SNAREs are incorporated into COPII vesicles by direct interaction with Sec24. In mammals, Sec24 isoforms recruit either Sec22b or the Q-SNARE complex comprising Syntaxin5, GS27, and Bet1. Analysis of immunoisolated COPII vesicles and intracellular localization of Sec24 isoforms indicates that all ER-Golgi SNAREs are present on the same vesicles.
Abstract
Secretory proteins are exported from the endoplasmic reticulum in COPII vesicles. SNARE proteins—core machinery for membrane fusion—are incorporated into COPII vesicles by direct interaction with Sec24. Here we report a novel mechanism for sorting of the ER–Golgi Q-SNAREs into COPII vesicles. Different mammalian Sec24 isoforms recruit either the R-SNARE Sec22b or the Q-SNAREs Syntaxin5, GS27, and Bet1. Syntaxin5 is the only Q-SNARE that directly interacts with Sec24C, requiring its “open” conformation. Mutation within the IxM cargo-binding site of Sec24C led to a drastic reduction in sorting of all three Q-SNAREs into COPII vesicles, implying their ER export as a preassembled complex. Analysis of immunoisolated COPII vesicles and intracellular localization of Sec24 isoforms indicate that all ER–Golgi SNAREs are present on the same vesicle. Combined with existing data, our findings yield a general concept of how Sec24 isoforms can recruit fusogenic SNARE subunits to keep them functionally apart and thus prime mammalian COPII vesicles for homotypic fusion.
INTRODUCTION
A key feature of eukaryotic cells is the presence of an elaborate endomembrane system that divides the cell into spatially and functionally separated compartments. These organelles are interconnected to each other via vesicular transport pathways maintaining their homeostasis and ensuring their specialized functions. Each trafficking step requires the orchestrated interplay of a distinct set of small GTP-binding proteins of the Arf and Rab family, coat proteins, tether proteins, and soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) proteins (reviewed in Bonifacino and Glick, 2004; Cai et al., 2007; Brocker et al., 2010; Faini et al., 2013).
SNAREs make up the core machinery for intracellular fusion events and were initially classified based on their distinct localization to either vesicle or target membrane as v- and t-SNAREs, respectively (Sollner et al., 1993). SNARE proteins are either tail-anchored membrane proteins or anchored to membranes via lipid modification. They are characterized by a 60– to 70–amino acid long stretch of heptad repeats generally referred to as the SNARE motif (Weimbs et al., 1997). A variety of N-terminal domains precede the SNARE motif, ranging from short, unstructured peptides to longin domains (e.g. in Sec22b, Ykt6, and VAMP7) or Habc domains (in Syntaxins) that are connected to the SNARE motif via a flexible linker region (reviewed in Brunger, 2005; Hong, 2005; Jahn and Scheller, 2006; Malsam et al., 2008). SNARE-mediated membrane fusion is driven by the formation of a four-helix coiled-coil bundle (known as a SNARE pin) of the heptad repeat SNARE motifs from a single v- and two or three t-SNAREs residing in opposing membranes (Sutton et al., 1998; Weber et al., 1998). Based on sequence and structural homologies and the presence of a characteristic amino acid (Arg [R] or Gln [Q]) in the central zero ionic layer (0-layer) of the heptad repeats, SNARE proteins were also classified as R-SNAREs or Q-SNAREs. Furthermore, it was proposed that fusion-competent SNARE complexes generally consist of one R-SNARE and three Q-SNAREs (Fasshauer et al., 1998). The R/Q-SNARE nomenclature is needed in particular for homotypic fusion events, in which the v/t-SNARE designation does not suffice to distinguish between SNARE complex subunits. SNARE proteins together with small GTP-binding proteins of the Rab family and tethering proteins contribute to compartmental specificity by guiding and regulating the different fusion events along the secretory pathway (reviewed in Cai et al., 2007). Therefore their intracellular distribution and activity has to be rigorously regulated.
COPII vesicles mediate export of newly synthesized proteins from the endoplasmic reticulum (ER; reviewed in Zanetti et al., 2012; D’Arcangelo et al., 2013; Adolf and Wieland, 2014). Their coat consists of the small GTP-binding protein secretion-associated and Ras-related protein (Sar1) and the heterooligomeric subcomplexes Sec23/24 and Sec13/31 (Barlowe et al., 1994). In mammalian cells, COPII vesicles are formed at specialized subdomains of the ER termed ER exit sites (ERES) or transitional ER (Orci et al., 1991; Bannykh et al., 1996; Hammond and Glick, 2000). The biogenesis of COPII vesicles is initiated by recruitment and activation of the small GTP-binding protein Sar1 via the guanine nucleotide exchange factor Sec12 (Nakano et al., 1988; d’Enfert et al., 1991; Barlowe and Schekman, 1993). The COPII coat is then assembled in a successive manner (Matsuoka et al., 1998). The inner coat subcomplex Sec23/24 is recruited to the ER via an interaction of Sec23 with Sar1 (Bi et al., 2002), which in turn recruits the outer coat subcomplex Sec13/31 (Bi et al., 2007).
Pioneering studies revealed the membrane-associated Sar1/Sec23/24 complex to be involved in the selection of transmembrane cargo proteins destined for ER export (Aridor et al., 1998; Kuehn et al., 1998). Subsequent genetic, biochemical, and structural studies identified Sec24 as the major cargo-binding subunit, with multiple cargo-binding sites (Pagano et al., 1999; Miller et al., 2002, 2003; Mossessova et al., 2003). Mammalian cells express four isoforms of Sec24 (Sec24A–D), which are believed to expand the cargo repertoire that can be selectively sorted into COPII vesicles (Pagano et al., 1999; Roberg et al., 1999; Peng et al., 2000; Shimoni et al., 2000; Miller et al., 2002; Mancias and Goldberg, 2007, 2008; Wendeler et al., 2007; Bonnon et al., 2010). All four Sec24 isoforms are ubiquitously expressed. Based on sequence homology, they were assigned to two subgroups, Sec24A/B and Sec24C/D (Pagano et al., 1999; Tang et al., 1999).
The R-SNARE Sec22p and the Q-SNAREs Sed5p, Bos1p, and Bet1p in Saccharomyces cerevisiae (Newman et al., 1990; Shim et al., 1991; Hardwick and Pelham, 1992; Lian and Ferro-Novick, 1993; Sacher et al., 1997), like their mammalian homologues Sec22b, Syntaxin5, GS27, and Bet1 (Dascher et al., 1994; Hay et al., 1997, 1998; Paek et al., 1997; Zhang et al., 1997; Xu et al., 2000), form a SNARE complex implicated in anterograde ER-to-ER–Golgi intermediate compartment (ERGIC)/Golgi transport.
Of note, the molecular mechanisms that underlie ER-to-Golgi SNARE sorting by COPII coat proteins are strikingly different in S. cerevisiae and mammalian cells. The S. cerevisiae Sec24 homologues Sec24p and Iss1p (both homologues of Sec24A/B in mammals) recruit all four ER-to-Golgi SNAREs into COPII vesicles. Sed5p, Bet1p, and Sec22p each has its own ER-export signal, which is recognized by an independent binding site within the Sec24 subunits termed the A-, B-, and C-sites, respectively (Kurihara et al., 2000; Miller et al., 2002, 2003, 2005; Mossessova et al., 2003). Consistently, Lst1p (homologue of Sec24C/D in mammals) was found to be present together with Sec24p on the same vesicular membrane (Shimoni et al., 2000)
In contrast, the mammalian SNAREs are sorted into COPII vesicles by either Sec24A/B or Sec24C/D. Short peptide motifs of the consensus sequence IxM within Syntaxin5 and GS27 serve for sorting of the Qa- and Qb-SNAREs, recognized by a specific binding site within Sec24C/D (Mancias and Goldberg, 2008). The R-SNARE Sec22b, on the other hand, is sorted into COPII vesicles via a conformational epitope formed when its N-terminal longin domain folds back onto its SNARE motif. This so-called “closed” conformation is recognized by a binding site molded in the interface between Sec23 and Sec24A/B (Mancias and Goldberg, 2007). Similarly, Bet1 was reported to interact with Sec24A/B via a short YxxCE sequence (Mancias and Goldberg, 2008), deduced from a previously identified LxxLE transport signal present in its S. cerevisiae homologue, Bet1p (Mossessova et al., 2003). Furthermore, in an independent study, the mammalian isoforms of the two Sec24 subclasses were proposed to localize to distinct ER exit sites and give rise to two subpopulations of COPII vesicles (Bonnon et al., 2010). This localization is difficult to reconcile with the reported SNARE specificities of the various Sec24 isoforms because the SNARE distribution reported would result in two types of vesicles incapable of undergoing homotypic fusion.
