The yeast transcription factor Gcn4 requires a ubiquitin ligase and the proteasome in order to function. Inhibiting proteasome function prevents the interaction of Gcn4 with target gene chromatin, and this activity is suppressed by inactivation of the ubiquitin-selective chaperone Cdc48. Thus proteolysis of Gcn4 is not required for its function.
Abstract
The ubiquitin–proteasome system (UPS) influences gene transcription in multiple ways. One way in which the UPS affects transcription centers on transcriptional activators, the function of which can be stimulated by components of the UPS that also trigger their destruction. Activation of transcription by the yeast activator Gcn4, for example, is attenuated by mutations in the ubiquitin ligase that mediates Gcn4 ubiquitylation or by inhibition of the proteasome, leading to the idea that ubiquitin-mediated proteolysis of Gcn4 is required for its activity. Here we probe the steps in Gcn4 activity that are perturbed by disruption of the UPS. We show that the ubiquitylation machinery and the proteasome control different steps in Gcn4 function and that proteasome activity is required for the ability of Gcn4 to bind to its target genes in the context of chromatin. Curiously, the effect of proteasome inhibition on Gcn4 activity is suppressed by mutations in the ubiquitin-selective chaperone Cdc48, revealing that proteolysis per se is not required for Gcn4 activity. Our data highlight the role of Cdc48 in controlling promoter occupancy by Gcn4 and support a model in which ubiquitylation of activators—not their destruction—is important for function.
INTRODUCTION
Regulated proteolysis by the ubiquitin (Ub)–proteasome system (UPS) is crucial for a myriad of processes, including control of gene transcription (Geng et al., 2012). In many cases, actions of the UPS in transcription follow the canonical paradigm in which ubiquitin ligases and the proteasome inhibit protein activity by reducing intracellular levels of target proteins. In the case of transcriptional activators, however, a counterintuitive scenario can play out in which components of the UPS stimulate the function of the activators they destroy. Transcription factors such as Gal4 (Muratani et al., 2005; Collins et al., 2009), Gcn4 (Lipford et al., 2005), and nuclear hormone receptors (Perissi et al., 2004), for example, have been shown to require their cognate Ub-ligases—and/or proteasomal proteolysis—for full activity, suggesting that Ub-mediated turnover of activator proteins promotes their function.
Precisely how ubiquitin-mediated destruction of an activator stimulates its activity is unclear. To account for the concept of how destruction of a protein could make it more active, we (Geng et al., 2012) and others (Lipford et al., 2005) hypothesized that the requirement for the UPS in transcription reflects the need for activators to cycle on chromatin to drive multiple rounds of transcriptional activation. We proposed that activator ubiquitylation occurs on promoter DNAs after transcription has initiated and that the modified activator remains bound to chromatin in an inactive configuration that must be cleared by the proteasome in order for subsequent rounds of activation to occur. This model assumes that activator ubiquitylation and proteolysis converge on a single mechanistic step in the activation process and that there is no difference between blocking activator ubiquitylation and inhibiting activator destruction. Moreover, this model was developed at a time when little was known about the role of factors that lie between ubiquitylation and proteolysis, such as complexes containing the Ub-dependent chaperone Cdc48 (Meyer et al., 2012), which extract ubiquitylated substrates from protein complexes for proteasomal destruction. The potential effect of Cdc48 on activator function is particularly intriguing, as Cdc48 was recently shown to mediate proteolysis-independent stripping of a Ub-fusion activator from its target DNA sites in vivo (Ndoja et al., 2014).
The purpose of this study was to challenge the assumption that ubiquitylation and proteolysis target the same step in activator function and ask how Cdc48 features in the control of a native Saccharomyces cerevisiae transcriptional activator, Gcn4. Gcn4 is induced in response to amino acid starvation and drives the expression of genes encoding amino acid biosynthetic enzymes (Hinnebusch, 2005). Like a majority of transcriptional activators, Gcn4 carries an overlapping transcriptional activation domain and degron (Geng et al., 2012) and is targeted for Ub-mediated proteolysis by the SCFCdc4 Ub ligase (Chi et al., 2001). Of importance, genetic inhibition of SCFCdc4 or inhibition of the proteasome reduces Gcn4 activity (Lipford et al., 2005), making Gcn4 an excellent focal point for our studies.
Here we confirm that full activity of Gcn4 is dependent on both SCFCdc4 and the proteasome but show that disrupting the function of each affects Gcn4 activity in different ways. We also show that inhibiting the proteasome blocks the ability of Gcn4 to bind target genes in vivo and that this phenotype is suppressed by mutations in Cdc48. These observations reveal that proteasomal proteolysis per se is not required for the function of Gcn4 and highlight the importance of Cdc48 in controlling the activity of a native yeast activator.
