Abstract
BACKGROUND
Approximately 75% of objective responses to anti–programmed death 1 (PD-1) therapy in patients with melanoma are durable, lasting for years, but delayed relapses have been noted long after initial objective tumor regression despite continuous therapy. Mechanisms of immune escape in this context are unknown.
METHODS
We analyzed biopsy samples from paired baseline and relapsing lesions in four patients with metastatic melanoma who had had an initial objective tumor regression in response to anti–PD-1 therapy (pembrolizumab) followed by disease progression months to years later.
RESULTS
Whole-exome sequencing detected clonal selection and outgrowth of the acquired resistant tumors and, in two of the four patients, revealed resistance-associated loss-of-function mutations in the genes encoding interferon-receptor–associated Janus kinase 1 (JAK1) or Janus kinase 2 (JAK2), concurrent with deletion of the wild-type allele. A truncating mutation in the gene encoding the antigen-presenting protein beta-2-microglobulin (B2M) was identified in a third patient. JAK1 and JAK2 truncating mutations resulted in a lack of response to interferon gamma, including insensitivity to its antiproliferative effects on cancer cells. The B2M truncating mutation led to loss of surface expression of major histocompatibility complex class I.
CONCLUSIONS
In this study, acquired resistance to PD-1 blockade immunotherapy in patients with melanoma was associated with defects in the pathways involved in interferon-receptor signaling and in antigen presentation. (Funded by the National Institutes of Health and others.)
Durable responses in metastatic cancers have been achieved with a variety of immunotherapies such as interleukin-2, adoptive cell transfer of tumor-infiltrating lymphocytes, antibodies that block cytotoxic T-lymphocyte–associated antigen 4 (CTLA4),1–5 and antibodies that block programmed death 1 (PD-1).6–10 However, in a recent study, approximately 25% of patients with melanoma who had had an objective response to PD-1 blockade therapy had disease progression at a median follow-up of 21 months.11
The mechanisms of immune-resistant cancer progression are mostly unknown. Previous studies involving humans examined the loss of beta-2- microglobulin as a mechanism of acquired resistance to several forms of cancer immunotherapy.12–14 In preclinical models, defects in the interferon signaling pathway have been proposed as a potential mechanism of cancer escape (insensitivity) to immunotherapy.15,16 In the current study, we assessed the effect of anti–PD-1 therapy on cancer genomic evolution, including acquired mutations in the genes affecting the interferon pathway and antigen-presentation pathway, in an effort to determine genetic mechanisms of acquired resistance to PD-1 blockade therapy.
METHODS
PATIENTS, RESPONSE ASSESSMENT, AND TUMOR BIOPSIES
Of 78 patients with metastatic melanoma who were treated with the anti–PD-1 antibody pembrolizumab at the University of California, Los Angeles (UCLA), 42 had an objective response, of whom 15 went on to have disease progression. Four of these 15 patients met all three selection criteria for this analysis. First, they must have had an objective tumor response while participating in a clinical trial with single-agent pembrolizumab.6,7,10,11 Tumor responses were evaluated at 12 weeks and confirmed 4 weeks later, and patients were assessed by imaging every 12 weeks thereafter with the use of both the Response Evaluation Criteria in Solid Tumors17 and the immune-related response criteria.18 Second, patients had to have late acquired resistance, defined as in situ recurrence or new lesion development, despite continuous dosing, after more than 6 months of tumor response. Third, patients had to have adequate biopsy material for whole-exome sequencing at two time points: before the initiation of pembrolizumab therapy and after disease progression. We processed tumor biopsy samples as described previously to perform pathological analyses, obtain DNA and RNA, and attempt to establish cell lines.19,20
IMMUNOHISTOCHEMICAL, IMMUNOFLUORESCENCE, WESTERN BLOT, AND FLOW-CYTOMETRIC ANALYSES
Immunohistochemical and immunofluorescence analyses19 as well as Western blot and flow-cytometric analyses21 were performed and analyzed as described previously. Full methods are included in the Supplementary Appendix, available with the full text of this article at NEJM.org.
GENETIC AND TRANSCRIPTIONAL-PROFILING ANALYSES
Whole-exome sequencing was performed at the UCLA Clinical Microarray Core with the use of the NimbleGen SeqCap EZ Human Exome Library, version 3.0 (Roche). Mutation calling was performed as described previously.22 Selected gene-expression profiling on interferon exposure was performed with the use of nCounter (NanoString Technologies). Whole-exome sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP076315.
