Abstract
Lipocalins, a widespread multifunctional family of small proteins (15–25 kDa) have been first described in eukaryotes and more recently in Gram-negative bacteria. Bacterial lipocalins belonging to class I are outer membrane lipoproteins, among which Blc from E. coli is the better studied. Blc is expressed under conditions of starvation and high osmolarity, conditions known to exert stress on the cell envelope. The structure of Blc that we have previously solved (V. Campanacci, D. Nurizzo, S. Spinelli, C. Valencia, M. Tegoni, C. Cambillau, FEBS Lett. 562 (2004) 183–188.) suggested its possible role in binding fatty acids or phospholipids. Both physiological and structural data on Blc, therefore, point to a role in storage or transport of lipids necessary for membrane maintenance. In order to further document this hypothesis for Blc function, we have performed binding studies using fluorescence quenching experiments. Our results indicate that dimeric Blc binds fatty acids and phospholipids in a micromolar Kd range. The crystal structure of Blc with vaccenic acid, an unsaturated C18 fatty acid, reveals that the binding site spans across the Blc dimer, opposite to its membrane anchored face. An exposed unfilled pocket seemingly suited to bind a polar group attached to the fatty acid prompted us to investigate lyso-phospholipids, which were found to bind in a nanomolar Kd range. We discuss these findings in terms of a potential role for Blc in the metabolism of lysophospholipids generated in the bacterial outer membrane.
Keywords: Lipocalins, Blc, Crystal structure, Lysophospholipids, Fatty acids
1. Introduction
The lipocalins are a widespread family of proteins identified initially in eukaryotes [1] and more recently in Gram-negative bacteria [2,3]. Lipocalins are carriers of lipophilic molecules whose functions, however, are often elusive and very diverse [4] (see also papers in Biochem. Biophys. Acta Vol. 1482). Retinol binding protein (RBP), which possess a well documented function and transport mechanism, has been the first lipocalin whose 3D structure has been solved [5]. Other lipocalins whose structures are known [6] include carriers of pheromones [7] and odorant molecules [8,9].
The lipocalin fold comprises an 8 stranded β-barrel followed by an α-helix at the C-terminus [6]. The number of disulfide bridges can vary from none to three in mammalian lipocalins. The amino acid sequences of lipocalins are very poorly conserved, with their most general sequence signature being the GXW motif at the N-terminus [1,6]. Sequence comparisons and evolutionary analyses of lipocalins have led to a classification in which ~14 clades have been identified [10]. Bacterial lipocalins belong to Clade 1 (the root clade) while the majority of eukaryotic lipocalins ranging from plants to invertebrates and mammals are found in the higher clades (2–14).
Many bacterial lipocalins are lipoproteins which contain a type 2 signal peptide allowing for export into the periplasm and anchoring in the inner leaflet of the outer membrane [2,3]. In the larger of the two identified groups, one or two cysteines are observed and no disulfide bridge is found. The first cysteine, located immediately after the signal peptide, is attached to the membrane anchor lipid forming the N-acyl-Ssn-1,2-diacylglycerylcysteine moiety at the N-terminus [2,3]. A smaller subgroup of bacterial lipocalins contains a putative disulfide bridge and its members have diverse predicted localizations in the cytoplasm, periplasm and the inner and outer membranes [2,3].
The paradigm for bacterial lipocalins is provided by Blc (Fig. 1) from Escherichia coli, the first bacterial lipocalin that has been identified [2] and whose 3D structure has been solved [11]. The structure of Blc at 1.8Å resolution revealed a fold similar to that of the moth bilin binding protein (BBP) [12] and the presence of an elongated and open cavity with the proper size to accommodate fatty acids or phospholipids. Blc is an outer-membrane bound protein, facing the periplasmic space. It is expressed during the stationary growth phase and under conditions of high osmolarity, during which the cell envelope suffers stress and requires maintenance [2,3].
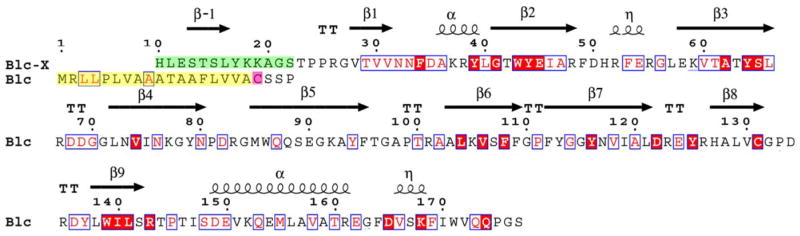
Fig. 1.
Sequence of the native Blc (Blc) and of the Blc construct used in this study (Blc-X). The N-terminus of Blc-X (green) replaces the N-terminus sequence in native Blc (yellow). The red filled boxes indicate identity with ApoD and Lazarillo, while the white ones indicate similarity. The secondary structure is represented as arrows and spirals (strands and helices) above the sequence. The cysteine linked to a membrane anchor in native Blc is identified in pink.
