Abstract
The outer membranes of Gram-negative bacteria are replete with integral membrane proteins that exhibit antiparallel β-barrel structures, but very few of these proteins function as enzymes. In Escherichia coli, only three β-barrel enzymes are known to exist in the outer membrane; these are the phospholipase OMPLA, the protease OmpT, and the phospholipid::lipid A palmitoyltransferase PagP, all of which have been characterized at the structural level. Structural details have also emerged for the outer membrane β-barrel enzyme PagL, a lipid A 3-O-deacylase from Pseudomonas aeruginosa. Lipid A can be further modified in the outer membrane by two β-barrel enzymes of unknown structure; namely, the Salmonella enterica 3′-acyloxyacyl hydrolase LpxR, and the Rhizobium leguminosarum oxidase LpxQ, which employs O2 to convert the proximal glucosamine unit of lipid A into 2-aminogluconate. Structural biology now indicates how β-barrel enzymes can function as sentinels that remain dormant when the outer membrane permeability barrier is intact. Host immune defenses and antibiotics that perturb this barrier can directly trigger β-barrel enzymes in the outer membrane. The ensuing adaptive responses occur instantaneously and rapidly outpace other signal transduction mechanisms that similarly function to restore the outer membrane permeability barrier.
Keywords: OMPLA, OmpT, PagP, PagL, LpxR, LpxQ
1. Introduction
Structural biology is now indicating how membrane-intrinsic β-barrel enzymes can function as sentinels that respond rapidly to perturbations in the outer membrane permeability barrier of Gram-negative bacteria. More generally, transmembrane signal transduction pathways control most adaptive responses toward environmental assaults on the integrity of the cell. An extracellular sensory transducer detects the environmental stimulus, which, upon binding, induces a conformational change that is communicated across the cytoplasmic membrane. The cytoplasmic components of the signal transduction pathway are thereby engaged to control gene expression and/or enzymatic activities that, ultimately, lead to an appropriate adaptive response. In the case of assaults on the integrity of the outer membrane, this general signal transduction mechanism can be troublesome when it is coupled to the expression of proteins that are themselves outer membrane components; these proteins are targeted for secretion through the cytoplasmic membrane and depend on extracytoplasmic chaperones and docking proteins in the outer membrane for their proper assembly.
Clearly, the outer membrane is first in line for external attack, but it is also last to receive any support from cellular utilities, which makes it vulnerable to potentially irreversible damage that might take effect before general signal transduction pathways can usher an adaptive response. Emerging evidence indicates that most β-barrel enzymes were selected to function in their own right as specialized sensory transducers, which are poised to respond instantaneously to environmental assaults on the bacterial outer membrane itself. General signal transduction pathways can respond gradually to looming danger by modulating the expression levels of these β-barrel enzymes. However, β-barrel enzymes can further act like air marshals at the cellular periphery by maintaining a low profile until the ultimate incursion on the outer membrane finally springs them into action. By expressing a small subset of β-barrel enzymes, the cell acquires extra protection for a membrane system at the furthest extremes of the cell’s centralized communication networks.
Here we discuss the structural biology of four β-barrel enzymes, each illustrating how their activity can be triggered to provide an appropriate adaptive response for the outer membrane in which they reside. The phospholipase OMPLA [1], the protease OmpT [2], the phospholipid::lipid A palmitoyltransferase PagP [3], and the lipid A 3-O-deacylase PagL [4] each represent β-barrel enzymes with established structure–function relationships that lend support to an emerging sentinel paradigm in outer membrane enzymology. In order to effectively illustrate their proposed roles as outer membrane sentinels, it is necessary to first discuss some basic principles of outer membrane biology.
2. Principles of outer membrane biology
2.1. Outer membrane β-barrel structure and assembly
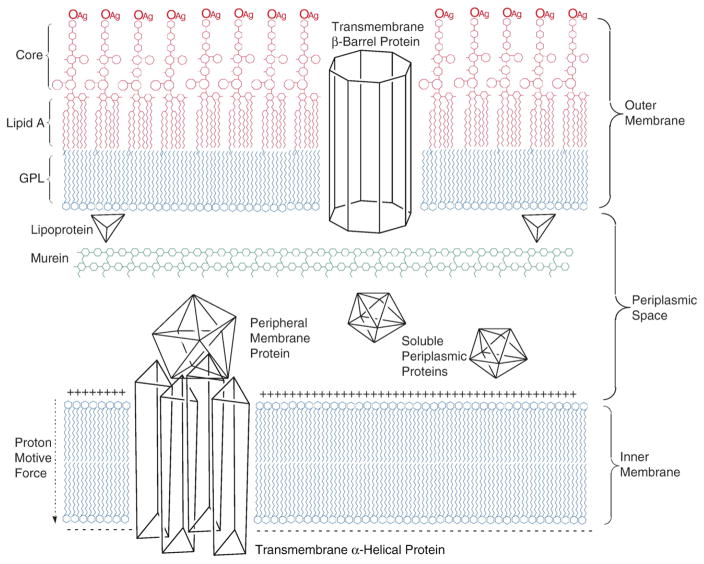
The Gram-negative cell envelope of Escherichia coli includes inner (cytoplasmic) and outer membrane systems, which are separated by the periplasmic space (Fig. 1). Whereas the cytoplasmic membrane is an energy-transducing glycerophospholipid bilayer structure that harbors transmembrane α-helical proteins, the outer membrane is a highly asymmetric bilayer with glycerophospholipids in the inner leaflet and lipopolysaccharides in the outer leaflet [5]. The lipopolysaccharides are tripartite molecules consisting of the hydrophobic and highly immunogenic anchor lipid A (endotoxin), the core oligosaccharide, which can be divided into the inner and outer core regions, and the O-antigen polysaccharide [6]. The peptidoglycan or murein provides a highly reticulated exoskeleton that determines cell shape and protects the cytoplasmic membrane from high internal osmotic pressure [7]. Bacterial lipoproteins can be anchored to the inner leaflet of the outer membrane by a lipid-modified amino terminal cysteine residue in order to provide a physical connection between the murein and the outer membrane [8–10]. The membrane-intrinsic proteins of the outer membrane are distinguished from their counterparts in the cytoplasmic membrane by an antiparallel β-barrel architecture [11,12].
Fig. 1.
Molecular organization of the Escherichia coli cell envelope. The outer membrane is an asymmetric bilayer with an inner leaflet of glycerophospholipids (GPL), and an outer leaflet of lipopolysaccharide, which can be divided into the lipid A, core oligosaccharide, and O-antigen (OAg) polysaccharide regions. The membrane-intrinsic proteins of the outer membrane are transmembrane β-barrels, while lipoproteins, anchored to the outer membrane inner leaflet, can provide a link with the murein exoskeleton. The energy-transducing inner membrane is a glycerophospholipid bilayer that supports the proton motive force and contains transmembrane α-helical proteins. The periplasmic space is the region between the inner and outer membranes and contains numerous globular proteins.
β-barrel membrane proteins are not normally found in the cytoplasmic membrane, although there is no physical reason why they could not assemble in that location. The β barrel structure is very effective at creating pores that would dissipate the proton motive force, suggesting that natural selection acts against their inner membrane localization. Indeed, several protein toxins act by forming β-barrel structures within energy-transducing membrane systems [13]. Compared to soluble globular domains, β-barrel membrane proteins appear to be inside out because they have a hydrophobic surface in contact with the lipid milieu of the membrane, and a more polar interior region. The relatively large extracellular loops and short periplasmic turns constitute the remaining hydrophilic surfaces [14]. The antiparallel β-barrel structure satisfies the requirement of providing continuous hydrogen bonding between the amide nitrogen proton and carbonyl oxygen of the peptide bonds, which are otherwise too polar to interact stably with the hydrocarbon chains in a lipid bilayer membrane [15].
Roughly 8 amino acids are minimally sufficient to span the membrane in the extended β-conformation, and half of these residues project into the polar interior region. These amphipathic β-strands can be much longer when they are significantly tilted in the membrane. Only those residues whose registration projects them into the lipid milieu of the membrane need to possess hydrophobic side-chains [11]. Consequently, β-barrel membrane proteins typically display an overall hydrophobicity that is intermediate between soluble globular proteins and transmembrane α-helical proteins. It is the outward exposure of their hydrophobic surfaces that calls for the use of detergents to keep properly folded β-barrel membrane proteins in solution, but detergents also promote folding by inducing polar groups to shield themselves internally. Many integral membrane β-barrel proteins can be purified in an unfolded state in guanidine or urea and refolded at high protein concentrations by dilution into detergent micelles, which greatly facilitates biochemical analysis [16].
β-barrel membrane proteins are normally transported through the inner membrane and, after cleavage of their amino-terminal secretory signal peptides, through the periplasmic space. A system of periplasmic chaperones and folding factors (SurA/Skp and DsbABCD) and an outer membrane docking complex (Omp85 β-barrel and YfgL/NlpB/YfiO/SmpA lipoproteins) are needed to assemble outer membrane β-barrel proteins in Gram-negative bacteria [17–23], and similar machinery appears to be needed to assemble β-barrel proteins in the outer membranes of mitochondria and plastids [24]. The extracytoplasmic accumulation of misfolded β-barrel proteins can be lethal in the absence of a key periplasmic protein DegP (HtrA), which functions both as a chaperone and a protease [25]. An extracellular function sigma factor σE directs RNA polymerase to transcribe genes like degP that are needed to respond to cell envelope stress [26]. The conserved carboxyl-terminal peptide sequences of misfolded β-barrel proteins can trigger the σE response by activating the periplasmic DegS protease, which initiates cleavage of the transmembrane protein RseA in order to release σE from inhibition at the cytoplasmic surface of the inner membrane [27]. This signal transduction pathway represents part of the so-called periplasmic stress response.
The periplasmic stress response also receives input from the CpxA/CpxR transmembrane phosphorylation cascade, which responds to misassembled subunits of a cell surface adhesive appendage known as the P-pilus [28,29]. CpxA/CpxR is representative of the two-component signal transduction pathways that depend on a membrane bound sensor kinase and cognate response regulator of transcription. The flux of phosphate between a phosphohistidine residue in the sensor kinase and a phosphoaspartate residue in the response regulator can be modulated by environmental conditions to control adaptive responses. Another example is provided by the EnvZ/OmpR transmembrane phosphorylation cascade, which is activated by osmotic stress exerted on the cell envelope, and modulates expression of a specific set of outer membrane channel proteins [30].
