Abstract
Bone marrow-derived skeletal stem/stromal cell (SSC) self-renewal and function are critical to skeletal development, homeostasis and repair. Nevertheless, the mechanisms controlling SSC behavior, particularly bone formation, remain ill-defined. Using knockout mouse models that target the zinc-finger transcription factors, Snail, Slug or Snail and Slug combined, a regulatory axis has been uncovered wherein Snail and Slug cooperatively control SSC self-renewal, osteoblastogenesis and bone formation. Mechanistically, Snail/Slug regulate SSC function by forming complexes with the transcriptional co-activators, YAP and TAZ, in tandem with the inhibition of the Hippo pathway-dependent regulation of YAP/TAZ signaling cascades. In turn, the Snail/Slug-YAP/TAZ axis activates a series of YAP/TAZ/TEAD and Runx2 downstream targets that control SSC homeostasis and osteogenesis. Together, these results demonstrate that SSCs mobilize Snail/Slug-YAP/TAZ complexes to control stem cell function.
Skeletal stem/stromal cells (SSCs) are found throughout tissues during embryogenesis and postnatal life1, 2. Though the multipotent potential of these cells is subject to debate, bone marrow-derived SSCs differentiate into osteoblasts, chondrocytes or adipocytes as well as hematopoietic stem cell-supportive stroma2. Interestingly, recent studies raise the possibility that SSCs express the transcriptional repressors, Snail and Slug (alternatively termed, Snail2)3. Though best known for their roles in orchestrating epithelial-mesenchymal transition (EMT) programs associated with development4–6, recent studies suggest that Snail and Slug impact stem cell functions via largely undefined mechanisms7–12. While screening mouse tissues for Snail and Slug expression, we unexpectedly discovered that both transcription factors are expressed in SSCs during pre- and post- natal states. Interestingly, targeting either Snail or Slug alone exerts only subtle effects on developmental programs. By contrast, dual knockout of both transcription factors markedly impairs SSC self-renewal, differentiation and bone formation. As such, we set out to define the means by which Snail and Slug co-regulate SSC function.
RESULTS
Snail/Slug Co-Dependent Regulation of SSC Proliferation and Differentiation
Using Snail/LacZ and Slug/LacZ knock-in mice11, 13, β-galactosidase activity is associated with cranial sutures, calvaria, long bones and cartilage (Fig. 1a,c, Supplementary Fig. 1a–e). Furthermore, in bone marrow, Snail-LacZ- as well as Slug-LacZ- knock-in mice display β-galactosidase activity in fibroblast-like cells (Fig. 1b,d). As such, SSCs were isolated by FACS, and both Snail and Slug mRNA expression confirmed in freshly sorted cells (Supplementary Fig. 1f). Furthermore, following the in vitro expansion of SSCs recovered from Snail-LacZ- or Slug-LacZ- knock-in mice, virtually all cells express β-galactosidase activity (Fig. 1e).
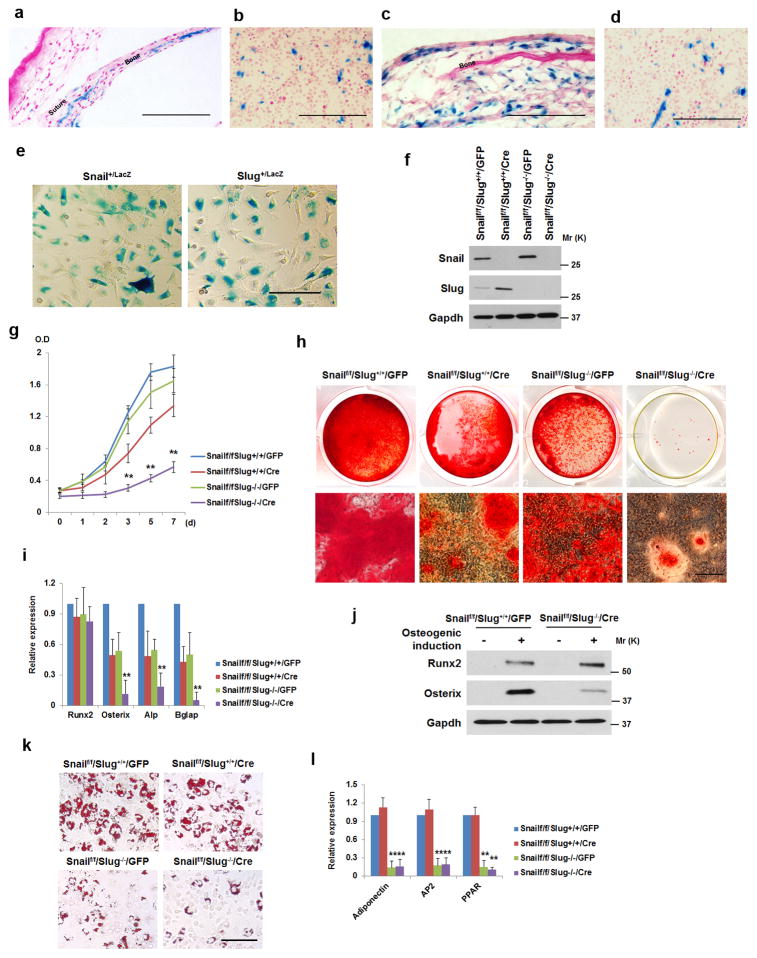
Figure 1. Snail/Slug Regulate SSC Proliferation and Differentiation.
a) LacZ expression in E17.5 Snail+/LacZ neonatal sagittal suture and parietal bone. Scale bar: 100 μm. Results are representative of 3 experiments performed.
b) LacZ expression in a sub-population of bone marrow cells in the femur of a 2-wk old Snail+/LacZ mouse. Scale bar: 100 μm. Results are representative of 3 experiments performed.
c) LacZ expression in Slug+/LacZ 7-d old sagittal suture and parietal bone. Scale bar: 100 μm. Results are representative of 3 experiments performed.
d) LacZ expression in a sub-population of bone marrow cells in the femur of a 2-wk old Slug+/LacZ mouse. Scale bar: 100 μm. Results are representative of 3 experiments performed.
e) LacZ expression in bone marrow-derived SSCs isolated from 4-wk old Snail+/LacZ or Slug+/LacZ mice. Scale bar: 100 μm. Results are representative of 3 experiments performed.
f) Western blot of Snail and Slug in SSCs isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice transduced with adeno-GFP or Cre expression vectors. Results are representative of 3 experiments performed.
g) Growth curve of SSCs isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice, and transduced with adeno-GFP or Cre expression vectors (mean ± s.d. n=3 independent experiments). **p<0.01; one-way ANOVA.
h) SSCs were isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. Cells were cultured under osteogenic conditions for 14 d, and stained with Alizarin Red S. Lower panels are magnified images of the upper panels. Scale bar: 100 μm. Results are representative of 3 experiments performed.
i) Relative mRNA expression of osteogenic markers in cultures from (h) (mean ± s.d., n=3 independent experiments). **p<0.01; one-way ANOVA.
j) SSCs were isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. After culture with or without osteogenic medium for 7 d, cell lysates were immunoblotted. Results are representative of 3 experiments performed.
k) SSCs were isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. After culture under adipogenic conditions for 7 d, cultures were stained with Oil Red O. Scale bar: 100 μm. Results are representative of 3 experiments performed.
l) Relative mRNA expression of adipogenic markers in cultures from (k) (mean ± s.d., n=3 independent experiments). **p<0.01; one-way ANOVA.
Next, bone marrow-derived SSCs (herein termed SSCs) were isolated from Snailf/f/Slug+/+ or Snailf/f/Slug−/− mice and transduced with adenoviral Cre- or a GFP control expression vector to yield either Snailf/f/Slug+/+, Snail−/−/Slug+/+, Snailf/f/Slug−/− or Snail−/−/Slug−/− progenitors. Consistent with studies demonstrating that Snail and Slug transcriptionally compensate for each other’s loss14, 15, deleting Snail up-regulates Slug expression while the loss of Slug expression induces Snail expression (Fig. 1f and Supplementary Fig. 1f). Under these conditions, deleting Slug or Snail alone yields only subtle effects on SSC proliferation (Fig. 1g). By contrast, in the absence of both Snail and Slug, SSC proliferation is decreased by ~75% with attendant losses in Ki67 expression (Fig. 1g and Supplementary Fig. 1g,h). Changes in stem cell-associated transcription factors, e.g., Nanog, Sox2 or Oct4, are not observed in double-null SSCs relative to controls. Further, while Snail and Slug can mediate anti-apoptotic effects16, Snail/Slug-deleted SSCs display no changes in apoptosis (Supplementary Fig. 1i).
To assess the roles of Snail and Slug in SSC differentiation, Snail/Slug-expressing cells (i.e., Snailf/f/Slug+/+/GFP), Snail-deleted cells (i.e., Snailf/f/Slug+/+/Cre), Slug-null cells (i.e., Snailf/f/Slug−/−/GFP) or double-nulls cells (i.e., Snailf/f/Slug−/−/Cre) were established as confluent cultures and induced to undergo osteogenesis. Over a 14 d culture period, the control Snailf/f/Slug+/+ SSCs differentiate into osteoblasts as assessed either by Alizarin Red S staining, the induction of the osteoblast markers, Runx2, Osterix, alkaline phosphatase (ALP) and Bglap2 at the mRNA level, or Runx2 and Osterix at the protein level (Fig. 1h–j). When compared to control SSCs, Snail- or Slug- deleted SSCs display only modest defects in mineralization or osteoblast commitment while maintaining their ability to up-regulate the master osteoblast transcription factor, Runx2 (Fig. 1h–j). By contrast, Snail/Slug-deficient SSCs display a complete defect in mineralization with commensurate losses in Osterix, ALP and Bglap2 expression (Fig. 1h,i) while Runx2 expression is fully retained (Fig. 1i,j), highlighting the fact that these cells maintain their ability to respond to osteogenic signals, whereas differentiation programs that operate downstream of Runx2 cannot be engaged. Significantly, a combined requirement for Snail and Slug in SSC differentiation is largely confined to osteoblastogenesis, i.e., when Snail, Slug or both Snail and Slug are targeted in SSCs and induced to undergo chondrogenesis, cartilage deposition as well as the expression of chondrogenic markers is are unaffected (Supplementary Fig. 1j). Alternatively, under adipogenic conditions, Snail-deleted SSCs generate lipid-rich adipocytes comparably to controls with normal expression of adipogenic markers (Fig. 1k,l). By contrast, and consistent with previous reports17, Slug-null SSCs are unable to mount an adipogenic response with no further adverse effects observed following the co-deletion of Snail/Slug (Fig. 1k,l). While these results demonstrate that osteoblast commitment in SSCs isolated from young mice (i.e., 4-wk old or younger) is dependent upon a dual requirement for Snail/Slug, recent studies suggest that SSCs arise from distinct populations in early versus late post-natal periods18, 19. Nevertheless, when SSCs were alternatively isolated from 3-month old mice, Snail and Slug regulate adult SSC function in a manner identical to SSCs isolated from young mice (Supplementary Fig. 1k,l).