RESULTS AND DISCUSSION
Full induction of Gcn4 target genes depends on SCFCdc4 and the proteasome
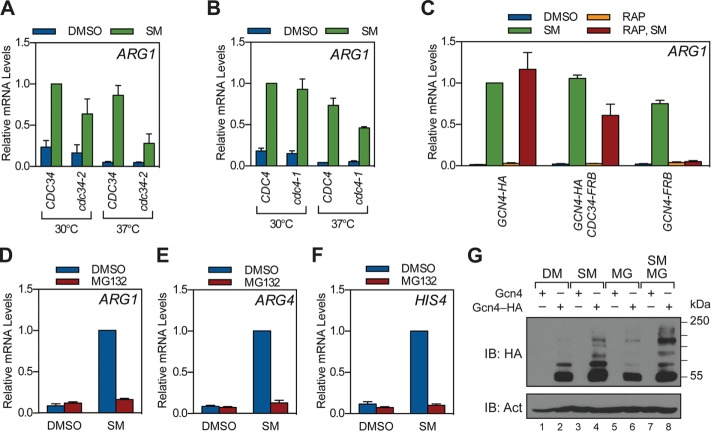
A previous study (Lipford et al., 2005) reported that activation of Gcn4 target genes is reduced by mutations in the Cdc34 and Cdc4 components of the SCFCdc4 Ub ligase (Chi et al., 2001), as well as by inhibition of proteasomal proteolysis. To confirm these findings, we monitored the effects of SCFCdc4 or proteasome perturbation on activation of Gcn4 target genes by sulfometuron methyl (SM), an agent that induces Gcn4 synthesis by blocking branched-chain amino acid production (Jia et al., 2000). Three sets of experiments were performed. First, we asked whether temperature-sensitive mutations in Cdc34 or Cdc4 (Patton et al., 1998) affect expression of the Gcn4 target gene ARG1. Here we found that shifting yeast to the restrictive temperature of 37°C reduced the expression of ARG1 in both the cdc34-2 (Figure 1A) and cdc4-1 (Figure 1B) strains. Second, we used the “anchor-away” technique (Haruki et al., 2008) to exclude Cdc34 from the nucleus via a rapamycin-induced interaction with a subunit of the ribosome. Here, treatment of the anchored Cdc34 strain with rapamycin reduced the expression of ARG1 (Figure 1C) to a level comparable to that observed in the cdc34-2 strain. Finally, we combined point mutations in the tryptic and caspase sites of the proteasome (Heinemeyer et al., 1997) with MG132-mediated inhibition of the chymotryptic site (Howard et al., 2012) to probe the effect of 20S proteasome inhibition on Gcn4 activity. Here, inhibition of proteasome activity blocked induction of ARG1 (Figure 1D), as well as of ARG4 (Figure 1E) and HIS4 (Figure 1F). Induction of Gcn4 protein by SM was not blocked by proteasome inhibition (Figure 1G), showing that this blockade is not at the level of Gcn4 synthesis. We did note, however, that proteasome inhibition promoted the accumulation of high–molecular weight Gcn4 species in total cell lysates, consistent with an increase in the level of ubiquitin-Gcn4 (Chi et al., 2001) or SUMO-Gcn4 (Rosonina et al., 2012) conjugates, or a combination of both. On the basis of these results, we conclude that induction of Gcn4 target genes is sensitive to mutations in SCFCdc4 and chemical-genetic inhibition of proteasomal proteolysis.
FIGURE 1:
Stimulation of Gcn4 target genes by SCFCdc4 and the proteasome. (A) CDC34 (W303-1a) and cdc34-2 (MT670) yeast were grown to log phase at 30°C in minimal medium and then shifted to 37°C or maintained at 30°C for 1 h as indicated. Strains were then treated with SM or DMSO for 1.5 h, at which time RNA was collected and ARG1 mRNA levels quantified by RT-qPCR. Relative mRNA levels for ARG1 were normalized to the CDC34 strain treated with SM at 30°C. n = 3. (B) As in A, except using CDC4 (W303-1a) and cdc4-1 (MT668) strains. n = 3. (C) Anchor-away strains expressing HA-tagged Gcn4 (GHY139), FRB-tagged Cdc34 (GHY149), or FRB-tagged Gcn4 (GHY145) were grown to log phase in minimal medium, treated with either DMSO or rapamycin for 1 h, and then further treated with either DMSO or SM for 1.5 h. RNA was collected, and ARG1 mRNA levels were measured by RT-qPCR, as in A. Relative mRNA levels for ARG1 were normalized to the GCN4-HA strain treated with SM at 30°C. n = 3. (D–F) Yeast bearing the pup1–T30A pre3–T20A mutations (GHY010) were grown to log phase in minimal medium and treated with either DMSO or MG132 for 1 h. Strains were then treated with SM or DMSO for 1.5 h, at which time RNA was collected and ARG1 (D), ARG4 (E), and HIS4 (F) mRNA levels quantified by RT-qPCR. Relative mRNA levels were then normalized to the SM-induced, DMSO-treated sample for each gene. n = 4. Error bars represent SEM. (G) Yeast expressing either native Gcn4 (GHY010) or HA-tagged Gcn4 (GHY025) were grown to log phase in minimal medium and treated with DMSO or MG132 for 1 h. Strains were then treated with SM or DMSO for 1.5 h, at which time protein was extracted, resolved by SDS–PAGE, and subject to Western blotting with antibodies against the HA epitope or β-actin (Act).
Proteasome inhibition disrupts the ability of Gcn4 to engage target gene chromatin
We next used chromatin immunoprecipitation (ChIP) to quantify the effect of Cdc4 or proteasome inactivation on binding of hemagglutinin (HA) epitope–tagged Gcn4 to target gene promoters. Consistent with a previous study (Lipford et al., 2005), Gcn4 binding to ARG1 was not different in the cdc4-1 strain versus its control strain at the restrictive temperature (Figure 2A). When we examined the effect of proteasome inhibition, however, we discovered that binding of Gcn4 to the ARG1 UAS was disrupted by proteasome inhibition (Figure 2B). The failure of Gcn4 to bind chromatin was accompanied by loss of binding of the TATA box–binding protein TBP to the ARG1 TATA box (Figure 2B) and was not restricted to ARG1, as we observed the same phenomenon at UAS elements in another 10 Gcn4 target genes (Figure 2C). Of importance, there was no obvious alteration in the cellular distribution of Gcn4 in response to proteasome inhibition (Figure 2D and Supplemental Figure S1), demonstrating that Gcn4 remained in the nucleus under these conditions but was unable to associate with target gene chromatin as measured by ChIP.
