Abstract
Membrane-intrinsic enzymes are embedded in lipids, yet how such enzymes interrogate lipid substrates remains a largely unexplored fundamental question. The outer membrane phospholipid:lipid A palmitoyltransferase PagP combats host immune defenses during infection and selects a palmitate chain using its β-barrel interior hydrocarbon ruler. Both a molecular embrasure and crenel in Escherichia coli PagP display weakened transmembrane β-strand hydrogen bonding to provide potential lateral routes for diffusion of the palmitoyl group between the hydrocarbon ruler and outer membrane external leaflet. Prolines in strands A and B lie beneath the dynamic L1 surface loop flanking the embrasure, whereas the crenel is flanked by prolines in strands F and G. Reversibly barricading the embrasure prevents lipid A palmitoylation without affecting the slower phospholipase reaction. Lys42Ala PagP is also a dedicated phospholipase, implicating this disordered L1 loop residue in lipid A recognition. The embrasure barricade additionally prevents palmitoylation of nonspecific fatty alcohols, but not miscible alcohols. Irreversibly barricading the crenel inhibits both lipid A palmitoylation and phospholipase reactions without compromising PagP structure. These findings indicate lateral palmitoyl group diffusion within the PagP hydrocarbon ruler is likely gated during phospholipid entry via the crenel and during lipid A egress via the embrasure.
The molecular mechanisms by which integral membrane enzymes select lipid substrates impact signal transduction and membrane biogenesis within all biological cells (1–3). Despite recent progress in the structural biology of integral membrane proteins (4), only a handful of known structures are enzymes of lipid metabolism. Along with the lipid cofactor-utilizing respiratory enzymes, only two other enzymes of eicosanoid biosynthesis (5–7) and a diacylglycerol kinase (8) represent determined subunit structures of bitopic transmembrane α-helical lipid metabolic enzymes. Another group of monotopic lipid enzyme structures engages the hydrophobic core of the bilayer using α- and/or β-motifs to bury into one membrane leaflet only (9, 10). Fundamental questions concerning lipid recognition by membrane enzymes remain largely unexplored (10), but robust β-barrel enzymes from the outer membranes of Gram-negative bacteria are providing intriguing answers (11–14).
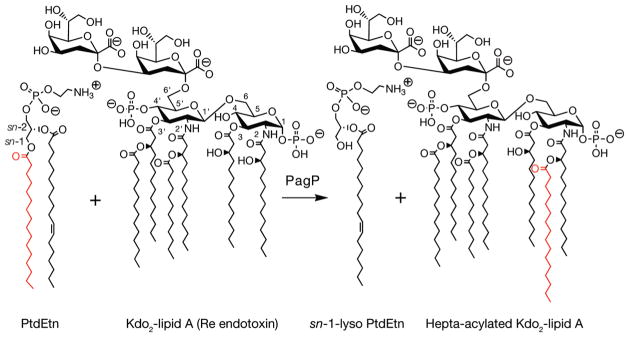
The outer membrane palmitoyltransferase PagP catalyzes transfer of palmitate from the sn-1 position of a phospholipid to the hydroxyl group of the (R)-3-hydroxymyristate chain at position 2 of lipid A (endotoxin) (Figure 1) (15, 16). PagP is a heat-stable 161-amino acid β-barrel enzyme, intrinsically lacking cysteine residues, which can fold reversibly in a defined detergent micellar enzymatic assay system (17). In Escherichia coli and related pathogenic Gram-negative bacteria, PagP combats host immune defenses by restoring the bacterial outer membrane permeability barrier (18–21) and attenuating endotoxin signaling through the host TLR4 inflammation pathway (22–25). The pagP gene is transcriptionally activated through the bacterial PhoP/PhoQ virulence signal transduction network in response to membrane-perturbing antimicrobial peptides (26), immunological agents PagP helps the bacteria to resist (18, 20, 27). PagP thus provides a target for the development of anti-infective agents, but it also provides a tool for synthesizing novel vaccine adjuvants and endotoxin antagonists (28).
Figure 1.
Reaction catalyzed by the phospholipid:lipid A palmitoyltransferase PagP. E. coli PagP transfers a palmitoyl group from the sn-1 position of a phospholipid, such as phosphatidylethanolamine (PtdEtn), to the free hydroxyl group of the N-linked (R)-3-hydroxymyristate chain on the proximal glucosamine unit of lipid A. One of the simplest lipid A acceptors for PagP in the outer membrane is known as Kdo2-lipid A or Re endotoxin.
The structure of E. coli PagP has been determined both by solution nuclear magnetic resonance (NMR)1 spectroscopy and by X-ray crystallography to reveal an eight-stranded antiparallel β-barrel preceded by an N-terminal amphipathic α-helix (29, 30). Like those of other β-barrel membrane proteins, the center of the PagP β-barrel is relatively rigid with an increasing mobility gradient as one moves toward the flexible outer loops. Remarkably, the PagP active site residues His33, Asp76, and Ser77, among others yet to be described, are localized to these dynamic extracellular loops. NMR studies indicate the presence of processes with time scales ranging from picoseconds to milliseconds in the L1 loop region where chain density could not be found in the X-ray analysis (29, 31). However, X-ray studies identified a single detergent molecule buried within the rigid β-barrel core, which corresponds to the binding site for the phospholipid sn-1 palmitate chain (30). These structural dynamics indicate PagP likely alternates between a relaxed state facilitating lipid substrate access from the membrane phase and a tense state organizing catalytic residues around the lipid substrates once bound.
PagP is exquisitely selective for a 16-carbon palmitate chain because its hydrocarbon ruler excludes lipid acyl chains varying in length by a solitary methylene unit (17, 30). The PagP hydrocarbon ruler is delineated by the detergent-binding hydrophobic pocket buried within the β-barrel structure (Figure 2) (30), andmutation of Gly88 can raise the hydrocarbon ruler floor to correspondingly shorten the selected acyl chain (17). The localization of the PagP hydrocarbon ruler within the outer membrane external leaflet must be rationalized with the asymmetric lipid distribution of the bacterial outer membrane. The external leaflet is lined by lipopolysaccharide (LPS) molecules anchored by their lipid A moiety, whereas the periplasmic leaflet is lined by phospholipid molecules (32, 33). Hydrophobic antibiotics and detergents freely permeate phospholipid bilayers, but they cannot penetrate the outer membrane because neighboring negatively charged LPS groups are ionically bonded by bridging divalent cations (34–36). PagP lies dormant in the outer membrane when lipid asymmetry restricts the phospholipid palmitoyl donor to the inner leaflet, but permeability defects involving aberrant external leaflet phospholipid migration directly trigger lipid A palmitoylation (21, 37–40).
Figure 2.