Snail/Slug Regulate SSC Function and Bone Formation
To determine whether the in vitro defects in SSC self-renewal and differentiation can be extended in vivo, we generated a conditional Snail-deleted mouse model in Slug wild-type or Slug-null backgrounds by crossing Snailf/f/Slug+/+ mice or Snailf/f/Slug−/− mice with Dermo1-Cre mice, wherein Cre is expressed in mesoderm-derived tissues, including SSCs20–22. While Snail conditional knockouts and Slug−/− mice are born in the expected ratios, and viable as well as fertile (see below), Snailf/f/Slug−/−/Dermo1-Cre mice die upon delivery [likely due to defective palate formation23]. Consequently, embryos from each of the crosses were harvested at E17.5 for analysis. Consistent with our in vitro data, deletion of either Snail or Slug (i.e., in Snailf/f/Slug+/+/Dermo1-Cre or Snailf/f/Slug−/− mice, respectively) results in only subtle changes in long bone ossification or cartilage structure (Fig. 2a and Supplementary Fig. 2b,d). Likewise, in the skull, where calvarial bone forms by intramembranous ossification, thereby bypassing the formation of a cartilaginous anlage24, only partial defects are observed in calvarial ossification or suture closure following targeting of Snail or Slug alone (Fig. 2b and Supplementary Fig. 2a,c). By contrast, in Snailf/f/Slug−/−/Dermo1-Cre mice, bone development is severely comprised with remarkable decreases in bone length and an almost complete loss of bone ossification (Fig. 2a). Further, in agreement with recent studies14, the columnar organization of proliferating chondrocytes is also misaligned (Supplementary Fig. 2e,f). Importantly, bone defects occur independently of cartilage development as bone formation and ossification are also undermined in the skulls of Snailf/f/Slug−/−/Dermo1-Cre mice (Fig. 2b), with cross-sections of E17.5 parietal plates highlighting an almost complete absence of calcified bone (Supplementary Fig. 2g). Further, the condensed mesenchymal layer, dominated by SSCs and osteoprogenitors25, is significantly thinner, consistent with a 60% decrease in Ki67 staining without changes in progenitor cell apoptosis (Fig. 2c–f and Supplementary Fig. 2h). Calvarial extracts recovered from Snailf/f/Slug−/−/Dermo1-Cre mice also display normal Runx2 expression in tandem with marked defects in Osterix, ALP and Bglap2 expression (Fig. 2g).
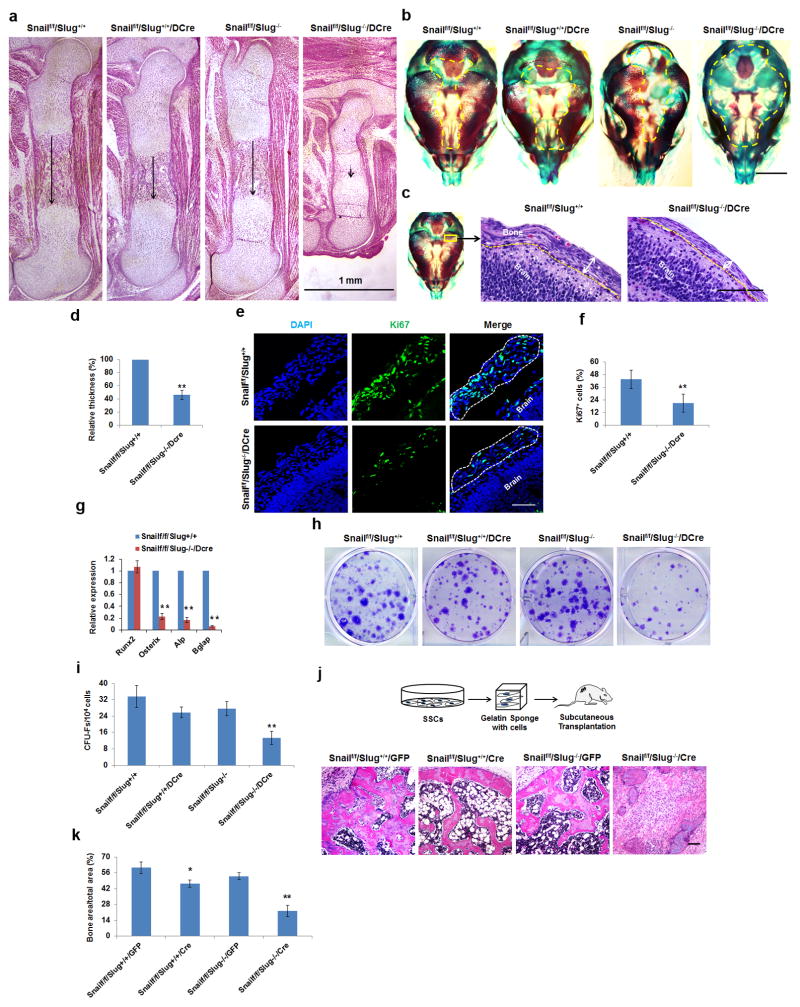
Figure 2. Skeletal and SSC-Associated Defects in Snail/Slug-Targeted Mice.
(a) Femur histology of E17.5 Snailf/f/Slug+/+, Snailf/f/Slug+/+/Dermo1-Cre Snailf/f/Slug−/− and Snailf/f/Slug−/−/Dermo1-Cre embryos. Scale bar: 1 mm. Results are representative of 5 experiments performed.
(b) Alizarin Red/Alcian Blue staining of skulls isolated from E17.5 Snailf/f/Slug+/+, Snailf/f/Slug+/+/Dermo1-Cre, Snailf/f/Slug−/− and Snailf/f/Slug−/−/Dermo1-Cre embryos. Scale bar: 500 μm. Results are representative of 5 experiments performed.
(c) Histology of E15 Snailf/f/Slug+/+ and Snailf/f/Slug+/+/Dermo1-Cre skulls across the parietal area. Scale bar: 100 μm. Results are representative of 5 experiments performed.
(d) Relative thickness of skull mesenchymal cell layers from (c) (mean ± s.d., n=5 mice). **p<0.01; unpaired t-test.
(e) Ki67 Immunofluorescence of parietal suture mesenchymal cell layers from E15 Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Dermo1-Cre skulls. Scale bar: 25 μm. Results are representative of 4 experiments performed.
(f) Quantification of Ki67-positive cells from (e) (mean ± s.d., n=4 mice). **p<0.01; unpaired t-test.
(g) Relative mRNA expression of osteogenic markers in E17.5 calvarial RNA extracts from Snailf/f/Slug+/+ versus Snailf/f/Slug−/−/Dermo1-Cre embryos (mean ± s.d., n=5 mice). **p<0.01; unpaired t-test.
(h) CFU-F generation from bone marrow cells isolated from E18.5 Snailf/f/Slug+/+, Snailf/f/Slug+/+/Dermo1-Cre, Snailf/f/Slug−/− and Snailf/f/Slug−/−/Dermo1-Cre mice. Results are representative of 5 experiments performed.
(i) CFU-F colony counts from (h) (mean ± s.d., n=5 mice). **p<0.01; one-way ANOVA.
(j) Schematic of the overall experimental design for in vivo implantation (upper panels).The lower panels show histology of tissues isolated from nude mice transplanted with Snailf/f/Slug+/+/GFP or Snailf/f/Slug−/−/Cre SSCs. Results are representative of 3 performed. Scale bar: 100 μm.
(k) Quantification bone formation in tissues from (j) (mean ± s.d., n=3 mice). **p<0.01, *p<0.05; one-way ANOVA.
To define the in vivo role of Snail/Slug in SSC homeostasis, colony-forming unit fibroblasts (CFU-Fs) assays were performed22, 26. Of note, dual deletion of Snail and Slug induces a severe loss of CFU-Fs in the embryonic bone of Snailf/f/Slug−/−/Dermo1-Cre mice relative to that found in the Snailf/f/Slug+/+/Dermo1-Cre, Snailf/f/Slug−/− mice or Snailf/f/Slug+/+ mice (Fig. 2h,i). The cell autonomous nature of the dual requirement for Snail and Slug is further confirmed following transplant of isolated Snail-deleted/Slug-null SSCs into nude mouse recipients. Whereas Snailf/f/Slug+/+/Cre SSCs or Snailf/f/Slug−/−/GFP SSCs generate bone-rich osteoids comparably to control Snailf/f/Slug+/+/GFP SSCs, the combined loss of Snail and Slug dramatically reduces bone formation (Fig. 2j,k).
