Abstract
The family of Shank scaffolding molecules (comprising Shank1, 2 and 3) are core components of the postsynaptic density (PSD) in neuronal synapses. Shanks link surface receptors to other scaffolding molecules within the PSD, as well as to the actin cytoskeleton. However, determining the function of Shank proteins in neurons has been complicated because the different Shank isoforms share a very high degree of sequence and domain homology. Therefore, to control Shank content while minimizing potential compensatory effects, a miRNA-based knockdown strategy was developed to reduce the expression of all synaptically targeted Shank isoforms simultaneously in rat hippocampal neurons. Using this approach, a strong (>75%) reduction in total Shank protein levels was achieved at individual dendritic spines, prompting an approximately 40% decrease in mushroom spine density. Furthermore, Shank knockdown reduced spine actin levels and increased sensitivity to the actin depolymerizing agent Latrunculin A. A SHANK2 mutant lacking the proline-rich cortactin-binding motif (SHANK2-ΔPRO) was unable to rescue these defects. Furthermore, Shank knockdown reduced cortactin levels in spines and increased the mobility of spine cortactin as measured by single-molecule tracking photoactivated localization microscopy, suggesting that Shank proteins recruit and stabilize cortactin at the synapse. Furthermore, it was found that Shank knockdown significantly reduced spontaneous remodelling of synapse morphology that could not be rescued by the SHANK2-ΔPRO mutant. It was concluded that Shank proteins are key intermediates between the synapse and the spine interior that, via cortactin, permit the actin cytoskeleton to dynamically regulate synapse morphology and function.
Keywords: dendritic spine, F-actin, postsynaptic density, rat, single-molecule tracking
Introduction
The actin cytoskeleton in neuronal spines forms a complex filamentous network required for a variety of neuronal functions (Frost et al., 2010a). Actin filaments and the complexes that promote actin dynamics are closely associated with the macromolecular complex of proteins that form the postsynaptic density (PSD) and retain amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)- and N-methyl-d-aspartate (NMDA)-type glutamate receptors that are critical for synaptic transmission and plasticity (Fifkova & Delay, 1982; Racz & Weinberg, 2004; Rostaing et al., 2006). Disrupting actin polymerization induces a rapid loss of synaptic scaffolding molecules and synaptic currents (Allison et al., 2000; Zhou et al., 2001; Kuriu et al., 2006; Duffney et al., 2013). Moreover, actin polymerization is highly active within 200 nm of the synapse (Frost et al., 2010b; Chazeau et al., 2014), and controls ongoing PSD restructuring and the subsynaptic distribution of AMPARs and scaffolding molecules (Kerr & Blanpied, 2012; MacGillavry et al., 2013), suggesting that actin polymerization actively coordinates synaptic function. However, although the actin cytoskeleton is critical for synaptic function, there is limited understanding of the molecular intermediates that permit efficient, spatially restricted control of actin polymerization over synaptic structure and function.
The family of Shank scaffolding molecules (Shank1, 2 and 3) are specifically expressed at excitatory synapses, and are among the core components of the PSD. Through multiple interaction domains, they link surface receptors to other scaffolding molecules within the PSD (Naisbitt et al., 1999; Tu et al., 1999), and the actin cytoskeleton via interactions with actin-regulatory proteins (Du et al., 1998; Boeckers et al., 2001; Park et al., 2003; Qualmann et al., 2004; Proepper et al., 2007; Haeckel et al., 2008; Han et al., 2013; Duffney et al., 2015). Furthermore, Shank proteins are generally found in the deeper layers of the PSD facing the spine interior (Valtschanoff & Weinberg, 2001), ideally positioned to control actin cytoskeleton–PSD interactions. Cortactin is a particularly attractive candidate to link synaptic Shank scaffold molecules to the actin cytoskeleton. Cortactin is an actin nucleation-promoting factor that recruits and activates the Arp2/3 (actin-related protein-2/3) complex (Uruno et al., 2001; Weaver et al., 2001), the molecular machinery essential for nucleating actin filament assembly. Thus, by recruiting cortactin to perisynaptic sites, Shank proteins could efficiently direct the dynamics of the actin cytoskeleton at the synapse.
However, determining the role of Shank proteins in neurons has been complicated because, in vitro, all three isoforms are commonly co-expressed simultaneously within individual hippocampal synapses (Grabrucker et al., 2011), and share a very high degree of sequence and domain homology, suggesting that individual isoforms can functionally compensate for each other. Furthermore, the sterile alpha motif (SAM) domains of Shanks confer oligomerization of individual Shanks (Naisbitt et al., 1999), but heterodimerization between different isoforms has also been suggested (Han et al., 2013). To test whether Shanks are required for controlling spine actin dynamics, a miRNA-based knockdown strategy was developed to simultaneously knock down most synaptically targeted Shank isoforms. Using this approach, the role of Shank proteins and their interaction with cortactin in spine actin dynamics was tested.
Materials and methods
All procedures used in the performance of this study were approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee, in accordance with NIH guidelines for the care and use of animals.
Neuronal cultures and transfections
Dissociated hippocampal cultures were prepared from embryonic day 18 rats of either sex as described previously (Frost et al., 2010b). For single-molecule imaging experiments, cells were plated on coverslips that had been cleaned by boiling in a 5 : 1 : 1 solution of filtered water, ammonium hydroxide and hydrogen peroxide for 3 h, rinsed in filtered water, ethanol and methanol, briefly flamed and coated overnight with poly-l-lysine (Sigma). Cells were transfected at 12–15 days in vitro (DIV) using Lipofectamine 2000 reagent (Invitrogen) and imaged 7 days later.
DNA constructs
For the mirShank construct miRNA sequences targeting Shank1 (mirShank1: 5′-ACAGACCAACCTGGATGAGAA-3′) and Shank3 (mirShank3: 5′-GGAAGTCACCAGAGGACAAGA-3′) were based on Grabrucker et al. (2011) and Verpelli et al. (2011), respectively. For Shank2 initially two miRNA sequences were selected (mir-Shank2 #1: 5′-AATCGATAGACAGCAGAATCT-3′ and mirShank2 #2: 5′-GGACTTGGATGAGGACTTTCT-3′) using the Invitrogen Block-IT miRNA design algorithm. Both sequences were found to efficiently reduce Shank2 immunoreactivity in neurons (~80%; data not shown), and mirShank2 #2, not targeting human SHANK2, was selected for the triple knockdown construct. These miRNA sequences were furthermore selected based on their ability to target most known rat isoforms of Shank1, 2 and 3 (isoform information based on UniProt IDs Q9WV48, Q9QX74 and Q9JLU4, respectively). The shorter isoform of Shank3 (isoform 3) is not targeted by the miRNA sequence, but this splice variant lacks a large portion of the C-terminus including the SAM domain that is required for synaptic targeting (Boeckers et al., 2005). However, because several alternative promoters have been described for the Shank3 gene (Jiang & Ehlers, 2013; Wang et al., 2014), it was decided here to use a miRNA sequence targeting the more C-terminal coding sequence of Shank3, to target most potential gene products. For Shank1, the miRNA sequence is targeted to the N-terminal region, and is predicted to not target potential shorter isoforms produced from a downstream alternative start site. Thus, the mirShank construct is predicted to target the majority of Shank isoforms that are synaptically localized. Scrambled versions of the targeting sequences were designed to have the same nucleotide composition, but to have no match with any known rat mRNA sequence: scrShank1: 5′-GAACTAGCGCAACATAAGAGC-5′; scrShank2: 5′-ATGGTATTCCGTTCGGAAGTG-3′; and scrShank3: 5′-GAACGGATAACCGAACGGAAG-3′. Oligos containing the 21 nucleotide target or scrambled sequence and the loop sequence (5′-GTTTTGGCCACTGACTGAC-3′) were annealed and cloned into pSM155-GFP (green fluorescent protein; kindly provided by G. Du, University of Texas, Houston, TX, USA; Du et al., 2006), which was digested with BsmBI leaving cohesive ends for the annealed miRNA oligos. To produce the triple-knockdown construct, first mirShank2 was digested with XbaI and MluI and ligated into the NheI and MluI sites of pSM155-mirShank1-GFP. Then, mirShank3 was digested with XbaI and MluI and ligated into the NheI and MluI sites of pSM155-mirShank1&2-GFP to make pSM155-mirShank1-3-GFP (hereafter referred to as mirShank-GFP). The triple scrambled construct was cloned similarly. To make pSM155-mirShank1-3-Cer3, the GFP coding sequence was replaced with mCerulean3 (Markwardt et al., 2011). All inserts were confirmed by DNA sequencing.
