Abstract
Quorum sensing (QS) is a microbial cell-to-cell communication process that relies on the production and detection of chemical signals called autoinducers (AIs) to monitor cell density and species complexity in the population. QS allows bacteria to behave as a cohesive group and coordinate collective behaviors. While most QS receptors display high specificity to their AI ligands, others are quite promiscuous in signal detection. How do specific QS receptors respond to their cognate signals with high fidelity? Why do some receptors maintain low signal recognition specificity? In addition, many QS systems are composed of multiple intersecting signaling pathways: what are the benefits of preserving such a complex signaling network when a simple linear ‘one-to-one’ regulatory pathway seems sufficient to monitor cell density? Here, we will discuss different molecular mechanisms employed by various QS systems that ensure productive and specific QS responses. Moreover, the network architectures of some well-characterized QS circuits will be reviewed to understand how the wiring of different regulatory components achieves different biological goals.
Keywords: intercellular communication, gene expression, group behavior, regulatory network, chemical signaling
This review focuses on the specificity and complexity of quorum-sensing circuits in both Gram-negative and Gram-positive bacterial species.
INTRODUCTION
Quorum sensing
Bacterial quorum sensing (QS) is a cell-to-cell communication process that relies on the production, secretion and detection of autoinducer (AI) signals to regulate gene expression in response to changes in population density. QS allows a group of bacterial cells to regulate their gene expression in unison, which is important for carrying out group behaviors such as bioluminescence production, biofilm formation, genetic exchange and virulence factor expression (Ng and Bassler 2009). Bacterial species depend on QS to regulate important cellular processes that are essential for surveillance, survival and adaptation to their changing environments (Bassler and Vogel 2013). By monitoring the accumulation of specific AIs, bacteria can track shifts in population density and species complexity in the vicinity and respond as a group accordingly (Fuqua and Greenberg 2002; Pappas, Weingart and Winans 2004; Novick and Geisinger 2008; Ng and Bassler 2009; Williams and Camara 2009; Ng et al. 2011).
QS systems have been identified in both Gram-negative and Gram-positive bacterial species. After synthesis, the signal is exported extracellularly, and its concentration increases proportionally to population density. When the concentration of the signal is above a certain threshold, the signal is detected by a QS receptor that elicits a downstream signal transduction cascade, triggering a high cell density gene expression program (Fig. 1). In Gram-negative bacteria, AI molecules are comprised of several chemical classes including acyl homoserine lactones (AHSLs), alkylquinolones, α-hydroxyketones and diffusible signal factor (fatty acid-like compounds) (Fig. 2). These signaling molecules are synthesized from common metabolites such as fatty acids, anthranilate and S-adenosylmethionine (SAM), either with a single signal synthase or through a series of enzymatic reactions (Fuqua and Greenberg 1998; Tiaden, Spirig and Hilbi 2010; Ryan et al. 2015). In contrast to the signals that regulate QS in Gram-negative bacteria, short oligopeptides are produced and detected in Gram-positive bacteria (Fig. 2). In most cases, the signal is synthesized as a longer peptide precursor, which is subsequently exported and modified upon its secretion by a dedicated transporter (Fig. 1C and D) (Kleerebezem et al. 1997; Pottathil and Lazazzera 2003; Lyon and Novick 2004; Dufour and Levesque 2013; Cook and Federle 2014; Monnet, Juillard and Gardan 2014).
Figure 1.
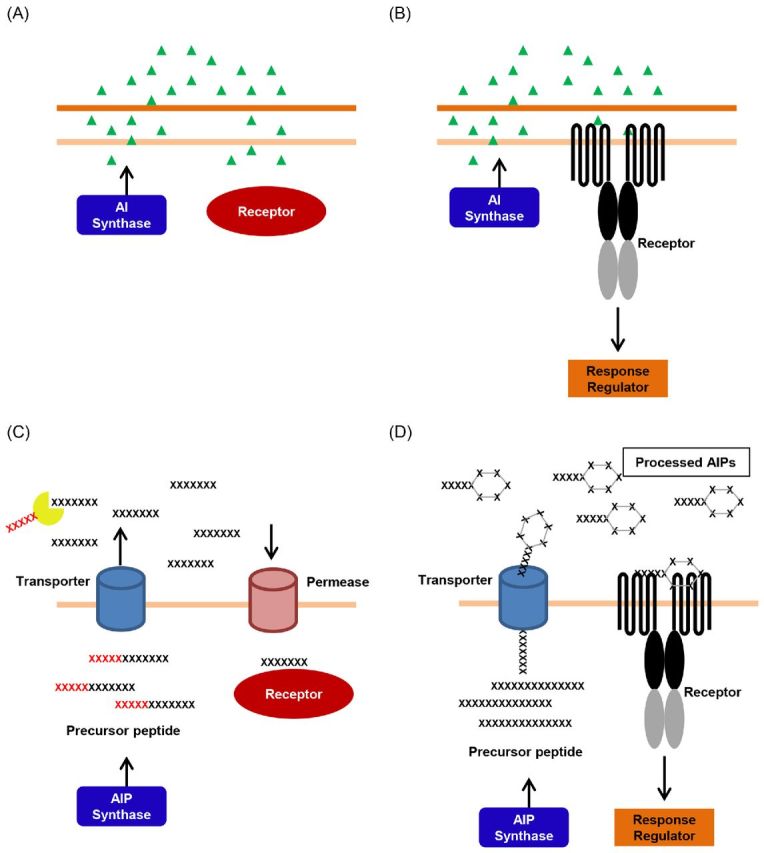
Basic QS circuit diagrams. (A) A Gram-negative one-component QS system. Autoinducer molecules are produced by the AI synthase, released into the extracellular environment, which are then diffused back into the cytoplasm where the QS receptor detects them, while also acting as a transcriptional regulator. (B) A Gram-negative two-component QS system. Autoinducer molecules are produced by the AI synthase, released into the extracellular environment, and are then detected by a transmembrane receptor. Detection of autoinducers triggers a phospho-relay that controls the downstream QS response. (C) A Gram-positive one-component QS system. Autoinducer peptides are produced by the AIP synthase and then released into the extracellular environment through a transporter, where they undergo proteolysis and are then transported back into the cytoplasm through a permease. In the cytoplasm, the modified AIP is detected by a QS receptor that also acts as a transcriptional regulator. (D) A Gram-positive two-component QS system. Autoinducer peptides are produced by the AIP synthase and released into the extracellular environment through a transporter where they undergo post-translational modifications and are then detected by a transmembrane receptor. Detection of autoinducer triggers a phospho-relay that controls the downstream QS response.
Figure 2.
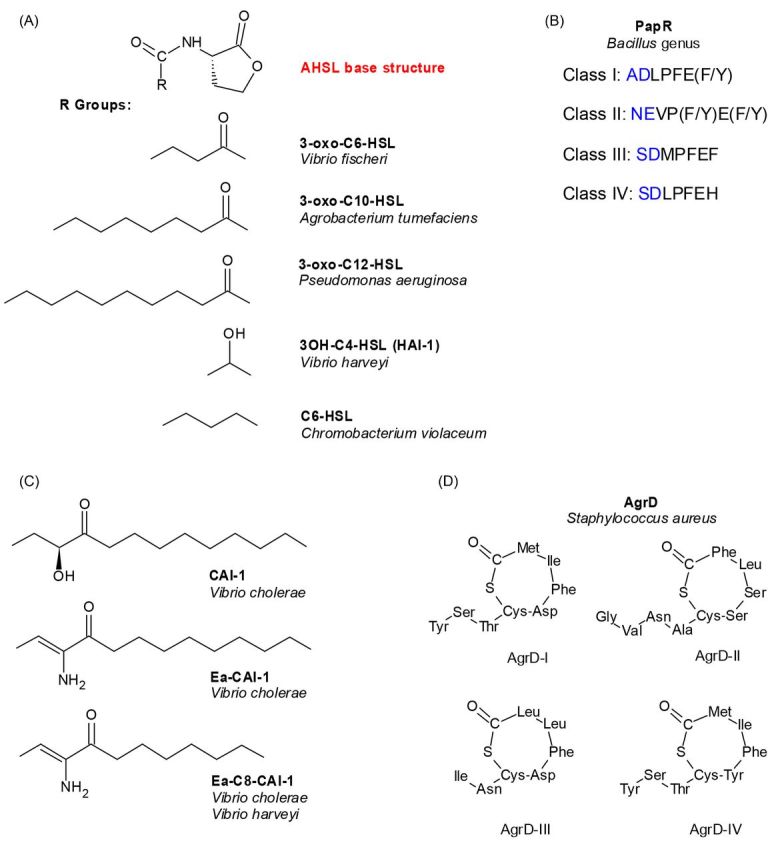
Structures of bacterial autoinducers. (A) Acyl-homoserine lactones (AHSLs) that are produced by various Gram-negative bacteria. Shown is the AHL base structure, plus various R groups that differ among species. (B) Small peptide autoinducers (AIPs) called PapR produced by Bacillus genus. Predicted physiologically-relevant heptapeptides are indicated by additional residues in blue. (C) CAI-1 and its related autoinducers produced by Vibrio species. (D) The four AgrD variants produced by Staphylococcus aureus.
