identify genetic variants that predispose an individual to nonresponse to or toxicity from drugs. The collective expertise of the network allows for synergy in developing methods and in populating a knowledgebase in pharmacogenomics. Research in the PGRN is necessarily diverse to allow for pharmacogenetic studies of drug therapies across multiple diseases. Thus, the impact of the PGRN on the field of pharmacogenetics is broad. In this overview, we have presented the goals and major findings of each of the groups in the PGRN and described the data and knowledge contained in the collaborative knowledge base, PharmGKB. The application of pharmacogenetics to clinical practice will advance only in the wake of strong, mechanistic research from multiple groups.104 Research in the PGRN has already contributed to and will continue to advance the field of pharmacogenetics.
Table 1.
Advantages of current asthma pharmacogenetic study design
| Technique | Advantage | How implemented |
|---|---|---|
| Screening algorithm | Helps address multiple comparisons issues | Screen using non-informative families before performing FBAT analysis |
| Family-based association test | Population stratification is not an issue | FBAT testing methodology using probands and parents |
| Principal components analysis | Increases statistical power | Each longitudinal outcome measure contributes to the observations |
| Replication population testing | Helps to assure generalizability and address false positives | Two population-based replication studies for each gene identified via screening, before confirmation |
| Functional studies | Extends association studies into the functional domain | Use of gene expression, in vitro, and animal models to test specific candidate genes and variants |
Acknowledgments
Sites in the Pharmacogenetics Research Network are supported by the following UO1 awards: GM61373, GM61390, GM74492, HL69757, GM61393, GM63340, HL65962, GM74518, GM61388, GM61374, DA20830, and HL65899.
Footnotes
CONFLICT OF INTEREST
D.M. Roden (GlaxoSmithKline, Pfizer, Inc., AstraZeneca, Abbott Laboratories, Novartis, 1st Genetic Trust), R.M. Krauss (Abbott Laboratories, AstraZeneca, Bristol-Myers Squibb, Merck & Co. Inc., Pfizer Inc., International Dairy Foods Association), M.J. Ratain (Prometheus, Genzyme Corp., Genentech); S.T. Weiss (Glaxo-Wellcome, Roche Pharmaceuticals, Millennium Pharmaceuticals, Genentech, Shering-Plough, Variagenics, Genome Therapeutics, Merck Frost). Stock ownership or operations (other than mutual funds): M.J. Ratain (Variagenics, Nuvelo, Applera). Grants received: D.M. Roden (1st Genetic Trusty), R.M. Krauss (King, Merck Schering Plough, Pfizer Inc.), S.T. Weiss (Glaxo-Wellcome, AstraZeneca, Pfizer). Patents received: M.J. Ratain (National Institutes of Health), M.V. Relling (National Institutes of Health).
References
- 1.Klein TE, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 2.Fields LE, et al. The burden of adult hypertension in the United States 1999–2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 3.Chobanian AV, et al. The seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: The JNC 7 Report. JAMA. 2003;289:2560–2571. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 4.Materson BJ, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [see comments] [published erratum appears in N Engl J Med 330, 1689 (1994)] [DOI] [PubMed] [Google Scholar]
- 5.Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, alpha-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16:834–839. doi: 10.1016/s0895-7061(03)01011-2. [DOI] [PubMed] [Google Scholar]
- 6.Frazier L, Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. Multilocus effects of the renin–angiotensin–aldosterone system genes on blood pressure response to a thiazide diuretic. Pharmacogenomics J. 2004;4:17–23. doi: 10.1038/sj.tpj.6500215. [DOI] [PubMed] [Google Scholar]
- 7.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. C825 T polymorphism of the G protein beta(3)-subunit and antihypertensive response to a thiazide diuretic. Hypertension. 2001;37(Part 2):739–743. doi: 10.1161/01.hyp.37.2.739. [DOI] [PubMed] [Google Scholar]
- 8.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. WNK1 kinase polymorphism and blood pressure response to a thiazide diuretic 10.1161/01.HYP.0000186240.81996.57. Hypertension. 2005;46:758–765. doi: 10.1161/01.HYP.0000186240.81996.57. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JA, et al. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 10.Mancia G, Omboni S, Parati G, Ravogli A, Villani A, Zanchetti A. Lack of placebo effect on ambulatory blood pressure. Am J Hypertens. 1995;8:311–315. doi: 10.1016/0895-7061(94)00250-F. [DOI] [PubMed] [Google Scholar]