Gram-negative bacterial outer membranes are built from diverse lipids and proteins. Lipid biosynthesis is restricted primarily to the inner (cytoplasmic) membrane. Many enzymes of lipid metabolism are integral membrane proteins, and among them only two structures are known. These are OMPLA [13] and PagP [14], both β-barrel proteins from the outer membrane of E. coli. In membrane metabolism, lipid trafficking events such as lateral diffusion and translocation between opposite leaflets are also essential processes, since the organization of lipids in the outer membrane is highly asymmetric; lipopolysaccharide (LPS) is located exclusively in the extracellular outer leaflet whereas phospholipids are normally restricted to the periplasmic inner leaflet. Although some trafficking events may occur spontaneously following chemical gradients or signals, some others may require a dedicated transporter, and Blc might be one of those. Two close eukaryotic neighbors of Blc, namely Lazarillo [15] and ApoD [16], are also believed to interact with membrane lipids [17].
In order to investigate the hypothesis that Blc might be a fatty acids/phospholipids/lysophospholipids carrier, we have performed binding experiments using tryptophan fluorescence quenching. We have also determined the crystal structure of Blc complexed to a fatty acid, vaccenic acid, and identified its dimeric functional organization. A likley role for Blc in the metabolism of lysophospholipids (LPLs) is indicated by the nanomolar affinity that Blc displays towards these compounds.
2. Materials and methods
2.1. Protein expression and purification
Sub-cloning and expression strategies used for our E. coli targets, including the blc gene, have already been described elsewhere [11,18]. Briefly, residues 1–22 in the native Blc have been replaced by the sequence MSYYHHHHHHLESTSLYKKAGS, coming from the Gateway pDEST17 vector [11], thus removing the N-terminal cysteine.
2.2. Tryptophan fluorescence quenching studies
Fatty acids and Phospholipids, PtdGro, PtdEtn and extracts, were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Pools of 100% methanolic solutions of the ligands were freshly prepared. Fluorescence quenching was measured using a Cary Eclipse (Varian) using a right angle configuration, at 20 °C by using 2.5-nm excitation and 10-nm emission bandwidths. The excitation wavelength was 280 nm and the emission spectra were measured between 290 and 540 nm. In all experiments the final methanol concentration in the cuvette was kept below 1%. Binding samples contained 1 μM protein in 10 mM Tris buffer, 25 mM NaCl, pH 8.0; ligands were used at concentrations between 0.02 and 22.5 μM. For the experiments with LPLs, the final Kd values were obtained with a protein concentration of 0.1 μM.
In order to estimate the affinity of the compounds for Blc, the fluorescence intensities at 348 nm, the maximum of emission, at increasing concentrations of quencher were plotted versus the quencher concentration. The Kd values and the standard errors were estimated by non-linear regression using Prism 3.02 (GraphPad software, Inc) following procedures already described [19].
2.3. Static light scattering study
Static light scattering experiments were performed with a Malvern Zetasizer nano S instrument (Malvern, UK). The protein concentration was varied between 1 and 20 mg/mL, and data were collected at 25 °C against a toluene standard.
2.4. Crystal structure determination
Crystals of the protein were grown at 20 °C using sitting drops and the nano-drops technology [20] followed by optimization. The Blc/vaccenic acid complex was prepared by adding vaccenic acid (molar ration 5:1) dissolved in methanol to Blc at 6–8 mg/mL in HEPES 5 mM, NaCl 150 mM, pH 7.5. A volume of 100 nL of the protein complex was mixed with 100 nL of precipitant solution consisting of sodium citrate 800–900 mM, sodium borate 50 mM, pH 7.0–7.5. Crystals grew within 1–3 days and proved to belong to the orthorhombic space group P212121 and are isostructural with the native Blc [11] (a = 57.9.0Å, b = 81.3Å, c = 89.0Å ) with two molecules per asymmetric unit (Vm = 2.6Å3/Da; 59 % solvent).
Data were collected on a single flash-cooled crystal (25% glycerol) on beamline ID14-EH4 at the European Synchrotron Radiation Facility (ESRF) in Grenoble (France) using an ADSC Quantum 4 CCD detector. A total of 180 images have been collected using a 1° oscillation range per image at Å wavelength. The data set were processed with MOSFLM and scaled using SCALA from the CCP4 [21] suite (Table 1). The isostructural crystals enabled the use of the native Blc model for initial phasing. Refinement was made with REFMAC [22] and model rebuilding with Turbo-Frodo [23]. Statistics for data collection and refinement are given in Table 1. Figures have been prepared with PyMol [24]. The coordinates have been deposited in the Protein Data Bank at RCSB (http://www.rcsb.org/pdb/) as entry 2ACO.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Resolution limits | 1.8–60.0/1.8–1.85 |
| % of data> 1σ (overall/last shell)a | 97.1 (89.1) |
| Overall I/σ (I) (overall/last shell)a | 13.7 (2.3) |
| R-merge (%) (overall/last shell)a | 8.9 (32.7) |
| Refinement | |
| Protein/ligand/solvent atoms | 2684/20/449 |
| Resolution limits (Å) | 1.8–60 |
| R-/R-free value (%) | 17.1/21.0 |
| r.m.s.d. on bonds (Å) and angles (°) | 0.011/1.3 |
| Mean B factor (Å2) protein | 15.2 |
| Vaccenic acid | 27.2 |
Last shell: 1.8–1.85Å.
