Abstract
The caudal migration of facial branchiomotor (FBM) neurons from rhombomere (r) 4 to r6 in the hindbrain is an excellent model to study neuronal migration mechanisms. Although several Wnt/Planar Cell Polarity (PCP) components are required for FBM neuron migration, only Celsr1, an atypical cadherin, regulates the direction of migration in mice. In Celsr1 mutants, a subset of FBM neurons migrates rostrally instead of caudally. Interestingly, Celsr1 is not expressed in the migrating FBM neurons, but rather in the adjacent floor plate and adjoining ventricular zone. To evaluate the contribution of different expression domains to neuronal migration, we conditionally inactivated Celsr1 in specific cell types. Intriguingly, inactivation of Celsr1 in the ventricular zone of r3-r5, but not in the floor plate, leads to rostral migration of FBM neurons, greatly resembling the migration defect of Celsr1 mutants. Dye fill experiments indicate that the rostrally-migrated FBM neurons in Celsr1 mutants originate from the anterior margin of r4. These data suggest strongly that Celsr1 ensures that FBM neurons migrate caudally by suppressing molecular cues in the rostral hindbrain that can attract FBM neurons.
Keywords: Hindbrain, Facial Branchiomotor Neuron, Neuronal migration, Celsr1
Introduction
Neuronal migration is a fundamental process in nervous system development, aiding in the establishment of complex neuronal architecture and underlying neural circuitry. All central nervous system neuron cell bodies undergo radial migration, usually along radial glial fibers, as they move from their birthplaces in the ventricular zone to their final locations near the pial surface. In addition, many types of neurons undergo tangential migration, which involves movement orthogonal to the radial direction within specific brain regions. In contrast to radial migration, which establishes distinct neuronal layers and is part of a neuron's maturation process, tangential migration helps generate neuronal diversity within a given brain region (Marín and Rubenstein, 2003; Valiente and Marín, 2010). The facial branchiomotor (FBM) neurons are a particularly intriguing neuron type that undergoes both radial and tangential migration. These neurons, which control facial and jaw movements, are born in rhombomere (r) 4 of the developing hindbrain. In most species studied, they migrate tangentially (caudally) into r6, followed by radial migration to form the facial motor nucleus near the pial surface of r6 (Chandrasekhar, 2004; Guthrie, 2007; Wanner et al., 2013).
Some, but not all, components of the non-canonical Wnt/planar cell polarity (PCP) pathway regulate tangential migration of FBM neurons in zebrafish and mice. While many PCP genes such as Celsr1-3, fzd3a, Pk1, Ptk7, Scrb, Vangl2, and Wnt5a are necessary for proper migration of FBM neurons into r6 (Carreira-Barbosa et al., 2003; Glasco et al., 2012; Mapp et al., 2010; Qu et al., 2010; Vivancos et al., 2009; Wada et al., 2006, 2005; Yang et al., 2014), other genes such as Dvl1-2 and glypican4/6 are not (Bingham et al., 2002; Glasco et al., 2012; Jessen et al., 2002; however, see also Davey et al., 2016), suggesting that caudal migration of FBM neurons is a PCP-independent process. Interestingly, one PCP gene, the atypical cadherin Celsr1, has been shown to regulate the directionality of FBM migration in mice (Glasco et al., 2012; Qu et al., 2010). In Celsr1-deficient mice, many FBM neurons migrate normally, yet a subset migrates rostrally into r2 instead of caudally into r6, completing their radial migration in r2. This reverse migration is not due to defects in neuronal specification or hindbrain patterning, suggesting that Celsr1 acts specifically to dictate the direction of migration, and unlike other Wnt/PCP genes, does not affect their ability to migrate (Qu et al., 2010). The Celsr1 mutant is the first mutant in zebrafish or mouse to exhibit a rostral migration phenotype, and provides a handle to investigate the mechanisms regulating directionality of neuronal migration.
Celsr1 is expressed in a dynamic fashion in the developing hindbrain, but is excluded from migrating FBM neurons (Formstone and Little, 2001; Qu et al., 2010; Shima et al., 2002; Tissir and Goffinet, 2006; Tissir et al., 2002). To understand how Celsr1 regulates the directionality of FBM neuron migration, we investigated the spatial requirement for Celsr1 function through tissue-specific knockouts by utilizing several tissue-specific Cre lines whose activities overlap with different aspects of the Celsr1 expression domain. Our data suggest strongly that Celsr1 functions in the ventricular zone of multiple rhombomeres to suppress the rostral migration of FBM neurons.
MATERIALS AND METHODS
Animals
Mouse colony maintenance and embryo collection were carried out using protocols approved by the Animal Care and Use Committee at the University of Missouri and the Animal Ethics Committee at the University of Louvain.
Mouse lines and genotyping
The Celsr1Crsh, Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (ROSA26mT/mG; gift from D. Cornelison, University of Missouri), Gt(ROSA)26Sortm1(EYFP)Cos (ROSA26-EYFP), Tg(Nkx6-2-icre)1Kess/SshiJ (Nkx6.2-Cre), and Tg(Isl1-EGFP)2Slp (SE1::GFP) mouse lines and genotyping protocols have been described previously (Curtin et al., 2003; Fogarty et al., 2007; Glasco et al., 2012; Muzumdar et al., 2007; Shirasaki et al., 2006; Soriano, 1999; Srinivas et al., 2001). Shhtm1(EGFP/cre)Cjt (ShhCre) mice (Harfe et al., 2004) were obtained from Jackson Laboratories (stock #005622) and genotyped via their recommended protocols. Tg(Hoxb1-cre)r4Mist (r4-Cre) mice were genotyped as previously described (Di Bonito et al., 2013). Egr2tm2(cre)Pch (Krox20Cre) mice (Voiculescu et al., 2000) were a gift from Susan Dymecki (Harvard University) and were genotyped with primers 5’-CTTTACACAGCATCGCCAAG-3’, 5’-TTGACCAGATGAACGGAGTG-3’, and 5’-ATCAGGACATAGCGTTGGCT-3’, the Krox20Cre and wild type alleles producing 500- and 659-bp bands, respectively. Sfrp1tm1Bsk (Bodine et al., 2004) and Sfrp2tm1Sato (Kobayashi et al., 2009) mutant mice were described previously. To obtain litters containing double mutant embryos, sFRP1+/−; sFRP2+/− adults were intercrossed, and embryos were genotyped as described (Misra and Matise, 2010).
The Celsr1 conditional mutant mouse line was described previously (Ravni et al., 2009). Genotyping assays to distinguish between Celsr1WT, Celsr1tm1Fati (Celsr1floxed, fl), and Celsr1tm1.1Fati (Celsr1KO, KO) alleles were designed by the University of Missouri-Columbia Research Animal Diagnostic and Investigative Laboratory (RADIL, now a part of IDEXX Laboratories). DNA was extracted from forelimb buds, which do not express Celsr1. The primer set 5’-CCACTCTGCTAACGGTAGG-3’ and 5’-GAAAGAGACTGTTGGTGAAGC-3’ produces a 596-bp band for the Celsr1fl allele and a 389-bp band for the Celsr1WT allele. The primer set 5’-CTCTGTTGACTTCTGACTGG-3’ and 5’-GAAAGAGACTGTTGGTGAGC-3’ produces a 3504-bp band for the Celsr1KO allele, and a 389-bp band for the Celsr1WT allele. Since the assay to detect the Celsr1fl allele does not amplify the Celsr1KO allele and vice versa, both assays were performed concurrently in order to identify mice with a Celsr1KO/fl genotype.
In situ hybridization and immunohistochemistry
Synthesis of digoxygenin-labeled probes (Celsr1, Islet1, Tbx20, Wnt5a) and whole mount in situ hybridization were carried out as described previously (Glasco et al., 2012; Qu et al., 2010; Song et al., 2006). Methods for demarcation of rhombomere boundaries were described previously (Glasco et al., 2012).
For immunohistochemistry, the following antibodies were used: mouse anti-Isl1 (1:500, DSHB 39.4d5), rabbit anti-GFP (1:500, Molecular Probes A11122), Alexa Fluor® 568 goat anti-mouse (1:250, Invitrogen A11004), and Alexa Fluor® 488 chicken anti-rabbit (1:250, Invitrogen A21441). 30-μm frozen tissue sections were prepared from OCT-embedded embryos on a Leica CM1900 cryostat, and slides were dried for 30 minutes at room temperature. Subsequent steps were carried out in a humidified chamber adapted from guidelines described elsewhere (Harlow and Lane, 2006a, 2006b, 2006c). The sections were permeabilized with PBT (1% Triton X-100 in PBS) three times for 10 minutes each. Non-specific binding was blocked with 10% filtered HIGS in PBT for one hour at room temperature. The diluted primary antibody was applied overnight at 4 °C. Following three ten-minute washes in PBT at room temperature, the sections were incubated in the diluted secondary antibody overnight at 4 °C. After three ten-minute PBT washes at room temperature, each slide was mounted in 175 μL of Mowiol mounting medium and imaged on a Zeiss LSM 510 confocal microscope. Images were processed using Zeiss software.