Taken together, our present state of knowledge on the transport of fusion-competent SNAREs does not allow us to answer the questions of 1) how homotypic fusion of ER-derived COPII vesicles can occur and 2) how the SNARE subunits can be kept in their fusogenic state during their transport.
In this study, we address seeming inconsistencies of previous studies by analyzing sorting of mammalian SNAREs in COPII vesicles in the context of vesicle formation. Similar to previous studies, we find that Sec24A/B recognize Sec22b, whereas Syntaxin5 and GS27 are selectively sorted by Sec24C/D. In addition, and in contrast to other reports, we find that Bet1 is specifically packaged into COPII vesicles as a subunit of a trimeric Q-SNARE complex that is recognized by a selective interaction of Sec24C/D with its Syntaxin5 subunit. Furthermore, by immunofluorescence and immunoprecipitation experiments, we find that Sec24 isoforms colocalize within ERES and consistently on COPII transport vesicles. Taken together, our data support a model in which isoform-specific sorting of R-SNARE and preassembled Q-SNAREs into the same population of COPII vesicles allow priming of these vesicles for homotypic fusion after coat disassembly.
RESULTS
The ER–Golgi R- and Q-SNAREs are differentially sorted by isoforms of the COPII coat subunit Sec24
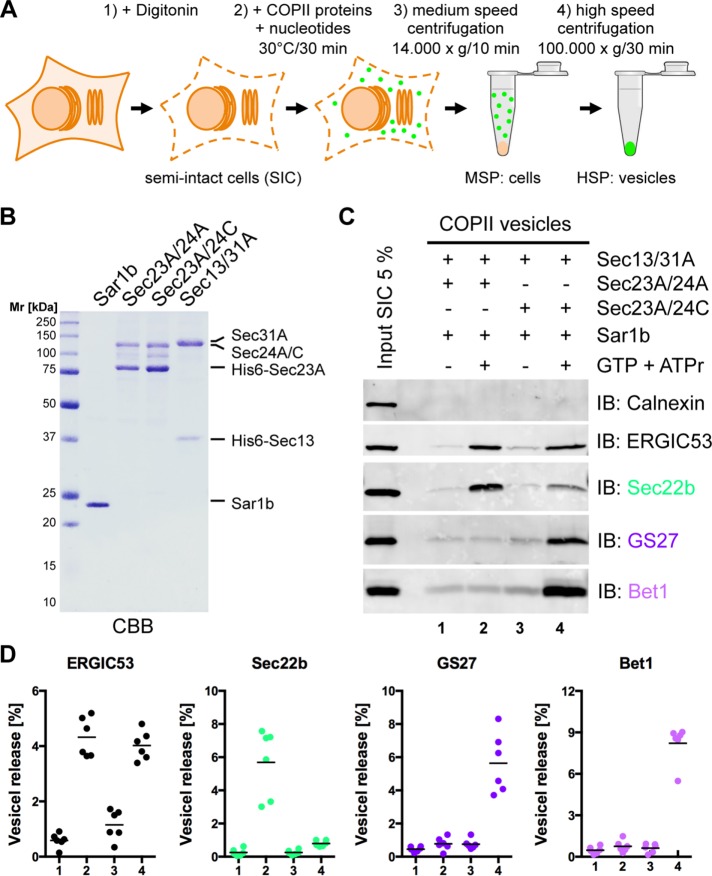
To investigate sorting of SNARE proteins into COPII-coated vesicles by human Sec24 isoforms, we used a well-established in vitro COPII vesicle reconstitution assay using digitonin-permeabilized cells as donor membranes (Figure 1A; Mancias and Goldberg, 2007, 2008; Adolf et al., 2013).
FIGURE 1:
The R-SNARE Sec22b and the Q-SNAREs GS27 and Bet1 are differentially sorted by isoforms of the COPII coat subunit Sec24. (A) Schematic of in vitro formation of COPII vesicles from semi-intact cells. (B) Recombinant purified hamster Sar1b (0.5 μg) expressed in E. coli, the inner COPII coat subcomplex His6-Sec23A/23A or His6-Sec23A/24C (1 μg), and the outer coat subcomplex His6-Sec13/31A (1.5 μg) expressed in Sf9 insect cells were affinity purified and separated by SDS–PAGE and stained with CBB. (C) Formation of COPII vesicles was reconstituted in vitro by incubation of semi-intact HeLa cells with Sar1b, Sec23A/24A, Sec23A/24C, or Sec13/31A in the presence of GTP and an ATP-regenerating system (ATPr) as indicated. Newly formed vesicles were separated from donor membranes by differential centrifugation. The vesicle fractions and 5% of SICs used for reconstitution were analyzed by Western blotting for the presence of the non–vesicle membrane marker calnexin and the ER-to-Golgi SNARE proteins GS27, Sec22b, and Bet1. (D) Amount of vesicle membrane marker ERGIC53 and ER-to-Golgi SNARE proteins GS27, Sec22b, and Bet1 in vesicle fractions quantified using Li-COR Image Studio software (n = 6; horizontal lines indicate the means).
To study isoform-specific cargo selection, we expressed Sec24A (representing the Sec24A/B subclass) or Sec24C (representing the Sec24C/D subclass) in complex with Sec23A, as well as the outer coat subcomplex Sec13/31, in insect cells. Preparations of the purified COPII coat proteins Sar1b, Sec23A/24A, Sec23A/24C, and Sec13/31A used in this study are depicted in Figure 1B. For reconstitution of COPII vesicles, semi-intact cells (SICs) were incubated with Sar1b, Sec23A/24A or Sec23A/24C, Sec13/31A, and GTP. Vesicle fractions were obtained by differential centrifugation and probed for the presence of various proteins by Western blotting (Figure 1C).
Clearly, vesicle formation was dependent on the presence of GTP (Figure 1C; compare lanes 1 and 3 with lanes 2 and 4, respectively). The ER-resident protein calnexin, which is not selected into COPII vesicles, was used as a control to assess specificity of the vesicle reconstitution assay (Figure 1C; compare Input with lanes 1–4). The cycling cargo adaptor protein ERGIC53, a Sec24 isoform–independent constituent of COPII vesicles (Mancias and Goldberg, 2007), served as a measure for the amount of vesicles generated. Based on the signal intensity of ERGIC53, similar amounts of COPII vesicles were generated when either the Sec23A/24A or Sec23A/24C coat subcomplex was used (Figure 1C; compare Input with lanes 2 and 4). Of importance, the R-SNARE Sec22b predominates in the vesicles formed with Sec24A, whereas the Q-SNAREs GS27 and Bet1 are mainly found in vesicles reconstituted with Sec24C (Figure 1C; compare lanes 2 and 4). A quantification of independent experiments is shown in Figure 1D. Sec22b is enriched sevenfold to eightfold in vesicles generated with the isoform Sec24A, and similarly, GS27 and Bet1 are enriched in Sec24C vesicles by a factor of approximately six to eight.
These results demonstrate that Sec24A and Sec24C discriminate between ER-to-Golgi R- and Q-SNAREs. Sec24A sorts only the R-SNARE Sec22b into COPII vesicles, whereas the Q-SNAREs Syntaxin5 and GS27 (Mancias and Goldberg, 2008), as well as the Qc-SNARE Bet1, are clients of Sec24C. These findings raised the questions of 1) how Bet1 is selectively sorted by the isoforms Sec24C/D and 2) whether in a cell, the ER-to-Golgi R-SNARE and Q-SNAREs are incorporated into distinct vesicles or the same vesicle.