In general, a specific set of AIs is produced and detected by each QS bacterial species; nonetheless, it is not unreasonable to assume that these species are exposed to non-cognate signal analogs produced by other species in the vicinity. How does a QS receptor detect and distinguish related AIs with similar structures? How do bacteria prevent premature induction of QS caused by signal fluctuations? Moreover, bacterial QS systems have likely evolved independently of one another, yet, there are elegant patterns within these circuits regarding signal detection and system architecture. Here, we will review how bacteria ensure steadfast QS signal transduction that only occurs upon detection of cognate signals with corresponding receptors. Specifically, we will discuss how several types of QS receptors have evolved to possess an exquisite and intrinsic specificity towards their cognate ligands. We will then consider strategies that maintain signal transduction specificity and fidelity in various QS systems and why multiple and distinct regulatory pathways are connected to form a complex signaling network.
LIGAND SPECIFICITY IN QS RECEPTORS
LuxR-type regulators
QS was first discovered as a regulatory mechanism of bioluminescence induction in the Gram-negative bacterium Vibrio fischeri, where light production is induced through a cell-density dependent activation of expression of the luciferase operon (Nealson, Platt and Hastings 1970; Nealson and Hastings 1979). This marine bacterium employs a LuxI-LuxR-type QS system to control light production, in which LuxR serves as both the cytoplasmic AI receptor and the transcriptional activator of the lux operon (Fig. 1A) (Engebrecht, Nealson and Silverman 1983; Engebrecht and Silverman 1984; Stevens, Dolan and Greenberg 1994; Schaefer et al. 1996a; Stevens and Greenberg 1997; Stevens et al. 1999). LuxI is the AI synthase, which catalyzes the production of N-(3-oxohexanoyl)-homoserine lactone (3-oxo-C6-HSL), the cognate signal for V. fischeri LuxR (Fig. 2A) (Eberhard et al. 1981; Engebrecht and Silverman 1984; Schaefer et al. 1996b).
LuxR and other similar regulators in the family are composed of two distinct domains: an N-terminal ligand binding domain (LBD) and a C-terminal DNA-binding domain (DBD) (Shadel, Young and Baldwin 1990; Slock et al. 1990; Choi and Greenberg 1991, 1992; Fuqua and Greenberg 1998). In the absence of AI, most LuxR-type proteins do not fold correctly and are rapidly degraded; however, the complex becomes stable upon ligand binding. In the V. fischeri LuxR, ligand (3-oxo-C6-HSL) binding also induces a conformational change that reveals the DBD of LuxR, therefore rendering it free to bind to the promoter of the lux operon and activate its transcription (Stevens, Dolan and Greenberg 1994; Hanzelka and Greenberg 1995). Homologs of the LuxI-LuxR QS system have been identified in many Gram-negative bacteria, including LasI-LasR (Passador et al. 1993; Pearson et al. 1994; Bottomley et al. 2007; Zou and Nair 2009), RhlI-RhlR (Ochsner et al. 1994; Brint and Ohman 1995; Ochsner and Reiser 1995; Pearson et al. 1995), QscR from Pseudomonas aeruginosa (Chugani et al. 2001), TraI-TraR from Agrobacterium tumefaciens (Fuqua and Winans 1994; Hwang et al. 1994; Vannini et al. 2002; Zhang et al. 2002) and CviR from Chromobacterium violaceum (McClean et al. 1997; Chen et al. 2011; Stauff and Bassler 2011). In these systems, AIs synthesized by the different signal synthases share a common homoserine lactone group, but differ in acyl chain length and modifications (Fig. 2A). Since these AIs are structurally similar and many QS species are capable of producing them, signal interference among non-cognate systems could lead to an unwanted, premature induction of the QS response. Thus, many QS bacteria have evolved their LuxR-type receptors with an exquisite specificity towards its cognate signal. On the other hand, some LuxR-type receptors display a more relaxed specificity and detect multiple ligands, such as CviR discussed later in this review (Swem et al. 2009; Chen et al. 2011). While the exact biological role for this promiscuous signal specificity is unknown, it is thought that the latter class of receptors can be used for inter-species signaling. Here, we use a few well-characterized LuxI-LuxR-type QS systems to illustrate the diversity in signal production and detection specificity.
AI binding is required for protein folding of some LuxR-type regulators
Agrobacterium tumefaciens employs a LuxI-LuxR-type QS system, called TraI-TraR, to regulate the transfer of the Ti plasmid from the bacterium to its plant host, ultimately causing tumor formation inside the host (Piper, Beck von Bodman and Farrand 1993; Fuqua and Winans 1994; Hwang et al. 1994; Christie 1997). Agrobacterium tumefaciens produces several AHSLs but the most abundant one is 3-oxo-C10-HSL (Fig. 2A), which is synthesized by TraI and is the cognate ligand of QS receptor TraR (Hwang et al. 1994; Zhu et al. 1998). Several unrelated AHSLs are also capable of activating TraR, however, these analogs are only active when present at high concentrations (Zhu et al. 1998). These findings suggest that TraR has evolved a very specific interaction with its cognate signal and is refractory to being activated by non-cognate signals in physiologically-relevant conditions. The 3D structure of TraR bound with its cognate AHSL ligand was the first reported among all LuxR-type receptors (Vannini et al. 2002; Zhang et al. 2002). The resolved structural model suggests that TraR requires its cognate ligand for proper folding, a characteristic shared with some of the other members of the LuxR family (Vannini et al. 2002; Zhang et al. 2002). The TraR protein is a symmetric homodimer, with each monomer comprised of a LDB and a DBD that are joined by a short linker region. Surprisingly, the 3-oxo-C10-HSL ligand is enclosed in a solvent-inaccessible region on the opposite side of the dimerization site of each monomer. The AI binding site is composed of aromatic and hydrophobic amino acids within each LBD. 3-oxo-C10-HSL interacts with TraR between the central β-sheet and α-helices α3, α4 and α5 to ensure that the signal remains bound and correctly-oriented (Vannini et al. 2002; Zhang et al. 2002). The invariant homoserine lactone portion of 3-oxo-C10-HSL interacts with multiple residues in the LBD, conserved among the majority of the LuxR-type receptors. While each AHSL signal is built upon a homoserine lactone core, the acyl chain of the AHSLs differs among the QS systems. These differences dictate how the molecule fits into the binding pocket of each LuxR-family receptor. For example, the acyl portion of the 3-oxo-C10-HSL interacts with TraR at residues Y53, L40, Y61, F62, some of which are not conserved. The fact that the AI binds within a buried hydrophobic pocket in the LBD suggests that AI binding plays a role in the correct folding and stabilization of the TraR protein and explains why TraR displays such high specificity for 3-oxo-C10-HSL (Zhu and Winans 1999, 2001; Vannini et al. 2002; Zhang et al. 2002).
While ligand binding is essential for TraR folding and stability, some LuxR proteins, such as LasR from P. aeruginosa and LuxR from V. fischeri, bind to their cognate ligands reversibly (Urbanowski, Lostroh and Greenberg 2004; Sappington et al. 2011). Indeed, low levels of active recombinant LasR can be detected in Escherichia coli grown without 3-oxo-C12-HSL, suggesting that LasR can fold into a functional conformation in the absence of signal through an unknown mechanism; however, this ligand-free form of LasR is very unstable (Sappington et al. 2011). Reversible AI binding allows the receptor to be inactivated via rapid dilution of ligand, which could be critical for the bacterium to switch behaviors from those of high cell density to those of low cell density. The crystal structure of the LBD of LasR bound to its cognate AI 3-oxo-C12-HSL provided further insight into how LasR detects its cognate AIs (Bottomley et al. 2007). Similar to TraR, LasR is a homodimer, in which one AI molecule binds to each monomer of the receptor in a solvent-inaccessible pocket comprised of a β-sheet and several α-helices (Bottomley et al. 2007). Likewise, LasR interacts with 3-oxo-C12-HSL by forming multiple hydrogen bonds in between several conserved LasR residues and the invariant homoserine lactone portion of the AI (Bottomley et al. 2007). However, the acyl chain of 3-oxo-C12-HSL is surrounded within a hydrophobic pocket in LasR by interacting with several non-conserved residues, possibly giving LasR the ability to distinguish 3-oxo-C12-HSL from other AHSLs (Bottomley et al. 2007).