FIGURE 2:
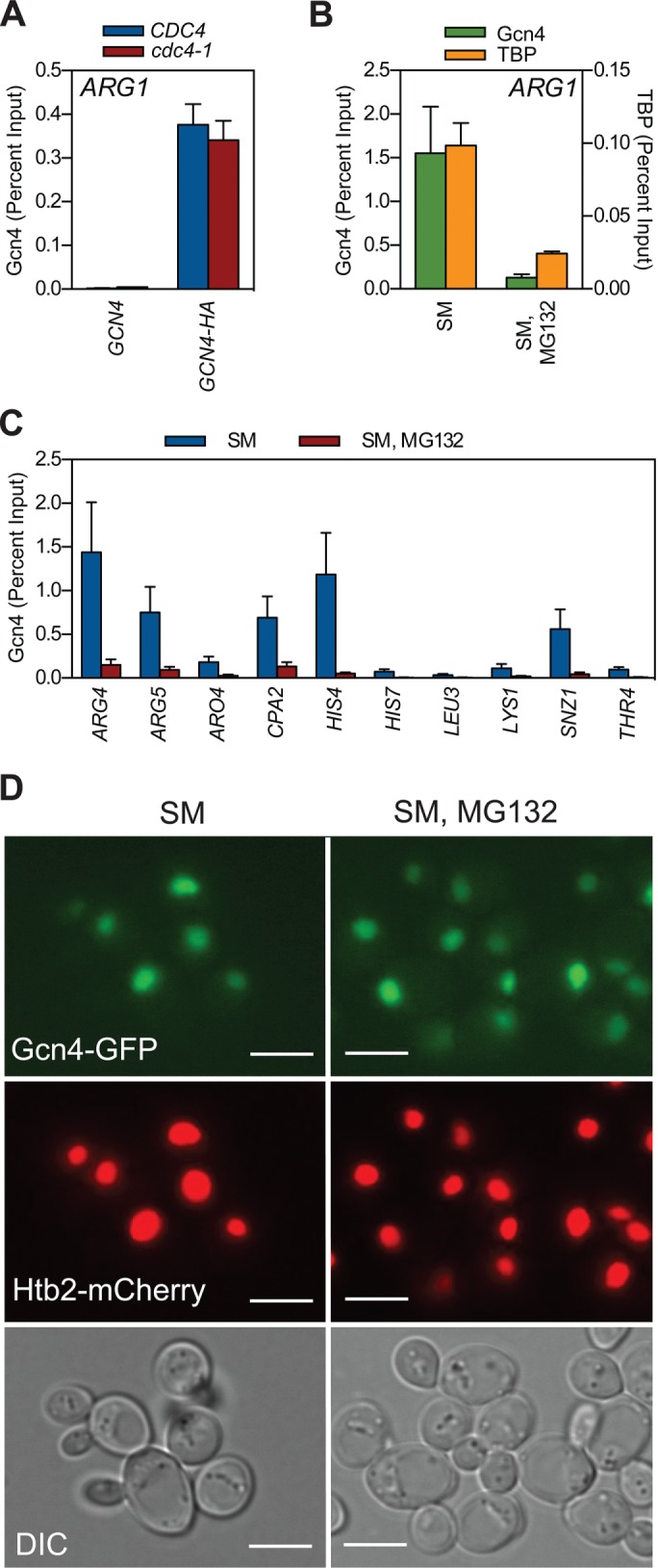
Inhibiting the proteasome impedes the ability of Gcn4 to bind target gene chromatin. (A) CDC4 (W303-1a), CDC4 GCN4-HA (GHY107), cdc4-1 (MT668), and cdc4-1 GCN4-HA (GHY107) strains were grown to log phase at 30°C in minimal medium, shifted to the restrictive temperature of 37°C for 1 h, and then induced with SM for an additional 1.5 h. At this time, ChIP was performed with an antibody against the HA-epitope tag. Coprecipitating ARG1 promoter DNA was quantified by qPCR, expressed relative to the percentage of input DNA. n = 3. (B) GCN4-HA (GHY025) yeast were grown to log phase at 30°C in minimal medium, treated with either DMSO or MG132 for 1 h, and induced with SM for 1.5 h. ChIP was performed using antibodies against the HA-epitope tag or TBP. Coprecipitating ARG1 promoter DNA was quantified by qPCR. n = 3. (C) As in B, except that coprecipitating DNAs from the anti-HA ChIP were quantified by qPCR, using primer pairs that amplify Gcn4-binding sites in the indicated genes. n = 3. Error bars represent SEM. (D) GCN4-GFP HTB2-mCherry (GHY339) yeast were grown to log phase at 30°C in minimal medium, treated with either DMSO or MG132 for 1 h, and induced with SM for 1.5 h. Samples were imaged using either fluorescence (top) or differential interference contrast microscopy (bottom). Scale bars, 5 μm.
Our finding that Gcn4 fails to associate with its cognate UAS elements when the proteasome is inhibited is at odds with a report by Lipford et al. (2005), who showed that Myc epitope–tagged Gcn4 robustly binds its target genes in the presence of MG132. This discrepancy raises the possibility that differential epitope tagging of Gcn4 is responsible for the disagreement between these studies. We therefore repeated our experiments with untagged Gcn4 using a polyclonal anti-Gcn4 antibody and with Myc-tagged Gcn4, as used in the Lipford et al. (2005) study. For untagged Gcn4 (Supplemental Figure S2A), we observed that proteasome inhibition reduced the level of Gcn4 binding at the ARG1 UAS to that observed in the uninduced state, mirroring the effect of proteasome inhibition on ARG1 gene induction (Figure 1D) and what we observed for HA-tagged Gcn4 (Figure 2B). When we examined Myc-tagged Gcn4, however, we found that proteasome inhibition had little if any effect on its binding to the ARG1 UAS (Supplemental Figure S2B). These results are consistent with the Lipford et al. (2005) study but also demonstrate that Myc tagging of Gcn4 changes the behavior of the protein—as measured by ChIP—in a way that does not recapitulate that of the native Gcn4 protein.
In sum, the contrasting effects of Cdc4 versus proteasome disruption on the ability of Gcn4 to bind DNA in vivo suggest that ubiquitylation and proteolysis influence distinct steps in Gcn4 function and that the profound effects of proteasome inhibition on Gcn4 target gene induction result from a failure of Gcn4 to stably bind chromatin when proteasome function is blocked.