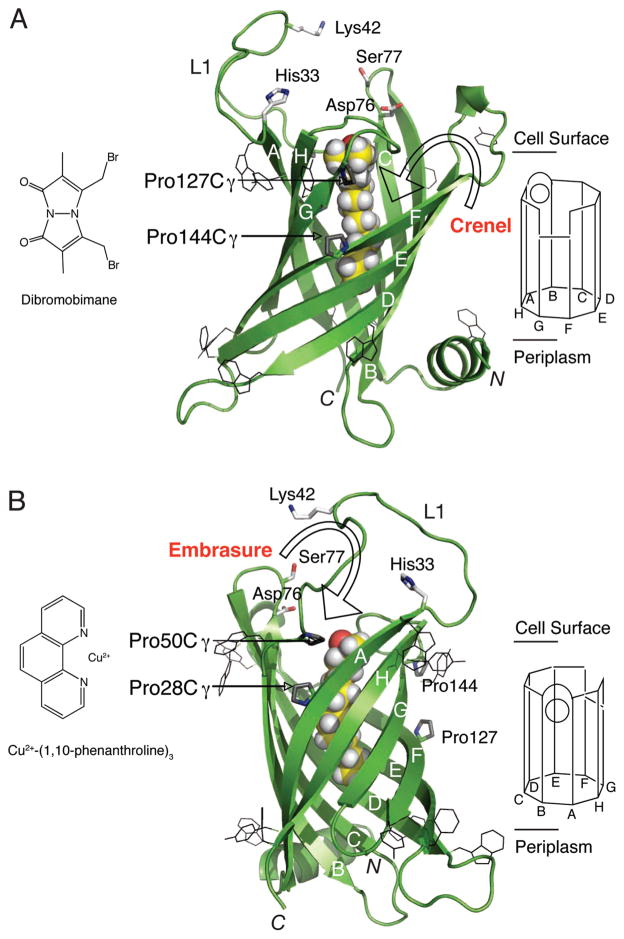
Lipid A palmitate chain selected by a β-barrel interior hydrocarbon ruler. PagP is an eight-stranded antiparallel β-barrel (green) with its hydrocarbon ruler delineated by an internalized LDAO detergent molecule (yellow), which is aligned with the external leaflet of the bacterial outer membrane. Aromatic belt residues in black wireframe representation define the membrane boundaries between the cell surface and periplasmic space. Pro → Cys substitutions flanking the PagP crenel (Pro127Cys/Pro144Cys) (A) and embrasure (Pro28Cys/Pro50Cys) (B) indicate cysteine sulfhydryl groups will localize near Cγ of the corresponding proline residues (gray). These prolines likely help to weaken local transmembrane β-strand hydrogen bonding in the outer membrane external leaflet to create the crenel and embrasure apertures shown schematically on the right. Cysteine oxidation of the PagP embrasure can be driven by copper phenanthroline, whereas the crenel cysteines can by alkylated by dibromobimane. The L1 loop and associated Lys42 are disordered in the PagP structure crystallized from LDAO (PDB entry 1THQ), but these elements were introduced and energy minimized in the model shown. His33, Asp76, and Ser77 were previously implicated in catalysis. The β-strands (A–H) are indicated in white letters on the structures and in black letters on the schematic diagrams. The amino (N) and carboxyl (C) termini are indicated for both structures.
Once a phospholipid molecule arrives in the outer membrane external leaflet, the mechanism by which the palmitoyl group transits between the membrane and PagP interior hydrocarbon ruler remains to be elucidated. A driving force for outer membrane β-barrel protein folding comes from hydrogen bonding between backbone amide proton and carbonyl oxygen atoms within the nonpolar membrane environment (41, 42). The interiors of typical β-barrelmembrane proteins are shielded from membrane lipids because of this continuous β-strand hydrogen bonding, and proline residues lacking an amide proton to donate a hydrogen bond are usually excluded from transmembrane β-strands (43). Nevertheless, proline residues are strikingly localized within the PagP transmembrane β-strands at two sites flanking Pro28 in strand A and Pro50 in strand B (Figure 2B), which creates a β-bulge beneath the dynamic L1 surface loop (29, 31), and flanking Pro127 in strand F and Pro144 in strand G (Figure 2A) (30). We refer to these external leaflet-exposed regions of weakened transmembrane β-strand hydrogen bonding in PagP as the embrasure, for the window between strands A and B, and as the crenel, for the indentation between strands F and G.
Although the embrasure and crenel provide potential routes for lateral lipid diffusion between the hydrocarbon ruler and outer membrane external leaflet, the PagP crystal structure indicates physical openings to facilitate lateral lipid exchange are blocked by amino acid side chains (30). The question of whether conformational changes gate lipid access or if an altogether different mechanism extrudes the palmitoyl group out of the membrane plane and over the β-barrel wall remains. Here we mutate prolines flanking the PagP embrasure and crenel to cysteines without compromising protein folding and activity in vitro. The influence on the PagP enzymology of physical barricades between the flanking cysteines is wholly consistent with a lateral diffusion lipid gating mechanism. These and other mutational and enzymological results support a model in which lipid acyl chains diffuse laterally between the biological membrane and the interior of an integral membrane lipid-metabolic enzyme.
EXPERIMENTAL PROCEDURES
DNA Manipulations and Protein Purification
Mutagenesis was performed using the primer sets listed in Table 1 and the pETCrcAHΔS vector as described previously (29). PagP was expressed using isopropyl β-D-thiogalactopyranoside (IPTG) induction in E. coli BL21(DE3) for purification and folding as previously reported (17). Protein samples precipitated from water prior to detergent folding were dissolved in a 50:50 solution of 1% formic acid and acetonitrile just prior to electrospray ionization mass spectrometry (ESI-MS). The sample concentration was maintained at 1 ng/μL and the sample injected directly onto a Waters/Micromass Q-TOF Ultima Global (a quadrupole time-of-flight) mass spectrometer. The spectra were reconstructed using the MassLynx 4.0 MaxEnt 1 module. Detergent-folded samples were precipitated for ESI-MS via addition of 1.25 mL of PagP at 1 mg/mL in 10 mM Tris-HCl (pH 8.0) and 0.1% lauryldimethylamine-N-oxide (LDAO) to 3.75 mL of 8 M guanidine hydrochloride (Gdn-HCl) and dialysis overnight against water. To avoid intermolecular cross-linking between Cys-carrying mutants during folding, precipitated samples were initially solubilized in 5 mL of 10 mM Tris-HCl (pH 8.0), 6 M Gdn-HCl, and 10 mM β-mercaptoethanol (βME) and heated at 50 °C for 2 h. The folding buffer also contained 5 mM βME (17). Folded proteins were concentrated through Ni2+ affinity chromatography and dialyzed against 10 mMTris-HCl (pH 8.0) and 0.1% LDAO to remove imidazole and βME. βME was not needed for Cys-carrying mutations after folding because the sulfhydryl groups were protected from air oxidation within the detergent micelle. All protein stock solutions were maintained at a concentration of 2 mg/mL determined using the Edelhoch method (44) or bicinchoninic acid assay (45).
Table 1.
Oligonucleotide Primers Used for Site-Directed Mutagenesis
| name | primer sequencea |
|---|---|
| Pro28Cys Forward | 5′-GATTTATATATTTGTGCCATCACCTGGCATGC-3′ |
| Pro28Cys Reverse | 5′-GCATGCCAGGTGATGGCACAAATATATAAATC-3′ |
| Pro50Cys Forward | 5′-CGCTATAACGAGCGATGCTGGGGTGGCGGTTTTGGC-3′ |
| Pro50Cys Reverse | 5′-GCCAAAACCGCCACCCCAGCATCGCTCGTTATAGCG-3′ |
| Pro127Cys Forward | 5′-CCCTCTCCCGGTTCTACTGTGCTTGGCCTCCGTGGG-3′ |
| Pro127Cys Reverse | 5′-CCCACGGAGGCCAAGCACAGTAGAACCGGGAGAGGG-3′ |
| Pro144Cys Forward | 5′-CAGATGACCTACATTTGCGGTACCTACAACAATGGC-3′ |
| Pro144Cys Reverse | 5′-GCCATTGTTGTAGGTACCGCAAATGTAGGTCATCTG-3′ |
| Lys42Ala Forward | 5′-CTTACGACAAAGAAGCAACCGATCGCTATAACGAG-3′ |
| Lys42Ala Reverse | 5′-CTCGTTATAGCGATCGGTTGCTTCTTTGTCGTAAG-3′ |
Mutagenized codons are underlined.