The majority of β-barrel membrane proteins function as channels or transporters of various substrates and usually possess large open pores within their interiors. These include non-selective channels known as porins, substrate-selective channels, and active transporters that derive energy from the inner membrane protein TonB [31]; the latter pores are plugged by residues contributed from an amino terminal domain that forms part of the ligand-binding site. Channels and transporters are all relatively large in possessing between 12 and 22 β-strands in order to create the space needed for transbilayer passage of substrates. However, several smaller outer membrane proteins possessing between 8 and 12 β-strands tend to have their interiors occluded by a network of mostly polar amino acid side-chains and solvent molecules. These proteins behave more like plugs in the outer membrane and tend to fulfill functions unrelated to transport, but including enzymatic functions, cell adhesion, and the binding of ligands such as detergents [32–35]. An intriguing exception is the 8-stranded β-barrel protein OmpW, which is implicated in the transport of small hydrophobic molecules [36].
All solved structures of β-barrel membrane proteins so far reveal an even number of antiparallel β-strands with the amino and carboxyl termini placed on the periplasmic side of the outer membrane. A notable exception that does not really violate this topological rule is seen in the autotransporter proteins, which display their amino terminal domains on the cell surface after threading them through the carboxyl terminal domains that form the β-barrel proper [37]. Another generally observed feature of β-barrel membrane proteins is the presence of aromatic belts composed of Trp, Tyr and Phe residues that demarcate the interfacial boundaries between the hydrophobic and aqueous domains on both sides of the outer membrane [11].
Outer membrane β-barrel proteins are most often formed from single polypeptide chains, which can undergo various levels of oligomerization, or the pore itself can sometimes be formed in segments supplied by multiple subunits. For example, many porins are 16-stranded β-barrels that assemble into trimers of three pore-forming subunits [38], whereas the TolC channel spans the outer membrane as a trimeric 12-stranded β-barrel in which each subunit contributes 4 β-strands to a single pore [39]. The fact that some 12-stranded β-barrels can form pores while others form plugs is a reflection of the role of β-strand tilt or shear angle in determining pore size [40,41]. Topological arguments indicate that the 8-stranded antiparallel β-barrel is likely the smallest design to be capable of spanning the outer membrane [42].
Nature’s other solution to provide continuous polypeptide hydrogen bonding is the transmembrane α-helix, which demands ~20 sequentially hydrophobic amino acid side-chains in order to span the membrane [15]. The greater hydrophobicity of transmembrane α-helical proteins dictates that their targeting to the inner membrane is essentially irreversible, a fact that explains why transmembrane α-helical proteins are not found in the outer membrane. One near-exception to this rule is seen in the structure of the Wza protein involved in the secretion of extracellular polysaccharides. Wza is an octamer that spans the outer membrane by assembling a single amphipathic α-helix in a segmental fashion to form an eight-fold symmetric pore structure [43]. Cationic amphipathic α-helices that function as antimicrobial peptides might form transient pores of similar structure as they cross the outer membrane en route to their target — the energy transducing inner membrane (see below). To date, no typically hydrophobic transmembrane α-helices have been described that remain stably integrated in the outer membrane.
2.2. Lipid trafficking to the outer membrane
The peptidoglycan or murein exoskeleton is sandwiched between the inner and outer membranes of Gram-negative bacteria and is believed to effectively block wholesale membrane exchange by vesicular trafficking [7]. Consequently, outer membrane biogenesis likely depends on soluble periplasmic lipid-transfer proteins and/or localized contact bridges between inner and outer membranes [19]. A well-characterized example is provided by the bacterial lipoproteins, which can serve to link the outer membrane with the peptidoglycan wall and are secreted with a distinctly lipidated amino terminus [8]. Nearly 100 lipoproteins are encoded in the E. coli genome and serve a multitude of functions in the biogenesis of the cell envelope. Roughly 90% of lipoproteins have their lipid anchors targeted to the inner leaflet of the outer membrane. An ATP-binding cassette transporter (LolCDE) disengages from the inner membrane external leaflet those lipoproteins lacking an inner membrane retention signal and delivers them into the jaws of a periplasmic chaperone (LolA). The LolA–lipoprotein complex is carried through the periplasmic space to an outer membrane docking protein (LolB), which itself is a lipoprotein and facilitates release of LolA concomitantly with integration of the lipoprotein payload into the outer membrane inner leaflet.
In contrast to lipoproteins, the precise mechanism by which other lipids are transported to the outer membrane is poorly understood [44]. Glycerophospholipids are known to freely exchange between the inner and outer membranes while the lipopolysaccharides are exported in a unidirectional manner [45]. The enzymes of the Kennedy pathway for glycerophospholipid biosynthesis require cytoplasmic cofactors and act on the cytoplasmic leaflet of the inner membrane [46]. Although various integral membrane proteins are reported to promote the rapid flip-flop of glycerophospholipids between the cytoplasmic and periplasmic leaflets of the inner membrane [47,48], the precise mechanism for reversibly transporting them between the inner and outer membranes remains a longstanding mystery. The ATP-binding cassette transporter MsbA is responsible for translocating the lipid A-core to the periplasmic surface of the inner membrane (see below), but its activity is also a required first step for the intermembrane exchange of glycerophospholipids in E. coli [49,50]. In Neisseria meningitidis, however, MsbA translocates lipid A-core independently of glycerophospholipid transport [51]. MsbA might be a dedicated flippase for lipid A-core that only secondarily affects glycerophospholipid transport in E. coli.
The lipopolysaccharide structure is assembled within three distinct subcellular compartments; namely, the cytoplasmic membrane, the periplasmic space, and the outer membrane. Like the Kennedy pathway enzymes, the primary lipopolysaccharide biosynthetic reactions require energy-rich precursors and are catalyzed by enzymes that reside within the cytoplasm or at the cytoplasmic surface of the inner membrane [6]. The Raetz pathway for lipid A biosynthesis was worked out primarily in E. coli and includes nine Lpx enzymes. The E. coli lipid A molecule is a β-1′,6-linked disaccharide of glucosamine, which is phosphorylated at positions 1 and 4′, and acylated with R-3-hydroxymyristate in ester linkage at positions 3 and 3′ and in amide linkage at positions 2 and 2′; the molecule is further acylated with lauroyl and myristoyl groups attached in acyloxyacyl linkage on the distal glucosamine unit (Fig. 2). The waa (rfa) operons encode the glycosyl transferase enzymes needed for step-wise assembly on lipid A of the core oligosaccharide, for which five distinct types can occur in E. coli [52]. Diverse operons control assembly of the various O-antigen polysaccharides. Although ~170 distinct O-antigen serotypes have been identified in E. coli alone, they are all assembled on the lipid carrier analog of dolichol known as bactoprenol [6]. The so-called rough lipopolysaccharide, or lipooligosaccharide, includes only the lipid A-core and is usually distinguished from the smooth lipopolysaccharide that additionally includes the polymerized O-antigen.
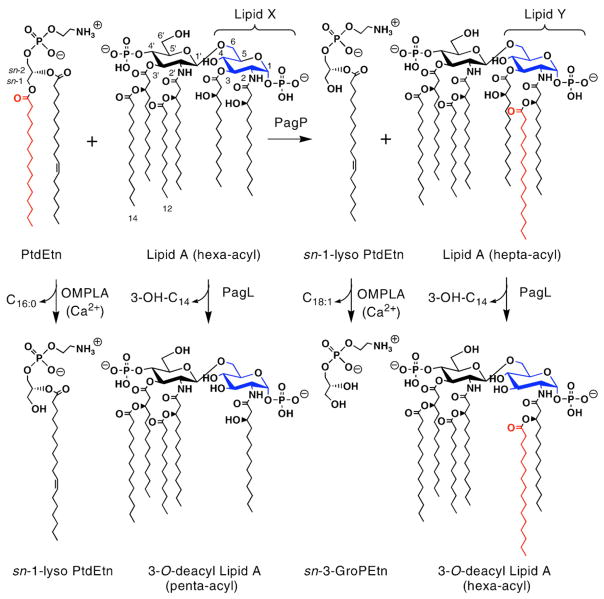
Fig. 2.
Outer membrane transformations of glycerophospholipids and lipid A. PagP catalyzes transfer of a palmitate chain (C16:0) from the sn-1 position of phosphatidylethanolamine (PtdEtn) to lipid A (endotoxin). Escherichia coli lipid A is a β-1′,6-linked disaccharide of glucosamine that is acylated with R-3-hydroxymyristate chains at the 2, 3, 2′, and 3′ positions, and phosphorylated at the 1 and 4′ positions. Acyloxyacyl linkages with laurate and myristate chains at the 2′ and 3′-positions, respectively, provide the constitutive hexa-acylated lipid A, which is a potent endotoxin. A regulated proportion of lipid A in E. coli contains a palmitate chain in acyloxyacyl linkage at position 2, which yields a hepta-acylated molecule that both is attenuated as an endotoxin and provides resistance to cationic antimicrobial peptides. PagL makes similar contributions by removing the R-3-hydroxymyristate chain at position 3 from either hexa-acyl or hepta-acyl lipid A. PagP can acylate the cytoplasmic monosaccharide lipid A precursor lipid X to produce lipid Y in vitro, and PagL can similarly deacylate lipids X and Y in vitro. The sn-1-lysophospholipid byproduct of the PagP reaction can be generated directly by the phospholipase activity of OMPLA when Ca2+ ions are available. OMPLA also displays lysophospholipase activity in vitro, which can release the sn-3-glycerophosphoethanolamine (sn-3-GroPEtn) polar head group from the remaining unsaturated fatty acyl chain (C18:1).