Syntaxin5 in its open conformation, but not Bet1, interacts with Sec24C
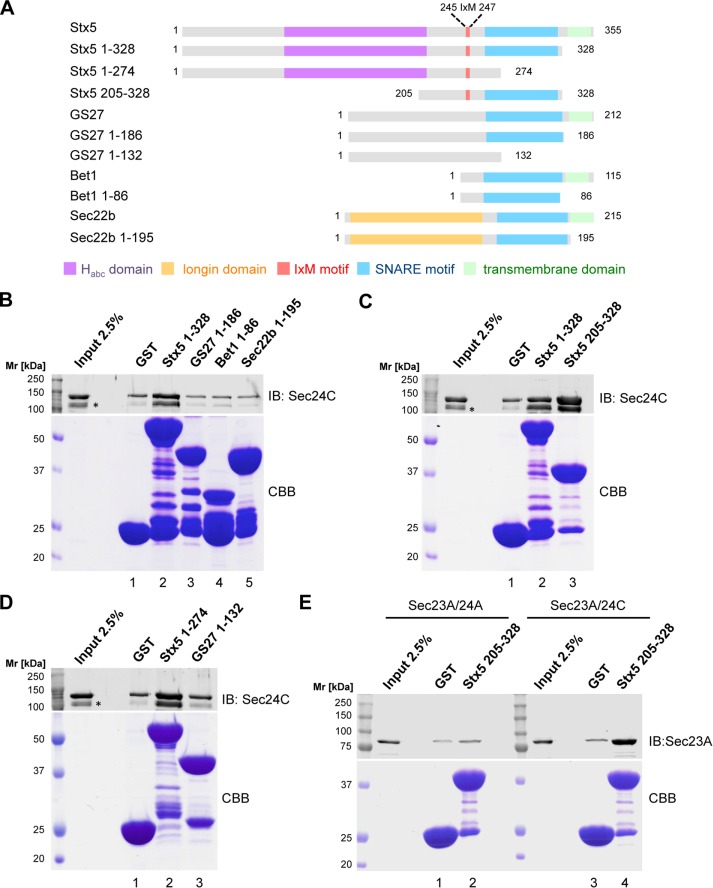
Syntaxin5 and its S. cerevisiae counterpart, Sed5p, as well as GS27 and its homologue, Bos1p, have structured N-terminal domains connected to their SNARE motifs via a flexible linker (Xu et al., 2000; Mossessova et al., 2003; Williams et al., 2004). Conversely, Bet1 is predicted to have a short, unstructured N-terminus preceding its SNARE motif. The domain organization of the various ER–Golgi SNAREs is depicted in Figure 2A.
FIGURE 2:
Syntaxin5 preferentially interacts with Sec24C in its open conformation. (A) Domain organization of human ER-to-Golgi SNARE and fusion proteins used in this study. The Habc domain of Syntaxin5 is depicted in purple, the longin domain of Sec22b in orange, the known IxM motif of Syntaxin5 in red, SNARE motifs in blue, and transmembrane domains (TMD) in green. (B–E) Direct interaction of the ER-to-Golgi SNARE fusion proteins was probed by GST pull-down assay. Sec23A/24A or Sec23A/24C bound to the SNARE-GST fusion proteins indicated and inputs of Sec23A/24A or Sec23A/24C (2.5%) were separated by SDS–PAGE. The lower part of the polyacrylamide gel containing SNARE-GST fusion proteins was stained with CBB, and the upper part was analyzed by Western blotting for the presence of Sec24C (B–D) or Sec23 (E). *N-terminally degraded recombinant Sec24C.
To investigate whether Bet1 is sorted into COPII vesicles by a direct interaction between the Qc-SNARE and the COPII coat subunit Sec24C, we expressed the entire cytoplasmic domains of the ER-to-Golgi SNAREs Syntaxin5, GS27, Bet1, and Sec22b fused to the N-terminus of glutathione S-transferase (GST) to mimic their natural orientation in a membrane and probed for binding to Sec24C. A schematic representation of the fusion proteins used is depicted in Figure 2A. A similar approach was used previously to identify the IxM cargo-sorting motifs in Syntaxin5 and GS27 (Mancias and Goldberg, 2008). Among the complete cytoplasmic domains of the ER-to-Golgi SNAREs, moderate but robust binding to Sec24C was detectable only for the Qa-SNARE Syntaxin5. By contrast, GS27, Bet1, and Sec22b fusion proteins did not show binding above background as determined with a GST control (Figure 2B).
Syntaxin family members are known to switch between “open” and “closed” conformations, which was first reported for Sytaxin1A and its yeast homologue, Sso1p. In the closed conformation, their N-terminal Habc domain is folded back to interact with the SNARE motif (Calakos et al., 1994; Fernandez et al., 1998; Nicholson et al., 1998; Dulubova et al., 1999; Fiebig et al., 1999; Misura et al., 2000; Munson et al., 2000). Syntaxin5 and Sed5p have similar N-terminal folded Habc domains, which also are believed to confer equilibrium between open and closed conformations (Xu et al., 2000; Yamaguchi et al., 2002; Williams et al., 2004). Of interest, the S. cerevisiae Syntaxin5 homologue, Sed5p, was reported to possess a higher binding affinity to Sec24p when in its open conformation (Mossessova et al., 2003). In light of these results, we further investigated the mechanism of binding of Syntaxin5 to Sec24C. To specifically study the open, form we analyzed binding to Sec24C of a truncated variant of Syntaxin5 that lacks the entire regulatory Habc domain. In pull-down experiments, we observed a markedly enhanced interaction of the open-conformation mimic of Syntaxin5 (Figure 2C). Similarly, to address whether lack of binding to Sec24C of the GS27 fusion protein comprising the entire cytoplasmic domain is due to the ability of the SNARE to adopt a closed conformation, we investigated Syntaxin5 and GS27 fusion constructs lacking their respective SNARE motifs. In line with the foregoing results described, binding to Sec24C was detectable only with the Syntaxin5 1-274-GST fusion protein (Figure 2D). To address the specificity of SNARE binding of Sec24 isoforms in the GST pull-down assay, we tested binding of either Sec23A/24A or Sec23A/24C to the Syntaxin5 205–328 fusion protein. To directly compare binding of the two subcomplexes, we analyzed pull downs for the presence of Sec23A. In line with results obtained from in vitro reconstitution of COPII vesicles (Figure 1), Syntaxin5 was found to interact directly with Sec24C but not with Sec24A (Figure 2E).
Taken together, our data imply that only the Qa-SNARE Syntaxin5 directly binds to the COPII coat subunit Sec24C, whereas GS27 and Bet1 do not directly interact with the COPII coat.
Mutation of the IxM-binding site within Sec24C leads to a sorting defect for all three Q-SNAREs
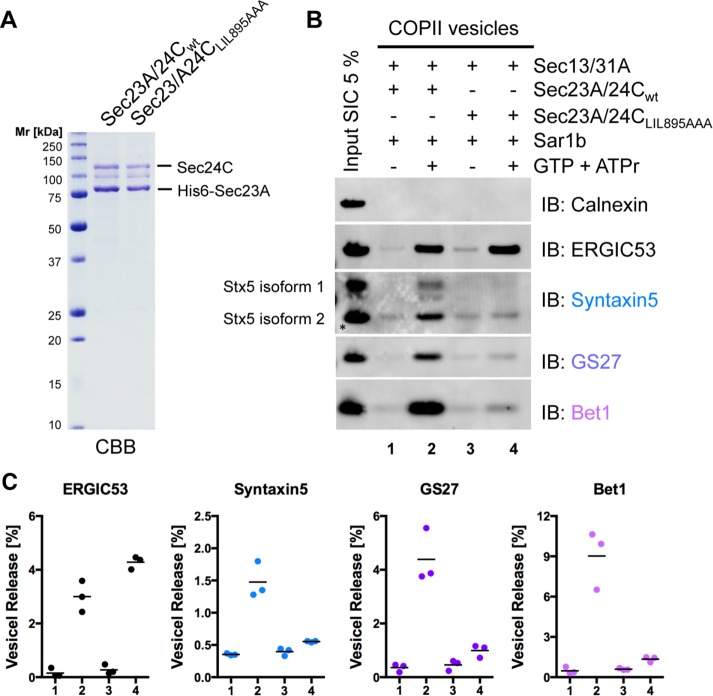
To further challenge this view, we took advantage of a previously described Sec24C mutant deficient in binding the IxM motif (Mancias and Goldberg, 2008). Sec24C LIL895AAA was expressed in complex with Sec23A in insect cells (Figure 3A) and used in the COPII vesicle budding assay.
FIGURE 3:
Mutation of the IxM-binding site within Sec24C affects sorting of the three ER–Golgi Q-SNAREs Syntaxin5, GS27, and Bet1. (A) Recombinant purified inner coat subcomplex His6-Sec23A/24C wild type or His6-Sec23A/24C LIL895AAA (1 μg) expressed in Sf9 insect cells was separated by SDS–PAGE and stained with CBB. (B) Formation of COPII vesicles was reconstituted in vitro by incubation of semi-intact HeLa cells with Sar1b, Sec23A/24C wild type, or Sec23A/24C LIL895AAA or Sec13/31A in the presence of GTP and an ATP-regenerating system (ATPr) as indicated. Newly formed vesicles were separated from donor membranes by differential centrifugation. The vesicle fractions and 5% of SICs used for reconstitution were analyzed by Western blotting for the presence of the non–vesicle membrane marker calnexin and the ER-to-Golgi Q-SNARE proteins Syntaxin5, GS27, and Bet1. (C) Amount of vesicle membrane marker ERGIC53 and ER-to-Golgi Q-SNARE proteins Syntaxin5, GS27, and Bet1 in vesicle fractions quantified using Li-COR Image Studio software (n = 3; horizontal lines indicate the means). The lower band of Syntaxin5 (isoform 2, marked by an asterisk) was used for quantification.