The rat Shank1-GFP expression plasmid was a gift from M. Sheng (Genentech, San Francisco, CA, USA). Rat Shank2-GFP and human mCherry-SHANK2 expression plasmids were kindly provided by S. Berkel (Institute of Human Genetics, Heidelberg, Germany; Berkel et al., 2012). To generate SHANK2 rescue constructs simultaneously expressing the three miRNAs and the miRNA-resistant SHANK2 sequence, the human SHANK2 sequence provided by S. Berkel was polymerase chain reaction (PCR) amplified and cloned into mirShank-GFP using InFusion cloning to make N-terminal GFP-tagged SHANK2-WT. The SHANK2-ΔSH3 mutant was generated by selective PCR amplification (SHANK2 amino acids 207–1463). The SHANK2-ΔPRO (removing the cortactin binding site in SHANK2, amino acids 1157–1164; Du et al., 1998) and P1035L mutants were made using QuickChange mutagenesis. Rat Shank3-GFP is a gift from P. Worley (Johns Hopkins University, Baltimore, MD, USA). The PSD-95 replacement plasmid is described in MacGillavry et al. (2013). SEP-GluA2 is a gift from R. Huganir (Johns Hopkins University, Baltimore, MD, USA). Lifeact- Ruby is a gift from R. Wedlich-Soldner (Max Planck Institute of Biochemistry, Martinsried, Germany). Cortactin-dsRED and cortactin- ΔSH3-dsRED are a gift from X. Zhan (University of Maryland School of Medicine, Baltimore, MD, USA). PA-GFP and mEos2-tagged cortactin and cortactin-ΔSH3 were made by replacing dsRED.
Western blot analysis
HEK cells transfected with indicated constructs were directly lysed in Laemmli sample buffer 48 h after transfection. Samples were boiled and run on a 4–20% gradient sodium dodecyl sulphate–polyacrylamide gel electrophoresis gel. Proteins were blotted on a nitrocellulose membrane and blocked with 5% (w/v) milk, 1% (v/v) Tween-20 in phosphate-buffered saline (PBS). Membranes were incubated with mouse anti-pan-Shank (UC Davis/NIH Neuromab Facility, clone N23B/4; 1 : 5000) and rabbit anti-GFP (eBiosciences; 1 : 1000) overnight at 4 °C, washed with PBS with 1% (v/v) Tween and incubated with alkaline phosphatase-conjugated secondary antibodies (Cell Signaling; 1 : 2000).
Immunocytochemistry
Hippocampal cultures transfected with indicated constructs were briefly washed in PBS, fixed in 4% (w/v) paraformaldehyde and 4% (w/v) sucrose in PBS for 10 min at room temperature (RT), and washed three times with PBS supplemented with 0.1 m glycine (PBS/Gly). Cells were then permeabilized and blocked in 0.1% (v/v) Triton X-100, 10% (v/v) normal goat serum (NGS) in PBS/Gly for 60 min at 37 °C. Primary antibodies were diluted in 0.1% (v/v) Triton X-100, 5% (v/v) NGS in PBS/Gly and incubated for 2 h at RT or overnight at 4 °C. Cells were washed three times with PBS/Gly, incubated with Alexa-488/561/647-conjugated secondary antibodies (Invitrogen) diluted 1 : 200 in 0.1% (v/v) Triton X-100, 5% (v/v) NGS in PBS/Gly for 60 min at RT, washed three times with PBS/Gly and mounted on glass microscope slides. Primary antibodies used were: mouse anti-pan-Shank (UC Davis/NIH Neuromab Facility; clone N23B/49; 1 : 400); mouse anti-Shank1 (UC Davis/NIH Neuromab Facility; clone N22/21; 1 : 400); mouse anti-Shank2 (UC Davis/NIH Neuromab Facility; clone N23B/6; 1 : 400); mouse anti-Shank3 (Neuromab Facility; clone N367/62; 1 : 400); rabbit anti-GFP (eBiosciences; 1 : 200); and rabbit anti-cortactin (Santa Cruz Biotechnology; H-191; 1 : 100).
Confocal imaging
All confocal images were acquired using a spinning disk confocal system (Andor Technology) consisting of a CSU-22 confocal (Yokagawa) and a Zyla 5.5 sCMOS camera (Andor Technology) mounted on an Olympus IX-81 inverted microscope with laser excitation (Coherent) and emission filters (Semrock). A 60 × objective (1.42 numerical aperture) oil-immersion objective with additional 1.6 × magnification in the light path and 1.2 × magnification placed between the confocal and the camera was used to yield a final 67-nm pixel size. Acquisition was controlled by iQ software (Andor). For live-cell experiments, cells were imaged in extracellular buffer (EB) containing (in mm): NaCl, 120; KCl, 3; CaCl2, 2; MgCl2, 2; glucose, 10; HEPES, 10; pH adjusted to 7.35 with NaOH. Latrunculin A (LatA) was applied as a 40 × solution (200 µm, diluted in EB from a 20 mm stock solution in dimethylsulphoxide (DMSO) to the imaging bath resulting in a final concentration of 5 µm (and a final concentration of 0.025% DMSO).
Spine morphology analysis
Neurons were transfected with indicated constructs at 14 DIV and fixed at 21 DIV. Cells were then stained for GFP, and GFP-positive neurons from at least three independent cultures were randomly selected and imaged. Maximum intensity projections of the confocal stacks were analysed using NeuronStudio (Rodriguez et al., 2008) by an observer blinded for the conditions. Spines were counted when their length was greater than 0.15 µm and less than 3.0 µm. Spines with a head to neck diameter ratio greater than 1.1 and a head diameter greater than or equal to 0.35 µm were considered mushroom spines. Spines with a length to head diameter ratio greater than 2.5 were considered thin spines. Spines with a head to neck diameter ratio smaller than 1.1 and a length to head diameter ratio smaller than 2.5 were considered stubby spines. For each condition, 13–23 neurons (2700–5000 spines) from at least three independent cultures were analysed. To normalize for variations in spine density between different cultures, for each condition spine density was normalized to the spine density in the control group of the corresponding culture. The control group consists of both GFP and scrShank-GFP transfected cells, no differences were found between these two groups.
Synapse morphology analysis
For PSD morphing experiments coverslips were warmed using an objective heater keeping the bath solution at ~34 °C. Z-stacks were acquired every minute over a 30-min time period. Maximum intensity projections were analysed in MetaMorph (Molecular Devices) using custom-written analysis journals essentially as described before (Blanpied et al., 2008b; Kerr & Blanpied, 2012). For each cell, 20–30 synapses were selected and digitally interpolated 4 × for analysis and display. Images of individual synapses were then background subtracted and segmented by applying a threshold set at 0.4 × the maximum fluorescence intensity within the synapse. Elliptical form (EF) measurements were smoothed over three time points, and the coefficient of variance (CV) was calculated with a running 10-min bin.
Single-molecule photoactivated localization microscopy (PALM) imaging and analysis
Cells expressing indicated constructs were imaged at RT in EB on an Olympus IX81 inverted microscope with a 100 × /1.45 TIRF oil immersion objective. Output from a set of diode lasers (Coherent) was directed to the rear of the microscope using a custom optical path. Molecules were simultaneously photoconverted and excited using 405 nm (<100 µW) and 561 nm (20 mW) illumination through oblique illumination to reduce background fluorescence. The acousto-optic tunable filter was controlled separately by a TTL timing source (AMPI Master-8) so that excitation pulse length could be set independently from the exposure time, reducing noise caused by molecular motion during the acquisition of each frame (Frost et al., 2012). Imaging was conducted at 1 Hz, with 100-ms laser pulses for 1800 frames. Fluorescence was detected by an iXon+ 897 EM-CCD camera (Andor Technology) placed after a 1.6 × magnifying optic, resulting in a pixel size of 100 nm. The Olympus ZDC2 feedback positioning system was used to maintain Z stability during imaging.
PALM stacks were exported as multilayer TIFF files and analysed offline using custom-written routines in MATLAB (Mathworks). Images were band-pass filtered and candidate peaks were localized by fitting an elliptical two-dimensional Gaussian function to a 9 × 9 pixel array surrounding the peak, as described previously (Frost et al., 2010b, 2013). Only molecules localized with a localization precision <25 nm (defined as in Thompson et al., 2002), EF <1.3 and emitting >100 photons were used for further analysis. Localized particles were tracked using available algorithms (http://physics.georgetown.edu/matlab/) such that particles appearing in consecutive frames separated by no more than 200 nm were collapsed in one track. Tracks consisting of four or more frames were used to calculate the instantaneous diffusion coefficient by fitting the slope of the corresponding mean-squared displacement (MSD) vs. elapsed time plot using linear fitting on the first three points and adding a value of 0 at MSD(0). The instantaneous diffusion coefficient Deff was then calculated using MSD = 4Defft. Tracks with a negative slope (<8%) were ignored. Based on the average y-intercept, the error e = ~15–20 nm was estimated using the function . Only tracks in spines were analysed by manually selecting a region of interest around spines identified in the scatter-plot of all the localized molecules. PALM images were rendered as density maps with pixels of 25 × 25 nm with intensity values relative to the number of localizations falling in that pixel.