Translocation of the lipid A-core and bactoprenol phosphate-O-antigen to the periplasmic surface of the inner membrane can be followed by polymerization of the O-antigen polysaccharide with its subsequent en-bloc ligation to the outer core. Although MsbA translocates lipid A-core, at least three distinct mechanisms for bactoprenol phosphate-O-antigen translocation have been reported [6]. At this periplasmic stage, several regulated partial modifications can occur on both lipid A and the inner core sugars [53]. Unlike the constitutive and cytoplasmic lipid A-core biosynthetic enzymes, which are widely conserved among Gram-negative bacteria, the regulated modification enzymes tend to be localized to the extracytoplasmic compartments and are distributed among particular groups of organisms. Modification enzymes located at the periplasmic face of the inner membrane largely control the addition or removal of polar groups, whereas those located in the outer membrane tend to control the addition or removal of fatty acyl-chains.
The recently discovered LptA/LptB proteins are implicated in transporting lipopolysaccharide across the periplasmic space for delivery to the inner leaflet of the outer membrane [54]. It is still undecided whether lipopolysaccharide transport is mediated by a soluble carrier or by localized contact bridges between the inner and outer membranes [19,55]. The lipopolysaccharide in the outer membrane next encounters the Imp β-barrel/RlpB lipoprotein complex, which is needed to display the lipopolysaccharide on the bacterial cell surface [56,57]. The E. coli outer membrane is an asymmetric lipid bilayer in which lipopolysaccharide exclusively lines the external leaflet while glycerophospholipids line the inner leaflet [5,31]. The asymmetric lipid organization provides a permeability barrier to hydrophobic antibiotics and detergents encountered in the natural and host environments. Divalent cations, especially Mg2+ and, to a lesser extent, Ca2+, are needed to maintain outer membrane lipid asymmetry by neutralizing negative charge repulsions that would otherwise occur between neighboring lipopolysaccharide molecules in the outer leaflet [58,59].
2.3. The outer membrane permeability barrier
The bridging of negatively charged groups by divalent cations between neighboring lipopolysaccharide molecules represents a double-edged sword for Gram-negative bacteria. Although the glycerophospholipid bilayer of the cytoplasmic membrane is intrinsically permeable to hydrophobic antibiotics and detergents, these same compounds are repelled on encountering Mg2+ ions bridged between the negatively charged inner core and lipid A sugars [5]. In the gastrointestinal tracts of mammals, enteric Gram-negative bacteria like E. coli and Salmonella encounter bile salts whose detergent action prevents their growth when the outer membrane structure is compromised [60]. While the majority of β-barrel proteins in the outer membrane allow exchange of essential nutrients and waste products, the asymmetric lipid organization helps to protect the cells from bile salts [31]. The outer membranes of these Gram-negative bacteria represent a major store for the divalent cations that are needed to maintain the permeability barrier.
This dependence on divalent cations can also be an Achilles’ heel for the bacterium. Under Mg2+-limited growth conditions, negatively charged groups begin to repel the neighboring lipopolysaccharide molecules and create space for glycerophospholipids that migrate from the outer membrane inner leaflet. Rafts of glycerophospholipid bilayers now mix among the lipopolysaccharides in the external leaflet. These locally symmetric bilayer rafts are freely permeable to hydrophobic antibiotics and detergents, which can now gain access to the periplasm while the outer membrane continues to retain the more polar periplasmic contents [5]. This model is based largely on permeability studies of E. coli cells treated briefly with EDTA [61], and likely explains the increased membrane permeability of imp mutants [57]. Some details might not apply in Pseudomonas aeruginosa, which is highly sensitive to EDTA [62], or in Neisseria species, where lipooligosaccharide is non-essential under certain conditions [63] and normal outer membrane lipid organization is not strictly asymmetric to begin with [64], but cell surface net negative charge appears to be a general property of Gram-negative outer membranes.
Nature exploits the outer membrane’s dependence on divalent cations, as is evidenced in the design principles for cationic antimicrobial peptides. Although capable of assuming an impressive diversity of structures, two common features are apparent in all antimicrobial peptides. Firstly, antimicrobial peptides take on a net positive charge and, secondly, they are capable of assuming an amphipathic structure that segregates polar and non-polar amino acid residues to opposite faces of the molecule [65,66]. In aqueous solution, antimicrobial peptides tend to remain unstructured, but their net positive charge electrostatically attracts them toward the negatively charged groups on the bacterial cell surface. As the dielectric constant of the medium gradually decreases, water molecules are stripped from the peptide bonds, which drives formation of hydrogen bonded secondary structures. At this stage, positively charged groups in the peptide likely displace some divalent cations from the outer membrane surface. The non-polar face of the induced amphipathic structure can now interact favorably with the hydrocarbon chains of the lipid bilayer, which facilitates the transit of the antimicrobial peptide into the periplasmic space. This model is aptly named the self-promoted uptake pathway [67].
The antimicrobial peptides are thought to kill bacteria primarily by disrupting the proton motive force in the cytoplasmic membrane, but it has recently emerged that some antimicrobial peptides likely target key cytoplasmic proteins as well [68]. In either case, antimicrobial peptides probably approach the cytoplasmic membrane in much the same manner as they approach the outer membrane because anionic glycerophospholipids are displayed on the external surface of bacterial cytoplasmic membranes. By contrast, eukaryotic cells actively sequester their anionic glycerophospholipids on the cytoplasmic surface of the plasma membrane. This lipid asymmetry, combined with the hydrophobic and van der Waals forces uniquely associated with the presence of cholesterol in eukaryotic membranes, has probably allowed the antimicrobial peptides to acquire selective toxicity towards bacteria [69]. In the host, antimicrobial peptides are not always benign as some can modulate the immune response [70].
Faced with cationic antimicrobial peptides in nature, it is no surprise that bacteria have acquired defensive measures to resist the threat [71]. In Gram-negative bacteria, the PhoP/PhoQ signal transduction pathway controls many important antimicrobial peptide resistance mechanisms [72]. The PhoQ protein is a transmembrane sensory transducer, which directs a patch of acidic amino acid residues in its periplasmic domain toward the anionic glycerophospholipids located at the external surface of the cytoplasmic membrane [73]. Mg2+ ions are thought to bridge the anionic lipid polar head groups with the acidic patch on the sensory transducer to maintain it in a repressed state [74]. Cationic antimicrobial peptides likely displace the bound Mg2+ ions and thus trigger a phosphorylation cascade that terminates in the activation of the PhoP transcription factor [75]. Mg2+ limitation similarly triggers the PhoP/PhoQ system in the absence of any challenge from antimicrobial peptides [76]. In Salmonella, PhoP/PhoQ activation triggers the PmrA/PmrB phosphorylation cascade via a posttranslational mechanism using an effector known as PmrD [77], but the pmrD gene is nonfunctional in E. coli [78]. PmrA/PmrB can also respond directly, as can PhoP/PhoQ [79], to mildly acidic growth conditions encountered in macrophage phagolysosomes [80], an environment that is already replete in both Mg2+ ions and antimicrobial peptides [81].
Among the consequences of PhoP/PhoQ activation is the expression of several lipid A modification enzymes. One PhoP/PhoQ-activated gene encodes the outer membrane β-barrel enzyme PagP, which contributes directly and indirectly to antimicrobial peptide resistance [82]. PagP introduces a palmitate chain into the lipid A molecule, which, by altering the hydrophobic and van der Waals forces in the outer membrane, is thought to block interactions with the non-polar faces of amphipathic antimicrobial peptides [83]. PagP also reduces the endotoxicity of lipid A, which otherwise contributes to the production of antimicrobial peptides by triggering the key host innate immune receptor TLR4 [82,84]. Another PhoP/PhoQ-activated gene encodes the outer membrane β-barrel enzyme PagL, which similarly contributes directly and indirectly to antimicrobial peptide resistance. PagL removes the R-3-hydroxymyristate chain at position 3 in lipid A [85], which can provide resistance to the antimicrobial peptide polymyxin B under certain conditions [86], and it also reduces lipid A signaling mediated by TLR4 [84].
PmrA/PmrB-dependent enzymes include those that incorporate phosphoethanolamine, derived from phosphatidylethanolamine [87,88], and 4-amino-4-deoxy-L-arabinose (L-Ara4N), derived from bactoprenol phosphate-L-Ara4N [89,90], into the lipid A phosphate groups presented at the periplasmic surface of the inner membrane. By incorporating positively charged groups into lipid A, it is thought that these modifications block the initial cell surface electrostatic interactions with antimicrobial peptides such as polymyxin B. Other strategies to alter cell surface charge include phosphate group removal [91,92], which can simultaneously provide polymyxin B resistance and attenuated lipid A endotoxicity [93,94], and the addition of a bactoprenol phosphate-activated galactosamine sugar [95]. Although the enzymes for these lipid A modifications might be subjected to different patterns of regulation, they similarly act at the periplasmic surface of the inner membrane [96]. Lipid A modification systems clearly endow bacterial cells with a coordinated response to antimicrobial peptides that not only blocks key steps in the self-promoted uptake pathway, but also attenuates antimicrobial peptide production by the host.
3. Sentinels of the bacterial outer membrane
Given the elaborate periplasmic stress response and PhoP/PhoQ signal transduction mechanisms, it would appear that the bacterial cell is well equipped to respond to environmental attacks on the integrity of its cell envelope. However, the structural biology of outer membrane β-barrel enzymes now suggests that a more rapid response can be achieved by directly triggering enzymatic activity in response to outer membrane lipid reorganization. In this regard, the signal transduction pathways can be seen to modulate the expression levels of effectors that are themselves designed to provide a complementary, but instantaneous, signal-response coupling mechanism.
3.1. The phospholipase OMPLA
The first outer membrane β-barrel enzyme to be characterized was the Ca2+-dependent phospholipase OMPLA. The activity was uncovered in the late 1960’s and its purification and association with the outer membrane fraction of E. coli was first reported in 1971 [97,98]. The association of a phospholipase with membranes was highly enigmatic because research at the time had focused its attention on snake venom phospholipases, which were known to be soluble toxins that attached at membrane surfaces to catalyze their dissolution. What business did the cell have in targeting a phospholipase to its own membrane systems? It should be remembered that this period coincided with the introduction of the fluid mosaic model of phospholipid bilayer membranes [99] and predated any knowledge of the correct structure for lipid A or the asymmetric lipid organization of the outer membrane [100–102].
