Abstract
Influenza A virus (IAV) assembles on the plasma membrane where viral proteins localize to form a bud encompassing the viral genome, which ultimately pinches off to give rise to newly formed infectious virions. Upon entry, the virus faces the opposite task—fusion with the endosomal membrane and disassembly to deliver the viral genome to the cytoplasm. There are at least four influenza proteins—hemagglutinin (HA), neuraminidase (NA), matrix 1 protein (M1), and the M2 ion channel—that are known to directly interact with the cellular membrane and modify membrane curvature in order to both assemble and disassemble membrane-enveloped virions. Here, we summarize and discuss current knowledge of the interactions of lipids and membrane proteins involved in the IAV replication cycle.
Influenza proteins shaping the plasma membrane during virion assembly
Hemagglutinin
Hemagglutinin (HA), a type I membrane glycoprotein, is synthesized as the precursor HA0. In all of the 18 currently known HA subtypes, the cytoplasmic tail, the transmembrane domain (TMD), the stem region, and the fusion peptide are the most conserved regions in the protein. HA S-acylation at the cytoplasmic tail [1], trimerization [2] and N-glycosylation [3] occur post-translationally in the endoplasmic reticulum. HA is delivered to the apical plasma membrane as a homotrimer via a secretory pathway sensitive to cholesterol removal [4]. HA0 is proteolytically cleaved to HA1 and HA2 peptides, which remain covalently bound by a disulfide bond. HA subtypes H5 and H7 are cleaved by furin-like serine proteases in the Golgi apparatus [5] while other subtypes are cleaved at the plasma membrane [6].
HA glycoproteins cluster on the plasma membrane [7,8] at a density consistent with that of influenza glycoproteins packed on the surface of released virions (about 1 glycoprotein/120 nm2) [9,10]. Thus, densely packed HA clusters are likely sites of virion budding. Patches from 40 nm (resolution limit of FPALM) up to several micrometers containing HAs were observed in living cells by fluorescence photoactivation localization microscopy (FPALM) and by negative stain immuno-electron microscopy (EM) on isolated membrane sheets retrieved from fibroblasts [7]. Interestingly, HA is neither static at the plasma membrane nor absolutely confined within a cluster since HA can diffuse laterally in and out of these clusters. The average HA diffusion coefficient in living cells, measured by fluorescence recovery after photobleaching (FRAP), is approximately 100x smaller (D ~0.07 μm2/s) compared to lipids [11], and the newer technique of live cell FPALM shows a large range of individual molecular mobilities that, on average, agree with the values determined by FRAP [12]. In addition, FPALM detected a subpopulation of confined molecules with a much smaller (~0.02 μm2/s) diffusion coefficient [12] consistent with the immobile fraction identified by FRAP.
The driving force behind the clustering of HA, and the role of HA clusters to promote bud formation has been controversial and needs re-evaluation. It has been hypothesized that HA clustering at the plasma membrane is driven by partitioning into liquid ordered (Lo) domains rich in cholesterol and sphingomyelin [8,13], based on the isolation of so-called detergent-resistant membranes (DRM), which were enriched in both cholesterol and WT (but not mutated) HA [1, 14 15, 16]. Phospholipid bilayers prepared from certain ratios of purified phospholipids and cholesterol can demix into co-existing but distinct membrane phases: Lo and liquid disordered (Ld) which differ in their degree of acyl chain packing. Since DRM and Lo phases are both moderately enriched in cholesterol it had been often assumed that DRM and the liquid ordered phases are identical. However, the ability of cold Triton X-100 to coalesce already existing domains [14] likely leads to protein rearrangements. Furthermore, cold Triton X-100 is able to induce phase separated domains in initially homogeneous membranes of the giant unilamellar vesicles (GUVs) [15]. Thus, the composition of DRM is not sufficiently reliable to accurately predict the composition of plasma membrane domains [16]. In accord with the hypothesis that lipid domain segregation drives HA clustering, the IAV membrane is ~1.4x enriched in sphinogolipids and ~1.1x in cholesterol in comparison with isolated apical membrane [17]. Furthermore, HA mobility and spacing is controlled by cholesterol in the plasma or viral membrane. HA mobility measured at room temperature increases ~1.6x after cholesterol depletion, arguing that the presence of cholesterol in the membrane retards HA mobility [18]. Interestingly, HA-HA spacing also decreases from ~ 9 nm to 8 nm after nearly complete (~90%) cholesterol depletion from the virus [19]. However, depleting cholesterol could just as easily promote gel phase (So) domains in mixed phospholipid bilayers, which are physically distinct from Lo. Thus, HA trimer decreased mobility as well as diminished HA-HA spacing might be caused by formation of the So domains. In disagreement with lipid domain segregation hypothesis driving HA clustering, imaging techniques have failed to unambiguously show HA residing in Lo domains and interacting with cholesterol at physiological temperature. Experiments monitoring FRET between HA TMD-YFP and a well-established raft marker, glycophosphatidylinositol (GPI) anchored CFP, showed that the two proteins clustered in a cholesterol dependent manner [20]. However, the behavior of GPI in cell membranes was recently revisited using micropatterning and single molecule tracking in living cells. These studies did not find that GPI-anchored proteins were targeted to artificially formed GPI-anchored GFP domains [21]. Other experiments have directly shown that HA does not reside in Lo domains: HA reconstituted in GUVs or HA present in giant plasma membrane vesicles (GPMVs) was found in the Ld domain or in both Lo and Ld [22,23]. Most of the studies of HA and cholesterol enriched domain association have relied on fluorescently tagged lipids which have inherently modified chemical structures and thus do not behave exactly as the unmodified lipids. In contradistinction, magic angle spinning (MAS) 1H NMR measures spectra gauche-trans isomerization and axial reorientation rate of lipid acyl chains and thus can detect Lo phase featuring a lower probability of gauche-trans isomerization without using any lipid probes. MAS NMR analysis revealed that the influenza viral envelope of intact virions, or influenza virion-extracted membranes, are composed of lipids whose ability to phase separate into Lo and Ld phases is dependent on temperature, with practically all the lipids in the disordered phase at physiological temperatures. In addition, no Lo phases were found at the higher temperatures experienced during fever by those suffering a bout of the flu, which can reach 41°C [24]. Furthermore, these experiments show that the viral proteins do not affect the phase behavior of these lipids.
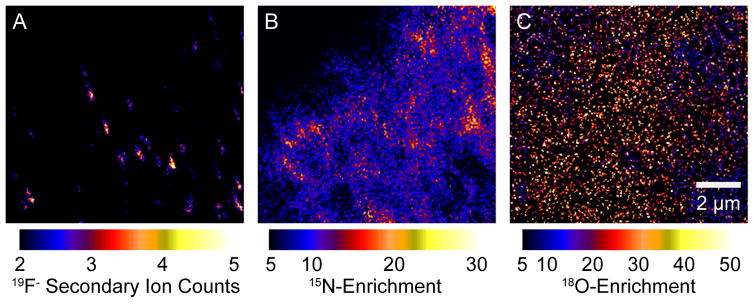
To directly determine the chemical composition of the HA-domains in the plasma membrane of cells expressing it, new imaging technology was used wherein a Cs beam serially scans a cell membrane allowing the mass determination of surface components (high-resolution secondary ion mass spectrometry (nano-SIMS)) with a lateral resolution of ~70 nm [25]. Isotope-labeled cholesterol and sphingomyelin were incorporated into fibroblasts stably expressing HA [26]. There were no detectable cholesterol domains in the cell plasma membrane, as cholesterol was evenly distributed over the plasma membrane with or without HA (Figure 1). Despite the lack of any cholesterol-enriched domains in the plasma membrane, there were readily detectable sphingomyelin domains discovered in these experiments. However, HA did not colocalize with these sphingomyelin domains.