Copper Phenanthroline Oxidation
Cu2+-(1,10-phenanthroline)3 [Cu(OP)3] can drive disulfide bond formation in membrane environments (46, 47). To obtain a final concentration of 10 mM Cu(OP)3 in 10 mL, 19.8 mg of 1,10-phenanthroline monohydrate was dissolved in 0.5 mL of ethanol and the volume was adjusted with 9.5mL of dH2O before 13.22 mg of Cu(II)SO4 was dissolved. The oxidation was conducted with 1 mM Cu(OP)3 at 4 °C for 2 h in a 15mL falcon tube using 5mL of 0.1mM PagP in 10 mMTris-HCl (pH 8.0) and 0.1% LDAO. The reaction was terminated by addition of 20 μL of 500 mM ethylenediaminetetraacetic acid (EDTA), and the mixture was dialyzed overnight against 10 mM Tris-HCl (pH 8.0) and 0.1% LDAO. Heating protein in the presence of 500 mM βME at 50 °C for 2 h reduced the disulfide bridge (48).
Dibromobimane Alkylation
Dibromobimane (bBBr) introduces a fluorophore across thiol groups lying within 5–7 A° of each other (49). To generate a 100 mM bBBr solution, 0.035 g of bBBr was dissolved in 1 mL of acetonitrile. PagP at 0.1 mM in 10mM Tris-HCl (pH8.0) and 0.1% LDAO was incubated at 37 °C for 1 h with 1mM bBBr. The reaction was terminated by addition of 5mM βME and the mixture dialyzed against 10mM Tris-HCl (pH 8.0) and 0.1% LDAO. Fluorescence spectra were recorded using a Varian Cary Eclipse fluorescence spectrophotometer. Protein samples in 10mM Tris-HCl (pH8.0) and 0.1% LDAO at a concentration of 0.50 mg/mL were excited at 393 nm, and emission spectra were recorded over the range of 400–600 nm.
Quantitation of Sulfhydryl Groups Using Ellman’s Reagent
Ellman’s reagent 5,5′-dithiobis(2-nitrobenzoate) (DTNB) was freshly dissolved in 50mM sodium acetate at 2mM and kept at 4 °C. Free thiols in PagP were reacted under either native (0.1% LDAO) or denaturing (6 M Gdn-HCl) conditions using 0.1 mM DTNB in 100 mM Tris-HCl (pH 8.0). After a 10 min incubation at room temperature, the absorbance at 412 nm was measured using a Varian Cary 50 Bio-UV-visible spectrophotometer and the free thiol concentration estimated using an extinction coefficient of 13600 M−1 cm−1 (50).
SDS–PAGE Analysis
The folded PagP samples were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using a noncommercial 16% Tris-glycine gel prepared under reducing and nonreducing conditions (51). Prior to the gel being loaded, samples were diluted 1:1 with Laemmli solubilization buffer with or without 10mM dithiothreitol (DTT) for reducing conditions and nonreducing conditions, respectively. The gel was run at 120 mV and stained using Coomassie Blue. After being stained for 2 h, the gel was destained overnight in 10% acetic acid. Each lane was loaded with 6 μg of PagP unheated or boiled at 100 °C for 10 min.
Circular Dichroism Spectroscopy
Samples analyzed by near- and far-ultraviolet (UV) circular dichroism (CD) were maintained at a concentration of 1.0 mg/mL (near-UV CD) or 0.3 mg/mL (far-UV CD) in 10 mMTris-HCl (pH 8.0) and 0.1% LDAO using a cuvette with a path length of 1 mm. The samples were analyzed using an Aviv 215 spectrophotometer, which was linked to a Merlin Series M25 Peltier device for temperature control. For each sample, three accumulations were averaged at a data pitch of 1 nm and a scanning speed of 10 nm/min. The temperature was maintained at 25 °C, and data sets were obtained from 200 to 260 nm for far-UV CD and 250–300 nm for near-UVCD. Thermal denaturation profiles were obtained by heating the samples from 20 to 100 °C at 218 nm with a temperature slope of 2 °C/min and a response time of 16 s.
Palmitoyltransferase Assays
Details for the preparation of [32P]Kdo2-lipid A, which carries two units of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo), for thin layer chromatography (TLC) assays have been previously described (17). Phospholipase assays were conducted using phosphatidylcholine (PtdCho) in a volume of 25 μL with di-[1-14C]-16:0-PtdCho to achieve a final concentration of 20 μM (4000 cpm/μL). The lipid was dried under a stream of N2(g) and dissolved in 22.5 μL of reaction buffer containing 0.1 M Tris-HCl (pH 8.0), 10 mM EDTA, and 0.25% n-dodecyl β-D-maltoside (DDM). The reactions were started via addition of 2.5 μL of PagP to give 1 μg/mL in the assay conducted at 30 °C. Reactions were terminated by adding 12.5 μL of the reaction mixture to 22.5 μL of a 1:1 CHCl3/MeOH mixture to generate a two-phase mixture from which 5 μL of the lower organic phase provided 10000 cpm for spotting onto a silica gel 60 TLC plate. The experiment was conducted for 5 h with the reaction being spotted once every hour. The TLC plates were developed in a CHCl3/MeOH/H2O (65:25:4, v/v) solvent system equilibrated in a sealed glass tank. The plates were exposed overnight to a Phosphor Imager screen and developed the following day with a Molecular Dynamics Typhoon 9200 Phosphor Imager. Reaction mixtures were supplemented with 10% (v/v) glycerol or 100 μM 16:0 and 14:0 monoglycerides where indicated. The phospholipase A2 control (4 milliunits/μL) was incubated with 10 mM CaCl2 in place of EDTA.
RESULTS
Proline residues were substituted with cysteine, both singly and in pairs, using site-directed mutagenesis with the primer sets shown in Table 1 and IPTG-inducible PagP expression plasmid pETCrcAHΔS (29). The expressed proteins possess a C-terminal hexahistidine tag and lack the N-terminal signal peptide to target PagP for secretion to the outer membrane. The proteins are expressed as insoluble aggregates, which can be dissolved in Gdn-HCl, purified by nickel affinity chromatography, and folded by dilution into the detergent LDAO (17). Mutations were confirmed both by DNA sequencing of the mutant plasmids and by ESI-MS of the purified proteins. ESI-MS performed on PagP and its derivatives (17, 52) yields experimental protein masses accurately matching theoretical predictions (Table 2). The presence of free thiol groups in PagP leads to the formation of an intermolecular disulfide-linked dimeric species when dilute formic acid is used to solubilize the precipitated samples prior to ESI-MS (17).
Table 2.