Awareness of the presence of OMPLA in the outer membrane provided plausible explanations for a number of phenomena that had been uncovered during fledgling studies of Gram-negative cell envelope biogenesis. Several E. coli mutants with defective outer membrane permeability barriers were found to exhibit elevated OMPLA activity [103,104]. Additionally, the secretion of colicin antibiotic proteins across the outer membrane was shown to depend on small bacteriolytic lipoproteins that were able to trigger OMPLA activity [105]. Colicins are encoded by extrachromosomal plasmid DNA molecules, which also encode a colicin immunity determinant [106]. Any cells in the population that jettison the colicin plasmid will likely be killed by the secreted colicin, which provides a kind of plasmid maintenance mechanism. Other exogenous assaults on the integrity of the outer membrane such as transfection of phage DNA [107], phage-induced lysis [108], spheroplast formation [109], heat shock [110], EDTA-treatment [111], and the action of antimicrobial peptides/proteins [112,113] have all been identified to trigger OMPLA.
It is difficult at first to envision how phospholipase activity provides a selective advantage towards most of the documented OMPLA triggers. As noted by Dekker, the constitutive expression of the enzyme suggested more of a housekeeping function [114]. The elucidation of the OMPLA crystal structure in 1999 served to localize the active site to the external leaflet of the outer membrane and indicated how activity could be modulated by a reversible dimerization mechanism [1]. When outer membrane lipid asymmetry is intact, OMPLA is proposed to assume an inactive monomeric structure. Events that promote the migration of glycerophospholipids into the external leaflet induce OMPLA dimerization, which leads to the formation of two active sites at the subunit interface where Ca2+ ions needed for catalysis are able to bind [115]. Perturbations of outer membrane lipid asymmetry that occur in the presence of divalent cations are most likely to trigger OMPLA, which would provide an appropriate adaptive response – restoration of the permeability barrier – by removing phospholipids from the external leaflet [114]. Although the fatty acid and lysophospholipid products have detergent-like properties that would be expected to destabilize the outer membrane, mechanisms for the active uptake and recycling of these compounds have been described [116–120].
The crystal structure of E. coli OMPLA shows how it hydrolyzes glycerophospholipids in the outer membrane (Fig. 3A). OMPLA consists of 12 antiparallel β-strands that fold into a transmembrane β-barrel. An intricate hydrogen-bonding network occludes the central cavity of OMPLA. The three active site residues, Asn156, His142, and Ser144, are organized on the exterior of the β-barrel in a catalytic triad that normally associates with the inert glucosamine backbone of lipid A in the outer leaflet [121,122]. The nucleophilic Ser144 of OMPLA can be irreversibly sulfonylated by the palmitate analog hexadecanesulfonyl fluoride. The crystal structure of sulfonylated OMPLA shows it to be a homodimer with Ca2+ contributing to the oxyanion holes that stabilize the tetrahedral intermediate during acylenzyme ester formation and hydrolysis [1,123–125]. Molecular dynamics investigations also suggest that increased stability around the active site in dimeric OMPLA is a consequence of the local ordering of water near the bound Ca2+ ions [126].
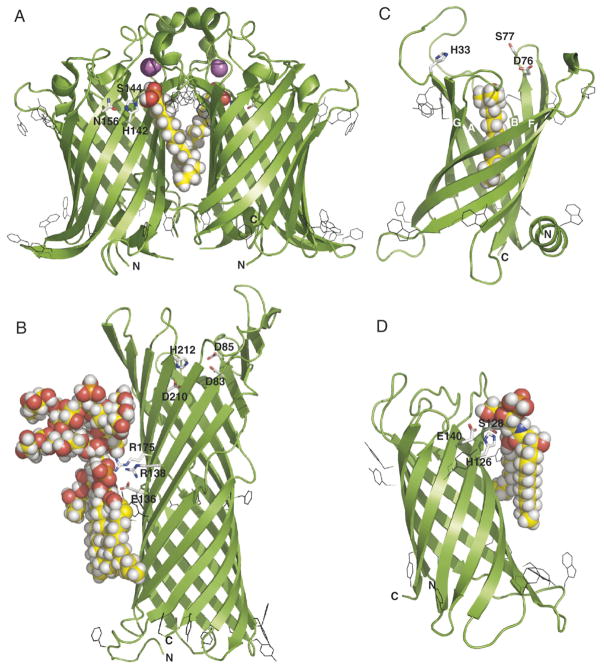
Fig. 3.
Structures of outer membrane β-barrel enzymes. (A) The outer membrane phospholipase OMPLA from E. coli with bound Ca2+ (purple) and hexadecanesulfonyl group (pdb: 1QD6). (B) The outer membrane protease OmpT from E. coli (pdb: 1I78); coordinates with bound lipooligosaccharide provided by Piet Gros and Lucy Rutten (Utrecht University). (C) The phospholipid::lipid A palmitoyltransferase PagP from E. coli with bound lauroyldimethylamine-N-oxide (pdb: 1THQ); coordinates with L1 loop introduced between β-strands A and B provided by Chris Neale and Régis Pomès (University of Toronto). (D) The outer membrane lipid A 3-O-deacylase from Pseudomonas aeruginosa (pdb 2ERV); coordinates with bound lipid X provided by Jan Tommassen and Lucy Rutten (Utrecht University).
OMPLA first selects a fatty acyl-chain at the sn-1 position of its glycerophospholipid substrate [127], but it can also hydrolyze the sn-2 acyl chain and will continue to hydrolyze acyl-chains from the lysophospholipid byproducts in vitro (Fig. 2). Snake venom phospholipases typically display positional selectivity in the glycerol backbone of their phospholipid substrates, but they are hard-pressed to reach into the bilayer and select particular fatty acyl-chain types. OMPLA is largely unspecific for phospholipid polar head groups, and recent thermodynamic studies of OMPLA dimerization have revealed that the formation of acyl-chain binding pockets at the subunit interface endows the enzyme with the ability to select fatty acyl-chains of 14 carbons or more in length [128,129].
The presence of OMPLA in Neisseria species is problematic because the outer membranes of these facultative intracellular pathogens exhibit considerable permeability toward hydrophobic compounds and likely display phospholipids on the cell surface [64]. Recent studies show that Neisseria OMPLA can induce bacteriolysis after prolonged growth. By releasing the cellular contents of a moribund subpopulation, it is proposed that this behavior altruistically sustains the remaining healthy cells in the population [130]. Although OMPLA activity is evident in most strains that cause gonorrhoeae and meningitis, some of the latter strains could be shown to contain premature stop codons in their OMPLA-encoding pldA genes. OMPLA activity is known to be associated with changes in cell surface structure that promote host–pathogen interactions in Campylobacter and Helicobacter infections [131–134], but its inactivation might also confer a selective advantage under certain pathogenic conditions in N. meningitidis.
3.2. The protease OmpT
An outer membrane protein of E. coli that was initially identified in polyacrylamide gels as protein a or 3B was later renamed OmpT because its expression was significantly reduced during growth at low temperature [135]. Proteolytic activity was first attributed to OmpT when outer membrane cleavage of the ferric enterobactin receptor protein was demonstrated in 1979 [136]. OmpT was subsequently recognized to be a nuisance for the purification of many recombinant proteins expressed in E. coli [137]. First purified in 1988, a striking feature of OmpT is its specificity for cleavage between paired basic amino acid residues [138]. The early identification of OmpT-deficient strains, such as E. coli BL21, provided a host strain to limit proteolysis during the expression and purification of recombinant proteins that is widely used today [137]. In most E. coli strains, the ompT gene is encoded by a cryptic prophage [139], but a homolog, ompP, can also be encoded by a conjugal F-plasmid that is sometimes present [140–142].
OmpT expression in E. coli can be regulated by the EnvZ/OmpR transmembrane phosphorylation cascade [135,143], which acts through the expression of two small RNA molecules OmrA/OmrB [144]. Control of OmpT is also exerted by PhoP/PhoQ, which, in E. coli, receives upstream signals from another transmembrane phosphorylation cascade EvgA/EvgS [145]. When rationalizing the teleonomic significance of regulatory signals acting on a gene, one should probably focus on the selective advantages conferred to the genetic element that encodes the gene [146]. In the case of a cryptic prophage, it is unclear if selection acted previously in favor of the phage or more recently in favor of the cell. Fortunately, a bona fide chromosomal gene encodes a Salmonella homolog of OmpT, known as PgtE, which is similarly governed by PhoP/PhoQ [147]. A logical function for PgtE, therefore, might be to confer antimicrobial peptide resistance on Salmonella. Indeed, the proteolytic specificity in OmpT homologs for paired basic residues appears to be ideally suited for the recognition of cationic antimicrobial peptides. OmpT provides resistance against protamine [148], a polycationic antimicrobial peptide known to disorganize outer membrane lipids, and PgtE is required for resistance to certain α-helical cationic antimicrobial peptides [147].
The crystal structure of E. coli OmpT shows how it degrades antimicrobial peptides presented at the bacterial cell surface (Fig. 3B). The enzyme is a 10-stranded antiparallel β-barrel that extends well above the external surface of the bilayer [2]. The active site is displayed within the external crevice formed by the OmpT surface-exposed loops. The catalytic groups include two pairings of Asp85 and Asp83 on one face, and His 212 and Asp210 on the other face. Although the X-ray structure was solved at insufficient resolution to visualize water molecules, arguments based on residue geometry and the enzyme’s pH optimum (pKa =6.2) suggested that a water molecule bridging His212 and Asp83 is delivered directly to the scissile peptide bond in the antimicrobial peptide substrate, and this model has since been substantiated by molecular dynamics simulations [149].
The OmpT active site organization appears to be stabilized by interactions on the β-barrel exterior with lipopolysaccharide, which is a required cofactor [150–152]. A motif of charged amino acids on the β-barrel exterior includes Arg175, Arg138, and Glu136, which are implicated in providing electrostatic binding interactions with charged groups in lipid A and the inner core [2,153]. A molecule of lipid A-core from its complex with the FhuA ferrichrome receptor [154] has been modeled onto the exterior of OmpT (Fig. 3B). This lipooligosaccharide molecule reaches toward, but not beyond, the active site cleft. The presence of the O-antigen polysaccharide in wild-type bacteria is expected to extend far beyond the surface of the OmpT active site and likely shields most proteins from OmpT-mediated digestion [155]. Those cationic peptides that interact with negatively charged groups in lipopolysaccharide are expected to penetrate through the O-antigen layer and suffer proteolytic degradation once they encounter OmpT [147]. The OmpT protein thereby captures only those cationic antimicrobial peptides that reveal its activity in the process of self-promoted uptake.
