MAPKs are catalytically and biologically active only when dually phosphorylated on a TEY motif. Mutations in the yeast MAPK Mpk1 are described that render it fully functional when mutated in its TEY motif and even when it carries a kinase-dead mutation.
Abstract
MAP kinases of the ERK family are conserved from yeast to humans. Their catalytic activity is dependent on dual phosphorylation of their activation loop’s TEY motif, catalyzed by MAPK kinases (MEKs). Here we studied variants of Mpk1, a yeast orthologue of Erk, which is essential for cell wall integrity. Cells lacking MPK1, or the genes encoding the relevant MEKs, MKK1 and MKK2, do not proliferate under cell wall stress, imposed, for example, by caffeine. Mutants of Mpk1, Mpk1(Y268C) and Mpk1(Y268A), function independently of Mkk1 and Mkk2. We show that these variants are phosphorylated at their activation loop in mkk1∆mkk2∆ and mkk1∆mkk2∆pbs2∆ste7∆ cells, suggesting that they autophosphorylate. However, strikingly, when Y268C/A mutations were combined with the kinase-dead mutation, K54R, or mutations at the TEY motif, T190A+Y192F, the resulting proteins still allowed mkk1∆mkk2∆ cells to proliferate under caffeine stress. Mutating the equivalent residue, Tyr-280/Tyr-261, in Erk1/Erk2 significantly impaired Erk1/2’s catalytic activity. This study describes the first case in which a MAPK, Erk/Mpk1, imposes a phenotype via a mechanism that is independent of TEY phosphorylation and an unusual case in which an equivalent mutation in a highly conserved domain of yeast and mammalian Erks causes an opposite effect.
INTRODUCTION
The extracellular-regulated kinases (Erks), encoded by ERK1 and ERK2, are serine/threonine kinases of the mitogen-activated protein kinase (MAPK) family. Erks modulate many cellular targets, primarily by phosphorylation of substrates (Haystead et al., 1992; Yang et al., 1998; Yoon and Seger, 2006; Cargnello and Roux, 2011; Michailovici et al., 2014) but also through noncatalytic processes (i.e., via protein–protein interactions; Rodriguez and Crespo, 2011). Erks also control transcription by direct binding to DNA (Hu et al., 2009; Rodriguez and Crespo, 2011). By modulating these proteins and genes, Erks regulate diverse cellular processes such as proliferation, differentiation, and cell survival (Boulton et al., 1991; Marshall, 1995; Avruch et al., 2001; Pearson et al., 2001; Shaul and Seger, 2007). They are overactive in most cancers and may acquire oncogenic properties themselves (Blume-Jensen and Hunter, 2001; Kohno and Pouyssegur, 2006; Roberts and Der, 2007; Samatar and Poulikakos, 2014; Smorodinsky-Atias et al., 2016). It is not clear whether the catalytic and the noncatalytic effects of Erks are both essential for the execution of their biological and pathological functions.
The three-dimensional structure of Erks is similar to that of most eukaryotic protein kinases (EPKs), but they contain two MAPK-specific domains, the MAPK insert and the C-terminal extension, whose functions are not fully known. As in most EPKs, the active site of Erks is stabilized and becomes functional only after activation and consequent conformational changes. Activation of Erks is achieved by dual phosphorylation of neighboring threonine and tyrosine residues (in a TEY motif) residing at the activation loop. This dual phosphorylation is catalyzed by MAPK kinases (MAPKKs; also known as MEKs or MKKs; Robbins et al., 1993; Marshall, 1994; Canagarajah et al., 1997; Cobb and Goldsmith, 2000; Pearson et al., 2001). Once activated, Erks interact with their substrates via two binding domains. One is known as the common docking (CD) domain and the other as the docking site for Erk FXF domain (DEF pocket). The CD domain is known to bind mainly cytoplasmic substrates, as well as upstream activators (MEKs), phosphatases, and probably other regulators (Tanoue et al., 2000; Tanoue and Nishida, 2003). The CD domain is not a mere docking motif, but is also a regulatory element. Proteins that bind it induce large conformational changes at the activation loop that expose the TEY motif to the solvent and assist activation (Zhou et al., 2006). The DEF pocket is a hydrophobic cavity found only in the conformation adopted by the dually phosphorylated Erk. It binds substrates, primarily transcription factors, that contain an FXF motif (Jacobs et al., 1999; Fantz et al., 2001; Galanis et al., 2001; Lee et al., 2004). It is not known whether proteins that bind the DEF pocket impose allosteric effects similarly to proteins that bind the CD domain.
In Erk2, mutations with interesting properties were reported in Tyr-261, which resides within the MAPK insert and serves as a central element of the DEF pocket. Mutating Tyr-261 to Ala has been shown to eliminate the binding of an array of DEF pocket substrates to Erk2 (Lee et al., 2004; Dimitri et al., 2005; Shin et al., 2010) and render the Erk2 protein incapable of inducing epithelial-to-mesenchymal transition in MCF10A cells (Shin et al., 2010). Mutating Tyr-261 to Asn impairs binding of MEK1 and causes nuclear localization of Erk2 (Robinson et al., 2002). The last-named study implies that Tyr-261 is involved not only in binding of substrates. Of interest, residues of the MAPK insert domain in the vicinity of Tyr-261 are critical for the DNA-binding activity of Erk2 (Hu et al., 2009). All of the foregoing studies suggest that mutations in Tyr-261 of the mammalian Erk2 have loss-of-function effects.
In an attempt to isolate molecules of the Erk family that are intrinsically active, that is, spontaneously active in a MEK-independent manner, we previously carried out a screen that took advantage of the yeast MAPK cascade known as the cell wall integrity pathway. The MAPK of this pathway is called MAPK 1 (Mpk1; also known as Slt2) and is one of the yeast orthologues of Erk. Yeast cells lacking components of the pathway have a weak cell wall and are sensitive to hypo-osmolarity and drugs such as calcofluor white and caffeine (Levin, 2005, 2011; Engelberg et al., 2014). In the screen, we looked for mutants of the Mpk1 protein that are able to function in cells lacking their MEKs, MKK1 and MKK2. Specifically, we looked for Mpk1 mutants that rescue mkk1∆mkk2∆ cells from caffeine stress (Levin-Salomon et al., 2008). Such mutants are by definition intrinsically active because they execute the downstream functions of Mpk1 in total absence of their direct activators. One of the mutants isolated was MPK1(R68S). Insertion of an equivalent mutation to Erk1 and Erk2, R84S and R65S, respectively, rendered them catalytically intrinsically active in vitro and spontaneously active in cell culture (Levin-Salomon et al., 2008; Smorodinsky-Atias et al., 2016; see Figure 1 for location of mutations within the MPK1, ERK1, and ERK2 sequences). Another mutant that was isolated in the screen, MPK1(Y268C), is mutated in the residue equivalent to Tyr-280 of Erk1 and Tyr-261 of Erk2 (Figure 1). In contrast to the case of the R68S/R84S/R65S mutations, mutating Tyr-280/261 in Erk1/2 to cysteine decreased their activity as assayed in vitro (Levin-Salomon et al., 2008). Namely, in Mpk1, the Y268C mutation had a dramatic gain-of-function effect, but the equivalent mutation in Erk1/2 seems to have a loss-of-function effect (Levin-Salomon et al., 2008).
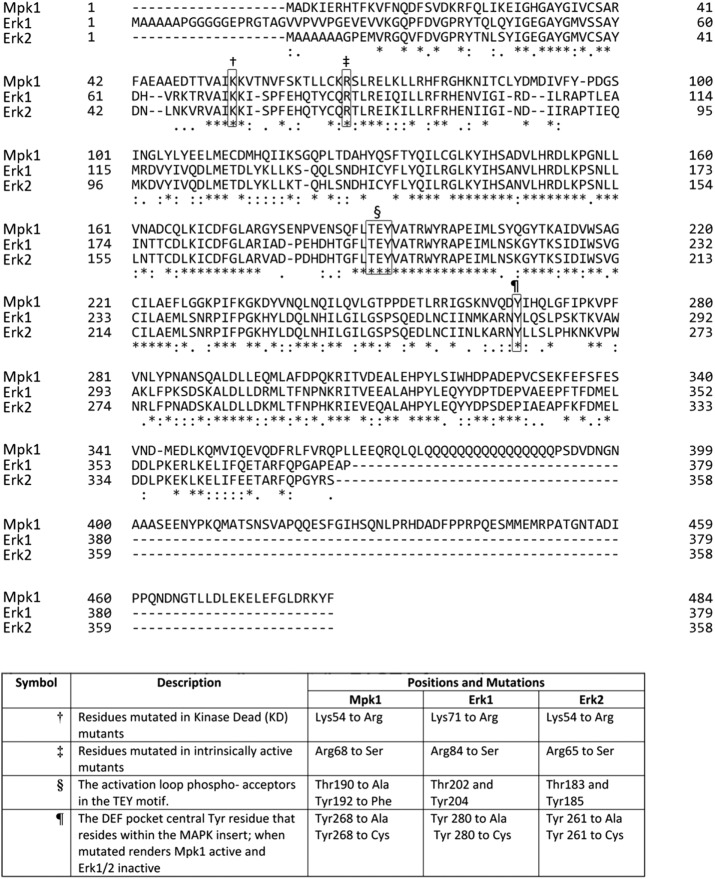
FIGURE 1:
Sequence alignment of the yeast Mpk1 and the mammalian Erk1 and Erk2 proteins. Locations of the residues that were mutated in this study are marked by symbols, and the concrete mutations are shown in the table.