- 11.Finkielman JD, Schwartz GL, Chapman AB, Boerwinkle E, Turner ST. Lack of agreement between office and ambulatory blood pressure responses to hydrochlorothiazide. Am J Hypertens. 2005;18:398–402. doi: 10.1016/j.amjhyper.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 12.Beitelshees AL, Zineh I, Yarandi HN, Pauly DF, Johnson JA. Discordant beta-blocker effects on clinic, ambulatory, and exercise hemodynamics in patients with hypertension. Pharmacotherapy. 2006;26:1247–1254. doi: 10.1592/phco.26.9.1247. [DOI] [PubMed] [Google Scholar]
- 13.Simon JA, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;15:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 14.Krauss RM, et al. Haplotypes in the HMGCoA reductase gene influence plasma LDL level and LDL response to statin in African Americans and Caucasians. Circulation Supplement II. 2005;112:725. (Abstract) [Google Scholar]
- 15.Chasman DI, et al. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 16.Zheng ZJ, Croft JB, Giles WH, Mensah GA. Sudden cardiac death in the United States, 1989–1998. Circulation. 2001;104:2158–2163. doi: 10.1161/hc4301.098254. [DOI] [PubMed] [Google Scholar]
- 17.Wattigney WA, Mensah GA, Croft JB. Increased atrial fibrillation mortality: United States, 1980–1998. Am J Epidemiol. 2002;155:819–826. doi: 10.1093/aje/155.9.819. [DOI] [PubMed] [Google Scholar]
- 18.Roden DM. Proarrhythmia as a pharmacogenomic entity: a critical review and formulation of a unifying hypothesis. Cardiovasc Res. 2005;67:419–425. doi: 10.1016/j.cardiores.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 19.Roden DM. Drug-induced prolongation of the QT Interval. N Engl J Med. 2004;350:1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 20.Mcanulty J, et al. A comparison of antiarrhythmic-drug therapy with implantable defibrillators in patients resuscitated from near-fatal ventricular arrhythmias. N Engl J Med. 1997;337:1576–1583. doi: 10.1056/NEJM199711273372202. [DOI] [PubMed] [Google Scholar]
- 21.Moss AJ, et al. Prophylactic implantation of a defibrillator in patients with myocardial infarction and reduced ejection fraction. N Engl J Med. 2002;346:877–883. doi: 10.1056/NEJMoa013474. [DOI] [PubMed] [Google Scholar]
- 22.The CAST Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med. 1989;321:406–412. doi: 10.1056/NEJM198908103210629. [DOI] [PubMed] [Google Scholar]
- 23.Chen QY, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 24.Bezzina C, et al. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 25.Yang P, Kupershmidt S, Roden DM. Cloning and initial characterization of the human cardiac sodium channel (SCN5A) promoter. Cardiovasc Res. 2004;61:56–65. doi: 10.1016/j.cardiores.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Yang P, et al. DNA polymorphisms with variant in vitro function are common in the cardiac sodium channel promoter. Circulation. 2004 [Google Scholar]
- 27.Simard C, Drolet B, Yang P, Kim RB, Roden DM. Polymorphism screening in the cardiac K+ channel gene KCNA5. Clin Pharmacol Ther. 2005;77:138–144. doi: 10.1016/j.clpt.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Drolet B, Simard C, Mizoue L, Roden DM. Human cardiac potassium channel DNA polymorphism modulates access to drug-binding site and causes drug resistance. J Clin Invest. 2005;115:2209–2213. doi: 10.1172/JCI23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKee SA, Sane DC, Deliargyris EN. Aspirin resistance in cardiovascular disease: a review of prevalence, mechanisms, and clinical significance. Thromb Haemost. 2002;88:711–715. [PubMed] [Google Scholar]
- 30.Eikelboom JW, Hankey GJ. Aspirin resistance: a new independent predictor of vascular events? J Am Coll Cardiol. 2003;41:966–968. doi: 10.1016/s0735-1097(02)03013-9. [DOI] [PubMed] [Google Scholar]
- 31.Gum PA, Kottke-Marchant K, Welsh PA, White J, Topol EJ. A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J Am Coll Cardiol. 2003;41:961–965. doi: 10.1016/s0735-1097(02)03014-0. [DOI] [PubMed] [Google Scholar]
- 32.Lau WC, et al. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation. 2004;109:166–171. doi: 10.1161/01.CIR.0000112378.09325.F9. [DOI] [PubMed] [Google Scholar]
- 33.Matetzky S, et al. Clopidogrel resistance is associated with increased risk of recurrent atherothrombotic events in patients with acute myocardial infarction. Circulation. 2004;109:3171–3175. doi: 10.1161/01.CIR.0000130846.46168.03. [DOI] [PubMed] [Google Scholar]
- 34.Gurbel PA, Bliden KP, Hiatt BL, O’Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 35.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 36.Malmstrom K, et al. Oral montelukast, inhaled beclomethasone and placebo for chronic asthmaA randomized controlled trial, Montelukast/Beclomethasone Study Group. Ann Intern Med. 1999;130:487–495. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- 37.Szefler SJ, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002;109:410–418. doi: 10.1067/mai.2002.122635. [DOI] [PubMed] [Google Scholar]