Proteasome inhibition does not act via the ArgR repressor or by modulating nucleosome occupancy
The simplest explanation for how proteasome inhibition disables the ability of Gcn4 to bind chromatin is that it induces a state on chromatin that is refractory to Gcn4 binding by either inducing an unstable repressor protein or perhaps altering nucleosome positioning. To address these possibilities, we first asked whether the ArgR repressor complex—which binds to DNA elements in the ARG promoters and inhibits their transcription (Messenguy et al., 1991)—mediates the effects of proteasome inhibition we observe. We found, however, that deletion of the gene encoding the Arg80 subunit of this complex (Dubois et al., 1987) had no detectable effect on the sensitivity of ARG1 to proteasome inhibition (Figure 3A), excluding the possibility that accumulation of this known repressor complex inhibits Gcn4 when proteasome function is blocked.
FIGURE 3:
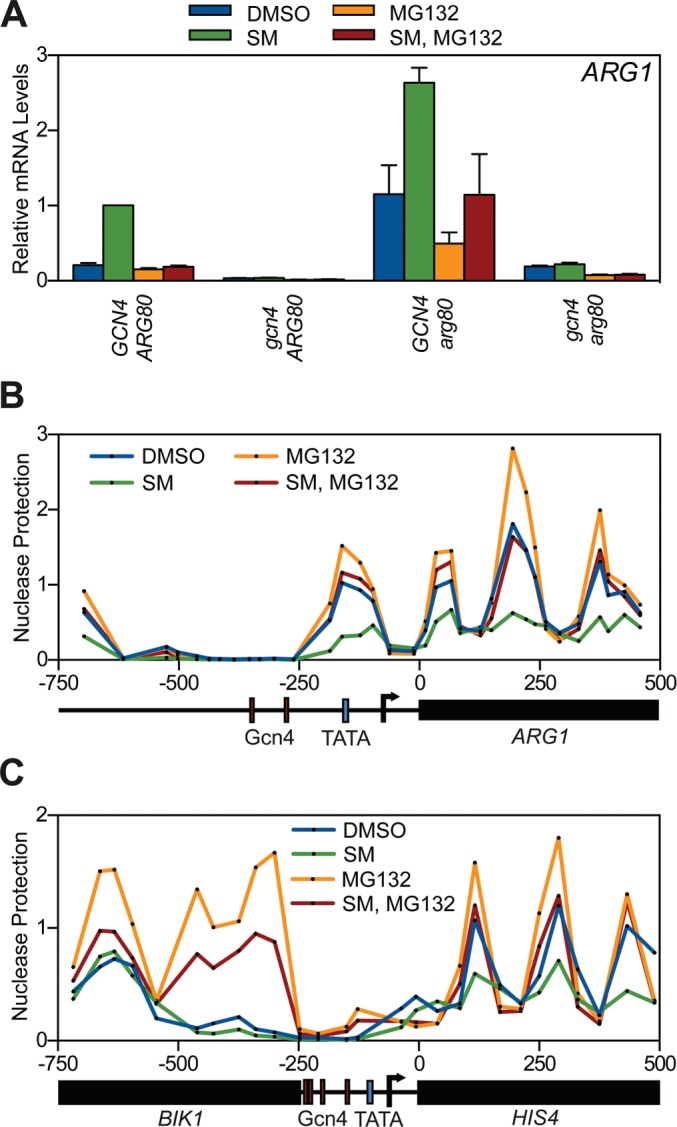
The effect of proteasome inhibition on Gcn4 activity is not mediated via the ArgR repressor or changes in nucleosome occupancy. (A) GCN4 ARG80 (GHY010), gcn4 ARG80 (GHY004), GCN4 arg80 (GHY081), and gcn4 arg80 (GHY079) strains were grown to log phase in minimal medium and treated with DMSO or MG132. After 1 h, strains were treated with DMSO or SM for 1.5 h and RNA harvested, and ARG1 mRNA levels were quantified by RT-qPCR. Each qPCR was normalized to the SM-treated GCN4 ARG80 sample. Error bars represent SEM. n = 4. (B) GCN4 yeast (GHY010) were grown to log phase at 30°C in minimal medium, treated with DMSO or MG132, and induced with SM or a DMSO control for an additional 1.5 h, and nucleosome occupancy was mapped by MNase digestion, coupled with tiled primer sets spanning ARG1. qPCR data were normalized to the signal from a GAL1-10 promoter-localized nucleosome. Data points represent an average of two independent experiments. (C) As in B, except monitoring nucleosome positioning surrounding the HIS4 locus. Data points represent an average of two independent experiments.
We next performed nucleosome scanning analysis (Sekinger et al., 2005) to ask whether proteasome inhibition alters nucleosome occupancy across the ARG1 (Figure 3B) and HIS4 (Figure 3C) genes. Consistent with a previous report (Crisucci and Arndt, 2012), we found that Gcn4-binding sites in the ARG1 (Figure 3B) and HIS4 (Figure 3C) promoters lack nucleosomes and that SM induction reduces the density of nucleosomes within the transcribed portions of ARG1 and HIS4, as expected when their transcription is induced. Proteasome inhibition increased the levels of nucleosomes across the ARG1 and HIS4 open reading frames—again consistent with a decrease in transcriptional activity—but had no detectable effect on nucleosome position or density across the “nucleosome-free” region bound by Gcn4. Of interest, nucleosome density at the HIS4-adjacent gene BIK1, which is not regulated by Gcn4, is markedly increased in response to MG132, demonstrating that proteasome function is involved in mediating some aspect of nucleosome dynamics that is yet to be described. In terms of Gcn4, however, these data exclude the notion that accumulation of nucleosomes in response to proteasome inhibition mediates the failure of Gcn4 to access its cognate sites on chromatin.