Figure 1. SIMS showing cholesterol, sphingomyelin clusters and HA clusters.
Images capturing the same region of the cell produced by spatial detection of secondary ions by mass spectrometry (SIMS, nanoSIMS implementation). Mouse fibroblast cells that stably express HA were metabolically labeled with 18O cholesterol and 15N sphingomyelin. Cells were chemically fixed and HA was labeled with an anti-HA antibody conjugated to fluorinated colloidal gold, which produces distinctive 19F− secondary ions. A) Locally high counts of 19F− secondary ions show HA clusters on the surface of the fibroblast. Sphingomyelin domains (B) and evenly distributed cholesterol (C) do not colocalize with HA clusters.
Although lipid (sphingomyelin) domains clearly form in the plasma membrane of fibroblasts, Lo and Ld phase separation might not be an unique mechanism of lipid domain formation. Importantly, depletion of cholesterol reorganizes the actin cytoskeleton [27], suggesting an alternative scenario in which HA clustering is facilitated by the actin cytoskeleton rather than Lo domain formation. Consistently, motion analysis revealed that HA preferentially moves on the plasma membrane in a quasi-linear fashion for short times [7], with the lattice of occupied spaces reminiscent of the actin cytoskeleton. Indeed, the mobility of individual GFP-labeled HA trimers decreased with increasing cytoskeletal actin density directly below the membrane along the path of diffusion. A study using FPALM imaging showed that actin associates with HA clusters, indicating that the cytoskeleton may drive HA clustering and control its lateral organization [12,28]. Actin has been shown to play a role in the formation of filamentous virions [29]. However, whether HA directly interacts with actin, in the presence of other viral proteins, in particular M1, remains to be investigated.
Transient expression of HA in 293T cells results in the release of HA containing vesicles, provoking the question of whether HA directly modulates plasma membrane curvature [30]. In the 293T cells, HA molecules covered most of the surface of the plasma membrane rather than being localized into patches. Further interest arose from an electron tomography (ET) study (where HA spikes are visible without antibody labeling) showing that expression of HA glycoproteins induces dramatic ruffling and plasma membrane vesiculation [31]. Interestingly, a large number of HA spikes were seen in modified membrane compartments in the cytoplasm [31], which might deliver HA prepackaged in quanta as a cluster, explaining the observed clustering on the surface of the membrane. HA induced membrane curvature and HA interaction with M1 layer may prevent or slow down HA mobility and its immediate dispersion from the cluster.
Neuraminidase
Neuraminidase (NA) is a type II transmembrane protein present on the viral surface as a tetramer together with HA. There are 11 known NA subtypes, all of which possess sialidase enzymatic activity, important to cleave the HA cellular binding receptor—sialic acid moieties—and facilitate the release of budded virions. The HA:NA ratio on the viral surface, which was found to be approximately 5:1 [9] is important for virus replication fitness since it maintains the right balance between receptor-binding and receptor-destroying capability of the virus. NA-lipid interactions have been far less explored in comparison to HA. Besides the well-characterized sialidase function, NA also seems to be able to modulate budding and release of virions. Expression of NA subtypes N1 and N2 in the absence of M1, M2, and HA results in production of NA containing particles, indicating that NA is able to induce membrane curvature [32,33]. In contrast to HA, NA containing particles are released more effectively and have a quasi-filamentous morphology [31–33]. In general, the filamentous morphology is typical for helical assemblies such as M1 matrix protein. Even without M1, NA forms particles with an extended filamentous morphology, indicating that NA glycoproteins favor a helical arrangement on the surface of the particle. The mechanism by which NA is able to induce curvature is not known, however a single Asp286 present in the virion-facing part of the NA globular domain is needed to confer this ability [33,34]. Interestingly, tetherin, an interferon-inducible antiviral host factor, abolishes the budding capability in NA N2 subtypes when Gly is present instead of the Asp286 residue [33,34]. This Asp286 residue might be involved in electrostatic interactions between individual NA spikes, leading to NA clusters typically found on the surface of the virion. The high lateral density of clustered NA together with NA’s conical molecular shape (with its large globular domain and long stem domain) might be responsible for inducing membrane curvature changes. At first glance, this idea is coordinate with the crowding hypothesis for the curvature effect of some scaffold proteins [35]. However, recent studies have shown that the crowding hypothesis works for disordered proteins and NA is well structured [36]. A coat hypothesis is possible, but the distribution of glycoproteins on the virion surface is more consistent with freely mobile molecules. We thus propose a crowded wedge hypothesis for membrane proteins with wedge-shaped ectodomains. Interestingly, NA preferentially occupies the ends of filamentous VLPs [31], areas with spherical curvature derived from the neck area of a budding virus that needs to develop more positive curvature to drive membrane scission. Thus, an NA cluster may self-assemble at the neck of a budding virion to promote its release, and the explanation for the lack of budding in the presence of the antiviral tetherin might be that tetherin modulates both NA’s preferential position to the spherical ends of virions and NA’s capability to change membrane curvature. More work is needed to investigate how much of a contribution NA makes to these processes.
Matrix protein 1
Matrix protein 1 (M1), a scaffold protein apposed to the membrane, is a pivotal interaction platform for influenza proteins. M1, which organizes in a helical manner [37], is a major determinant of IAV morphology through its ability to modulate membrane curvature. As a peripheral membrane protein M1 can reversibly bind to membranes through electrostatic interactions with negatively charged phospholipids such as phosphatidyl serine in the absence of other influenza proteins [38,39]. Neither the polybasic region nor the hydrophobic domains of M1 alone are required for membrane binding, thus M1 can bind membranes through multiple regions [40,41]. Recent studies combining atomic force microscopy and fluorescence correlation spectroscopy indicate that membrane binding itself is sufficient for M1 oligomerization [38,39,42].
In contradistinction to the spontaneous binding to negatively charged membranes described above, cellular expression of M1 in the absence of other influenza proteins leads to its accumulation in the nucleus, due to the lack of an inherent membrane targeting signal [43]. Interestingly, in the vaccinia virus-driven expression system, M1 was targeted to the membrane (possibly due to vaccinia background proteins) and released as filamentous particles in the absence of HA and NA, indicating that M1 itself induces membrane curvature and is able to promote budding and release when targeted to the plasma membrane [44]. In IAV infected cells either HA or NA are necessary for effective targeting of M1 to the plasma membrane [43,45,46]. In addition, M2 is also able to recruit M1 to the plasma membrane [43]. Removal of the cytoplasmic tail of HA, NA and M2 or removal of S-acylation on the cytoplasmic tail of the HA leads to decreased incorporation of M1 into virions, indicating that M1 interacts with HA and NA via cytoplasmic tails [43,47–49]. The structure of the complete M1 protein as well as the M1 layer organization is not fully known, much less in the context of these important tail interactions.