Interactions of two AHSL signals in Vibrio fischeri LuxR
Many QS bacteria produce multiple related AHSLs using different LuxI-type synthases. For example, aside from LuxI, V. fischeri carries another non-homologous AHSL synthase called AinS which produces C8-HSL. Together, these two AHSLs regulate bioluminescence production (Kuo, Callahan and Dunlap 1996; Hanzelka et al. 1999). C8-HSL is detected by a membrane-bound receptor AinR (we will discuss this type of AHSL receptor below). The ligand specificity of V. fischeri LuxR is somewhat stringent as several AHSL analogs such as 3-oxo-C5-HSL, 3-oxo-C8-HSL and 5-oxo-C6-HSL, are capable of activating lux expression through binding to LuxR in a heterologous E. coli host, but none of these analogs are as effective as the cognate signal 3-oxo-C6-HSL (Schaefer et al. 1996a). Although C8-HSL can bind to LuxR, the C8-HSL/LuxR complex is a weaker activator of the lux operon than the 3-oxo-C6-HSL/LuxR complex (Kuo, Callahan and Dunlap 1996; Schaefer et al. 1996a; Lupp et al. 2003). C8-HSL and 3-oxo-C6-HSL compete for the same binding site on LuxR and, therefore, high concentrations of C8-HSL could inhibit light production. Not surprisingly, this inhibition is suppressed by high doses of 3-oxo-C6-HSL (Schaefer et al. 1996a). Intriguingly, production of these two AHSLs varies among different V. fischeri isolates; some brighter strains that produce more luciferase, such as MJ1, secrete >1000-fold 3-oxo-C6-HSL and 5-fold less C8-HSL than other dimmer isolates (Boettcher and Ruby 1995). LuxR also displays only 75% identity among these different isolates. Directed evolution of LuxR that responds to C8-HSL but not 3-oxo-C6-HSL, reveals that residues both inside and outside of the LBD are responsible for this switch in ligand specificity (Collins, Arnold and Leadbetter 2005; Collins, Leadbetter and Arnold 2006; Hawkins et al. 2007). How different natural LuxR variants respond to these two competing AHSLs, and how polymorphisms among different LuxR proteins affects bioluminescence production in V. fischeri, remains unclear. Together, however, these findings strongly suggest that LuxR has evolved to respond to different AI compositions in order to achieve various biological goals.
Signal antagonism revealed in CviR bound with different ligands
CviR is a cytoplasmic QS receptor in C. violaceum, a bacterium known for the production of a purple pigment called violacein, along with biofilm formation and cyanide production (Stauff and Bassler 2011). The production of violacein is regulated by QS via CviR activation of the ‘vioABCD’ operon (McClean et al. 1997; August et al. 2000). CviR from different C. violaceum isolates displays various AHSL signal specificity. In strain ATCC 31532, the cognate signal for CviR is C6-HSL (Fig. 2A), which is synthesized by CviI. However, this CviR has promiscuous ligand specificity, as CviR can activate vioA transcription when bound to AHSLs with acyl chain lengths ranging from C4 to C8 (Swem et al. 2009). Although C4-HSL can induce maximum vioA expression, it requires a much higher signal concentration than cognate C6-HSL (Swem et al. 2009). In contrast, AHSLs with acyl chain lengths ranging from C10 to C14 are either weak agonists or are completely inactive. Indeed, C10-HSL is an effective CviR antagonist (Chen et al. 2011). Furthermore, CviR activity can also be antagonized by a synthetic molecule analogous to C6-HSL with two modifications: a phenoxy group at the end of the acyl chain and a homocysteine thiolactone ring in place of a homoserine lactone ring (Swem et al. 2009). Antagonism is further increased by the addition of a chlorine atom at the ‘para’ position on the phenoxy ring and by the removal of the methyl group at the ‘ortho’ position of the phenoxy ring (antagonist CTL). Additional antagonism is observed upon the replacement of sulfur in the thiolactone with an oxygen atom (antagonist CL).
When the structure of full-length CviR bound with antagonist chlorolactone (CL) was solved (Chen et al. 2011), it was discovered that the domain arrangement of the receptor diverges from the structure of TraR bound with its ligand. In the CviR/CL complex, the DBD is positioned underneath the LBD of the opposite monomer in a cross-domain structure. In this conformation, the DBD domains are far apart and incompatible with operator DNA binding. Moreover, bacterial two-hybrid studies showed that only C6-HSL allows CviR interaction with the RNA polymerase α-subunit (Chen et al. 2011).
Interestingly, residue M89 in the LBD exists in two separate orientations when different ligands are bound to CviR. This residue is typically buried when the C6-HSL agonist is bound. However, structures of the LBD alone bound to antagonists reveal that M89 acts as a gate that swings away from the binding pocket to accommodate larger HSLs such as C8-HSL, C10-HSL and CL. Activity of CviR bound to antagonist C10-HSL is restored when M89 is mutated to smaller residues (M89S or M89A), but not when M89 is mutated to residues that were similar in size or larger. CviR with mutation M89S or M89A bound to C10-HSL demonstrated an open conformation similar to CviR/C6-HSL complexes. The mutant CviR-C10-HSL could bind DNA, interact with RNA polymerase, and initiate vioA transcription (Chen et al. 2011).
Another C. violaceum strain (ATCC 12472) produces 3-OH-C10-HSL as its cognate AI and also responds to C10-HSL and CL, antagonists of the previously-studied CviR from another strain (ATCC31532). Interestingly, the CviR receptor from ATCC 12472 has a Ser residue at position 89, favoring a more open binding pocket that can bind C10-HSLs. However, a second amino acid change, N77Y, together with S89M, is necessary to switch ligand specificity for this CviR to sense C10-HSL and CL as antagonists (Chen et al. 2011). Thus, it appears that the two CviR receptors in these two C. violaceum strains have evolved to specifically detect the corresponding cognate AHSL signal. This series of structure-function analyses also gives important insight into how LuxR- type receptors discriminate structurally similar molecules and illustrate a possible antagonism mechanism for this important class of QS regulators.
Orphan (solo) LuxR-type receptors
While LuxR-type receptors and LuxI synthases are usually encoded in the same operon, some LuxR-type receptors are found to be orphans (or solos), meaning they have no genetically linked cognate AHSL synthases. These QS receptors are originally thought to respond only to AHSLs, however, it was recently found that these orphan LuxR-type proteins could respond to signals unrelated to AHSLs (Brachmann et al. 2013; Nguyen et al. 2015). Unsurprisingly, residues that are responsible for making contacts with AHSLs in other LuxR-type receptors are substituted in these orphan receptors. In the case of orphan regulators PluR and PauR, some of these substituted residues are essential for cognate signal sensing but not solely responsible for ligand-binding specificity (Brameyer and Heermann 2015).
Ligand specificity in Gram-positive RNPP-type receptors
A few years before Hastings and colleagues published their discovery of autoinduction of bioluminescence in V. fischeri (Nealson, Platt and Hastings 1970), a similar discovery was made in Streptococcus pneumoniae by Alexander Tomasz, in which the competence state of the bacterium can be induced by a hormone-like substance present in the culture medium (Tomasz 1965). Though this finding was not recognized as a QS-regulated behavior at the time, it is now known that a peptide-based QS system is involved in competence regulation (Havarstein, Coomaraswamy and Morrison 1995), and that this likely to be the first piece of evidence of cell–cell communication. In general, Gram-positive bacteria produce and detect short oligopeptides for QS regulation (Fig. 2B and D). These peptides are usually 5 to 34 amino acids in length and undergo critical modifications upon secretion into the extracellular space (Fig. 1C and D) (Kleerebezem et al. 1997; Pottathil and Lazazzera 2003; Lyon and Novick 2004; Dufour and Levesque 2013; Cook and Federle 2014; Monnet, Juillard and Gardan 2014). One family of this type of QS receptor is called the RNPP family, an acronym for the four receptors that are classified within this category: Rap, NprR, PlcR and PrgX (Declerck et al. 2007; Rocha-Estrada et al. 2010). For these systems, signal precursors are first synthesized as ∼40 amino acid peptides that contain a signal sequence for peptide export via either the general secretion system (Sec) or by dedicated ABC transporters (Jimenez and Federle 2014). The C-terminal region of these peptide signals contains ∼13 to 20 amino acids that are modified by proteolysis (Fig. 1C) (Pottathil and Lazazzera 2003; Declerck et al. 2007; Gohar et al. 2008; Rocha-Estrada et al. 2010). The modified peptides are then imported back into the cell through an oligopeptide permease for subsequent cytoplasmic receptor detection (Fig. 1C) (Perego 1997; Gominet et al. 2001). Rap is an aspartyl phosphate phosphatase and transcriptional activator protein (Rocha-Estrada et al. 2010; Parashar et al. 2011), while NprR, PlcR and PrgX are DNA-binding transcription factors (Wintjens and Rooman 1996; Aravind et al. 2005; Rocha-Estrada et al. 2010). NprR is a neutral protease regulator (Pottathil and Lazazzera 2003; Zouhir et al. 2013), PlcR is a phospholipase C regulator (Declerck et al. 2007; Gohar et al. 2008) and PrgX regulates plasmid conjugative transfer (Bae, Clerc-Bardin and Dunny 2000; Shi et al. 2005). Structural and mechanistic analyses of the RNPP-type receptors have been reviewed recently (Rocha-Estrada et al. 2010; Cook and Federle 2014). Here, we will use the well-characterized PlcR system to illustrate ligand specificity control in this family of regulators.