Bone Marrow-Directed Deletion of Snail/Slug Retards Postnatal Osteogenesis
The lethal phenotype or Snailf/f/Slug−/−/Dermo1-Cre-targeted mice precludes our ability to assess the role of Snail and Slug in regulating postnatal osteogenesis. Recently, however, Osterix-Cre has been shown to target bone marrow-associated stromal cells displaying SSC- as well as osteoprogenitor- like characteristics27–30. Hence, Osterix-Cre mice were crossed with Snailf/f/Slug+/− mice yielding Snailf/f/Slug+/+ (i.e., control) mice, Snailf/f/Slug+/+/Osterix-Cre (i.e., Snail-deleted) mice, Snailf/f/Slug−/− (Slug-null) mice and Snailf/f/Slug−/−/Osterix-Cre (i.e., Snail/Slug-targeted) mice. Mice from all four groups are born in the expected Mendelian rations and display normal lifespans. Whereas Snail-deleted and Slug-null mice are slightly smaller in stature than controls, the Snail/Slug-targeted mice reach only ~2/3 the size/weight of the controls (Fig. 3a). Further, though the Snail-deleted or Slug-null mice display a mild calvarial phenotype with retarded ossification and delayed suture fusion, bone development is strikingly compromised in the Snail/Slug-targeted mice (Fig. 3b). Femur cross-sections of Snail/Slug-targeted Osterix-Cre mice exhibit the most dramatic loss in bone mass, thickness and trabeculation (Fig. 3c–e). Consistent with our in vitro analyses, we also note a decrease in the proliferative potential, but not apoptotic rate, of bone surface-associated SSC/osteoprogenitors that is accompanied by a marked decrease in Osterix, ALP and Bglap2, but not Runx2, mRNA expression (Fig. 3f–h and Supplementary Fig. 3a,b). Likewise, a severe depletion of CFU-Fs is found in adult bone marrow of Snailf/f/Slug−/−/Osterix-Cre mice relative to the Snailf/f/Slug+/+/Osterix-Cre, Snailf/f/Slug−/− or Snailf/f/Slug+/+ mice (Supplementary Fig. 3c,d). Though bone loss can also be caused by unbalanced bone resorption24, differences in osteoclast counts are not detected (Supplementary Fig. 3e,f).
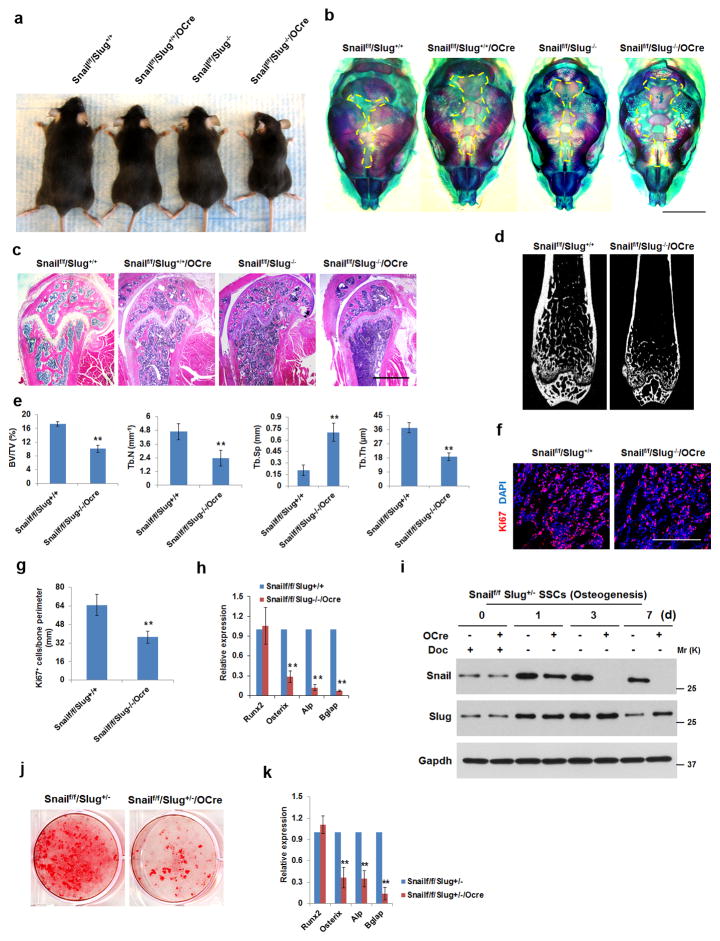
Figure 3. Osteogenic Defects in Osterix-Cre-Targeted Conditional Knockouts of Snail/Slug.
(a) Gross view of 3-month old Snailf/f/Slug+/+/Osterix-Cre, Snailf/f/Slug−/−, Snailf/f/Slug−/−/Osterix-Cre mice and a control littermate (Snailf/f/Slug+/+). Results are representative of 5 experiments performed.
(b) Alizarin Red/Alcian Blue staining of skulls isolated from neonatal Snailf/f/Slug+/+, Snailf/f/Slug+/+/Osterix-Cre, Snailf/f/Slug−/− and Snailf/f/Slug−/−/Osterix-Cre mice. Scale bar: 2 mm. Results are representative of 3 experiments performed.
(c) Femur histology of 3-month old Snailf/f/Slug+/+, Snailf/f/Slug+/+/Osterix-Cre, Snailf/f/Slug−/− and Snailf/f/Slug−/−/Osterix-Cre mice. Scale bar: 1 mm. Results are representative of 5 experiments performed.
(d) Microcomputed tomography of the proximal femur of 12-wk old Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Osterix-Cre mice. Results are representative of 10 experiments performed.
(e) Bone Mineral Density (BMD), Bone Volume/Tissue Volume (BV/TV), Trabecular Thickness (Tb.Th), Trabecular Number (Tb.N) and Trabecular Separation (Tb.Sp) were measured by microcomputed tomography in 12-wk old Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Osterix-Cre mice (mean ± s.d., n=10 mice). **p<0.01; unpaired t-test.
(f) Ki67 expression in proximal femurs isolated from neonatal Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Osterix-Cre mice. Scale bar: 100 μm. Results are representative of 5 experiments performed.
(g) Quantification of Ki67-positive cells from (f) (mean ± s.d., n=5 mice). **p<0.01; unpaired t-test.
(h) Relative mRNA expression of osteogenic markers in neonatal calvarial RNA extracts from Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Osterix-Cre mice (mean ± s.d., n=5 mice). **p<0.01; unpaired t-test.
(i) Western blot of Snail and Slug expression in SSCs isolated from Snailf/f/Slug+/− or Snailf/f/Slug+/−/Osterix-Cre mice cultured with doxycycline (Doc; 1.5 μg/ml) under control or osteogenic conditions for the indicated time periods. Results are representative of 3 experiments performed.
(j) SSCs from (i) were cultured under osteogenic conditions for 14 d and stained with Alizarin Red S. Results are representative of 3 experiments performed.
(k) Relative mRNA expression of osteogenic markers in cultures from (j) (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
Given the complex expression pattern of Osterix-Cre in transgenic mice27, 28, 30, we confirmed defects in Osterix-Cre-targeted SSC/osteoprogenitor function in vitro. As the Osterix-Cre transgene is tet-off regulated31, SSCs were isolated from Snailf/f mice maintained on doxycycline in a Slug heterozygous background (i.e., Slug+/− mice are indistinguishable from wild-type littermates while both Snailf/f/Slug+/−/Dermo1-Cre as well as Osterix-Cre mice display bone defects, albeit less severe than those observed in the Slug-null background). As expected, SSCs cultured in the presence of doxycycline co-express Snail and Slug comparably to controls (i.e., Snailf/f/Slug+/− SSCs). When doxycycline is removed from the media and osteogenesis induced (day 1), Snail protein levels fall to undetectable levels by culture day 3 while Slug proteins are induced relative to the heterozygous controls (Fig. 3i). Under these conditions, marked defects in osteoblastogenesis are again confirmed while early osteoblast commitment, reflected in Runx2 expression, is unaltered (Fig. 3j,k). Similar results are obtained when calvarial osteoprogenitors are recovered from Snailf/f/Slug−/− mice and transduced with adeno-Cre in vitro (Supplementary Fig. 3g,h). Thus, Snail and Slug are co-required during SSC as well as osteoprogenitor commitment to the bone lineage in vitro and in vivo.
Snail/Slug Promote SSC Proliferation and Osteogenesis via YAP/TAZ Activation
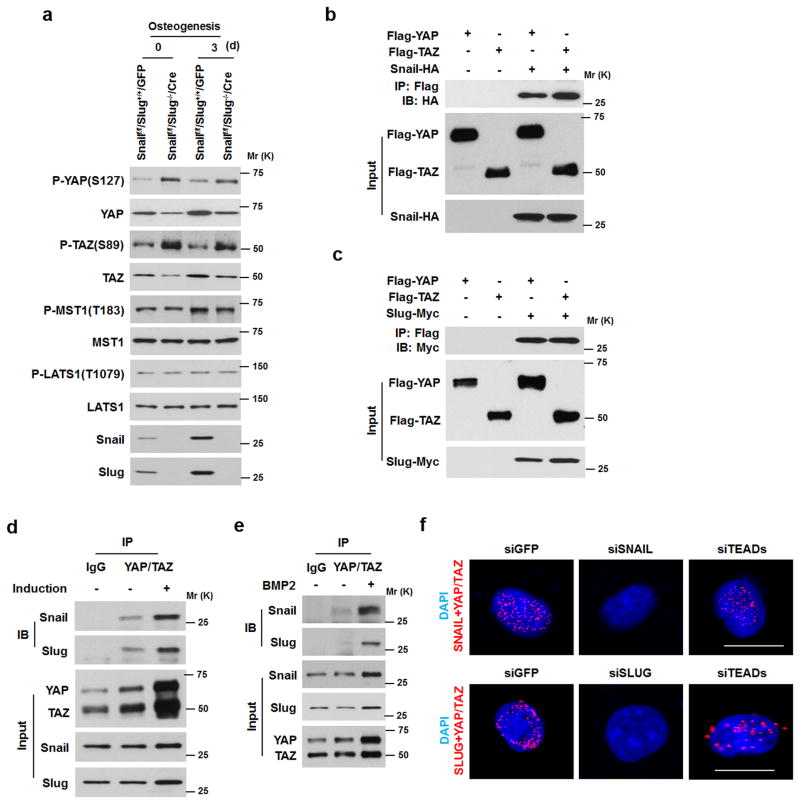
To date, a dual requirement for Snail and Slug has not been linked to the control of SSC proliferative responses or differentiation. Interestingly, the Hippo pathway components, YAP or TAZ, regulate stem cell self-renewal and differentiation programs32–35. Indeed, whereas control SSCs express YAP as well as TAZ at the protein level that each localize primarily to the nuclear compartment, Snail/Slug-deleted SSCs display significant decreases in YAP and TAZ protein levels with no changes in their relative mRNA levels (Fig. 4a,b and Supplementary Fig. 4a). Importantly, decreases in YAP/TAZ protein levels also correlate with lower expression of the YAP/TAZ targets, Ctgf and Ankrd132–35 (Supplementary Fig. 4b). Furthermore, while YAP and TAZ protein levels increase during SSC osteogenesis in tandem with increases in Snail and Slug levels, Snail/Slug-deleted SSCs continue to express lower protein levels of YAP and TAZ during differentiation (Fig. 4c). Likewise, osteoprogenitors undergoing osteoblast commitment with BMP-2 also increase YAP and TAZ protein levels in tandem with Snail and Slug (Fig. 4d,e and Supplementary Fig. 4c,d). By contrast, when both Snail and Slug are targeted, YAP and TAZ protein levels decrease (Fig. 4c,e). Similarly, SSCs/osteoprogenitors isolated from calvarial extracts of Snailf/f/Slug−/−/Dermo1-Cre mice or femurs of Snailf/f/Slug−/−/Osterix-Cre mice also exhibit decreases in YAP/TAZ protein levels (Fig. 4f,g and Supplementary Fig. 4e,f).