3. Results
3.1. Ligand binding studies using tryptophan fluorescence quenching
Fluorescence reporter molecules such as amino anthracene (AMA), NPS or 1-N-phenylnaphthylamine (NPN) have proved to be very useful with some lipocalins, such as odorant binding proteins (OBPs) [7,25–27]. Such reporters display a strong increase of fluorescence when accommodated in the hydrophobic ligand-binding environment of lipocalins known as the calyx [7,25]. Chasing these fluorescence reporters with ligands provide a fast and easy tool to estimate their true affinity for the host protein [19,25]. The three reporters tested with Blc, namely AMA, NPS or NPN, however, failed to display any fluorescence increase in the presence of the protein, indicating that they were unable to bind into its hydrophobic pocket. Visual inspection of the Blc structure indicated that among the four tryptophans present in the protein, two belong to the wall of the pocket (Trp139 and Trp43), while the other two (Trp86 and Trp171) are located at the external surface. Based on this observation, we thought that the two former tryptophans might be excellent binding reporters as their fluorescence would probably be quenched when adding a hydrophobic ligand. As expected, quenching of the Blc tryptophan fluorescence (max emission at 348 nm) occurred with most of the lipidic ligands (Table 2). We also assayed, as non-specific references, two “outlier” bulkier molecules, ampicillin and retinol. They both displayed Kd values higher than FAs or PLs, 5 and 20 μM, respectively.
Table 2.
Binding results, obtained with tryptophan quenching, of fatty acids, phospholipids and other molecules with Blc
| Fatty acids | Nr. carbons/chemical structure | Kd (μM) | Relative affinity |
|---|---|---|---|
| Myristic | 14
|
3.3 ± 0.3 | 0.8 |
| Palmitoleic | 16
|
3.1 ± 0.2 | 0.9 |
| Palmitic acid | 16
|
2.7 ± 0.2 | 1 |
| cis-Vaccenic | 18
|
2.5 ± 0.3 | 1.1 |
| Oleic | 18
|
2.0 ± 0.1 | 1.35 |
| Arachidonic | 20
|
1.8 ± 0.1 | 1.5 |
| Phospholipids | |||
| PtdGro | 4.7 ± 0.3 | 0.57 | |
| E.coli total mix | 2.5 ± 0.3 | 1.1 | |
| E.coli polar mix | 2.9 ± 0.4 | 0.9 | |
| Lysophospholipids | 1-Acyl-2-Hydroxy-sn-Glycero-3-Phosphoethanolamine | ||
| Oleoyl | 0.039 ± 0.006 | 70 | |
| Palmitoyl | 0.011 ± 0.001 | 245 | |
| Other molecules | |||
| Retinol |
|
20.2 ± 2 | 0.13 |
| Ampicillin |
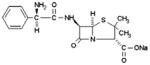
|
5.0 ± 0.3 | 0.54 |
Kd values are given in μM.
Since most, if not all, compounds were poorly soluble and had to be dissolved in methanol, we verified that the presence of small amounts of methanol did not significantly influence the binding. Among all ligands under study, fatty acids, ranging in acyl-chain length from C14 to C20, display good affinity for Blc. Binding of fatty acids, however, does not indicate a strong specificity as all the Kd values are in the narrow range of 3.3 μM (C14) and 1.8 μM (C20). A loose linear correlation is observed between the alkyl chain length and the Kd values, where the longer fatty acids yield the best affinity. The binding of three phospholipids was also studied. Pure phosphatidylglycerol (PtdGro) displayed a peculiar two range behavior: in the first range, quenching occurs as with fatty acids, and yields a Kd of 4.7 μM. However, upon increasing the concentration of PtdGro between 15 and 100 equiv. (with respect to the protein), another increase in fluorescence occurred at 316 nm, which reaches saturation at 50 equiv. We attributed this latter effect to surface tryptophans probably becoming involved in large PtdGro aggregates. Deconvolution of the signals indicates that the new band has its maximum at 308 nm. Total E. coli phospholipid extracts were also assayed with two extracts commercially available (Section 2). The total extract is composed of phosphatidylethanolamine (PtdEtn) (57.5%), PtdGro (15.1%), cardiolipin (9.8%) and a mix of non-documented compounds (17.6%). The “polar extract” contains 67%, 23.2% and 9.8% of the first three above mentioned products, respectively, and no undocumented compounds. Both extracts bind with constants identical within the experimental errors, 2.5 and 2.9 μM, respectively (Table 2), values comparable to those obtained with fatty acids.