Here we studied the characteristics of Mpk1 molecules mutated in Tyr-268, to understand the mechanism through which they escape the dependence on Mkk1/Mkk2 regulation. We report that Mpk1(Y268C) and Mpk1(Y268A) were phosphorylated at their activation loop not only in mkk1∆mkk2∆ cells, but also in mkk1∆mkk2∆pbs2∆ste7∆ cells, which lack all known yeast MEKs. It seems that Mpk1(Y268C) and Mpk1(Y268A) gained some degree of autophosphorylation capability. Furthermore, combining the mutations in Tyr-268 with the activating mutation R68S resulted in a synergistic effect, making Mpk1(R68S+Y268C) and Mpk1(R68S+Y268A) highly phosphorylated at their activation loop in mkk1∆mkk2∆ cells and highly capable of supporting the growth of these cells in the presence of caffeine. Unexpectedly, Mpk1 molecules carrying the Y268C/A mutation together with the kinase-dead mutation, K54R, or together with mutations at the activation loop phosphoacceptors, T190A and Y192F, which are crucial for Mpk1’s activation, could still rescue mkk1∆mkk2∆ cells. It seems that the Y268C/A mutations elevated both autocatalytic capability and the noncatalytic activity of Mpk1. The noncatalytic activity appeared to be responsible for most of the gain-of-function effect of the mutations and was sufficient, by itself, to support effective proliferation of mkk1∆mkk2∆ cells under cell wall stress.
We also measured the activity of the equivalent mutants of the mammalian Erk1 and Erk2 proteins in vitro in mammalian cells and yeast. In contrast to the effect of the mutations on Mpk1, mutating Tyr-280/Tyr-261 impaired both the catalytic and the biological capabilities of Erk1/2. Furthermore, when these mutations were combined with the R84S/R65S mutations, which render Erk1/Erk2 intrinsically active, the resulting double mutants, Erk1(R84S+Y280C/A) and Erk2(R65S+Y261C/A), showed reduced intrinsic activity, and even their MEK-induced activity was impaired. Accordingly, whereas Erk1(R84S) induces oncogenic transformation in NIH3T3 cells, Erk1(R84S+Y280C) and Erk1(R84S+Y280CA) do not. The destructive effect of the mutations at the Tyr-261 residue on Erk2 activity was also apparent when the mammalian Erk2 mutants were tested in yeast cells. We conclude that the function of Tyr-268/280/261 is not only to support substrate binding as part of the DEF pocket, but also to play regulatory roles in autophosphorylation and noncatalytic activities. These roles may be opposite in the yeast Mpk1 and the mammalian Erks.
RESULTS
Mutating Tyr-268 to Cys or Ala renders Mpk1 intrinsically active
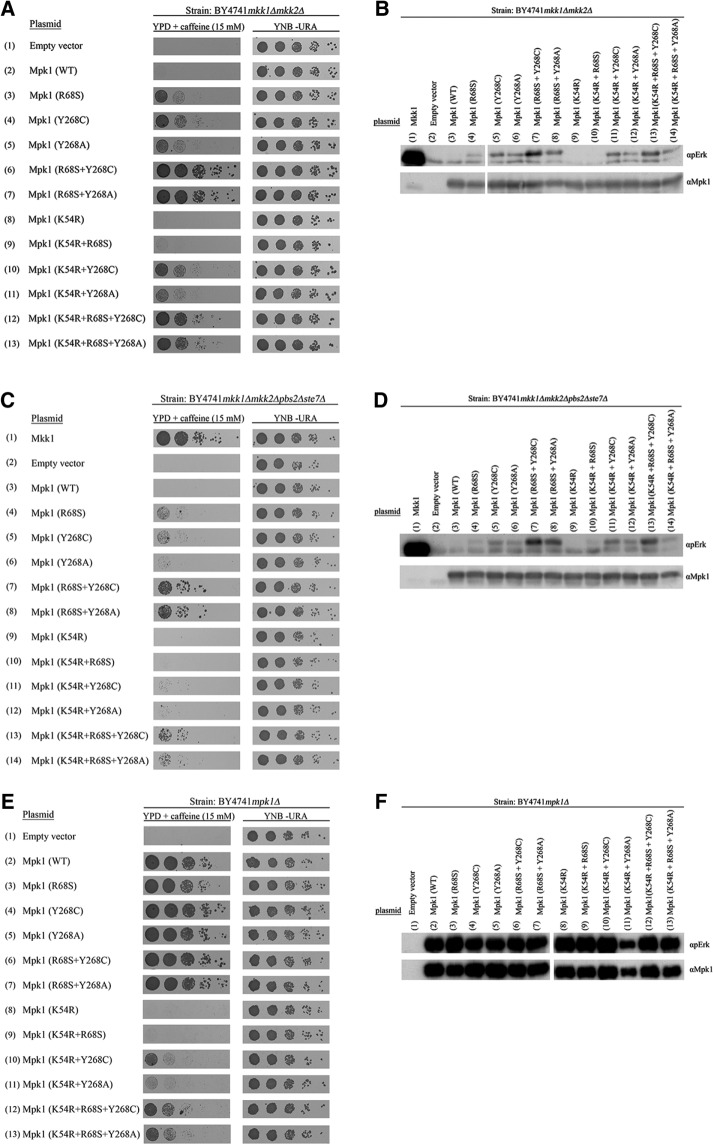
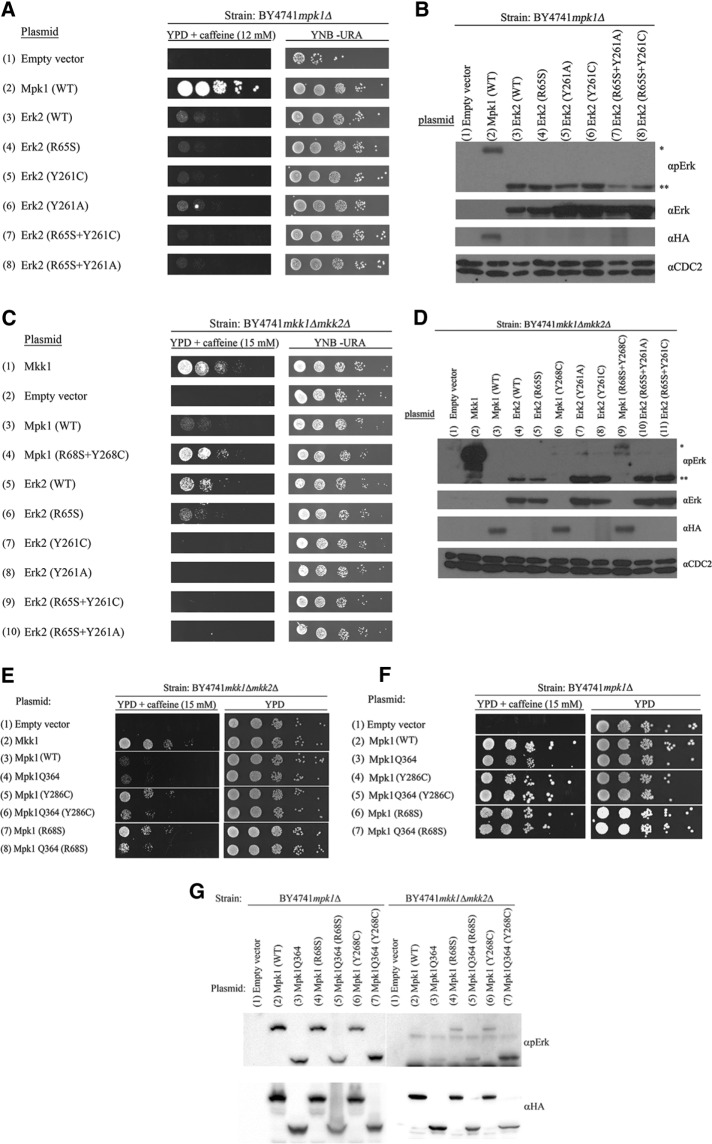
Several point mutations were found to render the yeast Mpk1 intrinsically (MEK independently) active (Levin-Salomon et al., 2008). Mpk1 molecules carrying any of these mutations allow growth of yeast cells lacking the Mpk1 activators Mkk1 and Mkk2 (mkk1∆mkk2∆ cells) in the presence of caffeine (Levin-Salomon et al., 2008; Figure 2A). These mutations can be defined therefore as gain-of-function mutations. One of these is the conversion of Tyr-268 to Cys (Levin-Salomon et al., 2008; Figure 2A, row 4). The Tyr-268 of Mpk1 is an orthologue of Tyr-261 of Erk2, which, when mutated to Ala or Asn, were described as loss-of-function mutations (Robinson et al., 2002; Lee et al., 2004; Dimitri et al., 2005; Shin et al., 2010). We thus tested whether mutating Tyr-268 of Mpk1 to Cys would render it intrinsically active, whereas mutating it to Ala would abolish its activity. We found, however, that Mpk1(Y268A) did allow mkk1∆mkk2∆ cells to proliferate in the presence of caffeine, although less efficiently than Mpk1(Y268C) (Figure 2A, row 5).
FIGURE 2:
The gain-of-function mutants Mpk1(Y268C) and Mpk1(Y268A) acquired the capability to spontaneously autophosphorylate their activation motif. (A) mkk1∆mkk2∆, (C) mkk1∆mkk2∆pbs2∆ste7∆, and (E) mpk1Δ cells expressing the indicated Mpk1 molecules were plated at five dilutions on plates containing YPD supplemented with 15 mM caffeine (left) or plates without caffeine (YNB(–URA); right). Western blot analysis with the indicated antibodies was performed on protein lysates prepared from (B) mkk1∆mkk2∆ cells, (D) mkk1∆mkk2∆pbs2∆ste7∆ cells, or (F) mpk1Δ cells expressing the indicated Mpk1 molecules.