Shigella flexneri is a close relative of E. coli that has acquired additional genetic determinants to enable actin-based intracellular motility during shigellosis. Part of this machinery includes an autotransporter protein VirG (IcsA), which displays a cell surface domain needed for actin polymerization at one bacterial pole. OmpT efficiently cleaves the VirG extracellular domain and thus limits intracellular motility [139]. It appears that ompT gene absence was a prerequisite in the acquisition of Shigella virulence from its E. coli ancestor [156]. Nevertheless, a Shigella virulence plasmid encodes another OmpT homolog, SopA (IcsP), which assists in the polar localization of the VirG protein by selectively removing it from other regions of the cell surface [157,158]. In Yersina pestis, the O-antigen-deficient causative agent of bubonic plague, a plasmid-encoded OmpT homolog, Pla, proteolytically activates the human proenzyme plasminogen and inactivates the antiprotease α2-antiplasmin in order to enhance bacterial migration through tissue barriers [159,160]. Although these different OmpT homologs clearly fulfill distinct physiological functions, their active site architectures are conserved; the so-called omptins are now widely recognized as a distinct proteolytic enzyme family [161].
3.3. The phospholipid::lipid A palmitoyltransferase PagP
PagP is only the third enzyme shown to be an integral outer membrane protein in E. coli [162]. PagP transfers a palmitate chain from the sn-1 position of a phospholipid to the hydroxyl group on the R-3-hydroxymyristate chain at position 2 of lipid A in the outer membrane (Fig. 2). The enzymology of palmitate addition to lipid A can be traced to the discovery of cytoplasmic monosaccharide lipid A precursors in 1983. Lipid X was shown to be a diacylglucosamine 1-phosphate bearing R-3-hydroxymyristoyl groups at positions 2 and 3, and lipid Y only differed from lipid X by the presence of a palmitoyl group in acyloxyacyl linkage at position 2 [163,164]. The discovery of a membrane-bound enzyme from E. coli that could generate lipid Y by transferring a palmitoyl group from the sn-1 position of a glycerophospholipid to lipid X was first reported in 1987 [165]. In 1997, several regulated covalent modifications of lipid A, including palmitoylation, were shown to be controlled by PhoP/PhoQ [166]. The gene pagP was then identified in a Salmonella mutant that exhibited both a deficiency in lipid A palmitoylation and a hypersensitivity to vertebrate cationic antimicrobial peptides [83]. A homolog in E. coli, known as crcA, had been identified previously among a cluster of genes involved in chromosome condensation and resistance to camphor vapors [167]. In the year 2000, the E. coli PagP (CrcA) protein was purified and established to be an outer membrane glycerophospholipid::lipid A palmitoyltransferase [162], thus demonstrating that the lipid A moiety of fully assembled lipopolysaccharide is the correct physiological substrate.
The structure and dynamics of E. coli PagP have been determined by both X-ray crystallography and nuclear magnetic resonance spectroscopy [3,168]. PagP is an 8-stranded antiparallel β-barrel with a short α-helix at its N-terminus and it sits in the membrane with the barrel axis tilted by roughly 25 degrees (Fig. 3C) [169]. The PagP palmitate-recognition pocket, known as the hydrocarbon ruler, resides within the interior of the β-barrel and is localized in the outer lipopolysaccharide-exposed region of the protein. A single molecule of the detergent lauroyl-dimethylamine-N-oxide serves to identify the position of the hydrocarbon ruler within the β-barrel interior. PagP is remarkable among lipid acyltransferases in its ability to select a single 16-carbon acyl-chain in its phospholipid donor. A cluster of buried aromatic amino acid side-chains that line the floor of the hydrocarbon ruler can undergo a sensitive exciton interaction, which has been exploited to probe PagP’s internal structure by circular dichroism spectroscopy [170]. Mutation and chemical modification at residue 88 can modulate acyl-chain selection with methylene unit precision provided that the exciton is not perturbed.
Two discontinuities in β-strand hydrogen bonding within the lipopolysaccharide-exposed region provide obvious routes for lateral access of lipids into the hydrocarbon ruler. The presence of conserved proline residues at key positions between β-strands A–B and F–G prevents the local formation of interstrand hydrogen bonds. Acting like crenellations in the turret of a medieval castle, lipids in the outer membrane external leaflet probably migrate laterally through these openings [3]. Although His33, Asp76, and Ser77 have been implicated in catalysis, no evidence supports an acylenzyme mechanism [168]. Catalysis most likely proceeds when both the lipid A and phospholipid substrates engage in a ternary complex with the enzyme [82]. PagP exists in two dynamically distinct states, but the solved structure represents an R-state that is inhibited by the bound detergent [171]. The catalytically competent T-state is largely determined by an ordering of residues in and around the L1 loop that connects β-strands A–B. Given that PagP displays slow phospholipase in addition to rapid lipid A palmitoyltransferase activities in vitro [172], and that L1 loop mutations can specifically block the lipid A palmitoyltransferase activity without affecting the phospholipase activity [82], it is likely that lipopolysaccharide approaches the hydrocarbon ruler between β-strands A–B; by default, the phospholipid approach is expected between β-strands F–G.
The implication that PagP depends on the aberrant migration of phospholipids into the external leaflet creates an important topological problem for the enzyme. PagP activity is insensitive to divalent cations and, conceivably, provides a specific response to those perturbations in outer membrane lipid asymmetry induced by Mg2+ limitation [162,165]. Under these conditions, transacylation to lipid A might provide a superior adaptive response compared with phospholipid hydrolysis, which is otherwise catalyzed exclusively by OMPLA, albeit inefficiently, in response to brief EDTA treatment [111]. By contrast, brief treatment of E. coli cells with EDTA induces lipid A palmitoylation instantaneously through a process that far outpaces induction of pagP gene expression, and occurs independently of de novo protein synthesis [173]. Lipid A palmitoylation induced by EDTA in vivo requires functional MsbA, which probably replenishes phospholipids in the outer membrane inner leaflet after they translocate into the external leaflet. E. coli imp mutants respond similarly by increasing lipid A palmitoylation [57] (unpublished observations). In E. coli O157:H7, a mutant deficient in lipid A myristoylation was reported recently to trigger palmitoylation of lipid A in the outer membrane [53]; palmitate addition could correct permeability defects associated with the absence of myristate and, surprisingly, exerted control over cytoplasmic enzymes involved in assembly of the core oligosaccharide. Emerging evidence has indicated that palmitoylated lipid A in the outer membrane can trigger the periplasmic stress response governed by σE [174]. It seems reasonable that palmitoylated lipid A subtypes and/or lysophospholipids, accumulated in the outer membrane in response to breaches in the permeability barrier, might adversely influence β-barrel membrane protein assembly and, thereby, trigger the periplasmic stress response.
As previously discussed, lipid A palmitoylation at position-2 attenuates endotoxin signaling through the host TLR4 pathway [84]. In more divergent organisms, where palmitate is attached on the distal glucosamine unit at position-3′, lipid A palmitoylation has been reported to accentuate the endotoxicity of lipid A [175,176], but this might reflect differences in the overall extent of acylation in addition to the different positional specificity. Combined with its contributions to antimicrobial peptide resistance [83], palmitoylated lipid A likely functions to help pathogenic bacteria evade the host immune response [82]. PagP homologues are now known to be necessary for disease causation in the respiratory pathogens Legionella pneumophila and Bordetella bronchiseptica [177,178]; in the latter organism, the pagP gene is needed to provide resistance to antibody-mediated lysis executed by the membrane-attack complex of the serum complement cascade [179]. Other PagP homologues are narrowly distributed among a group of pathogenic Gram-negative bacteria that often regulate pagP gene expression in order to meet the specific needs of their infectious lifestyles [82]. Mutational inactivation of PagP is observed in Y. pestis and B. pertussis, which, together with the aforementioned bacterium-specific genetic silencing of OMPLA and OmpT [130,139], reveals a theme that outer membrane β-barrel enzymes might be incompatible with certain pathogenic states.
3.4. The lipid A 3-O-deacylase PagL
The discovery of lipid A palmitoyltransferase activity in membranes from PhoP-constitutive mutants of Salmonella fortuitously revealed the activity of another enzyme that simultaneously removed an acyl chain from lipid A precursors (Fig. 2) [162]. The PhoP/PhoQ-activated gene pagL was subsequently established to encode the responsible enzyme [85]. PagL functions as a lipid A 3-O-deacylase and is localized to the outer membrane, but it shows no discernible activity under normal conditions. The presence of L-Ara4N in lipid A was shown to inhibit PagL and its activity in outer membranes only occurs when L-Ara4N is absent from lipid A [180]. Activated PagL confers polymyxin B resistance, which appears to compensate for sensitivity associated with the absence of L-Ara4N [86]. Lipid A 3-O-deacylation also attenuates lipid A endotoxicity and likely contributes to the evasion of immune defenses [84]. PagL is absent from E. coli, but, compared to PagP, its homologues are more widely distributed and not primarily restricted to pathogenic organisms [175,181]. Interestingly, loss of PagL function in P. aeruginosa has been observed during long-term adaptation to the cystic fibrosis airway [182].
The crystal structure of PagL from P. aeruginosa reveals an overall fold that is similar to PagP, and its active site likewise faces the outer surface of the outer membrane (Fig. 3D). PagL is similarly an 8-stranded antiparallel β-barrel and is strikingly tilted in the outer membrane [4]. However, PagL differs in the presentation of its lipid substrate to a distinct catalytic triad formed by Glu140, His126, and Ser128 on the β-barrel exterior. Energy minimization with the model substrate lipid X reveals that acyl-chains likely bind into hydrophobic grooves on the PagL β-barrel exterior. Although the enzyme normally encounters R-3-hydroxydecanoyl chains in its substrates in vivo, it also utilizes R-3-hydroxymyristoyl chains in vitro, which indicates that acyl-chains encountered on the β-barrel exterior cannot be measured with the same precision as performed by PagP [182]. The potential of PagL to dimerize at an interface between active sites suggests a possible mechanism to inhibit activity in the outer membrane when L-Ara4N is present. Whereas OMPLA is activated by Ca2+-dependent dimerization, a Ca2+-independent dimerization process is proposed to inactivate PagL [4].