Electrospray Ionization Mass Spectrometry of PagP Mutants
| PagP | theoretical mass (Da) (S–S dimer mass) | experimental mass (Da) (S–S dimer mass) |
|---|---|---|
| wild type | 20175.49 | 20176.97 ± 1.96 |
| Pro28Cys | 20181.51 (40361.02) | 20182.70 ± 1.31 (40362.89 ± 2.45) |
| Pro50Cys | 20181.51 (40361.02) | 20182.67 ± 1.46 (40360.88 ± 1.75) |
| Pro127Cys | 20181.51 (40361.02) | 20182.60 ± 1.21 (40361.99 ± 1.85) |
| Pro144Cys | 20181.51 (40361.02) | 20182.52 ± 1.11 (40362.21 ± 1.60) |
| Pro28Cys/Pro50Cys | 20187.54 (40371.08) | 20187.60 ± 1.53 (40372.55 ± 1.50) |
| Pro28Cys/Pro50Cys with Cu(OP)3 | 20185.52 | 20186.43 ± 1.90 |
| Pro127Cys/Pro144Cys | 20187.54 (40371.08) | 20187.83 ± 1.67 (40372.61 ± 1.70) |
| Pro127Cys/Pro144Cys with bBBr | 20375.75 | 20374.90 ± 2.10 |
| Lys42Ala | 20164.48 | 20163.92 ± 1.57 |
Barricading the PagP Embrasure and Crenel by Cysteine Cross-Linking
On the basis of the PagP structure previously crystallized from LDAO, we expected cysteine sulfhydryl groups in Pro → Cys double mutants to localize near Cγ of the corresponding proline residues (Figure 2) (30). On that basis, the embrasure sulfhydryl groups of Pro28Cys/Pro50Cys PagP should localize to a dynamic region of the protein (29, 31) within sufficient proximity to form an intramolecular disulfide bond. In contrast, the crenel sulfhydryl groups of Pro127Cys/Pro144Cys PagP should localize to a structured region separated by a gap of ~5 A° thus requiring a local polypeptide backbone repositioning to establish a 2.05 A ° disulfide bond (30). Interestingly, no spontaneous cysteine oxidation occurs in the detergent micelle environment for either folded Pro28Cys/Pro50Cys PagP or folded Pro127Cys/Pro144Cys PagP, and only in the former mutant could the reaction be driven forward by addition of the membrane permeable oxidizing reagent Cu(OP)3 (Figure 2B) (46, 47).
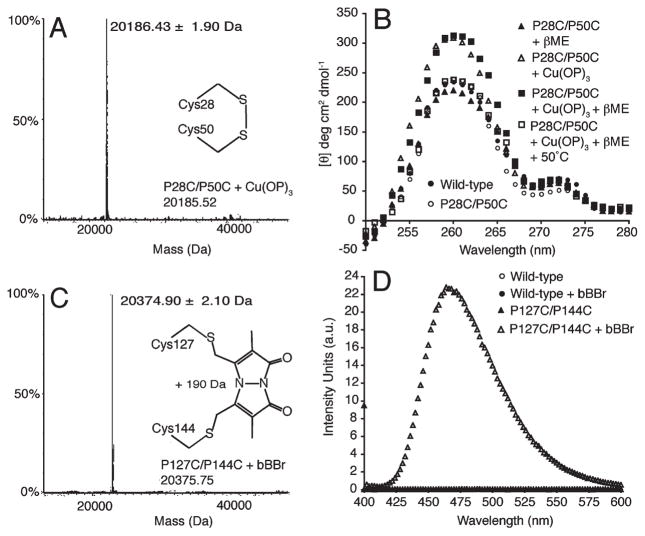
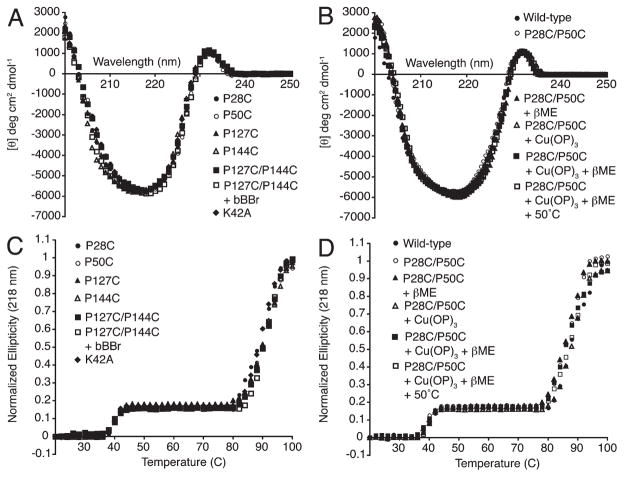
ESI-MS could not reliably measure the 2.02 Da decrease associated with PagP embrasure intramolecular disulfide bond formation (Table 2), but the full mass spectrum clearly excludes any intermolecular dimer formation following Cu(OP)3 oxidation (Figure 3A). To validate intramolecular disulfide bond formation in the folded protein, we took advantage of the intrinsically dissymmetric disulfide bond n → σ* transitions, which, together with contributions from interacting aromatic side chain π → π* transitions, can manifest near-UV CD ellipticity centered around 260 nm (53, 54). A dependence of the CD rotational strengths on the disulfide bond dihedral angle means either positive or negative ellipticity can manifest itself in the near UV (55–57). Intriguingly, PagP embrasure oxidation using Cu(OP)3 introduced positive ellipticity at 260 nm, and subsequent reduction by excess βME occurred only when combined with a 50 °C heat treatment (Figure 3B) (48). When PagP remained folded in 0.1% LDAO, free thiols flanking the PagP embrasure were unreactive to Ellman’s reagent DTNB (50). However, DTNB did react when the PagP embrasure mutant at 6.6 μM was denatured in 6 M Gdn-HCl, thus yielding ~2 molar equiv of sulfhydryl groups measured at 12.1 μM. Cu(OP)3 oxidation in LDAO proceeded to ~95% completion because subsequent DTNB reaction in Gdn-HCl yielded free thiols remaining in the PagP embrasure mutant at only 0.7 μM.
Figure 3.
Barricading the PagP embrasure and crenel by cysteine cross-linking. (A) ESI-MS identifies a single monomeric peak after oxidation of the PagP embrasure. (B) Near-UVCD identifies reversible oxidation and reduction of the PagP embrasure disulfide bond. (C) ESI-MS identifies a single monomeric peak after bimane alkylation of the PagP crenel. (D) Excitation at 393 nm reveals the emission spectrum for the bimane fluorophore.
Our failure to oxidize the folded PagP crenel sulfhydryl groups suggests insufficient conformational dynamics exist to bridge the ~5 A° gap. We opted instead to introduce a barricade using irreversible alkylation with the homobifunctional cross-linking reagent bBBr, which is ideally suited to bridge gaps of ~5 A° (49, 58–60). Intramolecular bimane cross-linking of the PagP crenel was validated by ESI-MS detection of the expected 190 Da increase (Table 2) with no intermolecular dimer formation (Figure 3C). Fluorescence imparted by bBBr bisalkylation was detected with an excitation at 393 nm resulting in a λmax of 466 nm (Figure 3D). The 477 nm λmax reported previously for soluble bBBr-modified proteins (49) suggests an 11 nm blue shift is associated with bimane cross-linking of the PagP crenel, possibly reflecting fluorophore localization within a more hydrophobic environment.