Mutation of Cdc48 suppresses the effects of proteasome inhibition on Gcn4 activity
Our data support the concept that ubiquitylation and proteolysis play distinct roles in controlling Gcn4 activity. One possibility is that ubiquitylation of chromatin-bound Gcn4 stimulates a step in the activation process but that once Gcn4 is ubiquitylated, it is unable to stably associate with chromatin. In this scenario, proteasome inhibition could force Gcn4 to accumulate off chromatin via the accrual of ubiquitylated Gcn4 species. Indeed, direct analysis of Gcn4–Ub conjugates via the His-Ub method (Yaglom et al., 1996) revealed that proteasome inhibition promotes a striking increase in the level of ubiquitylated Gcn4 protein (Figure 4A), suggesting that accumulation of Ub conjugates could underlie the defects in Gcn4 function we observe when the proteasome is inhibited.
FIGURE 4:
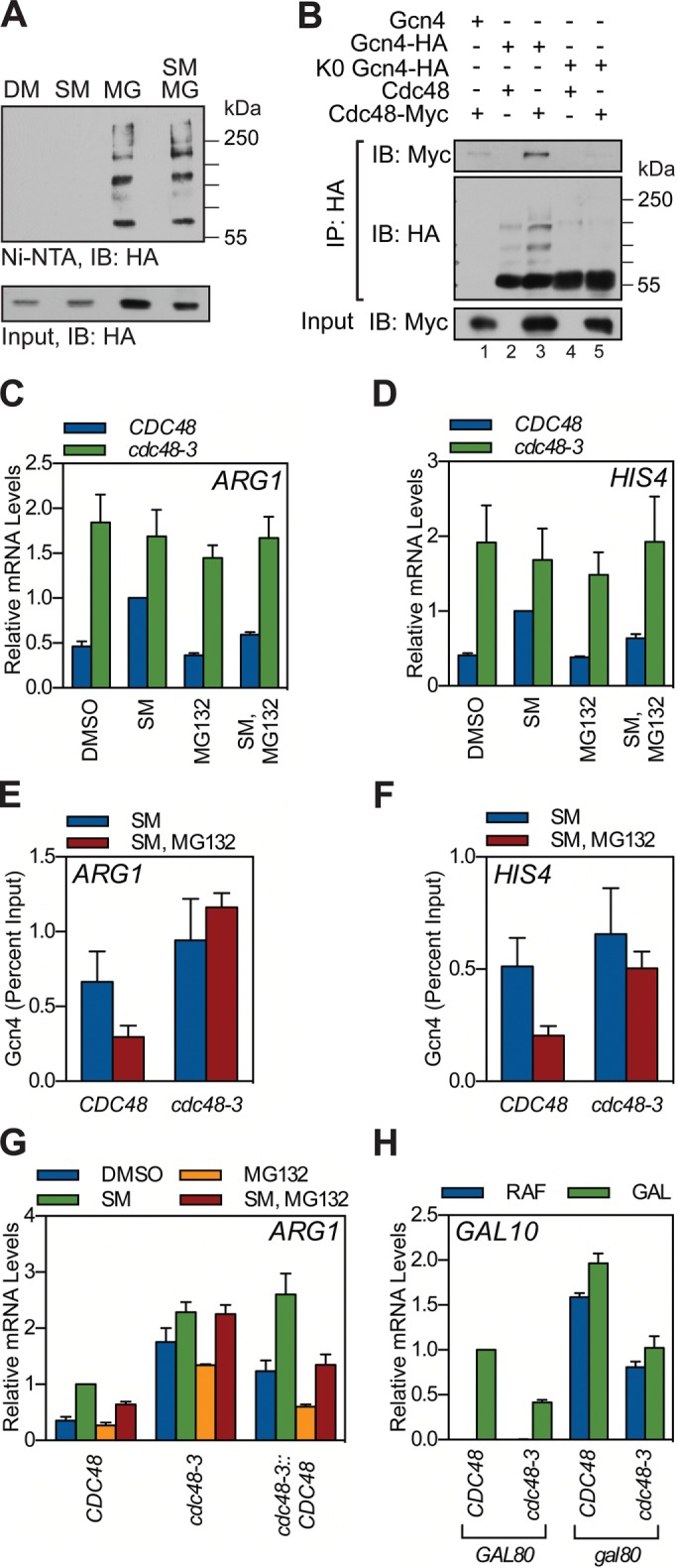
Mutation of Cdc48 suppresses the effect of proteasome inhibition on Gcn4 activity. (A) GCN4-HA (GHY356) yeast carrying a copper-inducible His-Ub expression plasmid (pUB221) were grown to log phase in minimal medium and treated with CuSO4 and DMSO or MG132 for 1 h. Yeast were induced with SM or DMSO for an additional 1.5 h, at which time protein lysates were collected under denaturing conditions. Ubiquitin-conjugates were captured by nickel-resin (Ni-NTA) chromatography, resolved by SDS–PAGE, and probed for HA-tagged Gcn4 protein by Western blotting. A sample of the input material to the nickel resin was also probed for HA-tagged Gcn4. IB, immunoblot. (B) GCN4 CDC48-MYC (GHY285), GCN4-HA CDC48 (GHY025), GCN4-HA CDC48-MYC (GHY287), K0-GCN4-HA CDC48 (GHY124), and K0-GCN4-HA CDC48-MYC (GHY293) yeast were grown to log phase at 30°C in minimal medium and treated with DMSO or MG132. After 1 h, Gcn4 was induced with SM for 1.5 h. Protein lysates were collected and Gcn4-HA immunoprecipitated (IP) via an anti-HA antibody, and IPs were probed with antibodies against the Myc-epitope (Cdc48) and HA-epitope (Gcn4) tags. A sample of the input material to the IP was also probed for Myc-tagged Cdc48. (C, D). CDC48 (RHY2455) and cdc48-3 (RHY2457) strains were grown to log phase at 30°C in minimal medium and treated with DMSO or MG132. After 1 h, strains were treated with DMSO or SM for 1.5 h, RNA harvested, and processed as described to measure mRNA levels from the ARG1 (C) and HIS4 (D) loci. n = 4. (E, F). CDC48 GCN4-HA (GHY116) and cdc48-3 GCN4-HA (GHY118) strains were grown to log phase at 30°C in minimal medium, treated with either DMSO or MG132 for 1 h, and induced with SM for 1.5 h. ChIP was then performed using antibodies against the HA-epitope tag. Coprecipitating ARG1 (E) or HIS4 (F) promoter DNAs were quantified by qPCR, expressed relative to the percentage of input DNA. n = 3. (G) CDC48 (RHY2455), cdc48-3 (RHY2457), and cdc48-3::CDC48 (GHY279) strains were grown and treated as described in C and RNA harvested, and RT-qPCR was performed to quantify ARG1 transcripts. n = 3. (H) CDC48 GAL80 (RHY2455), cdc48-3 GAL80 (RHY2457), CDC48 gal80 (GHY304), and cdc48-3 gal80 (GHY305) yeast were grown in raffinose medium and treated with water or 2% galactose for 1.5 h. RNA was collected and used for RT-qPCR for GAL10. Each qPCR was internally normalized to ACT1 and then normalized to the galactose-treated CDC48 GAL80 sample. Error bars represent SEM. n = 3.