The genetic screen that revealed the Mpk1 Y268C gain-of-function mutation also revealed another mutation, R68S, which provided the protein with an efficient autophosphorylation/autoactivation capability that readily explains its independence of MEKs (Levin-Salomon et al., 2008; Smorodinsky-Atias et al., 2016). If the R68S mutation renders Mpk1 intrinsically active by a different mechanism than the Y268C/A mutations, perhaps combining both mutations on the same Mpk1 molecule would result in a synergism that produced highly active Mpk1. Such an approach was very successful in obtaining superactive variants of Hog1 and p38 MAP kinases (Yaakov et al., 2003; Diskin et al., 2004; Askari et al., 2006, 2007). We thus generated mutants Mpk1(R68S+Y268C) and Mpk1(R68S+Y268A) and found that combining the mutations indeed had a synergistic effect, as manifested by inducing enhanced proliferation of mkk1∆mkk2∆ cells (Figure 2A, rows 6 and 7).
Mpk1(Y268C) and Mpk1(Y268A) acquire autophosphorylation capability
Because Tyr-268 is not known to be involved in Erk’s catalytic activity but instead in binding of substrates (as part of the DEF pocket) and MEK1 (Robinson et al., 2002; Dimitri et al., 2005; Shin et al., 2010), we presumed that the mechanism underlying the gain-of-function property of Mpk1(Y268C/A) involves changes in binding affinities and not necessarily changes in catalytic activation. Activation by phosphorylation of the TEY motif of Mpk1(Y268C/A) was thus not expected in mkk1∆mkk2∆ cells. We found, however, that Mpk1(Y268C) and Mpk1(Y268A) are phosphorylated at the activation loop in mkk1∆mkk2∆ cells (Figure 2B, lanes 5 and 6). Moreover, the double mutants Mpk1(R68S+Y268C) and Mpk1(R68S+Y268A) manifested even higher phosphorylation levels of the activation loop (Figure 2B, lanes 7 and 8). This phosphorylation, which rendered the kinases catalytically active, could be a result of increased affinity to a MAPKK other than Mkk1 or Mkk2. To examine this possibility, we expressed Mpk1(Y268C) and Mpk1(Y268A) in cells of the mkk1∆mkk2∆pbs2∆ste7∆ strain, which lack all four yeast MAPKKs (Levin-Salomon et al., 2009). mkk1∆mkk2∆pbs2∆ste7∆ cells expressing either Mpk1(Y268C) or Mpk1(Y268A) grew under caffeine stress (Figure 2C, rows 5 and 6, left). Of note, however, growth on caffeine of mkk1∆mkk2∆pbs2∆ste7∆ cells expressing the mutants was significantly less efficient than that of mkk1∆mkk2∆ cells expressing the same mutants (compare Figure 2, A and C). It could be, therefore, that Pbs2 and Ste7 play some role in the activation of the Mpk1 mutants. Alternatively, it could be that cells lacking the four MEK-encoding genes are weak and defective in other stress-response systems, so that the effect of knocking out PBS2 and STE7 on the Mpk1 mutants is indirect. In any case, the fact that Mpk1(Y268C) and Mpk1(Y268A) are functional to some extent in mkk1∆mkk2∆pbs2∆ste7∆ cells indicates that an important part of their activity is independent of any MAPKK. Furthermore, we found that both proteins were phosphorylated at their activation loop in mkk1∆mkk2∆pbs2∆ste7∆ cells as well (Figure 2D, lanes 5 and 6).
Thus the mutants are in fact catalytically activated to some degree in mkk1∆mkk2∆ and mkk1∆mkk2∆pbs2∆ste7∆ cells in a manner that does not involve any known MAPKK. Of importance, Mpk1(Y268C) and Mpk1(Y268A) did not lose the ability to be further phosphorylated and activated by MEKs when they were present, as mpk1∆ cells expressing Mpk1(Y268C) or Mpk1(Y268A) proliferated at a high rate on medium supplemented with caffeine (Figure 2E, rows 4 and 5), and the mutants were phosphorylated at higher levels than they were in mkk1∆mkk2∆ cells (Figure 2F, lanes 4 and 5). Because Mpk1(Y268C) and Mpk1(Y268A) were phosphorylated in mkk1∆mkk2∆pbs2∆ste7∆, their phosphorylation could possibly be achieved via autophosphorylation. To examine this idea, we constructed kinase-dead variants of Mpk1(Y268C) and Mpk1(Y268A) molecules by adding to them the kinase-dead mutation K54R. We confirmed that this mutation abolished the functionality of Mpk1 by showing that Mpk1(K54R) could not support proliferation of mpk1∆ cells under caffeine stress (Smorodinsky-Atias et al., 2016; Figure 2E, row 8). We expected loss of both biological activity and activation loop phosphorylation of Mpk1(Y268C) and Mpk1(Y268A). Totally unexpectedly, the Mpk1(K54R+Y268C) and Mpk1(K54R+Y268A) proteins, carrying the “kinase-dead” mutation, maintained activity and were able to promote growth of mkk1∆mkk2∆ cells (Figure 2A, rows 10 and 11) and mpk1∆ cells (Figure 2E, rows 10 and 11) in the presence of caffeine. Furthermore, these proteins were phosphorylated at their activation loop in cells lacking MEKs (Figure 2, B and D, lanes 11 and 12). In mkk1∆mkk2∆pbs2∆ste7∆ cells, the Mpk1(K54R+Y268C) and Mpk1(K54R+Y268A) proteins supported minimal growth on caffeine (Figure 2C, rows 11 and 12). Thus the K54R mutation did reduce the ability of Mpk1(Y268C) and Mpk1(Y268A) to support proliferation in the presence of caffeine, but this reduction was partial.
One explanation for the fact that proteins carrying kinase-dead mutations were still active is that in the Mpk1(Y268C) and Mpk1(Y268A) proteins, the kinase-dead mutation is not sufficient for abolishing the activity because the mutations in the 268 position enforce stabilization of the ATP-binding site, making Lys-54 dispensable for ATP binding, so that even without it, these proteins are capable of autophosphorylation and autoactivation. This notion is supported by the observation that Mpk1(K54R+Y268C) and Mpk1(K54R+Y268A) are phosphorylated in mkk1∆mkk2∆pbs2∆ste7∆ cells. Another possibility is that the K54R mutation significantly reduced the catalytic activity of Mpk1(Y268C) and Mpk1(Y268A), but they were still biologically active because catalysis is not critical for rescuing the cells from the caffeine stress. Instead, the mutants modulated the necessary downstream components primarily by noncatalytic means.
Mpk1(Y268C) and Mpk1(Y268A) function via a mechanism that does not require phosphorylation of the TEY motif
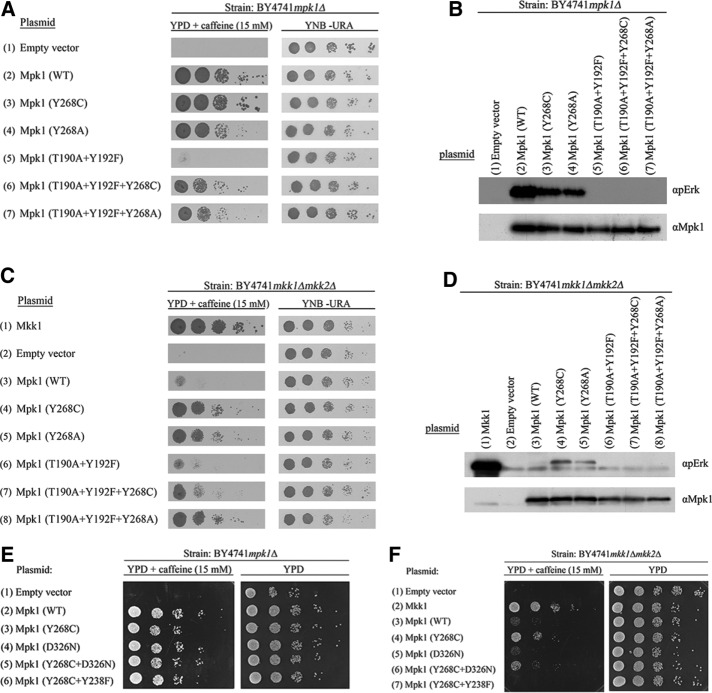
We currently cannot directly measure the intrinsic catalytic activity of the Mpk1 mutants to assess whether the K54R mutation abolished it because recombinant Mpk1 is not available and efforts we made to produce such proteins were not successful. We therefore took another approach to check the possibility that the unusual intrinsic biological activity of Mpk1(Y268C) and Mpk1(Y268A) does not require activation of their catalytic properties. We inserted into Mpk1(Y268C) and Mpk1(Y268A) mutations in their TEY motif (T190A and Y192F), so that they could not be catalytically activated at all, either by MEKs or via autophosphorylation. As expected, when expressed in mpk1∆ or mkk1∆mkk2∆ cells, Mpk1(T190A+Y192F) molecules did not support proliferation of these cells in the presence of caffeine (Figure 3, A, row 5, and C, row 6). However, Mpk1(T190A+Y192F+Y268C) and especially Mpk1(T190A+Y192F+Y268A) were able to rescue mpk1∆ and mkk1∆mkk2∆ cells from caffeine stress (Figure 3, A, rows 6 and 7, and C, rows 7 and 8; note controls in Figure 3, B and D), although less efficiently than the parental mutants Mpk1(Y268C) and Mpk1(Y268A). This suggests that the ability of the Mpk1 variants carrying mutations in Tyr-268 to execute biological functions is only partially dependent on activation of catalysis. The mutants function quite efficiently without catalytic activation by phosphorylation of the TEY motif. These results, together with the results with the proteins carrying the K54R+Y268C/A mutations, strengthen the possibility that mutations in Tyr268 stabilize a conformation that promotes biological functions of the protein, primarily via noncatalytic activities.