Embrasure and Crenel Barricades Do Not Compromise PagP Structural Integrity
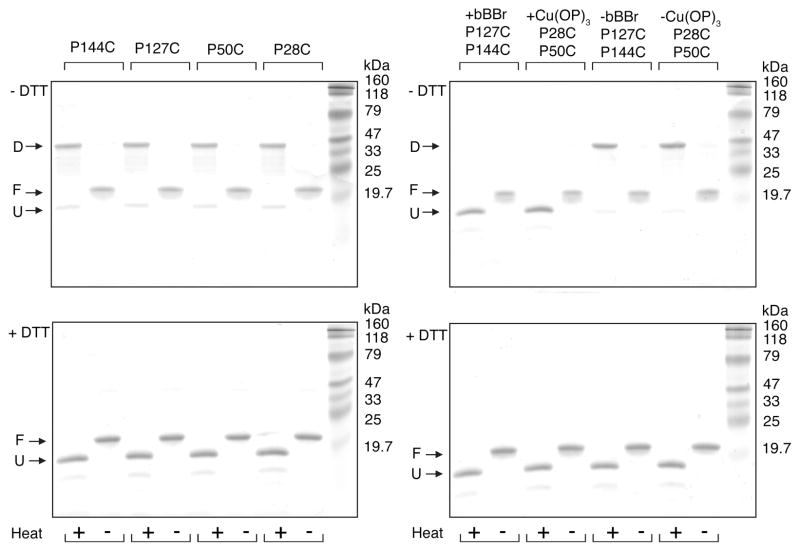
SDS–PAGE provides a facile means of evaluating the effectiveness of PagP folding in vitro. PagP folded in LDAO can be diluted into SDS where the β-barrel structure is maintained until heating induces a denatured state dominated by α-helical structure (17). When resolved by SDS–PAGE, the folded β-form of PagP migrates faster in some gel systems (15, 17) and slower in others (61, 62), but quantitative folding manifests a single band regardless of the gel system employed. According to this criterion, the PagP embrasure and crenel Pro → Cys mutants were all completely folded, as were the double mutants before and after chemical treatments with Cu(OP)3 and bBBr (Figure 4). Under nonreducing conditions, only the unfolded α-forms possessing free sulfhydryl groups were capable of migrating as disulfide-linked dimers. Consistent with our observed Cu(OP)3 requirement to drive embrasure disulfide bond formation in LDAO (Figure 3B), the sulfhydryl groups in the folded β-form PagP Pro → Cys mutants were all protected from spontaneous air oxidation (Figure 4), possibly due to shielding within a more hydrophobic micellar milieu.
Figure 4.
SDS–PAGE of PagP embrasure and crenel Pro→Cys single mutants and the double mutants with and without heat denaturation and under dithiothreitol (DTT) reducing or nonreducing conditions. The folded β-form (F) and the heat-denatured α-form (U) are monomeric species, whereas an intermolecular disulfide bond links the dimeric species (D).
The PagP far-UV CD spectrum provides a measure of secondary structure content and thermal stability (17, 53), but also a measure of tertiary structural integrity because the hydrocarbon ruler floor is coupled to a rare signature in CD known as an aromatic exciton couplet (63). The exciton is a consequence of excited state delocalization between Tyr26 and Trp66 aromatic π → π* transitions, which in their folded geometry near the hydrocarbon ruler floor interact to manifest positive ellipticity at 232 nm together with an equivalent enhancement of the negative ellipticity at 218 nm. The 218 nm signal in PagP thus results from the super positioning of the negative exciton band upon the stronger Cotton effect for the polypeptide n → π* transition, which also arises at 218 nm due to the PagP β-barrel structure (17). The exciton contribution at 218 nm is thermally sensitive because it disappears in response to heating above 40 °C, whereas the n → π* transition is lost only at temperatures above 80 °C (Figure 5). Protein aggregation occurring at temperatures above 80 °C precludes reversible folding in this LDAO detergent system, but the midpoint of 88 °C corresponds to a true melting temperature established previously by differential scanning calorimetry (17). Importantly, the embrasure and crenel Pro → Cys mutants, and the double mutants before and after chemical treatments with Cu(OP)3 and bBBr, all display far-UV CD spectra and 218 nm thermal unfolding profiles indistinguishable from those of wild-type PagP (Figure 5). Furthermore, the intrinsic thermal stability of the PagP β-barrel satisfactorily protects the enzyme from denaturation during 50 °C βME reduction of the oxidized embrasure (Figure 5B,D). We conclude that PagP tertiary structural integrity and stability have not been compromised by the embrasure and crenel Pro → Cys substitutions, by the reversible oxidation or reduction of the embrasure, or by the irreversible alkylation of the crenel.
Figure 5.
Far-UV CD spectroscopy performed on PagP embrasure and crenel Pro → Cys mutants, before and after embrasure oxidation or reduction and crenel alkylation, including the Lys42Ala substitution. Wavelength scans are shown in panels A and B. Thermal melts are shown in panels C and D.
Lys42Ala PagP Is a Dedicated Phospholipase in Vitro
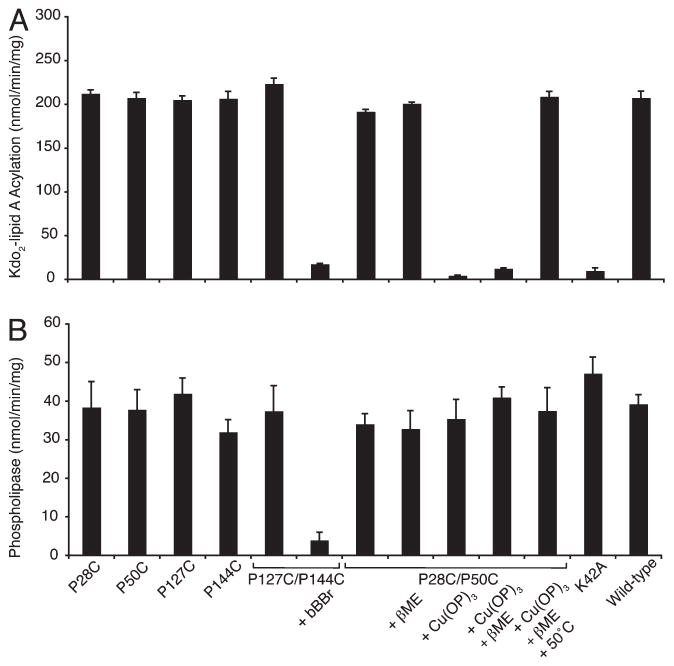
PagP phospholipid:lipid A palmitoyltransferase activity is monitored in vitro using a defined detergent micellar enzymatic assay with TLC separation of radioactive lipid products (17). The inhibitory LDAO detergent used during PagP folding is exchanged by dilution into DDM to support enzymatic activity. PagP is highly selective for a palmitoyl group at the sn-1-position in a glycerophospholipid, but largely unspecific for the polar head group (15). We employ the palmitoyl donor di-16:0-PtdCho and the palmitoyl acceptor [32P]Kdo2-lipid A. In the absence of a lipid A acceptor, a slow phospholipase reaction occurring at a rate that is 20% of that of lipid A palmitoylation can also be monitored in vitro using di-[1-14C]-16:0-PtdCho (Figure 6) (64). Although three PagP amino acid residues were shown previously by mutagenesis to be essential for in vitro lipid A palmitoylation (29), a subsequent survey of all conserved cell surface-exposed ionizable functional groups identified 11 such residues (unpublished results). Uniquely, the Lys42Ala substitution revealed the only PagP mutant to function in vitro as a dedicated phospholipase (Figure 6). Lys42Ala PagP was purified and its structural integrity validated here (Tables 1 and 2 and Figure 5A,C) because it remains the only known PagP specificity determinant for lipid A recognition. Lys42 localization in the disordered L1 surface loop thus identifies the PagP embrasure as a likely locus for the access of lipid A to the hydrocarbon ruler.