We attempted to challenge this notion by producing a form of Gcn4 that cannot be ubiquitylated. We engineered alanine-substitution mutations in Gcn4 (Supplemental Figure S3A) that block phosphorylation events required for ubiquitylation by SCFCdc4 (Chi et al., 2001). Unfortunately, this form of Gcn4 still accumulated a single Ub-conjugated form in response to proteasome inhibition (Supplemental Figure S3B) and remained sensitive to proteasome inhibition (Supplemental Figure S3, C and D). We also constructed a version of Gcn4 in which all 23 lysine residues are simultaneously mutated to arginine (K0). Although this mutation blocked the ubiquitylation of Gcn4 (Supplemental Figure S3E), we were unable to detect binding of K0 Gcn4 to the UAS of ARG1 by ChIP (Supplemental Figure S3F) and thus did not pursue this mutant further.
Instead, we considered the possibility that the Gcn4–Ub conjugates that accumulate in response to proteasome inhibition are removed from chromatin by Cdc48, as previously reported for a Ub-fusion activator (Ndoja et al., 2014). Coimmunoprecipitation assays demonstrated that Gcn4 interacts with Cdc48 (Figure 4B) and that this interaction is disrupted by the K0 mutation in Gcn4, which disrupts ubiquitylation (Figure 4B). Moreover, a temperature-sensitive mutant of CDC48 (cdc48-3; Sato and Hampton, 2006), assayed at the semipermissive temperature, suppressed the effects of proteasome inhibition on transcription at both ARG1 and HIS4 (Figure 4, C and D), as well as restored binding of Gcn4 to these genes (Figure 4, E and F). Suppression of the effects of proteasome inhibition on Gcn4 activity in the cdc48-3 strain is due to the mutations in CDC48, as expression of wild-type Cdc48 in this strain background restored proteasome sensitivity (Figure 4G). The ability of mutations in Cdc48 to suppress the effects of proteasome inhibition supports the concept that proteasome inhibition drives the accumulation of Gcn4–Ub conjugates that are prevented from stably binding chromatin by Cdc48.
In addition to suppressing defects associated with proteasome inhibition, we observed that the cdc48-3 mutation increased levels of Gcn4 target gene activity in the absence of MG132 (Figure 4, C and D). This result contrasts with our report that Cdc48 stimulates the activity of the yeast activator Gal4 (Bonizec et al., 2014). To confirm that Gcn4 and Gal4 are affected differently by the cdc48-3 mutation, we examined Gal4 activity in the cdc48-3 mutant strain; we also examined the effect of GAL80 deletion in this context, which has been argued to be a point of Gal4 regulation by the UPS (Ang et al., 2012). Supporting our previous study (Bonizec et al., 2014), mutation of Cdc48 reduced activation of the GAL10 locus (Figure 4H), whereas the GAL80 deletion did not affect the sensitivity of GAL10 transcription to the cdc48-3 mutation. Thus, despite many parallels in how Gcn4 and Gal4 are regulated by the UPS, the effect of Cdc48 on the function of these two activators is different. Further work will be required to determine the mechanistic basis for the differences between Gcn4 and Gal4 in terms of the role that Cdc48 plays in their function.
Conclusion
In this study, we confirmed the importance of an intact UPS to the ability of Gcn4 to activate transcription of select target genes but made the surprising discovery that ubiquitylation and proteasomal destruction appear to control distinct steps in the activation process. On one hand, disrupting the activity of the SCFCdc4 Ub ligase reduces the expression of Gcn4 target genes via a process that is most likely downstream of target gene recognition by Gcn4, as previously suggested (Lipford et al., 2005). On the other hand, inhibition of the proteasome inhibits Gcn4 target gene activation at a step that is apparently upstream of chromatin recognition by Gcn4 and in a manner that can be suppressed by a mutation in the Ub-selective chaperone Cdc48. Although we have been unable to test whether the actions of SCFCdc4 and the proteasome in this context are mediated directly on Gcn4, the simplest conclusion from these data is that ubiquitylation of Gcn4 stimulates two distinct processes, promoting the inherent transcriptional activity of the protein while at the same time limiting activation by triggering the Cdc48-mediated extraction of Gcn4 from target gene promoters. In this way, disruption of SCFCdc4 reduces the level of Gcn4-target gene transcription without affecting Gcn4-promoter binding, whereas proteasome inhibition promotes the accumulation of ubiquitylated Gcn4, which is then either stripped off chromatin or prevented from stably binding once it dissociates (Supplemental Figure S4).
