Abstract
CRISPR/Cas9 system has been used widely in animals and plants to direct mutagenesis. To date, no such method exists for fish somatic cell lines. We describe an efficient procedure for genome editing in the Chinook salmon Oncorhynchus tshawytscha CHSE. This cell line was genetically modified to firstly overexpress a monomeric form of EGFP (cell line CHSE-E Geneticin resistant) and additionally to overexpress nCas9n, a nuclear version of Cas9 (cell line CHSE-EC, Hygromycin and Geneticin resistant). A pre-validated sgRNA was produced in vitro and used to transfect CHSE-EC cells. The EGFP gene was disrupted in 34.6 % of cells, as estimated by FACS and microscopy. The targeted locus was characterised by PCR amplification, cloning and sequencing of PCR products; inactivation of the EGFP gene by deletions in the expected site was validated in 25 % of clones. This method opens perspectives for functional genomic studies compatible with high-throughput screening.
Keywords: CRISPR/Cas9, CHSE-214, EGFP, FACS, Genome editing, nCas9n
Introduction
The CRISPR/Cas (clustered regularly interspaced palindromic repeats/CRISPR-associated) system was initially discovered in the genome of Escherichia coli (Ishino et al. 1987), and later found in about 40 % of bacteria, and a great majority of Archae (Horvath and Barrangou 2010). It functions as a bacterial protection system with memory directed against bacteriophages whereby double-stranded DNA molecules from viral origin are cleaved (Horvath and Barrangou 2010). The mechanism was characterised in Streptococcus, where the endonuclease Cas9 binds two RNA molecules, a CRISPR (cr)RNA targeting a complementary sequence on the viral genome, and a trans-activating (tra)crRNA acting as a transactivator (Jinek et al. 2012; Deltcheva et al. 2011). The isolation and characterisation of the Cas9 protein enabled to establish a method for genome editing in a wide range of organisms generating knockout models very efficiently and very quickly using small “guide RNA molecules” sgRNA in which cr and tracrRNA are fused (Ran et al. 2013; Sander and Joung 2014; Hsu et al. 2014). This system has been used in a number of species including fish, to generate lines of site-directed mutated animals by egg manipulation: zebrafish (Hruscha et al. 2013; Hwang et al. 2013; Kimura et al. 2014; Jao et al. 2013; Irion et al. 2014), Atlantic salmon (Edvardsen et al. 2014) or tilapia (Li et al. 2014). However, while CRISPR/Cas9-based methods have been used for genome editing in mammalian cell lines, to date, no such a system had been set up in somatic fish cell lines. This communication presents an efficient method to generate CRISPR/Cas9 knockout in the Chinook salmon Oncorhynchus tshawytscha cell line, CHSE-214.
Material and Methods
The CHSE-214 (referred as CHSE) cell line (ATCC CRL1681) was genetically engineered to overexpress a monomeric, cytosolic form of EGFP (mEGFP) using a method described previously by Lester et al. (2012) with the plasmid pmEGFP-N1 kindly provided by Prof. Reich (Chen and Reich 2010), conferring resistance to Geneticin G418. The resulting cell line, named CHSE-E, was maintained in CHSE culture medium supplemented with 500 μg/ml G418 (ThermoFisher Scientific).
A eukaryotic expression plasmid encoding the protein nCas9n, a nuclear version of the Cas9 from Streptococcus pyogenes, was constructed by subcloning the HindIII/XbaI fragment of pCS2-nCas9n (a gift from Wenbiao Chen, Addgene plasmid # 47929) (Jao et al. 2013; Edvardsen et al. 2014) into pcDNA3.1-Hyg (ThermoFisher Scientific) to obtain the plasmid pcDNA3.1-Hyg-nCas9n. The CHSE-E cell line (passage 30) was genetically engineered using this plasmid as described previously by Lester et al. (2012) to generate the CHSE-EC cell line, and cultivated in CHSE-E medium supplemented with 30 μg/ml Hygromycin B (ThermoFisher Scientific). The nCas9n transcript levels were measured by qPCR as described previously by Collet and Lester (2011) using primers nCas9nF 5′-CGACGCAAATCTGGATAAGG-3′ and nCas9nR 5′-GATCAGTGTAGCGTCCAGCA-3′.