Vesicle fractions generated with Sec24C wild type and Sec24C LIL895AAA were analyzed for the presence of the non–vesicle marker calnexin as well as for the COPII vesicle markers ERGIC53 and the three ER–Golgi Q-SNARE proteins. Vesicle reconstitutions with Sec24C wild type and Sec24C LIL895AAA yielded similar amounts of vesicles deduced form the abundance of ERGIC53 (Figure 3B; compare lanes 2 and 4). Strikingly, not only were Syntaxin5 and GS27 lost from vesicle fractions generated with Sec24C LIL895AAA, as has been described (Mancias and Goldberg, 2008), but so was the Qc-SNARE Bet1 (Figure 3B; compare lanes 2 and 4). Quantification of independent reconstitution experiments confirmed consistent depletion of all three Q-SNAREs (Figure 3C). Taking the results together, binding of Sec24C to Syntaxin5 only and the loss of all Q-SNAREs in vesicles made with Sec24C LIL895AAA strongly imply that Syntaxin5, GS27, and Bet1 are sorted as a preassembled complex via the previously described IxM cargo-sorting motif present in Syntaxin5.
The R-SNARE Sec22b and the Q-SNAREs Syntaxin5, GS27, and Bet1 are incorporated into the same vesicle
To form a fusion-competent trans-SNARE complex, the prevailing concept stipulates that the R-SNARE Sec22b must reside in one membrane and the corresponding Q-SNAREs Syntaxin5, GS27, and Bet1 in an opposing membrane (Fasshauer et al., 1998; Xu et al., 2000). Of interest, it has been proposed that isoforms of the two subclasses Sec24A/B and Sec24C/D reside in different ER subdomains and give rise to COPII vesicles coated homogeneously with Sec24 isoforms from one subclass (Bonnon et al., 2010).
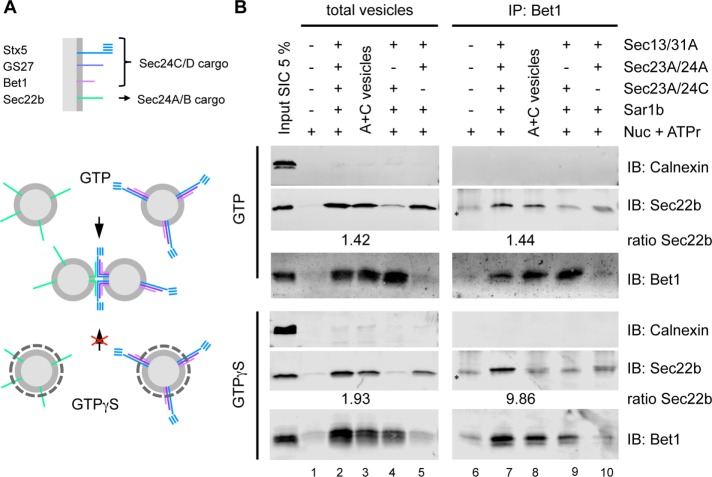
The SNARE selectivity of Sec24 isoforms as reported earlier would allow such a segregation of Sec24A/B and Sec24C/D and consequently of their respective cargo proteins into distinct vesicles. To test this hypothesis experimentally, we analyzed whether the R-SNARE and the Q-SNAREs can be found on the same vesicles. To this end, we adapted a previously reported method to immunoisolate COPII vesicles (Muniz et al., 2001). Vesicles reconstituted in the presence of GTP rapidly lose their coat after membrane scission, whereas vesicles reconstituted with the poorly hydrolyzable GTP analogue GTPγS remain coated and therefore cannot undergo homotypic fusion, as schematically depicted in Figure 4A. Hence, for immunoisolation of COPII vesicles, we performed the standard in vitro budding assay in the presence of either GTP or GTPγS, which allowed us to discriminate between segregation of SNARE proteins into distinct vesicles and vesicles that are fused/tethered to each other. After medium-speed centrifugation, the COPII vesicle preparations were divided. From one fraction, vesicles were directly harvested by high-speed centrifugation to assess the total amount of vesicles (Figure 4B, left, total vesicles). To isolate Q-SNARE–containing vesicles, the other fraction was immunoprecipitated with anti-Bet1 antibodies (Figure 4B, right, IP:Bet1). Both samples were subsequently analyzed by Western blotting for presence of the R-SNARE Sec22b and the Q-SNAREs (Bet1). COPII vesicles were reconstituted with either Sec24 isoform alone (Figure 4B, lanes 4/5 and 9/10) or with a combination of both (Figure 4B, lanes 2 and 7). Similarly, vesicles were generated separately with either isoform, mixed after medium-speed centrifugation, and further incubated for 10 min at 30°C to allow tethering/fusion (Figure 4B, lanes 3 and 8, A+C vesicles).
FIGURE 4:
The R-SNARE Sec22b and the Q-SNAREs Bet1 are present on the same vesicle membrane. (A) Schematic representation of homotypic COPII vesicle fusion. COPII vesicles reconstituted in the presence of GTP rapidly lose their coat after vesicle membrane scission, whereas vesicles reconstituted with the poorly hydrolyzable GTP analogue GTPγS remain coated and hence cannot undergo homotypic COPII vesicle fusion. (B) Formation of COPII vesicles was reconstituted in vitro by incubation of semi-intact HeLa cells with Sar1b, Sec23A/24A, or Sec23A/24C, Sec13/31A, and GTP or GTPγS plus an ATP-regenerating system (ATPr) as indicated and separated from donor membranes by medium-speed centrifugation. Vesicle fractions were analyzed by Western blotting for the presence of the non–vesicle membrane marker calnexin and the ER-Golgi SNARE proteins Sec22b and Bet1 as indicated. (Asterisks in B indicate immunostaining of immunoglobulin light chains from immunoprecipitated antibodies.)
The same isoform preference for sorting of the SNARE proteins Sec22b and Bet1 was observed irrespective of whether GTP (Figure 4B, top) or the nonhydrolyzable analogue GTPγS (Figure 4B, bottom) was used for COPII vesicle reconstitution (Figure 4B; compare lanes 4 and 5). As expected, whenever Sec24C was present in the various reconstitution reactions, Bet1 was found in the corresponding vesicles (Figure 4B, lanes 2–4). Technically, it is of note that Bet1-containing vesicles could be immunoisolated irrespective of whether they were fusion competent (reconstituted with GTP) or fusion incompetent/coated (reconstituted with GTPγS; Figure 4B, lanes 7–9).
The Sec24A-specific cargo Sec22b was found in fusion-competent vesicles reconstituted with Sec24A and Sec24C in the same reaction (Figure 4B, lane 7, top) and similarly when vesicles generated separately with Sec24A or Sec24C were mixed together (Figure 4B, lane 8, top). This indicates that the mixed vesicles generated in the presence of GTP were able to tether or fuse with each other. In contrast, when reconstitutions were performed with GTPγS, Sec22b was predominantly found in vesicles made in the presence of both Sec24 isoforms (Figure 4B, lane 7, bottom).
We quantified the Sec22b signals and formed their ratio for vesicles prepared in the presence of both coats divided by the corresponding signals of mixed vesicles. Whereas under GTP conditions the ratios between pelleted and immunoprecipitated vesicles were unchanged, under GTPγS conditions, the ratio of the immunoprecipitated sample was about fivefold higher than the respective ratio of the input (total vesicles; Figure 4B; compare ratios in the top and bottom of lanes 2/3 with 7/8).
Taken together, these results show that, when both Sec24A and Sec24C are present, R- and Q-SNAREs are packaged into the same vesicle population, indicating that Sec24 isoforms cooperate in the biogenesis of individual vesicles and thus have to be present on the same ERES/vesicles.