Two LPLs have been chosen to complete the panel of putative ligands: oleoyl-sn-2-lyso-PtdEtn (LOP) and palmitoylsn-2-lyso-PtdEtn (LPP). Contrary to the “flat” range of Kd constants obtained with FAs or PLs, LPLs exhibited much higher affinities. LPP has 70 times higher affinity than oleic acid, while LPP has 245 times higher affinity than palmitic acid. Interestingly, while the longer acyl-chain and double bond in oleic acid gave a slightly favourable effect on the affinity compared to palmitic acid (ratio 1.35), LOP binds 5 times less well than LPP. This may reveal a much more constrained binding for LPLs than for fatty acids.
3.2. Crystal structure with vaccenic acid
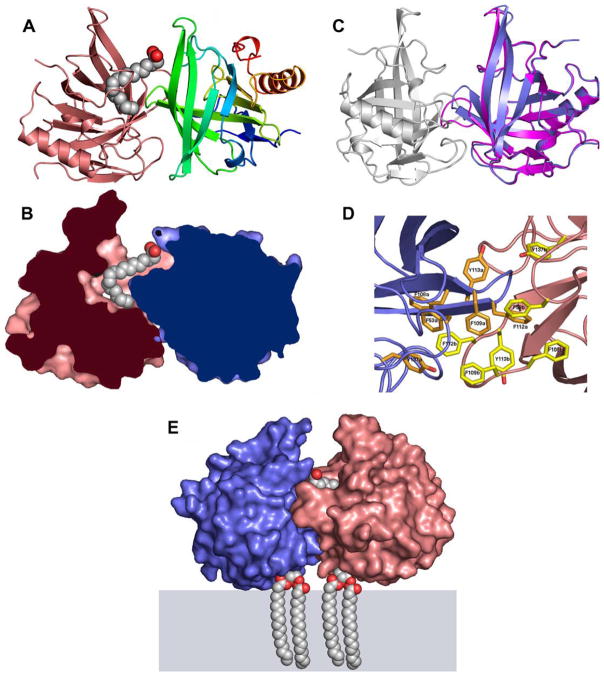
We have co-crystallized Blc in the presence of vaccenic acid, and collected data at 1.8Å resolution. All attempts to crystallize Blc-LPLs complexes were unsuccessful, and vaccenic acid has provided the best diffracting crystals. The structure is isostructural with that of the native Blc. Two monomers of Blc are contained in the asymmetric unit. Blc monomer has a typical lipocalin fold consisting of a β-barrel with eight anti-parallel strands and an α-helix at the C-terminus. Inside the cavity of monomer B, a well defined elongated electron density could easily accommodate a vaccenic acid molecule, while monomer A was empty. The molecule is bent at the position of the cis-double bond (Z11–12). Careful examination of monomers A and B relationship revealed that they interact tightly together: the interaction covers 786Å2 and 825Å2 of water accessible surface (WAS) area, on monomer A and B, respectively, upon binding (Fig. 2). The total WAS area of each monomer being 7800Å2, the buried WAS area represents ~10% of the total surface, a value indicating that dimerisation is not a crystallization artefact [28]. For comparison, these values of interacting surface area are comparable to those observed in immunoglobulin fragments/protein complexes (Table 3A) [29]. The interaction between the two monomers was present and essentially identical in the original native structure, but it escaped our observation and is not described in [11].
Fig. 2.
X-ray structure of Blc in complex with vaccenic acid. (A) Electon density map (2fo–fc) contoured in the binding site of Blc with vaccenic acid (red sticks) inside. (B) Ribbon view of Blc with vaccenic acid inside (spheres). Monomer B (left) is pink; monomer A (right) is rainbow colored from N-(blue) to C-terminus (red). (C) Compact view of Blc sliced at the level of the inside cavity, illustrating shape complementarity of both monomers and of vaccenic acid. Same orientation as in (B); vaccenic acid in spheres; monomer B (left) is pink; monomer A (right) is blue. (C) View of Blc dimer after superposition of monomers A and B (blue and pink) showing the structural difference involving strands 6 and 7 and the loop in between. (D) Inter-monomers hydrophobic core. Monomer B (right) is pink with yellow side-chains; monomer A (right) is blue with orange side-chains. (E) Compact view of the Blc dimer attached to the membrane anchor lipid forming a S-sn-1,2-diacylglycerylcysteine moiety attached to the first cysteine (computer model). Monomer B (right) is pink and monomer A (right) is blue; vaccenic acid and lipidic anchors are represented as spheres.
Table 3.