Pre-validated sgRNA targeting EGFP was produced as described previously by Hwang et al. (2013) using DR274-eGFP (a gift from Filippo Del Bene; Addgene plasmid #61051; Auer et al. 2014). The targeted site for EGFP was GGCGAGGGCGATGCCACCTA starting at position 100 (within the ORF and relative to START codon) followed by the Protospacer adjacent motif (PAM) CGG. The sgRNA was produced in vitro using the Promega T7 transcription kit, purified using TRIzol (ThermoFisher Scientific), treated with RNAse-free DNase (ThermoFisher Scientific) and quantified by spectrometry (Nanodrop, Labtech International). CHSE-EC cells (passage 5) were transfected with sgRNA using the same conditions as established for plasmids for CHSE cells and described previously by Collet and Lester (2011), i.e., 500 ng sgRNA was used per 10 μl cell suspension at approx. 107 cells/ml. Twenty four (24) hours after passage, an analysis by FACS was performed alongside the parental CHSE and CHSE-E cell lines, acting as negative and positive controls for EGFP. Cells were trypsined with 1x trypsin (ThermoFisher Scientific), washed in 1x HSBSS (ThermoFisher Scientific), centrifuged for 10 min at 400g, resuspended in IF Media (PBS supplemented with 2 % FCS and 0.05 % sodium azide (Sigma-Aldrich)) and analysed using an Accuri®C6 analyser (BD Biosciences). A total of 50,000 events were collected from the live cell gate and data analysed using CFlow®Plus software.
From the same population of sgRNA-transfected CHSE-EC cells analysed by FACS, genomic DNA was purified using the DNeasy kit (Qiagen). A fragment containing the 20 nucleotide EGFP genomic target was amplified using the primers mEGFP-F 5′-ACGGCCACAAGTTCAGC-3′ and mEGFP-R 5′CGTCGCCGATGGGGGTGTTCT-3′. PCR was carried out using the FastStart High Fidelity PCR system (Roche) according to the manufacturer’s instructions. The PCR product was cloned using pGEM-T easy (Promega) according to the manufacturer’s instructions and sequenced (VWR sequencing services). Sequencher software (Gene Codes Corporation) was used to analyse chromatogram and detect indel mutations.
Results
CHSE-E displayed an intense fluorescence under an EVOSfl inverted fluorescent microscope (Advanced Microscopy Group) after more than 30 passages (Fig. 1a). The cloning process used in CHSE and CHSE-E was previously described by Collet and Lester (2011) and consisted in a small number of clonal cells (<500 cells) isolated, subcultured in a 25-cm2 flask and passaged at least 4 times in 75-cm2 flasks. Therefore, it is reasonable to assume that the quantity of the residual plasmid had become insignificant relative to the integrated transgene. PCR analysis of cDNA made from RNA purified from the CHSE-EC clonal cell line showed a strong expression of nCas9n when compared to mock cDNA produced by omitting reverse transcriptase (data not shown). In addition, the 192 nucleotide Cas9 fragment could be amplified by PCR directly from genomic DNA purified from CHSE-EC, indicating that at least one copy of the plasmid should be integrated in the genome of the CHSE-EC cell line (data not shown).
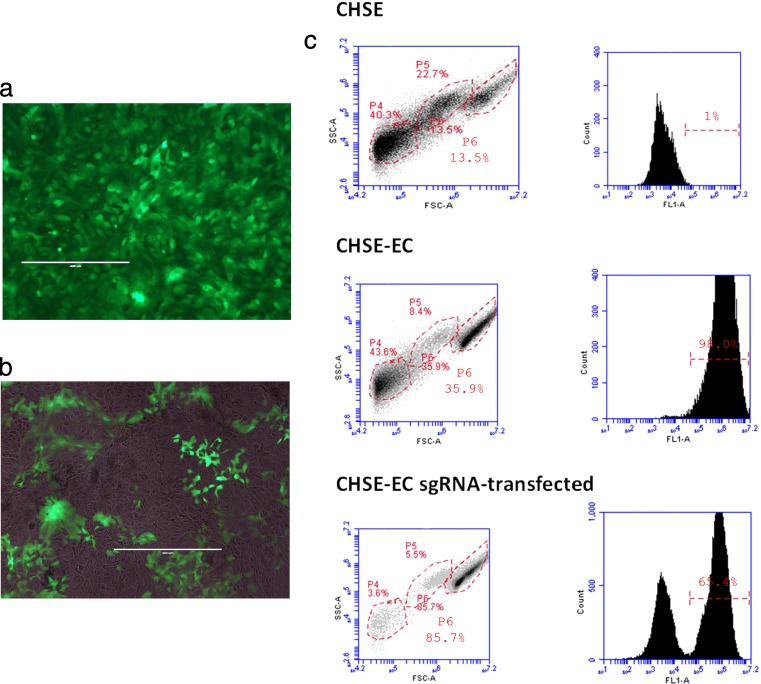
Fig. 1.