PlcR is a cytoplasmic QS receptor known as a phospholipase C regulator in the Gram-positive Bacillus genus, including B. anthracis, B. cereus, B. thuringiensis, and other Bacillus species (Lereclus et al. 1996; Declerck et al. 2007). The peptide ligand of PlcR is called PapR. There are four PlcR/PapR classes within the B. cereus group, identified as PlcRI/PapRI (LPFE(F/Y)), PlcRII/PapRII (VP(F/Y)E(F/Y)), PlcRIII/PapRIII (MPFEF) and PlcRIV/PapRIV (LPFEH) (Fig. 2B) (Slamti and Lereclus 2002, 2005). The PapR variations are generally in the first and last residues of the pentapeptide (the longer, physiologically-relevant forms of these peptides will be discussed below), giving rise to species ligand specificity (Slamti and Lereclus 2002, 2005). The X-ray crystal structure of B. thuringiensis PlcRI bound with PapR (LPFEF) revealed that PlcRI is a symmetrical, dimeric protein composed of an N-terminal DBD and a C-terminal LBD, which also serves as the dimerization site between the two monomers (Declerck et al. 2007). The LBD is comprised of five tetratricopeptide repeats (TPR) (Declerck et al. 2007; Grenha et al. 2013). TPR domains are known for their protein–protein and protein–peptide interactions (Blatch and Lassle 1999) and the two PlcR monomers meet at their respective TPR domains which also contain the PapR-binding sites. Within this binding site, the backbone of the PapR ligand forms hydrogen bonds with several residues of PlcRI. Specially, the PapR glutamic acid binds to Y275 of the TPR helix and also to residues K87 and K89, which function as the gatekeepers to select for PlcR/PapR binding only, while the K197 residue interacts with the PapR C-terminus (Declerck et al. 2007; Bouillaut et al. 2008). The two PapR phenylalanines, which are not always present in PapR peptides in other Bacillus species, stabilize the peptide by inserting it into a hydrophobic cleft between α-helices 5 and 7 of PlcR. The processed PapR in all four pentapeptide classes has a proline residue that is predicted to fit the peptide into the binding pocket within the TPR domain of PlcR (Bouillaut et al. 2008). After the structure of B. thuringiensis PlcR was determined, it was found that the more biologically-relevant PapR sequence is ADLPFEF instead of the shortened pentapeptide LPFEF (Bouillaut et al. 2008). The additional two amino acids in the heptapeptide were predicted to fit into the continuation of the groove of the PapR-binding site in PlcRI, and also account for receptor-ligand specificity (Bouillaut et al. 2008; Rocha-Estrada et al. 2010). Since there is some PapR similarity, cross-talk does exist, but PlcRI in B. thuringiensis was shown to be highly activated by its cognate PapR heptapeptide and to a much lesser extent by the other three heptapeptides, while PlcRII and III are more promiscuous (Bouillaut et al. 2008). The positioning of the TPR is thought to be influenced by the binding of the ligand, which subsequently affects dimerization and positioning of the DBD to DNA (Declerck et al. 2007; Grenha et al. 2013). Remarkably, the structures of PlcR in B. thuringiensis and PgrX in Enterococcus faecalis are extremely similar, even where the peptide signal binds to the receptor, yet these proteins have completely different functions. DNA binding is non-existent in PlcR unless bound to the PapR peptide, while PgrX binds to DNA sans signal (Declerck et al. 2007; Bouillaut et al. 2008).
Specificity in two-component Gram-positive peptide QS systems
In addition to cytoplasmic peptide receptors, Gram-positive bacteria also employ membrane-bound receptors for QS communication. These peptide QS receptors usually are histidine kinases that transfer a phosphate to cytoplasmic response regulators (Hoch and Silhavy 1995; Inouye and Dutta 2003; Simon, Crane and Crane 2007), which either initiate or repress transcription of a particular set of genes (Fig. 1D). While the cytoplasmic portions of these membrane-bound receptors belong to the histidine kinase family and share homology, little homology exists in their LBDs, which determine ligand specificity (Magnuson, Solomon and Grossman 1994; Pestova, Havarstein and Morrison 1996; Miller and Bassler 2001; Novick and Geisinger 2008; Ng and Bassler 2009). Since these receptors are membrane-bound, structural analysis is difficult. Here, we will use AgrC as an example to illustrate the mechanisms used by this type of receptor to differentiate between related peptide signals.
Staphylococcus aureus is a clinically important human pathogen that uses a two-component accessory gene regulator (Agr) system as the primary QS circuit. Agr controls the expression of several virulence factors that allow this bacterium to persist in almost any human tissue (Novick and Geisinger 2008; Le and Otto 2015). This system is expressed from two divergently transcribed loci: the agrBDCA operon (RNAII) and RNAIII (Novick et al. 1995; Novick and Geisinger 2008). The agrC gene encodes the AgrC transmembrane QS histidine kinase receptor, while agrA encodes the cytoplasmic response regulator protein (Lina et al. 1998; Peng et al. 1988). The agrD gene encodes the 46–47 amino acid peptide precursor AgrD, which is further processed by AgrB, a transmembrane endopeptidase, to a 7 to 9 amino acid peptide that contains a conserved thiolactone ring formed between the sulfur atom from a cysteine residue to the C-terminus of the autoinducer peptide (AIP) that is required for its activity (Fig. 1D) (Ji, Beavis and Novick 1995, 1997; Novick et al. 1995; Mayville et al. 1999; Zhang and Ji 2004; Novick and Geisinger 2008). RNAIII is a non-coding RNA that modulates expression of other regulators (Geisinger et al. 2006; Boisset et al. 2007; Novick and Geisinger 2008; Gupta, Luong and Lee 2015).
The Agr QS system is conserved among staphylococci, but variations within AgrD and the C-terminus of AgrB result in different mature AIPs. Moreover, polymorphism is observed in the extracellular LBD of AgrC among different Staphylococci strains. Binding of cognate AgrD peptide to AgrC sensor kinase leads to the activation of kinase activity of the receptor, while binding of non-cognate AgrD peptide leads to receptor kinase inhibition (Ji, Beavis and Novick 1997; Mayville et al. 1999; Otto et al. 2001; Olson et al. 2014; Le and Otto 2015).
Staphylococcus aureus Agr systems can be classified into four different groups based on variations in the agrB, agrD and agrC genes, with even more variations found in other species as well (Otto et al. 1999; Jarraud et al. 2000; Lyon and Novick 2004; Novick and Geisinger 2008). All AgrD AIPs have distinctly defined sequences with a conserved cysteine in the fifth position from the C-terminus that serves as the site of the thiolactone bond with the C-terminal carboxyl end of the peptide. AgrD-I is an octapeptide (YSTCDFIM) whose sequence varies from AgrD-IV (YSTCYFIM) with a single residue change; AgrD-II is a nonapeptide with the sequence (GVNACSSLF) and AgrD-III is a heptapeptide (INCDFLL) (Fig. 2D) (Jarraud et al. 2000; Lyon et al. 2002; Novick and Geisinger 2008). AgrD AIP is found in all known strains of Staph. intermedius and contains a serine residue in place of the cysteine residue to form a cyclic lactone instead (Bannoehr et al. 2007).
AIP specificity appears to correlate to diseases caused by the different Staph. aureus groups in the host. Each of the four Staph. aureus groups has a characteristic biological consequence: Group I is linked to enterotoxin disease, Group II is linked to early vancomycin-resistant strains, Groups II and III are linked to endocarditis, Group III is linked to menstrual toxic shock syndrome, and Group IV is linked to exfoliative disease (Jarraud et al. 2000, 2002; Sakoulas et al. 2002; Novick and Geisinger 2008). Using AgrC chimeras and a battery of AIP molecules, it was determined that the AIP makes two interactions with the AgrC protein. The first is a hydrophobic interaction between AgrC and the C-terminal residues on the AIP. The second interaction is more sequence specific, lending to specificity of the receptor for the AIP. For example, it was shown that the tail region of AgrD-II is required for AgrC-II activation and a mutant peptide with a single residue change (GVAACSSLF) is an inhibitor of all four AgrC classes (Mayville et al. 1999). The exocyclic residue N in AgrD-II, and endocyclic residues D and Y in AgrD-I and AgrD-IV, respectively, are critical for cognate AgrC receptor activation. Also, additional residues added to the N-terminal end of AgrD-III abolish receptor activation (Lyon et al. 2002). Taken together, these studies have shown that not only that the sequence of the AgrD AIP important for either recognition or inhibition of AgrC, but that stereochemistry can also alter activity.
The AgrC receptor has an N-terminal transmembrane sensor domain plus a C-terminal histidine kinase domain. The C-terminal domain is highly conserved among staphylococci; however, the N-terminal sensor domain is as divergent as agrD and agrB (Wright et al. 2004; Novick and Geisinger 2008). Mutagenesis studies of AgrC concluded that AgrD discrimination likely occurs in the second extracellular loop in the sensor domain between helices three and four. Interestingly, switching five residues that differed in this region between AgrC-I and AgrC-IV switched the specificity of the cognate AgrD AIP (Wright et al. 2004). For AgrC-IV, residue T101 is crucial for activation, and making T101A, V107S and I116S mutations are enough to change the specificity of AgrC-IV to that of AgrC-I (Chen, Tsou and Chen 2009). In contrast, mutating AgrC-I residue Y100 to F and I171 to K broadened the specificity of AgrC-I greatly (Geisinger et al. 2008; Jensen et al. 2008; Novick and Geisinger 2008; Geisinger, Muir and Novick 2009). AgrC-I residue I171 was also deemed a critical inhibitory residue by non-cognate AgrD AIPs (Novick and Geisinger 2008).