FUNCTIONAL STUDIES
Patient-derived and previously established human melanoma cell lines were used to analyze recognition by T-cell receptor transgenic T cells23 with the use of in vitro coculture assays that detect antigen-induced release of interferon-γ assessed by enzyme-linked immunosorbent assay. Cell- proliferation and growth-inhibition assays were performed with the use of an automated live-cell imaging system (IncuCyte, Essen BioScience) with or without exposure to interferons. Full methods are described in the Supplementary Appendix.
STUDY OVERSIGHT
Data generated and collected by the study investigators were analyzed by the last author, who vouches for the completeness and accuracy of the data, analyses, and reported results. Summaries of the clinical protocol have been reported by Hamid et al.6 and Robert et al.10
STATISTICAL ANALYSIS
Student’s t-test and a two-way analysis of variance were used for cell-culture experiments, with Dunnett’s correction applied for multiple comparisons with untreated controls.
RESULTS
CLINICAL COURSE AND IMMUNE INFILTRATES
We analyzed paired tumor samples from four (nonconsecutive) selected patients with meta-static melanoma who had had a relapse while receiving PD-1–inhibition therapy with pembrolizumab (Tables S1 and S2 in the Supplementary Appendix). All four patients met objective criteria for a partial response,17,18 though with slightly different kinetics (Fig. 1, and Figs. S1, S2, and S3 in the Supplementary Appendix). The mean time to relapse was 624 days (range, 419 to 888). The baseline biopsy samples were obtained just before the initiation of pembrolizumab therapy in Patients 2, 3, and 4, whereas for Patient 1, the only available baseline biopsy sample was obtained before an earlier course of therapy with the BRAF inhibitor vemurafenib. The baseline biopsy samples from Patients 1, 2, and 3 showed preexisting CD8 T-cell infiltrates at the invasive margin that colocalized with programmed death ligand 1 (PD-L1) expression on surrounding macrophages and melanoma cells (Fig. 1B, and Figs. S1B and S2B in the Supplementary Appendix). The biopsy samples obtained at the time of response in Patients 2, 3, and 4 showed a marked increase in intratumoral CD8 T-cell infiltrates (Figs. S1C, S2C, and S3C and Table S3 in the Supplementary Appendix; no biopsy sample during therapy was available for Patient 1). At the time of relapse, all four biopsy samples showed CD8 T-cell infiltration and PD-L1 expression concentrated at the tumor margins again (Fig. 1C and Figs. S1D, S2D, and S3D in the Supplementary Appendix). Multiplex immunofluorescence assays revealed that melanoma cells at the time of relapse in Patients 1 and 2 were negative for PD-L1 even when directly adjacent to T cells, whereas macrophages and stromal cells were positive for PD-L1.
Figure 1. Clinical Pattern of Acquired Resistance to Anti–Programmed Death 1 (PD-1) Therapy in Patient 1.
In Panel A, computed tomographic images show a melanoma small-bowel metastasis at baseline, at the time of maximum response, and at the time of an in situ relapse after a year of minimal residual disease; the red dots in the graph on the right indicate these three time points. Immunohistochemical staining and multiplexed immunofluorescence analysis showed abundant CD8 T-cell infiltrates and programmed death ligand 1 (PD-L1) expression at baseline and again at the time of relapse at the tumor margin in Panels B and C, respectively. In the immunofluorescence images, red indicates PD-L1, yellow CD8 T cells, light blue the melanoma marker S100 (cytoplasmic staining), and dark blue the melanoma marker Sox10 (nuclear staining).
GENETIC CHANGES IN RELAPSE BIOPSY SAMPLES
The pattern of a strong initial response, long dormancy, and rapid late progression led us to hypothesize that relapse in these patients resulted from immune-mediated clonal selection and tumor outgrowth.24 To identify mutations that might confer immune resistance, we extracted DNA from bulk-tumor biopsy samples or early-passage primary cell lines (Table S2 in the Supplementary Appendix) and performed whole-exome sequencing to compare baseline and matched relapsed tissues. We achieved a median coverage of 149×, and the percent of tumor cells (as compared with stromal cells) was more than 40% in all samples (Table S2 in the Supplementary Appendix). Nonsynonymous mutations for all samples are shown in Table S4 in the Supplementary Appendix.