List of interactions within Blc dimer (A) and between Blc and vaccenic acid (B) as illustrated by the WAS (Å2) area buried in the interaction
| Res | Nr | WAS | Res | Nr | WAS | ||
|---|---|---|---|---|---|---|---|
| A | |||||||
| GLY | 21 | A | 11 | LYS | 19 | B | 6 |
| ARG | 52 | A | 33 | ALA | 20 | B | 54 |
| PHE | 53 | A | 28 | GLY | 21 | B | 53 |
| ARG | 67 | A | 15 | LEU | 22 | B | 8 |
| ASP | 69 | A | 20 | PRO | 25 | B | 54 |
| SER | 89 | A | 4 | HIS | 51 | B | 4 |
| GLU | 90 | A | 2 | ARG | 52 | B | 59 |
| LYS | 92 | A | 53 | PHE | 53 | B | 57 |
| TYR | 94 | A | 42 | VAL | 106 | B | 6 |
| LYS | 105 | A | 64 | PHE | 108 | B | 27 |
| SER | 107 | A | 1 | PHE | 109 | B | 83 |
| PHE | 108 | A | 10 | PRO | 111 | B | 73 |
| PHE | 109 | A | 89 | PHE | 112 | B | 138 |
| GLY | 110 | A | 40 | TYR | 113 | B | 36 |
| PRO | 111 | A | 84 | GLY | 114 | B | 24 |
| PHE | 112 | A | 100 | PRO | 133 | B | 37 |
| TYR | 113 | A | 85 | ASP | 134 | B | 43 |
| GLY | 114 | A | 13 | ASP | 136 | B | 7 |
| PRO | 133 | A | 28 | TYR | 137 | B | 51 |
| ASP | 134 | A | 18 | TRP | 139 | B | 3 |
| TYR | 137 | A | 46 | Total | 823 | ||
| Total | 786 |
| Nr | WAS | ||
|---|---|---|---|
| B | |||
| HIS | 51 | B | 9 |
| ARG | 52 | B | 9 |
| PHE | 53 | B | 51 |
| GLU | 54 | B | 4 |
| ASN | 76 | B | 13 |
| SER | 89 | B | 20 |
| VAL | 106 | B | 2 |
| PHE | 108 | B | 43 |
| PHE | 109 | B | 19 |
| GLY | 114 | B | 1 |
| Total | B | 171 | |
| ARG | 52 | A | 40 |
| PHE | 53 | A | 2 |
| PHE | 109 | A | 16 |
| PRO | 111 | A | 10 |
| PHE | 112 | A | 21 |
| Total | A | 89 |
The columns A and B refer to the Blc monomer involved in the interaction B.
The oligomerization state of Blc, alone or in complex with palmitoleic acid, was investigated using static light scattering. In both cases, Blc appears as a dimer of 43.1 ± 2.3 kDa and 38 ± 3 kDa, respectively. This confirms the crystallographic data that Blc dimerisation is an intrinsic property of the protein. It is interesting to note that such an asymmetric binding pocket (as shown in Fig. 2A and B) in a homodimer is maintained even without a ligand present. Furthermore, binding of two ligands to the Blc dimer is impossible due to steric clashes between them.
The comparison of subunits A and B indicate significant conformational differences that we think are necessary for dimerisation (Fig. 2C). In order to fit within the other monomer, strands 6 and 7, and particularly the loops between them, move up to 5Å in one monomer relative to the other (Fig. 2C). The dimer interface involves in large part these loops, and other residues among which the aromatics Tyr113, Tyr137, and several surface-exposed phenylalanines including Phe53 A and B, Phe108 A and B, Phe109 A and B and Phe112 A and B together form an inter-dimer hydrophobic core (Fig. 2D, Table 3A). The presence of many exposed phenylalanines is an uncommon feature at the surface of a monomeric globular protein, and has to be regarded as a hallmark of protein-protein or protein-lipid interactions. Both subunits are involved in the binding site of Blc, accounting for the stoichiometry of one vaccenic acid molecule per dimer (Fig. 2A and B). The vaccenic acid interacts with both subunits and covers 89Å2 and 171Å2 of the WAS area of monomers A and B, respectively (Table 3B).
It should be stressed that Blc is anchored to membranes by a covalently attached lipid modification at the amino terminus of the protein (Fig. 1). The manner in which the lipid-anchor of each monomer is arranged relative to the other is a critical issue. Our Blc construct has the signal peptide and the first four residues (CSSP) removed and replaced by the Gateway ATTB1 sequence (Fig. 1) [11,30]. We have modelled into our dimeric structure the four original residues of Blc, as well as the major part of the anchoring lipid, S-sn-1,2-diacylglycerylcysteine, that we included at each N-terminal cysteine. This model reveals that the lipid-anchors of each of the 2 monomers are close to each other in the dimer, and are located on the same face in a geometry compatible with the insertion of the dimer into the membrane (Fig. 2E). The binding site for vaccenic acid is located opposite to the membrane insertion site where it is expected to face the periplasmic space (Fig. 2E).
4. Discussion
Using intrinsic fluorescence quenching, Blc has been shown to bind several FAs and PLs with μM affinities. In contrast, Blc binds weakly to retinol and does not bind fluorescent reporters such as AMA, amino-naphthalene sulfonate (ANS) or NPN. Ampicillin was found to bind Blc, but with slightly lower affinity than FAs. Blc exhibits, therefore, some preference for FAs and PLs over other compounds, the Kd differences being less than a factor 2 (Table 2). LPLs, in contrast, bind Blc two orders of magnitude better than the corresponding FAs or PLs (Table 2). Interestingly, while the Kds ratio oleoyl-LPL/oleic acid is 51, the Kds ratio for palmitoyl-LPL/ palmitic acid is 5 times larger. It seems that binding of the LPL polar headgroup is modulated by the FA aliphatic chain length.