The foregoing scenario is thematically similar to the previously proposed “licensing” (Salghetti et al., 2001) and “Ub-clock” (Wu et al., 2007) models, in that activator ubiquitylation sets an inherent limitation on the functional lifetime of transcriptional activators but differs profoundly in the role of proteasomal proteolysis in this process. Indeed, the ability to suppress the effect of proteasome inhibition on Gcn4 activity by inactivation of Cdc48 strongly implies that proteasomal proteolysis per se cannot play a positive role in Gcn4-mediated transcriptional activation, irrespective of whether Gcn4 or some other factor is the relevant substrate. Instead, the effects of proteasome inhibition on Gcn4 must be indirect, perhaps a result of the accumulation of ubiquitylated Gcn4 species, as we propose. A revised view of how the proteasome features in transcriptional activation is more consistent with work from the Laney (Wilcox and Laney, 2009) and Yao (Ndoja et al., 2014) laboratories, which pinpoint Cdc48-mediated extraction of ubiquitylated activators as a limiting point in the activation process, and implies that the widespread and intimate relationship between activation and destruction elements in transcription factors (Salghetti et al., 2000; Geng et al., 2012), if anything, reflects the functional importance of activator ubiquitylation over destruction.
MATERIALS AND METHODS
Yeast manipulations
Yeast strains are described in Supplemental Table S1. Gene deletions were performed via homologous recombination using PCR-amplified auxotrophic markers or antibiotic resistance genes from plasmids, as indicated. Epitope tagging of endogenous loci was performed similarly (Knop et al., 1999; Sheff and Thorn, 2004). mCherry tagging of endogenous Htb2 was performed as described except using an mCherry-tagging cassette amplified from SWY5678 genomic DNA (Lord et al., 2015). Strains carrying the 3T2S-GCN4 allele were generated through deletion of endogenous GCN4 with the URA3 cassette followed by insertion of PCR-amplified 3T2S-GCN4 through homologous recombination. PCR genotyping was used to confirm all genomic manipulations. Replacement of the cdc48-3 allele for wild-type CDC48 was performed through homologous recombination of a PCR fragment encoding wild-type CDC48 and spanning the sites of the two mutations in cdc48-3 (P257L, R387K). Colonies were selected for ability to grow at the restrictive temperature of 37°C, and restoration of the wild-type CDC48 sequence was confirmed by PCR amplification of the locus and Sanger DNA sequencing. For anchor-away strains, epitope tagging of GCN4 and deletion of PDR5 were performed as described. CDC34 and GCN4 were tagged with FKBP12-rapamycin-binding domain (FRB) as described for epitope tagging except using an FRB-tagging construct (Haruki et al., 2008). A high-efficiency yeast transformation protocol was used for genomic manipulations and introduction of plasmids (Gietz and Schiestl, 2007). Supplemental Table S2 lists primer sequences.
RNA isolation and analysis
For Gcn4-based experiments, yeast were grown overnight in yeast extract/peptone/adenine/dextrose (YPAD), washed with sterile water, and diluted to an OD600 of 0.3 in minimal medium (0.67% yeast nitrogen base without amino acids and 2% dextrose, supplemented with amino acids as appropriate). Yeast were grown for 5 h and treated with 0.5 μg/ml SM (or dimethyl sulfoxide [DMSO] control) before RNA was harvested by the hot-phenol method (Leung et al., 2011). If proteasome inhibition was part of the experiment, yeast were grown for 4 h and treated with 50 μM MG132 (or DMSO control) plus 0.004% SDS (Liu et al., 2007) for 1 h before SM induction. For anchor-away experiments, yeast were grown for 4 h in minimal medium and treated with 1.0 μg/ml rapamycin (or DMSO control) for 1 h before SM induction. For Gal4-based experiments, yeast were grown overnight in YPAD, washed with sterile water, and diluted to an OD600 of 0.3 in CSM-RAF medium (0.67% yeast nitrogen base without amino acids, 2% raffinose) supplemented with CSM. After 5 h, cultures were induced with 2% galactose for 90 min before RNA was isolated. In all cases, mRNA levels were quantified by reverse-transcription quantitative PCR (RT-qPCR) as described (Leung et al., 2011) and normalized to an ACT1 primer pair. Supplemental Table S2 lists primer sequences.
Western blotting, coimmunoprecipitation, and ubiquitylation assays
For Western blotting, yeast pellets were resuspended in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 10 mM Imidazole) to a final OD600 of ∼150, and proteins were extracted by bead beating. For each protein preparation, 20 μg was ethanol precipitated to remove guanidine, resolved by SDS–PAGE, and transferred to nitrocellulose membranes. Membranes were probed using the appropriate antibodies: anti-HA–horseradish peroxidase (HRP; 12013819001; Roche, Basel, Switzerland), anti-c-Myc-HRP (11814150001; Roche), or anti–β-actin (ab8224; Abcam, Cambridge, MA). For coimmunoprecipitation assays, cell pellets from 100-ml cultures were resuspended in 800 μl of yeast lysis buffer (10 mM Na2HPO4/NaH2PO4, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40, 50 mM NaF, 0.1 mM Na3VO4, 1× Roche Complete, EDTA-free Protease Inhibitor Cocktail; 0.4 mg/ml Pefabloc SC, 50 μM MG132, 2 mg/ml iodoacetamide, and 200 μM 1,10-phenanthroline). Cells were lysed by bead beating at 4°C for 40 s, followed by incubation in ice water for 2 min, for a total of five times. Cell lysate was collected and cleared by centrifugation. Immunoprecipitation reactions were performed using the anti-HA antibody 12CA5 as described (Daulny et al., 2008). Ubiquitylation assays were performed using the His-tagged Ub method essentially as described (Daulny et al., 2008). Copper-inducible, His-tagged Ub was expressed from the plasmid pUB221 (Yaglom et al., 1996).