FIGURE 3:
Mpk1(Y268C) and Mpk1(Y268A) rescue mpk1∆ and mkk1∆mkk2∆ cells even when mutated in their TEY motif. (A) mpk1Δ and (C) mkk1∆mkk2∆ cells expressing the indicated Mpk1 molecules were plated at five dilutions on plates containing YPD supplemented with 15 mM caffeine (left) or plates without caffeine (YNB(–URA); right). Western blot analysis with the indicated antibodies was performed on protein lysates prepared from (B) mpk1∆ and (D) mkk1∆mkk2∆ cells expressing the indicated Mpk1 molecules. (E, F) Mpk1(Y268C+D326N) and Mpk1(Y268C+Y238F) rescue mpk1∆ (E) but not mkk1∆mkk2∆ (F) cells.
The observation that TEY phosphorylation was not required for the functionality of Mpk1(Y268C) and Mpk1(Y268A) led us to test whether intact binding sites of regulators and substrates are required. This was done by combining the Y268C mutation with the D326N mutation (equivalent to the sevenmaker mutation, which is known to reduce binding to the CD domain; Bott et al., 1994; Brunner et al., 1994) or with the Y238F mutation, which is important for the function of the DEF pocket (Lee et al., 2004). Mpk1(Y268C+D326N) and Mpk1(Y268C+Y238F) were not able to rescue mkk1∆mkk2∆ cells but did rescue mpk1∆ cells from caffeine stress (Figure 3, E and F). Thus the Mpk1(Y268C) does require intact binding domains to function when Mkk1/2 are absent. Because the effect of the Y238F was more dramatic than that of the D326 mutation (Figure 3F), it seems that the DEF pocket is more critical than the CD domain. The ability of the Mpk1(Y268C+D326N) and Mpk1(Y268C+Y238F) mutants to rescue mpk1∆ cells, which do possess Mkk1/2, may suggest that activation loop phosphorylation makes the CD domain and the DEF pocket rigidly stable, and hence D326 or Y238 can be mutated without significant effects on the activity. In mkk1∆mkk2∆ cells, stabilization of these structural elements relies on the Y268C mutation alone, and it is therefore more sensitive to mutagenesis.
Insertion of equivalent mutations to mammalian Erk1 and Erk2 imposes loss-of-function effects on their catalytic activity and biological functions
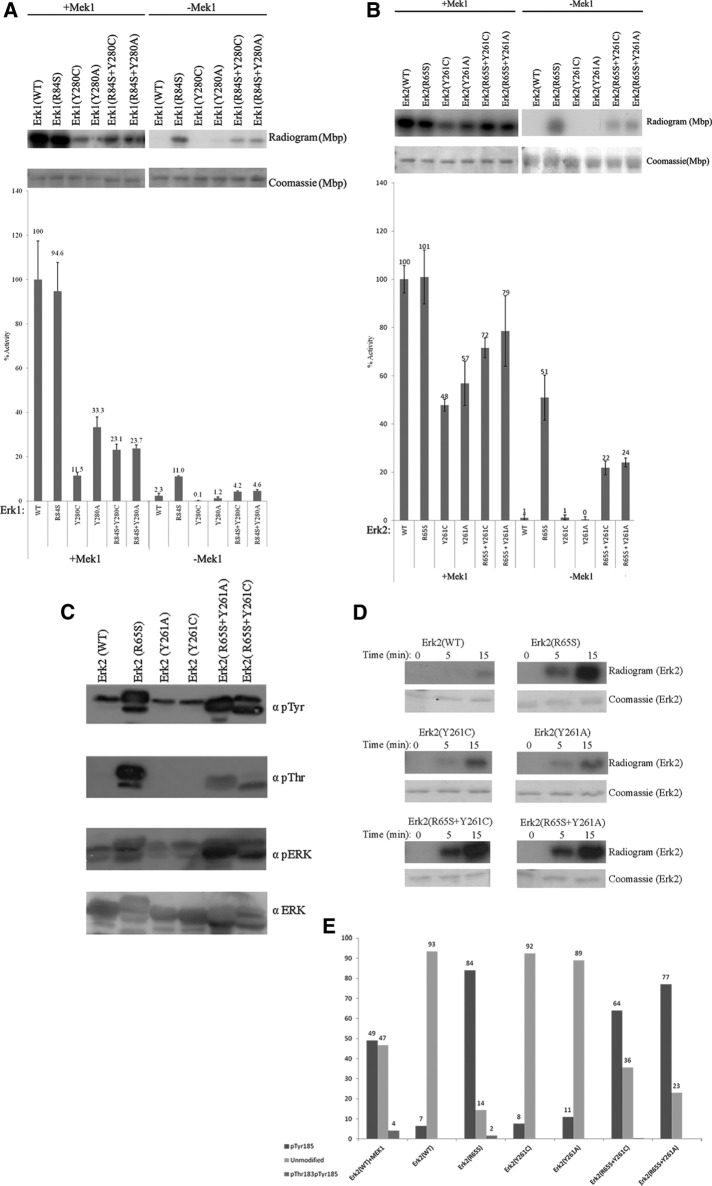
A large number of yeast proteins have orthologues in mammals, and in many cases, structure–function and regulatory aspects are faithfully conserved (Engelberg et al., 2014). As a result, many interesting mutations have similar effects on the yeast and mammalian orthologues (Engelberg et al., 1989, 2014). This is particularly common in functionally interchangeable proteins such as Mpk1 and Erk2. Indeed, the R68S mutation that was identified in Mpk1 as a gain-of-function mutation has similar effects on the mammalian Erk1 and Erk2 proteins (Levin-Salomon et al., 2008; Smorodinsky-Atias et al., 2016). However, inserting mutations equivalent to the Y268C/A mutations of MPK1 to Erk1 and Erk2 did not create MEK independence. Furthermore, not only did Erk1(Y280C/A) and Erk2(Y261C/A) manifest no intrinsic activity, but their MEK-dependent activity was lower than that of Erk1(WT) and Erk2(WT), as assayed in vitro with purified proteins (Levin-Salomon et al., 2008; Figure 4, A and B). MEK1-phosphorylated Erk1(Y280C) showed only 11.5% of the activity manifested by MEK1-phosphorylated Erk1(WT) (Figure 4A), and MEK1-phosphorylated Erk2(Y261C) showed only 48% of the activity manifested by MEK1-phosphorylated Erk2(WT) (Figure 4B). Similarly, MEK1-phosphorylated Erk1(Y280A) and Erk2(Y261A) proteins showed only 33.3 and 57% of the activity of MEK1-phosphorylated Erk1/2(WT), respectively (Figure 4, A and B).
FIGURE 4:
Mutating Tyr-280/261 in the mammalian Erk1/2 proteins has loss-of-function effects on their catalytic properties. (A, B) Catalytic activity of purified recombinant Erk1/2 carrying mutations at Tyr-280/Tyr-261 was monitored with or without preincubation with active MEK1 using [γ-32p]ATP and MBP as substrates. Reaction mixtures were spotted on filter papers and quantified. Activity of MEK1-activated Erk2(WT) was defined as 100%. In parallel, a sample from each reaction was subjected to SDS–PAGE, stained with Coomassie brilliant blue, and exposed to x-ray film. (C) Purified recombinant Erk2 proteins were subjected to Western blot analysis using antibodies that react with Erk1/2 proteins phosphorylated at their TEY motif (αpErk), antibodies that react with phosphorylated Tyr (αpTyr) or Thr (αpThr) residues, and antibodies that react with Erk (αErk). (D) The autophosphorylation capabilities of the purified Erk2 variants were assessed by incubating the proteins in a kinase assay mixture with [γ-32p]ATP and no other substrate. Reactions were terminated at the indicated time points, separated by SDS–PAGE, stained with Coomassie brilliant blue, and exposed to an x-ray film. (E) Summary of results obtained from LC-MS/MS analysis of the indicated purified recombinant proteins. LC-MS/MS analysis was performed on proteins that underwent an autophosphorylation reaction.
We further tested the effect of combining the Tyr-280/Tyr-261 mutations with the R84S/R65S mutation. Whereas equivalent combinations lead to a significant synergism in Mpk1 (Figure 2A, rows 6 and 7), combining these mutations in Erks had a negative effect. Mutating the Tyr-280/Tyr-261 residues in Erk1(R84S) and Erk2(R65S) reduced both the intrinsic catalytic activity and the MEK-dependent activity of these active variants, as assayed in vitro with myelin basic protein (MBP) as a substrate (Figure 4, A and B). We also tested whether Erk2(Y261A) and Erk2(Y261C) gained any autophosphorylation capabilities. Western blot analysis with anti–phospho-Thr and anti–phospho-Tyr showed that these proteins, just like Erk2(WT), have a negligible autophosphorylation capability (Figure 4C). Direct measurement of autophosphorylation capability as assessed by incorporation of a 32PO4 group from [γ-32p]ATP confirmed this notion (Figure 4D). Finally, liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of these mutants showed that ∼90% of Erk2(Y261C/A) and Erk2(WT) molecules were unmodified at their TEY motif. On the other hand, relevant peptides derived from Erk2(R65S) and Erk2(R65S+Y261C/A) were mainly phosphorylated on the Tyr-185 residue (Figure 4E). Erk2(R65S) was also phosphorylated, although at a low level (2% of the peptides), on Thr-183, and this level was even lower in Erk2(R65S+Y261C/A) (Figure 4E).
In summary, the very same mutations that endowed gain-of-function properties (catalytic and biological) on Mpk1 were destructive for the catalytic activity of Erk1 and Erk2.