JAK MUTATIONS WITH CONCURRENT LOSS OF HETEROZYGOSITY AT RELAPSE
We found strong evidence that the relapsed tumors were closely genetically related to their baseline counterparts, despite up to 2 years between biopsies. In the case of Patients 1 and 2, of 1173 and 240 nonsynonymous mutations, respectively, originally identified in the baseline sample, 92.5% and 95.8% were also seen in the resistant tumor (Fig. 2A, and Fig. S4 in the Supplementary Appendix). The relapsing tumors also contained the same chromosomal loss-of-heterozygosity events as the baseline tumors, and all differences were due to further loss in the relapse samples. In the relapse biopsy samples from both patients, we identified new homozygous loss-of-function mutations in the kinases associated with the interferon-receptor pathway, with a Q503* nonsense mutation in the gene encoding Janus kinase 1 (JAK1) in Patient 1 (Fig. 2A and 2B) and a F547 splice-site mutation in the gene encoding Janus kinase 2 (JAK2) in Patient 2 (Fig. S4 in the Supplementary Appendix). RNA sequencing showed that the JAK2 splice-site mutation caused intron inclusion, producing an in-frame stop codon 10 bp after exon 12 (Fig. S5 in the Supplementary Appendix). Therefore, both mutations were upstream of the kinase domains and probably truncated the protein or caused nonsense-mediated decay. Neither mutation was seen at baseline in the exome sequencing reads, by Sanger sequencing, or by targeted amplicon resequencing (Fig. S6 in the Supplementary Appendix).
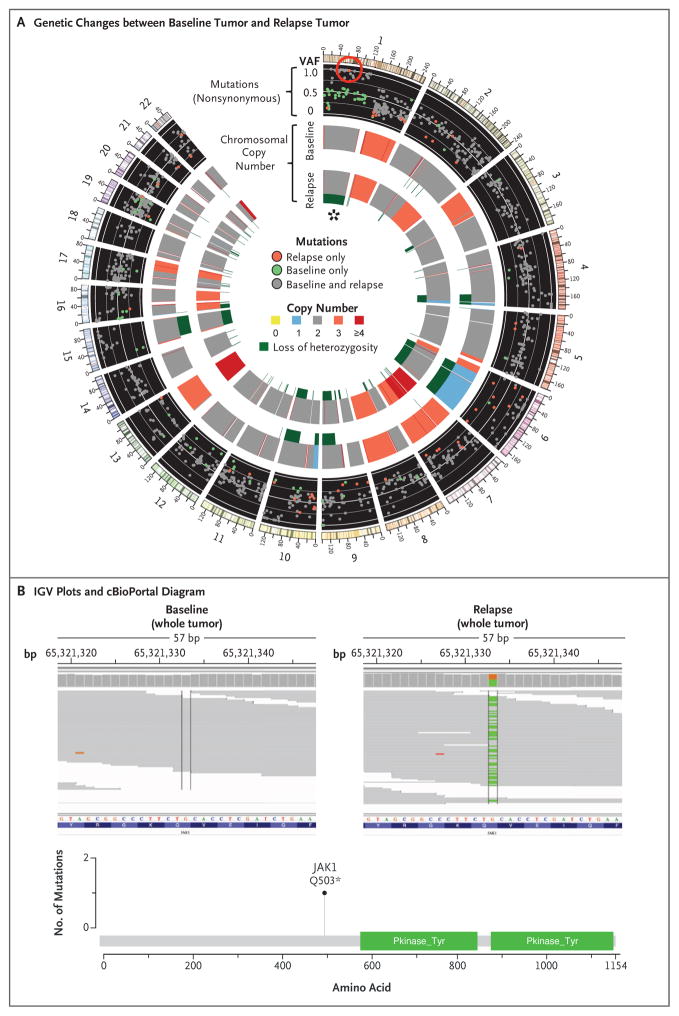
Figure 2. Acquired JAK1 Loss-of-Function Mutation at Relapse, with Accompanying Loss of Heterozygosity.
In Panel A, a Circos plot25 of Patient 1 shows differences in whole-exome sequencing between the pre-pembrolizumab and post-relapse biopsies. The red circle highlights a new, high-allele-frequency, relapse-specific mutation in the gene encoding Janus kinase 1 (JAK1) in the context of chromosomal loss of heterozygosity (asterisk). Each wedge represents a chromosome. In the outer track (black background), each point represents a nonsynonymous mutation, with most detected in both biopsy samples (gray) rather than at relapse only (red) or baseline only (green). The y-axis position indicates the variant allele frequency (VAF) at relapse, unless baseline-specific. The middle and inner tracks show copy-number status for the baseline and relapse biopsy, respectively; dark green in the subtrack indicates loss of heterozygosity. In Panel B, Integrative Genomics Viewer (IGV) plots (top) show that the JAK1 Q503* nonsense mutation is relapse-specific, and the cBioPortal26 diagram (bottom) shows that the JAK1 mutation is upstream of the kinase domains.