Neuronal tracing with lipophilic dyes
Protocols previously described (Fritzsch et al., 2005; Qu et al., 2010) were adapted for anterograde labeling of hindbrain neurons. Small pieces of eyebrow hairs were soaked in liquid NeuroVue dyes (Molecular Targeting Technologies) and placed into the appropriate positions of intact embryo hindbrains. NeuroVue Jade (508 nm), Maroon (667 nm), and Red (588 nm) were used in this study. After dye diffusion was visually confirmed, hindbrains were dissected out, flat-mounted in 100% glycerol, and stored at 4 °C until imaging. Images were captured on a Leica SP5 confocal microscope and processed using Leica software.
Results
Celsr1 functions non-cell autonomously, and independently of the floor plate, to regulate FBM neuron migration
We showed previously that a subset of FBM neurons migrates rostrally into r3 and r2 in Celsr1Crsh/+ and Celsr1KO/KO embryos (Qu et al., 2010). Interestingly, Celsr1 is primarily expressed in midline tissues, and does not appear to be expressed in FBM neurons (Qu et al., 2010). Consistent with this, conditional inactivation of Celsr1 in FBM neurons did not result in rostral migration, as seen in whole animal knockouts, suggesting strongly that Celsr1 functions non-cell autonomously to suppress the rostral migration of FBM neurons.
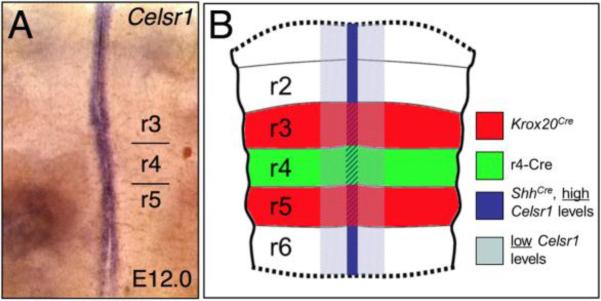
Celsr1 is expressed in midline tissues adjacent to the migrating FBM neurons (Fig. 1A), including the ventral aspects of the ventricular zone at all axial levels in the hindbrain (Qu et al., 2010). It is robustly expressed in floor plate cells throughout the period of FBM neuron migration (Formstone et al., 2010; Qu et al., 2010). Since floor plate cells play a critical role in FBM neuron migration in mice and zebrafish (Sittaramane et al., 2013), we wondered whether Celsr1 may act in the floor plate to prevent the rostral migration of FBM neurons. We tested this hypothesis by specifically inactivating Celsr1 in floor plate cells using the ShhCre (Fig. 1B) and Celsr1floxed mouse lines.
Figure 1. Overlap of the Celsr1 expression pattern with tissue-specific Cre driver domains.
(A), Flat-mounted E12 hindbrain processed for Celsr1 in situ hybridization (ISH). Celsr1 is expressed in the floor plate at all axial levels. B, Overview of the tissue-specific Cre lines used in this study showing overlap of the Cre domains with Celsr1 expression. “High” and “low” refer to the relative levels of Celsr1 expression in the floor plate and ventricular zone, respectively.
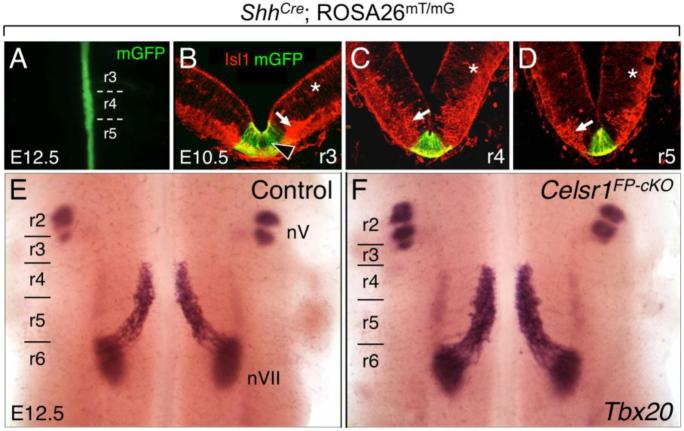
To confirm that Cre activity was restricted to floor plate cells, we crossed ShhCre mice (Harfe et al., 2004) with ROSA26mT/mG reporter mice (Muzumdar et al., 2007) to generate ShhCre; ROSA26mT/mG embryos expressing membrane-targeted EGFP in tissues with active Cre recombinase. In addition to GFP immunostaining on sections (to label Cre-expressing cells), we examined the expression of the motor neuron marker Islet1 (Isl1) to provide landmarks along the dorsal-ventral and rostral-caudal axes. At the onset of FBM neuron migration (E10.5) and later (E12.5), Cre-mediated GFP expression was robust in floor plate cells at all axial levels of the hindbrain (Fig. 2A-D) (n=6; 3 whole mount, 3 sectioned). Next, we generated Celsr1 floor plate conditional mutants (FP-cKO) using a breeding strategy designed to produce two FP-cKO genotypes, ShhCre; Celsr1KO/fl and ShhCre; Celsr1fl/fl (Fig. S1). All embryos were assayed for FBM neuron migration defects by Tbx20 in situ hybridization (ISH). In FP-cKO embryos, FBM neurons migrated only in the caudal direction in all embryos tested (n=14) (Figs. 2E, F and 5D), demonstrating that Celsr1 does not function in the floor plate to suppress rostral migration of FBM neurons.
Figure 2. Disruption of Celsr1 function in the floor plate does not affect FBM neuron migration.
A, E, F, Dorsal views of E12.5 embryos visualized live for GFP (A) or processed for Tbx20 ISH (E, F). GFP expression along the midline in floor plate cells in a ShhCre; ROSA26mT/mG embryo (A) labels the expression domain of Cre used for the floor plate-specific knockout of Celsr1 function (FP-cKO). B-D, 30-μm coronal sections of an E10.5 wild type hindbrain processed for anti-GFP (green) and anti-Isl1 (red) immunostaining to mark Cre expression (GFP), and FBM neurons (arrow), respectively. The red background signal (asterisks) is due to mTomato expression in all tissues of the ROSA26mT/mG mouse where Cre is not active. At all axial levels, Cre activity is restricted to floor plate cells (arrowhead). E, F, In control embryos (ShhCre; Celsr1fl/+ or ShhCre; Celsr1KO/+, 8/8 embryos), Tbx20 labels trigeminal BM neurons (nV) in r2 and r3, and FBM neurons (nVII) migrating caudally from r4 to r6 (E). FBM neurons also migrate normally in FP-cKO embryos (F) (ShhCre; Celsr1KO/fl or ShhCre; Celsr1fl/fl, 14/14 embryos).
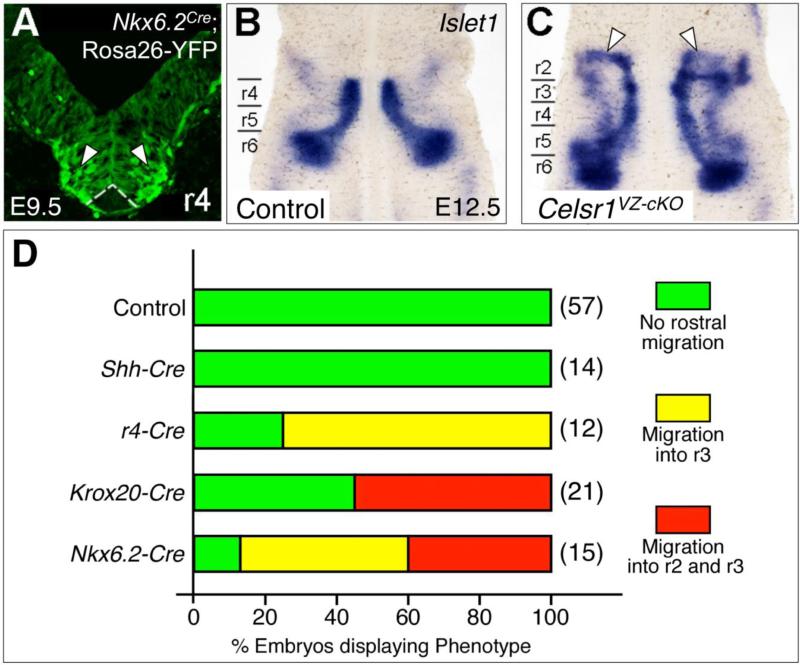
Figure 5. Conditional Celsr1 inactivation in the Nkx6.2 expression domain generates a similar rostral migration phenotype to Celsr1 knockout.