Statistics
Statistical significance was tested using a Student’s t-test when comparing two groups. Statistical significance was tested with a one-way anova followed by a Bonferroni’s or Tukey multiple comparison test when more than two groups were compared. A P-value below 0.05 was considered significant. In all figures, *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
Simultaneous knockdown of Shank1, -2 and -3 in hippocampal neurons disrupts spine morphology
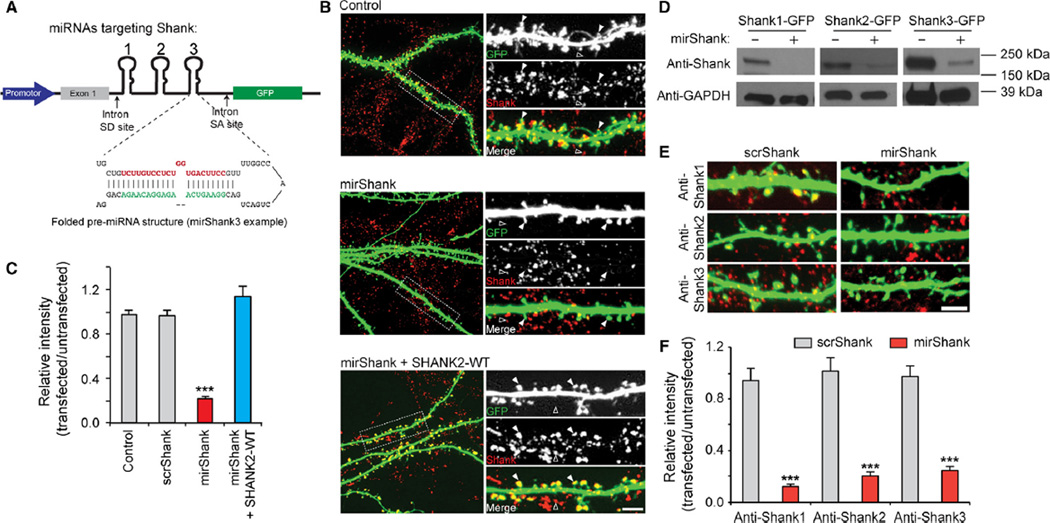
To simultaneously knockdown the major Shank isoforms, for each Shank family member, one 21-nt targeting sequence was selected that is shared by most of its potential splice variants. Each of these three sequences was then cloned in tandem in an artificial miRNA expression cassette designed to drive the expression of one long primary RNA transcript, which is then efficiently spliced to form three individual miRNA short-hairpins and a mRNA coding for EGFP to mark transfected cells (Du et al., 2006; Fig. 1A). First, to measure the efficiency of this construct HEK cells were co-transfected with either Shank1, Shank2 or Shank3 together with the triple-knockdown construct (referred to as mirShank hereafter) and protein expression examined using Western blotting. Expression of all three individual Shank isoforms was significantly reduced in cells expressing mirShank (Fig. 1B). Second, to examine the level of knockdown at neuronal synapses, cultured hippocampal neurons were transfected with mirShank-GFP and immunostaining performed using an antibody recognizing all three Shank isoforms. Expression of a control GFP-expressing plasmid, or a vector expressing scrambled versions of the Shank miRNAs (scrShank) did not change Shank levels, but a strong (>75%) reduction in total Shank protein levels was found at individual spines in mirShank-GFP transfected neurons compared with untransfected neighbouring neurons, which was restored by co-expression of a miRNA-resistant human SHANK2 sequence (mean ± SEM normalized intensity relative to untransfected cells for GFP control: 0.98 ± 0.04; scrShank: 0.97 ± 0.04; mirShank: 0.22 ± 0.02; mirShank + SHANK2-WT: 1.14 ± 0.09; n = 7–18 neurons per group; F2,51 = 70.07, P < 0.0001, one-way anova; Fig. 1C and D). Thus, in this condition most synaptic Shank isoforms were reduced while the relative levels of synaptic Shank2 increased. Here SHANK2 was chosen for all rescue experiments throughout the rest of this work because this isoform is expressed in the majority of mature excitatory synapses of hippocampal neurons (Grabrucker et al., 2011). While it was not practical to evaluate expression levels in all cells in the following experiments, it is possible that Shank2 expression in the rescue condition sometimes exceeds the endogenous protein level. Finally, staining for individual Shank isoforms confirmed that mirShank-GFP expression efficiently reduced levels of Shank1, Shank2 and Shank3 at individual spines (mean ± SEM normalized intensity relative to untransfected cells for anti-Shank1: 0.13 ± 0.01; anti-Shank2: 0.20 ± 0.03; anti-Shank3: 0.24 ± 0.04; F1,24 = 181.3, P < 0.0001, two-way anova; Fig. 1E and F). To test the specificity of these antibodies, COS7 cells were transfected with Shank1-GFP, Shank2-GFP or Shank3-GFP, and stained with anti-Shank1, anti-Shank2 or anti-Shank3. It was found that each of the antibodies only recognized the correct family member, with no observable cross-reactivity (Supporting Information).
Fig 1.
Simultaneous knockdown of Shank1, Shank2 and Shank3 in hippocampal neurons. (A) Schematic overview of the triple-knockdown vector driving the simultaneous expression of three miRNA sequences, targeting Shank1, -2 and -3, and GFP from artificial introns. The miRNA expression cassette is placed in between synthetic splice donor (SD) and acceptor sites (SA) based on the human globin gene to facilitate splicing of the synthetic miRNA component from the GFP mRNA component. The individual miRNAs are cleaved in the nucleus to form single, folded miR155-based miRNA precursors, which are exported to the cytoplasm and further processed to form mature miRNAs. Separate processing of the miRNAs and the GFP mRNA promotes both the efficient knockdown of the target genes and efficient expression of GFP. (B) Western blot analysis of HEK cells transfected with rat Shank1-GFP, Shank2-GFP or Shank3-GFP together with pSM155-GFP or mirShank-GFP. (C) Confocal images of cultured hippocampal neurons transfected with GFP (green), scrShank-GFP, mirShank-GFP or mirShank-GFP and SHANK2-WT-mCherry stained with an antibody recognizing all Shank isoforms (pan-Shank/Alexa647; red). Arrowheads indicate Shank clusters in transfected cells (filled arrowheads) and untransfected cells (open arrowheads). Scale bar: 3 µm. (D) Quantification of relative pan-Shank immunoreactivity in spines of GFP, scrShank-GFP, mirShank-GFP and mirShank-GFP + SHANK2-WT-mCherry rescue transfected neurons compared with untransfected neighbouring neurons. (E) Confocal images of cultured hippocampal neurons transfected with scrShank-GFP or mirShank-GFP (green) stained with antibodies recognizing individual Shank isoforms (red). Scale bar: 3 µm. (F) Quantification of relative Shank1, -2 or -3 immunoreactivity in spines of scrShank-GFP and mirShank-GFP transfected neurons. Data in bar graphs are represented as mean ± SEM. ***P < 0.001, anova with post hoc Bonferroni multiple comparison test.