The loss-of-function effects of mutating Tyr-280/Tyr-261 in Erk1/2 are also manifested in mammalian and yeast cells
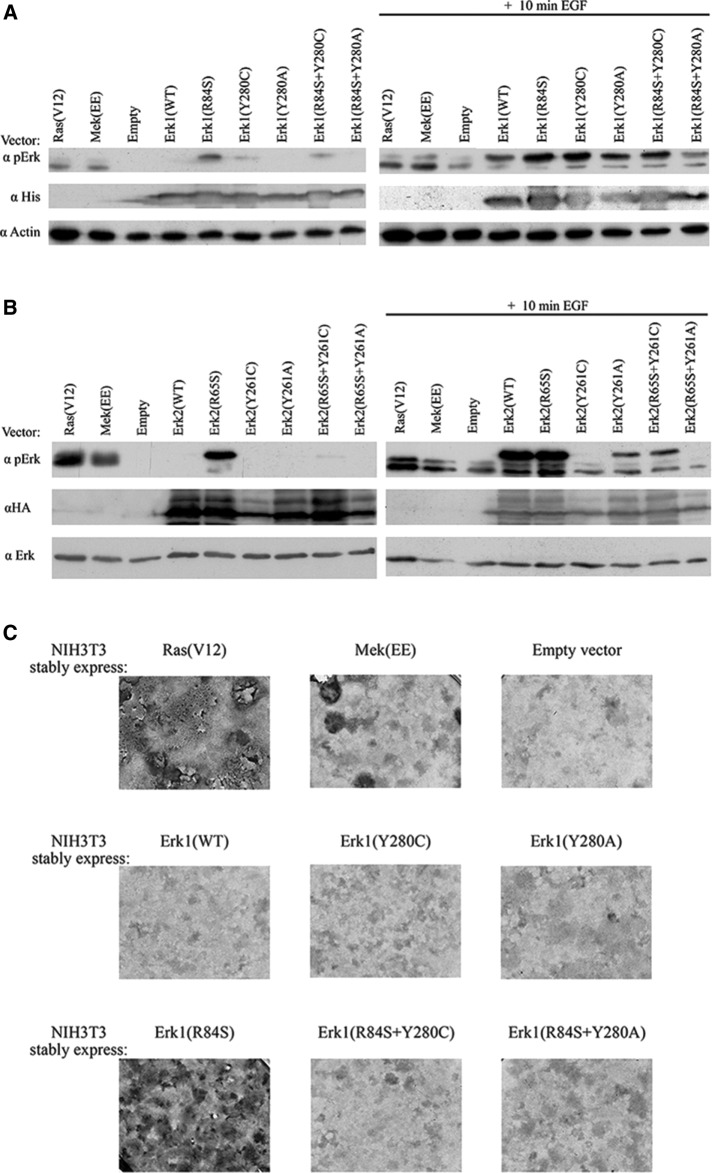
Whereas the loss-of-function effects of the Tyr-280/261 mutations in Erk1/2 were observed in vitro using purified proteins, the gain-of-function effects of the equivalent mutations in Mpk1 were manifested in vivo in yeast cells. The possibility remains, therefore, that the Erk mutant would also manifest gain-of-function properties in living cells of mammals or yeast. However, when we expressed the Erk1 and Erk2 mutants in HEK293T cells, we found that, as in the in vitro assays, the proteins carrying a mutation in Y280/Y261 had no intrinsic autophosphorylation ability and that proteins mutated in Y261 had a significantly lower phosphorylation level even after exposure to EGF (compared with that of Erk2(WT)). Moreover, mutating these residues in Erk1(R84S) and Erk2(R65S) abolished the spontaneous activity of these active variants in HEK293T cells (Figure 5, A and B). We also tested all mutants in NIH3T3 cells. This experiment is of particular importance because we recently observed that Erk1(R84S) can transform NIH3T3 cells (Smorodinsky-Atias et al., 2016), raising the question of whether combining the Y280C and Y280A mutations would increase its oncogenic capabilities (analogous to the effect of combining these mutations in Mpk1; Figure 2A, rows 6 and 7) or instead impair these capabilities, as suggested by the in vitro assays (Figure 4, A and B). The results of this assay showed that Erk1(R84S+Y280C) was not capable of inducing oncogenic transformation in NIH3T3 cells (Figure 5C), demonstrating that the loss-of-function effect of the Y280C/A mutations is manifested in living cells too.
FIGURE 5:
Mutating Tyr280/Tyr-261 in Erk1/2 causes loss-of-function effects on their biological properties in mammalian cells. (A, B) pCEFL vectors carrying cDNAs encoding the indicated Erk1 (A) or Erk2 (B) molecules, a control empty vector, or vectors encoding Ha-Ras(V12) or Mek(EE) were introduced into HEK293 cells. At 48 h posttransfection, cells were exposed to 50 ng/ml EGF for 10 min and harvested. Protein lysates prepared from these cells were analyzed by Western blot analysis, using antibodies that specifically recognize phospho-Erk or the relevant tag (His-Erk1 or HA-Erk2). (C) NIH3T3 cells transfected with the indicated vectors were allowed to grow in the presence of G-418 to high density and reach confluence. Continuation of proliferation and appearance of foci was monitored. Cells were fixed and stained with crystal violet 4 wk after transfection. We estimate that the following number of foci appear when 1 μg of plasmid is introduced into the cells: ∼500 foci with a plasmid carrying Ha-ras(V12), ∼60 with a plasmid carrying ERK1(R84S), ∼10 with a plasmid carrying MEK(EE), and 0 with plasmids carrying ERK1(R84S + Y280C) or ERK1(R84S + Y280A).
The foregoing results further strengthen the notion that the same mutations in Mpk1 and Erk1/2 in a highly conserved domain have opposite effects on the proteins. A possible explanation for this paradoxical observation is that the cellular environment affects the behavior of the mutants in addition to the effect of the mutation itself. It could be, for example, that within the yeast cell, Mpk1(Y268C) and Mpk1(Y268A) manifest gain-of-function properties because they are associated with some allosteric effector(s). Therefore the mammalian Erks carrying these mutations may also show gain-of-function effects in yeast. Because the mammalian Erk2 can functionally replace Mpk1 to some extent, allowing mpk1∆ cells to proliferate, although slowly, in the presence of caffeine (Levin-Salomon et al., 2009; Figure 6A, row 3), we could directly test whether Erk2(Y261C) and Erk2(Y261A) can also support proliferation of mpk1∆ cells on caffeine. We found that Erk2(Y261C) and Erk2(Y261A) did support some proliferation of the cells in the presence of caffeine, similar to Erk2(WT) (Figure 6A, rows 5 and 6), and were efficiently phosphorylated at their activation loop when expressed in mpk1∆ cells (Figure 6B, lanes 5 and 6). In the course of these experiments, however, we found that mammalian Erk2(WT) could also rescue mkk1∆mkk2∆ cells, probably because in yeast, it is autoactivated spontaneously (Levin-Salomon et al., 2009; Figure 6C, row 5). However, Erk2(Y261C) and Erk2(Y261A) could not rescue these cells, although they were phosphorylated on their TEY motif (Figure 6, C, rows 7 and 8, and D, lanes 7 and 8). These results demonstrate that the Y261C/A mutations in Erk2 caused loss-of-function effects within the yeast cells too, although they were autophosphorylated. We concluded that the opposite effect of the mutation on Mpk1 and Erk1/2 is inherent to the molecules.
FIGURE 6:
Mutating Tyr-280/Tyr-261 in Erk1/2 causes loss-of-function effects on their activity in yeast cells. (A) mpk1Δ or (C) mkk1∆mkk2∆ yeast cells expressing the indicated mammalian Erk2 molecules were plated at five dilutions on plates containing YPD supplemented with 15 mM caffeine (left) or plates without caffeine (YNB(–URA); right). Western blot analysis with the indicated antibodies was performed on protein lysates prepared from (B) mpk1∆ cells or (D) mkk1∆mkk2∆ cells expressing the indicated Erk2 molecules. In B and D, asterisk marks the position of Mpk1 and double asterisks mark the position of Erk2. (E–G) When inserted into a truncated Mpk1 molecule that is very similar to Erk2 (Mpk1Q364), the Y268C and the R68S mutations do not render it intrinsically active but also do not abolish its standard activity.
What could explain the inherent difference between Mpk1 and Erk1/2 with respect to the Y261/268 residue? An obvious difference between the proteins is the presence of a longer C-terminal tail in Mpk1, which makes this protein 126 residues longer than Erk2 (Figure 1). Removal of this tail, generating Mpk1Q364, makes a Mpk1 molecule that is very similar to Erk2 and is still biologically functional (see Figure 6 in Levin-Salomon et al., 2009). We thus wondered about the effect of the Y268C mutation on the truncated Mpk1 molecule. Insertion of the mutation to Mpk1Q364 allowed the resulting protein, Mpk1Q364(Y268C), to rescue mkk1∆mkk2∆ but significantly less efficiently than Mpk1(Y268C) (Figure 6E), indicating that in the absence of the tail, only low Mkk1/2-independent activity is induced by the Y268C. Thus, somewhat similar to the case of Erks, the Y268C mutation is much less effective on the short, Erk-like, Mpk1 protein. Of interest, as a control, we also inserted the R68S mutation to Mpk1(Q364) and saw that, similarly to the Mpk1Q364(Y268C) protein, Mpk1Q364(R68S) is also less efficient in rescuing mkk1∆mkk2∆ cells than MPK1(R84S) (Figure 6, E and F). It is therefore impossible to be conclusive on whether the long C-terminus of Mpk1 is partially responsible for the different effect of the Y268C/261C mutation on the Mpk1 and Erk proteins.
DISCUSSION
Enzymes possess unusual capabilities to enhance the rates of catalysis of specific reactions. The rates of a catalyzed reaction are 5–17 orders of magnitude higher than the rates of a noncatalyzed reaction (Kraut, 1988; Miller and Wolfenden, 2002).