Matrix protein 2
Matrix protein 2 (M2) is a transmembrane homotetramer. Each 97 amino acid alpha-helical monomer is composed of an N-terminal ectodomain, a TMD and an amphipathic helix at the cytoplasmic tail, which is post-translationally modified by palmitoylation [50,51]. Immunofluorescence data indicate that while M2 is abundant in the plasma membrane of infected or transfected cells, fewer copies of M2 are incorporated into released virions (compared to HA or NA). Estimates made based on quantitative Western blot analysis revealed that there are 14–68 molecules of M2 per virion [52] in contrast to ~300 HA and ~ 50 NA per spherical virion with a diameter of 120 nm as shown by cryo-ET [9].
Many proteins featuring amphipathic helices with alternating hydrophobic and positively charged surfaces are able to induce or sense changes in membrane curvature by lateral wedge-like insertion into the bilayer [53]. Accordingly, in vitro studies showed that wild-type M2 reconstituted into GUV or the M2 amphipathic helix added into preformed GUV were able to induce blebbing of GUVs [54]. Thus, it has been proposed that the M2 amphipathic helix is able to drive membrane scission by inducing negative membrane curvature. Consistently, a high-resolution small angle X-ray scattering (SAXS) study showed that full length as well as the cytoplasmic domain of M2 reconstituted into liposomes are able to restructure the membrane bilayer (lamellar phase) into non-bilayer structures (cubic phase) [55]. Membrane reorganization from bilayer to non-bilayer is typically observed during membrane fusion and scission. M2 induced GUV blebbing was observed only at low cholesterol levels (<17 mol%) [54], suggesting that cholesterol might be an inhibitor of M2 driven membrane scission. However, cholesterol is a lipid with negative spontaneous monolayer curvature (J0 <0) and is also able to induce cubic phases [56]. Thus, contrary to the data on M2 induced GUV blebbing [54], the presence of cholesterol in the membrane likely supports the M2 induced negative membrane curvature. This is consistent with an NMR study which showed that the presence of cholesterol increases the ability of the M2 TMD with amphipathic helix to induce negative curvature [57]. In addition, another study showed that the M2 induced cubic phases were observed only in the presence of high dioleoylphosphatidyl ethanolamine content (>60 mol%) [55], which has a spontaneous negative curvature (J0 = −0.399 nm−1) comparable to cholesterol (J0 = −0.494 nm−1) [58]. In contrast, in a recent NMR study the M2 TMD with amphipathic helix was able to induce curvature in the absence of lipids with high spontaneous curvature [59]. Thus, so far whether cholesterol or other lipids with negative spontaneous curvature are regulating the capability of M2 to induce membrane scission remains elusive.
It is unlikely that in cells M2 scission is regulated only by cholesterol content since in such a scenario M2 would likely induce membrane scission after its synthesis in the endoplasmic reticulum where cholesterol is low. However, Rab11 was shown to control M2 delivery to the plasma membrane [54] and a recent study showed that IAV infection increases cholesterol content in Rab11 positive recycling endosomes, as indicated by increased filipin staining, which binds to cholesterol [60]. It is plausible that an elevated cholesterol concentration inhibits M2 propensity to induce negative membrane curvature to prevent premature scission during trafficking to the plasma membrane.
Several studies have been conducted to investigate whether M2 directly interacts with cholesterol and partitions into Lo domains. UV Cross-linking of M2 with [3H] photocholesterol and detection of [3H] cholesterol associated with M2 suggest that M2 can bind directly to cholesterol [61,62] through a putative cholesterol recognition amino acid consensus (CRAC) motif [63]. However, not all influenza strains posses a CRAC consensus motif in M2, e.g. the filamentous Udorn strain [61]. Moreover, mutation of both CRAC and palmitoylation motifs do not impede virus growth [64]. The M2 amphipathic helix was shown to avoid cholesterol enriched Lo phase in GUV experiments and clustered into the boundary between the two phases [54], suggesting that M2 scission takes place at the interface of phase separated membrane. However, another domain partitioning study using GPMVs showed that M2 is found in both Lo and Ld domains with no accumulation at the domain interface [62]. Removal of the M2 palmitoylation motif leads to a weak preference for the Ld domain. Since palmitoylation is reversible, palmitoylation has been proposed as a switch controlling M2 lateral distribution [62]. Thus, whether M2 is indeed localized to the Lo – Ld interface and what is the driving force behind this localization is not clear. Imaging HA clusters by FPALM and nano-SIMS in the presence and absence of M2 proteins will bring additional information on M2 lateral distribution in the cellular plasma membrane.
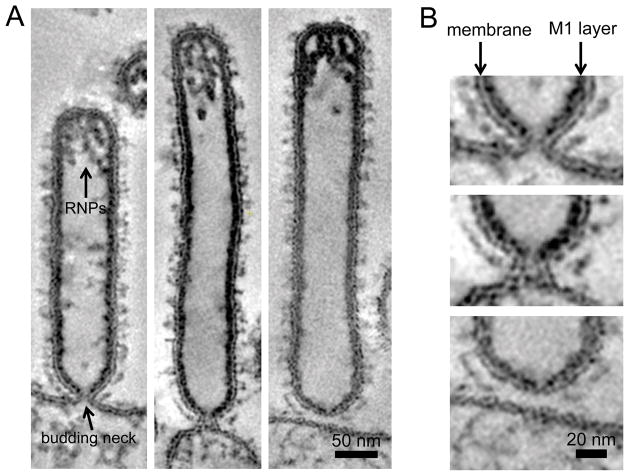
The M2 mediated membrane scission hypothesis was also tested in cells infected by IAV lacking M2. Thin-section EM analyses of IAV lacking M2 show virions that look like beads-on-a-string, a phenotype suggestive of scission impairment. However, studies investigating the M2 driven membrane scission of the budding virions at the plasma membrane are complicated by the fact that M2 also plays an important role in virion morphology. Deletion of M2 abolishes the typical filamentous morphology of the Udorn virus, [65–67] and results in the beads-on-a-string viruses, which do not seem to contain a complete M1 layer. Thus, the absence of the M1 layer might be responsible for this phenotype rather than the absence of M2 driven scission [54,66,67]. In fact, the M2 cytoplasmic tail and M1 are known to interact and removal of the M2 cytoplasmic tail reduces M1 incorporation [68]. ET studies on the viruses displaying beads-on-a-string phenotype are needed to conclude whether the connections between viruses are real budding necks or a result of improper M1 layer assembly. In the system using plasmid driven VLP, which is closer to the real scenario of infected cells than reconstituted systems using GUVs, M2 is dispensable for HA, NA and M1 release [30]. Thus, whether M2 is strictly needed for scission is still not clear. Additional experiments using VLPs without M2 or M2 deletion mutant viruses analyzed in three-dimensions by ET are needed to understand whether M2 is needed for the formation of budding necks or solely for the final scission by changing budding neck diameter. It is plausible that the local M1:M2 ratio in the virion determines whether M1 assembles as a layer with cylindrical or spherical curvature, which have different helical pitches [37]. A straightforward suggestion is that the assembly of M1 layer with a spherical curvature drives both formation of the budding neck and membrane scission. Consistently, the budding virion does not have a complete M1 layer in the area of the budding neck (Figure 2). Complete assembly of the M1 layer found in the rear end of the budding virion could provide sufficient energy to drive membrane scission. Thin-section EM showed that budding virions, which accumulate at late stages of budding at the plasma membrane are connected to the plasma membrane via budding necks, indicating that either M1 layer closure or membrane scission is a rate-limiting step.