4. β-Barrel enzymes of unknown structure
At least two other β-barrel enzymes of unknown structure have been identified. LpxR removes the 3′-acyloxyacyl unit in lipid A and was recently identified in outer membranes of Salmonella and other important enteric pathogens [183]. The enzyme requires Ca2+ as a cofactor and is not regulated by PhoP/PhoQ. The function of LpxR is unknown, but alteration of the lipid A acylation pattern probably affects lipid A endotoxicity and/or antimicrobial peptide resistance. The activity of LpxR cannot be detected under normal growth conditions, indicating that its activity is latent. It seems likely that LpxR will function together with OMPLA in response to membrane perturbations when divalent cations are replete. Another lipid A modification that requires Ca2+ as a cofactor is the EptB-catalyzed incorporation of phosphoethanolamine into a key inner core sugar at the periplasmic surface of the inner membrane [88]. EptB also requires Ca2+ in the growth medium for expression [184], but LpxR expression, like that of OMPLA, appears to be constitutive.
Rhizobium species establish a symbiotic relationship with the root cells of certain plants and are responsible for nitrogen fixation. These Gram-negative bacteria fail to trigger the host innate immune response partly because they express a detoxified lipid A. The conserved Raetz pathway enzymes are largely intact, but additional extracytoplasmic enzymes serve to remodel the lipid A structure [53]. Among the observed structural variations is a two-step modification of the proximal glucosamine sugar. First, the phosphate group is removed by the lipid A 1-phosphatase LpxE, which acts at the periplasmic surface of the inner membrane [91,96]. Second, oxidation of the glucosamine aldehyde yields the open chain sugar 2-aminogluconate. The responsible oxidase is the EDTA-sensitive and O2-dependent outer membrane enzyme LpxQ [185,186]. Based on the foregoing discussion, it might be tempting to speculate that LpxQ behaves, like the other outer membrane β-barrel enzymes, as a sentinel triggered by perturbations to the outer membrane structure. However, the LpxQ reaction proceeds efficiently under normal conditions when it encounters 1-dephosphorylated lipid A in the outer membrane. It is unknown if the 2-aminogluconate moiety is compatible with the transport of lipid A across the periplasmic compartment, but LpxQ might be a terminal biosynthetic enzyme that is restricted to the outer membrane simply because its activity is incompatible with lipid A transport. While the majority of outer membrane β-barrel enzymes are most easily comprehended in terms of the sentinel hypothesis, LpxQ underscores the fact that it need not necessarily apply to them all. It will certainly be interesting to learn how LpxQ uses O2 to catalyze lipid A oxidation in the outer membrane environment.
5. Conclusions
The structure and function of β-barrel enzymes indicates that they have been adapted to serve as sentinels in the outer membrane. By functioning as sensory transducers whose activity is triggered by perturbations to outer membrane lipid organization, β-barrel enzymes provide a first line of defense against external attack. Centralized signal transduction networks that function to maintain the outer membrane permeability barrier can modulate the expression levels of these outer membrane sentinels, but the expansive outer membrane assembly pathway places huge demands for speed in responding to external assaults. The sentinels likely provide the instantaneous responses needed to outpace the centralized signal transduction pathways. Additionally, the discovery of outer membrane β-barrel enzymes has provided useful probes to monitor the biogenesis of the Gram-negative cell envelope. By following the modification of cell surface lipids catalyzed by β-barrel enzymes, insights into outer membrane assembly and function that were not previously possible are now dramatically changing our perspective on Gram-negative cell envelope structure. The importance of β-barrel enzymes in bacterial pathogenesis testifies that knowledge of their structure-function relationships might also lead to novel treatments for bacterial infections.
Acknowledgments
We thank Piet Gros, Lucy Rutten, Jan Tommassen, Chris Neale, and Régis Pomès for providing structure coordinates. This work was supported by CIHR operating grant MOP-43886.
Abbreviations
- L-Ara4N
4-amino-4-deoxy-L-arabinose
- GroPEtn
glycerophosphoethanolamine
- GPL
glycerophospholipid
- OAg
O-antigen
- PtdEtn
phosphatidylethanolamine
References
- 1.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 2.Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, Gros P. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 2001;20:5033–5039. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, de Haan A, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Ann Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita S, Matsuyama S, Tokuda H. Lipoprotein trafficking in Escherichia coli. Arch Microbiol. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- 9.Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta. 1975;415:335–377. doi: 10.1016/0304-4157(75)90013-1. [DOI] [PubMed] [Google Scholar]
- 10.Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 11.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 12.Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 13.Montoya M, Gouaux E. Beta-barrel membrane protein folding and structure viewed through the lens of alpha-hemolysin. Biochim Biophys Acta. 2003;1609:19–27. doi: 10.1016/s0005-2736(02)00663-6. [DOI] [PubMed] [Google Scholar]
- 14.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 15.White SH, Ladokhin AS, Jayasinghe S, Hristova K. How membranes shape protein structure. J Biol Chem. 2001;276:32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 16.Buchanan SK. Beta-barrel proteins from bacterial outer membranes: structure, function and refolding. Curr Opin Struck Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 17.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 18.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Ann Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 19.Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 21.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 25.Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 26.Alba BM, Gross CA. Regulation of the Escherichia coli sigma E-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 27.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. Omp peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its Pdz domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 28.Hung DL, Raivio TL, Jones CH, Silhavy TJ, Hultgren SJ. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 2001;20:1508–1518. doi: 10.1093/emboj/20.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 30.Pratt LA, Hsing W, Gibson KE, Silhavy TJ. From acids to OsmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P. Crystal structure of neisserial surface protein a (NspA), a conserved outer membrane protein with vaccine potential. J Biol Chem. 2003;278:24825–24830. doi: 10.1074/jbc.M302803200. [DOI] [PubMed] [Google Scholar]
- 33.Prince SM, Achtman M, Derrick JP. Crystal structure of the OpcA integral membrane adhesin from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2002;99:3417–3421. doi: 10.1073/pnas.062630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 35.Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J Mol Biol. 2000;298:273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 36.Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 37.Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss MS, Wacker T, Weckesser J, Welte W, Schulz GE. The three-dimensional structure of porin from Rhodobacter capsulatus at 3 Å resolution. FEBS Lett. 1990;267:268–272. doi: 10.1016/0014-5793(90)80942-c. [DOI] [PubMed] [Google Scholar]
- 39.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 40.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 41.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. II. The observed structures. J Mol Biol. 1994;236:1382–1400. doi: 10.1016/0022-2836(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 42.Gouaux E. Roll out the barrel. Nat Struct Biol. 1998;5:931–932. doi: 10.1038/2804. [DOI] [PubMed] [Google Scholar]
- 43.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huijbregts RP, de Kroon AI, de Kruijff B. Topology and transport of membrane lipids in bacteria. Biochim Biophys Acta. 2000;1469:43–61. doi: 10.1016/s0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 45.Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem. 1977;252:7405–7412. [PubMed] [Google Scholar]
- 46.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 47.Kol MA, de Kruijff B, de Kroon AI. Phospholipid flip-flop in biogenic membranes: what is needed to connect opposite sides. Semin Cell Dev Biol. 2002;13:163–170. doi: 10.1016/s1084-9521(02)00044-7. [DOI] [PubMed] [Google Scholar]
- 48.Kol MA, van Dalen A, de Kroon AI, de Kruijff B. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J Biol Chem. 2003;278:24586–24593. doi: 10.1074/jbc.M301875200. [DOI] [PubMed] [Google Scholar]
- 49.Doerrler WT, Reedy MC, Raetz CR. An Escherichia coli mutant defective in lipid export. J Biol Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 50.Doerrler WT, Gibbons HS, Raetz CR. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 51.Tefsen B, Bos MP, Beckers F, Tommassen J, de Cock H. MsbA is not required for phospholipid transport in Neisseria meningitidis. J Biol Chem. 2005;280:35961–35966. doi: 10.1074/jbc.M509026200. [DOI] [PubMed] [Google Scholar]
- 52.Heinrichs DE, Yethon JA, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 53.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Ann Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J Biol Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- 56.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci U S A. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler M, Osborn MJ. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 59.Coughlin RT, Tonsager S, McGroarty EJ. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry. 1983;22:2002–2007. doi: 10.1021/bi00277a041. [DOI] [PubMed] [Google Scholar]
- 60.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 61.Leive L. The barrier function of the Gram-negative envelope. Ann NY Acad Sci. 1974;235:109–129. doi: 10.1111/j.1749-6632.1974.tb43261.x. [DOI] [PubMed] [Google Scholar]
- 62.Watt SR, Clarke AJ. Role of autolysins in the EDTA-induced lysis of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1994;124:113–119. doi: 10.1111/j.1574-6968.1994.tb07270.x. [DOI] [PubMed] [Google Scholar]