A, 30-μm coronal section of an E9.5 ROSA26-EYFP; Nkx6.2-Cre hindbrain in r4, imaged to directly visualize YFP fluorescence, which indicates the expression domain of Cre used for the ventricular zone-specific knockout of Celsr1 function (VZ-cKO). Nkx6.2-Cre is expressed in the motor neuron progenitor domain at all axial levels (arrowheads), and is excluded from the floor plate (dotted lines). B-C, Dorsal views of E12.5 hindbrains processed for Isl1 ISH. In a control embryo (Nkx6.2-Cre; Celsr1fl/+, 11/11 embryos), Isl1-expressing FBM neurons migrate caudally from r4 to r6 (B). In VZ-cKO embryos (C), (Nkx6.2-Cre; Celsr1fl/fl), significant numbers of FBM neurons (arrowheads) migrate rostrally into r3 and r2 in a majority of embryos (13/15 embryos). D, Quantitation of rostral migration phenotypes in various tissue-specific Celsr1 knockouts. In Krox20-Cre and Nkx6.2-Cre embryos, embryos displaying the r2 migration phenotype also have some FBM neurons in r3.
Celsr1 partly functions in rhombomere 4 to suppress rostral neuronal migration
Since Celsr1 function in the floor plate is dispensable, we reasoned that its expression in the ventricular zone in r4, where the FBM neurons are born, could play a role. Therefore, we conditionally inactivated Celsr1 in r4 and its derivatives using the r4-Cre mouse line (Di Bonito et al., 2013). To confirm Cre expression within r4, we crossed r4-Cre mice (Fig. 1B) with ROSA26mT/mG reporter mice and immunostained for Isl1 and GFP at E10.5. Cre-mediated GFP was localized exclusively to r4-derived cells, including FBM neurons along their migration route in r4-r6 (Fig. 3A-D) (n=9; 2 whole mount, 7 sectioned). Notably, Cre-mediated GFP expression was robust throughout the neuroepithelium of r4, and was tightly restricted to this rhombomere. The only cells expressing GFP outside r4 in E10.5-12.5 wild type embryos were located in r5, likely representing the caudally migrating stream of FBM neurons (Fig. 3A, D).
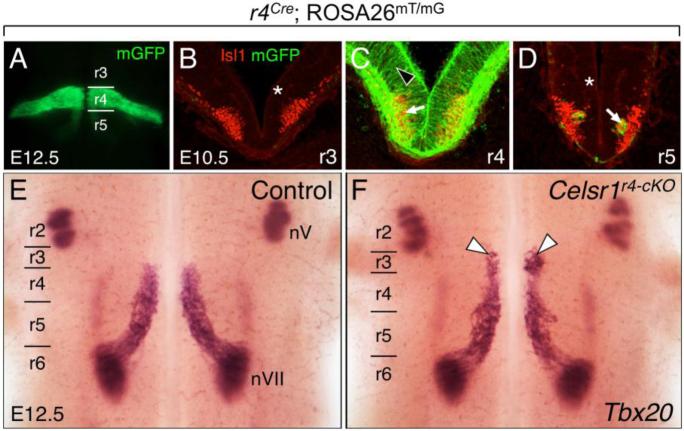
Figure 3. Some FBM neurons migrate rostrally into rhombomere 3 (r3) following disruption of Celsr1 function in r4.
A, E, F, Dorsal views of E12.5 embryos visualized live for GFP (A) or processed for Tbx20 ISH (E, F). GFP expression in r4 in a r4-Cre; ROSA26mT/mG embryo (A) labels the expression domain of Cre used for the r4-specific knockout of Celsr1 function (r4-cKO). BD, 30-μm coronal sections of an E10.5 wild type hindbrain processed for anti-GFP (green) and anti-Isl1 (red) immunostaining to mark r4-Cre expression (GFP), and FBM neurons (arrow), respectively. The light red background signal (asterisks) is due to mTomato expression in all tissues of the ROSA26mT/mG mouse where Cre is not active. Cre activity is restricted to tissues within r4 (black arrowhead). The GFP-expressing cells in D (arrow) are the earliest caudally-migrating FBM neurons originating in r4. E, F, In control embryos (r4-Cre; Celsr1fl/+ or r4-Cre; Celsr1KO/+, 20/21 embryos), Tbx20 labels trigeminal BM neurons (nV) in r2 and r3, and FBM neurons (nVII) migrating caudally from r4 to r6 (E). A small but significant number of FBM neurons (arrowheads) migrates rostrally into r3 in a majority of r4-cKO embryos (F) (r4-Cre; Celsr1KO/fl or r4-Cre; Celsr1fl/fl, 9/12 embryos).
In Celsr1 r4-cKO embryos, a subset of Tbx20+ve FBM neurons migrated rostrally instead of caudally (Figs. 3E-F and 5D) (9/12 embryos), as seen in Celsr1Crsh/+ (also referred to as Celsr1Crsh) and Celsr1KO/KO embryos (also referred to as Celsr1KO). Interestingly, these aberrantly migrating FBM neurons were found exclusively in r3, typically in clusters near the r3/r4 boundary. Therefore, although FBM neurons migrated rostrally in r4-cKO embryos, it was less severe than the Celsr1Crsh and Celsr1KO mutant phenotypes, where FBM neurons migrate two rhombomeres in the rostral direction and settle in r2 adjacent to the trigeminal motor nuclei (Qu et al., 2010). These data suggest that Celsr1 function in r4 contributes to the ability of Celsr1 to suppress rostral migration of FBM neurons.
Celsr1 functions in rhombomeres 3 and 5 to regulate FBM neuron migration
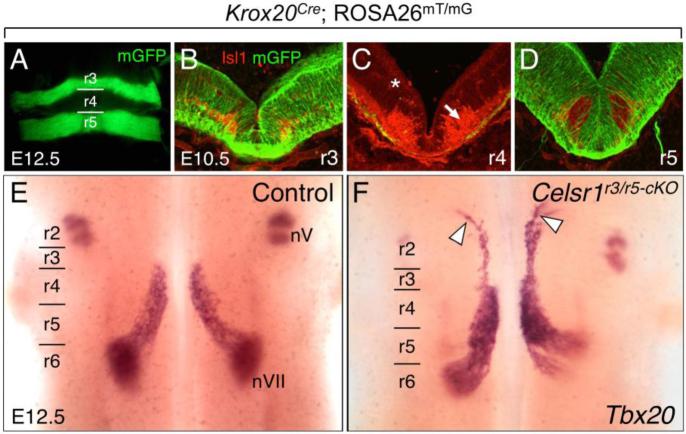
Since r4-specific inactivation generated only a partial rostral migration phenotype, we tested whether Celsr1 inactivation in more anterior rhombomeres like r3 could generate a similar phenotype. Since an r3-specific Cre driver is not available, we employed the Krox20Cre knock-in mouse line, where Cre recombinase driven by the endogenous Krox20 promoter is expressed in rhombomeres 3 and 5 (Voiculescu et al., 2000). To confirm Cre expression in r3 and r5, we crossed Krox20Cre mice (Fig. 1B) with ROSA26mT/mG reporter mice and immunostained for Isl1 and GFP at E10.5. Cre-mediated GFP expression was seen at all dorsal-ventral levels of the neuroepithelium in r3 and r5 (Fig. 4A-D) (5 whole mount, 5 sectioned). Notably, Cre-mediated GFP was excluded from r4-derived FBM neurons (Isl1+ve) migrating caudally within r5 (Fig. 4D).
Figure 4. Some FBM neurons migrate rostrally following inactivation of Celsr1 in r3 and r5.