Figure 6.
PagP phospholipid:lipid A palmitoyltransferase and phospholipase activity measured for PagP embrasure and crenel Pro → Cys mutants, before and after embrasure oxidation or reduction and crenel alkylation, including the Lys42Ala substitution. Specific activity for the palmitoyltransferase reaction is shown in panel A and that for the phospholipase reaction in panel B.
Lateral Lipid Diffusion through the PagP Embrasure and Crenel
The PagP embrasure and crenel double mutants, together with each single Pro→Cys mutant, all display wild-type lipid A palmitoylation and phospholipase activity (Figure 6). Exposure to βME does not stimulate PagP activity, but closing the embrasure by Cu(OP)3 oxidation eliminates lipid A palmitoylation without affecting phospholipase activity, thus recapitulating Lys42Ala mutant behavior. Subsequent exposure of the oxidized embrasure to excess βME fully restores lipid A palmitoylation (Figure 6), but only after 50 °C heat treatment needed to reduce the embrasure disulfide bond (Figure 3B). Barricading the crenel with bBBr inhibits both lipid A palmitoylation and phospholipase reactions (Figure 6) without compromising PagP structure (Figure 5A,C), thus implicating the crenel as a locus for glycerophospholipid discrimination. The retention of slow phospholipid hydrolysis after embrasure closure indicates water molecule access remains unimpeded. PagP does not function as a phopholipase in vivo where it is continually exposed to LPS (28), but in vitro phospholipid hydrolysis implies the existence of a “hydrophilic funnel” to channel water from bulk solvent to the active site as proposed for other membrane-intrinsic lipid enzymes employing a soluble cosubstrate (10).
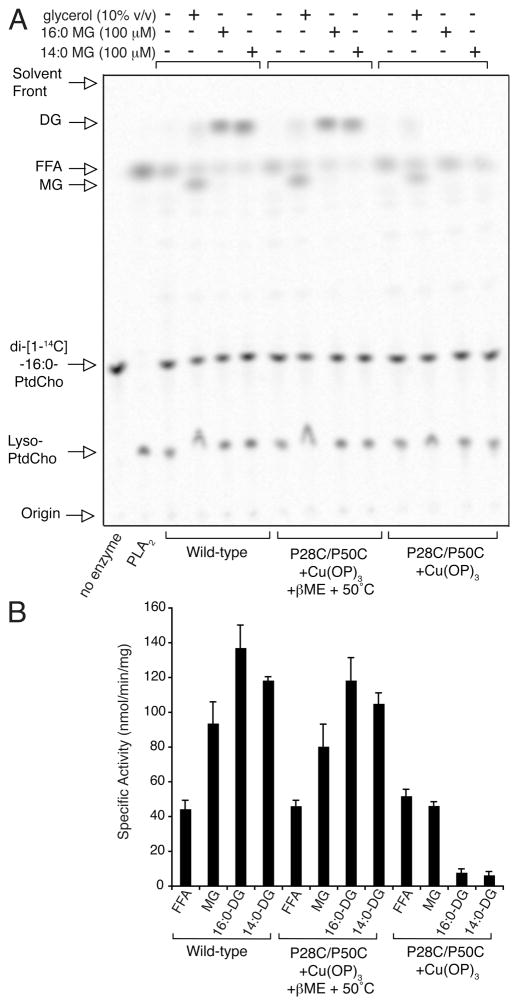
To further validate this model, we took advantage of findings PagP can palmitoylate various nonspecific miscible and fatty alcohols in vitro (64). Addition of methanol, ethanediol, propanediol, or glycerol to the phospholipase reaction results in production of the corresponding palmitoyl esters. The fatty alcohols Triton X-100 and various monoglycerides are also palmitoylated when added to the phospholipase reaction mixture, and the palmitoylation of lysophospholipids can result in palmitoyl group exchange between different classes of phospholipids in vitro (64). We reasoned embrasure closure should distinguish palmitoyl group transfer to acceptors arriving via a lateral route versus a hydrophilic funnel. Using glycerol and its palmitoylated or myristoylated monoglycerides as acceptors, we demonstrate palmitoylation of only the fatty alcohols is completely impeded by embrasure closure (Figure 7). Palmitoyl group transfer to water remains unimpeded, whereas embrasure closure renders glycerol just 2-fold less effective as a palmitoyl acceptor. These findings indicate phospholipid donor palmitoyl groups within the PagP hydrocarbon ruler can be resolved slowly by water ormiscible alcohols arriving from the bulk solvent phase in vitro, but either lipid A or nonspecific fatty alcohol acceptors are more rapidly engaged via a strict lateral route taken through the embrasure.
Figure 7.
PagP palmitoylation of nonspecific miscible and lipid alcohols. (A) Representative TLC of the phospholipase reaction supplemented with and without glycerol and its palmitoylated (16:0) or myristoylated (14:0) monoglycerides (MG). Phospholipid hydrolysis liberates palmitate as the free fatty acid (FFA), which is indicated by reaction with phospholipase A2 (PLA2). Addition of glycerol generates 16:0 MG, whereas addition of 16:0 or 14:0 MG generates the corresponding diglycerides (DG). (B) Specific activities for phospholipid hydrolysis (FFA), glycerol palmitoylation (MG), and monoglyceride palmitoylation (16:0 DG and 14:0 DG) for wild-type PagP are compared to those of PagP having the embrasure either oxidized or reduced.
DISCUSSION
Amphiphilic detergents possessing both polar and nonpolar functional groups often disrupt lipid bilayer structural features essential for the normal function of integral membrane lipid metabolic enzymes, and many such enzymes prove to be recalcitrant toward functional detergent extraction from the membrane environment. The PagP detergent micellar enzymatic assay faithfully replicates key conditions in vivo because enzyme activity is triggered by lipid redistribution associated with a breach in the outer membrane permeability barrier (21, 37–40). Detergent micelles clearly accommodate PagP and both of its natural substrates (15, 17), but the enzyme’s exposure to local polar and nonpolar environments also matches those anticipated to exist in vivo. Shielding introduced cysteine sulfhydryl groups spanning the embrasure and crenel from oxidation only in the folded state (Figure 4), combined with blue shifting of the crenel-spanning bimane fluorophore (Figure 3D), indicates these membrane-intrinsic regions experience a hydrophobic environment also in the micellar milieu. The expected membrane surface exposure of the active site also faithfully anticipates the bulk solvent access revealed by slow phospholipid hydrolysis observed in vitro (Figures 6 and 7). Since bacterial protein secretion is generally coupled to disulfide redox enzymology (65), periplasmic chaperones (66), outer membrane β-barrel docking machinery (67, 68), and dedicated outer membrane lipid transport systems (39, 69), addressing PagP lateral lipid diffusion in vivo is a considerable challenge. By reconstituting this basic process in vitro, we now provide a first validation for the PagP crenel and embrasure as bona fide structural adaptations for controlling lateral lipid diffusion between the outer membrane external leaflet and the β-barrel interior hydrocarbon ruler.