Specificity in two-component Gram-negative QS receptors
While the LuxI-LuxR-type system is common among Gram-negative QS bacteria, some species use membrane-bound receptors for AHSL detection (Fig. 1B). For instance, V. harveyi responds to 3OH-C4-HSL, or HAI-1 (Fig. 2A), exclusively using a membrane-bound histidine kinase receptor LuxN (Cao and Meighen 1989; Bassler et al. 1993). HAI-1 is made by synthase LuxM, which shares little homology to LuxI (Bassler et al. 1993; Bassler, Wright and Silverman 1994; Freeman, Lilley and Bassler 2000; Ng and Bassler 2009). AHSLs with longer acyl tails, or lacking a hydroxyl group at the C3 position of the acyl tail, do not induce QS in V. harveyi, suggesting that LuxN is specific for HAI-1 (Ke, Miller and Bassler 2015). Furthermore, HSLs with an eight or longer carbon acyl chain can outcompete HAI-1 as antagonists with increasing potency in accordance with increasing acyl chain length, even without the C3 hydroxyl group (Ke, Miller and Bassler 2015).
It is predicted that LuxN contains nine transmembrane helices, and the HAI-1-binding site lies between helices four and seven (Swem et al. 2008). Mutations in LuxN revealed that residue H210 is responsible for recognizing the C3 hydroxyl moiety on the HAI-1 AI but is not important for determining acyl chain length (Ke, Miller and Bassler 2015). Mutations in LuxN residue pair L166H/N176D renders LuxN unable to detect any HAI-1 molecules, while mutations in residue pairs G147D/Y194H, S184I/S230P and T159I/Y193K broadens the specificity of LuxN to detect longer acyl chains up to C10 (Ke, Miller and Bassler 2015). More specifically, LuxN residue L166 discriminates HSLs for ones that have four carbon acyl chains, and decreasing the size of the side chain on residue L166 allowed longer acyl chains to be detected. Taken together, LuxN residues H210 and L166 provide simple, yet rigorous discrimination for the cognate HAI-1, leading to efficient and informative signaling that is not easily interrupted.
Although related to V. fischeri and V. harveyi, V. cholerae does not produce or detect any AHSLs. Instead, this human pathogen possesses a CqsS/CqsA QS system which is conserved in many other Vibrio species (Miller et al. 2002; Henke and Bassler 2004; Higgins et al. 2007; Ng and Bassler 2009). CqsA is the synthase for the signal CAI-1, which is detected by the membrane-bound receptor CqsS. CAI-1 was first purified and identified as (S)-3-hydroxytridecan-4-one (Fig. 2C) (Higgins et al. 2007). However, CAI-1 is not directly synthesized by CqsA. Instead, CqsA uses decanoyl-CoA and SAM to make the precursor enamino-CAI-1 (Ea-CAI-1), which is further metabolized into CAI-1. Surprisingly, CqsA can also use octanoyl-CoA to make EA-C8-CAI-1; and both Ea-CAI-1and Ea-C8-CAI-1 are potent agonists of CqsS (Fig. 2C) (Higgins et al. 2007; Kelly et al. 2009; Ng et al. 2011; Wei et al. 2011).
Since CqsS is predicted to have six transmembrane helices, structural studies are very difficult, if not impossible. Thus, an orthogonal chemical genetic approach was developed to identify key residues important for CAI-1 and CqsS interactions. Specifically, an array of CAI-1 analogs with defined modifications was synthesized and used to screen for CqsS mutants that would respond to these molecules. Using this approach, it was revealed that the highly conserved residues W104 and S107 located in the fourth transmembrane helix serve to distinguish between the hydroxyl and amino moieties at the C3 position on CAI-1 (Ng et al. 2010). Two chemically modified CAI-1 analogs were used to probe CqsS specificity for tail length and head group. Phenyl-CAI-1 has a bulky modification on the head group of CAI-1 and functions as a CqsS antagonist, while C8-CAI-1 has a shorter tail length of 8 carbons instead of the 10 carbon tail length seen with CAI-1 and functions as a weak CqsS agonist. By screening for CqsS mutants that could use these analogs but at the same time were insensitive to the native CAI-1 ligand, it was discovered that CqsS residue F162 is important for CAI-1 head group recognition, while residue C170 determines CAI-1 tail length preference (Ng et al. 2010). Intriguingly, C170 is only present in the V. cholerae CqsS receptor, whereas other CqsS receptors such as the one in V. harveyi, have a phenylalanine residue at this position (Ng et al. 2010, 2011). This observation infers that other Vibrio species recognize CAI-1 molecules with a C8 tail instead of the longer C10 tail, as the phenylalanine in the corresponding position to V. cholerae C170 would prevent binding of longer tailed CAI-1 molecules. A more extensive study on ligand specificity revealed that V. cholerae mutant CqsS C170F can only detect Ea-C8-CAI-1 and C8-CAI-1, but not CAI-1 or Ea-CAI-1 (Ng et al. 2011). As expected, V. harveyi CqsS only recognizes Ea-C8-CAI-1 and does not respond to CAI-1 with a C10 tail (Ng et al. 2011). Mutating V. harveyi CqsS residue F175 (the corresponding position to V. cholerae C170) to a cysteine relaxes the specificity of V. harveyi CqsS, and the mutant receptor is able to detect CAI-1 and Ea-CAI-1 as well as Ea-C8-CAI-1. Interestingly, V. harveyi only makes CAI-1 molecules with C8 tails due to high substrate specificity of its CqsA synthase (Ng et al. 2011). Thus, signal production and signal detection in the CqsA/CqsS system in these two Vibrio species have co-evolved. While it is likely that both receptors emerged from a common ancestor, the divergence in specificity of these two Vibrio QS systems could be due to selective advantages that require relaxed specificity in V. cholerae and more stringent requirements in V. harveyi.
COMPLEXITY IN QS NETWORK ARCHITECTURE
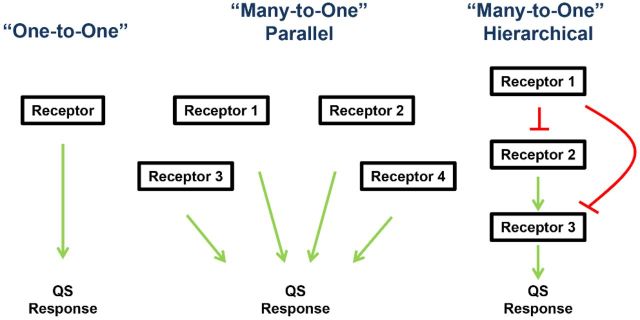
As illustrated above, intrinsic receptor ligand specificity plays an important role in determining the final QS output in response to cognate and non-cognate signals. This is especially important when certain QS bacteria employ a ‘one-to-one’ QS network configuration, in which the overall QS response is solely controlled by a single receptor responding to a single signal (Fig. 3). Yet, it is not uncommon for different QS signal transduction pathways to be inter-connected into a complex QS network. A few of these bacterial QS systems that have a parallel (or ‘many-to-one’) or hierarchical configuration will be examined below (Fig. 3). We will discuss how varying architectures are advantageous for achieving different biological goals.
Figure 3.
Different QS network configurations. In a ‘One-to-One’ system, a single receptor controls the entire QS response. In a ‘Many-to-One’ parallel circuit, information contained in multiple autoinducers are integrated together to control the QS response. In a ‘Many-to-One’ hierarchical system, many QS receptors are connected in a signaling cascade in which the downstream receptor activity is controlled by the upstream receptors. Arrows and T-bar denote hypothetical activation and repression pattern, respectively.
Parallel QS systems in Vibrio species
Parallel network architectures that utilize a phospho-relay signal transduction mechanism are common in Vibrio QS systems. While V. fischeri contains a LuxI-LuxR-type QS system, V. cholerae and V. harveyi do not utilize this type of receptor for AI detection. QS networks in these two species are composed of multiple histidine kinase receptors converging to control a single regulator that governs the downstream QS response (Fig. 1B). Although network architecture is similar between the two Vibrio species, the exact identities of the receptors used differ (Miller et al. 2002; Zhu et al. 2002; Henke and Bassler 2004; Lenz et al. 2004; Long et al. 2009). In V. harveyi, three QS receptors LuxN, LuxPQ and CqsS, which are all transmembrane histidine kinases, convey their AI information to LuxO through LuxU (Bassler, Wright and Silverman 1994; Freeman, Lilley and Bassler 2000; Henke and Bassler 2004; Neiditch et al. 2005, 2006; Higgins et al. 2007; Swem et al. 2008). In contrast, V. cholerae possesses four QS receptors, three of which are transmembrane histidine kinases, LuxPQ, CqsS and CqsR, while the fourth is predicted to be a cytoplasmic receptor VpsS (Miller and Bassler 2001; Miller et al. 2002; Waters and Bassler 2005; Ng and Bassler 2009; Jung, Chapman and Ng 2015). It should be noted that, in addition to the LuxI-LuxR system, V. fischeri also carries membrane-bound LuxPQ and AinN receptors for QS gene regulation (Kuo, Callahan and Dunlap 1996; Schaefer et al. 1996a; Lupp et al. 2003). Kinase activity of all of these receptors predominates when AI concentration is low; LuxO is activated by phosphorylation, resulting in a low cell density gene expression program. When AI concentration is high, receptor phosphatase activity predominates and LuxO becomes inactive, resulting in a high cell density gene expression program.