- 38.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 39.Tantisira KG, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 40.Tantisira KG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci USA. 2004;101:18099–18104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenyl cyclase type 9 in asthma: interaction between β-agonist and corticosteroid pathways. Hum Mol Genet. 2005;14:1671–1677. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 42.Lima JJ, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Steen K, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet. 2005;37:683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 44.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. Using the noninformative families in family-based association tests: a powerful new testing strategy. Am J Hum Genet. 2003;73:801–811. doi: 10.1086/378591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 46.Murphy A, et al. Genomic screening in family-based association testing. BMC Genet. 2005;6(Suppl 1):S115. doi: 10.1186/1471-2156-6-S1-S115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med. 2000;343:1054–1063. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 48.Weiss ST, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006;6:311–326. doi: 10.1038/sj.tpj.6500387. [DOI] [PubMed] [Google Scholar]
- 49.Malaiyandi V, et al. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychol. 2006;11:400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 50.Lerman C, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79:600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 51.Benowitz NL, Swan GE, Jacob P, Lessov-Schlaggar CN, Tyndale RT. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80:457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Kuehl P, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 53.Givens RC, et al. CYP3A5 genotype predicts renal CYP3A activity and blood pressure in healthy adults. J Appl Physiol. 2003;95:1297–1300. doi: 10.1152/japplphysiol.00322.2003. [DOI] [PubMed] [Google Scholar]
- 54.Thompson EE, et al. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, et al. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics. 2001;11:555–572. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 56.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 57.Lamba J, et al. MDR1 genotype is associated with hepatic cytochrome P450 3A4 basal and induction phenotype. Clin Pharmacol Ther. 2006;79:325–338. doi: 10.1016/j.clpt.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 58.Lan LB, Dalton JT, Schuetz EG. Mdr1 limits CYP3A metabolism. in vivo Mol Pharmacol. 2000;58:863–869. doi: 10.1124/mol.58.4.863. [DOI] [PubMed] [Google Scholar]
- 59.Maitland ML, et al. Comparative genomics analysis of human sequence variation in the UGT1A gene cluster. Pharmacogenomics J. 2006;6:52–62. doi: 10.1038/sj.tpj.6500351. [DOI] [PubMed] [Google Scholar]
- 60.Gupta E, et al. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- 61.Gupta E, Wang X, Ramirez J, Ratain MJ. Modulation of glucuronidation of SN-38, the active metabolite of irinotecan by valproic acid and phenobarbital. Cancer Chemother Pharmacol. 1997;39:440–444. doi: 10.1007/s002800050595. [DOI] [PubMed] [Google Scholar]
- 62.Iyer L, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101:847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iyer L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–47. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 64.Innocenti F, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382–1388. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 65.Dolan ME, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 66.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 67.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 68.Liu W, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 69.Liu W, et al. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–1012. [PubMed] [Google Scholar]
- 70.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 71.Rocha JC, et al. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105:4752–4758. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Relling MV, et al. Pharmacogenetic risk factors for osteonecrosis of the hip among children with leukemia. J Clin Oncol. 2004;22:3930–3936. doi: 10.1200/JCO.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 73.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–844. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 74.French D, et al. Global gene expression as a function of germline genetic variation. Hum Mol Genet. 2005;14:1621–1629. doi: 10.1093/hmg/ddi170. [DOI] [PubMed] [Google Scholar]