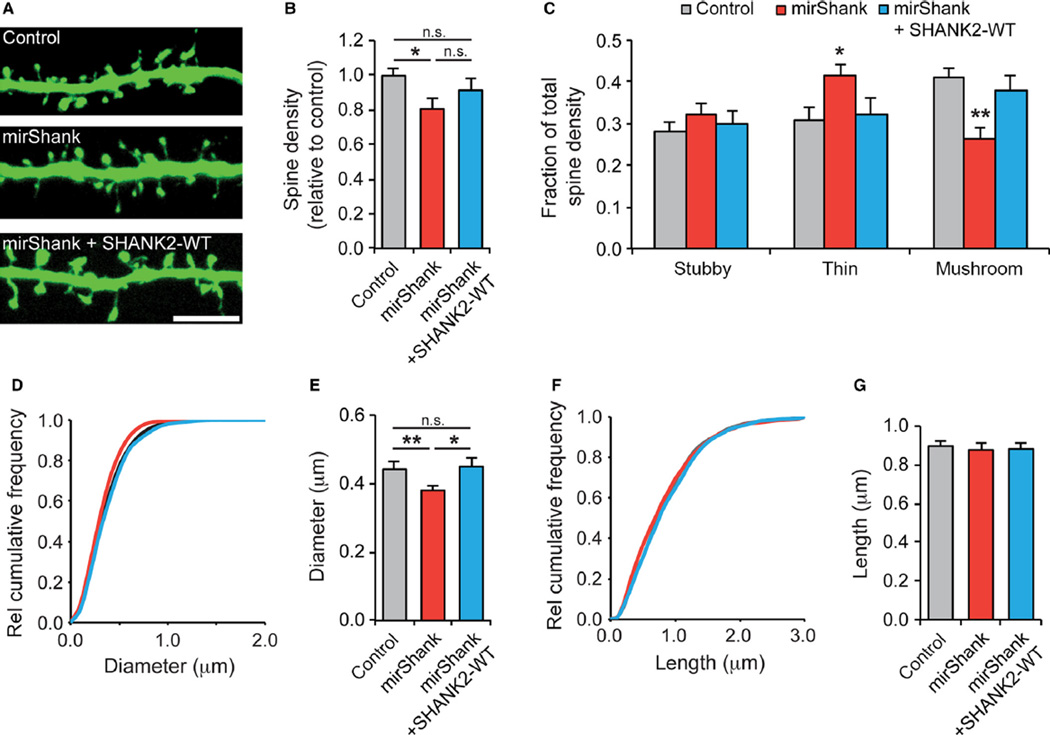
Numerous studies have established that all three Shank isoforms play important roles in the regulation of dendritic spine morphology (Roussignol et al., 2005; Sala et al., 2005; Haeckel et al., 2008; Hung et al., 2008; Grabrucker et al., 2011; Peca et al., 2011; Verpelli et al., 2011; Berkel et al., 2012; Durand et al., 2012; Schmeisser et al., 2012), and knockout of either Shank2 (Schmeisser et al., 2012) or Shank3 (Peca et al., 2011) reduces spine numbers and the frequency of spontaneous miniature excitatory postsynaptic synaptic potentials. To measure the effect of knockdown of all three isoforms of Shank simultaneously on spine morphology, cultured hippocampal neurons were transfected with mirShank-GFP at 14 DIV and fixed 7 days later at 21 DIV. Neurons were stained with anti-GFP to enhance contrast, and maximum intensity projections of confocal images were analysed to quantify the density and morphology of dendritic spines (Fig. 2A). An overall 20% decrease in total spine density in mirShank neurons was found, which was only partially rescued by co-expression of SHANK2 (mean ± SEM number of spines per 10 µm dendrite for control: 7.3 ± 0.5; mirShank: 5.9 ± 0.5; and mirShank + SHANK2-WT: 6.3 ± 0.6; n = 13–23 neurons; F2,54 = 3.56, P = 0.035, one-way anova; Fig. 2B). Next, all spines were classified into the three main morphological types: stubby; thin; and mushroom. It was found that while control and rescue neurons had a higher fraction of mushroom (41%) than stubby (28%) and thin (31%) spines, mirShank neurons had a relatively high fraction of thin spines (42%) and a relatively low fraction of mushroom spines (26%; Fig. 2C). In terms of absolute spine density (Table 1), it appeared that mushroom spines were the primary target of Shank knockdown. Consistent with this, a significant reduction was found in spine head diameter in Shank knockdown neurons (mean ± SEM head diameter control: 0.44 ± 0.02 µm; mirShank: 0.38 ± 0.02 µm; and mirShank + SHANK2-WT: 0.45 ± 0.03 µm; F2,54 = 6.57, P = 0.0028, one-way anova; Fig. 2D and E), but no change in spine length (mean ± SEM head diameter control: 0.90 ± 0.03 µm; mirShank: 0.88 ± 0.04 µm; and mirShank + SHANK2-WT: 0.88 ± 0.03 µm; F2,54 = 2.11, P = 0.13, one-way anova). Together, these data confirm that expression of the mirShank construct specifically and efficiently reduces the expression of all three Shank isoforms at synapses of cultured hippocampal neurons and disrupts spine morphology.
Fig 2.
Shank knockdown alters spine morphology. (A) Example images of dendrites from control, mirShank and mirShank + SHANK2-WT rescue neurons stained for GFP. Scale bar: 5 µm. (B) Bar graph summary of average spine density normalized to control. (C) Bar graph of the fraction of thin, stubby and mushroom spines of the total number of spines for control, mirShank and mirShank+SHANK2-WT rescue neurons. (D) Relative cumulative distribution plot for spine head diameter for control, mirShank and mirShank+SHANK2-WT rescue neurons. (E) Bar graph of mean spine head diameter. (F) Relative cumulative distribution plot for spine length for control, mirShank and mirShank+SHANK2-WT rescue neurons. (G) Bar graph of spine length. Data in bar graphs are represented as mean±SEM. *P < 0.05, **P < 0.01, one-way anova with post hoc Bonferroni multiple comparison test.
Table 1.
Spine morphology measurements in Shank triple knockdown neurons
| Control | mirShank | mirShank + SHANK2-WT rescue | |
|---|---|---|---|
| Total spine density per 10 µm dendrite (mean ± SEM) | 7.31 ± 0.53 | 5.96 ± 0.50 | 6.33 ± 0.63 |
| Stubby spine density per 10 µm dendrite (mean ± SEM) | 1.98 ± 0.13 | 1.75 ± 0.14 | 1.69 ± 0.11 |
| Thin spine density per 10 µm dendrite (mean ± SEM) | 2.38 ± 0.33 | 2.43 ± 0.27 | 2.13 ± 0.37 |
| Mushroom spine per 10 µm dendrite density (mean ± SEM) | 2.94 ± 0.24 | 1.78 ± 0.24 | 2.51 ± 0.35 |
| Head diameter in µm (mean ± SEM) | 0.44 ± 0.02 | 0.38 ± 0.02 | 0.45 ± 0.03 |
| Spine length in µm (mean ± SEM) | 0.90 ± 0.03 | 0.88 ± 0.04 | 0.88 ± 0.03 |
| n (spines/cells/cultures) | 4946/23/3 | 3567/21/3 | 2734/13/3 |
Shank maintains stability of the spine actin cytoskeleton via its C-terminal cortactin binding site
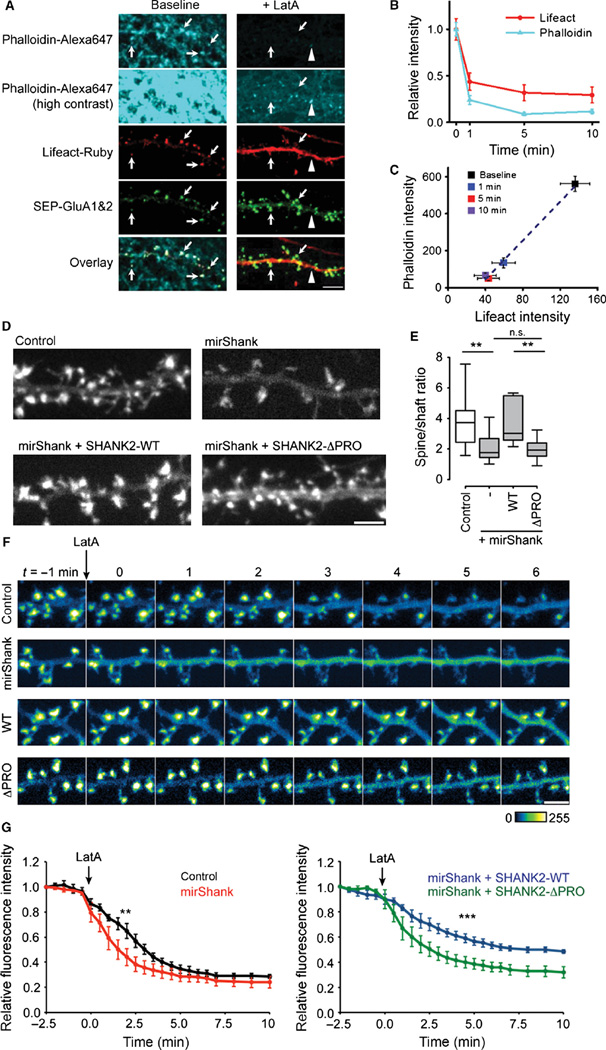
Overexpression of Shank promotes actin polymerization in heterologous cells (Durand et al., 2012) and the accumulation of F-actin in neuronal spines (Sala et al., 2001; Durand et al., 2012; Han et al., 2013), but it is unknown whether Shanks are required for maintaining the integrity of the actin cytoskeleton in mature spines. To test if Shank knockdown alters spine actin content, F-actin was labelled in live cells with LifeAct-Ruby, a small (17 amino-acid) recombinant F-actin probe fused to the red-fluorescent protein Ruby (Riedl et al., 2008). LifeAct signal was specifically enriched in dendritic spines where F-actin is generally most abundant, was highly correlated with intensity of phalloidin staining in fixed cells and showed a similar time course and degree of loss of fluorescence after application of the actin depolymerizering agent LatA (Fig. 3A–C), confirming that LifeAct specifically labels F-actin. To measure the relative spine F-actin content, the ratio of LifeAct intensity in spines compared with the dendritic compartment was quantified to normalize for differences in expression levels between neurons. In control neurons, the LifeAct signal was ~four times higher in spines than in dendrites (mean enrichment ± SEM control: 3.9 ± 0.5; n = 15 neurons); however, expression of mirShank significantly decreased the levels of LifeAct-Ruby signal in spines (2.2 ± 0.3; n = 15 neurons). Re-expression of full-length miRNA-resistant SHANK2 fully restored F-actin content to control levels 3.7 ± 0.4; n = 12 neurons; Fig. 3D and E), confirming that Shank proteins are required to maintain levels of F-actin in spines. Interestingly, while a previous study suggested that the interactions of actin-binding proteins with the N-terminal ANK domain of Shank are required to mediate over-expression-induced increase in F-actin content (Durand et al., 2012), it was found that SHANK2, a Shank isoform that naturally lacks the ANK domain, could still rescue the observed decrease in spine F-actin. It was therefore tested whether interactions with the cortactin binding motif in the C-terminal proline-rich region of SHANK2 are required to regulate spine actin content. To test this, a SHANK2 mutant lacking the 7-amino-acid-long cortactin-binding motif (SHANK2-ΔPRO) was re-expressed in mirShank neurons. This mutant was still efficiently targeted to spines (data not shown) but, compared with wild-type SHANK2, did not rescue the loss of spine F-actin (2.0 ± 0.2; n = 12 neurons; F3,45 = 5.47, P = 0.0027, one-way anova; Fig. 3D and E). Thus, Shank-mediated maintenance of the spine actin cytoskeleton requires its C-terminal cortactin binding motif.