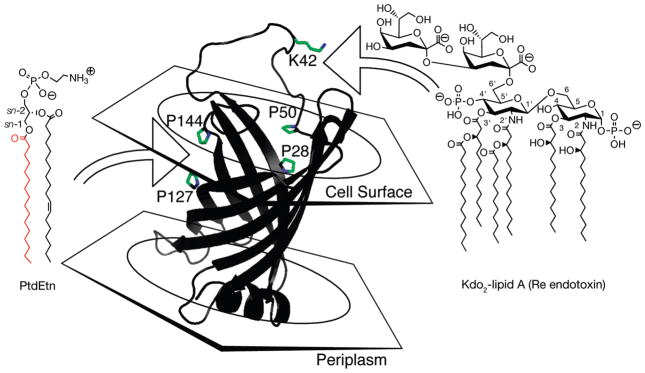
We propose a model in which lateral palmitoyl group diffusion within the PagP hydrocarbon ruler is gated during phospholipid entry via the crenel and during lipid A egress via the embrasure (Figure 8). A nonsequential acyl–enzyme mechanism would be obligatory if only a single lipid gateway existed for PagP hydrocarbon ruler access, but the presence of two such gateways indicates a sequential ternary complex mechanism is equally plausible (70). The crenel and embrasure represent distinctly different regions in terms of protein structural dynamics, perhaps reflecting known differences in PagP substrate selectivity. The PagP crenel is highly structured according to X-ray crystallography (30) and incapable of spanning the ~5 A° gap with an intramolecular disulfide bond (Figures 2A and 3C,D), but we also expect it to be stringently selective for a phospholipid palmitoyl group (Figure 6), with sn-1-palmitoyl phosphatidic acid previously demonstrated to provide the simplest of the known efficient palmitoyl donors (15). In contrast, NMR studies reveal the PagP embrasure to be highly dynamic (29, 31) and remarkably unselective insofar as it was previously demonstrated to recognize multiple lipid A substructures in vitro, including the most basic diacylglucosamine 1-phosphate moiety (lipid X) (15). This dynamic embrasure region is also clearly responsible for the palmitoylation of nonspecific lipid alcohols (Figure 7). After a conformational ordering in and around the L1 loop induces a catalytically competent PagP state, as revealed by prior relaxation–dispersion NMR investigations (31), the single embrasure-spanning Lys42 might suffice in vitro as the sole specificity determinant to neutralize negative charge on Kdo2-lipid A (Figure 8).
Figure 8.
Model for lateral lipid diffusion between the PagP β-barrel interior hydrocarbon ruler and the outer membrane external leaflet. Donor phospholipid palmitoyl groups diffuse laterally into the hydrocarbon ruler through the crenel flanked by prolines 144 and 127. Lipid A acceptors diffuse laterally into the hydrocarbon ruler through the embrasure flanked by prolines 28 and 50. The ε-amino group of lysine 42 in the dynamic L1 surface loop flanking the embrasure likely provides a single lipid A specificity determinant capable of neutralizing negative charge on Kdo2-lipid A in vitro. Disordered residues, including lysine 42 in the L1 loop region, were introduced and energy minimized in the PagP model shown.
We have previously argued the LPS-replete outer membrane external leaflet represents an environment of overly low selective pressure to demand stringent lipid A selectivity from PagP (28), but selective pressure likely acts to protect the phospholipid palmitoyl donor from unproductive hydrolysis. An induced-fit substrate binding mechanism might provide not only a means of shielding substrates from water molecules (71) but also a means of gating lipid access between the membrane and hydrocarbon ruler. The aforementioned ordering of residues in and around the L1 surface loop occurs only in a detergent system known to support PagP activity (31). The crystal structure in the inhibitory LDAO (30) likely represents a latent state before PagP is triggered into an active state by perturbations to outer membrane lipid asymmetry (11). Additional structural data derived from PagP in novel detergent systems (62) might eventually reveal the mechanisms by which lipid substrates and/or their analogues induce the conformational changes governing both catalysis and hydrocarbon ruler gating.
Although highly conserved or invariant proline residues flank the PagP crenel and embrasure, we successfully managed to fold all of the Pro→Cys mutants in vitro (Figure 4). Prolines flanking the crenel are invariant in all PagP homologues so far identified, but those flanking the embrasure are not conserved in the most divergent species, perhaps reflecting known lipid A structural variations (16, 28). Proline residues are clearly not essential to enable PagP hydrocarbon ruler lateral lipid diffusion, but the weakened transmembrane β-strand hydrogen bonding is likely critical. The outer membrane β-barrel transporter FadL channels long chain fatty acids from the bulk solvent directly into the outer membrane external leaflet through another embrasure structure displaying weakened hydrogen bonding, but achieved instead by a proline-independent shifting of the local β-strand registration (72). If LPS received by the outer membrane protein LptD (Imp) (69, 73) proves to be transported through its β-barrel interior, then lateral lipid diffusion during LPS delivery into the outer membrane external leaflet could conceivably require channeling through a crenel structure. Our findings thus have general significance for outer membrane biogenesis processes in bacteria, chloroplasts, and mitochondria (74, 75).
Acknowledgments
We thank Eileen Lo and Konrad Szymanski for conducting pilot studies on K42A PagP, Joel Moktar for assistance with site-directed mutagenesis, and Chris Neale for modelling the PagP L1 surface loop.
Footnotes
This work was supported by CIHR Operating Grant MOP-84329 to R.E.B.
Abbreviations: βME, β-mercaptoethanol; CD, circular dichroism; Cu-(OP)3, Cu2+-(1,10-phenanthroline)3; Kdo, 3-deoxy-D-manno-oct-2-ulosonic acid; bBBr, dibromobimane; DTNB, 5,5′-dithiobis(2-nitrobenzoate); DTT, dithiothreitol; DDM, n-dodecyl β-D-maltoside; ESI-MS, electrospray ionization mass spectrometry; EDTA, ethylenediaminetetraacetic acid; Gdn-HCl, guanidine hydrochloride; IPTG, isopropyl β-D-thiogalactopyranoside; LDAO, lauryldimethylamine-N-oxide; LPS, lipopolysaccharide; NMR, nuclear magnetic resonance; PDB, Protein Data Bank; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; SDS–PAGE, sodiumdodecyl sulfate–polyacrylamide gel electrophoresis; TLC, thin layer chromatography; UV, ultraviolet.