Sec24A and Sec24C colocalize within the same ER exit sites
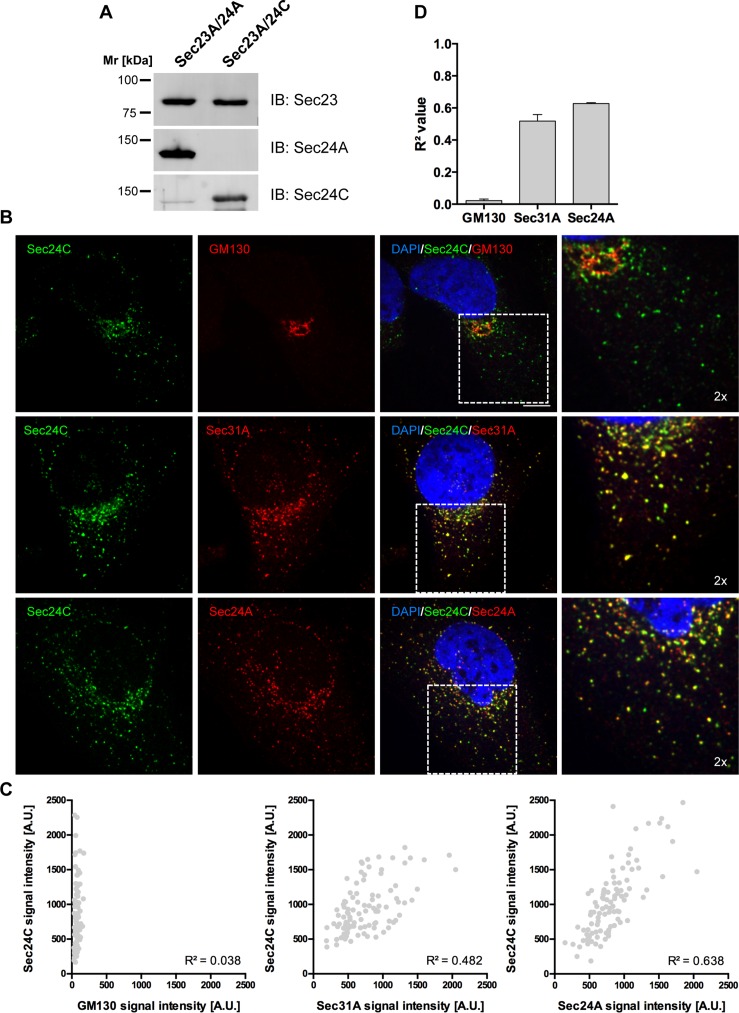
We further challenged our biochemical data by analyzing the intracellular localization of Sec24A and Sec24C in HeLa cells via immunofluorescence microscopy. To this end, we used antibodies that specifically recognize distinct Sec24 isoforms. To assess isoform specificity, we separated purified, recombinant Sec23A/24A and Sec23A/24C subcomplexes by SDS–PAGE and analyzed them by Western blotting. The antibodies directed against Sec24A and Sec24C showed barely any cross-reactivity (Figure 5A).
FIGURE 5:
Sec24A and Sec24C colocalize on the same ER exit sites in HeLa cells. (A) Specificity of Sec24-isoform antibodies. Equal amounts of recombinant Sec23A/24A and Sec23A/24C were separated by SDS–PAGE and analyzed by Western blotting with the antibodies indicated. (B) HeLa cells were methanol fixed and double stained with antibodies directed against the inner coat subunit Sec24C (rabbit anti-Sec24C) and the Golgi matrix protein GM130 (mouse anti-GM130, top), the outer coat subunit Sec31A (mouse anti-Sec31A, middle), or the inner coat subunit Sec24A (goat anti-Sec24A, bottom). Rabbit Igs were visualized with Alexa 488–labeled anti-rabbit IgGs, whereas mouse and goat IgGs were visualized with Alexa 546–labeled anti-mouse and anti-goat IgGs, respectively. Samples were embedded with ProLong Gold Antifade with DAPI and examined by confocal laser scanning microscopy. Confocal z-stacks were acquired on a Nikon ERS-6 spinning-disk CLSM, and z-stakes were deconvolved with Huygens. Shown is a single slice of the representative z-stacks (scale bar, 10 μm). Right, enlargements of the boxed regions in overlays (2×). (C) Quantification of colocalization of Sec24C with GM130, Sec31A, and Sec24A. Scatter plot of fluorescence intensities measured for Sec24C with GM130 (left), Sec31A (middle), and Sec24A (right) in individual ERES of a single cell shown in B. (D) Correlation coefficients (R2) derived from quantifications of ∼100 ERES from three individual cells (means ± SD).
To investigate whether endogenous Sec24 isoforms are segregated into different ER exit sites and consequently distinct COPII-coated vesicles, we fixed HeLa cells and coimmunolabeled them with anti-Sec24C antibody in combination with an antibody against Golgi matrix protein GM130, the outer COPII coat subunit Sec31A, or Sec24A. Single slices of representative z-stacks are depicted in Figure 5B.
Barely any colocalization was detected between GM130 and Sec24C (Figure 5B, top). When the localization of Sec24C was compared with outer COPII coat component Sec31A, a high level of colocalization was observed (Figure 5B, middle). Comparison of Sec24C with Sec24A resulted in a similar picture (Figure 5B, bottom). The degree of colocalization observed in punctuated ERES was even slightly higher in cells immunostained for Sec24C and Sec24A than with cells stained with Sec24C and Sec31A.
For a statistical evaluation of immunofluorescence images, ERES were randomly selected, and fluorescent signals for both channels were evaluated as previously described (Iwasaki et al., 2015). Adjacent signals for Sec24C were blotted against the corresponding signals of GM130, Sec31A, and Sec24A, and R2 values were obtained by linear regression (Figure 5C). Correlation coefficients derived from quantification of roughly 100 ERES in three different cells per condition are shown in Figure 5D. The highest correlation was obtained for colocalization of Sec24C and Sec24A, clearly indicating that there is no strict segregation of Sec24 isoforms into distinct ERES. This finding is consistent with our biochemical results that show coexistence of Sec24A- and Sec24C-specific cargo within the same vesicle.
DISCUSSION
SNARE proteins play a pivotal role in vesicle-targeting and fusion events in eukaryotic cells. Therefore their intracellular localization and coherent sorting into nascent transport vesicles are strictly regulated. In an attempt to reconcile proposed models for sorting of the ER-to-Golgi SNARE proteins, we investigated their sorting by mammalian Sec24 isoforms, as well as the intracellular localization of these COPII coat subunits. We found that the two subclasses of the mammalian Sec24 isoforms discriminate between the ER-to-Golgi R-SNARE Sec22b and the three Q-SNAREs Syntaxin5, GS27, and Bet1.
Using an established in vitro COPII vesicle reconstitution assay to monitor the requirement of individual Sec24 isoforms for sorting of ER-to-Golgi SNAREs, we show that Sec22b is specifically recognized by Sec24A, whereas Syntaxin5, GS27, and Bet1 are clients of Sec24C. On one hand, these results agree with the previously described preference of Sec24A/B for the R-SNARE Sec22b and of Sec24C/D for the Qa-SNARE Syntaxin5 and the Qb-SNARE GS27 (Mancias and Goldberg, 2007, 2008). On the other hand, earlier studies proposed that the mammalian Qc-SNARE Bet1 is sorted into COPII vesicles by Sec24A/B via a YxxCE motif deduced form the LxxLE motif of its S. cerevisiae homologue, Bet1p (Mancias and Goldberg, 2008). In yeast, the LxxLE motif binds to the so-called B-site, which also recognizes diacidic DxE motifs (Miller et al., 2003; Mossessova et al., 2003). However, the Sec24A/B preference for mammalian Bet1 was not tested in the context of COPII vesicle formation. Instead, a peptide of Bet1 comprising the 25YxxCE29 motif was soaked into a crystal of Sec24A (Mancias and Goldberg, 2008).
Because IxM sorting motifs have been identified in both Syntaxin5 and GS27 (Mancias and Goldberg, 2008), we initially speculated that Bet1 is also selectively incorporated into COPII vesicles by direct interaction with Sec24C/D, mediated by a yet-to-be-identified cargo-sorting motif. We tested this possibility with GST pull-down experiments. Strikingly, we were not able to detect a direct interaction between Sec23A/24C and GS27 or Bet1. Only the Qa-SNARE Syntaxin5 displayed moderate but robust binding to Sec23A/24C. Of particular interest was the observation that the strongest interaction between Syntaxin5 and Sec24C was detected with a minimal construct comprising only the linker region (containing the IxM motif previously identified) and the SNARE motif but lacking the entire N-terminal regulatory domain. Hence it is the open conformation of Syntaxin5 that binds Sec24C. This is consistent with the idea that its yeast homologue, Sed5p, binds to Sec24p in its open conformation via a YxxxNPF motif that is not accessible when the N-terminal Habc domain is folded back onto its own SNARE motif (Mossessova et al., 2003). We further excluded the possibility that the lack of binding observed of the GS27 construct comprising the entire cytoplasmic domain is a result of the adoption of a similar closed conformation. Consistent with our observation, a lack of binding of Bet1 to Sec23A/24C was previously reported (Kim et al., 2005).