The JAK2 mutation was the only homozygous mutation (adjusted variant allele frequency, >85%) of 76 new nonsynonymous mutations in Patient 2, and the JAK1 mutation was 1 of only 3 homozygous mutations among 53 new mutations in Patient 1 (Table S5 in the Supplementary Appendix). To become homozygous, both JAK mutations were acquired in the context of a copy-number–neutral nondisjunction event, resulting in loss of the wild-type chromosome and duplication of the mutated allele. This is seen clearly in Patient 1: at relapse, chromosome 1p (containing JAK1) showed a decrease in minor-allele frequencies for germline single-nucleotide polymorphisms relative to baseline (Fig. S7 in the Supplementary Appendix), was missing 36 heterozygous baseline mutations (presumably on the lost allele), and contained 20 mutations (presumably on the amplified allele) that became homozygous (adjusted variant allele frequency, >85%, with change of >35 percentage points from baseline). A similar loss-of-heterozygosity event occurred for chromosome 9 in Patient 2 (Fig. S8 and Table S5 in the Supplementary Appendix). Together, these data suggest that the tumors resistant to anti–PD-1 are a relatively homogenous population derived directly from the baseline tumor and that acquisition of the JAK mutations was an early founder event before clonal selection and relapse despite the fact that the mutation was not detected in pre-treatment tumor tissue.
FUNCTIONAL EFFECTS OF JAK2 MUTATION
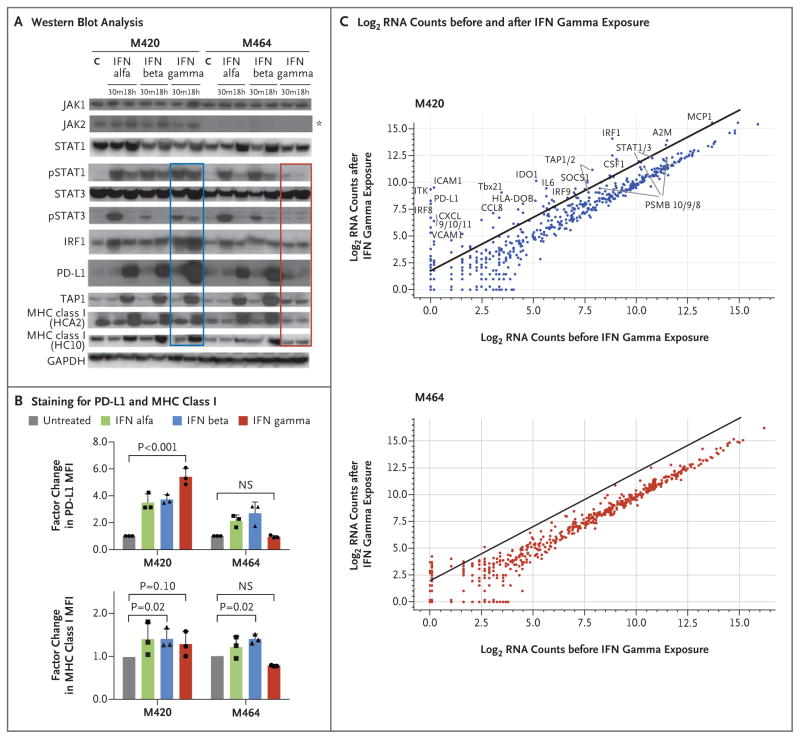
To assess the functional consequences of the observed JAK mutations, we focused on the JAK2 mutation from Patient 2 using two cell lines established at baseline (M420, wild-type JAK2) and at the time of relapse (M464, JAK2 F547 splice-site mutation). Whole-exome sequencing confirmed that the original bulk tumor was well represented by M464 (Fig. S9 in the Supplementary Appendix). Western blot analysis showed that the baseline cell line responded to interferon alfa, beta, and gamma with the expected signal transduction, including an increase in signal transducer and activator of transcription 1 (STAT1) and interferon regulatory factor (IRF) expression, STAT1 phosphorylation (pSTAT1), and the production of downstream interferon targets such as PD-L1, transporter associated with antigen processing 1 (TAP1), and major histocompatibility complex (MHC) class I (Fig. 3A). However, the cell line from the progressing lesion showed a total loss of JAK2 protein (Fig. 3A), resulting in a lack of response to interferon gamma, without change in sensitivity to interferon alfa or beta. This was true of the pSTAT1 response (Fig. 3A) and the expression of PD-L1 and MHC class I molecules (Fig. 3A and 3B). The progressing cell line also failed to upregulate a wider panel of interferon-induced transcripts involved in antigen presentation and T-cell chemotaxis (Fig. 3C, and Table S6 in the Supplementary Appendix). Together, these data indicate a total loss of functional response to interferon gamma and are consistent with JAK2 being required for signaling through the interferon-γ receptor, as opposed to the interferon-α/β receptor, which uses TYK2 and JAK1.27–29
Figure 3. Loss of Interferon Gamma–Induced Signaling and Gene-Expression Changes through Acquired JAK2 Mutation.