Figure 2. Influenza A virus budding neck.
Slices of dual-axis electron tomograms capturing A/Udorn/72 virus budding from MDCK cells at 22 hours post infection. A) Filamentous virions containing vRNPs are connected to the cell surface by a budding neck. B) The M1 layer is not complete in the area of the budding neck, which has an inner diameter of approximately 10 nm. Both M1 and M2 are important determinants of IAV morphology. The M1:M2 ratio may control the M1 layer helical pitch and thus may trigger M1 layer transition from cylindrical to spherical M1 layer. Closure of the M1 layer may drive membrane scission as indicated by increased constriction of the neck in proximity to the M1 layer (arrowhead).
Influenza virus entry and membrane fusion
HA mediated membrane fusion
IAV is a pleiomorphic virus whose size and shape seems to be a determinant of the initial cell entry pathway. IAV with predominantly spherical particles enter the host cell by receptor-mediated endocytosis whereas IAV with predominantly long filamentous virions enter through macropinocytosis [69,70]. In both cases, the sialic acid moieties of the glycoproteins serve as binding receptor recognized by the HA1 domain of the HA trimer. Human IAV with HA subtypes H2 and H3 preferentially bind to the N-acetylneuraminic acid-α2,3-galactose (NeuAcα2,6Gal) linkage while most avian IAV with subtypes such as H5 and H7 recognize the NeuAcα2,3Gal linkage [71]. The virus is delivered into the late endosome where membrane fusion occurs and the viral ribonucleoproteins (vRNPs) are released into the cytoplasm. The HA trimer is a type I fusion protein with its hydrophobic fusion peptides tacked onto the interface of the HA2 domains [72]. From the standpoint of the phospholipids, membrane fusion is thermodynamically neutral, since one begins and ends with all phospholipids in bilayers. The chief thermodynamic consideration is the rather large energy barrier due to (i) hydration repulsion which increases exponentially as the two repelling negatively charged surfaces of the membranes approach each other [73] and (ii) the hydrophobic effect resisting the exposure of hydrophobic surfaces at contact. Fusion proteins are able to (i) decrease the distance between two apposing phospholipid bilayers below 2 nm overcoming some of the formidable hydration force, (ii) dimple membranes into point contacts to minimize the surface area needed to be in close contact, and (iii) locally disrupt the structure of the target membrane by fusion peptide insertion, which results in increased local membrane curvature and partial exposure of hydrophobic acyl chains [74]. HA irreversibly changes its conformation in the presence of H+, beginning by dissociation of the HA1 domains and exposing the fusion peptide forming the predicted (but not yet directly observed) extended HA intermediate that can bridge the viral and endosomal membrane. Subsequently, the extended HA intermediate folds back into the so-called post-fusion conformation, whose structure was determined by X-ray crystallography [75], bringing the two apposing membranes together. The structure of the post-fusion intermediate also indicated that the HA TMDs and the HA fusion peptides are drawn together, however, the exact position of these two hydrophobic domains in the fused membranes are missing in the structure due to limitations of X-ray crystallography. Cryo-ET proved to be an essential technique in elucidating the structural details of viral membrane fusion intermediates. A recent cryo-ET study of the HA spikes at low pH revealed two different HA conformational stages with decreasing electron density in the membrane proximal domain and overall shortening of the spike, presumably resulting from HA1 domain release and HA2 rearrangements [76]. HA spikes at low pH are hard to discern by cryo-ET as individual entities, presumably due to dissociated HA1 domains, which stay attached to the HA2 domain. The thinner HA2 stem domain of the low pH modified HA was discerned by cryo-ET after trypsin treatment, which cleaves most of the HA1 domain [37,77]. Cryo-ET with recently developed detectors directly detecting electrons and phase plate technology will likely provide a tool to confirm the existence and deliver the structure of the extended “pre-hairpin” HA intermediate, which is a cornerstone of the prevailing membrane fusion model.
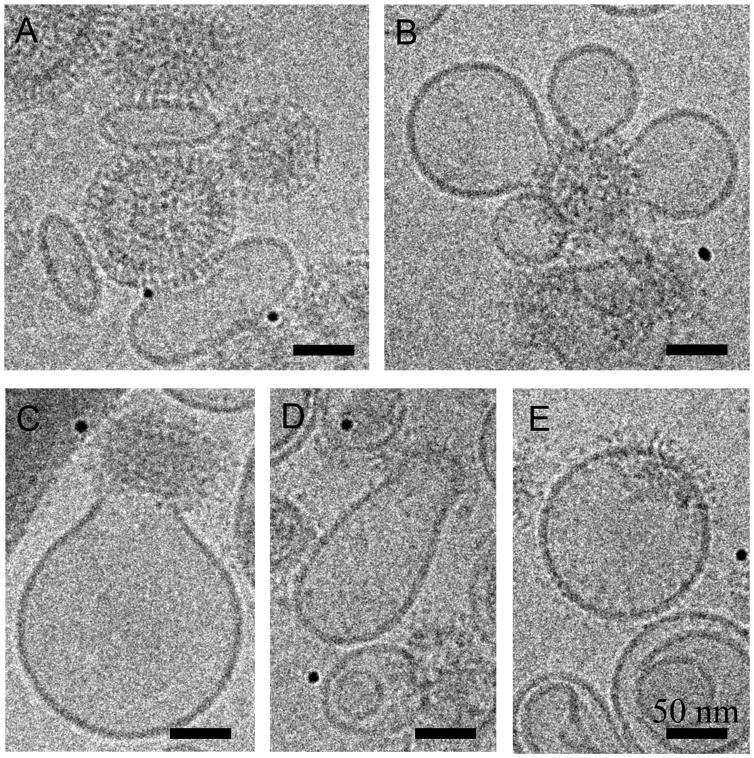
There is good evidence that membrane fusion occurs in steps through short-lived lipidic intermediates with local free energy minima as predicted by theory [78]. In the hemifusion intermediate the two opposing membranes are locally fused but an aqueous pore is not formed. This essential membrane fusion intermediate is well documented by both experimental and theoretical studies for HA and other viral or cellular fusion machineries. Mutations in the fusion peptide which is located at the N termini of the HA2 domain [79] or replacement of the HA TMD domain with a GPI anchor (GPI-HA) [80] were found to arrest this stage, suggesting that both fusion peptide and TMD play a crucial role in fusion pore initiation. Although two different hemifusion structures, the hemifusion stalk (HS) and the hemifusion diaphragm (HD), were theoretically predicted [78] their existence remains to be confirmed by direct visualization. According to the prevailing model, HS can either remodel into a fusion pore or expand into HD, which can form a pore upon breakage [78]. However, theoretical studies using membrane mechanics or molecular dynamics indicate that extension of a stalk into an HD is energetically permitted only for membranes composed of lipids with substantial spontaneous negative curvature [81] or by formation of a small pore at the rim of the HD, respectively [82]. Thus, it is more likely that the fusion pore is directly formed from HS, omitting HD. HD was recently visualized by cryo-EM in the SNARE liposome reconstituted system and the concomitant fluorescence dequenching kinetics experiment suggested that HD transform into a fusion pore, albeit with slow kinetics [83]. Cryo-EM and cryo-ET can be used to study lipidic intermediates and products of IAV and liposome fusion triggered at low pH (Figure 3). Cryo-ET of X31 IAV in the presence of a liposomes at low pH revealed locally curved membrane “dimples” and funnel-like structures, presumably membrane intermediates of the fusion pore [84]. The same study also showed that liposomal membrane transiently opens and is subsequently inserted into the viral membrane. However, no gangliosides were added into the liposomes as HA receptors and the high defocus values used to collect cryo-ET precluded obtaining a detailed structure of the lipidic intermediates in more physiological conditions.