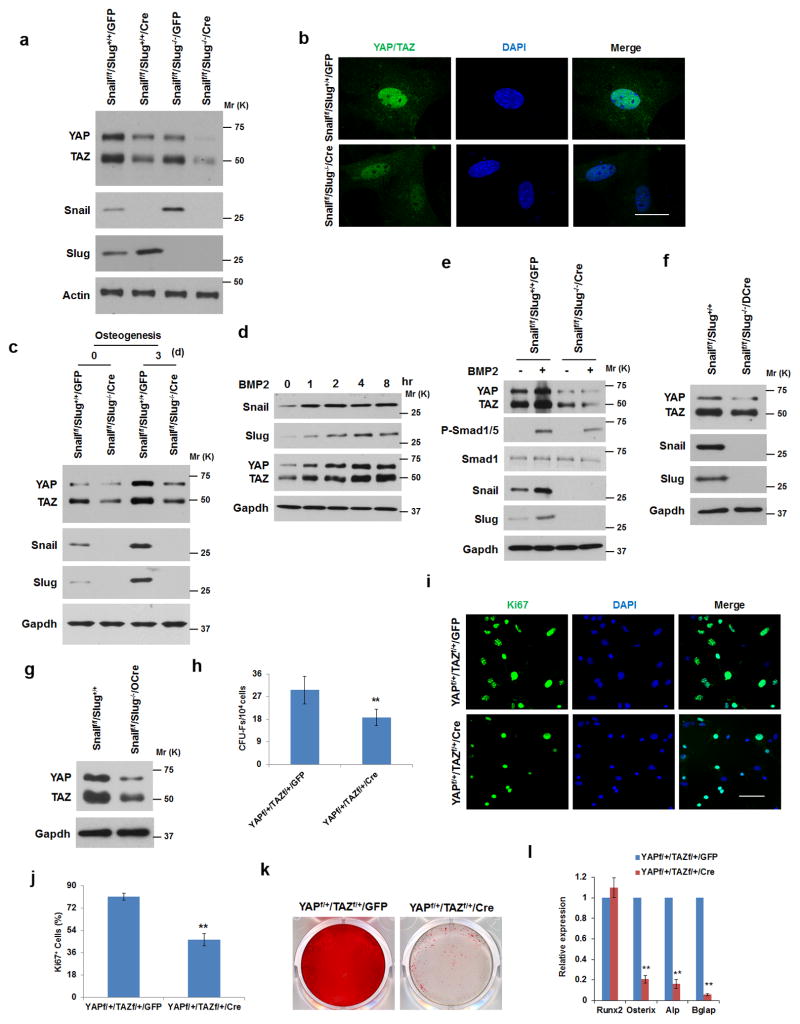
Figure 4. Snail/Slug Regulate YAP/TAZ Levels in SSCs.
(a) YAP and TAZ expression levels in SSCs isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. Results are representative of 3 experiments performed.
(b) YAP and TAZ localization in Snailf/f/Slug+/+ and Snailf/f/Slug−/− SSCs following transduction with adeno-LacZ or Cre. Nuclei were counterstained with DAPI (blue). Scale bar: 10 μm. Results are representative of 3 experiments performed.
(c) YAP and TAZ expression levels in Snailf/f/Slug+/+/GFP and Snailf/f/Slug−/−/Cre SSCs cultured under osteogenic conditions as indicated. Results are representative of 3 experiments performed.
(d) BMP2 induces Snail/Slug and YAP/TAZ levels in osteoblast progenitors. Calvarial osteoblast progenitors were isolated from E18 mice embryos, treated with 100 ng/ml BMP2 and protein expression monitored by Western blot. Results are representative of 3 experiments performed.
(e) Deletion of Snail/Slug in osteoblast progenitors blunts the BMP2-induced up-regulation of YAP/TAZ. Calvarial osteoblast progenitors were isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. Cells were then treated with 100 ng/ml BMP2 for 2 h, and protein expression monitored by Western blot. Results are representative of 3 experiments performed.
(f) YAP and TAZ expression levels in E17.5 Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Dermo1-Cre calvarial lysates. Results are representative of 5 experiments performed.
(g) YAP and TAZ expression levels in E17.5 Snailf/f/Slug+/+ and Snailf/f/Slug−/−/Osterix-Cre femur lysates. Results are representative of 5 experiments performed.
(h) CFU-F colony count from bone marrow cells isolated from YAPf/+/TAZf/+ mice following transduction with lentivirus GFP or Cre expression vectors (mean ± s.d., n=5 independent experiments). **p<0.01; unpaired t-test.
(i) Ki67 expression in SSCs isolated from YAPf/+/TAZf/+ mice following transduction with adeno-GFP or Cre. Scale bar: 50 μm. Results are representative of 3 experiments performed.
(j) Quantification of Ki67-positive cells from (i) (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
(k) Alizarin Red S staining of SSCs isolated from YAPf/+/TAZf/+ mice following transduction with adeno-GFP or Cre and cultured under osteogenic conditions for 14 d. Results are representative of 3 experiments performed.
(l) Relative mRNA expression of osteogenic markers in cultures from (k) (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
To determine whether decreases in YAP/TAZ protein levels affect SSC self-renewal and differentiation in a manner that phenocopies the Snail/Slug-deleted state, SSCs were isolated from YAPf/+/TAZf/+ mice and transduced with adenoviral-GFP or Cre expression vectors. As predicted, YAP and TAZ protein levels decrease in hemizygous SSCs (without affecting either Snail/Slug protein levels or their subcellular localization) with coordinate decreases in SSC proliferation and CFU-F formation (Fig. 4h–j and Supplementary Fig. 4g,h). Likewise, when YAP/TAZ-hemizygote SSCs are cultured under osteogenic conditions, mineralization is blocked (Fig. 4k). As observed in Snail/Slug double-null mice, Runx2 expression is up-regulated normally in YAP−/+/TAZ−/+ SSCs, while Osterix, ALP and Bglap2 are repressed (Fig. 4l). Similar changes are not observed in YAP−/+ SSCs or TAZ−/+ SSCs. Interestingly, transient expression of Snail in YAPf/+/TAZf/+ osteoprogenitors partially rescues both proliferative and osteogenic activities (Supplementary Fig. 4i–k).
Snail/Slug–YAP/TAZ Complexes Regulate the Hippo Pathway
YAP/TAZ stability is regulated by the Hippo cascade-associated kinases, MST1 and MST2 as well as the Lats kinases, LATS1 and LATS2, that ultimately control YAP/TAZ phosphorylation, subcellular localization and proteasomal degradation34–37. While Snail/Slug did not affect either MST1 or LATS1 protein levels or phosphorylation-dependent activation states, the combined absence of Snail and Slug increases phospho-YAP and phospho-TAZ levels with consequent losses in total YAP/TAZ levels – both prior to, and during, osteogenesis (Fig. 5a). Consistent with increased YAP/TAZ phosphorylation levels, binding interactions between Lats1/2 and YAP/TAZ are increased in Snail/Slug-deleted SSCs (Supplementary Fig. 5a). The half-lifes of YAP/TAZ proteins also decrease in Snail/Slug-deleted osteoprogenitors, while, by contrast, YAP/TAZ protein levels increase when proteasome activity is inhibited (Supplementary Fig. 5b–d). As such, we considered a model wherein Snail/Slug form complexes with YAP/TAZ that directly or indirectly protect the transcriptional co-activators from proteasomal degradation. Indeed, following co-expression of epitope-tagged YAP or TAZ with epitope-tagged Snail or Slug, interacting complexes between Snail and YAP, Snail and TAZ, Slug and YAP and Slug and TAZ, are detected by co-immunoprecipitation in COS-1 cells as well as by in situ proximity ligation assay in the C3H10T1/2 progenitor cell line (Fig. 5b,c and Supplementary Fig. 5e). Further, endogenous complexes between Snail/Slug and YAP/TAZ are found in SSCs as well as calvarial osteoprogenitors undergoing osteogenesis (Fig. 5d–f and Supplementary Fig. 5f–h). Complementing these findings, formation of nuclear YAP/TAZ-Snail/Slug complexes occurs independently of the expression of TEAD, the dominant co-transcriptional activator of YAP/TAZ function34–36. (Fig. 5d–f and Supplementary Fig. 5f–h).
Figure 5. Snail/Slug-Dependent Regulation of YAP/TAZ Stability.
(a) Western blot of p-YAP, p-TAZ, p-Lats1 and p-Mst1 in SSCs isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre. Results are representative of 3 experiments performed.
(b) Association of YAP/TAZ with Snail was detected by immunoprecipitation in Cos-1 cells transfected with Flag-YAP, Flag-TAZ and Snail-HA, respectively, as indicated. Results are representative of 3 experiments performed.
(c) Association of YAP/TAZ with Slug was detected by immunoprecipitation in Cos-1 cells transfected with Flag-YAP, Flag-TAZ and Slug-Myc, respectively, as indicated. Results are representative of 3 experiments performed.
(d) Associations between endogenous YAP/TAZ and Snail/Slug were detected in SSCs cultured in the absence or presence of osteogenic medium. Results are representative of 3 experiments performed.
(e) BMP2 increases Snail/Slug-YAP/TAZ complex formation. Calvarial osteoblast progenitors isolated from E17.5 mice were treated with 100 ng/ml BMP2 for 2 h. Endogenous Snail/Slug-YAP/TAZ complexes were detected by immunoprecipitation. Results are representative of 3 experiments performed.