In Panel A, Western blot analysis of lysates from cell lines M420 (Patient 2, baseline) and M464 (Patient 2, relapse) shows Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling events and downstream target induction after either 30 minutes (m) or 18 hours (h) of exposure to interferon (IFN) alfa, beta, or gamma (C indicates untreated control). Janus kinase 2 (JAK2) protein expression was absent in the relapse cell line (asterisk), and M464 failed to phosphorylate intermediate signaling components STAT1 and STAT3 or to up-regulate interferon-response targets TAP1, PD-L1, and major histocompatibility complex (MHC) class I after treatment specifically with interferon gamma (red box), as compared with intact signaling in M420 (blue box). There was no change in response to interferon alfa or beta. As shown in Panel B, a lack of response to interferon gamma exposure was also seen in surface staining for PD-L1 and MHC class I by flow cytometry. Each point represents an independent experiment, T bars represent standard deviations, and P values are for a two-way analysis of variance with Dunnett’s correction. MFI denotes mean fluorescent intensity, and NS not significant. Panel C shows log2 RNA counts of expression for 790 immune-related genes on exposure to interferon gamma or vehicle control. The baseline cell line M420 (top) showed up-regulation of many interferon-stimulated genes (line represents an increase by a factor of 4), whereas the JAK2 mutated progression cell line M464 (bottom) lacked a similar response.
LOSS OF INTERFERON GAMMA–INDUCED GROWTH ARREST THROUGH ACQUIRED JAK MUTATIONS
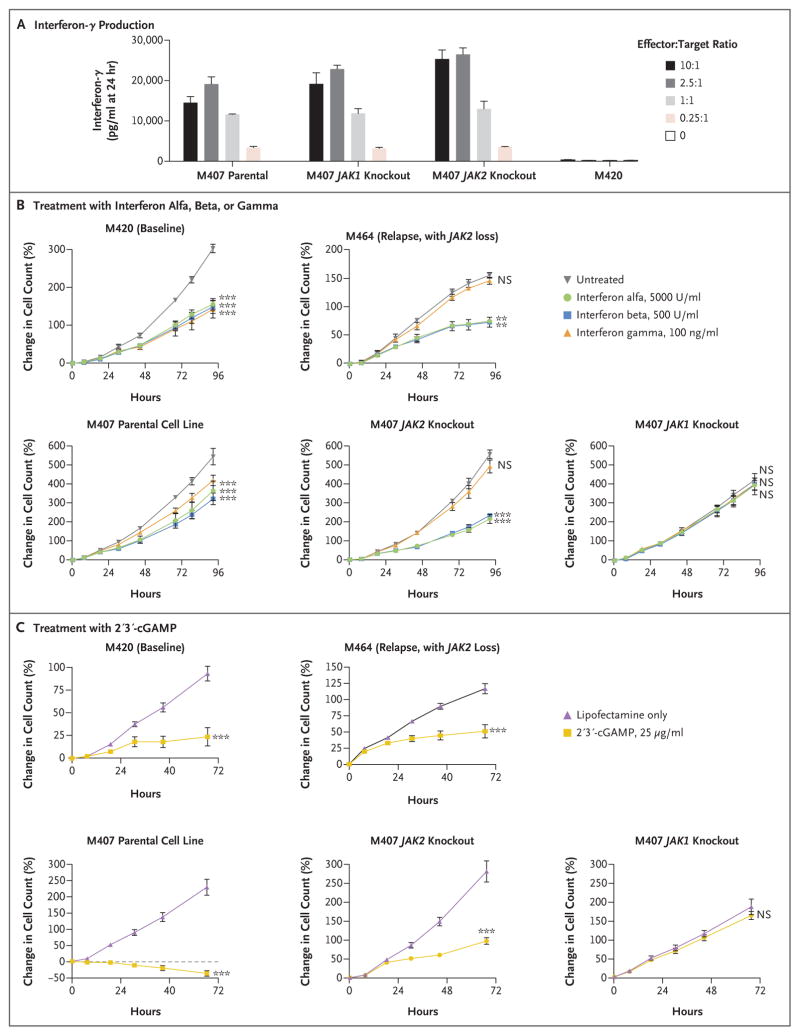
We hypothesized that inactivating JAK mutations may result in a functional advantage for the progressive tumors because the lack of interferon signaling either decreased antigen presentation or allowed escape from interferon-induced inhibition of growth. In addition to using M420 and M464, we engineered the human melanoma cell line M407 by means of the CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 approach to create sublines without expression of JAK1 or JAK2 (Figs. S10 and S11 in the Supplementary Appendix). These created truncating mutations analogous to those from Patients 1 and 2, and M407 is positive for HLA-A*02:01 and expresses the cancer–testis antigen NY-ESO-1, which allowed us to model T-cell recognition using T cells genetically modified to express an NY-ESO-1–specific T-cell receptor.23 M407 and both JAK-loss sublines were equally recognized by NY-ESO-1–specific T cells, leading to high levels of interferon-γ production (Fig. 4A).