The crystal structure of Blc in complex with vaccenic acid reveals a stoichiometry of one ligand per dimer. Blc dimerisation seems to be biologically relevant, since the WAS area buried in the dimer is larger than the accepted threshold, comparable to those observed in immunoglobulins/proteins complexes. Dimerisation of another lipocalin, bovine OBP (bOBP), by domain swapping was observed in the crystal structure. In this case, however, a ligand is bound by monomer [25,31]. The exchange of the α-helix in bOBP was made possible by the absence of a disulfide bridge and resulted in that case only in the stabilisation of the protein. The different arrangement of the Blc dimer, driven by the unusual number of hydrophobic Phe groups exposed to the surface and possibly by a few movements of surface loops, results in two major advantages: it creates a larger binding site bridging two subunits, and it orients the polarity of the protein so as to promote the insertion into the membrane on one side and the binding function on the opposite side.
The trafficking of lipids between the inner and outer membranes of the Gram-negative bacterial cell envelope is a subject of intensive current interest. Significant recent progress has been made in the study of lipid export and import across both the inner and outer membranes. The ATP-binding cassette transporter MsbA accelerates the export of both phospholipids and LPS across the inner membrane [32,33], while the outer membrane protein Imp is needed for the display of LPS on the extracellular surface of the outer membrane [34]. Additionally, mechanisms for the uptake of fatty acids across both the outer and inner membranes are now well established [35]. However, the mechanisms for transperiplasmic movement of lipids between the inner and outer membrane systems remain largely unknown.
Pathways for the rapid exchange of glycerophospholipids between the two membranes and for the unidirectional export of LPS in Gram-negative bacteria were first revealed by the ground-breaking studies of Jones and Osborn [36,37]. In eukaryotic cells, lipid trafficking has been documented to proceed by lipid transfer proteins or by vesiculation mechanisms. In Gram-negative bacteria, vesiculation models seem less likely given the structural barrier provided by the peptidoglycan exoskeleton, which is sandwiched between the two membranes [38]. Bacterial lipoproteins clearly depend on a specific transfer protein during outer membrane biogenesis, suggesting that other lipids are transported by similar mechanisms [39]. Lipocalins represent an important class of lipid transfer proteins, and Blc, while a bacterial lipoprotein anchored in the inner leaflet of the outer membrane, is a lipocalin of unknown function.
Given the high affinity of Blc for LPL’s, it seems likely that Blc may fulfil a role in cell envelope LPL transport. Although LPL’s are key inner membrane intermediates of phospholipid metabolism, we do not know of any evidence to indicate that LPL’s are exported to the outer membrane. However, Rock and colleagues have established that exogenously supplied LPL’s can be taken up by deep-rough LPS mutants and converted by reacylation into glycerophospholipids using inner membrane-associated enzymes [40,41]. To date, no accessory factors needed for LPL uptake have been identified. In wild-type cells, at least two enzymes can generate LPL’s in the outer membrane, namely, the phospholipase OMPLA [13] and the lipid A palmitoyltransferase PagP [14,42–45]. Both enzymes preferentially generate the sn-1 LPL regioisomers, but these are known to spontaneously rearrange into the more stable sn-2 LPL’s [46] shown here to bind Blc with high affinity in our fluorescence quenching studies.
In eukaryotic cells, sn-2 LPL’s are generated by phospholipases that mobilize arachidonic acid for eicosanoid-mediated signal transduction. Recently, the sn-2 LPL products have been shown to function as potent biological mediators in their own right [47]. Mammalian apolipoprotein D and Drosophila lazarillo are the closest eukaryotic homologous of Blc and the only eukaryotic lipocalins that, like Blc, are anchored to lipid membranes. The ligand for lazarillo is unknown, but apolipoprotein D binds a variety of ligands including arachidonic acid [48]. Recently, functional studies on ApoD and lazarillo clearly pointed to their essential role in lipid metabolism [49,50]. Perhaps the high affinity of Blc for LPL’s may help shed some light on the enigmatic functions of lazarillo and apolipoprotein D.
Acknowledgments
We thank the structural genomics team of the AFMB laboratory, Louis Reese and Nicolas Babeault for technical assistance. This study was supported by the French Ministry of Industry (grant ASG) and the Marseille-Nice Genopole. Work in the laboratory of REB was supported by Canadian Institutes of Health Research operating Grant MOP-43886.