On the basis of the Sec24 isoform–specific sorting of the SNARE proteins and the finding that only Syntaxin5 directly interacts with Sec24C, we hypothesized that GS27 and Bet1 are incorporated into COPII vesicles as a preassembled Q-SNARE complex via Syntaxin5. This hypothesis is further supported by the observation that mutations within the IxM cargo-binding site of Sec24C led to a severe defect in sorting of all three ER-to-Golgi Q-SNAREs into COPII vesicles. These two independent lines of evidence argue strongly against an alternative interpretation that GS27 and Bet1 are recruited into COPII vesicles via an additional, unknown protein that competes with their natural partner, Syntaxin5, for binding to the same Sec24 cargo-binding site.
A coupled transport mechanism similar to the one we propose for the mammalian ER-to-Golgi SNAREs has been proposed for the S. cerevisiae ER-to-Golgi SNAREs Sed5p, Bos1p, and Sec22p (Mossessova et al., 2003). Sed5p is selectively sorted into COPII vesicles by binding of its YxxxNPF transport signal to the so-called A-site within Sec24p (Mossessova et al., 2003; Miller et al., 2005). However, only Sed5p was depleted from COPII vesicles generated with a disrupted A-site variant of Sec24p, arguing against a coupled ER export mechanism of an Sed5p, Bos1p, Sec22p SNARE complex (Miller et al., 2003, 2005). Mutations within the R-SNARE Sec22p that disrupt SNARE complex formation but not ER export corroborate this view and led the authors to propose that the yeast Sec24 proteins have a preference for individual ER-to-Golgi SNARE proteins (Liu et al., 2004).
The sorting preference reported here of Sec24A/B for the R-SNARE Sec22b or of Sec24C/D for the preassembled Q-SNARE complex is consistent with the prevailing view that in both mammalian cells and S. cerevisiae, the R-SNAREs have to reside on one membrane, whereas the Q-SNAREs form the cognate SNARE complex on the opposing membrane (Xu et al., 2000; Furukawa and Mima, 2014). The preference of the mammalian Sec24A/B for the closed conformation of the R-SNARE Sec22b (Mancias and Goldberg, 2007) strengthens this view.
Of note, whereas the role of the subunits of the ER–Golgi SNARE complex are conserved between mammalian cells and S. cerevisiae, the organization of the early secretory pathway in yeast and mammals is strikingly different. Mammalian COPII vesicles homotypically fuse with each other after membrane scission/uncoating to form the ERGIC/vesicular tubular cluster (VTC; Xu and Hay, 2004). In contrast, COPII vesicles in S. cerevisiae fuse directly with cis-Golgi compartments to deliver their cargo content, as shown most recently by superresolution microscopy (Kurokawa et al., 2014). Accordingly, in yeast, Sed5p is required for ER-to-Golgi transport only on the cis-Golgi acceptor compartment but not on vesicular membranes (Cao and Barlowe, 2000), whereas its mammalian homologue, Syntaxin5, is required for the formation of the ERGIC (Rowe et al., 1998). Thus the sorting preference of the mammalian Sec24 isoforms for the fusogenic forms of the ER–Golgi SNARE subunits would prime COPII vesicles for homotypic fusion to promote the formation of an ERGIC/VTC.
Immunoisolation of COPII vesicles and analysis of their Sec24 isoform-specific cargo proteins show that the various Sec24 isoforms are found on the same population of ER-derived vesicles. Colocalization of endogenous Sec24 isoforms corroborates this view. These observations raise the important question of how the R-SNARE and the Q-SNAREs are prevented from forming a premature cis-SNARE complex when present on the same vesicle membrane. Our results suggest that fusion competence is preserved by segregation of SNARE subunits as clients of different Sec24 isoforms. In such a scenario, the inner coat proteins would scaffold the SNARE proteins until uncoating and keep Sec22b in its inactive conformation.
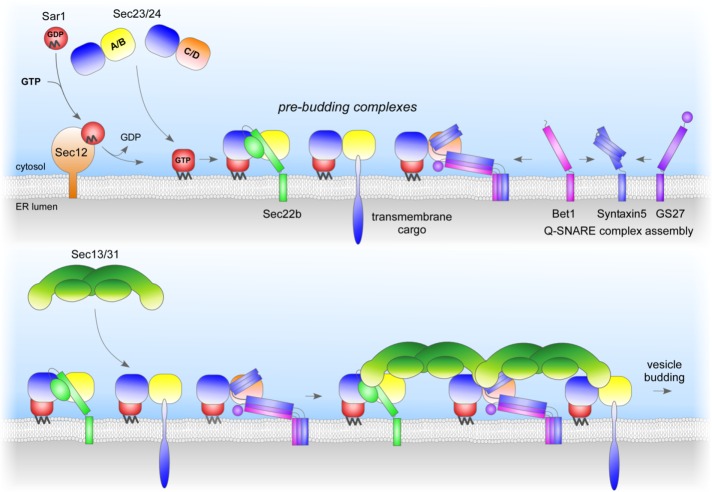
A model of Sec24 isoform–dependent SNARE sorting in mammals is given in Figure 6. Sec23/24 is recruited to ER membranes by Sar1-GTP. Cargo proteins are recruited into nascent COPII vesicles by Sec24 isoforms that discriminate between the fusogenic states of the ER-to-Golgi SNARE proteins. Sec24A/B selectively recruit the R-SNARE Sec22b when it is not engaged in a SNARE complex, whereas the three Q-SNAREs are sorted into vesicles as a preassembled complex via a direct interaction between Sec24C/D with an IxM motif presented by Syntaxin5. The mammalian Sec24 isoforms colocalize on ERES in living cells and hence promote sorting of their isoform-specific cargo proteins into the same vesicles. The resulting pre–budding complexes then recruit the outer coat layer Sec13/31 to form a bud that eventually undergoes membrane scission to become released as a COPII vesicle.
FIGURE 6:
Model for Sec24 isoform–dependent sorting into COPII vesicles of the preassembled ER-to-Golgi Q-SNARE complex Sytaxin5/GS27/Bet1. On membrane recruitment of the inner coat subcomplex Sec23/Sec24 via the small GTP-binding protein Sar1, the Sec24 subunit binds to either the R-SNARE Sec22b (Sec24A/B) via a conformational epitope or the Q-SNARE complex Syntaxin5/GS27/Bet1 (Sec24C/D). The ER-to-Golgi Q-SNARE complex binds to a conserved binding site within Sec24C or Sec24D via the motif IxM present in the flexible linker region between the Habc domain and the SNARE motif of its subunit Syntaxin5. The different Sec24 isoforms are not segregated to distinct ERES, and hence their cargo proteins are present on the same vesicle membranes.
Besides ER-to-Golgi SNARE proteins, some other cargo proteins are exported from the ER in a Sec24 isoform–specific manner (Farhan et al., 2007; Bonnon et al., 2010; Merte et al., 2010; Sucic et al., 2011), whereas many are likely to be unknown. Further studies are required to gain a comprehensive knowledge of cargo sorting. Ultimately, this will help to unravel the molecular basis of human disorders associated with mutations in Sec24 isoforms (Yang et al., 2013; Garbes et al., 2015).
MATERIALS AND METHODS
Antibodies
The following antibodies were used: Sec31A (32/Sec31A, 612350) and GM130 (35/GM130, 610822), both from BD Bioscience (San Jose, CA); calnexin (ab75801) and Sec22L (ab116676), both from Abcam (Cambridge, MA); Sec23 (E19, sc-12107), Sec24A (N-14, sc-169279), ERGIC53 (C-6, sc-365158), Syntaxin5 (B-8, sc-365124), GS27 (25, sc-135932), Sec22b (29-F7, sc-101267), and Bet1 (17, sc-136390), all from Santa Cruz Biotechnology (Dallas, TX); and Sec24C (SZ505), a kind gift from Randy Schekman (University of California, Berkeley, CA). For immunofluorescence and Western blot, the following secondary antibodies were used: goat anti-rabbit Alexa Fluor 488, donkey anti-goat Alexa Fluor 546, goat anti-mouse Alexa Fluor 546, donkey anti-goat Alexa Fluor 680, goat anti-rabbit Alexa Fluor 680, and goat anti-mouse Alexa Fluor 680, all from Invitrogen (Carlsbad, CA).