(f) In situ proximity ligation assay detection of endogenous Snail/YAP/TAZ and Slug/YAP/TAZ interactions in human SSCs. siRNAs: siGFP, siSNAIL, siSLUG or siTEADs(1–4) were transfected, respectively. Nuclei were counterstained with DAPI (blue). The detected interaction sites are marked by fluorescent dots (red). Scale bar: 5μm. Results are representative of 3 experiments performed.
Snail/Slug–YAP/TAZ Complexes Regulate Transcriptional Activity
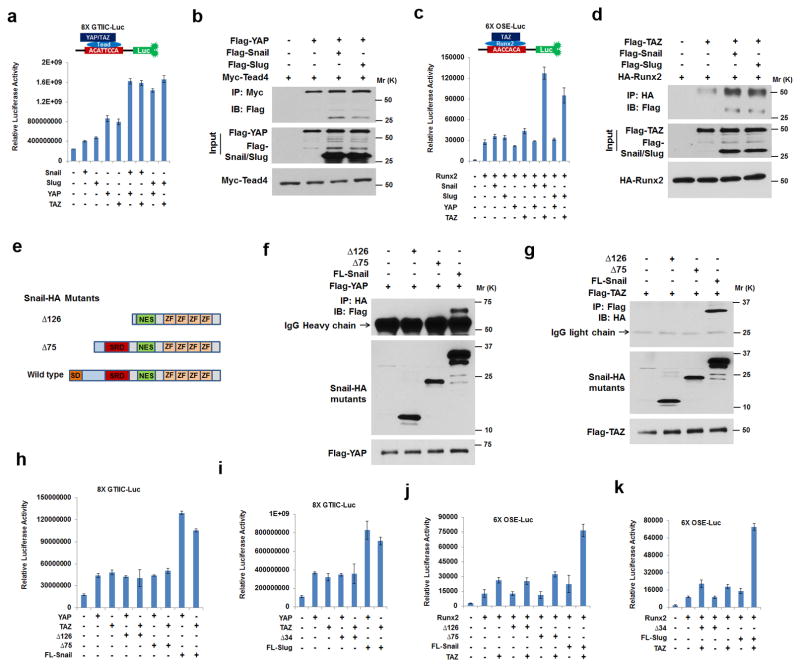
YAP and TAZ can regulate transcriptional events by forming complexes with TEAD transcription factors (i.e., TEAD1–4)34–36. Given the ability of Snail and Slug to interact with YAP/TAZ, we sought to determine whether YAP/TAZ-dependent TEAD transcriptional activity is modulated by Snail/Slug complex formation. As such, TEAD-luciferase reporter activity (i.e., 8X GTIIC-Luc)38 was assessed in cells transfected with wild-type YAP or TAZ in the absence or presence of Snail/Slug. As expected, YAP as well as TAZ increase the activity of the TEAD reporter with only subtle effects exerted by Snail or Slug alone (presumably due to interactions with endogenous YAP/TAZ) (Fig. 6a). When YAP or TAZ is expressed in tandem with Snail or Slug, however, reporter activity is increased significantly with binding interactions between YAP/Snail or YAP/Slug and TEAD confirmed by co-immunoprecipitation (Fig. 6a,b). As YAP or TAZ can also associate with Runx2, and Runx2/TAZ complexes play key roles in controlling downstream Runx2 signaling cascades33, 37, 39, the modulation of transcriptional activity was assessed with a Runx2-responsive luciferase reporter construct (i.e., 6X OSE-Luc)40. YAP, a previously described Runx2 transcriptional inhibitor39, is unable to support 6X OSE-Luc reporter activity in the presence of either Snail or Slug. By contrast, though Runx2 reporter activity is moderately up-regulated by TAZ alone, the combination of TAZ and either Snail or Slug induces a synergistic effect where both Snail and Slug are found in association with TAZ in Runx2 complexes (Fig. 6c,d).
Figure 6. Mapping of Snail/Slug-YAP/TAZ Binding Interactions and Regulation of Transcriptional Activity.
(a) 8XGTIIC luciferase activity was determined in Cos-1 cells co-transfected with YAP, TAZ, Snail or Slug (mean ± s.d., n=3 independent experiments).
(b) Snail/Slug binding to YAP/Tead4 complexes. Epitope-tagged Snail, Slug, YAP and Tead4 were co-transfected into Cos-1 cells, lysates immunoprecipitated with anti-Myc antibody (Tead4 tagged with Myc) and immunoblotted. Results are representative of 3 experiments performed.
(c) 6XOSE luciferase activity was monitored in Cos-1 cells co-transfected with Runx2, Snail, Slug, YAP or TAZ (mean ± s.d., n=3 independent experiments).
(d) Snail/Slug incorporation into TAZ/Runx2 complexes. Epitope-tagged Snail, Slug, YAP or Runx2 were co-transfected into Cos-1 cells, lysates immunoprecipitated with anti-HA antibody (Runx2 tagged with HA) and samples immunoblotted. Results are representative of 3 experiments performed.
(e) Schematic of HA-tagged Snail mutants.
(f) YAP interacts with the SNAG domain of Snail. Cos-1 cells were co-transfected with the indicated Snail-HA or mutant constructs in tandem with Flag-YAP. Cell lysates were subjected to anti-HA immunoprecipitation, and YAP in the complexes determined by anti-Flag immunoblotting. Results are representative of 3 experiments performed.
(g) TAZ interacts with the SNAG domain of Snail. Cos-1 cells were co-transfected with the indicated Snail-HA or mutant constructs in tandem with Flag-TAZ. Cell lysates were subjected to anti-Flag immunoprecipitation, and wild-type Snail or Snail mutants in the complexes assessed following anti-HA immunoblotting. Results are representative of 3 experiments performed.
(h) 8XGTIIC luciferase activity was measured in Cos-1 cells following co-transfection with YAP, TAZ, Snail or Snail mutants (mean ± s.d., n=3 independent experiments).
(i) 8XGTIIC luciferase activity was determined in Cos-1 cells co-transfected with YAP, TAZ, Slug or Slug mutants (mean ± s.d., n=3 independent experiments).
(j) 6XOSE luciferase activity was determined in Cos-1 cells co-transfected with Runx2, TAZ, Snail or Snail mutants (mean ± s.d., n=3 independent experiments).
(k) 6XOSE luciferase activity was monitored in Cos-1 cells co-transfected with Runx2, TAZ, Slug or Slug mutants (mean ± s.d., n=3 independent experiments).
As neither the TEAD nor OSE reporter constructs contain definable Snail- or Slug- binding sites, we mapped the Snail/Slug-YAP/TAZ protein domains that mediate complex formation. YAP and TAZ can be divided into TEAD binding (TB), WW, SH3 binding (SH3), transcriptional activation (TA) and PDZ binding (PB) domains (Supplementary Figure 6a,d)34, 35. Following expression of a series of deletion mutants of YAP and TAZ in combination with either epitope-tagged, wild-type Snail or Slug, the dominant YAP/TAZ domains responsible for Snail/Slug binding can be localized to the WW domains of YAP and TAZ (Supplementary Fig. 6a–f). Conversely, deletion mutants of Snail and Slug identify the conserved N-terminal repressor motifs (termed the extended SNAG domain4), as the key binding elements mediating interactions with the WW domains (Fig. 6e–g and Supplementary Fig. 6g–i). Confirming the importance of these interactions, SNAG domain-deletion mutants of Snail or Slug no longer support the YAP/TAZ-dependent cooperative effects on TEAD or Runx2 transcriptional activity (Fig. 6h–k).
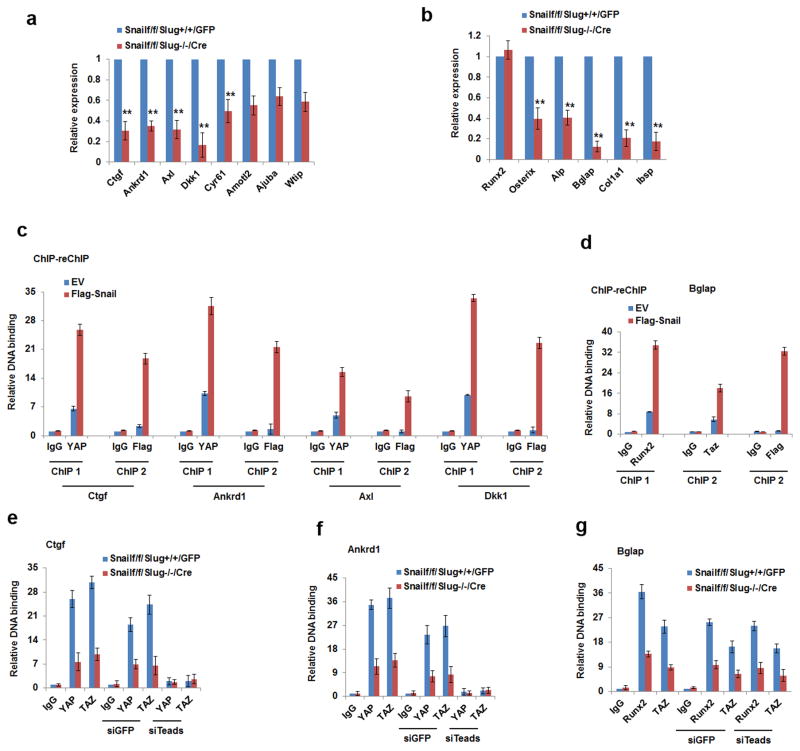
Given the characterization of Snail/Slug-YAP/TAZ complexes, we examined the ability of Snail/Slug to control the expression of YAP/TAZ/TEAD- and Runx2- downstream gene targets. Co-deletion of Snail/Slug significantly decreases the expression of the YAP/TAZ/TEAD targets, Ctgf, Ankrd1, Axl, Dkk1 and Cyr6141–43, while simultaneously inhibiting expression of the Runx2 target genes, Osterix, Alp, Osteocalcin, Col1a1 and Ibsp44–46 (Fig. 7a,b). Further, when Snail is re-expressed at physiologic levels in Snail/Slug-null osteoblast progenitors, chromatin immunoprecipitation (ChIP) analysis confirms the presence of both YAP and Snail in the promoter regions of Ctgf, Ankrd1, Axl and Dkk1 (Fig. 7c and Supplementary Fig. 7a). Similarly, a sequential ChIP for Runx2 followed by anti-Flag-Snail (or Flag-Slug) re-ChIP documents the ability of Runx2 and Snail/Slug to co-occupy the Bglap2 promoter (Fig. 7d and Supplementary Fig. 7b,c). Finally, endogenous Snail/Slug increases YAP/TAZ binding to the promoters of the YAP/TAZ/TEAD-target genes, Ankrd1 and Ctgf, in a TEAD-dependent manner (Fig. 7e,f and Supplementary Fig. 7d), while simultaneously increasing TAZ binding to Runx2 in the Bglap promoter in a TEAD-independent fashion (Fig. 7g).