Figure 4. Loss of Interferon Gamma– Induced Growth Arrest through Acquired JAK Mutations.
In Panel A, the M407 parental cell line as well as the M407 JAK1-knockout and JAK2-knockout sublines were recognized by NY-ESO-1–specific, HLA-A*02:01–restricted T cells, as assessed by interferon-γ production after 24 hours of in vitro coculture. M420 is negative for HLA-A*02:01 and served as a negative control. In Panel B, cell lines M420 and M407 showed growth inhibition in response to direct in vitro treatment with interferon alfa, beta, or gamma (left), whereas the JAK2-deficient counterpart M464 and the M407 JAK2 knockout were insensitive specifically to interferon gamma (middle). The M407 JAK1 knockout was insensitive to all three interferons (right). In Panel C, treatment with 2′3′-cGAMP (cyclic guanosine monophosphate–adenosine monophosphate), a direct cytosolic agonist of the stimulator of interferon genes (STING), was able to produce growth arrest in all cell lines, regardless of JAK2 status, yet had no effect in M407 with JAK1 knockout. Growth curves represent the percent change in the number of melanoma cells over time as measured by IncuCyte continuous live-cell imaging in one of three independent experiments. I bars in Panels A, B, and C indicate standard deviations for three replicate wells. Three asterisks indicate P<0.001 and two asterisks P<0.01 for the percent change in growth with the treatment shown at the 72-hour end point as compared with the untreated control, with Dunnett’s multiple-comparison correction applied in Panel B. NS denotes not significant.
When cultured in recombinant interferon alfa, beta, or gamma, the M420 and M407 parental cell lines showed interferon-induced growth inhibition in a dose-dependent manner (Fig. S12 in the Supplementary Appendix). However, both the JAK2-deficient M464 cell line (from Patient 2 at relapse) and the M407 JAK2-knockout subline were insensitive specifically to interferon gamma–induced growth arrest, yet remained sensitive to type I interferons alfa and beta; in contrast, the M407 JAK1-mutated subline was resistant to all three interferons (Fig. 4B). This is again consistent with the specific association of JAK2 with the interferon-γ receptor and the common use of JAK1 by all three interferon receptors.27–29 As an orthogonal test of these effects, we treated our cell lines with 2′3′-cGAMP (cyclic guanosine monophosphate–adenosine monophosphate); this di-nucleotide, which is produced in response to cytosolic double-stranded DNA, directly activates the stimulator of interferon genes (STING) and leads to interferon-β production through activation of interferon regulatory factor 3 (IRF-3).30 After 2′3′-cGAMP treatment, we observed growth arrest in all cell lines independent of JAK2 status but no effect in the JAK1-knockout subline (Fig. 4C). Therefore, the JAK1 and JAK2 loss-of-function mutations did not decrease in vitro T-cell recognition but selectively blocked the interferon-γ signaling that leads to cell-growth inhibition, which for JAK2 loss could be corrected by type I pathway activation or a STING agonist.
FUNCTIONAL EFFECTS OF MUTATION IN THE GENE ENCODING BETA-2-MICROGLOBULIN (B2M)
In Patient 3, whole-exome sequencing of the baseline and progressive lesions showed a 4-bp S14 frame-shift deletion in exon 1 of the beta-2-microglobulin component of MHC class I as 1 of only 24 new relapse-specific mutations and the only such mutation that was homozygous (Fig. S13A and S13B in the Supplementary Appendix). Immunohistochemical analysis for MHC class I heavy chains revealed loss of outer-membrane localization as compared with adjacent stroma or the baseline tumor, even though diffuse intracellular staining indicated continued production of MHC class I molecules (Fig. S14 in the Supplementary Appendix). This finding is in line with the role of beta-2-microglobulin in proper MHC class I folding and transport to the cell surface, and its deficiency has long been recognized as a genetic mechanism of acquired resistance to immunotherapy.12–14 Both the baseline and relapse biopsy samples were negative for MHC class II expression (Fig. S14 in the Supplementary Appendix), which suggests a lack of compensatory MHC upregulation.