This power underlies the ability of enzymes to carry out the processes of life. It is becoming apparent, however, that enzymes also affect the fate of cells and organisms via mechanisms that do not use their catalytic capabilities. For example, various enzymes that function in signal transduction cascades have an additional activity, mainly as DNA-binding proteins involved in transcriptional regulation (Alepuz et al., 2001; Zeitlinger et al., 2003; Pokholok et al., 2006; Hu et al., 2009; Rodriguez and Crespo, 2011). Other enzymes may function as scaffold molecules (Pbs2, Ste5, Mek, PI3K) or stabilizers of large protein complexes (Cdc25) in addition to their catalytic activities (Engelberg et al., 1990; Posas and Saito, 1997; Posas et al., 1998; Iyer et al., 2005; Taylor et al., 2013). Noncatalytic activities have also been proposed for Mpk1 and its pseudokinase paralogue, Mlp1. These enzymes activate the transcription regulator Swi4/Swi6 (SBF; Kim et al., 2008). Because Mlp1 does not have any phosphotransfer activity, it is clear that its effect on SBF is mediated solely via the interaction with Swi4, one of the subunits that form SBF. Similarly, Mpk1 molecules carrying the kinase-dead (K54R) mutation are also capable of activating SBF (Kim et al., 2008). Of importance, although the catalytic activity of Mpk1 and Mlp1 is not required for SBF activation, both proteins must be phosphorylated to execute it. Mpk1 also functions as a transcriptional elongation factor via its interaction with one of the subunits of the Paf1C elongation complex (Kim and Levin, 2011).
Here we showed that Mpk1 molecules carrying a mutation in Tyr-268 executed the entire downstream biological function, using mechanisms that do not require activation of catalysis or the catalytic Lys-54 residue. It could be therefore that the noncatalytic activities of Mpk1, such as activation of SBF, transcription elongation, and activities not yet defined, are sufficient to allow proliferation under cell wall stress. This notion readily explains why these mutants are independent of MEKs. The possibility remains that Mpk1(Y268C/A+K54R) and Mpk1(Y268C/A+T190A+Y192F) still possess some residual catalytic activity that contributes to their biological activity. It is difficult to detect low levels of catalytic activity and unequivocally determine whether it is involved in inducing the biological phenotype. Based on the known effects of the K54R and the T190A+Y192F mutations on the protein’s catalytic properties, we believe that the existence of residual catalytic properties is an unlikely explanation. Further, given the destructive effect of the mutations in Tyr-280/261 of Erk1/2, it seems inconceivable that the Y268C/A mutations would enforce on Mpk1 a catalytically active conformation in which K54, T190, and Y192 are dispensable. We believe that it is likelier that Mpk1(Y268C/A+K54R) and Mpk1(Y268C/A+T190A+Y192F) are inactive as kinases or manifest at most marginal phosphotransfer activities. It should be emphasized that although Mpk1(Y268C/A+K54R) and Mpk1(Y268C/A+T190A+Y192F) molecules are biologically active, they rescue mpk1∆ and mkk1∆mkk2∆ cells less efficiently than Mpk1(Y268C/A). This implies that their catalytic activity does play a role in their biological effect. Thus the power of the mutants relies on a combination of catalytic and noncatalytic effects. Their noncatalytic activity suffices to allow certain growth rates on caffeine, and only when combined with the catalytic activity is the full effect of the mutations manifested.
Although eukaryotic protein kinases have been thoroughly studied, critical structure–function aspects of their activity and regulation are still not understood. In the case of the Erk molecules, these matters have been addressed comprehensively via classical biochemistry and genetic approaches, crystal structures, nuclear magnetic resonance, and hydrogen–deuterium exchange MS analysis (Zhang et al., 1994; Canagarajah et al., 1997; Robinson et al., 2002; Lee et al., 2004; Kinoshita et al., 2008; Peti and Page, 2013). These studies pointed to the importance of Tyr-261 in Erk2. Tyr-261, together with other residues, undergoes a significant reorientation upon activation/phosphorylation of Erk2 (Canagarajah et al., 1997; Lee et al., 2004) and functions as one of the foundations of the hydrophobic cavity that serves as the binding domain of substrates that possess a DEF motif (Robinson et al., 2002; Lee et al., 2004; Shin et al., 2010). In addition, an intact Tyr-261 is essential for MEK1 binding (Robinson et al., 2002).
Here we added new roles to this residue, not in binding of substrates and regulators, but in two other aspects of Erk’s functions. One is an important role in catalysis per se, and the other is a critical role in suppressing the noncatalytic activity of Mpk1. The latter notion is strongly supported by the finding that mutating Tyr-268 rendered Mpk1 intrinsically active, independent of intact K54, T190, and Y192. Its role in catalytic activity was disclosed by the observation that in mammalian Erk1/2, mutating Tyr-280/261 resulted in proteins with a significantly reduced phosphotransfer capability toward MBP. An intact Tyr-280/261 was further found to be critical for the MEK-independent catalytic activity of Erk1(R84S) and Erk2(R65S) in vitro and in cell lines. It is thus critical for both MEK-independent (autophosphorylation-mediated) and MEK-dependent catalysis. We believe that the effect on MBP phosphorylation observed in these mutants is a direct effect on catalytic properties and not a result of reduced binding to the substrate, because MBP does not have a DEF motif. The equivalent mutations had a dramatic effect on Mpk1 activation as well, but it was a positive effect, as reflected by the increased autophosphorylation activity of Mpk1(Y268C) and Mpk1(Y268A) in yeast cells.
A model that can consolidate all observations, at least for Mpk1, may suggest that binding of substrates or other proteins (e.g., Mek1) to Tyr-268 imposes allosteric changes in several domains: 1) the activation loop, 2) the catalytic sites (analogous to the effect of binding of substrates to the CD domain; Zhou et al., 2006), and 3) domains important for noncatalytic activities. Mutating Tyr-268 may partially mimic such allosteric effects.
Mpk1 and mammalian Erks are highly conserved, structurally and functionally (Herskowitz, 1995; Gustin et al., 1998; Posas et al., 1998; Widmann et al., 1999; van Drogen and Peter, 2001; Levin-Salomon et al., 2009). In particular, five of the residues that form the Erks’ DEF pocket, (Tyr-231, Leu-235, Met-197, Leu-198, and Tyr-261) are identical in Mpk1, and the sixth residue, Leu-232, is Val in Mpk1. Thus this domain would be expected to execute similar functions. It is therefore not clear why the equivalent mutations have opposite effects on Mpk1 and Erk1/2.
MATERIALS AND METHODS
Yeast strains and media
The Saccharomyces cerevisiae strains used in this study were mkk1Δ/mkk2Δ (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YPL140c::kanMX4; mkk1::LEU2; Levin-Salomon et al., 2008), mpk1Δ (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YHR030c::kanMX4; obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis, Frankfurt, Germany), and pbs2Δste7Δmkk1Δmkk2Δ (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YJL128c::kanMX4; ste7::LEU2; mkk1::HygromycinB; mkk2::HIS3; Levin-Salomon et al., 2009). Yeast cultures were maintained on YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose with or without 15 mM caffeine) or on the synthetic medium YNB(–URA) (0.17% yeast nitrogen base without amino acids and NH4(SO4)2, 0.5% ammonium sulfate, 2% glucose, and 40 mg/l adenine, histidine, tryptophan, lysine, leucine, and methionine).
Cell cultures and transfection procedures
NIH3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 0.044 M sodium hydrogen carbonate, streptomycin, and penicillin (Beit-Haemek, Israel). HEK293T cells were cultured in the same medium supplemented with 10% FBS, 0.017 M sodium hydrogen carbonate, 1 mm sodium pyruvate, streptomycin, and penicillin. All cells were incubated at 37ºC and 5% CO2. HEK293T cells were plated 24 h before transfection at 1.5 × 104 cells per 3-cm plate or 8 × 104 cells per 10-cm plate. Medium was changed to half the volume ∼2 h before transfection. HEK293T cells were transfected with 2 μg of cDNA (pCEFL plasmids) for 24 h using the calcium phosphate method (Kingston et al., 2003), washed with phosphate-buffered saline, and provided with fresh medium with or without 50 ng/ml EGF for 10 min. Then cells were lysed by addition of 80-μl sample buffer containing 10% glycerol, 3% SDS, 0.2 M Tris, pH 6.8, 5% β-mercaptoethanol, and phenol blue dye, followed by 10 min of boiling at 100°C. NIH3T3 cells were transfected using TransIT-X2 System (Mirus, Madison, WI) according to the manufacturer’s protocol.
Foci formation assay
Approximately 1.5 × 104 NIH3T3 cells were plated on a 3-cm plate, transfected with the relevant plasmid, and selected for the presence of the plasmid by incubation with 1 mg/ml G-418. Four weeks after transfection, cells were fixed with 100% MeOH for 20 min, stained with 4% crystal violet dye for 5 min, and photographed.
Plasmids
Plasmids carrying the mutants ERK1(R84S), ERK1(Y280C), ERK2(R65S), ERK2(Y261C), MPK1(R68S), MPK1(Y268C), MPK1(K54R), and MPK1(K54R+R68S), as well as the short MPK1(Q364), have been described (Levin-Salomon et al., 2008; Smorodinsky-Atias et al., 2016). New mutations were introduced as to be described. For mammalian expression systems, pCEFL vector (Invitrogen, Carlsbad, CA) containing ERK1(WT), ERK1(R84S), ERK2(WT), or ERK2(R65S) with ERK1 N-terminally tagged with the hexahistidine (6His) and ERK2 N-terminally tagged with the hemagglutinin (HA) sequence (YPYDVPDYA) served as the template for site-directed mutagenesis for the mutations ERK1(Y280A), ERK2(Y261A), ERK1(R84S+Y280C/A), and ERK2(R65S+Y261C/A) using the Stratagene QuikChange kit (Agilent), according to manufacturer’s instructions. All cDNAs were verified via DNA sequencing. Primers used for the mutagenesis are shown in Table 1. The pET15b (Addgene) vectors containing the Erk variants were used for protein expression and purification.