Figure 3. Cryo-EM projections of IAV driven membrane fusion with liposomes containing gangliosides as a binding receptor of HA.
A) IAV (V) binds to the liposomes (L) containing gangliosides at pH 7. HA spikes are clearly discernable. B–E) IAV induced liposomal membrane modulation at pH 5.5. HA spikes do not appear as single entities after undergoing conformational changes and dissociation of HA1 domains. B and C) Liposomal membrane is deformed pointing towards the virus. D and E) Liposomal and IAV membrane are continuous. E) The M1 layer dissociates and the fusion product becomes spherical.
Electrophysiological recordings of membrane fusion occurring between fibroblasts expressing HA and red blood cells using whole-cell patch clamp technology and time-resolved capacitance measurements showed that the initial fusion pore attains a diameter of 1–2 nm in less than a millisecond [85]. Interestingly, the initial fusion pore undergoes multiple cycles of open-closed transitions with kinetics on the order of seconds, presumably oscillating between hemifusion and an open small fusion pore, an intermediate termed flickering pore (89). There is good evidence that formation of the fusion pore requires more than one HA trimer. Cell-cell fusion assays with varying HA densities showed a temporal lag phase preceding onset of the HA mediated membrane fusion. The sigmoidal shape of the curve relating the lag-phase and HA density infer that there is positive cooperativity required between a minimum of 3 HA to form a fusion pore [86]. Recently developed single virion kinetic assays utilizing dual labeled virions and dextran cushion-supported membrane bilayers have allowed the study of hemifusion and fusion pore intermediates in great detail [87]. Analysis of lipid fluorescence quenching alleviated upon hemifusion revealed that exposure of the fusion peptide is a rate-limiting step of the pathway leading to hemifusion. Furthermore, the study suggests that HA cooperativity takes place in the lag phase prior to the onset of membrane fusion and is likely required at the stage of the extended HA intermediates to bring the membranes together in concerted action [88]. In addition, the fusion kinetics depend on the lipid composition of the target membrane suggesting that endosomal levels of cholesterol may play a role in permissiveness for fusion [89].
Electrophysiological studies also indicate a second stage of fusion pore widening after the initial HA-mediated fusion pore forms (89). Fusion pore expansion requires additional HA [90]; contradicting results exist about the role of HA palmitoylation at the cytoplasmic tail in fusion pore expansion. Hemifusion, monitored by fluorescent lipid demixing, does not depend on HA palmitoylation. However, in some HA subtypes such as H1 (A/USSR/77) [91], H2 (A/Japan/305/57) [92] and H7 [93] but not H3 (A/Aichi/2/68) [94] and H3 (A/Udorn/72) [48], the substitution of the three cysteines in the cytoplasmic tail leads to formation of a small fusion pore unable to expand as inferred from the aqueous demixing assay, which showed that the pore is permeable only to small fluorescent molecules like calcein and carboxyfluorescein (hydrodynamic diameter 0.61 nm) but not to 10kDa dextran (hydrodynamic diameter 4.6 nm). Conductance measurements of fusion pores induced by H3 (A/Udorn/72) detected that the removal of palmitoyls reduced the flickering of the fusion pore but did not influence the pore enlargement kinetics in comparison to wild-type HA [95]. It is unlikely that the source of discrepancies is solely due to differences in HA subtype amino acid sequences since the cytoplasmic tail and the positions of the cysteines is highly conserved. More likely, the differences arise from using different expression systems and fusion assays. Therefore, a study that would test several HA subtypes in parallel by different fusion assays is needed.
Membrane fusion and cellular factors
Is HA bound to a target-bound receptor the minimal requirement for productive membrane fusion (when productivity is defined as resulting in infectious virion replication)? Although decades of accumulated evidence has shown that HA alone can drive the fusion of both synthetic and cellular membranes (including liposomes and cells), cellular proteins are nevertheless involved and can either enhance or block HA-mediated membrane fusion. Recent data utilizing genome-wide RNA interference (RNAi) screens found that cathepsin W, a cysteine protease present in the late endosomes, is important for IAV membrane fusion in late endosomes [96]. Knock down of CD81, a tetraspanin residing in both plasma and endosomal membrane known to be a hepatitis C virus co-receptor, resulted in reduced numbers of IAV fusion events [97]. In contrast, interferon-induced transmembrane proteins (IFITMs) were found to curtail the entry of several enveloped viruses including IAV. IFITMs recruitment to the endosomal membrane increases cholesterol concentration and alters membrane curvature. A study employing single virion fusion assays inside the cells showed that IFITM3 is able to inhibit the transition from hemifusion to a fusion pore [98].
IAV uncoating during entry
M1 and M2 proteins play crucial roles in the successful release of vRNPs from the virion during viral entry. The M2 ion channel allows transport of H+ and K+ [99] to the interior of the virion, a process which can be inhibited by adamantane-based drugs. Acidification of the IAV interior is necessary for both vRNP release and disassembly of the M1 layer from the virion membrane to allow fusion pore expansion. An acid bypass-fusion assay wherein the X31 virus was acid-pretreated before fusion implies that gradual rather than abrupt acidification in the endosome enhances vRNP and M1 layer dissociation [99,100]. Although no changes of the M1 layer inside the virions were obvious by cryo-EM at pH 6, atomic force microscopy (AFM) captured decreased stiffness of the virions, indicating that already at mild acidification the M1 layer undergoes restructuralization [100]. Interestingly, the stiffness of the virion was recovered to original values when the pH was adjusted back to pH 7, suggesting that the initial changes of M1 disassembly occurring at pH 6 are reversible. At more acidic pH the overall morphology of the virion changes dramatically and the M1 layer disassembles irreversibly [100]. Cryo-ET showed thinning of the M1 layer and formation of M1 aggregates usually accumulated in the proximity of the membrane at pH 5 [76,101]. Another cryo-EM study analyzing the filamentous virions of the Udorn strain after low pH treatment revealed M1 multilayered coils with a typical pitch of 11 nm dissociated from the viral membrane, indicating that M1 disassembly occurs through more organized intermediates [37].
M1 layer uncoating in vivo is not only pH dependent but is also facilitated by a plethora of cellular factors including dynein/dynactin, myosin II, MTs, actin and takes advantage of the aggresome machinery by mimicking misfolded proteins [102].
IAV alternative entry pathway
Interestingly, recent findings show an alternative entry pathway, which does not seem to involve HA mediated fusion. IAV infected cells show increased formation of long tubular structures referred to as tunneling nanotubules (TNTs) that allow the transfer of the vRNPs to neighboring cells [103]. Influenza induced TNTs contain HA and actin but no microtubules. Since tubular structures connecting cells have been reported as a means of communication and nutrition exchange between cells, it is plausible that IAV hijacks the cellular machinery underlying TNT formation. Whether HA or other viral protein is directly involved in formation and fusion of TNTs with the neighboring cells is yet to be revealed.