Plasmids
The pFBDM transfer vector (Berger et al., 2004) and the plasmids pFBDM-His6-Sec23A/Sec24A and pFBDM-His6-Sec13/Sec31A (Adolf et al., 2013) have been described previously. For generation of the pFBDM-hexahistidine (His6)-Sec23A/Sec24C construct, His6-Sec23A was excised with XmaI and NcoI from pFBDM-His6-Sec23A/Sec24A and inserted into the respective restriction sites of pFBDM. The cDNA of human Sec24C was obtained as I.M.A.G.E. clone from Source BioScience (Nottingham UK), amplified by PCR with the primers 5′-AAA AAG CGG CCG CAT GAA CGT CAA CCA GTC AG-3′ and 5′-AAA AAT CTA GAT TAG CTC AGT AGC TGC-3′, digested with NotI and XbaI, and inserted into the respective restriction sites of pFBDM-His6-Sec23A. The pFBDM-His6-Sec23A/Sec24C LIL895AAA construct was generated by site-directed mutagenesis. For the pET29a-SNARE-GST fusion constructs, GST was amplified by PCR from pGEX-2T (GE Healthcare, Little Chalfont, United Kingdom) with the primers 5′-AAA AAA CCA TGG GAT CC CCT ATA CTA GG-3′ and 5′-AAA AAA CTC GAG TCA ATC CGA TTT TGG AGG-3′, digested with NcoI and XhoI, and inserted into respective restriction sites of pET29a (Novagen). Rat Syntaxin5 1–328 (5′-AAA AAA CAT ATG ATC CCG CGG AAA-3′ and 5′-AAA AAA GGT ACC TGA CTG GAA GTA CTT-3′), rat Syntaxin5 205–328 (5′-AAA AAA CAT ATG AGG AAC CGT CGG GAA-3′ and 5′-AAA AAA GGT ACC TGA CTG GAA GTA CTT-3′), rat Syntaxin5 1–274 (5′-AAA AAA CAT ATG ATC CCG CGG AAA-3′ and 5′-AAA AAA GGT ACC CTG CAT GGT GTC TGC-3′), human GS27 1–186 (5′-AAA AAA CAT ATG GAT CCC CTG TTC-3′ and 5′-AAA AAA GGT ACC AGC CCG CTT CTC GAT-3′), human GS27 1–132 (5′-AAA AAA CAT ATG GAT CCC CTG TTC-3′ and 5′-AAA AAA GGT ACC TTT CTG GAG GGA GGA-3′), and human Bet1 1–86 (5′-AAA AAA CAT ATG AGG CGT GCA GGC-3′ and 5′-AAA AAA GGT ACC CTT CAG TTT GCC CAT-3′) were amplified by PCR with the primer pairs indicated, digested with NdeI and KpnI, and inserted into the respective restriction sites of pET29a-GST.
Protein expression and purification
The COPII coat subcomplexes His6-Sec23A/24A, His6-Sec23A/24C, and His6-Sec13A/31A were expressed in Sf9 insect cells and purified as described previously (Adolf et al., 2013). GST- tagged hamster Sar1b was expressed in Escherichia coli BL21 (DE3) pLysS (Thermo Fisher Scientific, Waltham, MA) and purified as described previously (Kim et al., 2005). SNARE-GST fusion constructs were expressed in E. coli BL21 (DE3) at 25°C for 3 h after induction with 0.5 mM isopropyl-β-d-thiogalactoside and purified on glutathione Sepharose 4 fast flow (GE Healthcare). Before snap freezing, buffer was exchanged to 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.2 (KOH), 200 mM KCl, 5 mM MgCl2, 0.02% (vol/vol) monothioglycerol, and 10% (wt/vol) glycerol on PD-10 columns (GE Healthcare).
In vitro COPII vesicle formation from semi-intact cells
HeLa ACC57 cells (DSMZ, Braunschweig, Germany) were grown at 37°C with 5% CO2 in alpha-MEM (Sigma-Aldrich, St. Louis, MO) with 10% fetal bovine serum. Preparation of semi-intact HeLa cells (Mancias and Goldberg, 2007) and the COPII budding assay were carried out as described previously (Adolf et al., 2013). When Sec22b and GS27 were to be detected in the same assay, the standard COPII budding reaction was scaled up twofold, and 40% of the vesicle fractions were analyzed by quantitative Western blotting.
SNARE-GST binding assay
For the GST binding assay, 50% (vol/vol) slurry of glutathione Sepharose 4 fast was mixed 1:1 with 50% (vol/vol) slurry of Sepharose CL-4B (Sigma-Aldrich). GST or SNARE-GST fusion proteins (100–300 μg, to obtain equal amounts of fusion protein binding) were incubated with 20 μl of the 50% (vol/vol) mixed slurry in assay buffer (25 mM HEPES, pH 7.2 [KOH], 150 mM KOAc, 2 mM MgOAc, 0.02% [vol/vol] monothioglycerol) in a total volume of 1 ml for 1 h at 4°C. Beads were washed three times with 750 μl of assay buffer and subsequently incubated with 20 μg of His6-Sec23A/24C in assay buffer in a total volume of 500 μl for 1 h at 4°C. Subsequently, glutathione Sepharose beads were washed three times with 750 μl of assay buffer to remove unbound material, and bound His6-Sec23A/24C was eluted with SDS–PAGE sample buffer. Input fractions of His6-Sec23A/24C (3 and 5%) and 40% of samples were analyzed by SDS–PAGE and Coomassie brilliant blue (CBB) staining (GST and GST-fusion proteins) and Western blotting (Sec24C or Sec23A).
Immunoisolation of COPII vesicle
For immunoprecipitation of COPII vesicles, the standard COPII vesicle budding reaction was scaled up threefold. After medium-speed centrifugation at 14.000 × g for 10 min at 4°C, the COPII vesicle–containing supernatants, generated with only Sec24A, Sec24C, a mixture of both isoforms, or a sample containing a 1:1 mixture of homotypic vesicles (twofold reaction of Sec24A and Sec24C vesicles each), were incubated for an additional 10 min at 30°C. After the short incubation to allow vesicle fusion/intervesicle tethering, one-third of each sample was harvested by ultracentrifugation at 100,000 × g for 10 min at 4°C. The remainder of each sample was adjusted to 1 ml containing 1 μg of anti-Bet1 antibody in assay buffer (20 mM HEPES, pH 7.2 [KOH], 150 mM KOAc, 2 mM MgOAc) supplemented with 5% (wt/vol) bovine serum albumin (BSA) and incubated for 14–16 h at 4°C. Subsequently, 30 μl of a 50% (vol/vol) slurry of protein A Sepharose 4 Fast Flow was added, and the incubation was prolonged for further 6 h at 4°C. Subsequently, beads were washed five times with 1 ml of assay buffer supplemented with 0.1% (wt/vol) BSA, and bound material was eluted with SDS–PAGE sample buffer. Input fractions of semi-intact cells (5 μg), and vesicle fractions corresponding to 75% of the high-speed pellets (HSP) and 60% of the immunoisolated COPII vesicles (IP: Bet1) were analyzed by Western blotting.
Immunofluorescence microscopy
For immunofluorescence, HeLa ACC57 cells were seeded on coverslips 24 h before immunostaining, fixed with ice-cold methanol for 5 min, and subsequently blocked with 5% (wt/vol) BSA in phosphate-buffered saline (PBS) for 15 min at room temperature. Sequential incubation with primary and fluorescently labeled secondary antibodies was carried out in 5% (wt/vol) BSA in PBS for 30 min at room temperature. The Sec24C primary antibodies were detected with Alexa 488–labeled anti-rabbit immunoglobulins (Igs), whereas GM130 (mouse), Sec31A (mouse), and Sec24A (goat) primary antibodies were stained with Alexa 546–labeled Igs raised in the indicated hosts. Samples were embedded in ProLong Gold Antifade with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Images of z-stacks were acquired on a Nikon ERS-6 spinning-disk confocal microscope with a 63× objective and deconvolved with Huygens. Quantification of colocalization was preformed as described (Iwasaki et al., 2015). Briefly, ∼100 ERES were randomly selected per cell, and fluorescence intensity in both channels was quantified with ImageJ (National Institutes of Health, Bethesda, MD).