Fig 3.
Shank proteins maintain spine F-actin levels. (A) Example images of dendrites expressing LifeAct-Ruby (red) and SEP-GluA1 and 2 (green) before and after LatA treatment fixed and stained with phalloidin-Alexa647 (Cyan) displayed at moderate and high contrast. (B) Quantification of LifeAct and phalloidin signal 1, 5 and 10 min after LatA treatment, plotted as mean±SEM. (C) Scatterplot of phalloidin intensity vs. LifeAct intensity (mean±SEM) compared at different time points after LatA treatment. (D) Example images of LifeAct-Ruby in dendrites from control, mirShank, mirShank+SHANK2-WT and mir-Shank+SHANK2-ΔPRO rescue neurons. Scale bar: 2.5 µm. (E) Boxplot of median spine/dendrite ratio of LifeAct-Ruby signal intensity. **P < 0.01, one-way anova with post hoc Bonferroni multiple comparison test. (F) Confocal images of LifeAct-Ruby signal. Arrow indicates time of application of 5 µm LatA. Scale bar: 2 µm. (G) Line graph of LifeAct-Ruby fluorescence (mean ± SEM) measured in spines over time before and after application of 5 µm LatA (indicated by the arrow). **P < 0.01, ***P < 0.001, one-way anova with post hoc Bonferroni multiple comparison test.
These results indicate that the loss of Shank reduces the stability of the actin cytoskeleton in spines. To address this more directly in live cells, the real-time loss of LifeAct-Ruby intensity was measured in spines after treatment with LatA. In control neurons, application of LatA induced a rapid decrease in LifeAct signal from spines, which reached plateau levels 8 min after application, consistent with previous experiments (Kerr & Blanpied, 2012). In spines of Shank knockdown neurons, a significantly faster loss of Lifeact signal after LatA treatment was observed, confirming that the actin cytoskeleton in these spines was less stable (Fig. 3F and G). However, expression of SHANK2-WT on the knockdown background dramatically slowed the loss of LifeAct signal and reduced the net loss of LifeAct signal. In contrast, expression of the SHANK2-ΔPRO mutant did not slow down the LatA-induced loss of F-actin (Fig. 3F and G). A two-way anova revealed a main effect of time (F22,368 = 115.1, P < 0.0001), condition (F3,368 = 67.98, P < 0.0001), and a significant interaction effect between condition and time (F66,368 = 1.59, P = 0.0041). Thus, Shank proteins are required to maintain a stable F-actin network in spines, which in part relies on interactions with its C-terminal cortactin binding motif.
Spine retention of cortactin requires its interaction with Shank
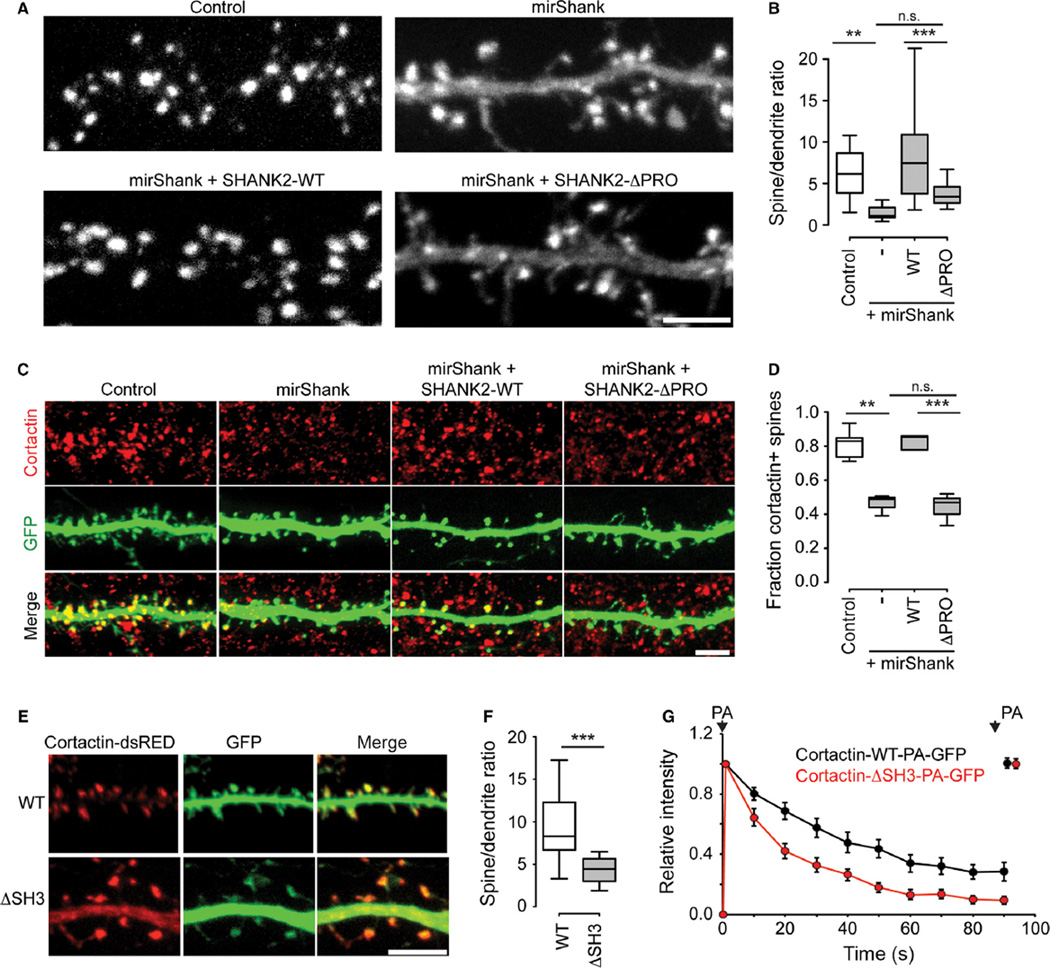
Cortactin is a spine-enriched actin nucleation factor shown to directly interact with actin, Shank2 and Shank3 (Du et al., 1998; Naisbitt et al., 1999); however, previous studies suggested that the targeting of cortactin to spines is primarily dependent on its interaction with the actin cytoskeleton (Hering & Sheng, 2003). Here, to test if Shank additionally helps to recruit and anchor cortactin to spines, the distribution of dsRED-tagged cortactin expressed in control and mirShank neurons was measured. While in control neurons cortactin-dsRED was almost exclusively enriched in dendritic spines, in Shank knockdown neurons cortactin-dsRED was found more evenly distributed throughout the dendritic shaft and spines (Fig. 4A). Quantification of the relative intensity of cortactin-dsRED in spines compared with dendrites confirmed a significant loss of spine enrichment in Shank knockdown cells, which was rescued by re-expression of full-length SHANK2 (mean enrichment ± SEM control: 6.0 ± 0.7; mirShank: 1.6 ± 0.3; mirShank + SHANK2-WT: 8.8 ± 1.6; n = 15 neurons).
Fig 4.
Spine cortactin retention is controlled by Shank proteins. (A) Cortactin-dsRED expression in dendrites from control, mirShank, mirShank + SHANK2-WT and mirShank + SHANK2-ΔPRO rescue neurons. Scale bar: 5 µm. (B) Boxplot of median spine/dendrite ratio of cortactin-dsRED fluorescence intensity in control, mirShank, mirShank + SHANK2-WT and mirShank + SHANK2-ΔPRO rescue neurons. (C) Confocal images of neurons transfected stained with GFP, mirShank, mirShank + GFP-SHANK2-WT and mirShank + GFP-SHANK2-ΔPRO (green), and stained for endogenous cortactin (red). Scale bar: 5 µm. (D) Quantification of the percentage of cortactin-positive spines for the indicated conditions. (E) Confocal image of dendrites from neurons expressing wild-type (WT) or SH3 deletion mutant (ΔSH3) cortactin-dsRED (red) and GFP (green). Scale bar: 5 µm. (F) Quantification of the relative fluorescence intensity in spines for the indicated conditions. (G) Average fluorescence intensity measured in spines over time for wild-type cortactin-PA-GFP (black trace) and cortactin-ΔSH3-PA-GFP (red trace) after 2-photon photoactivation (PA). The signal was normalized to fluorescence intensity after the first photoactivation step and plotted as mean±SEM. Spines were exposed to a second photoactivation step at the end of the imaging period. **P < 0.01, ***P < 0.001, one-way anova with post hoc Bonferroni multiple comparison test.