Abbreviations
- AMA
amino anthracene
- ANS
amino-naphthalene sulfonate
- LPS
lipopolysaccharide
- NPN
1-N-phenylnaphthylamine
- FA
fatty acid
- PL
phospholipid
- LPL
lysophospholipid
- PtdGro
phosphatidylglycerol
- PtdEtn
phosphatidylethanolamine
- LOP
oleoyl-sn-2-lyso-PtdEtn
- LPP
palmitoyl-sn-2-lyso-PtdEtn
- WAS
water accessible surface
- SLS
static light scattering
References
- 1.Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 2.Bishop RE, Penfold SS, Frost LS, Holtje JV, Weiner JH. Stationary phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein D. Implications for the origin of lipocalins. J Biol Chem. 1995;270:23097–23103. doi: 10.1074/jbc.270.39.23097. [DOI] [PubMed] [Google Scholar]
- 3.Bishop RE. The bacterial lipocalins. Biochim Biophys Acta. 2000;1482:73–83. doi: 10.1016/s0167-4838(00)00138-2. [DOI] [PubMed] [Google Scholar]
- 4.Akerstrom B, Flower DR, Salier JP. Lipocalins: unity in diversity. Biochim Biophys Acta. 2000;1482:1–8. doi: 10.1016/s0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- 5.Newcomer ME, Jones TA, Aqvist J, Sundelin J, Eriksson U, Rask L, Peterson PA. The three-dimensional structure of retinol-binding protein. Embo J. 1984;3:1451–1454. doi: 10.1002/j.1460-2075.1984.tb01995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flower DR. Experimentally determined lipocalin structures. Biochim Biophys Acta. 2000;1482:46–56. doi: 10.1016/s0167-4838(00)00147-3. [DOI] [PubMed] [Google Scholar]
- 7.Ramoni R, Vincent F, Grolli S, Conti V, Malosse C, Boyer FD, Nagnan-Le Meillour P, Spinelli S, Cambillau C, Tegoni M. The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein. J Biol Chem. 2001;276:7150–7155. doi: 10.1074/jbc.M010368200. [DOI] [PubMed] [Google Scholar]
- 8.Spinelli S, Ramoni R, Grolli S, Bonicel J, Cambillau C, Tegoni M. The structure of the monomeric porcine odorant binding protein sheds light on the domain swapping mechanism. Biochemistry. 1998;37:7913–7918. doi: 10.1021/bi980179e. [DOI] [PubMed] [Google Scholar]
- 9.Ramoni R, Vincent F, Ashcroft AE, Accornero P, Grolli S, Valencia C, Tegoni M, Cambillau C. Control of domain swapping in bovine odorant-binding protein. Biochem J. 2002;365:739–748. doi: 10.1042/BJ20011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez G, Ganfornina MD, Sanchez D. Evolution of the lipocalin family as inferred from a protein sequence phylogeny. Biochim Biophys Acta. 2000;1482:35–45. doi: 10.1016/s0167-4838(00)00151-5. [DOI] [PubMed] [Google Scholar]
- 11.Campanacci V, Nurizzo D, Spinelli S, Valencia C, Tegoni M, Cambillau C. The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding. FEBS Lett. 2004;562:183–188. doi: 10.1016/S0014-5793(04)00199-1. [DOI] [PubMed] [Google Scholar]
- 12.Huber R, Schneider M, Mayr I, Muller R, Deutzmann R, Suter F, Zuber H, Falk H, Kayser H. Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution. J Mol Biol. 1987;198:499–513. doi: 10.1016/0022-2836(87)90296-8. [DOI] [PubMed] [Google Scholar]
- 13.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 14.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. Embo J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganfornina MD, Sanchez D, Bastiani MJ. Lazarillo, a new GPI-linked surface lipocalin, is restricted to a subset of neurons in the grasshopper embryo. Development. 1995;121:123–134. doi: 10.1242/dev.121.1.123. [DOI] [PubMed] [Google Scholar]
- 16.Davis RaVJE. Comprehensive Biochemistry. Elsevier; Amsterdam: 1996. [Google Scholar]
- 17.Breustedt DA, Schonfeld DL, Skerra A. Comparative ligand-binding analysis of ten human lipocalins. Biochim Biophys Acta. 2006;1764:161–173. doi: 10.1016/j.bbapap.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Vincentelli R, Bignon C, Gruez A, Canaan S, Sulzenbacher G, Tegoni M, Campanacci V, Cambillau C. Medium-scale structural genomics: strategies for protein expression and crystallization. Acc Chem Res. 2003;36:165–172. doi: 10.1021/ar010130s. [DOI] [PubMed] [Google Scholar]
- 19.Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A, Cambillau C, Breer H, Tegoni M. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J Biol Chem. 2001;276:20078–20084. doi: 10.1074/jbc.M100713200. [DOI] [PubMed] [Google Scholar]
- 20.Sulzenbacher G, Gruez A, Roig-Zamboni V, Spinelli S, Valencia C, Pagot F, Vincentelli R, Bignon C, Salomoni A, Grisel S, Maurin D, Huyghe C, Johansson K, Grassick A, Roussel A, Bourne Y, Perrier S, Miallau L, Cantau P, Blanc E, Genevois M, Grossi A, Zenatti A, Campanacci V, Cambillau C. A medium-throughput crystallization approach. Acta Crystallogr D Biol Crystallogr. 2002;58:2109–2115. doi: 10.1107/s0907444902013938. [DOI] [PubMed] [Google Scholar]