We could not find defined genetic alterations in Patient 4 that had clear potential to result in acquired resistance to T cells, but cancer cells in the baseline and relapse biopsy samples did not express PD-L1 despite proximity to T cells and PD-L1–expressing stroma (Fig. S3D in the Supplementary Appendix). These findings suggest possible nongenetic mechanisms of altered expression of interferon-inducible genes.16
DISCUSSION
With the approval of PD-1 checkpoint-blockade agents for the treatment of patients with melanoma, lung cancer, and other cancers, it is anticipated that cases of late relapse after initial response will increase. Understanding the molecular mechanisms of acquired resistance by focused comparison of biopsy samples from paired baseline and relapsing lesions may open options for the rational design of salvage combination therapies or preventive interventions and may guide mechanistic biomarker studies for the selection of patients, before the initiation of treatment, who are unlikely to have a response.
Tumor-infiltrating T cells are the effectors that kill cancer cells during PD-1 blockade therapy.19,31 We found it striking that after intratumoral CD8 T-cell infiltration during active response, CD8 T cells were usually still present and abundant at the time of relapse, though they were restricted to the tumor margin. This observation suggested to us that the T cells were no longer able to exert their cytotoxic activity, because of either a lack of tumor antigen recognition and activation or a loss of sensitivity to their effector molecules by the cancer cells. The general possibilities are loss of mutational or shared tumor antigens that are recognized by T cells, loss of antigen-presenting machinery components (e.g., beta-2-microglobulin and HLA),12–14 tumor-cell–induced or myeloid-cell–induced inactivation of T-cell signaling,32,33 or insensitivity to the pro-apoptotic effects of toxic granules (e.g., perforin and granzymes), death receptors (e.g., Fas and tumor necrosis factor–related apoptosis-inducing ligand [TRAIL]), or interferons.34 Any of these escape mechanisms would be hypothesized to be fostered by the selective pressure of CD8 attack, which would be particularly active during the new round of immunoediting35 that is unleashed after PD-1 blockade.
The inactivation of JAK1 or JAK2, as noted in two of the patients, may be particularly advantageous to cancer cells in the context of anti–PD-1 therapy as compared with other immunotherapies. The interferon-induced adaptive expression of PD-L1, which allows the cancer to inactivate adjacent CD8 T cells,36 would be of no use after the PD-1–PD-L1 interaction is blocked by therapeutic antibodies. We propose that without this benefit, the advantage for cancer cells tilts toward abolishing interferon signaling in order to avoid the detrimental increase in antigen presentation and direct antiproliferative effects.27 Although we identified inactivating mutations in JAK1 and JAK2, which are receptor-level signaling bottlenecks, interferon insensitivity through other means — such as epigenetic silencing of interferon-signaling components as previously documented in lung-cancer and prostate-cancer cell lines15,16 or increased expression of negative regulators37 — might lead to the same end. We also documented one case of beta-2-microglobulin inactivation, which corroborates a previously described mechanism of acquired resistance to cancer immunotherapy in humans through loss of this shared component of all human MHC class I molecules that is required for CD8 T-cell recognition.12–14
In conclusion, the nearly identical mechanism of acquisition, functional consequence, and evidence of clonal selection for JAK1 or JAK2 mutations in two independent cases with a similar clinical course of acquired resistance suggests that resistance to interferon gamma contributes to immune resistance and escape. This genetic alteration of immune resistance joins the previously described loss of B2M in decreasing immune-cell recognition of cancer cells, leading to acquired resistance to cancer immunotherapy. Although we have identified four cases and worked out a potential mechanism of resistance in three of them, additional cases will need to be closely examined to assess the generalizability of these findings.