To test whether this loss of cortactin was a direct effect of the loss of the Shank–cortactin interaction, next SHANK2-ΔPRO was re-expressed, and it was found that compared with wild-type SHANK2, this mutant only partially rescued the loss of spine cortactin enrichment (mean ± SEM: 3.6 ± 0.4; n = 14 neurons; F3,54 = 10.84, P < 0.0001, one-way anova; Fig. 4A and B). A SHANK2 mutant that lacks the SH3 domain (SHANK2-ΔSH3) or a mutant that is unable to bind Homer (SHANK2-P1035L) both rescued the loss of spine cortactin similarly as SHANK2-WT (mirShank + SHANK2-ΔSH3: 7.0 ± 1.8; mirShank + SHANK2-P1035L: 9.3 ± 1.5; n = 5 and 8 neurons; P ≫ 0.05, data not shown). Furthermore, to test whether endogenous levels of cortactin are also reduced in Shank knockdown neurons, transfected neurons were stained for cortactin, and a significant reduction in the fraction of spines with detectable levels of cortactin was found in mirShank neurons (mean fraction ± SEM control: 0.81 ± 0.03; mirShank: 0.46 ± 0.04; mirShank + SHANK2-WT: 0.82 ± 0.06; mirShank + SHANK2-ΔPRO: 0.44 ± 0.06; n = 3–6 neurons; F3,13 = 17.37; P < 0.0001; one-way anova; Fig. 4C and D). These results indicate that any potential recruitment of the small pool of remaining endogenous Shank isoforms by expression of replacement Shank2 constructs was not sufficient to rescue cortactin distribution. Note that because cortactin staining can also be found in axons and other cellular compartments than spines, here the fraction of spines positive for cortactin staining rather than the ratio of staining intensity in transfected compared with untransfected neurons was quantified.
The observed loss of cortactin from spines could be an indirect effect of the loss of spine F-actin in mirShank neurons. Therefore, it was tested whether a cortactin mutant that lacks the SH3 domain (cortactin-ΔSH3), and is therefore unable to bind Shank (Naisbitt et al., 1999), but can still bind actin, would be targeted to spines as efficiently as wild-type cortactin in control neurons. Indeed, it was found that although this mutant is still targeted to spines as described before (Hering & Sheng, 2003), spine enrichment is significantly decreased compared with wild-type cortactin (ratio for cortactin-WT: 9.4 ± 1.3; cortactin-ΔSH3: 4.6 ± 0.8; n = 11 neurons; t20 = 3.26; P = 0.0040; unpaired t-test; Fig. 4E and F). To further test whether cortactin requires its C-terminal SH3 domain for its stability at mature spines, wild-type cortactin or the cortactin-ΔSH3 mutant tagged with photoactivatable GFP were expressed and the decay rate of fluorescence was measured after photoactivation. Consistent with the observed reduction in spine levels, a significantly faster decay of cortactin-ΔSH3 fluorescence compared with wild-type cortactin was found (Fig. 4G). Together these data support that, although actin binding is important for spine localization of cortactin, its interaction with synaptic Shank proteins additionally determines its retention and stability in spines.
Shank regulates mobility of cortactin within spines
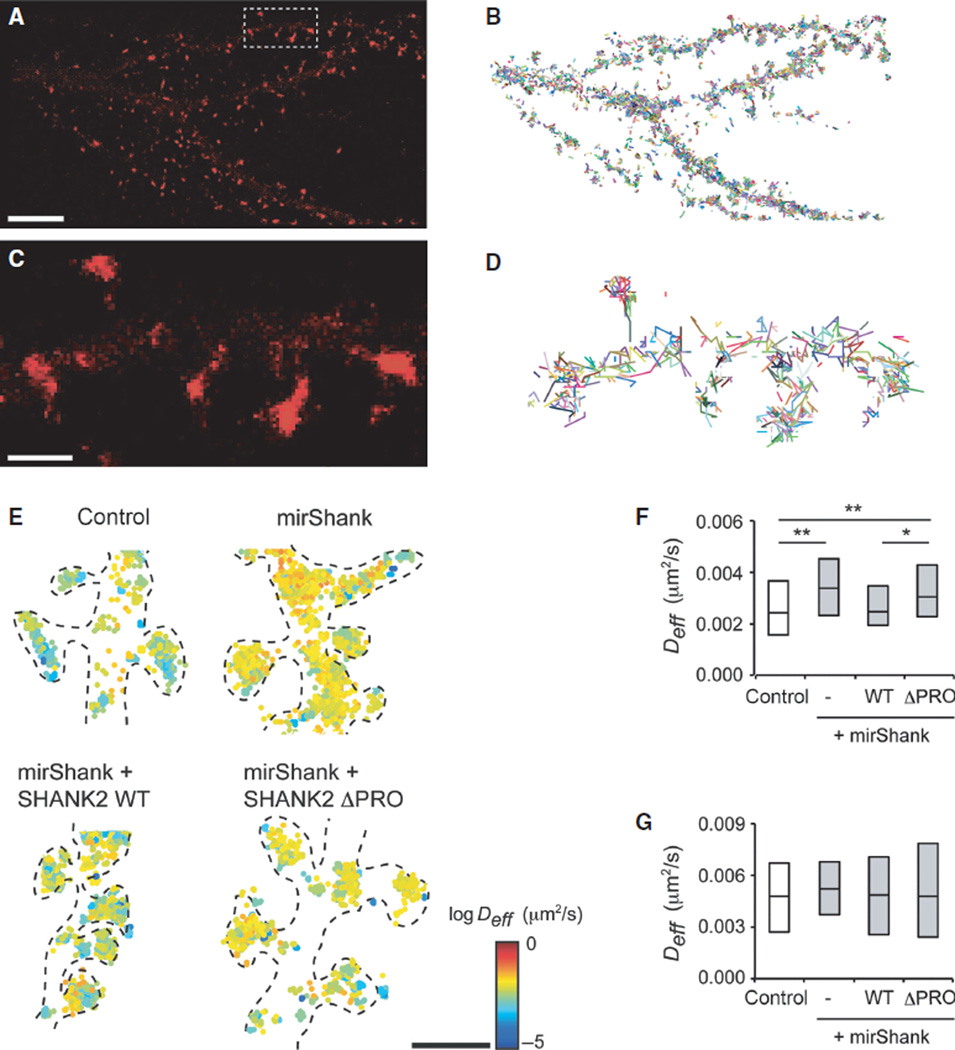
Based on these findings, it was predicted that interactions with Shank proteins determine not only the localization but also the mobility of cortactin within spines. To measure the mobility of cortactin directly within individual spines of live neurons, single-molecule tracking PALM of cortactin tagged with the photoconvertible fluorophore mEos2 was used. Individual molecules were sparsely activated and tracked over consecutive frames to construct a super-resolved map of cortactin mobility in individual spines. Individual cortactin molecules are likely to exist in the cell either in a freely mobile state when unbound and cytosolic, or in a less mobile state when bound to actin filaments or other regulatory molecules like Shank (Lu et al., 2014). To selectively track bound cortactin molecules in spines while preventing the detection of freely moving molecules, low-frequency imaging (1 Hz) with pulsed excitation (100 ms) was used (Frost et al., 2010b, 2012). Using these imaging parameters, single molecules were imaged and tracked at high density and precision (Fig. 5A–D). Consistent with the distribution of cortactin measured by confocal microscopy, it was found that the majority of localized molecules were enriched in spines (Fig. 5A and C). Molecules appearing in consecutive frames were then compiled into trajectories, and the MSDs between time points were calculated for tracks persisting for more than three frames. The instantaneous diffusion coefficient Deff was then estimated by fitting the slope of the MSD vs. time curve using a least-square linear fit on the first four time points. Single cortactin molecules in spines moved at relatively low diffusive rates (mean Deff: 0.0031 ± 0.0002 µm2/s; n = 97 spines from nine neurons), reflecting that for a large fraction of cortactin molecules, mobility is restricted by interactions with binding partners within spines. In contrast, cortactin molecules tracked within dendritic shafts were much sparser and moved significantly faster (mean Deff: 0.0050 ± 0.0005 µm2/s; n = 38 dendritic segments from six neurons) than the spine cortactin population (Fig. 5E–G).
Fig 5.
Cortactin mobility in spines is regulated by Shank proteins. (A) Density map PALM for cortactin-mEos2 in control neuron. Scale bar: 5 µm. (B) Individual tracks of cortactin-mEos2. (C and D) Enlarged images of density map (C) and tracks (D) of the box in (A). Scale bar: 1 µm. (E) Scatterplots of tracked cortactin-mEos2 molecules, colour-coded for their diffusion coefficient, in control, mirShank, mirShank + SHANK2-WT and mirShank+SHANK2-ΔPRO rescue neurons. Scale bar: 2.5 µm. (F and G) Boxplot of median diffusion in spines (F) and dendrites (G) of control, mirShank, mirShank+SHANK2-WT and mirShank + SHANK2-ΔPRO rescue neurons. *P < 0.05, **P < 0.01, one-way anova with post hoc Bonferroni multiple comparison test.