- 21.CCP4 & 4, C.C.P.N. The CCP4 suite: programs for crystallography. Acta Cryst D. 1994;50:760–766. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Roussel A, Cambillau C. The TURBO-FRODO graphics package. Silicon graphics Geometry Partners Directory; Mountain View, USA: 1991. [Google Scholar]
- 24.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: ( http://www.pymol.org) [Google Scholar]
- 25.Vincent F, Ramoni R, Spinelli S, Grolli S, Tegoni M, Cambillau C. Crystal structures of bovine odorantbinding protein in complex with odorant molecules. Eur J Biochem. 2004;271:3832–3842. doi: 10.1111/j.1432-1033.2004.04315.x. [DOI] [PubMed] [Google Scholar]
- 26.Briand L, Nespoulous C, Perez V, Remy JJ, Huet JC, Pernollet JC. Ligand-binding properties and structural characterization of a novel rat odorant-binding protein variant. Eur J Biochem. 2000;267:3079–3089. doi: 10.1046/j.1432-1033.2000.01340.x. [DOI] [PubMed] [Google Scholar]
- 27.Marchese S, Pes D, Scaloni A, Carbone V, Pelosi P. Lipocalins of boar salivary glands binding odours and pheromones. Eur J Biochem. 1998;252:563–568. doi: 10.1046/j.1432-1327.1998.2520563.x. [DOI] [PubMed] [Google Scholar]
- 28.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 29.Desmyter A, Spinelli S, Payan F, Lauwereys M, Wyns L, Muyldermans S, Cambillau C. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J Biol Chem. 2002;277:23645–23650. doi: 10.1074/jbc.M202327200. [DOI] [PubMed] [Google Scholar]
- 30.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 31.Tegoni M, Ramoni R, Bignetti E, Spinelli S, Cambillau C. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nat Struct Biol. 1996;3:863–867. doi: 10.1038/nsb1096-863. [DOI] [PubMed] [Google Scholar]
- 32.Doerrler WT, Gibbons HS, Raetz CR. MsbAdependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 33.Reyes CL, Chang G. Structure of the ABC transporter MsbA in complex with ADP. vanadate and lipopolysaccharide. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 34.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dirusso CC, Black PN. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. J Biol Chem. 2004;279:49563–49566. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 36.Jones NC, Osborn MJ. Interaction of Salmonella typhimurium with phospholipid vesicles. Incorporation of exogenous lipids into intact cells. J Biol Chem. 1977;252:7398–7404. [PubMed] [Google Scholar]
- 37.Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem. 1977;252:7405–7412. [PubMed] [Google Scholar]
- 38.Huijbregts RP, de Kroon AI, de Kruijff B. Topology and transport of membrane lipids in bacteria. Biochim Biophys Acta. 2000;1469:43–61. doi: 10.1016/s0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 39.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Hsu L, Jackowski S, Rock CO. Uptake and acylation of 2-acyl-lysophospholipids by Escherichia coli. J Bacteriol. 1989;171:1203–1205. doi: 10.1128/jb.171.2.1203-1205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu L, Jackowski S, Rock CO. Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J Biol Chem. 1991;266:13783–13788. [PubMed] [Google Scholar]
- 42.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. Embo J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang PM, Bishop RE, Kay LE. The integral membrane enzyme PagP alternates between two dynamically distinct states. Proc Natl Acad Sci USA. 2004;101:9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia W, Zoeiby AE, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 46.Pluckthun A, Dennis EA. Acyl and phosphoryl migration in lysophospholipids: importance in phospholipid synthesis and phospholipase specificity. Biochemistry. 1982;21:1743–1750. doi: 10.1021/bi00537a007. [DOI] [PubMed] [Google Scholar]
- 47.Anliker B, Chun J. Lysophospholipid G protein-coupled receptors. J Biol Chem. 2004;279:20555–20558. doi: 10.1074/jbc.R400013200. [DOI] [PubMed] [Google Scholar]
- 48.Morais Cabral JH, Atkins GL, Sanchez LM, Lopez-Boado YS, Lopez-Otin C, Sawyer L. Arachidonic acid binds to apolipoprotein D: implications for the protein’s function. FEBS Lett. 1995;366:53–56. doi: 10.1016/0014-5793(95)00484-q. [DOI] [PubMed] [Google Scholar]
- 49.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein d leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–679. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez D, Lopez-Arias B, Torroja L, Canal I, Wang X, Bastiani MJ, Ganfornina MD. Loss of glial lazarillo, a homolog of apolipoprotein d, reduces lifespan and stress resistance in Drosophila. Curr Biol. 2006;16:680–686. doi: 10.1016/j.cub.2006.03.024. [DOI] [PubMed] [Google Scholar]