- 75.Cheok MH, et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003;34:85–90. doi: 10.1038/ng1151. [Erratum in: Nat. Genet. 34, 231 (2003)] [DOI] [PubMed] [Google Scholar]
- 76.Holleman A, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 77.Lugthart S, et al. Identification of genes associated with chemotherapy cross resistance and treatment response in childhood acute lymphoblastic leukemia. Cancer Cell. 2005;7:375–386. doi: 10.1016/j.ccr.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Cheng Q, et al. Karyotypic abnormalities create discordance of germline genotype and cancer cell phenotypes. Nat Genet. 2005;37:878–882. doi: 10.1038/ng1612. [DOI] [PubMed] [Google Scholar]
- 79.Stearns V, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Nat Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 80.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharm Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 81.Johnson MD, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 82.Lim YC, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–512. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 83.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 84.Goetz MP, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 85.Goetz MP, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res and Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 86.Zhang W, et al. Expression of drug pathway proteins is independent of tumour type. J Pathol. 2006;209:213–219. doi: 10.1002/path.1955. [DOI] [PubMed] [Google Scholar]
- 87.Freimuth RR, et al. Polymorphism discovery in 51 chemotherapy pathway genes. Hum Mol Genet. 2005;14:3595–3603. doi: 10.1093/hmg/ddi387. [DOI] [PubMed] [Google Scholar]
- 88.Yu J, et al. DNA repair pathway profiling and microsatellite instability in colorectal cancer. Clin Cancer Res. 2006;12:5104–5111. doi: 10.1158/1078-0432.CCR-06-0547. [DOI] [PubMed] [Google Scholar]
- 89.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 90.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 91.Kroetz DL, et al. Sequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) gene. Pharmacogenetics. 2003;13:481–494. doi: 10.1097/00008571-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 92.Leabman MK, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Urban TJ, et al. Functional genomics of membrane transporters in human populations. Genome Res. 2006;16:223–230. doi: 10.1101/gr.4356206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heils A, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 95.Smeraldi E, Serretti A, Artioli P, Lorenzi C, Catalano M. Serotonin transporter gene-linked polymorphic region: possible pharmacogenetic implications of rare variants. Psychiatr Genet. 2006;16:153–158. doi: 10.1097/01.ypg.0000218611.53064.a0. [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Weinshilboum RM. Thiopurine S-methyltransferase (TPMT) pharmacogenetics: insights, challenges and future directions. Oncogene Rev. 2006;25:1629–1638. doi: 10.1038/sj.onc.1209372. [DOI] [PubMed] [Google Scholar]
- 97.Weinshilboum R, Wang L. Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin Pharmacol Ther. 2004;75:253–258. doi: 10.1016/j.clpt.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 98.Salavaggione OE, Wang L, Wiepert M, Yee VC, Weinshilboum RM. Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet Genomics. 2005;15:801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- 99.Martin YN, et al. Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet Genomics. 2006;16:265–277. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- 100.Hildebrandt MA, et al. Genetic diversity and function in the human cytosolic sulfotransferases. Pharmacogenomics J. doi: 10.1038/sj.tpj.6500404. epub, June 27, 2001. [DOI] [PubMed] [Google Scholar]
- 101.Tai H-L, Krynetski EY, Schuetz EG, Yanishevski Y, Evans WE. Enhanced proteolysis of thiopurine S-methyltransferase (TPMT) encoded by mutant alleles in humans (TPMT*3A, TPMT*2): mechanisms for the genetic polymorphism of TPMT activity. Proc Natl Acad Sci USA. 1997;94:6444–6449. doi: 10.1073/pnas.94.12.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003;13:555–564. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
- 103.Wang L, et al. Human thiopurine S-methyltransferase (TPMT) pharmacogenetics: variant misfolding and aggresome formation. Proc Natl Acad Sci USA. 2005;102:9394–9399. doi: 10.1073/pnas.0502352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roden DM, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]