Next, to test whether the retention and stabilization of cortactin in spines is regulated by interactions with Shank molecules, the mobility of cortactin-mEos2 in neurons co-transfected with mirShank was tracked. Indeed, cortactin molecules were localized at high density in both spines and dendrites of Shank knockdown neurons, and moved significantly faster in knockdown spines than in control spines (mean Deff mirShank: 0.0040 ± 0.0002 µm2/s; n = 93 spines from nine neurons). Importantly, this was rescued by re-expression of SHANK2-WT, but not by re-expression of the SHANK2-ΔPRO mutant (mirShank + SHANK2-WT: 0.0032 ± 0.0001 µm2/s; n = 122 spines from eight neurons; mirShank + SHANK2-ΔPRO: 0.0042 ± 0.0002 µm2/s; n = 79 spines from six neurons; F3,330 = 4.96, P = 0.0022, one-way anova; Fig. 5E–G). The mobility of cortactin in the dendritic compartment was not changed in any of these conditions (mirShank: 0.0053 ± 0.0004 µm2/s; mirShank + SHANK2-WT: 0.0049 ± 0.0007 µm2/s; mirShank + SHANK2-ΔPRO: 0.0053 ± 0.0007 µm2/s; n = 34–40 dendritic segments from six–seven neurons; Fig. 5G). It was thus concluded that the mobility of cortactin within spines is largely controlled by its direct interaction with Shank proteins.
Shank–cortactin interactions are required for spontaneous morphing of synapses
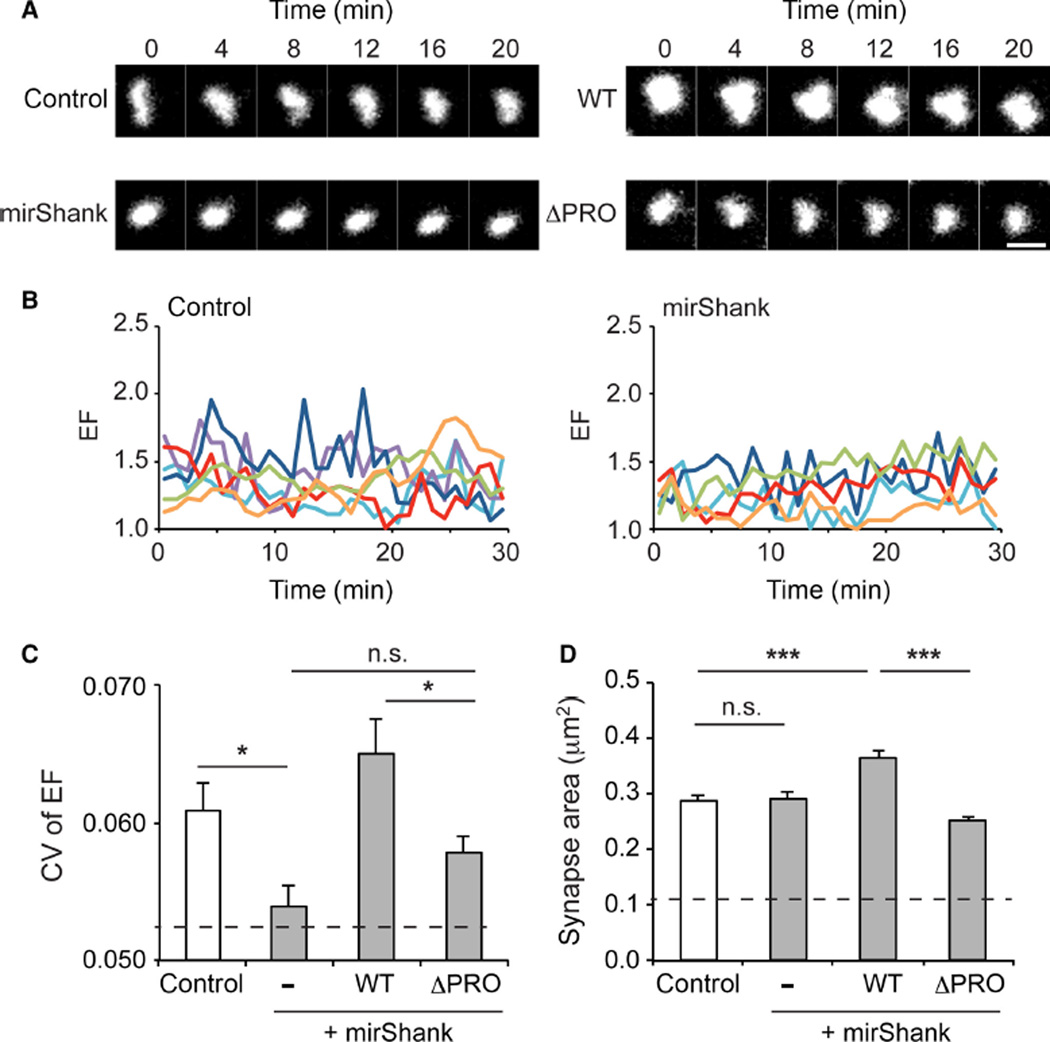
Previous observations showed that the morphology of individual synapses undergoes continuous remodelling, which heavily relies on ongoing actin dynamics (Blanpied et al., 2008a; Kerr & Blanpied, 2012). Thus, based on the results to this point, it was predicted that the reduced spine actin stability in Shank knockdown neurons and the loss of the interaction of cortactin with the PSD would reduce the spontaneous morphing of synapses. To test this, PSD morphology changes were measured in control or Shank knockdown neurons using a PSD-95 molecular replacement strategy to fluorescently label synapses (MacGillavry et al., 2013). As shown before, the EF (length/width) of individual PSDs marked with shrPSD-95-mCherry varied continuously, dynamically changing shape on a minute time-scale (Fig. 6A and B). To quantify and compare the level of PSD reshaping between control and mirShank neurons, the CV of the EF was calculated in 10-min time-bins. Interestingly, it was found that this measure of morphological dynamics was significantly reduced in Shank knockdown neurons (mean CV ± SEM control: 0.061 ± 0.002; n = 89 synapses from four neurons; mirShank: 0.054 ± 0.001; n = 118 synapses from five neurons; Fig. 6C), and was almost as low as the CV measured for 100-nm fluorescent beads immobilized to a glass coverslip (0.052 ± 0.003; n = 24 beads; Fig. 6C, dashed line). Re-expression of SHANK2-WT rescued this defect and even showed a slight, but not significant, enhancement in CV (0.065 ± 0.002; n = 80 synapses from three neurons) compared with control neurons, while the SHANK2-ΔPRO mutant only partially rescued the morphological dynamics of PSDs (0.058 ± 0.002; n = 155 synapses from five neurons; F3,468 = 10.91; P < 0.0001, one-way anova).
Fig 6.
Spontaneous morphing of synapses is regulated by Shank. (A) Examples of time-lapse imaging of individual synapses marked by shrPSD95-mCherry. Scale bar: 1 µm. (B) Individual traces of elliptical form (EF) over time for postsynaptic densities (PSDs) in control (left) and mirShank (right) neurons. (C) Bar graph of the mean CV of EF for the different groups. Dashed line indicates average CV measured for fluorescent 100-nm beads. (D) Bar graph of mean synapse area for the different groups. Error bars represent SEM, *P < 0.05, ***P < 0.001, one-way anova with post hoc Bonferroni multiple comparison test.
A potential reduction in PSD area could have prevented the detection of shape changes using the imaging parameters. However, no change was found in the measured area of the PSDs that were analysed between control and mirShank neurons (mean area control: 0.29 ± 0.01 µm2; mirShank: 0.29 ± 0.01 µm2), and it was found that the area of 100-nm fluorescent beads appeared much smaller (0.12 ± 0.001 µm2) than the area of synapses in mirShank neurons, excluding this possibility. Interestingly, however, it was found that the average area of PSDs in SHANK2-WT, but not SHANK2-ΔPRO, expressing neurons was significantly larger than in control and mutant neurons (mirShank + SHANK2-WT: 0.36 ± 0.01 µm2; mirShank + SHANK2-ΔPRO: 0.25 ± 0.01 µm2; F3,390 = 10.51, P < 0.0001, one-way anova; Fig. 6D), confirming previous observations (MacGillavry et al., 2013). Together, these results demonstrate that the stability of the spine actin cytoskeleton maintained by Shank–cortactin interactions is required for ongoing morphological dynamics of individual synapses.