Acknowledgments
We thank Jonathan Goldberg (Memorial Sloan Kettering Cancer Center) and Martin Lowe (University of Manchester, Manchester, United Kingdom) for cDNAs of ER-to-Golgi SNARE proteins, Randy Scheckman (University of California, Berkeley, CA) for Sec24 isoform–specific antibodies, and Ulrike Engel and Christian Ackerman (Nikon Imaging Center, Heidelberg, Germany) for help with the ERS-6 CLSM and Huygens. We are grateful to Thomas Söllner and Julien Béthune (Heidelberg University Biochemistry Center) for critical reading and discussion of the manuscript and all members of the Wieland lab for discussions, comments, and support. This work was supported by German Research Council Grant SFB 638, A10 to F.T.W.
Abbreviations used:
- COPII
coat protein complex II
- ER
endoplasmic reticulum
- ERES
ER exit sites
- ERGIC
ER-Golgi intermediate compartment
- SIC
semi-intact cells
- SNARE
N-ethylmaleimide–sensitive factor attachment protein receptor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0229) on July 13, 2016.
REFERENCES
- Adolf F, Herrmann A, Hellwig A, Beck R, Brugger B, Wieland FT. Scission of COPI and COPII vesicles is independent of GTP hydrolysis. Traffic. 2013;14:922–932. doi: 10.1111/tra.12084. [DOI] [PubMed] [Google Scholar]
- Adolf F, Wieland FT. Small G proteins: Arf family GTPases in vesicular transport. In: Wittinghofer A, editor. Ras Superfamily Small G Proteins: Biology and Mechanisms 2: Transport. Cham, Switzerland: Springer International Publishing; 2014. pp. 181–214. [Google Scholar]
- Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonnon C, Wendeler MW, Paccaud JP, Hauri HP. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J Cell Sci. 2010;123:1705–1715. doi: 10.1242/jcs.062950. [DOI] [PubMed] [Google Scholar]
- Brocker C, Engelbrecht-Vandre S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–R952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Calakos N, Bennett MK, Peterson KE, Scheller RH. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Cao X, Barlowe C. Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with acceptor membranes. J Cell Biol. 2000;149:55–66. doi: 10.1083/jcb.149.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo JG, Stahmer KR, Miller EA. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim Biophys Acta. 2013;1833:2464–2472. doi: 10.1016/j.bbamcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Matteson J, Balch WE. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J Biol Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Enfert C, Barlowe C, Nishikawa S, Nakano A, Schekman R. Structural and functional dissection of a membrane glycoprotein required for vesicle budding from the endoplasmic reticulum. Mol Cell Biol. 1991;11:5727–5734. doi: 10.1128/mcb.11.11.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faini M, Beck R, Wieland FT, Briggs JA. Vesicle coats: structure, function, and general principles of assembly. Trends Cell Biol. 2013;23:279–288. doi: 10.1016/j.tcb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Farhan H, Reiterer V, Korkhov VM, Schmid JA, Freissmuth M, Sitte HH. Concentrative export from the endoplasmic reticulum of the gamma-aminobutyric acid transporter 1 requires binding to SEC24D. J Biol Chem. 2007;282:7679–7689. doi: 10.1074/jbc.M609720200. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Furukawa N, Mima J. Multiple and distinct strategies of yeast SNAREs to confer the specificity of membrane fusion. Sci Rep. 2014;4:4277. doi: 10.1038/srep04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbes L, Kim K, Riess A, Hoyer-Kuhn H, Beleggia F, Bevot A, Kim MJ, Huh YH, Kweon HS, Savarirayan R, et al. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am J Hum Genet. 2015;96:432–439. doi: 10.1016/j.ajhg.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond AT, Glick BS. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Yorimitsu T, Sato K. Distribution of Sec24 isoforms to each ER exit site is dynamically regulated in Saccharomyces cerevisiae. FEBS Lett. 2015;589:1234–1239. doi: 10.1016/j.febslet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem. 2005;280:7758–7768. doi: 10.1074/jbc.M411091200. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kurihara T, Hamamoto S, Gimeno RE, Kaiser CA, Schekman R, Yoshihisa T. Sec24p and Iss1p function interchangeably in transport vesicle formation from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:983–998. doi: 10.1091/mbc.11.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Okamoto M, Nakano A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat Commun. 2014;5:3653. doi: 10.1038/ncomms4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JP, Ferro-Novick S. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell. 1993;73:735–745. doi: 10.1016/0092-8674(93)90253-m. [DOI] [PubMed] [Google Scholar]
- Liu Y, Flanagan JJ, Barlowe C. Sec22p export from the endoplasmic reticulum is independent of SNARE pairing. J Biol Chem. 2004;279:27225–27232. doi: 10.1074/jbc.M312122200. [DOI] [PubMed] [Google Scholar]
- Malsam J, Kreye S, Sollner TH. Membrane fusion: SNAREs and regulation. Cell Mol Life Sci. 2008;65:2814–2832. doi: 10.1007/s00018-008-8352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, Ginty DD. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol. 2010;12:41–46. doi: 10.1038/ncb2002. supp. 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Miller EA, Liu Y, Barlowe C, Schekman R. ER-Golgi transport defects are associated with mutations in the Sed5p-binding domain of the COPII coat subunit, Sec24p. Mol Biol Cell. 2005;16:3719–3726. doi: 10.1091/mbc.E05-03-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Muniz M, Morsomme P, Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Meda P, Holcomb C, Moore HP, Hicke L, Schekman R. Mammalian Sec23p homologue is restricted to the endoplasmic reticulum transitional cytoplasm. Proc Natl Acad Sci USA. 1991;88:8611–8615. doi: 10.1073/pnas.88.19.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek I, Orci L, Ravazzola M, Erdjument-Bromage H, Amherdt M, Tempst P, Sollner TH, Rothman JE. ERS-24, a mammalian v-SNARE implicated in vesicle traffic between the ER and the Golgi. J Cell Biol. 1997;137:1017–1028. doi: 10.1083/jcb.137.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A, Letourneur F, Garcia-Estefania D, Carpentier JL, Orci L, Paccaud JP. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7840. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- Peng R, De Antoni A, Gallwitz D. Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J Biol Chem. 2000;275:11521–11528. doi: 10.1074/jbc.275.15.11521. [DOI] [PubMed] [Google Scholar]
- Roberg KJ, Crotwell M, Espenshade P, Gimeno R, Kaiser CA. LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J Cell Biol. 1999;145:659–672. doi: 10.1083/jcb.145.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- Sacher M, Stone S, Ferro-Novick S. The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J Biol Chem. 1997;272:17134–17138. doi: 10.1074/jbc.272.27.17134. [DOI] [PubMed] [Google Scholar]
- Shim J, Newman AP, Ferro-Novick S. The BOS1 gene encodes an essential 27-kD putative membrane protein that is required for vesicular transport from the ER to the Golgi complex in yeast. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R. Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J Cell Biol. 2000;151:973–984. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sucic S, El-Kasaby A, Kudlacek O, Sarker S, Sitte HH, Marin P, Freissmuth M. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J Biol Chem. 2011;286:16482–16490. doi: 10.1074/jbc.M111.230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Tang BL, Kausalya J, Low DY, Lock ML, Hong W. A family of mammalian proteins homologous to yeast Sec24p. Biochem Biophys Res Commun. 1999;258:679–684. doi: 10.1006/bbrc.1999.0574. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeler MW, Paccaud JP, Hauri HP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AL, Ehm S, Jacobson NC, Xu D, Hay JC. rsly1 binding to syntaxin 5 is required for endoplasmic reticulum-to-Golgi transport but does not promote SNARE motif accessibility. Mol Biol Cell. 2004;15:162–175. doi: 10.1091/mbc.E03-07-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167:997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Joglekar AP, Williams AL, Hay JC. Subunit structure of a mammalian ER/Golgi SNARE complex. J Biol Chem. 2000;275:39631–39639. doi: 10.1074/jbc.M007684200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Sudhof TC. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- Yang XY, Zhou XY, Wang QQ, Li H, Chen Y, Lei YP, Ma XH, Kong P, Shi Y, Jin L, et al. Mutations in the COPII vesicle component gene SEC24B are associated with human neural tube defects. Hum Mutat. 2013;34:1094–1101. doi: 10.1002/humu.22338. [DOI] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wong SH, Tang BL, Xu Y, Peter F, Subramaniam VN, Hong W. The mammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in vesicular transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1997;139:1157–1168. doi: 10.1083/jcb.139.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]