A, E, F, Dorsal views of E12.5 embryos visualized live for GFP (A) or processed for Tbx20 ISH (E, F). GFP expression in r3 and r5 in a Krox20Cre; ROSA26mT/mG embryo (A) labels the expression domain of Cre used for the r3/r5-specific knockout of Celsr1 function (r3/r5-cKO). B-D, 30-μm coronal sections of an E10.5 wild type hindbrain processed for anti-GFP (green) and anti-Isl1 (red) immunostaining to mark Cre expression (GFP), and FBM neurons (arrow), respectively. The light red background signal (asterisk) is due to mTomato expression in all tissues of the ROSA26mT/mG mouse where Cre is not active. In r3 (B) and r5 (D), Cre activity is ubiquitous, including in floor plate cells. In r4 (C), Cre is inactive except for a few cells along the pial surface, which was seen in all sections (5/5 embryos). E, F, In control embryos (Krox20Cre; Celsr1fl/+ or Krox20Cre; Celsr1KO/+, 18/18 embryos), Tbx20 labels trigeminal BM neurons (nV) in r2 and r3, and FBM neurons (nVII) migrating caudally from r4 to r6 (E). A small but significant number of FBM neurons migrate rostrally into r2 in several r3/r5-cKO embryos (F, white arrowheads) (Krox20Cre; Celsr1KO/fl or Krox20Cre; Celsr1fl/fl, 3/12 embryos).
Next, we generated Celsr1 r3/r5 conditional mutant embryos (r3/r5-cKO) with the Krox20Cre; Celsr1KO/fl or Krox20Cre; Celsr1fl/fl genotypes, and assayed for FBM neuron migration defects by Tbx20 ISH. In some Celsr1 r3/5-cKO embryos (3/12), a subset of Tbx20+ve FBM neurons migrated rostrally into r2 instead of caudally into r6 (Fig. 4F), similar to Celsr1Crsh and Celsr1KO mutants (Qu et al., 2010). In several r3/r5-cKO embryos (9/12), some caudal migration defects were seen, where FBM neurons exhibited dorsolateral migration in r5 instead of r6 (Fig. 4F). To rule out the possibility that the FBM neuron migration defects in r3/r5-cKO embryos might be due to Krox20Cre activity in the male germ line (Fig. S1), we implemented an alternate breeding strategy using Krox20Cre; Celsr1KO/+ males and Celsr1fl/fl females to generate Krox20Cre; Celsr1KO/fl (r3/r5-cKO) embryos. In most r3/r5-cKO embryos (8/9; Fig. 5D), FBM neurons migrated rostrally (data not shown), confirming that the phenotype resulted from the conditional deletion of Celsr1 in r3/r5.
Together, these results indicate that loss of Celsr1 function in r3/r5 can generate all of the migration defects seen in Celsr1Crsh and Celsr1KO embryos, albeit at a lower frequency. However, rostral migration defects are seen in >90% of Celsr1Crsh and Celsr1KO mutants (Qu et al., 2010; unpublished data), indicating that Celsr1 function in tissues other than r3 (and/or r5) normally contributes to the suppression of rostral migration. Given our data for a contribution of r4 in this process (Fig. 3), an attractive possibility is that loss of Celsr1 function in the ventricular zone at all axial levels from r2 to r5, but excluding the floor plate, is sufficient to generate the migration defects seen in Celsr1KO/KO mutants. We tested this hypothesis by conditional inactivation of Celsr1 using an Nkx6.2-Cre driver, which is expressed in the appropriate tissues.
Celsr1 functions in the ventricular zone along the ventral midline of the hindbrain to regulate FBM neuron migration
We deleted Celsr1 in Nkx6.2-expressing cells using Nkx6.2-Cre mice (Fogarty et al., 2007). Throughout FBM neuron specification (E9.5) and at the onset of their migration (E10.5), Nkx6.2 is expressed broadly in the neural tube at all axial levels of the hindbrain (Moreno-Bravo et al., 2010; Qiu et al., 1998) but is mostly absent from the floor plate (Fig. 5A). Thereafter (E11.5), Nkx6.2 expression is largely restricted to pre-migratory FBM neurons in r4 (Pattyn et al., 2003).
As predicted, deletion of Celsr1 in Nkx6.2-expressing cells generated the rostral migration phenotype seen in Celsr1KO/KO mutants and Celsr1Crsh/+ embryos. In many Nkx6.2-Cre; Celsr1KO/fl embryos (6/15; 40%), a subset of Isl1+ve FBM neurons migrated rostrally into r2 instead of caudally into r6 (Fig. 5B, C). In other embryos (7/15; 47%), a subset of FBM neurons also migrated rostrally, albeit to a lesser extent, into r3. Overall, 87% of Nkx6.2-Cre embryos contained FBM neurons migrating rostrally into r2 and/or r3, which is similar to the fraction (~93%) of Celsr1Crsh and Celsr1KO embryos exhibiting rostral migration defects (Qu et al., 2010; unpublished data). The summarized data from the various conditional knockouts (Fig. 5D) clearly demonstrate the importance of Celsr1 function in the ventricular zone in the anterior hindbrain up to rhombomere 5 for suppressing rostral migration of FBM neurons.
Rostral migration of FBM neurons in Celsr1Crsh embryos may result from the loss of local guidance cues
We wondered why, in Celsr1 mutants, only a subset of FBM neurons migrates rostrally, while the majority undergoes normal caudal migration. One possibility is that loss of Celsr1 function leads to failure of PCP and loss of cell polarity in pre-migratory FBM neurons in r4 (even though they do not express Celsr1) such that they migrate either rostrally or caudally in a random fashion. In this case, rostrally migrating FBM neurons would be expected to originate at various positions, spanning the rostral-caudal extent of r4. Alternatively, since disruption of Celsr1 function in r2 to r5 facilitates rostral migration of FBM neurons, this phenotype may result from the loss of local cues that normally act at the r3/r4 boundary to suppress rostral migration of the FBM neurons. In this case, rostrally migrating FBM neurons would be predicted to originate preferentially from the anterior aspects of r4 adjacent to the r3/r4 boundary.
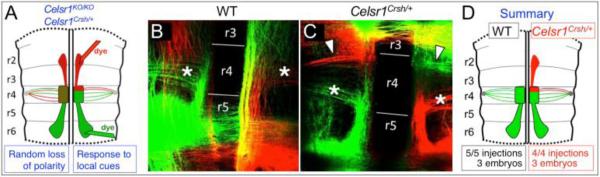
To distinguish between these putative mechanisms, we placed NeuroVue lipophilic dyes into the regions of the hindbrain neuroepithelium corresponding to both the normally-migrating (caudal) and aberrantly-migrating (rostral) populations of FBM neurons in Celsr1Crsh mutants. By E9.5, FBM axons extend to the r4 exit point at various rostralcaudal levels corresponding to their soma positions within r4, generating a fan shape (Fritzsch et al., 1993; Rossel and Capecchi, 1999; Schneider-Maunoury et al., 1997). Therefore, through anterograde labeling of migrating FBM neurons from outside of r4, we could infer their starting positions within r4 prior to migration (Fig. 6A).
Figure 6. The rostrally migrating FBM neurons in Celsr1Crsh/+ embryos originate in the rostral portion of r4, adjacent to the r3/r4 boundary.
A, Schematic of a flat-mounted E12.5 hindbrain indicating the dye application sites within the rostral (NeuroVue Maroon, red) and caudal (NeuroVue Jade, green) migrating streams of FBM neurons to anterogradely label the FBM axons exiting in r4. Two hypotheses (blue boxes) and predicted phenotypes are shown. If the rostral migration phenotype of Celsr1 mutants results from a loss of polarity in FBM neurons in r4 (left side), a newborn FBM neuron in any location within r4 (mixed red/green stipples) could potentially migrate rostrally, resulting in a mixed collection of FBM axons exiting from r4. However if rostrally migrating neurons arise exclusively from the anterior margin of r4 in response to guidance cues unmasked in the mutants (right side), the FBM axons extended by the rostrally and caudally migrating neurons would be clearly segregated in r4. B-C, Flat-mounted views of dye-labeled WT and Celsr1Crsh/+ hindbrains. To maximize data collection from each embryo, the migratory streams were labeled on both sides (caudal left-green; rostral left-red; caudal right-red; rostral right-green). In a WT embryo (B), dye application in r5/r6 anterogradely labels FBM axons (asterisks) spanning the anterior-posterior extent of r4. Dye application in r2/r3 does not label any axon projections into the r4 exit points. In a Celsr1Crsh/+ embryo (C), dye application in r5/r6 anterogradely labels FBM axons in r4 (asterisks). Importantly, dye application in r2/r3 anterogradely labels FBM axons projecting toward the r4 exit points, extending along the anterior margin of r4 (arrowheads). Axons crossing the midline in r5 (B) and r3 (C) are commissural axons that were inadvertently labeled in these particular embryos, and are not relevant to the rostral migration defect. D, Schematic summarizing the results shown in B and C.