TABLE 1:
Primers sequences used in site-directed mutagenesis reactions.
| Primer name | Primer sequence |
|---|---|
| ERK1(Y280C)_F | 5′-GAGACTGTAGGCAGTTTCGGGCCTTCATG-3′ |
| ERK1(Y280C)_R | 5′-CATGAAGGCCCGAAACTGCCTACAGTCTC-3′ |
| ERK2(Y261C)_F | 5′-GCTAGAAACTGTTTGCTTTCTCTCCCGCAC-3′ |
| ERK2(Y261C)_R | 5′-GTGCGGGAGAGAAAGCAAACAGTTTCTAGC-3′ |
| ERK1(Y280A)_F | 5′-GAAGCCCCGAAACGCCCTACAGTCTCTGCCC-3′ |
| ERK1(Y280A)_R | 5′-GGGCAGAGACTGTAGGGCGTTTCGGGCCTTC-3′ |
| ERK2(Y261A)_F | 5′-GCTAGAAACGCTTTGCTTTCTCTCCCGCAC-3′ |
| ERK2(Y261A)_R | 5′-GTGCGGGAGAGAAAGCAAAGCGTTTCTAGC-3′ |
| MPK1(Y268A)_F | 5′-GGTTCTAAAAATGTTCAGGACGCCATACATCAATTAGG-3′ |
| MPK1(Y268A)_R | 5′-CCTAATTGATGAATGGCGTCCTGAACATTTTTAGAACC-3′ |
| MPK1(T190A+Y192F)_F | 5′- CAGTCAATTTTTGGCGGAGTTCGTGGCCACTAGATGG-3′ |
| MPK1(T190A+Y192F)_R | 5′-CCATCTAGTGGCCACGAACTCCGCCAAAAATTGACTG-3′ |
| MPK1(D326N)_F | 5′-CTATATGGCATGATCCAGCTAACGAACCTGTGTGTAGTG-3′ |
| MPK1(D326N)_R | 5′-CACTACACACAGGTTCGTTAGCTGGATCATGCCATATAG-3′ |
| MPK1(Y238F)_F | 5′-CTTCAAAGGAAAGGATTTCGTTAATCAATTGAATC-3′ |
| MPK1(Y238F)_R | 5′-GATTCAATTGATTAACGAAATCCTTTCCTTTGAAG-3′ |
For expression of Mpk1 and Erk2 variants in yeast, we used the pAES426 vector (Friedmann et al., 2006). pAES426-HA-MPK1(Y268A), pAES46-HA-MPK1(R68S+Y268A), pAES426-HA-MPK1(T190A+Y192F), pAES426-HA-MPK1(Y268C+D326N), pAES426-HA-MPK1(Y268C+Y238F), pAES426-HA-MPK1Q364(Y268C), and pAES426-HA-MPK1Q364(R68S) were obtained via site-directed mutagenesis using the Stratagene QuikChange kit, according to manufacturer’s instructions. Primers used for the mutagenesis are shown in Table 1. pAES426-HA-MPK1(WT) and pAES426-HA-MPK1(R68S), respectively, served as templates (Levin-Salomon et al., 2008). pAES426-HA-MPK1(R68S+Y268C) was obtained by subcloning the gene segment carrying the Y268C mutation from pAES426-HA-MPK1(Y268C) (Levin-Salomon et al., 2008) into pAES426-HA-MPK1(R68S) (Levin-Salomon et al., 2008) via restriction/ligation using the SacI and NotI restriction enzymes. pAES426-HA-MPK1(K54R+Y268C), pAES426-HA-MPK1(K54R+Y268A), pAES426-HA-MPK1(K54R+R68S+Y268C), and pAES426-HA-MPK1(K54R+R68S+Y268A) were constructed by restriction/ligation of the K54R-mutation containing region from pAES426-HA-MPK1(K54R) (Smorodinsky-Atias et al., 2016) into pAES426-HA-MPK1(Y268C), pAES426-HA-MPK1(Y268A), pAES426-HA-MPK1(R68S+Y268C), or pAES426-HA-MPK1(R68S+Y268A), respectively, using the SalI and SacI restriction enzymes. pAES426-HA-MPK1(T190A+Y192F) was obtained via site-directed mutagenesis using the Stratagene QuikChange kit as well. pAES426-HA-MPK1(WT) served as the template. Primers used for the mutagenesis are shown in Table 1. pAES426-HA-MPK1(T190A+Y192F+Y268C) and pAES426-HA-MPK1(T190A+Y192F+Y268A) were constructed by restriction/ligation of the T190A+Y192F–mutation containing region from pAES426-HA-MPK1(T190A+Y192F) into pAES426-HA-MPK1(Y268C) and pAES426-HA-MPK1(Y268A), respectively, using SalI and SacI restriction enzymes.
For expressing mammalian ERK2 in yeast, pAES426-HA-ERK2 vectors were constructed by subcloning the HA-ERK2 gene, either WT or mutant, from pCEFL-based plasmids (described here earlier; Smorodinsky-Atias et al., 2016) into pAES426 (Friedmann et al., 2006) vector via restriction/ligation using the HindIII and NotI restriction enzymes.
Spot assay of yeast cells
Yeast cultures were grown in YNB(–URA) liquid medium to mid logarithmic phase (OD600 of 0.5). Five serial 10-fold dilutions were prepared, starting at an OD600 of 0.4 (approximate concentrations 107, 106, 105, 104, and 103 cells/ml). We spotted 5 μl from each dilution at 30ºC for 6 d on YPD plates supplemented with 15 mM caffeine or plates containing YNB(–URA) medium.
Preparation of yeast cell lysates
Cell cultures (12 ml) were grown on YNB(–URA) to an OD600 of 0.4–0.6. Cultures were pelleted and resuspended in 10 ml of 20% trichloroacetic acid. Samples were pelleted again and resuspended in 200 μl of 20% trichloroacetic acid at room temperature (25°C). We added 400 mg of glass beads and vortex-mixed each sample twice for 8 min. Supernatants were transferred to new Eppendorf tubes, and glass beads were rinsed twice with 200 μl of 5% trichloroacetic acid. After centrifugation, pellets were resuspended in 100 μl of 2× Laemmli sample buffer, followed by addition of 50 μl of 1 M Tris base. Samples were vortexed for 30 s and boiled for 3 min before centrifugation. The supernatant was used.
Western blotting
We separated 100 ng of recombinant proteins, 30 μg of protein lysates prepared from mammalian cells, or 45 μg of lysates prepared from yeast cells by SDS–PAGE and subsequently transferred them to a nitrocellulose membrane. After incubation of the membrane with the appropriate antibodies (Table 2), specific proteins were visualized using an enhanced chemiluminescence detection reagent.
TABLE 2:
Antibodies used in this study.
| Antibody name | Manufacturer |
|---|---|
| Anti–phospho-Erk | Cell Signaling, Boston, MA (4377) |
| Anti-Erk | Cell Signaling (9102) |
| Anti-Mpk1 | Santa Cruz Biotechnology, Dallas, TX (SC6803) |
| Anti-polyhistidine | Sigma Aldrich (H1029) |
| Anti-actin | MP Biomedical, Santa Ana, CA (691000) |
| Anti-CDC2 | Santa Cruz Biotechnology (SC-53) |
| Anti-HA | Roche, Basel, Switzerland (11-867-423001) |
Protein expression and purification
Erk variants were expressed and purified as previously described (Levin-Salomon et al., 2008).
Paper-spotted kinase assay
To activate the recombinant Erk variants, 1 μg of a purified recombinant 6His-tagged Erk protein and 10 ng of recombinant active MEK1 (14-429; Upstate Biotechnology, Lake Placid, NY) were used. The final reaction conditions were 100 mM NaCl, 35 mM Tris-Cl, pH 8, 15 mM MgCl2, 5 mM 2-glycerolphosphate, 0.2 mM Na3VO4, 0.02 mM dithiothreitol (DTT), 1 mM ethylene glycol tetraacetic acid, and 100 μM ATP. The activation reactions proceeded for 30 min at 30°C and were terminated by placement on ice.
The kinase assays were initialized by the addition of 45 μl of reaction mixture to 5 μl of 0.04 μg/μl activated Erk enzyme. The final reaction conditions were 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 8.0, 0.1 mM benzamidine, 10 mM MgCl2, 25 mM 2-glycerolphosphate, 1 mM Na3VO4, 0.1 mM dithiothreitol, 0.5 μg/μl MBP (M1891; Sigma-Aldrich, St. Louis, MO), 0.1 mM ATP, and 0.1 μCi of [γ-32P]ATP. The kinase reactions proceeded for 15 min at 30°C and were terminated by the addition of 50 μl of 0.5 M EDTA, pH 8 (250 mM final), and placement on ice. After reaction termination, aliquots of 85 μl from each well were spotted onto 3 × 3–cm Whatman No. 3MM paper squares and briefly air-dried. Each square was rinsed four times with 10% trichloroacetic acid and 3% sodium pyrophosphate (10 ml/square) for 1.5 h (each time) and twice with 100% ethanol (4 ml/square) for 20 min each time and air-dried. The radioactivity of each square was counted using a scintillation counter running a 32P Cherenkov program. Experimental points were triplicates. In addition, Laemmli sample buffer was added to 15 μl of each reaction. All samples were boiled at 100°C for 5 min and separated on 15% SDS–PAGE. The gel was exposed to film.