Distinct information contained within each signal in Vibrio QS Systems
Although it seems perplexing that LuxO is controlled by multiple redundant receptors, it is possible that each signal could be translated into a particular output response. In other words, each detected signal may have its own meaning (Bassler, Wright and Silverman 1994; Miller et al. 2002; Henke and Bassler 2004; Ng et al. 2011). For example, V. harveyi detects three AIs HAI-1, CAI-1 and AI-2, using receptors LuxN, CqsS and LuxPQ, respectively. These systems are thought to be used for intra-species, intra-genus, and inter-species communication (Federle and Bassler 2003; Henke and Bassler 2004; Ng et al. 2011). HAI-1 is exclusively made and detected by V. harveyi. CAI-1 and its related molecules are made and detected by many Vibrio species. As discussed above, CAI-1 with an 8- or 10-carbon tail length regulates V. cholerae CqsS kinase activity, while CAI-1 with an 8-carbon tail length controls V. harveyi CqsS (Ng et al. 2011). Thus, CAI-1 with an 8-carbon tail length may be used for inter-Vibrio signaling, while CAI-1 with a 10-carbon tail length could be a V. cholerae-specific QS signal. AI-2 is made by a variety of bacterial species carrying the gene luxS and therefore LuxPQ could be used for Vibrios to enumerate the abundance of surrounding microbial species (Bassler, Wright and Silverman 1994; Surette, Miller and Bassler 1999; Chen et al. 2002; Miller et al. 2002; Ng et al. 2011; Pereira, Thompson and Xavier 2013). Although the signals that regulate the recently-identified receptors CqsR and VpsS are unknown, it is likely that these two receptors also detect small chemical molecules that are unique from one another (Jung, Chapman and Ng 2015). As a result, specific information contained in each signal may allow for subtle behavioral changes that are more suitable for adapting to certain environmental niches.
Parallel inputs prevent premature QS induction in Vibrio cholerae
In V. cholerae, the loss of three of the four QS receptor histidine kinase activities has little effect on V. cholerae colonization of animal hosts. Thus, any one of its four QS receptors acting alone is sufficient to regulate gene expression in response to cell density (Jung, Chapman and Ng 2015). In contrast, mutants lacking all four QS receptors do not colonize the infant mouse host. Even though having four receptors seems redundant, V. cholerae inhabits vastly different environmental niches and it may encounter a variety of different bacterial species that produce their own array of signaling molecules. In contrast to V. harveyi, where QS can be induced by addition of a single AI such as HAI-1 (Bassler et al. 1993; Bassler, Wright and Silverman 1994; Mok, Wingreen and Bassler 2003; Henke and Bassler 2004), it was shown that excess CAI-1 added to V. cholerae does not induce QS prematurely (Jung, Chapman and Ng 2015). Based on these findings, it is thought that these receptors may act together to prevent a premature committed responses caused by signal perturbations or, possibly, analogous molecules associated with environmental changes and noise (Fig. 3) (Ng et al. 2011, 2012; Jung, Chapman and Ng 2015). Previous studies have shown that molecules with structures drastically different from CAI-1 could control CqsS activity (Ng et al. 2012), suggesting that there are possibilities for decoy molecules acting alone on a single QS receptor. It should be noted that CqsS of V. harveyi has higher ligand detection specificity than that of V. cholerae and is less likely to be affected by signal perturbation (Ng et al. 2011). Therefore, even though these Vibrio species utilize parallel signaling networks, their biological goals achieved are different. The V. harveyi QS system is proposed to function as a ‘coincidence detector’ (Mok, Wingreen and Bassler 2003; Henke and Bassler 2004), while the V. cholerae redundant receptors function together in parallel to resist signal perturbations. Taken together, this particular circuit architecture could be crucial in maintaining population-wide expression of QS genes, and it could also function in preventing premature commitment to high cell density gene expression until the cells are ready and when the response is appropriate.
Hierarchical QS systems in Pseudomonas aeruginosa
In addition to parallel signal transduction pathways, different network configurations have been identified in other QS systems (Fig. 3). For instance, the P. aeruginosa QS network adopts a unique, hierarchical architecture that consists of two intimately-connected pathways: the LasI-LasR system which makes and detects 3-oxo-C12-HSL, and the RhlI-RhlR system which makes and detects C4-HSL (Whitehead et al. 2001; Fuqua and Greenberg 2002). Together, these two QS systems regulate expression of >300 genes involved in virulence factor production and biofilm formation in this pathogen (Schuster et al. 2003; Schuster, Urbanowski and Greenberg 2004; Wagner, Gillis and Iglewski 2004; Wei and Ma 2013). While each of these two QS systems regulates a specific set of genes, there exists some overlap. This is because the LasI-LasR circuit is hierarchically positioned to regulate expression of the RhlI-RhlR circuit. LasR, in the presence of 3-oxo-C12-HSL, activates expression of rhlI and rhlR (Latifi et al. 1996; Pesci et al. 1997; Medina et al. 2003a,b; Gilbert et al. 2009). This network arrangement allows temporal expression of different sets of genes in the P. aeruginosa QS regulon (Schuster et al. 2003; Smith and Iglewski 2003; Schuster, Urbanowski and Greenberg 2004; Venturi 2006). Additionally, the orphan QscR regulator functions as a negative regulator to repress LasI and RhlI expression (Chugani et al. 2001). QscR responds to 3-oxo-C12-HSL and a variety of AHSLs. Therefore, through QscR, the QS response of P. aeruginosa could be modulated by its own AHSL feedback or by the presence of other AHSL-producing species (Williams et al. 2000; Chugani et al. 2001; Ledgham et al. 2003; Parsek and Greenberg 2005; Waters and Bassler 2005; Lee, Lequette and Greenberg 2006; Lequette et al. 2006; Mattmann and Blackwell 2010).
In addition to network hierarchy, the two P. aeruginosa AHSL ligands have different decaying rates, and P. aeruginosa behaves differently with different combinatorial (non-additive) concentrations of the two AIs (Cornforth et al. 2014). These researchers suggest that signal concentration threshold gates correspond to different responses, such that bacteria can infer both social (cell density) and physical (mass-transfer) environments. Accordingly, by utilizing multiple signals and different combinatorial signal concentrations, personal environments are more fully resolved and result in more complex gene expression patterns within individual cells (Cornforth et al. 2014; Drees et al. 2014). Finally, there has been an increasing evidence that nutritional cues within infection environments can alter this complex hierarchy (Welsh and Blackwell 2016).
Competition between two related QS regulatory Rgg systems
Signal antagonism is also incorporated into complex QS systems to dampen output responses. For instance, Streptococcus pyogenes regulates its QS response through four different Rgg regulators. Each Rgg receptor presumably responds to a specific peptide signal called SHP (short hydrophobic peptide) and XIP (SigX inducing peptide) (Ibrahim et al. 2007a,b; Fontaine et al. 2010; Mashburn-Warren, Morrison and Federle 2010, 2012; Chang et al. 2011; Fleuchot et al. 2011). Of particular interest is the antagonistic action discovered between the Rgg2 and Rgg3 proteins, which respectively respond to SHP2 and SHP3 peptide pheromones (Chang et al. 2011; LaSarre, Chang and Federle 2013). While Rgg2 is a transcriptional activator, Rgg3 represses the same target genes. Both regulators bind to a conserved DNA motif present in the promoters of these target genes, including the genes encoding SHP2 and SHP3. Since they share binding sites, only one Rgg protein may bind to the same promoter. Additionally, SHP concentration can skew binding to favor Rgg2. Rgg2 binding induces QS, while Rgg3 binding results in QS repression by monopolizing binding sites essential for SHP production (Lasarre, Aggarwal and Federle 2013; Aggarwal et al. 2014). Taken together, Rgg3 binding antagonizes Rgg2 binding and, therefore, may protect against premature induction of the QS circuit in S. pyogenes (Aggarwal et al. 2014).
Newly acquired receptors and facultative cheating
A recent study provided insight on the selective advantage of utilizing multiple receptors for the QS response (Travisano and Velicer 2004; Even-Tov et al. 2016). The central hypothesis of this work is that strains that have acquired new QS receptors are selected via facultative cheating. Cheaters are individuals that do not contribute to the group but reap the benefits of the cooperative members of the group. From the perspective of QS, cheaters are strains that do not respond to the AI signals due to various mechanisms such as missing a functional QS system. When present in low number within a population, cheaters could exhibit higher fitness under certain growth conditions (Travisano and Velicer 2004; West et al. 2006; Popat et al. 2012). Moreover, cheating can be either obligate or facultative. In the latter case, facultative cheaters will cooperate with their own kind but cheat when they are in a group of genetically dissimilar individuals (Travisano and Velicer 2004). It is predicted that evolved strains with multiple QS systems are less cooperative than their ancestral strains that have fewer QS receptors because the concentration of the cognate signal of the new receptor is relatively low. However, these new, evolved strains will cooperate in a homogenous population with cells bearing the same number of receptors (Travisano and Velicer 2004; Even-Tov et al. 2016). Facultative cheating in these strains with higher number of QS receptors enable these individuals to invest less (e.g. producing less public goods) but maintain the same level of fitness in the presence of its ancestor and yet remain cooperative when the population becomes clonal. Based on this model, in a parallel QS circuit, the newly-introduced receptor must be able to repress its QS response in the absence of cognate signal (such as the QS receptors in the Vibrio QS system), while receptors that positively regulate QS could potentially be favored in a hierarchical network (Even-Tov et al. 2016).