Figure 7. Snail/Slug-YAP/TAZ Complexes Regulate Gene Expression.
(a) Snail/Slug regulates YAP/TAZ/TEAD targeted genes. mRNA expression of YAP/TAZ/TEAD-direct targets in calvarial osteoblast progenitors isolated from Snail1f/f/Slug+/+ or Snail1f/f/Slug−/− mice and transduced with adeno-GFP or Cre expression vectors (mean ± s.d., n=3 independent experiments). **p<0.01, unpaired t-test.
(b) Relative mRNA expression of Runx2 and its downstream genes were assessed in calvarial osteoblast progenitors isolated from Snail1f/f/Slug+/+ or Snail1f/f/Slug−/− mice and transduced with adeno-GFP or Cre expression vectors, cultured under osteogenic conditions (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
(c) Co-occupation of YAP and Snail in the YAP/TAZ/TEAD targeted promoters. ChIP experiments using anti-YAP antibody were performed in Snail/Slug double null osteoblast progenitors re-expressed with Flag-Snail and then re-ChIPed with anti-Flag antibody. Results are shown as the fold-enrichment relative to IgG IP controls (mean ± s.d., n=3 independent experiments).
(d) Co-occupation of Runx2, TAZ and Snail in the Bglap2 promoter. ChIP experiments using anti-Runx2 antibody were performed in Snail/Slug double null osteoblast progenitors re-expressed with Flag-Snail under osteogenic culture condition and re-ChIPed with anti-TAZ or anti-Flag antibody. Results are shown as fold-enrichment relative to IgG IP controls (mean ± s.d., n=3 independent experiments).
(e) Snail and Slug increase YAP/TAZ/TEADs binding to the Ctgf promoter. ChIP experiments using anti-YAP or anti-TAZ were performed in SSC lysates isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice that were transduced with adeno- GFP or Cre, and then transfected with siGFP or siTEADs(1–4). Results are presented as the fold-enrichment relative to IgG IP controls (mean ± s.d., n=3 independent experiments).
(f) Snail and Slug increases YAP/TAZ/TEADs binding to the Ankrd1 promoter. ChIP experiments using anti-YAP or anti-TAZ were performed in SSCs isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre followed by transfection with siGFP or siTEADS(1–4). Results are presented as fold-enrichment relative to IgG IP controls (mean ± s.d., n=3 independent experiments).
(g) Snail and Slug increase Runx2 binding to the Bglap2 promoter. ChIP experiments using anti-Runx2 or anti-TAZ were performed in lysates of calvarial osteoblast progenitors isolated from Snailf/f/Slug+/+ and Snailf/f/Slug−/− mice and transduced with adeno-GFP or Cre followed by transfection with siGFP or siTEADs(1–4). Results are presented as fold-enrichment relative to IgG IP controls (mean ± s.d., n=3 independent experiments).
Snail/Slug–YAP/TAZ Complexes Control SSC Function
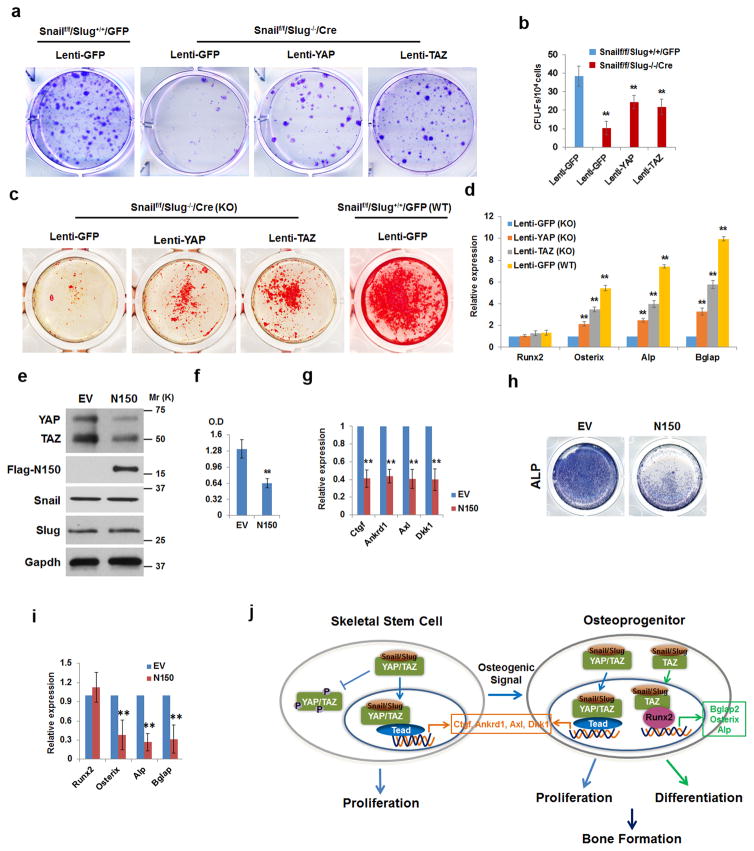
The ability of Snail/Slug to promote YAP/TAZ protein stability and transcriptional activity led us to postulate that i) over-expressing wild-type YAP or TAZ should only partially rescue defects in Snail/Slug-deleted SSCs and ii) a dominant-negative mutant that prevents binding interactions between endogenous Snail/Slug and YAP/TAZ would inhibit SSC function. Indeed, whereas Snail/Slug-deleted SSCs display defects in CFU-F formation, cells overexpressing wild-type YAP or TAZ only display a partially rescued phenotype (Fig. 8a,b and Supplementary Fig. 8a). Likewise, overexpressing YAP and TAZ only partially reverses defects in the osteogenic potential of Snail/Slug-deleted SSCs (Fig. 8c,d). To block binding interactions between YAP/TAZ and Snail/Slug, we constructed a Snail-deletion mutant that retains the SNAG domain, but deletes the Snail zinc-finger domains that support its nuclear import and DNA-binding activities47, 48 (Supplementary Fig. 8b). As expected, the Snail mutant, termed N150, binds YAP/TAZ (Supplementary Fig. 8c), blocks the formation of YAP/TAZ-Snail/Slug complexes and inhibits the cooperative effect of wild-type Snail/Slug and YAP or TAZ on either TEAD- or OSE- reporter activities, as well as the ability of YAP to occupy the Ctgf promoter or Rnx2 to bind to the Bglap2 promoter (Supplementary Fig. 8d–h). When the Snail N150 mutant is transfected into C3H10T1/2 cells, YAP and TAZ levels decrease in the predicted fashion with no effect on Snail/Slug levels (Fig. 8e). More importantly, SSC proliferation as well as the expression of YAP/TAZ/TEAD target genes are inhibited while osteogenesis, despite normal Runx2 induction, is blocked in tandem with decreased expression of Runx2 downstream targets (Fig. 8f–i). Hence, inhibiting Snail/Slug-YAP/TAZ complex formation alone recapitulates the phenotypic defects observed in Snail/Slug-null or YAP/TAZ hemizygous cells, thereby supporting a model wherein the formation of Snail/Slug-YAP/TAZ complexes control SSC proliferation and differentiation (Fig. 8j).
Figure 8. A Snail/Slug-YAP/TAZ Axis Regulates SSC Function.
(a) CFU-F generation from Snailf/f/Slug+/+/GFP or Snailf/f/Slug−/−/Cre SSCs that were transduced with lentiviral GFP, YAP or TAZ constructs. Results are representative of 5 experiments performed.
(b) CFU-F colony counts from (a) (mean ± s.d., n=5 independent experiments). **p<0.01; one-way ANOVA.
(c) Alizarin Red S staining of Snailf/f/Slug−/−/Cre or Snailf/f/Slug+/+/GFP SSCs transduced with lentiviral GFP, YAP or TAZ, and cultured under osteogenic conditions for 14 d. Results are representative of 3 experiments performed.
(d) Relative mRNA expression of osteogenic markers (Runx2, Osterix, Alp or Bglap2) in cultures from (c) (mean ± s.d., n=3 independent experiments). **p<0.01; one-way ANOVA.
(e) Immunoblotting of YAP/TAZ, Snail/Slug and mutant Snail (N150). C3H10T1/2 cells were transfected with mutant Snail (N150) or empty vector (EV). Results are representative of 3 experiments performed.
(f) Mutant Snail (N150) inhibits proliferation of C3H10T1/2 cells. Proliferative response in cells from (e) were assayed by XTT assay (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
(g) Relative mRNA expression of YAP/TAZ/TEAD targets was assessed in cells from (e) (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
(h) Mutant Snail (N150) inhibits osteogenesis of C3H10T1/2 cells. Cells from (e) were cultured under osteogenic conditions for 7 d. Osteogenesis was monitored by ALP staining.
(i) Relative mRNA expression of osteogenic markers (Runx2, Osterix, Alp and Bglap2) was assessed in cells from (h) (mean ± s.d., n=3 independent experiments). **p<0.01; unpaired t-test.
(j) Schematic model of Snail/Slug in regulating SSC proliferation and osteogenic differentiation.
Discussion
Skeletal SSCs comprise an heterogeneous assortment of mesoderm- and neural crest- derived progenitors that play distinct and age-specific roles in osteoblastogenesis 1, 2, 18, 19, 25, 49–51. Though effects of Snail/Slug on stem cell function are normally assigned to epithelial cell populations, we demonstrate herein that SSCs – despite their mesenchymal origin – co-express these factors, emphasizing their ability to function outside of classic EMT-related programs. While recent reports have proposed major roles for Snail alone in osteoblastogenesis under in vitro conditions3, 52, 53, neither Snail- nor Slug- targeted mice display remarkable skeletal phenotypes.