Autophosphorylation assay
One microgram of a purified recombinant 6His-tagged Erk protein was incubated in Erk activating buffer lacking MEK1. The autophosphorylation reactions proceeded for 5 and 15 min at 30°C and were terminated by placement on ice and addition of Laemmli sample buffer. All samples were boiled at 100°C for 5 min and separated on 10% SDS–PAGE. The gel was exposed to film.
Capillary LC/MS-MS analysis for identification of phosphorylation sites
One microgram of protein from each autophosphorylated Erk sample was trypsin digested and desalted on disposable reversed-phase column (C18 Micro Tip Column; Harvard Apparatus, Holliston, MA). The samples were first brought to a concentration of 8 M urea and 100 mM ammonium bicarbonate. Next 45 mM DTT was added to a final concentration of 2.6 mM, and the samples were incubated for 30 min at 60°C with mild agitation. The samples were cooled to room temperature, and 150 mM iodoacetamide was added to final concentration of 8.8 mM for 30 min at room temperature in the dark. After a fourfold dilution (to 2 M urea) with water, trypsin was added at a ratio of 1:50 (trypsin to protein), and the samples were incubated overnight at 37°C. A second dose of trypsin was given after the overnight digestion, and the samples were left to continue the proteolysis for an additional 4 h at 37°C. The samples were then acidified with 2 μl of 0.1% trifluoroacetic acid (TFA) to stop the reaction. Peptide desalting was carried out by C18 Micro Tip Columns (Harvard Apparatus). The Tips were washed with 80% acetonitrile (ACN) and 0.1% TFA (elution solution), followed by 0.1% TFA (desalting solution). The samples were loaded in 0.1% TFA on the C18 microcolumns, and the flowthrough fractions were kept. The tryptic peptides were desalted with 0.1% TFA and eluted with 80% ACN and 0.1% TFA. Next the samples were completely dried by vacuum centrifugation and resuspended in 0.1% formic acid for LC-MS/MS on an OrbitrapXL (Thermo Fisher Scientific, Waltham, MA).
Acknowledgments
We thank Michal Bell for critically reading and editing the manuscript. This study was supported by the Israel Science Foundation (Center of Excellence Grants 180/09 and 1772/13 and personal grant 593/15), the Bi-National US-Israel Science Foundation (Grant 2009116), the Israel Cancer Research Fund, and the Singapore National Research Foundation under its HUJ-NUS partnership program at the Campus for Research Excellence and Technology Enterprise (CREATE).
Abbreviations used:
- Erk
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- MKK
mitogen-activated protein kinase kinase
- Mpk1
mitogen-activated protein kinase 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0167) on July 13, 2016.
REFERENCES
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- Askari N, Diskin R, Avitzour M, Capone R, Livnah O, Engelberg D. Hyperactive variants of p38alpha induce, whereas hyperactive variants of p38gamma suppress, activating protein 1-mediated transcription. J Biol Chem. 2007;282:91–99. doi: 10.1074/jbc.M608012200. [DOI] [PubMed] [Google Scholar]
- Askari N, Diskin R, Avitzour M, Yaakov G, Livnah O, Engelberg D. MAP-quest: could we produce constitutively active variants of MAP kinases. Mol Cell Endocrinol. 2006;252:231–240. doi: 10.1016/j.mce.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Avruch J, Khokhlatchev A, Kyriakis JM, Luo Z, Tzivion G, Vavvas D, Zhang XF. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bott CM, Thorneycroft SG, Marshall CJ. The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett. 1994;352:201–205. doi: 10.1016/0014-5793(94)00958-9. [DOI] [PubMed] [Google Scholar]
- Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yancopoulos GD. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- Brunner D, Oellers N, Szabad J, Biggs WH, 3rd, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MH, Goldsmith EJ. Dimerization in MAP-kinase signaling. Trends Biochem Sci. 2000;25:7–9. doi: 10.1016/s0968-0004(99)01508-x. [DOI] [PubMed] [Google Scholar]
- Dimitri CA, Dowdle W, MacKeigan JP, Blenis J, Murphy LO. Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr Biol. 2005;15:1319–1324. doi: 10.1016/j.cub.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Diskin R, Askari N, Capone R, Engelberg D, Livnah O. Active mutants of the human p38alpha mitogen-activated protein kinase. J Biol Chem. 2004;279:47040–47049. doi: 10.1074/jbc.M404595200. [DOI] [PubMed] [Google Scholar]
- Engelberg D, Perlman R, Levitzki A. Transmembrane signalling in Saccharomyces cerevisiae. Cell Signal. 1989;1:1–7. doi: 10.1016/0898-6568(89)90015-6. [DOI] [PubMed] [Google Scholar]
- Engelberg D, Perlman R, Levitzki A. Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: state of the art after 25 years. Cell Signal. 2014;26:2865–2878. doi: 10.1016/j.cellsig.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Engelberg D, Simchen G, Levitzki A. In vitro reconstitution of cdc25 regulated S. cerevisiae adenylyl cyclase and its kinetic properties. EMBO J. 1990;9:641–651. doi: 10.1002/j.1460-2075.1990.tb08156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantz DA, Jacobs D, Glossip D, Kornfeld K. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J Biol Chem. 2001;276:27256–27265. doi: 10.1074/jbc.M102512200. [DOI] [PubMed] [Google Scholar]
- Friedmann Y, Shriki A, Bennett ER, Golos S, Diskin R, Marbach I, Bengal E, Engelberg D. JX401, A p38alpha inhibitor containing a 4-benzylpiperidine motif, identified via a novel screening system in yeast. Mol Pharmacol. 2006;70:1395–1405. doi: 10.1124/mol.106.022962. [DOI] [PubMed] [Google Scholar]
- Galanis A, Yang SH, Sharrocks AD. Selective targeting of MAPKs to the ETS domain transcription factor SAP-1. J Biol Chem. 2001;276:965–973. doi: 10.1074/jbc.M007697200. [DOI] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haystead TA, Dent P, Wu J, Haystead CM, Sturgill TW. Ordered phosphorylation of p42mapk by MAP kinase kinase. FEBS Lett. 1992;306:17–22. doi: 10.1016/0014-5793(92)80828-5. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Truman AW, Levin DE. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol. 2008;28:2579–2589. doi: 10.1128/MCB.01795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Rose JK. Calcium phosphate transfection. Curr Protoc Mol Biol. 2003 doi: 10.1002/0471142727.mb0901s63. Chapter 9, Unit 9.1. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yoshida I, Nakae S, Okita K, Gouda M, Matsubara M, Yokota K, Ishiguro H, Tada T. Crystal structure of human mono-phosphorylated ERK1 at Tyr204. Biochem Biophys Res Commun. 2008;377:1123–1127. doi: 10.1016/j.bbrc.2008.10.127. [DOI] [PubMed] [Google Scholar]
- Kohno M, Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann Med. 2006;38:200–211. doi: 10.1080/07853890600551037. [DOI] [PubMed] [Google Scholar]
- Kraut J. How do enzymes work. Science. 1988;242:533–540. doi: 10.1126/science.3051385. [DOI] [PubMed] [Google Scholar]
- Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Salomon V, Kogan K, Ahn NG, Livnah O, Engelberg D. Isolation of intrinsically active (MEK-independent) variants of the ERK family of mitogen-activated protein (MAP) kinases. J Biol Chem. 2008;283:34500–34510. doi: 10.1074/jbc.M806443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Salomon V, Maayan I, Avrahami-Moyal L, Marbach I, Livnah O, Engelberg D. When expressed in yeast, mammalian mitogen-activated protein kinases lose proper regulation and become spontaneously phosphorylated. Biochem J. 2009;417:331–340. doi: 10.1042/BJ20081335. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Michailovici I, Harrington HA, Azogui HH, Yahalom-Ronen Y, Plotnikov A, Ching S, Stumpf MP, Klein OD, Seger R, Tzahor E. Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development. 2014;141:2611–2620. doi: 10.1242/dev.107078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BG, Wolfenden R. Catalytic proficiency: the unusual case of OMP decarboxylase. Annu Rev Biochem. 2002;71:847–885. doi: 10.1146/annurev.biochem.71.110601.135446. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- Robinson FL, Whitehurst AW, Raman M, Cobb MH. Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J Biol Chem. 2002;277:14844–14852. doi: 10.1074/jbc.M107776200. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Crespo P. Working without kinase activity: phosphotransfer-independent functions of extracellular signal-regulated kinases. Sci Signal. 2011;4:re3. doi: 10.1126/scisignal.2002324. [DOI] [PubMed] [Google Scholar]
- Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smorodinsky-Atias K, Goshen-Lago T, Goldberg-Carp A, Melamed D, Shir A, Mooshayef N, Beenstock J, Karamansha Y, Saadon ID, Livnah O, et al. Intrinsically active variants of Erk oncogenically transform cells and disclose unexpected autophosphorylation capability that is independent of TEY phosphorylation. Mol Biol Cell. 2016;27:1026–1039. doi: 10.1091/mbc.E15-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E. Molecular recognitions in the MAP kinase cascades. Cell Signal. 2003;15:455–462. doi: 10.1016/s0898-6568(02)00112-2. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Shaw A, Hu J, Meharena HS, Kornev A. Pseudokinases from a structural perspective. Biochem Soc Trans. 2013;41:981–986. doi: 10.1042/BST20130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Peter M. MAP kinase dynamics in yeast. Biol Cell. 2001;93:63–70. doi: 10.1016/s0248-4900(01)01123-6. [DOI] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- Yaakov G, Bell M, Hohmann S, Engelberg D. Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol Cell Biol. 2003;23:4826–4840. doi: 10.1128/MCB.23.14.4826-4840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]