Population heterogeneity
The phrase ‘phenotypic heterogeneity’ describes phenotypic variations observed between individual bacterial cells in what was thought to be a genetically homogenous population (Davidson and Surette 2008). In V. harveyi, the presence of multiple QS systems renders the population more homogenous in a laboratory batch culture (Plener et al. 2015). Potentially, different combinations of AI abundances could lead to heterogeneity of the QS response in V. harveyi. Although QS is thought to promote unison behavior, diversification in QS output responses could be beneficial and necessary for the group's survival and well-being (Anetzberger, Schell and Jung 2012). Variation of gene expression profiles in a given population likely depends on the micro-environments encountered by different cells in the community such as stratification within a biofilm (Vlamakis et al. 2008; Bischofs et al. 2009). For example, Bacillus subtilis cells that are motile, biofilm-producing and sporulating localize to specific niches within a biofilm structure, and this localization and ratio of each cell type is temporally and spatially dynamic within the population (Vlamakis et al. 2008). Cell-to-cell heterogeneity has also been reported in Listeria monocytogenes biofilm communities (Garmyn et al. 2011) and in deep tissue infected with Yersinia pseudotuberculosis (Davis, Mohammadi and Isberg 2015). Specialized differentiated populational behaviors may allow for cells to react better to changes in their environmental surroundings (Bischofs et al. 2009). Therefore, these multiple signal transduction systems are crucial to integrate environmental signals based on spatiotemporal community structure, which differs among neighboring subpopulations, as is seen in biofilm structures (Bischofs et al. 2009). Within these biofilm communities, varying AI concentrations could represent different stages of biofilm development, whereby multiple systems are used for a multi-developmental program, where different genes are regulated at differential expression levels (Mehta et al. 2009).
CONCLUSIONS
It is fascinating to consider the evolution of the vastly different bacterial QS systems. Even in related species with similar QS network architectures, the functional goals of these signaling pathways could be vastly different. On the other hand, recurring patterns and characteristics including ligand specificity, evolution and the inter-connection of multiple pathways are observed in unrelated systems. All in all, bacteria have adopted various elegant mechanisms to resist signal antagonism and perturbations. In the last few decades, we have uncovered a vast knowledge about the components and basic concepts in building a QS system; yet, there are many more questions that remain: what governs the transition from a simple ancestral QS system to a complex QS network that we observe in modern bacterial species today? Do different environmental niches determine QS ligand specificity or QS network architecture? How do micro-environments influence QS behavior in a bacterial community? Will and can QS networks become a focus for therapeutic development? We hope that continued research on these topics will provide answers to these important questions.
Acknowledgments
We apologize to colleagues whose work was not cited here due to space limitations.
FUNDING
This work is supported by [R01AI121337].
Conflict of interest. None declared.
REFERENCES
- Aggarwal C, Jimenez JC, Nanavati D, et al. Multiple length peptide-pheromone variants produced by Streptococcus pyogenes directly bind Rgg proteins to confer transcriptional regulation. J Biol Chem. 2014;289:22427–36. doi: 10.1074/jbc.M114.583989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anetzberger C, Schell U, Jung K. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiology. 2012;12:209. doi: 10.1186/1471-2180-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, et al. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–62. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- August PR, Grossman TH, Minor C, et al. Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J Mol Microb Biotech. 2000;2:513–9. [PubMed] [Google Scholar]
- Bae T, Clerc-Bardin S, Dunny GM. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297:861–75. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- Bannoehr J, Ben Zakour NL, Waller AS, et al. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol. 2007;189:8685–92. doi: 10.1128/JB.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B, Vogel J. Bacterial regulatory mechanisms: the gene and beyond. Curr Opin Microbiol. 2013;16:109–11. doi: 10.1016/j.mib.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, et al. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–86. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Bischofs IB, Hug JA, Liu AW, et al. Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. P Natl Acad Sci USA. 2009;106:6459–64. doi: 10.1073/pnas.0810878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–9. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J Bacteriol. 1995;177:1053–8. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Gene Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Muraglia E, Bazzo R, et al. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282:13592–600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Bouillaut L, Perchat S, Arold S, et al. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008;36:3791–801. doi: 10.1093/nar/gkn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann AO, Brameyer S, Kresovic D, et al. Pyrones as bacterial signaling molecules. Nat Chem Biol. 2013;9:573–8. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- Brameyer S, Heermann R. Specificity of Signal-Binding via Non-AHL LuxR-Type Receptors. PLoS One. 2015;10:e0124093. doi: 10.1371/journal.pone.0124093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–63. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–6. [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, et al. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Swem LR, Swem DL, et al. A strategy for antagonizing quorum sensing. Mol Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LC, Tsou LT, Chen FJ. Ligand-receptor recognition for activation of quorum sensing in Staphylococcus aureus. J Microbiol. 2009;47:572–81. doi: 10.1007/s12275-009-0004-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–9. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Choi SH, Greenberg EP. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. P Natl Acad Sci USA. 1991;88:11115–9. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Greenberg EP. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992;174:4064–9. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–94. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. P Natl Acad Sci USA. 2001;98:2752–7. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CH, Arnold FH, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol Microbiol. 2005;55:712–23. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol. 2006;24:708–12. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev. 2014;38:473–92. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Popat R, McNally L, et al. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. P Natl Acad Sci USA. 2014;111:4280–4. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson CJ, Surette MG. Individuality in bacteria. Annu Rev Genet. 2008;42:253–68. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- Davis KM, Mohammadi S, Isberg RR. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe. 2015;17:21–31. doi: 10.1016/j.chom.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck N, Bouillaut L, Chaix D, et al. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. P Natl Acad Sci USA. 2007;104:18490–5. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Reiger M, Jung K, et al. A modular view of the diversity of cell-density-encoding schemes in bacterial quorum-sensing systems. Biophys J. 2014;107:266–77. doi: 10.1016/j.bpj.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Levesque CM. Bacterial behaviors associated with the quorum-sensing peptide pheromone ('alarmone') in streptococci. Future Microbiol. 2013;8:593–605. doi: 10.2217/fmb.13.23. [DOI] [PubMed] [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, et al. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–9. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–81. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. P Natl Acad Sci USA. 1984;81:4154–8. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Tov E, Omer Bendori S, Valastyan J, et al. Social evolution selects for redundancy in bacterial quorum sensing. PLoS Biol. 2016;14:e1002386. doi: 10.1371/journal.pbio.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Bassler BL. Interspecies communication in bacteria. Journal of clinical investigation. 2003;112:1291–9. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuchot B, Gitton C, Guillot A, et al. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol. 2011;80:1102–19. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- Fontaine L, Boutry C, de Frahan MH, et al. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 2010;192:1444–54. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Lilley BN, Bassler BL. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000;35:139–49. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–9. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Bio. 2002;3:685–95. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmyn D, Gal L, Briandet R, et al. Evidence of autoinduction heterogeneity via expression of the Agr system of Listeria monocytogenes at the single-cell level. Appl Environ Microb. 2011;77:6286–9. doi: 10.1128/AEM.02891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Adhikari RP, Jin R, et al. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–48. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- Geisinger E, George EA, Chen J, et al. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP. Agr receptor mutants reveal distinct modes of inhibition by Staphylococcal autoinducing peptides. P Natl Acad Sci USA. 2009;106:1216–21. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KB, Kim TH, Gupta R, et al. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–85. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohar M, Faegri K, Perchat S, et al. The PlcR virulence regulon of Bacillus cereus. PLoS One. 2008;3:e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gominet M, Slamti L, Gilois N, et al. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol. 2001;40:963–75. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- Grenha R, Slamti L, Nicaise M, et al. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. P Natl Acad Sci USA. 2013;110:1047–52. doi: 10.1073/pnas.1213770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Luong TT, Lee CY. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. P Natl Acad Sci USA. 2015;112:14036–41. doi: 10.1073/pnas.1509251112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelka BL, Greenberg EP. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol. 1995;177:815–7. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelka BL, Parsek MR, Val DL, et al. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181:5766–70. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. P Natl Acad Sci USA. 1995;92:11140–4. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins AC, Arnold FH, Stuermer R, et al. Directed evolution of Vibrio fischeri LuxR for improved response to butanoyl-homoserine lactone. Appl Environ Microb. 2007;73:5775–81. doi: 10.1128/AEM.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–14. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Pomianek ME, Kraml CM, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–6. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- Hoch J, Silhavy T. Two-Component Signal Transduction. Washington, DC: ASM Press; 1995. [Google Scholar]
- Hwang I, Li PL, Zhang L, et al. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. P Natl Acad Sci USA. 1994;91:4639–43. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M, Guillot A, Wessner F, et al. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J Bacteriol. 2007a;189:8844–54. doi: 10.1128/JB.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M, Nicolas P, Bessieres P, et al. A genome-wide survey of short coding sequences in streptococci. Microbiology. 2007b;153:3631–44. doi: 10.1099/mic.0.2007/006205-0. [DOI] [PubMed] [Google Scholar]
- Inouye M, Dutta R. Histidine Kinases in Signal Transduction. London: Academic; 2003. [Google Scholar]