In considering complementary pathways that intersect with the Snail/Slug-dependent transcriptional programs operative in SSCs, YAP/TAZ, like Snail/Slug, undergo cytosolic-nuclear translocation, are regulated by phosphorylation-dependent β-TRCP-mediated ubiquitination, and interface with canonical Wnt as well as TGFβ signaling5, 34, 35, 37. Extending these regulatory interactions, we find Snail/Slug forms binary complexes with YAP/TAZ that not only modulate YAP/TAZ protein levels, but also the expression of YAP/TAZ-targeted genes. Further studies demonstrated that the extended SNAG domains of Snail/Slug (that recruit chromatin modifying enzymes critical for transcriptional repression) mediate binding interactions with the YAP/TAZ WW domains5, 35, 37. WW domains classically interact with protein partners via embedded PPxY motifs35, 37, 54, but the extended SNAG domains of neither Snail nor Slug include this sequence. Interestingly, the Snail and Slug extended SNAG domains contain a conserved PxY motif that potentially participates in YAP/TAZ binding4.
Recent studies demonstrate that genome-wide, YAP/TAZ DNA-binding regions include a small subset of promoters with most YAP/TAZ/TEAD sites localized to distal enhancers that associate with target promoters through chromatin looping in a context-specific fashion 41–43. As such, the regulatory effects mediated by Snail/Slug-YAP/TAZ complexes likely extend beyond the targets that we have characterized, though it is noteworthy that several of the affected genes identified herein regulate osteogenesis55–57. Given the widespread expression of Snail/Slug as well as YAP/TAZ in both epithelial as well as mesenchymal cell populations, we posit that the interacting transcriptional networks outlined here may prove equally important to stem cell function in a variety of normal and neoplastic states.
Methods (Online Only)
Mice
Mice with Snailflox/flox alleles and Snail+/LacZ have been generated in our laboratory and are maintained on a C57BL/6 genetic background1, 2. YAPf/f/TAZf/f and Slug+/− mice have been previously developed3, 4. Slug+/LacZ mice were obtained from T. Gridley and maintained on a C57BL/6 background5. Dermo1-Cre knock-in mice 6 and Osterix-GFP-Cre transgenic mice7 were obtained from the Jackson Laboratory (Maine, USA). Nu/Nu mice were from Charles River Laboratories International, Inc. All mouse work was performed with IACUC approval and in accordance with a protocol approved by University of Michigan Institutional Animal Care & Use Committee.
CFU-F Assay
Fetal limb skeletal tissues were dissected from embryos, cut into small pieces and digested with 3 mg/ml collagenase (type I; Sigma) for 30 min at 37°C with constant agitation. Postnatal femur and tibia specimens were similarly treated, but digested with 3 mg/ml collagenase for 60 min at 37°C with constant agitation. Following enzymatic digestion, the skeletal preparations were filtered through a 40 μm cell strainer, the cell suspensions pelleted, washed and resuspended 8. For fibroblast colony-forming unit (CFU-F) assays, 5×105 mouse bone marrow cells were plated into individual wells of 6-well plates, and 3 d later, the unattached cells removed by washing, and the adherent cells further cultured for 7–10 d. The total number of colonies formed was determined after the samples were fixed and stained with crystal violet. Colonies with more than 50 cells were scored as CFU-Fs9.
Quantitative RT-PCR
RNA purified from embryonic, neonatal skull, femurs or cell cultures was used for cDNA synthesis and amplification by real-time PCR according to the manufacturer’s instructions (Power SYBR Green, Applied Biosystems). The primer information was provided in Supplementary Table 1.
Cell Culture
4–6-wk old mice femur and tibia specimens were dissected and cleaned from surrounding tissue, cut into pieces and digested with 3 mg/ml collagenase for 60 min at 37°C with constant agitation. Single-cell suspensions were collected after passing through a 40 μm cell strainer (BD Bioscience) and incubated in tissue culture-grade plastic dishes with DMEM supplemented with 10% fetal bovine serum. After 2 d of culture, cells were washed twice with PBS and the adherent cells further cultured in DMEM supplemented with 10% fetal bovine serum. The adherent colonies were then sorted by flow cytometry with antibodies to Sca-1, CD29, CD45 and CD11b as described9, 10. To further deplete hematopoietic cell populations from the cultures, cell suspensions were purified with a mouse hematopoietic progenitor cell enrichment set (BD Biosciences), and hematopoietic cell lineages captured with BD IMag Streptavidin Particles Plus-DM magnetic nanoparticles according to the manufacturer’s instructions. The purified cells isolated from CFU-Fs were used as SSCs.
For culture of calvarial mesenchymal progenitor cells, calvaria were harvested from embryo skulls and digested in 0.1% collagenase and 0.2% dispase solution11. Mesenchymal progenitor cells isolated from digested calvaria were cultured in alpha-Minimum Essential Medium supplemented with 10% FBS.
Human mesenchymal stem cells (Lonza), C3H/10T1/2 and MC3T3-E1 cells (ATCC) were cultured according to the manufacturer’s instructions. Human mesenchymal stem cells were authenticated by cell surface markers and cell differentiation assay. C3H/10T1/2 and MC3T3-E1 cells were confirmed by cell differentiation assay. All cell lines were routinely tested for mycoplasma contamination and were negative. None of the cell lines used in this study is present in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample.
Differentiation of SSCs In Vitro
SSCs were induced with osteogenic medium [50 μM ascorbic acid, 10 nM dexamethasone, 10 mM β-glycerolphosphate and 2% fetal bovine serum (FBS) in low-glucose DMEM] or adipogenic medium [1 μM dexamethasone, 50 μM indomethacin, 500 nM IBMX, 5 μg/ml insulin and 2% FBS in low-glucose DMEM]12. For chondrogenesis, micro-mass cultures were performed as described13 with SSCs induced with a STEMPRO Chondrogenesis Differentiation Kit (Invitrogen). Differentiated cells were stained for Alizarin Red S to detect calcium deposits in osteogenic cultures, Oil Red O for detecting lipid deposits during adipogenesis, and Safranin O (cartilage mucopolysaccharides staining)/Fast Green for monitoring chondrogenesis14.
Histology
Embryonic, neonatal or adult skeletal tissues were dissected, fixed (4 h at 4°C for embryonic and neonatal bones, and overnight at 4°C for adult bones) in 4% paraformaldehyde. Adult bones were decalcified in EDTA. The skeletal specimens were embedded in OCT compound or paraffin, and sectioned (10 μm for cryostat sections and 5 μm for paraffin sections). Sections were stained with Hematoxylin/Eosin (H&E) according to standard procedures while LacZ activity was analyzed in frozen sections 14. Femur osteoclasts were identified by staining for tartrate resistant acid phosphatase (TRAP) using a leukocyte acid phosphatase assay kit (Sigma-Aldrich). The number of TRAP-positive cells per millimeter of trabecular bone perimeter was quantified in the secondary spongiosa 14.
Microcomputed Tomography
Tibiae and femora from adult mice were fixed in 4% paraformaldehyde overnight at 4°C, and stored in 70% ethanol. Femurs were scanned in water at an 18 μm isotropic voxel resolution using Explore Locus SP (GE Healthcare Pre-Clinical Imaging, London, ON, Canada), and calibrated three-dimensional images reconstructed. Bone morphometric variables were analyzed, including Bone Mineral Density (BMD), Bone Volume/Tissue Volume (BV/TV), Trabecular Number (Tb.N), Trabecular Separation (Tb.Sp) and Trabecular Thickness (Tb.Th) as described 14.
In Vivo Assay of SSCs Osteogenic Capacity
For in vivo osteogenic differentiation, 2×106/ml mouse SSCs were seeded into gelatin sponges that were implanted subcutaneously into Nu/Nu mice. The specimens were dissected from the mouse recipients and analyzed 4 wk later as described 14.
Skeleton Analysis
Skulls collected at E17.5 or P0 were skinned, and fixed in 95% ethanol. Skull tissues were stained with Alizarin Red S and Alcian Blue as described 15.
Proliferation Assay
Cell were seeded at 5×103 cells/well in 96-well plates and proliferation determined using a XTT Cell Viability Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Alternatively, cells were seeded on glass cover slips, incubated overnight, fixed and immunostained for the cell proliferation marker, Ki67. Total cell number was evaluated by DAPI staining. The ratio of Ki67 positive cells to the total cell number was determined.
Immunostaining, In Situ PLA and Imaging
Cells cultured on glass surfaces were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized and incubated with primary antibodies at 4°C. For frozen sections, permeabilization and antibody incubations were performed under identical conditions as described above. All primary antibodies were used at a 1:200 dilution. For in situ PLA, protein-protein interactions between Snail/Slug and YAP/TAZ were detected with secondary proximity probes (Anti-Rabbit Plus and Anti-Mouse Minus) according to the Duolink In Situ Fluorescence User Guide (Sigma-Aldrich). Images were captured with a Leica confocal microscope. The antibody information was provided in the Supplementary Table 2.