References
- 1.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins, and Membranes. Elsevier; Amsterdam: 2002. [Google Scholar]
- 3.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 5.Martinez Molina D, Wetterholm A, Kohl A, McCarthy AA, Niegowski D, Ohlson E, Hammarberg T, Eshaghi S, Haeggstrom JZ, Nordlund P. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson AD, McKeever BM, Xu S, Wisniewski D, Miller DK, Yamin TT, Spencer RH, Chu L, Ujjainwalla F, Cunningham BR, Evans JF, Becker JW. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 7.Ago H, Kanaoka Y, Irikura D, Lam BK, Shimamura T, Austen KF, Miyano M. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature. 2007;448:609–612. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 8.van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 10.Forneris F, Mattevi A. Enzymes without borders: Mobilizing substrates, delivering products. Science. 2008;321:213–216. doi: 10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
- 11.Bishop RE. Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim Biophys Acta. 2008;1778:1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, de Haan A, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutten L, Mannie JP, Stead CM, Raetz CR, Reynolds CM, Bonvin AM, Tommassen JP, Egmond MR, Trent MS, Gros P. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium. Proc Natl Acad Sci USA. 2009;106:1960–1964. doi: 10.1073/pnas.0813064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 15.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan MA, Neale C, Michaux C, Pomes R, Prive GG, Woody RW, Bishop RE. Gauging a hydrocarbon ruler by an intrinsic exciton probe. Biochemistry. 2007;46:4565–4579. doi: 10.1021/bi602526k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilione MR, Pishko EJ, Preston A, Maskell DJ, Harvill ET. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect Immun. 2004;72:2837–2842. doi: 10.1128/IAI.72.5.2837-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston A, Maxim E, Toland E, Pishko EJ, Harvill ET, Caroff M, Maskell DJ. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol Microbiol. 2003;48:725–736. doi: 10.1046/j.1365-2958.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 20.Robey M, O’Connell W, Cianciotto NP. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AE, Kim SH, Liu F, Jia W, Vinogradov E, Gyles CL, Bishop RE. PagP activation in the outer membrane triggers R3 core oligosaccharide truncation in the cytoplasm of Escherichia coli O157:H7. J Biol Chem. 2008;283:4332–4343. doi: 10.1074/jbc.M708163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki K, Ernst RK, Miller SI. 3-O-Deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through toll-like receptor 4. J Biol Chem. 2004;279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 23.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 24.Muroi M, Ohnishi T, Tanamoto K. MD-2, a novel accessory molecule, is involved in species-specific actions of Salmonella lipid A. Infect Immun. 2002;70:3546–3550. doi: 10.1128/IAI.70.7.3546-3550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanamoto K, Azumi S. Salmonella-type heptaacylated lipid A is inactive and acts as an antagonist of lipopolysaccharide action on human line cells. J Immunol. 2000;164:3149–3156. doi: 10.4049/jimmunol.164.6.3149. [DOI] [PubMed] [Google Scholar]
- 26.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 27.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 28.Bishop RE. The lipid A palmitoyltransferase PagP: Molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 29.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBOJ. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang PM, Bishop RE, Kay LE. The integral membrane enzyme PagP alternates between two dynamically distinct states. Proc Natl Acad Sci USA. 2004;101:9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol. 2006;1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 35.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leive L. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun. 1965;21:290–296. doi: 10.1016/0006-291x(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 37.Jia W, Zoeiby AE, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 38.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 42.Gao J, Bosco DA, Powers ET, Kelly JW. Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity substantially stabilizes proteins. Nat Struct Mol Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li SC, Goto NK, Williams KA, Deber CM. α-Helical, but not β-sheet, propensity of proline is determined by peptide environment. Proc Natl Acad Sci USA. 1996;93:6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 45.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 46.Loo TW, Bartlett MC, Clarke DM. Val133 and Cys137 in transmembrane segment 2 are close to Arg935 and Gly939 in transmembrane segment 11 of human P-glycoprotein. J Biol Chem. 2004;279:18232–18238. doi: 10.1074/jbc.M400229200. [DOI] [PubMed] [Google Scholar]
- 47.Lee GF, Lebert MR, Lilly AA, Hazelbauer GL. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl cross-linking in vivo. Proc Natl Acad Sci USA. 1995;92:3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinrikson RL. The selective S-methylation of sulfhydryl groups in proteins and peptides with methyl-p-nitrobenzenesulfonate. J Biol Chem. 1971;246:4090–4096. [PubMed] [Google Scholar]
- 49.Kim JS, Raines RT. Dibromobimane as a fluorescent crosslinking reagent. Anal Biochem. 1995;225:174–176. doi: 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]
- 50.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 51.Bishop RE, Weiner JH. Overproduction, solubilization, purification and DNA-binding properties of AmpR from Citrobacter freundii. Eur J Biochem. 1993;213:405–412. doi: 10.1111/j.1432-1033.1993.tb17775.x. [DOI] [PubMed] [Google Scholar]
- 52.Whitelegge JP, le Coutre J, Lee JC, Engel CK, Prive GG, Faull KF, Kaback HR. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woody RW. Circular dichroism. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 54.Sreerama N, Manning MC, Powers ME, Zhang JX, Goldenberg DP, Woody RW. Tyrosine, phenylalanine, and disulfide contributions to the circular dichroism of proteins: Circular dichroism spectra of wild-type and mutant bovine pancreatic trypsin inhibitor. Biochemistry. 1999;38:10814–10822. doi: 10.1021/bi990516z. [DOI] [PubMed] [Google Scholar]
- 55.Bewley TA. Optical activity of disulfide bonds in proteins. 1 Studies on plasmin modified human somatotropin. Biochemistry. 1977;16:209–215. doi: 10.1021/bi00621a008. [DOI] [PubMed] [Google Scholar]
- 56.Kurapkat G, Kruger P, Wollmer A, Fleischhauer J, Kramer B, Zobel E, Koslowski A, Botterweck H, Woody RW. Calculations of the CD spectrum of bovine pancreatic ribonuclease. Biopolymers. 1997;41:267–287. doi: 10.1002/(SICI)1097-0282(199703)41:3<267::AID-BIP3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 57.Bewley TA. Optical activity of disulfide bonds in proteins: Studies on human choriomammotropin and bovine pituitary somatotropin. Biochemistry. 1977;16:4408–4414. doi: 10.1021/bi00639a013. [DOI] [PubMed] [Google Scholar]
- 58.Loo TW, Clarke DM. Identification of residues within the drug-binding domain of the human multidrug resistance P-glycoprotein by cysteine-scanning mutagenesis and reaction with dibromobimane. J Biol Chem. 2000;275:39272–39278. doi: 10.1074/jbc.M007741200. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharjee H, Rosen BP. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J Biol Chem. 1996;271:24465–24470. doi: 10.1074/jbc.271.40.24465. [DOI] [PubMed] [Google Scholar]
- 60.Wu J, Voss J, Hubbell WL, Kaback HR. Site-directed spin labeling and chemical crosslinking demonstrate that helix V is close to helices VII and VIII in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1996;93:10123–10127. doi: 10.1073/pnas.93.19.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huysmans GH, Radford SE, Brockwell DJ, Baldwin SA. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP. J Mol Biol. 2007;373:529–540. doi: 10.1016/j.jmb.2007.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michaux C, Pomroy NC, Prive GG. Refolding SDS-denatured proteins by the addition of amphipathic cosolvents. J Mol Biol. 2008;375:1477–1488. doi: 10.1016/j.jmb.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 63.Grishina IB, Woody RW. Contributions of tryptophan side chains to the circular dichroism of globular proteins: Exciton couplets and coupled oscillators. Faraday Discuss. 1994:245–262. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 64.Bishop RE, Raetz CR. Fortieth Interscience Conference on Antimicrobial Agents and Chemotherapy; Toronto, ON. 2000. [Google Scholar]
- 65.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 66.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 67.Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 69.Sperandeo P, Deho G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 70.Segel IH. Enzyme kinetics: Behavior and analysis of rapid equilibrium and steady state enzyme systems. Wiley; New York: 1975. [Google Scholar]
- 71.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: The long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walther DM, Rapaport D, Tommassen J. Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: Evolutionary conservation and divergence. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0029-z. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: The role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]