- Jarraud S, Lyon GJ, Figueiredo AM, et al. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–22. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarraud S, Mougel C, Thioulouse J, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–41. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RO, Winzer K, Clarke SR, et al. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–9. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–30. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. P Natl Acad Sci USA. 1995;92:12055–9. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JC, Federle MJ. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol. 2014;4:127. doi: 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SA, Chapman CA, Ng WL. Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog. 2015;11:e1004837. doi: 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Miller LC, Bassler BL. Determinants governing ligand specificity of the Vibrio harveyi LuxN quorum-sensing receptor. Mol Microbiol. 2015;95:127–42. doi: 10.1111/mmi.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RC, Bolitho ME, Higgins DA, et al. The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA. Nat Chem Biol. 2009;5:891–5. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Quadri LE, Kuipers OP, et al. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- Kuo A, Callahan SM, Dunlap PV. Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol. 1996;178:971–6. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasarre B, Aggarwal C, Federle MJ. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. MBio. 2013;3:e00333-12. doi: 10.1128/mBio.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B, Chang JC, Federle MJ. Redundant group a streptococcus signaling peptides exhibit unique activation potentials. J Bacteriol. 2013;195:4310–8. doi: 10.1128/JB.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Foglino M, Tanaka K, et al. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–46. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- Le KY, Otto M. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol. 2015;6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgham F, Ventre I, Soscia C, et al. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–9. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Lee JH, Ledgham F, et al. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–70. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D, Agaisse H, Gominet M, et al. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J Bacteriol. 1996;178:2749–56. doi: 10.1128/jb.178.10.2749-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Jarraud S, Ji G, et al. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–62. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Urbanowski M, Greenberg EP, et al. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. 2003;50:319–31. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;25:1389–403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Lyon GJ, Wright JS, Muir TW, et al. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry. 2002;41:10095–104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- McClean KH, Winson MK, Fish L, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(Pt 12):3703–11. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–16. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J Bacteriol. 2012;194:4589–600. doi: 10.1128/JB.00830-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattmann ME, Blackwell HE. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J Org Chem. 2010;75:6737–46. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayville P, Ji G, Beavis R, et al. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. P Natl Acad Sci USA. 1999;96:1218–23. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina G, Juarez K, Diaz R, et al. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology. 2003a;149:3073–81. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- Medina G, Juarez K, Valderrama B, et al. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J Bacteriol. 2003b;185:5976–83. doi: 10.1128/JB.185.20.5976-5983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Long T, et al. Information processing and signal integration in bacterial quorum sensing. Mol Syst Biol. 2009;5:325. doi: 10.1038/msb.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, et al. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–14. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 2003;22:870–81. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnet V, Juillard V, Gardan R. Peptide conversations in Gram-positive bacteria. Crit Rev Microbiol. 2014 doi: 10.3109/1040841X.2014.948804. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–22. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Miller ST, et al. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–18. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Perez L, Cong J, et al. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Perez LJ, Wei Y, et al. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79:1407–17. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Wei Y, Perez LJ, et al. Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. P Natl Acad Sci USA. 2010;107:5575–80. doi: 10.1073/pnas.1001392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y, Nguyen NX, Rogers JL, et al. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. MBio. 2015;6:02429-14. doi: 10.1128/mBio.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Novick RP, Projan SJ, Kornblum J, et al. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–58. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Koch AK, Fiechter A, et al. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–54. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. P Natl Acad Sci USA. 1995;92:6424–8. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ME, Todd DA, Schaeffer CR, et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol. 2014;196:3482–93. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M, Echner H, Voelter W, et al. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:1957–60. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M, Sussmuth R, Vuong C, et al. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–62. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- Pappas KM, Weingart CL, Winans SC. Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signalling. Mol Microbiol. 2004;53:755–69. doi: 10.1111/j.1365-2958.2004.04212.x. [DOI] [PubMed] [Google Scholar]
- Parashar V, Mirouze N, Dubnau DA, et al. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS Biol. 2011;9:e1000589. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Passador L, Cook JM, Gambello MJ, et al. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–30. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. P Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Passador L, Iglewski BH, et al. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. P Natl Acad Sci USA. 1995;92:1490–4. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HL, Novick RP, Kreiswirth B, et al. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–72. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. P Natl Acad Sci USA. 1997;94:8612–7. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–81. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- Pesci EC, Pearson JP, Seed PC, et al. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–32. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–62. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–50. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Plener L, Lorenz N, Reiger M, et al. The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J Bacteriol. 2015;197:1747–56. doi: 10.1128/JB.02544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat R, Crusz SA, Messina M, et al. Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci. 2012;279:4765–71. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci. 2003;8:d32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- Rocha-Estrada J, Aceves-Diez AE, Guarneros G, et al. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biot. 2010;87:913–23. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- Ryan RP, An SQ, Allan JH, et al. The DSF Family of Cell-Cell Signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 2015;11:e1004986. doi: 10.1371/journal.ppat.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G, Eliopoulos GM, Moellering RC, Jr, et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Ch. 2002;46:1492–502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappington KJ, Dandekar AA, Oinuma K, et al. Reversible signal binding by the Pseudomonas aeruginosa quorum-sensing signal receptor LasR. MBio. 2011;2:e00011–11. doi: 10.1128/mBio.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Hanzelka BL, Eberhard A, et al. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996a;178:2897–901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Val DL, Hanzelka BL, et al. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. P Natl Acad Sci USA. 1996b;93:9505–9. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, et al. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–79. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. P Natl Acad Sci USA. 2004;101:15833–9. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Young R, Baldwin TO. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J Bacteriol. 1990;172:3980–7. doi: 10.1128/jb.172.7.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Brown CK, Gu ZY, et al. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. P Natl Acad Sci USA. 2005;102:18596–601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Crane B, Crane A. Two-Component Signaling Systems. San Diego: Academic; 2007. [Google Scholar]
- Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002;21:4550–9. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti L, Lereclus D. Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group. J Bacteriol. 2005;187:1182–7. doi: 10.1128/JB.187.3.1182-1187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slock J, VanRiet D, Kolibachuk D, et al. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J Bacteriol. 1990;172:3974–9. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J Clinical Invest. 2003;112:1460–5. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff DL, Bassler BL. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol. 2011;193:3871–8. doi: 10.1128/JB.05125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Dolan KM, Greenberg EP. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. P Natl Acad Sci USA. 1994;91:12619–23. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Fujita N, Ishihama A, et al. Involvement of the RNA polymerase alpha-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J Bacteriol. 1999;181:4704–7. doi: 10.1128/jb.181.15.4704-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Greenberg EP. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–62. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. P Natl Acad Sci USA. 1999;96:1639–44. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, O'Loughlin CT, et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol Cell. 2009;35:143–53. doi: 10.1016/j.molcel.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, et al. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–73. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Hilbi H. Bacterial gene regulation by alpha-hydroxyketone signaling. Trends Microbiol. 2010;18:288–97. doi: 10.1016/j.tim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Tomasz A. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature. 1965;208:155–9. doi: 10.1038/208155a0. [DOI] [PubMed] [Google Scholar]
- Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends Microbiol. 2004;12:72–8. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186:631–7. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gargioli C, et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–91. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, et al. Control of cell fate by the formation of an architecturally complex bacterial community. Gene Dev. 2008;22:945–53. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Gillis RJ, Iglewski BH. Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeruginosa. Vaccine. 2004;22(Suppl 1):S15–20. doi: 10.1016/j.vaccine.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev B. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14:20983–1005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Perez LJ, Ng WL, et al. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem Biol. 2011;6:356–65. doi: 10.1021/cb1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh MA, Blackwell HE. Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem Biol. 2016;23:361–9. doi: 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, et al. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Whitehead NA, Barnard AM, Slater H, et al. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–91. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Williams P, Camara M, Hardman A, et al. Quorum sensing and the population-dependent control of virulence. Philos T Roy Soc B. 2000;355:667–80. doi: 10.1098/rstb.2000.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintjens R, Rooman M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- Wright JS, III, Lyon GJ, George EA, et al. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. P Natl Acad Sci USA. 2004;101:16168–73. doi: 10.1073/pnas.0404039101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ji G. Identification of a staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J Bacteriol. 2004;186:6706–13. doi: 10.1128/JB.186.20.6706-6713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RG, Pappas KM, Brace JL, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–4. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- Zhu J, Beaber JW, More MI, et al. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. P Natl Acad Sci USA. 2002;99:3129–34. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Winans SC. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. P Natl Acad Sci USA. 1999;96:4832–7. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. P Natl Acad Sci USA. 2001;98:1507–12. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem Biol. 2009;16:961–70. doi: 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhir S, Perchat S, Nicaise M, et al. Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR. Nucleic Acids Res. 2013;41:7920–33. doi: 10.1093/nar/gkt546. [DOI] [PMC free article] [PubMed] [Google Scholar]