SiRNAs, Constructs, Gene Expression and Reporter Assays
Human or mouse TEF-1, TEF-3, TEF-4 and TEF-5 (TEAD1–4) siRNAs, and human SNAIL or SLUG siRNAs were obtained from Santa Cruz Technology. To silence gene expression, PepMute siRNA Transfection Reagent (SignaGen Laboratories) was used for all siRNAs transfection. Flag-Snail, Snail-HA, Slug-Myc, Flag-YAP, Flag-TAZ, and 8xGTIIC were obtained from Addgene. HA-Runx2 and 6xOSE reporter constructs were provided by G. Stein 16. Snail, Slug, YAP and TAZ mutants were constructed by PCR. Where indicated, Flag-YAP and Flag-TAZ were cloned into pLentilox-IRS-GFP lentiviral vectors and used to transduce SSCs, with the expression of the exogenous proteins confirmed by Western blot. For gene transfection, COS-1 cells or osteoblast progenitors were transfected with Lipofectamine 2000 (Invitrogen). For reporter assays, COS-1 cells were transfected with expression vectors and either 8xGTIIC or 6xOSE reporters. Luciferase activity in cell lysates was determined as previously described 9.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was carried out by using the Imprint Chromatin Immunoprecipitation Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, cells were harvested and resuspended in fresh culture medium and then crosslinked with 1% formaldehyde (for 10 min), and quenched with 0.125 M glycine for 5 min at room temperature. Harvested cells were washed twice with ice-cold PBS and then resuspended in nuclei preparation buffer. The nuclear pellet was harvested and resuspended in shearing buffer and sonicated on ice until the sheared DNA was approximately 200–1000 bp in size. The samples were then centrifuged at 16,000 × g for 10 min at 4°C to remove debris, and the supernatants diluted 1:1 in dilution buffer. The prepared material was then used for protein/DNA immunoprecipitation. Antibodies directed against YAP, TAZ, Runx2 or Flag, normal control mouse IgG or rabbit IgG (Cell Signaling) were pre-bound to the assay wells and immunoprecipitation reactions carried out with chromatin extracts. Five per cent of the chromatin extract was set aside for input. After the immunoprecipitation, cross-link reversal was carried out and the precipitated DNA purified. DNA was quantitated by real-time PCR analysis. All ChIP signals were normalized to the input, and relative fold-enrichment compared to IgG controls.
For ChIP-reChIP, chromatin isolated from cells expressing Flag–Snail or Slug was incubated with anti-YAP or anti–Runx2 antibody overnight at 4 °C. Following washing, the immunoprecipitated complexes were incubated with protein A/G magnetic beads (Thermo) and then eluted by incubating beads in lysis buffer with purified GST-YAP or HA-Runx2 proteins, respectively for 2 h at 4 °C. The GST-YAP fusion protein was purified by GST-pulldown assay by using the pGEX-KG-GST-YAP construct (Addgene). While HA-Runx2 was purified from 293 cells transfected with a HA-Runx2 expression vector following incubation with anti-HA magnetic beads (ThermoFisher) and elution with HA peptide (Sigma). The eluted chromatin was diluted 1:3 with lysis buffer, supplemented with 1% Triton X-100 and incubated with normal mouse IgG, anti-Flag or anti-TAZ antibodies overnight at 4 °C. Following immunoprecipitation, cross-link reversal was carried out and the precipitated DNA purified. DNA was quantitated by real-time PCR analysis. The antibody information was provided in the Supplementary Table 2, and the primer information was provided in Supplementary Table 1.
Immunoprecipitation and Immunoblotting
Embryonic, neonatal skeleton homogenates or cell lysates were prepared using RIPA buffer for Western blotting. For immunoprecipitation, embryonic, neonatal skeleton homogenates or cell lysate were prepared using a lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100 and 1 mM EDTA. The lysates were incubated with antibody and Dynabeads Protein G (Life Technologies). Following, immunoprecipitation, the immunocomplexes were analyzed by Western blotting 17. The antibody information was provided in the Supplementary Table 2.
Statistics and Reproducibility
Statistical analysis was performed with the Student’s t-test or by one-way ANOVA. *: p<0.05, **: p<0.01. All tests are two-sided with all experiments performed 3 or more times. In each experiment, the sample size is determined on the basis of our prior knowledge of the variability of experimental output. In animal experiments, the gender, age of animals were matched and a sample size of 4–10 mice per group allows us to detect the difference in bone differentiation and proliferation markers, microCT and bone formation parameters, and in CFU-Fs counting with the confidence of 90%. No randomization was performed in all the experiments. The researchers were not blinded during experiments and outcome assessment. For in vitro experiments analyses, in most of the cases three or more independent experiments were performed and each with three or more replicates. The corresponding information has been included in the figure legends. The difference between means of different experimental groups was analyzed by using two-tailed unpaired Student’s t-test noted in the respective legends. t-tests were performed with the assumption of equal variation accordingly. F-tests were performed to compare variation within different groups. The statistical information was further provided in Supplementary Table 3.
Data Availability
Statistics source data for Figure 1g,i,l; Figure 2d,f,g,i,k; Figure 3e,g,h,k; Figure 4h,j,l; Figure 7a,b; Figure 8b,d,f,g,i; Supplementary Figure 1f,h,j,k,l; Supplementary Figure 2f; Supplementary Figure 3b,d,f,h; Supplementary Figure 4a,b,d,j,k are provided in Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
We thank E. N. Olson (UTSW) for providing YAP+/−/TAZ+/− mice. This work was supported by a grant from the Breast Cancer Research Foundation (S.J.W.). Work performed in this study was also supported by NIH Grant R01-1AR065524 (S.J.W.) and by P01-CA093900 (ETK).
Footnotes
Note: Supplementary Information is available in the online version of the paper.
Author Contributions
Y.T. Designed and performed experiments, analyzed data and wrote the paper
T.F. Designed and performed experiments
E.T.K. Designed and performed experiments
X-Y. L. Designed and performed experiments
S.J.W. Oversaw project, designed experiments, analyzed data and wrote the paper
References
- 1.Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Bianco P, Robey PG. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batlle R, et al. Snail1 controls TGF-beta responsiveness and differentiation of mesenchymal stem cells. Oncogene. 2013;32:3381–3389. doi: 10.1038/onc.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrallo-Gimeno A, Nieto MA. Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 2009;25:248–252. doi: 10.1016/j.tig.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 6.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 7.Desgrosellier JS, et al. Integrin alphavbeta3 drives slug activation and stemness in the pregnant and neoplastic mammary gland. Dev Cell. 2014;30:295–308. doi: 10.1016/j.devcel.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvay K, et al. Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelium. EMBO J. 2015;34:1319–1335. doi: 10.15252/embj.201490881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang WL, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y, et al. Snail1-dependent control of embryonic stem cell pluripotency and lineage commitment. Nat Commun. 2014;5:3070. doi: 10.1038/ncomms4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oram KF, Carver EA, Gridley T. Slug expression during organogenesis in mice. Anat Rec A Discov Mol Cell Evol Biol. 2003;271:189–191. doi: 10.1002/ar.a.10027. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Gridley T. Compensatory regulation of the Snai1 and Snai2 genes during chondrogenesis. J Bone Miner Res. 2013;28:1412–1421. doi: 10.1002/jbmr.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Gridley T. The SNAI1 and SNAI2 proteins occupy their own and each other’s promoter during chondrogenesis. Biochem Biophys Res Commun. 2013;435:356–360. doi: 10.1016/j.bbrc.2013.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vega S, et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Mancera PA, et al. Adipose tissue mass is modulated by SLUG (SNAI2) Hum Mol Genet. 2007;16:2972–2986. doi: 10.1093/hmg/ddm278. [DOI] [PubMed] [Google Scholar]
- 18.Worthley DL, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Isolation of murine bone marrow derived mesenchymal stem cells using Twist2 Cre transgenic mice. Bone. 2010;47:916–925. doi: 10.1016/j.bone.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, et al. MT1-MMP-dependent control of skeletal stem cell commitment via a beta1-integrin/YAP/TAZ signaling axis. Dev Cell. 2013;25:402–416. doi: 10.1016/j.devcel.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 24.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9:e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizoguchi T, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Osterix-cre labeled progenitor cells contribute to the formation and maintenance of the bone marrow stroma. PLoS One. 2013;8:e71318. doi: 10.1371/journal.pone.0071318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 32.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 33.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 34.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 36.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 38.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 39.Zaidi SK, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein C, et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galli GG, et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishio Y, et al. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 46.Harada H, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 47.Mingot JM, Vega S, Maestro B, Sanz JM, Nieto MA. Characterization of Snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. J Cell Sci. 2009;122:1452–1460. doi: 10.1242/jcs.041749. [DOI] [PubMed] [Google Scholar]
- 48.Choi S, et al. Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin beta. Acta Crystallogr D Biol Crystallogr. 2014;70:1050–1060. doi: 10.1107/S1399004714000972. [DOI] [PubMed] [Google Scholar]
- 49.Chan CK, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isern J, et al. The neural crest is a source of mesenchymal stem cells with specialized hematopoietic stem cell niche function. eLife. 2014;3:e03696. doi: 10.7554/eLife.03696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Frutos CA, et al. Snail1 controls bone mass by regulating Runx2 and VDR expression during osteoblast differentiation. EMBO J. 2009;28:686–696. doi: 10.1038/emboj.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SJ, et al. The transcription factor snail regulates osteogenic differentiation by repressing Runx2 expression. Bone. 2010;46:1498–1507. doi: 10.1016/j.bone.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 54.Wang W, et al. Defining the protein-protein interaction network of the human hippo pathway. Mol Cell Proteomics. 2014;13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivkovic S, et al. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacDonald BT, et al. Bone mass is inversely proportional to Dkk1 levels in mice. Bone. 2007;41:331–339. doi: 10.1016/j.bone.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 58.Rowe RG, et al. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol. 2009;184:399–408. doi: 10.1083/jcb.200810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu K, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, et al. Inhibition of Sca-1-positive skeletal stem cell recruitment by alendronate blunts the anabolic effects of parathyroid hormone on bone remodeling. Cell Stem Cell. 2010;7:571–580. doi: 10.1016/j.stem.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anjos-Afonso F, Bonnet D. Isolation, culture, and differentiation potential of mouse marrow stromal cells. Curr Protoc Stem Cell Biol. 2008;Chapter 2(Unit 2B):3. doi: 10.1002/9780470151808.sc02b03s7. [DOI] [PubMed] [Google Scholar]
- 63.Murdoch AD, et al. Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold-free cartilage. Stem Cells. 2007;25:2786–2796. doi: 10.1634/stemcells.2007-0374. [DOI] [PubMed] [Google Scholar]
- 64.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 65.Tang Y, Liu Z, Zhao L, Clemens TL, Cao X. Smad7 stabilizes beta-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J Biol Chem. 2008;283:23956–23963. doi: 10.1074/jbc.M800351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistics source data for Figure 1g,i,l; Figure 2d,f,g,i,k; Figure 3e,g,h,k; Figure 4h,j,l; Figure 7a,b; Figure 8b,d,f,g,i; Supplementary Figure 1f,h,j,k,l; Supplementary Figure 2f; Supplementary Figure 3b,d,f,h; Supplementary Figure 4a,b,d,j,k are provided in Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.