Concluding remarks and open questions
IAV proteins interplay in a concerted fashion, modulating host membrane curvature during both budding and entry. HA mediates membrane fusion during entry and M2 is hypothesized to drive membrane scission during budding (Figure 4). M1 protein controls IAV membrane rigidity and serves as a major budding driving force when targeted to the plasma membrane by HA, NA and M2. NA localizes in the vicinity of the budding neck where it may contribute to membrane curvature changes and membrane scission.
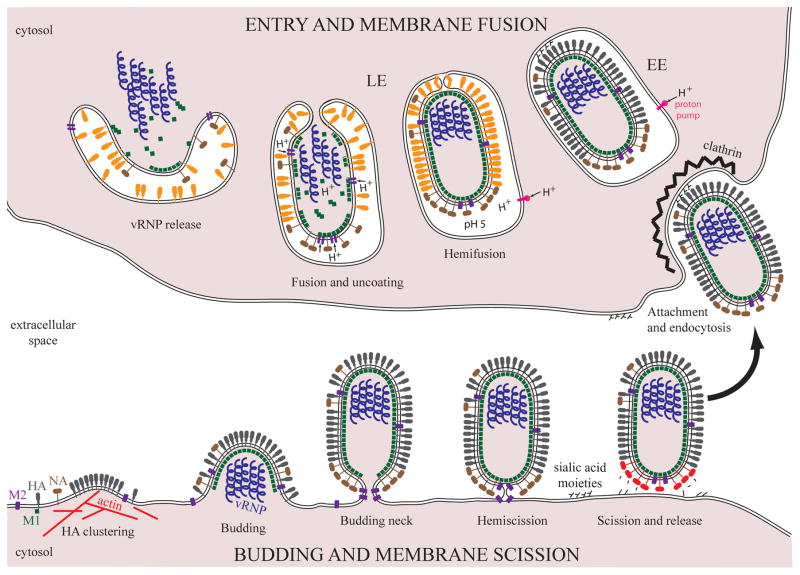
Figure 4. Cell membrane remodeling during IAV budding and entry.
IAV budding begins by formation of an HA cluster driven by the remodeling of the actin cytoskeleton at the plasma membrane (bottom left). The viral genome composed of 8 segments (vRNP) is incorporated into a nascent bud. M1 polymerizes into a layer with a helical arrangement underneath the plasma membrane providing the driving force for budding. Once the vRNP are incorporated, the plasma membrane is constricted into a budding neck at the rear end of the budding virion. NA concentrated around the budding neck cleaves off the sialic acid moieties (red NA) from the surface of the infected cells to prevent re-entry. M2 is thought to perform membrane scission, a process with a high-energy barrier, through an intermediate called hemiscission. Hemiscission proceeds to full scission, separating both leaflets of the viral and plasma membrane. The newly formed virion is released and can attach to the sialic acid moieties present on the plasma membrane of another host cell (arrow). Spherical virions are endocytosed by the clathrin-mediated pathway. Filamentous virions enter by macropinocytosis (not shown). In the early endosome (EE), the vacuolar proton pump gradually decreases the pH. HA changes conformation (orange HA) at ~pH 5 in the late endosome (LE) and mediates membrane fusion, a process with a high-energy barrier, through an intermediate called hemifusion. The M2 protein conducts protons into the virion interior, triggering the disassembly of the M1 layer and vRNP release. The initial HA mediated fusion pore expands and vRNP are released into the cytoplasm of the host cell.
A majority of imaging studies on HA clustering and HA-lipid association report results performed in the absence of viral infection or other influenza proteins. Thus, they are likely to be irrelevant to viral assembly. In infected cells, M1, which interacts with HA and assembles into a 4 nm thick layer underlying the membrane, likely influences HA mobility or actin association, but it does not affect lipid phase behavior. The question of how M1 and other influenza proteins influence HA clustering needs investigation. Importantly, super-resolution light microscopy studies need augmentation from 3D imaging with highest axial resolution possible in order to appropriately assess the contribution of membrane curvature in the HA clusters. Development of electron-dense tags, which would specifically label cholesterol or sphingomyelin would be extremely useful to fully exploit the potential of cryo-ET to study lipid domains in three dimensions at high resolution, such as the fascinating sphingomyelin-rich but cholesterol neutral domains discovered in the search for rafts.
The M1 matrix is the major determinant of IAV shape. The helical arrangement of the M1 layer in the filamentous or spherical particles differs. What determines the helical arrangement of the M1 layer? Is the helical rearrangement responsible for initiation and closure of the M1 layer in the filamentous virions? Additional cryo-EM studies elucidating the M1 structure at high resolution in the spherical and filamentous virions and at the ends of the filamentous virions would greatly facilitate our understanding of morphology and assembly of IAV virions. Since the M2 protein has been shown to play a crucial role in particle morphology, it is important to elucidate whether the local M1:M2 ratio may be a switch between helical arrangements determining whether the particle will be spherical or filamentous.
IAV, unlike other enveloped viruses, does not hijack the endosomal sorting complex required for transport (ESCRT) for scission of the budding membranous neck at the final stage of budding. Instead, M2 protein has been shown to perform membrane scission. Is M2 protein responsible for formation of a budding neck or is it only responsible for scission at the site of the budding neck? The most intriguing question is why M2 protein is dispensable for influenza VLP release, which are structurally similar to the real virus. Further structural studies utilizing VLP produced in the absence of the M2 protein will provide the answer to this contradiction.
NA expression in transfected cells results in formation and release of quasi-filamentous particles. In addition, NA localizes at spherically curved areas of the influenza VLP. This indicates that NA is able to modulate or respond to membrane curvature in a different way than HA and might be involved in the assembly and release of the IAV beyond its well-known enzymatic activity. Given that NA is found at the rear end of virions, does NA’s ability to modulate or sense curvature play a role in budding neck formation or scission?
HA mediated membrane envelope fusion with the endosomal membrane is thought to occur via membranous intermediates such as hemifusion, whose structure remains to be elucidated. In addition, future cryo-ET studies are needed to understand the conformational changes of HA driving membrane fusion. In particular, it is necessary to confirm the existence of the HA extended structure, which is the basis for the prevailing model of HA mediated membrane fusion.
Acknowledgments
We thank to Mary L. Kraft for kindly providing the nano-SIMS figure and Samuel T. Hess for useful suggestions. We are grateful to members of the Zimmerberg lab and to Fredric S. Cohen for critical reading of the manuscript. We sincerely apologize to all scientists whose important work could not be cited here due to space restrictions. This work was supported by the Division of Intramural Research of the NICHD, in the Intramural Research Program of the NIH.