In E12.5 WT embryos, dye application in the caudally migrating stream in r5/r6 anterogradely labeled FBM axons projecting to the r4 exit point from various points spanning the rostral-caudal extent of r4 (5 injections; 3 embryos) (Fig. 6B). Dye application into r2/r3 did not label any axons extending to the r4 exit point. The medial longitudinal fascicle and reticular formation were consistently labeled in all embryos, since they run parallel to the floor plate at all axial levels. Dye application into the caudally migrating stream of Celsr1Crsh/+ embryos consistently labeled a smaller number of FBM axons projecting to the r4 exit point (Fig. 6C). Importantly, dye application into the rostrally migrating stream in r2/r3 labeled FBM axons extending to the r4 exit point (4 injections; 3 embryos). Furthermore, the two axon populations were completely segregated (no mixing), with the rostrally labeled axons originating entirely near the r3/r4 boundary, and the caudally labeled axons originating from the central and caudal aspects of r4 (Fig. 6C, D). These results are consistent with our predictions based on putative underlying mechanisms (Fig. 6A), and suggest that rostral migration of FBM neurons in Celsr1 mutants is a response of mutant neurons at the r3/r4 boundary to local (short range) attractive cues that are normally suppressed in wild type embryos (Fig. 7).
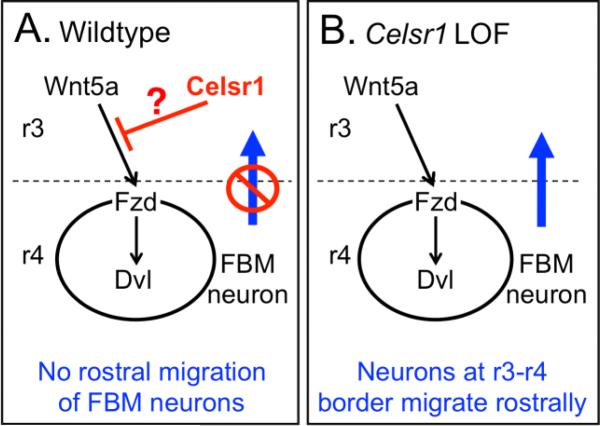
Figure 7. Model of Celsr1 function in repressing rostral migration of FBM neurons.
We propose that Wnt5a, which is expressed along the midline in r3 and more anterior regions of the hindbrain, can act as a chemoattractive cue to facilitate the migration of FBM neurons located in anterior r4, adjacent to the r3/r4 boundary, rostrally into r3 and r2. A, In a wild type hindbrain, Celsr1 expression in the ventricular zone inhibits Wnt5a-dependent activation through an unknown mechanism (?), blocking the rostral migration of FBM neurons into r3. B, In a Celsr1-deficient hindbrain, the inhibition is relieved and FBM neurons migrate rostrally into r3 toward the Wnt5a source.
We also noted differences between embryos in the number and location of crossing axons (reticulo- and vestibulo-bulbar projections in rhombomeres 2-6; Straka et al., 2014) (Fig. 5B, C). However, there was no reproducible difference between WT and Celsr1Crsh/+ embryos, suggesting that the observed differences are likely due to variability in NeuroVue dye application and extent of anterograde labeling. However, we cannot rule out that some crossing axons may be differentially affected in Celsr1 mutants.
Potential roles for Wnt signaling in generating the rostral migration phenotype of Celsr1 mutants
Since the rostrally migrating subset of FBM neurons originates from the anterior part of r4, adjacent to the r3/r4 boundary, we wondered whether these FBM neurons may be responding to a chemoattractant in r3 whose expression/function is unmasked by Celsr1 inactivation (Fig. 7). Wnt5a, a Wnt/PCP ligand, is expressed in a rhombomere-specific fashion, and can serve as a chemoattractant for caudally migrating FBM neurons (Vivancos et al., 2009). In E11.5 control embryos, Wnt5a was expressed strongly in the hindbrain neuroepithelium posterior to r5, virtually absent from r4, and strongly expressed in the floor plate anterior to the r3/r4 boundary (Fig. S2A, C) (Vivancos et al., 2009). Wnt5a expression was unaffected in r3/r5-cKO or r4-cKO hindbrains (Fig. S2B, D), indicating that the rostral migration phenotype in Celsr1 mutants is not caused by ectopic or increased expression of Wnt5a.
While Wnt5a expression is not affected, its activity in the anterior hindbrain may be unmasked in Celsr1 mutants, and induce chemotactic migration of FBM neurons into r2 and r3 (Fig. 7). Vivancos et al. (2009) showed that FBM neurons can respond to and migrate toward Wnt5a-soaked beads placed laterally in r4, consistent with a role for Wnt5a in directing caudal migration of FBM neurons. Interestingly, we found that Wnt5a-soaked beads placed in r3 in wild type hindbrain explants could induce the rostral migration of FBM neurons from r4 (Fig. S3), suggesting that excess Wnt5a can overcome putative inhibitory factors that normally suppress rostral migration. We hypothesized that two Wnt-binding proteins sFRP1 and sFRP2 (secreted frizzled-related proteins 1 and 2) act as putative inhibitory factors (Bodine et al., 2004; Bovolenta et al., 2008; Jones and Jomary, 2002; Kobayashi et al., 2009; Lei et al., 2006; Leimeister et al., 1998; Misra and Matise, 2010). The two genes are expressed in partially overlapping patterns (Fig. 8A, B and Fig. S4), with sFRP1 showing strongest expression medially and sFRP2 more laterally. sFRP1 is also expressed weakly along the midline in r3 and r2 (Fig. S4), overlapping with Celsr1 and Wnt5a expression domains. If sFRPs act in r3 to block Wnt5a activity (in a Celsr1-dependent fashion) and suppress rostral migration of FBM neurons, we predicted that loss of sFRP function would recapitulate the migration defect of Celsr1 mutants.
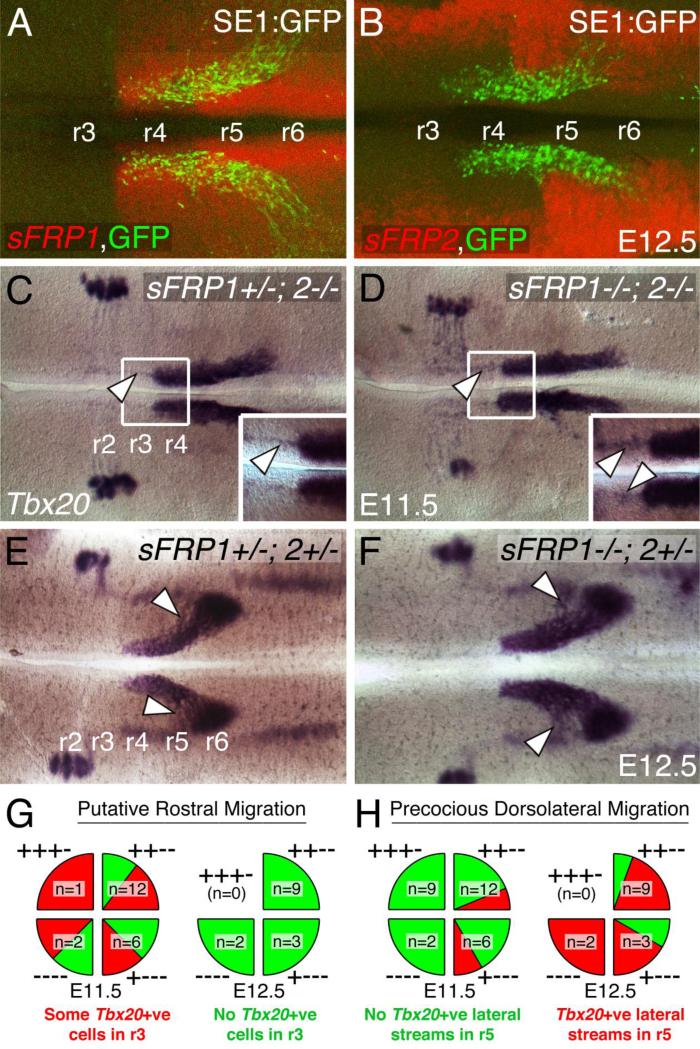
Figure 8. Wnt antagonist genes sFRP1/2 modulate caudal migration of FBM neurons.