Fluorescent monolayer of the CHSE-EC (passage 5) cell line before (a) and after (b) transfection with the sgRNA targeting the EGFP locus. Bright field and fluorescence images have been overlaid in B in order to visualise non-fluorescent cells. Intensities of each channel have been adjusted for maximum visibility. FACS analysis of CHSE, CHSE-EC before and after sgRNA transfection (c): left panel: flow cytometry gating scheme (FSC-A, x-axis; SSC-A, y-axis). Populations P4, P5 and P6 were distinguished. Right panel: GFP fluorescence intensity of the P6 gated population (FL1-A, x-axis), number of cells (y-axis) and % of gated fluorescent cells are indicated
Following transfection, a proportion of cells on the monolayer had lost its fluorescence as assessed by simple microscopy observation (Fig. 1a, b). During FACS analysis, the viable cells were gated (region P6, Fig. 1c) and analysed for their fluorescence. Cell populations from the regions 4 and 5 were composed mainly of dead cells. The sgRNA-transfected CHSE-EC cells were composed of two populations, with levels of fluorescence corresponding to EGFP− CHSE (35 %) and EGFP+ CHSE-E (65 %), respectively (Fig. 1c, bottom panel). These results suggested that transfection with sgRNA resulted in a decreased level of EGFP in 35 % of the CHSE-EC cells.
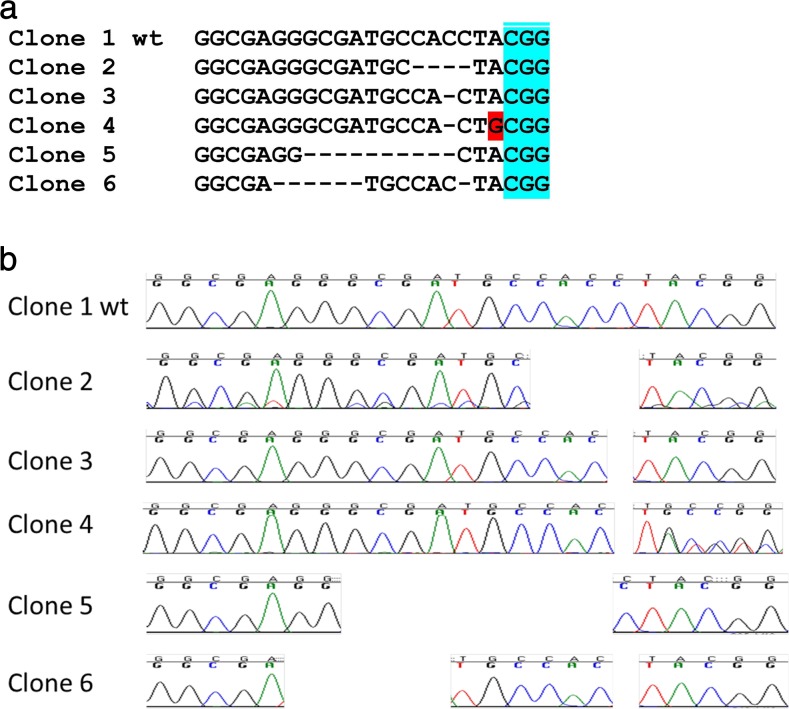
The genomic region around the targeted locus was sequenced from a PCR product obtained from genomic DNA purified from the sgRNA-transfected CHSE-EC cells. Out of 24 clones sequenced, 18 were showing 5 different mutated patterns and 6 were wild type (Fig. 2).
Fig. 2.