- 63.Steeghs L, de Cock H, Evers E, Zomer B, Tommassen J, van der Ley P. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 2001;20:6937–6945. doi: 10.1093/emboj/20.24.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lysko PG, Morse SA. Neisseria gonorrhoeae cell envelope: permeability to hydrophobic molecules. J Bacteriol. 1981;145:946–952. doi: 10.1128/jb.145.2.946-952.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Epand RM, Shai Y, Segrest JP, Anantharamaiah GM. Mechanisms for the modulation of membrane bilayer properties by amphipathic helical peptides. Biopolymers. 1995;37:319–338. doi: 10.1002/bip.360370504. [DOI] [PubMed] [Google Scholar]
- 66.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 67.Hancock RE, Falla T, Brown M. Cationic bactericidal peptides. Adv Microb Physiol. 1995;37:135–175. doi: 10.1016/s0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 68.Hancock RE, Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 69.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 70.Scott MG, Rosenberger CM, Gold MR, Finlay BB, Hancock RE. An alpha-helical cationic antimicrobial peptide selectively modulates macrophage responses to lipopolysaccharide and directly alters macrophage gene expression. J Immunol. 2000;165:3358–3365. doi: 10.4049/jimmunol.165.6.3358. [DOI] [PubMed] [Google Scholar]
- 71.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 72.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 74.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 75.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 76.Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 77.Kox LF, Wosten MM, Groisman EA. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winfield MD, Groisman EA. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A. 2004;101:17162–17167. doi: 10.1073/pnas.0406038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 80.Gibbons HS, Kalb SR, Cotter RJ, Raetz CR. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol Microbiol. 2005;55:425–440. doi: 10.1111/j.1365-2958.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 81.Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller S, Finlay BB, Gros P, Grinstein S. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol Biol Cell. 2006;17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 83.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 84.Kawasaki K, Ernst RK, Miller SI. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through toll-like receptor 4. J Biol Chem. 2004;279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 85.Trent MS, Pabich W, Raetz CR, Miller SI. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 86.Kawasaki K, China K, Nishijima M. Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica. J Bacteriol. 2007;189:4911–4919. doi: 10.1128/JB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee H, Hsu FF, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reynolds CM, Kalb SR, Cotter RJ, Raetz CR. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 89.Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CR. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J Biol Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- 90.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 91.Karbarz MJ, Kalb SR, Cotter RJ, Raetz CR. Expression cloning and biochemical characterization of a Rhizobium leguminosarum lipid A 1-phosphatase. J Biol Chem. 2003;278:39269–39279. doi: 10.1074/jbc.M305830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, McGrath SC, Cotter RJ, Raetz CR. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J Biol Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CR. Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proc Natl Acad Sci U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Persing DH, Coler RN, Lacy MJ, Johnson DA, Baldridge JR, Hershberg RM, Reed SG. Taking toll: lipid A mimetics as adjuvants and immunomodulators. Trends Microbiol. 2002;10:S32–S37. doi: 10.1016/s0966-842x(02)02426-5. [DOI] [PubMed] [Google Scholar]
- 95.Wang X, Ribeiro AA, Guan Z, McGrath SC, Cotter RJ, Raetz CR. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CR. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Scandella CJ, Kornberg A. A membrane-bound phospholipase A1 purified from Escherichia coli. Biochemistry. 1971;10:4447–4456. doi: 10.1021/bi00800a015. [DOI] [PubMed] [Google Scholar]
- 98.Bell RM, Mavis RD, Osborn MJ, Vagelos PR. Enzymes of phospholipid metabolism: Localization in the cytoplasmic and outer membrane of the cell envelope of Escherichia coli and Salmonella typhimurium. Biochim Biophys Acta. 1971;249:628–635. doi: 10.1016/0005-2736(71)90144-1. [DOI] [PubMed] [Google Scholar]
- 99.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 100.Imoto M, Kusumoto S, Shiba T, Naoki H, Iwashita T, Rietschel ET, Wollenweber HW, Galanos C, Luderitz O. Chemical-structure of Escherichia-coli lipid-A — linkage site of acyl-groups in the disaccharide backbone. Tetrahedron Lett. 1983;24:4017–4020. [Google Scholar]
- 101.Takayama K, Qureshi N, Mascagni P. Complete structure of lipid A obtained from the lipopolysaccharides of the heptoseless mutant of Salmonella typhimurium. J Biol Chem. 1983;258:12801–12803. [PubMed] [Google Scholar]
- 102.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 103.Audet A, Nantel G, Proulx P. Phospholipase A activity in growing Escherichia coli cells. Biochim Biophys Acta. 1974;348:334–343. doi: 10.1016/0005-2760(74)90213-6. [DOI] [PubMed] [Google Scholar]
- 104.Michel GP, Starka J. Phospholipase A activity with integrated phospholipid vesicles in intact cells of an envelope mutant of Escherichia coli. FEBS Lett. 1979;108:261–265. doi: 10.1016/0014-5793(79)81224-7. [DOI] [PubMed] [Google Scholar]
- 105.Pugsley AP, Schwartz M. Colicin E2 release: lysis, leakage or secretion? Possible role of a phospholipase. EMBO J. 1984;3:2393–2397. doi: 10.1002/j.1460-2075.1984.tb02145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taketo A. Sensitivity of Escherichia coli to viral nucleic acid. 8. Idiosyncrasy of Ca2+-dependent competence for DNA. J Biochem (Tokyo) 1974;75:895–904. doi: 10.1093/oxfordjournals.jbchem.a130463. [DOI] [PubMed] [Google Scholar]
- 108.Cronan JE, Jr, Wulff DL. A role for phospholipid hydrolysis in the lysis of Escherichia coli infected with bacteriophage t4. Virology. 1969;38:241–246. doi: 10.1016/0042-6822(69)90365-1. [DOI] [PubMed] [Google Scholar]
- 109.Patriarca P, Beckerdite S, Elsbach P. Phospholipases and phospholipid turnover in Escherichia coli spheroplasts. Biochim Biophys Acta. 1972;260:593–600. doi: 10.1016/0005-2760(72)90008-2. [DOI] [PubMed] [Google Scholar]
- 110.de Geus P, van Die I, Bergmans H, Tommassen J, de Haas G. Molecular cloning of pldA, the structural gene for outer membrane phospholipase of E. coli K12. Mol Gen Genet. 1983;190:150–155. doi: 10.1007/BF00330338. [DOI] [PubMed] [Google Scholar]
- 111.Hardaway KL, Buller CS. Effect of ethylenediaminetetraacetate on phospholipids and outer membrane function in Escherichia coli. J Bacteriol. 1979;137:62–68. doi: 10.1128/jb.137.1.62-68.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weiss J, Beckerdite-Quagliata S, Elsbach P. Determinants of the action of phospholipases A on the envelope phospholipids of Escherichia coli. J Biol Chem. 1979;254:11010–11014. [PubMed] [Google Scholar]
- 113.Wright GC, Weiss J, Kim KS, Verheij H, Elsbach P. Bacterial phospholipid hydrolysis enhances the destruction of Escherichia coli ingested by rabbit neutrophils. Role of cellular and extracellular phospholipases. J Clin Invest. 1990;85:1925–1935. doi: 10.1172/JCI114655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dekker N. Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 115.Dekker N, Tommassen J, Lustig A, Rosenbusch JP, Verheij HM. Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J Biol Chem. 1997;272:3179–3184. doi: 10.1074/jbc.272.6.3179. [DOI] [PubMed] [Google Scholar]
- 116.Dirusso CC, Black PN. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. J Biol Chem. 2004;279:49563–49566. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 117.McIntyre TM, Bell RM. Escherichia coli mutants defective in membrane phospholipid synthesis: binding and metabolism of 1-oleoylglycerol 3-phosphate by a plsB deep rough mutant. J Bacteriol. 1978;135:215–226. doi: 10.1128/jb.135.1.215-226.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hsu L, Jackowski S, Rock CO. Uptake and acylation of 2-acyl-lysophospholipids by Escherichia coli. J Bacteriol. 1989;171:1203–1205. doi: 10.1128/jb.171.2.1203-1205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hellion P, Landry F, Subbaiah PV, Proulx P. The uptake and acylation of exogenous lysophosphatidylethanolamine by Escherichia coli cells. Can J Biochem. 1980;58:1381–1386. doi: 10.1139/o80-187. [DOI] [PubMed] [Google Scholar]
- 120.Homma H, Nishijima M, Kobayashi T, Okuyama H, Nojima S. Incorporation and metabolism of 2-acyl lysophospholipids by Escherichia coli. Biochim Biophys Acta. 1981;663:1–13. doi: 10.1016/0005-2760(81)90189-2. [DOI] [PubMed] [Google Scholar]
- 121.Kingma RL, Fragiathaki M, Snijder HJ, Dijkstra BW, Verheij HM, Dekker N, Egmond MR. Unusual catalytic triad of Escherichia coli outer membrane phospholipase A. Biochemistry. 2000;39:10017–10022. doi: 10.1021/bi000786d. [DOI] [PubMed] [Google Scholar]
- 122.Snijder HJ, Van Eerde JH, Kingma RL, Kalk KH, Dekker N, Egmond MR, Dijkstra BW. Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn–His interaction in the catalytic triad. Protein Sci. 2001;10:1962–1969. doi: 10.1110/ps.17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Snijder HJ, Dijkstra BW. Bacterial phospholipase A: structure and function of an integral membrane phospholipase. Biochim Biophys Acta. 2000;1488:91–101. doi: 10.1016/s1388-1981(00)00113-x. [DOI] [PubMed] [Google Scholar]
- 124.Kingma RL, Snijder HJ, Dijkstra BW, Dekker N, Egmond MR. Functional importance of calcium binding sites in outer membrane phospholipase A. Biochim Biophys Acta. 2002;1561:230–237. doi: 10.1016/s0005-2736(02)00351-6. [DOI] [PubMed] [Google Scholar]
- 125.Snijder HJ, Kingma RL, Kalk KH, Dekker N, Egmond MR, Dijkstra BW. Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase a from Escherichia coli. J Mol Biol. 2001;309:477–489. doi: 10.1006/jmbi.2001.4675. [DOI] [PubMed] [Google Scholar]
- 126.Baaden M, Meier C, Sansom MS. A molecular dynamics investigation of mono and dimeric states of the outer membrane enzyme OMPLA. J Mol Biol. 2003;331:177–189. doi: 10.1016/s0022-2836(03)00718-6. [DOI] [PubMed] [Google Scholar]
- 127.Horrevoets AJ, Hackeng TM, Verheij HM, Dijkman R, de Haas GH. Kinetic characterization of Escherichia coli outer membrane phospholipase A using mixed detergent-lipid micelles. Biochemistry. 1989;28:1139–1147. doi: 10.1021/bi00429a031. [DOI] [PubMed] [Google Scholar]
- 128.Stanley AM, Chuawong P, Hendrickson TL, Fleming KG. Energetics of outer membrane phospholipase A (OMPLA) dimerization. J Mol Biol. 2006;358:120–131. doi: 10.1016/j.jmb.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 129.Stanley AM, Treubrodt AM, Chuawong P, Hendrickson TL, Fleming KG. Lipid chain selectivity by outer membrane phospholipase A. J Mol Biol. 2007;366:461–468. doi: 10.1016/j.jmb.2006.10.055. [DOI] [PubMed] [Google Scholar]