Supplementary Material
Acknowledgments
Supported in part by National Institutes of Health (NIH) grants R35 CA197633, P01 CA168585, 1U54 CA199090, and R01 CA170689 (to Dr. Ribas). Dr. Ribas was supported by the Ressler Family Foundation, the Dr. Robert Vigen Memorial Fund, the Grimaldi Family Fund, the Samuels Family Fund, the Ruby Family Fund, and the Garcia-Corsini Family Fund. Drs. Ribas and Schumacher were supported by a Stand Up To Cancer–Cancer Research Institute Cancer Immunology Dream Team Translational Research Grant (SU2C-AACR-DT1012); Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. Mr. Zaretsky is part of the UCLA Medical Scientist Training Program supported by NIH training grant GM08042. Dr. Shin was supported by the Oncology (5T32CA009297-30), Dermatology (5T32AR058921-05), and Tumor Immunology (5T32CA009120-39) training grants and a Tower Cancer Research Foundation Grant. Dr. Hu-Lieskovan was supported by a Young Investigator Award and a Career Development Award from the American Society of Clinical Oncology, a Tower Cancer Research Foundation Grant, and a Dr. Charles Coltman Fellowship Award from the Hope Foundation. Dr. Homet Moreno was supported in part by the Rio Hortega Scholarship (08/142) from Hospital 12 de Octubre, Madrid. Dr. Torrejon was supported by a grant from the Spanish Society of Medical Oncology for Translational Research in Reference Centers. Dr. Hugo was supported by the American Skin Association. Dr. Graeber was supported by a Melanoma Research Alliance Established Investigator Award (20120279) and an American Cancer Society Research Scholar Award (RSG-12-257-01-TBE). Dr. Lo was supported by the Steven C. Gordon Family Foundation, the Wade F.B. Thompson/Cancer Research Institute Clinic and Laboratory Integration Program Grant, the Ressler Family Foundation, the Grimaldi Family Fund, and the Ian Copeland Melanoma Fund.
We thank the staff of the Translational Pathology Core Laboratory and Rongqing Guo and Wang Li from UCLA for blood and biopsy processing; Xinmin Li, Ling Dong, Janice Yoshizawa, and Jamie Zhou from the UCLA Clinical Microarray Core for sequencing expertise; and members of Dr. Ribas’s laboratory, Pia Kvistborg and Marit van Buuren from the Netherlands Cancer Institute, Jordan Freeman from FLX Bio, and Leticia Corrales and Thomas Gajewski from the University of Chicago for useful discussions.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Atkins MB, Kunkel L, Sznol M, Rosenberg SA. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J Sci Am. 2000;6(Suppl 1):S11–4. [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yang JC, Sherry RM, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prieto PA, Yang JC, Sherry RM, et al. CTLA-4 blockade with ipilimumab: long-term follow-up of 177 patients with metastatic melanoma. Clin Cancer Res. 2012;18:2039–47. doi: 10.1158/1078-0432.CCR-11-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eroglu Z, Kim DW, Wang X, et al. Long term survival with cytotoxic T lymphocyte-associated antigen 4 blockade using tremelimumab. Eur J Cancer. 2015;51:2689–97. doi: 10.1016/j.ejca.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schadendorf D, Hodi FS, Robert C, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33:1889–94. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti–PD-1) in melanoma. N Engl J Med. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert C, Ribas A, Wolchok JD, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384:1109–17. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 8.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–30. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 10.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–32. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 11.Ribas A, Hamid O, Daud A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315:1600–9. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 12.Restifo NP, Marincola FM, Kawakami Y, Taubenberger J, Yannelli JR, Rosenberg SA. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst. 1996;88:100–8. doi: 10.1093/jnci/88.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Urso CM, Wang ZG, Cao Y, Tatake R, Zeff RA, Ferrone S. Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in B2m gene expression. J Clin Invest. 1991;87:284–92. doi: 10.1172/JCI114984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sucker A, Zhao F, Real B, et al. Genetic evolution of T-cell resistance in the course of melanoma progression. Clin Cancer Res. 2014;20:6593–604. doi: 10.1158/1078-0432.CCR-14-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan DH, Shankaran V, Dighe AS, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556–61. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn GP, Sheehan KC, Old LJ, Schreiber RD. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res. 2005;65:3447–53. doi: 10.1158/0008-5472.CAN-04-4316. [DOI] [PubMed] [Google Scholar]
- 17.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Wolchok JD, Hoos A, O’Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–20. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- 19.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atefi M, Avramis E, Lassen A, et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20:3446–57. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins PF, Morgan RA, Feldman SA, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–24. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 25.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–91. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 28.Müller M, Briscoe J, Laxton C, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. 1993;366:129–35. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 29.Watling D, Guschin D, Müller M, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–70. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 30.Corrales L, Gajewski TF. Endogenous and pharmacologic targeting of the STING pathway in cancer immunotherapy. Cytokine. 2016;77:245–7. doi: 10.1016/j.cyto.2015.08.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finke JH, Zea AH, Stanley J, et al. Loss of T-cell receptor zeta chain and p56lck in T-cells infiltrating human renal cell carcinoma. Cancer Res. 1993;53:5613–6. [PubMed] [Google Scholar]
- 33.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor mi-croenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–64. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 35.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 36.Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 2015;5:915–9. doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fish EN, Platanias LC. Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res. 2014;12:1691–703. doi: 10.1158/1541-7786.MCR-14-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.