Indel mutations isolated from genomic DNA purified from the sgRNA-transfected CHSE-EC cells. Sequence alignment in the sgRNA target (a) and corresponding chromatograms (b)
Discussion
To our knowledge, this is the first description of a successful method to generate KO in somatic cell lines from non-mammalian vertebrates. We have genetically engineered the cell line CHSE-EC expressing EGFP and nCas9n using CRISPR/Cas 9-based genome editing to knockout the integrated EGFP gene. An early step of verification by PCR and sequencing can detect and estimate the frequency of deletion mutants, which can be followed by isolation of clonal cell lines by flow sorting or low-density seeding. It allows fast and efficient knockout of genome targets in a single round of transfection with a specific sgRNA easily produced in vitro.
One clone (clone 6, Fig. 2) showed deletions in two sites whereas Auer et al. (2014) did not detect any such events at the targeted site when editing the genome in zebrafish embryos using the same sgRNA. Although the number of clones analysed is low, this may reflect slight differences in the way Cas9 operates between salmon and zebrafish cells, possibly related to different temperatures (23 °C in the present study, 28 °C for zebrafish). The efficiency measured in the present study (approx. 35 %) is related to the editing of a single haplotype. Efficient knockout of a given endogenous gene would require the mutation of the target on two haplotypes which may be performed at lower efficiency with the present method. However, many factors can influence the knockout efficiency, and it is likely that the delivery of sgRNA to the nucleus of CHSE-EC is one of the main limiting factors rather than the number of haplotypes to target, in which case the efficiency of a double disruption should be comparable to the one assessed here for EGFP. As a matter of fact, the CRISPR/Cas9 system has been used efficiently to target several alleles in hexaploid species such as bread wheat (Wang et al. 2014) or other multiple target species (see for review Kim and Kim 2014). In consequence, we are confident that the frequency of knockout provided by this method on two haplotypes would be higher than a few per cent.
Non-fluorescent CHSE-C and ASK-C (Atlantic Salmon Kidney, Salmo salar) cell lines are currently being engineered for genome editing in both salmon species to provide material for high-efficiency CRISPR/Cas9 genome editing. This method has the potential to be applied to any endogenous target and to provide a cost-effective functional genomic tool in fish compatible with high-throughput screening.
Acknowledgments
This project was carried out during CD’s BBSRC Eastbio PIPS placement at Marine Scotland. The authors are grateful to Dr. Milena Monte (University of Aberdeen) for the help with the FACS analysis. The authors wish to thank Dr. Filippo Del Bene (Neuronal Circuit Development, Institut Curie) and Dr. Wenbiao Chen (School of Medicine, Vanderbilt University) for the Addgene plasmids, #61051 and #47929, respectively, and Prof. Nancy C. Reich Marshall (Department of Molecular Genetics and Microbiology, Stony Brooks University) for the plasmid pmEGFP-N1.
Abbreviations
- CRISPR
Clustered regularly interspaced short palindromic repeat
- Cas
CRISPR-associated
- PAM
Protospacer adjacent motif
- qPCR
Quantitative polymerase chain reaction
- CHSE
CHinook salmon embryo
- FACS
Fluorescence-activated cell sorting
- crRNA
CRISPR RNA
- tracrRNA
Trans-activating crRNA
- G418
Geneticin
References
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-C, Reich NC. Live cell imaging reveals continuous STAT6 nuclear trafficking. J Immunol. 2010;185:64–70. doi: 10.4049/jimmunol.0903323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet B, Lester K. Establishment of a Chinook salmon cell line with an inducible gene expression system. In Vitro Cell Dev Biol Anim. 2011;47:695–697. doi: 10.1007/s11626-011-9466-8. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A. Targeted mutagenesis in Atlantic Salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS One. 2014;9:e108622. doi: 10.1371/journal.pone.0108622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Krauss J, Nüsslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. PNAS. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S-I. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester K, Hall M, Urquhart K, Gahlawat S, Collet B. Development of an in vitro system to measure the sensitivity to the antiviral Mx protein of fish viruses. J Virol Methods. 2012;182:1–8. doi: 10.1016/j.jviromet.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Li M, Yang H, Zhao J, Fang L, Shi H, Li M, Sun Y, Zhang X, Jiang D, Zhou L, Wang D. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics. 2014;197:591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]