References
- 1.Kordyukova LV, Serebryakova MV, Baratova LA, Veit M. S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J Virol. 2008;82:9288–92. doi: 10.1128/JVI.00704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatu U, Hammond C, Helenius A. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 1995;14:1340–8. doi: 10.1002/j.1460-2075.1995.tb07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mir-Shekari SY, Ashford DA, Harvey DJ, Dwek RA, Schulze IT. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J Biol Chem. 1997;272:4027–36. doi: 10.1074/jbc.272.7.4027. [DOI] [PubMed] [Google Scholar]
- 4.Keller P, Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J Cell Biol. 1998;140:1357–67. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas S, Yin SR, Blank PS, Zimmerberg J. Cholesterol promotes hemifusion and pore widening in membrane fusion induced by influenza hemagglutinin. J Gen Physiol. 2008;131:503–13. doi: 10.1085/jgp.200709932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 7.Hess ST, Gould TJ, Gudheti MV, Maas SA, Mills KD, Zimmerberg J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc Natl Acad Sci U S A. 2007;104:17370–5. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–27. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 9.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–7. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasilewski S, Calder LJ, Grant T, Rosenthal PB. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine. 2012;30:7368–73. doi: 10.1016/j.vaccine.2012.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippov A, Oradd G, Lindblom G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J. 2003;84:3079–86. doi: 10.1016/S0006-3495(03)70033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudheti MV, et al. Actin mediates the nanoscale membrane organization of the clustered membrane protein influenza hemagglutinin. Biophys J. 2013;104:2182–92. doi: 10.1016/j.bpj.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. [DOI] [PubMed] [Google Scholar]
- 14.Pathak P, London E. Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation. Biophys J. 2011;101:2417–25. doi: 10.1016/j.bpj.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casadei BR, Domingues CC, de Paula E, Riske KA. Direct visualization of the action of Triton X-100 on giant vesicles of erythrocyte membrane lipids. Biophys J. 2014;106:2417–25. doi: 10.1016/j.bpj.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lichtenberg D, Goni FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci. 2005;30:430–6. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Gerl MJ, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol. 2012;196:213–21. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shvartsman DE, Kotler M, Tall RD, Roth MG, Henis YI. Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J Cell Biol. 2003;163:879–88. doi: 10.1083/jcb.200308142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domanska MK, Dunning RA, Dryden KA, Zawada KE, Yeager M, Kasson PM. Hemagglutinin Spatial Distribution Shifts in Response to Cholesterol in the Influenza Viral Envelope. Biophys J. 2015;109:1917–24. doi: 10.1016/j.bpj.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scolari S, Engel S, Krebs N, Plazzo AP, De Almeida RF, Prieto M, Veit M, Herrmann A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J Biol Chem. 2009;284:15708–16. doi: 10.1074/jbc.M900437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevcsik E, Brameshuber M, Folser M, Weghuber J, Honigmann A, Schutz GJ. GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat Commun. 2015;6:6969. doi: 10.1038/ncomms7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson SA, Stinson BM, Go MS, Carmona LM, Reminick JI, Fang X, Baumgart T. Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim Biophys Acta. 2010;1798:1427–35. doi: 10.1016/j.bbamem.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaus J, et al. Hemagglutinin of influenza virus partitions into the nonraft domain of model membranes. Biophys J. 2010;99:489–98. doi: 10.1016/j.bpj.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polozov IV, Bezrukov L, Gawrisch K, Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat Chem Biol. 2008;4:248–55. doi: 10.1038/nchembio.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraft ML, Klitzing HA. Imaging lipids with secondary ion mass spectrometry. Biochim Biophys Acta. 2014;1841:1108–19. doi: 10.1016/j.bbalip.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Wilson RL, Frisz JF, Klitzing HA, Zimmerberg J, Weber PK, Kraft ML. Hemagglutinin clusters in the plasma membrane are not enriched with cholesterol and sphingolipids. Biophys J. 2015;108:1652–9. doi: 10.1016/j.bpj.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M, Northup N, Marga F, Huber T, Byfield FJ, Levitan I, Forgacs G. The effect of cellular cholesterol on membrane-cytoskeleton adhesion. J Cell Sci. 2007;120:2223–31. doi: 10.1242/jcs.001370. [DOI] [PubMed] [Google Scholar]
- 28.Gunewardene MS, Subach FV, Gould TJ, Penoncello GP, Gudheti MV, Verkhusha VV, Hess ST. Superresolution imaging of multiple fluorescent proteins with highly overlapping emission spectra in living cells. Biophys J. 2011;101:1522–8. doi: 10.1016/j.bpj.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts PC, Compans RW. Host cell dependence of viral morphology. Proc Natl Acad Sci U S A. 1998;95:5746–51. doi: 10.1073/pnas.95.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–23. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chlanda P, Schraidt O, Kummer S, Riches J, Oberwinkler H, Prinz S, Krausslich HG, Briggs JA. Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology. J Virol. 2015;89:8957–66. doi: 10.1128/JVI.00592-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai JC, Chan WW, Kien F, Nicholls JM, Peiris JS, Garcia JM. Formation of virus-like particles from human cell lines exclusively expressing influenza neuraminidase. J Gen Virol. 2010;91:2322–30. doi: 10.1099/vir.0.019935-0. [DOI] [PubMed] [Google Scholar]
- 33.Yondola MA, Fernandes F, Belicha-Villanueva A, Uccelini M, Gao Q, Carter C, Palese P. Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J Virol. 2011;85:2480–91. doi: 10.1128/JVI.02188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leyva-Grado VH, Hai R, Fernandes F, Belicha-Villanueva A, Carter C, Yondola MA. Modulation of an ectodomain motif in the influenza A virus neuraminidase alters tetherin sensitivity and results in virus attenuation in vivo. J Mol Biol. 2014;426:1308–21. doi: 10.1016/j.jmb.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stachowiak JC, et al. Membrane bending by protein-protein crowding. Nat Cell Biol. 2012;14:944–9. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 36.Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun. 2015;6:7875. doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A. 2010;107:10685–90. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batishchev OV, et al. pH-Dependent Formation and Disintegration of the Influenza A Virus Protein Scaffold to Provide Tension for Membrane Fusion. J Virol. 2015 doi: 10.1128/JVI.01539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruigrok RW, Barge A, Durrer P, Brunner J, Ma K, Whittaker GR. Membrane interaction of influenza virus M1 protein. Virology. 2000;267:289–98. doi: 10.1006/viro.1999.0134. [DOI] [PubMed] [Google Scholar]
- 40.Kretzschmar E, Bui M, Rose JK. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology. 1996;220:37–45. doi: 10.1006/viro.1996.0283. [DOI] [PubMed] [Google Scholar]
- 41.Thaa B, Herrmann A, Veit M. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology. 2009;383:150–5. doi: 10.1016/j.virol.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Hilsch M, Goldenbogen B, Sieben C, Hofer CT, Rabe JP, Klipp E, Herrmann A, Chiantia S. Influenza A matrix protein M1 multimerizes upon binding to lipid membranes. Biophys J. 2014;107:912–23. doi: 10.1016/j.bpj.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D, Harmon A, Jin J, Francis DH, Christopher-Hennings J, Nelson E, Montelaro RC, Li F. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J Virol. 2010;84:4673–81. doi: 10.1128/JVI.02306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Puertas P, Albo C, Perez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding. J Virol. 2000;74:11538–47. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu T, Ye Z. Restriction of viral replication by mutation of the influenza virus matrix protein. J Virol. 2002;76:13055–61. doi: 10.1128/JVI.76.24.13055-13061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Z, Robinson D, Wagner RR. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J Virol. 1995;69:1964–70. doi: 10.1128/jvi.69.3.1964-1970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–19. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen BJ, Takeda M, Lamb RA. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J Virol. 2005;79:13673–84. doi: 10.1128/JVI.79.21.13673-13684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–7. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugrue RJ, Belshe RB, Hay AJ. Palmitoylation of the influenza A virus M2 protein. Virology. 1990;179:51–6. doi: 10.1016/0042-6822(90)90272-s. [DOI] [PubMed] [Google Scholar]