A-F, Flat-mounted hindbrains (left is rostral) processed for ISH with sFRP1 (A), sFRP2 (B) and Tbx20 (C-F). The distribution of GFP-expressing FBM neurons in A, B (from SE1::GFP embryos) helps to delineate rhombomere boundaries. A, At E12.5, sFRP1 is strongly expressed caudal to the r3/r4 boundary in the entire neuroepithelium. Although not apparent in fluorescent ISH, sFRP1 is also expressed weakly in r3 and r2. B, sFRP2 is weakly expressed in medial regions, and strongly expressed along the lateral margins of the hindbrain at all axial levels (r3-r6). C-D, In E11.5 hindbrains, when 3 or more copies of sFRP1/2 are removed, a small number of Tbx20-expressing cells (arrowheads) appear to migrate rostrally into r3 (see insets). There are no defects in the caudally migrating streams of FBM neurons. E-F, In E12.5 hindbrains, when 2 or more copies of sFRP1/2 are removed, the caudal migration streams frequently contain ectopic streams indicating precocious dorsolateral migration (arrowheads). Interestingly, there are no rostral migration defects. G-H, Summary data showing distribution of migration defects among various sFRP1/2 compound mutants at E11.5 and E12.5. The four quarters in each “pie” show the breakdown of two phenotypes (red and green sectors) in the indicated sFRP1; sFRP2 genotypes. For example, ++−− corresponds to sFRP1+/+; sFRP2−/−. G, While over 50% of E11.5 mutants (12/21 embryos) contain some rostrally migrating Tbx20+ve cells, these cells are no longer evident in E12.5 mutants (14/14 embryos). H, While less than 20% of E11.5 mutants (5/29 embryos) contain precocious lateral streams of Tbx20+ve cells, most E12.5 mutants (11/14 embryos) contain such streams.
Therefore, we examined FBM neurons by Tbx20 ISH in sFRP compound mutants lacking two or more copies of these genes (Fig. 8C-H). Weak streams of Tbx20+ve putative FBM neurons were seen in r3 in over 50% of E11.5 mutants (Fig. 8C, D, G; 12/21 embryos), but the “rostral migration” phenotype was much weaker than seen in Celsr1 mutants. No Tbx20+ve cells were seen in these locations in any E12.5 mutant (Fig. 8E, F, G; 0/14 embryos), suggesting that sFRPs play a minor role, if any, in suppressing the migration of FBM neurons into r3. Interestingly, lateral streams of FBM neurons in r5, indicative of precocious dorsolateral migration, were seen in sFRP compound mutants, sometimes at E11.5 (Fig. 8H, 5/29 embryos) and frequently at E12.5 (Fig. 8E, F, H; 11/14 embryos), suggesting that sFRP1 and 2 help establish the medial pathway of the caudal migratory stream.
Discussion
The caudal migration of FBM neurons in the developing hindbrain is a tightly regulated process involving diverse signaling pathways, including non-canonical Wnt/PCP signaling. Though several genes are required for the caudal migration of FBM neurons (Chandrasekhar, 2004; Song, 2007; Wanner et al., 2013), our studies indicated for the first time that some genes like Celsr1 function to suppress rostral migration (Glasco et al., 2012; Qu et al., 2010). Our conditional knockout studies demonstrate that Celsr1 function is required in a relatively small expression domain, the Nkx6.2+ve ventricular zone spanning rhombomeres 2-5, to suppress rostral migration of FBM neurons. This analysis provides a starting point for elucidating the molecular mechanisms that control the directionality of FBM neuron migration.
No discernable function for floor plate-derived Celsr1 in FBM neuron migration
Given the strong expression of Celsr1 in floor plate cells (Qu et al., 2010), it was surprising to find that loss of Celsr1 function in the floor plate did not affect FBM neuron migration. Celsr1 is expressed in the floor plate at the onset of migration, and persists even after its downregulation at the ventricular surface. However, a minor role for Celsr1 in the floor plate cannot be ruled out for two reasons. First, in both r3/r5-cKO and r4-cKO embryos, which exhibit partially overlapping rostral migration phenotypes, Celsr1 function is inactivated in the entire neuroepithelium of specific rhombomeres including the floor plate. Second, the weaker ability of FBM neurons in Nkx6.2-Cre (VZ-cKO) embryos to migrate into r2 (Fig. 5) compared to that in Celsr1Crsh and Celsr1 KO embryos (Qu et al., 2010) may reflect a functional role for Celsr1 in floor plate cells in anterior rhombomeres. Therefore, although the floor plate itself plays an important role in regulating FBM neuron migration (Sittaramane et al., 2013; however see also Davey et al., 2016), there is only a minor role, if any, for floor plate-derived Celsr1 in this process. Importantly, floor plate-derived Celsr1 may regulate other floor plate-dependent processes such as neural tube closure (Doudney and Stanier, 2005; Greene et al., 1998; Murdoch et al., 2001), orientation of floor plate primary cilia (Borovina et al., 2010; Okada et al., 2005), guidance of commissural axons (Serafini et al., 1996), migration of inferior olivary, lateral reticular, and external cuneate nuclei (de Diego et al., 2002), or the contralateral projection of olivocochlear efferent neurons (Fritzsch et al., 1993). While there were no obvious neural tube closure defects in Celsr1 floor plate cKO embryos, potential roles for Celsr1 in these processes have not been directly examined.
Celsr1 suppresses rostral migration of FBM neurons
Our analysis of Celsr1 mutants (Qu et al., 2010) suggests that FBM neurons possess the ability to migrate in either the rostral or caudal direction, but that Celsr1 normally mediates directionality cues that steer them caudally. Rhombomere specification and patterning occur normally in Celsr1 mutants indicating that the reversed migration is not a consequence of aberrant rhombomere boundary formation (Qu et al., 2010). One possibility is that loss of Celsr1 function results in the random orientation of FBM neurons in rhombomere 4 due to loss of tissue polarity in the Celsr1-expressing cells in their environment, causing FBM neurons to migrate rostrally or caudally in random fashion. In this case, one would expect about 50% of FBM neurons to migrate rostrally out of r4; however, in all Celsr1 mutants analyzed, the rostrally migrating stream contains substantially fewer neurons. While functional redundancy between Celsr1 and related genes may account for this “weak” rostral migration phenotype, another plausible explanation is that it is the consequence of the inactivation of Celsr1-dependent local cues that normally suppress rostral migration. Given that Wnt5a is expressed in a gradient in rhombomeres 5-7 and plays a role in caudal migration of FBM neurons (Vivancos et al., 2009), it might play a similar role to induce rostral migration in Celsr1 mutants. Consistent with this, Wnt5a is expressed in midline tissues in the rostral hindbrain up to r3, overlapping in expression with Celsr1 (Fig. S2). Furthermore, loss of Dvl2 function in Celsr1Crsh/+ embryos suppresses the rostral FBM neuron migration phenotype (Glasco et al., 2012), suggesting that the ability to migrate rostrally involves activation of the Wnt/Dvl signaling pathway. Based on these observations, we propose that Wnt5a in r3 can potentially act as a chemoattractant to induce FBM neurons to migrate rostrally from r4 (Fig. 7A). Further, we suggest that in the wild type hindbrain, Celsr1 functions to suppress the activity of Wnt5a in inducing neuronal migration. In Celsr1 mutants, this suppression is relieved, resulting in rostral migration (Fig. 7B). Since the Wnt-mediated chemoattraction would decrease with increasing distance from the source in r3, this model also explains why the rostrally migrating FBM neurons in Celsr1 mutants originate exclusively from near the r3/r4 boundary (Fig. 6), where chemoattraction would be the strongest. Consistent with this model, placement of numerous Wnt5a-soaked beads in r3 in wild type hindbrain explants results in rostral migration of FBM neurons (Fig. S3), suggesting that excess Wnt5a can overcome Celsr1-mediated suppression.
Although a model invoking the loss of local cues at the r3-r4 boundary adequately explains the rostral migration phenotype of Celsr1 mutants, and is consistent with several independent observations discussed above, the differing phenotypes between r3/r5-cKO and r4-cKO embryos suggest that the polarity of Celsr1-expressing cells in r3 may also be important. When Celsr1 is inactivated solely in r4 (r4-cKO embryos), a subset of FBM neurons migrates rostrally into r3, but not further into r2 (Fig. 3). In this case, Celsr1 is not inactivated in r3, which may represent a non-permissive (polarized?) environment that prevents FBM neurons from migrating further rostrally into r2. In r3/r5-cKO embryos, however, FBM neurons may be able to migrate rostrally into r2, since inactivation of Celsr1 in the cells adjacent to the FBM neurons in r3 may represent a permissive (non-polarized) environment.