- 130.Bos MP, Tefsen B, Voet P, Weynants V, van Putten JP, Tommassen J. Function of neisserial outer membrane phospholipase A in autolysis and assessment of its vaccine potential. Infect Immun. 2005;73:2222–2231. doi: 10.1128/IAI.73.4.2222-2231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grant KA, Belandia IU, Dekker N, Richardson PT, Park SF. Molecular characterization of pldA, the structural gene for a phospholipase a from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect Immun. 1997;65:1172–1180. doi: 10.1128/iai.65.4.1172-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bukholm G, Tannaes T, Nedenskov P, Esbensen Y, Grav HJ, Hovig T, Ariansen S, Guldvog I. Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand J Gastroenterol. 1997;32:445–454. doi: 10.3109/00365529709025079. [DOI] [PubMed] [Google Scholar]
- 133.Tannaes T, Bukholm IK, Bukholm G. High relative content of lysophospholipids of Helicobacter pylori mediates increased risk for ulcer disease. FEMS Immunol Med Microbiol. 2005;44:17–23. doi: 10.1016/j.femsim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 134.Tannaes T, Dekker N, Bukholm G, Bijlsma JJ, Appelmelk BJ. Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect Immun. 2001;69:7334–7340. doi: 10.1128/IAI.69.12.7334-7340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rupprecht KR, Gordon G, Lundrigan M, Gayda RC, Markovitz A, Earhart C. OmpT: Escherichia coli K12 structural gene for protein a (3b) J Bacteriol. 1983;153:1104–1106. doi: 10.1128/jb.153.2.1104-1106.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fiss EH, Hollifield WC, Jr, Neilands JB. Absence of ferric enterobactin receptor modification activity in mutants of Escherichia coli K12 lacking protein a. Biochem Biophys Res Commun. 1979;91:29–34. doi: 10.1016/0006-291x(79)90578-3. [DOI] [PubMed] [Google Scholar]
- 137.Grodberg J, Dunn JJ. OmpT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sugimura K, Nishihara T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: Identity of protease VII and OmpT. J Bacteriol. 1988;170:5625–5632. doi: 10.1128/jb.170.12.5625-5632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M, Sasakawa C. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the OmpT and KcpA loci. Mol Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 140.Kaufmann A, Stierhof YD, Henning U. New outer membrane-associated protease of Escherichia coli K12. J Bacteriol. 1994;176:359–367. doi: 10.1128/jb.176.2.359-367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matsuo E, Sampei G, Mizobuchi K, Ito K. The plasmid F OmpP protease, a homologue of OmpT, as a potential obstacle to E. coli-based protein production. FEBS Lett. 1999;461:6–8. doi: 10.1016/s0014-5793(99)01418-0. [DOI] [PubMed] [Google Scholar]
- 142.Hwang BY, Varadarajan N, Li H, Rodriguez S, Iverson BL, Georgiou G. Substrate specificity of the Escherichia coli outer membrane protease OmpP. J Bacteriol. 2007;189:522–530. doi: 10.1128/JB.01493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lundrigan M, Earhart CF. Reduction in three iron-regulated outer membrane proteins and protein a by the Escherichia coli K12 perA mutation. J Bacteriol. 1981;146:804–807. doi: 10.1128/jb.146.2.804-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 145.Eguchi Y, Okada T, Minagawa S, Oshima T, Mori H, Yamamoto K, Ishihama A, Utsumi R. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J Bacteriol. 2004;186:3006–3014. doi: 10.1128/JB.186.10.3006-3014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dawkins R. The extended phenotype: The long reach of the gene. Oxford University Press; Oxford; New York: 1983. [Google Scholar]
- 147.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. 1998;180:4002–4006. doi: 10.1128/jb.180.15.4002-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baaden M, Sansom MS. OmpT: Molecular dynamics simulations of an outer membrane enzyme. Biophys J. 2004;87:2942–2953. doi: 10.1529/biophysj.104.046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kramer RA, Zandwijken D, Egmond MR, Dekker N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2000;267:885–893. doi: 10.1046/j.1432-1327.2000.01073.x. [DOI] [PubMed] [Google Scholar]
- 151.Kramer RA, Brandenburg K, Vandeputte-Rutten L, Werkhoven M, Gros P, Dekker N, Egmond MR. Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2002;269:1746–1752. doi: 10.1046/j.1432-1327.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 152.Brandenburg K, Garidel P, Schromm AB, Andra J, Kramer A, Egmond M, Wiese A. Investigation into the interaction of the bacterial protease OmpT with outer membrane lipids and biological activity of OmpT:lipopolysaccharide complexes. Eur Biophys J. 2005;34:28–41. doi: 10.1007/s00249-004-0422-3. [DOI] [PubMed] [Google Scholar]
- 153.Ferguson AD, Welte W, Hofmann E, Lindner B, Holst O, Coulton JW, Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 154.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 155.Kukkonen M, Suomalainen M, Kyllonen P, Lahteenmaki K, Lang H, Virkola R, Helander IM, Holst O, Korhonen TK. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol Microbiol. 2004;51:215–225. doi: 10.1046/j.1365-2958.2003.03817.x. [DOI] [PubMed] [Google Scholar]
- 156.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 157.Egile C, d’Hauteville H, Parsot C, Sansonetti PJ. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997;23:1063–1073. doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- 158.Shere KD, Sallustio S, Manessis A, D’Aversa TG, Goldberg MB. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol Microbiol. 1997;25:451–462. doi: 10.1046/j.1365-2958.1997.4681827.x. [DOI] [PubMed] [Google Scholar]
- 159.Sodeinde OA, Goguen JD. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to OmpT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kukkonen M, Lahteenmaki K, Suomalainen M, Kalkkinen N, Emody L, Lang H, Korhonen TK. Protein regions important for plasminogen activation and inactivation of alpha2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol Microbiol. 2001;40:1097–1111. doi: 10.1046/j.1365-2958.2001.02451.x. [DOI] [PubMed] [Google Scholar]
- 161.Kukkonen M, Korhonen TK. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int J Med Microbiol. 2004;294:7–14. doi: 10.1016/j.ijmm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 162.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Takayama K, Qureshi N, Mascagni P, Anderson L, Raetz CR. Glucosamine-derived phospholipids in Escherichia coli. Structure and chemical modification of a triacyl glucosamine 1-phosphate found in a phosphatidylglycerol-deficient mutant. J Biol Chem. 1983;258:14245–14252. [PubMed] [Google Scholar]
- 164.Takayama K, Qureshi N, Mascagni P, Nashed MA, Anderson L, Raetz CR. Fatty acyl derivatives of glucosamine 1-phosphate in Escherichia coli and their relation to lipid A. Complete structure of a diacyl Glcn-1-P found in a phosphatidylglycerol-deficient mutant. J Biol Chem. 1983;258:7379–7385. [PubMed] [Google Scholar]
- 165.Brozek KA, Bulawa CE, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A membrane-bound enzyme that transfers a palmitoyl residue from a glycerophospholipid to lipid X. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 166.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes PhoP–PhoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 167.Hu KH, Liu E, Dean K, Gingras M, DeGraff W, Trun NJ. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Evanics F, Hwang PM, Cheng Y, Kay LE, Prosser RS. Topology of an outer-membrane enzyme: measuring oxygen and water contacts in solution NMR studies of PagP. J Am Chem Soc. 2006;128:8256–8264. doi: 10.1021/ja0610075. [DOI] [PubMed] [Google Scholar]
- 170.Khan MA, Neale C, Michaux C, Pomes R, Prive GG, Woody RW, Bishop RE. Gauging a hydrocarbon ruler by an intrinsic exciton probe. Biochemistry. 2007;46:4565–4579. doi: 10.1021/bi602526k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hwang PM, Bishop RE, Kay LE. The integral membrane enzyme PagP alternates between two dynamically distinct states. Proc Natl Acad Sci U S A. 2004;101:9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bishop RE, Raetz CR. Fortieth Interscience Conference on Antimicrobial Agents and Chemotherapy; Toronto, ON, Canada. 2000; 2000. Vol. abstr. 2290. [Google Scholar]
- 173.Jia W, Zoeiby AE, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 174.Tam C, Missiakas D. Changes in lipopolysaccharide structure induce the sigma E-dependent response of Escherichia coli. Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- 175.Geurtsen J, Steeghs L, Hamstra HJ, Ten Hove J, de Haan A, Kuipers B, Tommassen J, van der Ley P. Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect Immun. 2006;74:5574–5585. doi: 10.1128/IAI.00834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 177.Robey M, O’Connell W, Cianciotto NP. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Preston A, Maxim E, Toland E, Pishko EJ, Harvill ET, Caroff M, Maskell DJ. Bordetella bronchiseptica PagP is a bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol Microbiol. 2003;48:725–736. doi: 10.1046/j.1365-2958.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 179.Pilione MR, Pishko EJ, Preston A, Maskell DJ, Harvill ET. PagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect Immun. 2004;72:2837–2842. doi: 10.1128/IAI.72.5.2837-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kawasaki K, Ernst RK, Miller SI. Inhibition of Salmonella enterica serovar typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J Bacteriol. 2005;187:2448–2457. doi: 10.1128/JB.187.7.2448-2457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Geurtsen J, Steeghs L, Hove JT, Ley PV, Tommassen J. Dissemination of lipid A deacylases (PagL) among Gram-negative bacteria: identification of active-site histidine and serine residues. J Biol Chem. 2005;280:8248–8259. doi: 10.1074/jbc.M414235200. [DOI] [PubMed] [Google Scholar]
- 182.Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, Trent MS, Miller SI. The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J Bacteriol. 2006;188:191–201. doi: 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CR, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium LpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kanipes MI, Lin S, Cotter RJ, Raetz CR. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J Biol Chem. 2001;276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- 185.Que-Gewirth NL, Karbarz MJ, Kalb SR, Cotter RJ, Raetz CR. Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ. J Biol Chem. 2003;278:12120–12129. doi: 10.1074/jbc.M300379200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Que-Gewirth NL, Lin S, Cotter RJ, Raetz CR. An outer membrane enzyme that generates the 2-amino-2-deoxy-gluconate moiety of Rhizobium leguminosarum lipid A. J Biol Chem. 2003;278:12109–12119. doi: 10.1074/jbc.M300378200. [DOI] [PMC free article] [PubMed] [Google Scholar]