Chromatin immunoprecipitation
For each reaction, 100-ml cultures of yeast were processed as described for RNA isolation and analysis and cross-linked with 1% formaldehyde, and ChIP was performed as described (Geng and Tansey, 2012). The 12CA5 anti-HA antibody, 9E10 anti-Myc antibody, anti-Gcn4 antibody (sc-50443; Santa Cruz Biotechnology, Dallas, TX), or anti-TBP antibody (a gift from P. A. Weil, Vanderbilt University, Nashville, TN), was used to immunoprecipitate chromatin as indicated. Coprecipitating DNAs were quantified by qPCR using primer sets that amplify TATA box–proximal Gcn4-binding sites in each gene (for detection of Gcn4–promoter interaction) or the TATA box of the ARG1 gene. In each case, signal is calculated as equivalent to percentage of the input to the ChIP reaction. Primer sequences are listed in Supplemental Table S2.
Fluorescence microscopy
Yeast cultures were processed as described for RNA isolation and analysis and transferred to a glass slide for imaging. Images were acquired with a standard microscope (BX50; Olympus, Center Valley, PA) equipped with a motorized stage (model 999000; Ludl), UPlanF1 100×/numerical aperture 1.30 oil immersion objective, and digital charge-coupled device camera (Orca-R2; Hamamatsu, Hamamatsu City, Japan). Image manipulations were performed using ImageJ software.
Micrococcal nuclease protection assay
The micrococcal nuclease (MNase) protection assay was performed as described (Crisucci and Arndt, 2012). Briefly, 185-ml yeast cultures were processed as described for RNA isolation and analysis and cross-linked with a final concentration of 2% formaldehyde while being shaken at 30°C for 30 min. Formaldehyde was quenched with glycine, cells collected, and spheroplasts prepared by Zymolyase treatment. Spheroplasts (300 μl) were treated with increasing concentrations of MNase (10107921001; Roche) at 0, 1.0, 2.5, 5.0, 10, or 20 U MNase at 37°C for 45 min on a nutator. Reactions were stopped by addition of SDS (1% final concentration) and EDTA (10 mM final concentration), DNA purified by phenol-chloroform extraction, and recovered by ethanol precipitation. The efficiency of each MNase digestion was determined using a qPCR primer set flanking a region in the GAL1-10 upstream activating sequence (UAS) protected from MNase digestion by a nucleosome and a primer set flanking a region in the GAL1-10 UAS not protected from MNase digestion. Samples with MNase digestion efficiency of ∼95% were used for subsequent qPCR analysis using tiled primer sets. Primer sequences are listed in Supplemental Table S2.
Supplementary Material
Acknowledgments
We thank R. Adams, K. Arndt, L. Burns, R. Deshaies, D. Finley, M. Funk, R. Hampton, S. Lorey, J. MacGurn, and P. A. Weil for reagents and experimental assistance. We thank R. Deshaies, A. Weissmiller, P. A. Weil, and S. Wenzel for advice and guidance. This work is supported by National Institutes of Health Grant GM067728 and Vanderbilt Ingram Cancer Center Support Grant P30CA68485.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- RT-qPCR
reverse-transcription quantitative PCR
- Ub
ubiquitin
- UPS
ubiquitin–proteasome system.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0192) on July 6, 2016.
REFERENCES
- Ang K, Ee G, Ang E, Koh E, Siew WL, Chan YM, Nur S, Tan YS, Lehming N. Mediator acts upstream of the transcriptional activator Gal4. PLoS Biol. 2012;10:e1001290. doi: 10.1371/journal.pbio.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizec M, Herissant L, Pokrzywa W, Geng F, Wenzel S, Howard GC, Rodriguez P, Krause S, Tansey WP, Hoppe T, et al. The ubiquitin-selective chaperone Cdc48/p97 associates with Ubx3 to modulate monoubiquitylation of histone H2B. Nucleic Acids Res. 2014;42:10975–10986. doi: 10.1093/nar/gku786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Lipford JR, Deshaies RJ, Tansey WP. Gal4 turnover and transcription activation. Nature. 2009;461:E7. doi: 10.1038/nature08406. discussion E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisucci EM, Arndt KM. Paf1 restricts Gcn4 occupancy and antisense transcription at the ARG1 promoter. Mol Cell Biol. 2012;32:1150–1163. doi: 10.1128/MCB.06262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci USA. 2008;105:19649–19654. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Bercy J, Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987;207:142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- Geng F, Tansey WP. Similar temporal and spatial recruitment of native 19S and 20S proteasome subunits to transcriptionally active chromatin. Proc Natl Acad Sci USA. 2012;109:6060–6065. doi: 10.1073/pnas.1200854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Howard GC, Collins GA, Tansey WP. Letter to the editor. Chemical-genetic strategy for inhibiting proteasome function in Saccharomyces cerevisiae. Yeast. 2012;29:93–94. doi: 10.1002/yea.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia MH, Larossa RA, Lee JM, Rafalski A, Derose E, Gonye G, Xue Z. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Leung A, Cajigas I, Jia P, Ezhkova E, Brickner JH, Zhao Z, Geng F, Tansey WP. Histone H2B ubiquitylation and H3 lysine 4 methylation prevent ectopic silencing of euchromatic loci important for the cellular response to heat. Mol Biol Cell. 2011;22:2741–2753. doi: 10.1091/mbc.E11-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158. doi: 10.2144/000112389. 160, 162. [DOI] [PubMed] [Google Scholar]
- Lord CL, Timney BL, Rout MP, Wente SR. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J Cell Biol. 2015;208:729–744. doi: 10.1083/jcb.201412024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E, Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991;11:2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Ndoja A, Cohen RE, Yao T. Ubiquitin signals proteolysis-independent stripping of transcription factors. Mol Cell. 2014;53:893–903. doi: 10.1016/j.molcel.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [correction published in Genes Dev (1998). 12, 3144] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev. 2012;26:350–355. doi: 10.1101/gad.184689.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Hampton RY. Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast. 2006;23:1053–1064. doi: 10.1002/yea.1407. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.