Mechanism of Celsr1-mediated suppression of rostral migration of FBM neurons
While our genetic and bead-explant data suggest strongly that Celsr1 normally suppresses rostral migration, likely by antagonizing Wnt function (Fig. 7), potential mechanisms remain obscure. In Celsr1 mutants, even a small change in the Wnt concentration gradient could in principle affect only the rostral-most FBM neurons in r4, and induce them to migrate rostrally. Alternatively, Celsr1-mediated changes in the adhesion properties of the ventricular zone in r3 (adjacent to the rostral-most FBM neurons) may modify the local “non-migratory” environment, causing the rostral-most FBM neurons to migrate into r3. Expression profiling using RNA-seq of hindbrain fragments from wild type and Celsr1Crsh embryos have identified candidate secreted molecules and receptors that are being validated.
Independently of this analysis, we investigated potential roles for secreted Frizzled related proteins (sFRPs) due to their well-documented roles in antagonizing Wnt function, and their expression patterns in the developing hindbrain (Bodine et al., 2004; Bovolenta et al., 2008; Jones and Jomary, 2002; Kobayashi et al., 2009; Lei et al., 2006; Leimeister et al., 1998; Misra and Matise, 2010). If sFRPs were mediating the suppressive effect of Celsr1 on Wnt5a, we would predict that FBM neurons would migrate rostrally in sFRP mutants. While there was a weak transient effect reminiscent of rostral migration in sFRP1/2 compound mutants (Fig. 8), it is unlikely that the putative inhibition of Wnt activity by Celsr1 is mediated through sFRPs. However, we did observe a strong effect on caudal migration in sFRP mutants, consistent with a role in modulating Wnt5a activity. In Wnt5a−/− hindbrains, caudally migrating FBM neurons frequently undergo precocious dorsolateral migration, resulting in the formation of ectopic streams in r5 (Vivancos et al., 2009), and suggesting that when Wnt signaling is reduced, FBM neurons may prematurely respond other cues mediating dorsolateral migration. Interestingly, precocious dorsolateral streams are also seen in sFRP compound mutants (Fig. 8E, F), in which Wnt signaling is likely to be enhanced. These results suggest that Wnt signaling must be maintained in an optimal range to prevent precocious dorsolateral migration. In addition, the exclusion of sFRP gene expression from the FBM migratory pathway (Fig. 8A, B) suggests that sFRPs may help establish a medial Wnt5a-enriched pathway that is permissive for FBM neuron migration.
We provide here novel insight into the cellular mechanism by which the atypical cadherin Celsr1 regulates the direction of migration of FBM neurons. Our data suggest strongly that Celsr1 functions in a relatively small expression domain in the ventricular zone, rather than in a more prominent floor plate domain, to suppress rostral migration. We propose that Celsr1 suppresses rostral migration by antagonizing a Wnt5a chemoattractive source in the anterior hindbrain. How Celsr1 mediates its putative effects on Wnt activity, and whether this mechanism is conserved between mice and zebrafish are important issues for further study.
Supplementary Material
Highlights.
Celsr1 regulates the directionality of mouse facial branchiomotor neuron migration.
Celsr1 functions specifically in the ventricular zone of rhombomeres 3-5.
In Celsr1 mutants, rostrally-migrated neurons originate from anterior rhombomere 4.
Celsr1 may suppress attractive cues to prevent inappropriate rostral migration.
Acknowledgements
We thank members of the Chandrasekhar lab for discussion and help with animal care. We thank Mi-Ryoung Song (Gwangju Institute of Science and Technology) for the Wnt5a cDNA.
Funding
This work was supported by an NIH predoctoral fellowship 1F31NS063513 (D.G.), Queen Elisabeth Medical Foundation (F.T.), and NIH grant NS040449 (A.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.M.G. designed and performed all experiments except those indicated, analyzed the data, and co-wrote the manuscript. W.P. performed the sFRP experiments, and the Wnt bead experiments, which L.R. initiated and standardized. Y.Q., A.M.G. and F.T. performed the Nkx6.2 cKO experiments. K.M. provided some sFRP mutant embryos, and both sFRP knockout lines. M.D.B. and M.S. generated and provided the r4-Cre line before publication. B.F. performed the NeuroVue labeling experiments with assistance from D.M.G. All authors commented on and contributed to manuscript revisions. A.C. conceived the project, supervised the experiments and co-wrote the manuscript.
References
- Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. The Zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev. Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. doi:10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine PVN, Zhao W, Kharode YP, Bex FJ, Lambert A-J, Goad MB, Gaur T, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol. Endocrinol. 2004;18:1222–1237. doi: 10.1210/me.2003-0498. doi:10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. doi:10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. doi:10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Concha ML, Takeuchi M, Ueno N, Wilson SW, Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. doi:10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning Heads: Development of Vertebrate Branchiomotor. Neurons. Dev. Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. doi:10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM, Nolan PM, Steel KP, Brown SDM, Gray IC, Murdoch JN. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 2003;13:1129–33. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- de Diego I, Kyriakopoulou K, Karagogeos D, Wassef M. Multiple influences on the migration of precerebellar neurons in the caudal medulla. Development. 2002;129:297–306. doi: 10.1242/dev.129.2.297. [DOI] [PubMed] [Google Scholar]
- Davey CF, Mathewson AW, Moens CB. PCP Signaling between Migrating Neurons and their Planar-Polarized Neuroepithelial Environment Controls Filopodial Dynamics and Directional Migration. PLoS Genet. 2016;12:e1005934. doi: 10.1371/journal.pgen.1005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonito M, Narita Y, Avallone B, Sequino L, Mancuso M, Andolfi G, Franzè AM, Puelles L, Rijli FM, Studer M. Assembly of the Auditory Circuitry by a Hox Genetic Network in the Mouse Brainstem. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003249. doi:10.1371/journal.pgen.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudney K, Stanier P. Epithelial cell polarity genes are required for neural tube closure. Am. J. Med. Genet. C. Semin. Med. Genet. 2005;135C:42–47. doi: 10.1002/ajmg.c.30052. doi:10.1002/ajmg.c.30052. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marín O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. doi:10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone CJ, Little PFR. The flamingo-related mouse Celsr family (Celsr1-3) genes exhibit distinct patterns of expression during embryonic development. Mech. Dev. 2001;109:91–94. doi: 10.1016/s0925-4773(01)00515-9. doi:10.1016/S0925-4773(01)00515-9. [DOI] [PubMed] [Google Scholar]
- Formstone CJ, Moxon C, Murdoch J, Little P, Mason I. Basal enrichment within neuroepithelia suggests novel function(s) for Celsr1 protein. Mol. Cell. Neurosci. 2010;44:210–222. doi: 10.1016/j.mcn.2010.03.008. doi:10.1016/j.mcn.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Christensen MA, Nichols DH. Fiber pathways and positional changes in efferent perikarya of 2.5- to 7-day chick embryos as revealed with DiI and dextran amines. J. Neurobiol. 1993;24:1481–1499. doi: 10.1002/neu.480241104. doi:10.1002/neu.480241104. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Muirhead K. a., Feng F, Gray BD, Ohlsson-Wilhelm BM. Diffusion and imaging properties of three new lipophilic tracers, NeuroVue Maroon, NeuroVue Red and NeuroVue Green and their use for double and triple labeling of neuronal profile. Brain Res. Bull. 2005;66:249–258. doi: 10.1016/j.brainresbull.2005.05.016. doi:10.1016/j.brainresbull.2005.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasco DM, Sittaramane V, Bryant W, Fritzsch B, Sawant A, Paudyal A, Stewart M, Andre P, Cadete Vilhais-Neto G, Yang Y, Song M-R, Murdoch JN, Chandrasekhar A. The mouse Wnt/PCP protein Vangl2 is necessary for migration of facial branchiomotor neurons, and functions independently of Dishevelled. Dev. Biol. 2012;369:211–222. doi: 10.1016/j.ydbio.2012.06.021. doi:10.1016/j.ydbio.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene NDE, Gerrelli D, Van Straaten HWM, Copp AJ. Abnormalities of floor plate, notochord and somite differentiation in the loop-tail (Lp) mouse: A model of severe neural tube defects. Mech. Dev. 1998;73:59–72. doi: 10.1016/s0925-4773(98)00029-x. doi:10.1016/S0925-4773(98)00029-X. [DOI] [PubMed] [Google Scholar]
- Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8:859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. doi:10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Preparing frozen tissue sections for immunostaining. CSH Protoc. 2006a;2006 doi: 10.1101/pdb.prot4328. doi:10.1101/pdb.prot4328. doi:10.1101/pdb.prot4328. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Binding antibodies to tissue sections. CSH Protoc. 2006b;2006 doi: 10.1101/pdb.prot4331. doi:10.1101/pdb.prot4331. doi:10.1101/pdb.prot4331. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Mounting samples in gelvatol or mowiol. CSH Protoc. 2006c;2006 doi: 10.1101/pdb.prot4461. doi:10.1101/pdb.prot4461. doi:10.1101/pdb.prot4461. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Topczewski J, Bingham S, Sepich DS, Marlow F, Chandrasekhar A, Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. doi:10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. doi:10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Luo M, Zhang Y, Wilkes DC, Ge G, Grieskamp T, Yamada C, Liu T-C, Huang G, Basson CT, Kispert A, Greenspan DS, Sato TN. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat. Cell Biol. 2009;11:46–55. doi: 10.1038/ncb1811. doi:10.1038/ncb1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q, Jeong Y, Misra K, Li S, Zelman AK, Epstein DJ, Matise MP. Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev. Cell. 2006;11:325–337. doi: 10.1016/j.devcel.2006.06.013. doi:10.1016/j.devcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Bach A, Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech. Dev. 1998;75:29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- Mapp OM, Wanner SJ, Rohrschneider MR, Prince VE. Prickle1b mediates interpretation of migratory cues during zebrafish facial branchiomotor neuron migration. Dev. Dyn. 2010;239:1596–1608. doi: 10.1002/dvdy.22283. doi:10.1002/dvdy.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Rubenstein JLR. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. doi:10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Misra K, Matise MP. A critical role for sFRP proteins in maintaining caudal neural tube closure in mice via inhibition of BMP signaling. Dev. Biol. 2010;337:74–83. doi: 10.1016/j.ydbio.2009.10.015. doi:10.1016/j.ydbio.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Moreno-Bravo JA, Perez-Balaguer A, Martinez S, Puelles E. Dynamic expression patterns of Nkx6.1 and Nkx6.2 in the developing mes-diencephalic basal plate. Dev. Dyn. 2010;239:2094–2101. doi: 10.1002/dvdy.22327. doi:10.1002/dvdy.22327. [DOI] [PubMed] [Google Scholar]
- Murdoch J, Doudney K, Paternotte C, Copp A, Stanier P. Severe neural tube defects in the loop-tail mouse result from mutations of loopin, a novel transmembrane protein. 2001;10:2593–2601. doi: 10.1093/hmg/10.22.2593. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. doi:10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Okada Y, Takeda S, Tanaka Y, Belmonte JCI, Hirokawa N. Mechanism of nodal flow: A conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. doi:10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Sander M, Ericson J. Complementary roles for Nkx6 and Nkx2 class proteins in the establishment of motoneuron identity in the hindbrain. Development. 2003;130:4149–4159. doi: 10.1242/dev.00641. doi:10.1242/dev.00641. [DOI] [PubMed] [Google Scholar]
- Qiu M, Shimamura K, Sussel L, Chen S, Rubenstein JLR. Control of anteroposterior and dorsoventral domains of Nkx-6.1 gene expression relative to other Nkx genes during vertebrate CNS development. Mech. Dev. 1998;72:77–88. doi: 10.1016/s0925-4773(98)00018-5. doi:10.1016/S0925-4773(98)00018-5. [DOI] [PubMed] [Google Scholar]
- Qu Y, Glasco DM, Zhou L, Sawant A, Ravni A, Fritzsch B, Damrau C, Murdoch JN, Evans S, Pfaff SL, Formstone C, Goffinet AM, Chandrasekhar A, Tissir F. Atypical cadherins Celsr1-3 differentially regulate migration of facial branchiomotor neurons in mice. J. Neurosci. 2010;30:9392–9401. doi: 10.1523/JNEUROSCI.0124-10.2010. doi:10.1523/JNEUROSCI.0124-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravni A, Qu Y, Goffinet AM, Tissir F. Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J. Invest. Dermatol. 2009;129:2507–2509. doi: 10.1038/jid.2009.84. doi:10.1038/jid.2009.84. [DOI] [PubMed] [Google Scholar]
- Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–5040. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Seitanidou T, Charnay P, Lumsden a. Segmental and neuronal architecture of the hindbrain of Krox-20 mouse mutants. Development. 1997;124:1215–1226. doi: 10.1242/dev.124.6.1215. [DOI] [PubMed] [Google Scholar]
- Serafini T, Colamarino S. a., Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. doi:10.1016/S0092-8674(00)81795-X. [DOI] [PubMed] [Google Scholar]
- Shima Y, Copeland NG, Gilbert DJ, Jenkins N. a., Chisaka O, Takeichi M, Uemura T. Differential expression of the seven-pass transmembrane cadherin genes Celsr1-3 and distribution of the Celsr2 protein during mouse development. Dev. Dyn. 2002;223:321–332. doi: 10.1002/dvdy.10054. doi:10.1002/dvdy.10054. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Lewcock JW, Lettieri K, Pfaff SL. FGF as a target-derived chemoattractant for developing motor axons genetically programmed by the LIM code. Neuron. 2006;50:841–53. doi: 10.1016/j.neuron.2006.04.030. doi:10.1016/j.neuron.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Sittaramane V, Pan X, Glasco DM, Huang P, Gurung S, Bock A, Li S, Wang H, Kawakami K, Matise MP, Chandrasekhar A. The PCP protein Vangl2 regulates migration of hindbrain motor neurons by acting in floor plate cells, and independently of cilia function. Dev. Biol. 2013;382:400–412. doi: 10.1016/j.ydbio.2013.08.017. doi:10.1016/j.ydbio.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M-R, Shirasaki R, Cai C-L, Ruiz EC, Evans SM, Lee S-K, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. doi:10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR. Moving cell bodies: Understanding the migratory mechanism of facial motor neurons. Arch. Pharm. Res. 2007;30:1273–1282. doi: 10.1007/BF02980268. doi:10.1007/BF02980268. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. doi:10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. doi:10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka H, Fritzsch B, Glover JC. Connecting ears to eye muscles: evolution of a 'simple' reflex arc. Brain Behav Evol. 2014;83:162–175. doi: 10.1159/000357833. [DOI] [PubMed] [Google Scholar]
- Tissir F, De-Backer O, Goffinet a. M., Lambert de Rouvroit C. Developmental expression profiles of Celsr (Flamingo) genes in the mouse. Mech. Dev. 2002;112:157–160. doi: 10.1016/s0925-4773(01)00623-2. doi:10.1016/S0925-4773(01)00623-2. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Expression of planar cell polarity genes during development of the mouse CNS. Eur. J. Neurosci. 2006;23:597–607. doi: 10.1111/j.1460-9568.2006.04596.x. doi:10.1111/j.1460-9568.2006.04596.x. [DOI] [PubMed] [Google Scholar]
- Valiente M, Marín O. Neuronal migration mechanisms in development and disease. Curr. Opin. Neurobiol. 2010;20:68–78. doi: 10.1016/j.conb.2009.12.003. doi:10.1016/j.conb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Vivancos V, Chen P, Spassky N, Qian D, Dabdoub A, Kelley M, Studer M, Guthrie S. Wnt activity guides facial branchiomotor neuron migration, and involves the PCP pathway and JNK and ROCK kinases. Neural Dev. 2009;4:7. doi: 10.1186/1749-8104-4-7. doi:10.1186/1749-8104-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiculescu O, Charnay P, Schneider-Maunoury S. Expression pattern of a Krox-20/Cre knock-in allele in the developing hindbrain, bones, and peripheral nervous system. genesis. 2000;26:123–126. doi: 10.1002/(sici)1526-968x(200002)26:2<123::aid-gene7>3.0.co;2-o. doi:10.1002/(SICI)1526-968X(200002)26:2<123::AID-GENE7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Wada H, Iwasaki M, Sato T, Masai I, Nishiwaki Y, Tanaka H, Sato A, Nojima Y, Okamoto H. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 2005;132:2273–2285. doi: 10.1242/dev.01810. doi:10.1242/dev.01810. [DOI] [PubMed] [Google Scholar]
- Wada H, Tanaka H, Nakayama S, Iwasaki M, Okamoto H. Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development. 2006;133:4749–4759. doi: 10.1242/dev.02665. doi:10.1242/dev.02665. [DOI] [PubMed] [Google Scholar]
- Wanner SJ, Saeger I, Guthrie S, Prince VE. Facial motor neuron migration advances. Curr. Opin. Neurobiol. 2013;23:943–50. doi: 10.1016/j.conb.2013.09.001. doi:10.1016/j.conb.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Bassuk AG, Stricker S, Fritzsch B. Prickle1 is necessary for the caudal migration of murine facial branchiomotor neurons. Cell Tissue Res. 2014;357:549–561. doi: 10.1007/s00441-014-1925-6. doi:10.1007/s00441-014-1925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.