- 51.Veit M, Klenk HD, Kendal A, Rott R. The M2 protein of influenza A virus is acylated. J Gen Virol. 1991;72(Pt 6):1461–5. doi: 10.1099/0022-1317-72-6-1461. [DOI] [PubMed] [Google Scholar]
- 52.Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–72. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–7. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J Am Chem Soc. 2013;135:13710–9. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tenchov BG, MacDonald RC, Siegel DP. Cubic phases in phosphatidylcholine-cholesterol mixtures: cholesterol as membrane “fusogen”. Biophys J. 2006;91:2508–16. doi: 10.1529/biophysj.106.083766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang T, Cady SD, Hong M. NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide. Biophys J. 2012;102:787–94. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kollmitzer B, Heftberger P, Rappolt M, Pabst G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/C3SM51829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T, Hong M. Investigation of the curvature induction and membrane localization of the influenza virus M2 protein using static and off-magic-angle spinning solid-state nuclear magnetic resonance of oriented bicelles. Biochemistry. 2015;54:2214–26. doi: 10.1021/acs.biochem.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawaguchi A, Hirohama M, Harada Y, Osari S, Nagata K. Influenza Virus Induces Cholesterol-Enriched Endocytic Recycling Compartments for Budozone Formation via Cell Cycle-Independent Centrosome Maturation. PLoS Pathog. 2015;11:e1005284. doi: 10.1371/journal.ppat.1005284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder C, Heider H, Moncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur Biophys J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- 62.Thaa B, Levental I, Herrmann A, Veit M. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem J. 2011;437:389–97. doi: 10.1042/BJ20110706. [DOI] [PubMed] [Google Scholar]
- 63.Thaa B, Siche S, Herrmann A, Veit M. Acylation and cholesterol binding are not required for targeting of influenza A virus M2 protein to the hemagglutinin-defined budozone. FEBS Lett. 2014;588:1031–6. doi: 10.1016/j.febslet.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 64.Thaa B, Tielesch C, Moller L, Schmitt AO, Wolff T, Bannert N, Herrmann A, Veit M. Growth of influenza A virus is not impeded by simultaneous removal of the cholesterol-binding and acylation sites in the M2 protein. J Gen Virol. 2012;93:282–92. doi: 10.1099/vir.0.038554-0. [DOI] [PubMed] [Google Scholar]
- 65.Roberts KL, Leser GP, Ma C, Lamb RA. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol. 2013;87:9973–82. doi: 10.1128/JVI.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roberts PC, Lamb RA, Compans RW. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998;240:127–37. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 67.Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J Virol. 2010;84:5078–88. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–70. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jimenez V, Scholte F, Garcia-Sastre A, Rottier PJ, de Haan CA. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossman JS, Leser GP, Lamb RA. Filamentous influenza virus enters cells via macropinocytosis. J Virol. 2012;86:10950–60. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito T, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–73. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 73.Parsegian VA, Fuller N, Rand RP. Measured work of deformation and repulsion of lecithin bilayers. Proc Natl Acad Sci U S A. 1979;76:2750–4. doi: 10.1073/pnas.76.6.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lague P, Roux B, Pastor RW. Molecular dynamics simulations of the influenza hemagglutinin fusion peptide in micelles and bilayers: conformational analysis of peptide and lipids. J Mol Biol. 2005;354:1129–41. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 75.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 76.Fontana J, Cardone G, Heymann JB, Winkler DC, Steven AC. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J Virol. 2012;86:2919–29. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruigrok RW, Wrigley NG, Calder LJ, Cusack S, Wharton SA, Brown EB, Skehel JJ. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5:41–9. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci U S A. 2001;98:7235–40. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiao H, Armstrong RT, Melikyan GB, Cohen FS, White JM. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–69. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–91. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 81.Kozlovsky Y, Chernomordik LV, Kozlov MM. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys J. 2002;83:2634–51. doi: 10.1016/S0006-3495(02)75274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Risselada HJ, Smirnova Y, Grubmuller H. Free energy landscape of rim-pore expansion in membrane fusion. Biophys J. 2014;107:2287–95. doi: 10.1016/j.bpj.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. Elife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spruce AE, Iwata A, Almers W. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc Natl Acad Sci U S A. 1991;88:3623–7. doi: 10.1073/pnas.88.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–69. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci U S A. 2008;105:15382–7. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. Elife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Domanska MK, Wrona D, Kasson PM. Multiphasic effects of cholesterol on influenza fusion kinetics reflect multiple mechanistic roles. Biophys J. 2013;105:1383–7. doi: 10.1016/j.bpj.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leikina E, Mittal A, Cho MS, Melikov K, Kozlov MM, Chernomordik LV. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J Biol Chem. 2004;279:26526–32. doi: 10.1074/jbc.M401883200. [DOI] [PubMed] [Google Scholar]
- 91.Sakai T, Ohuchi R, Ohuchi M. Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J Virol. 2002;76:4603–11. doi: 10.1128/JVI.76.9.4603-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Naeve CW, Williams D. Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 1990;9:3857–66. doi: 10.1002/j.1460-2075.1990.tb07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner R, Herwig A, Azzouz N, Klenk HD. Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J Virol. 2005;79:6449–58. doi: 10.1128/JVI.79.10.6449-6458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steinhauer DA, Wharton SA, Wiley DC, Skehel JJ. Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology. 1991;184:445–8. doi: 10.1016/0042-6822(91)90867-b. [DOI] [PubMed] [Google Scholar]
- 95.Melikyan GB, Jin H, Lamb RA, Cohen FS. The role of the cytoplasmic tail region of influenza virus hemagglutinin in formation and growth of fusion pores. Virology. 1997;235:118–28. doi: 10.1006/viro.1997.8686. [DOI] [PubMed] [Google Scholar]
- 96.Edinger TO, Pohl MO, Yanguez E, Stertz S. Cathepsin W Is Required for Escape of Influenza A Virus from Late Endosomes. MBio. 2015;6:e00297. doi: 10.1128/mBio.00297-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He J, Sun E, Bujny MV, Kim D, Davidson MW, Zhuang X. Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog. 2013;9:e1003701. doi: 10.1371/journal.ppat.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stauffer S, Feng Y, Nebioglu F, Heilig R, Picotti P, Helenius A. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J Virol. 2014;88:13029–46. doi: 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li S, Sieben C, Ludwig K, Hofer CT, Chiantia S, Herrmann A, Eghiaian F, Schaap IA. pH-Controlled two-step uncoating of influenza virus. Biophys J. 2014;106:1447–56. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fontana J, Steven AC. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J Virol. 2013;87:5621–8. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Banerjee I, et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science. 2014;346:473–7. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 103.Roberts KL, Manicassamy B, Lamb RA. Influenza A virus uses intercellular connections to spread to neighboring cells. J Virol. 2015;89:1537–49. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]