Discussion
The Shank family of synaptic scaffolding molecules is of key interest because it is uniquely positioned to control the interaction between glutamate receptors at the postsynaptic membrane and the actin cytoskeleton in the spine head. However, because the different Shank family members share a high degree of sequence and domain homology and are co-expressed within individual synapses, it is difficult to study the requirement of individual Shanks for synaptic processes in the absence of within-family compensatory effects. Here, a specific and efficient miRNA-based knockdown approach was established to simultaneously reduce the expression of all three Shank family members in synapses of mature hippocampal neurons. This molecular toolset was employed to test whether the family of Shank proteins is required for maintaining the integrity and dynamics of the actin cytoskeleton in mature spines. A combination of live-cell confocal and single-molecule imaging techniques was used to demonstrate that the Shank family is a central organizer of the synaptic actin cytoskeleton. Specifically, it was shown that the interaction of SHANK2 with the actin nucleation factor cortactin is critical for maintaining the stability of existing actin filaments in mature spines and for the dynamic morphing of synapses. These findings imply that beyond their role as stable scaffolding molecules within the PSD, Shank proteins are key intermediates between the synapse and the spine interior that direct actin-based force via cortactin to regulate synapse morphology and function.
Extending previous studies that found that overexpression of individual Shank isoforms is sufficient to increase spine actin content (Sala et al., 2001; Durand et al., 2012; Han et al., 2013), here a significant reduction in F-actin was found in Shank knockdown spines, indicating that Shank proteins are required to actively maintain the spine actin network. Furthermore, the loss of F-actin triggered by LatA application was accelerated in Shank knockdown neurons, suggesting that the remaining actin cytoskeleton was more prone to depolymerization. On the other hand, miRNA-resistant SHANK2 was sufficient to rescue F-actin levels, and greatly increased the resistance of the actin network to destabilization by LatA; even 10 min after LatA application, a large fraction of spine F-actin remained. These results clearly indicate that Shanks have a robust influence on the integrity of the spine actin cytoskeleton.
Interestingly, it was found that a SHANK2 mutant unable to bind the actin nucleation-promoting factor cortactin was severely impaired in rescuing these defects, suggesting that the ability of Shanks to recruit cortactin is important for regulation of the spine actin cytoskeleton. Indeed, it was found that the loss of actin from Shank knockdown spines was paralleled by a marked loss of cortactin spine enrichment. While molecular replacement with full-length SHANK2 completely restored spine cortactin levels, the SHANK2 mutant lacking the cortactin binding site could not, indicating that this is a direct effect of the elimination of the Shank–cortactin interaction. Moreover, removing cortactin’s C-terminal SH3 domain, responsible for its interaction with Shank (Naisbitt et al., 1999), resulted in a similar loss of cortactin levels from spines, and greatly reduced its stability as measured by two-photon activation experiments, further confirming that Shank proteins are necessary for retaining cortactin in dendritic spines. Thus, although it has been suggested that cortactin retention in spines is primarily regulated via its interactions with the actin cytoskeleton and does not require its C-terminal SH3 domain (Hering & Sheng, 2003), strong evidence was found that its interaction with Shank significantly contributes to its localization in spines. Moreover, single-molecule tracking showed that Shank knockdown greatly increased the overall mobility of cortactin in spines, further supporting a model where Shank proteins in the PSD are necessary for retaining and stabilizing cortactin in spines. Interestingly, while the proline-rich sequence that is responsible for the interaction with cortactin is conserved between Shank2 and Shank3, Shank1 seems to lack this sequence (Naisbitt et al., 1999) and has never been reported to interact with cortactin, suggesting functional diversity among Shank family members. Apart from Shanks, cortactin-binding protein 2 (CTTNBP2) was also shown to interact with and stabilize cortactin in dendritic spines (Chen & Hsueh, 2012), indicating that multiple synaptic proteins contribute to the regulation of cortactin spine retention.
Immunogold EM localization suggests that the majority of cortactin resides in the core of the spine head with a relatively small fraction associated with the PSD (Racz & Weinberg, 2004), consistent with proteomic analysis (Husi et al., 2000; Peng et al., 2004). Here, mapping the mobility of cortactin using single-molecule tracking PALM revealed a homogenous population of fairly immobile cortactin molecules throughout the spine head, consistent with a largely stable pool of cortactin bound to the actin cytoskeleton. Also, recent in vitro single-molecule studies demonstrated that actin-bound cortactin is essentially static, and has a strong preference for accumulating at actin branch points (Helgeson & Nolen, 2013). Together, these data suggest that whereas only a minority of cortactin is PSD-associated at any point, transient interactions with Shank at the PSD are required for overall retention of the large pool of spine cortactin.
Apart from cortactin, Shank can interact with several other actin nucleation-promoting factors. For instance, Abp1 (actin-binding protein-1), which like cortactin is found at sites of active actin assembly in heterologous cells (Kessels et al., 2000), can also bind Shank and regulate spine morphology (Qualmann et al., 2004; Haeckel et al., 2008). In contrast to cortactin, however, Abp1 does not directly interact with the Arp2/3 complex, but promotes Arp2/3 activity by releasing the auto-inhibition of the neural Wiskott–Aldrich syndrome protein (N-WASP; Kim et al., 2000; Pinyol et al., 2007). Interestingly, on its own, cortactin only weakly activates the Arp2/3 complex but, together with activated WASP, cortactin synergistically enhances Arp2/3 activity (Weed et al., 2000; Uruno et al., 2001; Weaver et al., 2001; Helgeson & Nolen, 2013). Moreover, Abi-1 (Abelson interacting protein-1), another Shank interacting protein involved in synapse formation (Proepper et al., 2007), is part of the WAVE (WASP family verprolin homologous protein) complex, and is able to determine its localization and activity (Gautreau et al., 2004; Innocenti et al., 2004; Leng et al., 2005). Thus, Shank proteins might serve as an efficient scaffolding platform bringing together cortactin, N-WASP, WAVE and Arp2/3 to cooperatively promote actin nucleation close to the synapse.
The recent findings that Shank proteins can also directly interact with the Arp2/3 and WAVE complexes (Han et al., 2013) and that cortactin can bind directly to WAVE2 (Han et al., 2014) further strengthen this model. Moreover, a recent super-resolution imaging study found that the Arp2/3 complex is preferentially immobilized in regions close to the PSD, and that members of the WAVE complex are even more tightly associated with the PSD, consistent with the idea that the PSD is a central organizer of actin nucleation (Chazeau et al., 2014). Such an organization very much resembles the zonula adherens found in adhesive junctions of epithelial cells, where E-cadherin recruits Arp2/3 via cortactin as well as other nucleation promoting factors such as WAVE2 and N-WASP to seed the localized assembly of actin filaments (Verma et al., 2012; Han et al., 2014). This type of organization suggests a general model of coincident regulation of actin assembly, where multiple upstream signals are integrated to control actin nucleation with great temporal and spatial precision.
The recruitment and regulation of actin regulating proteins by Shank suggest that the force generated by local actin polymerization and branching can directly alter synaptic organization. Indeed, actin dynamics in spines is known to promote ongoing changes in PSD morphology (Blanpied et al., 2008a; Kerr & Blanpied, 2012), and it was demonstrated here that these actin-driven PSD dynamics require the Shank–cortactin interaction. Interestingly, the distribution of Shank within the PSD is heterogeneous and characterized by distinct nanoclusters of elevated protein density (MacGillavry et al., 2013). This suggests further that the Shank-guided distribution of force application may be relatively focal, controlling protein organization within the bounds of the PSD on the nanometer scale. Consistent with this idea, it was found previously that depolymerization of the actin cytoskeleton with LatA acutely disrupts the maintenance of nanoscale scaffolding domains in the PSD (MacGillavry et al., 2013) as well as the subsynaptic distribution of AMPARs (Kerr & Blanpied, 2012; MacGillavry et al., 2013). Because receptor positioning within the PSD contributes to establishing synaptic strength (Freche et al., 2011; MacGillavry et al., 2011; Nair et al., 2013), these effects may directly and acutely regulate synaptic transmission.
Even more broadly, activity-induced spine actin remodelling underlies many forms of synaptic plasticity (Matus, 2000; Bosch & Hayashi, 2012). Indeed, it was recently shown that long-term potentiation (LTP) induction using glutamate uncaging triggers the rapid translocation into spines of many actin regulators that are likely responsible for the concurrent, rapid spine expansion (Bosch et al., 2014). In contrast, however, both LTP induction using theta-burst stimulation in hippocampal slices and NMDA application to induce synaptic depression in hippocampal cultures have been shown to induce a translocation of cortactin out of spines, into the dendritic shaft (Hering & Sheng, 2003; Iki et al., 2005; Seese et al., 2012). Thus, while substantial work remains to define these mechanisms clearly, the Shank–cortactin interaction provides an attractive mechanism by which neuronal activity (and other signalling events) can trigger acute reorganization of the synaptic cytoskeleton. Indeed, the transient loss and replenishment of Shank itself from the synapse spine in a brief window following LTP induction (Steiner et al., 2008) is consistent with it playing a key role in linking regulation of spine morphology with plasticity of receptor number and pattern in the PSD during this critical window of plasticity.
The miRNA-based molecular approach to reduce the expression of all three Shank family members simultaneously proved to be a powerful method to unveil critical functions of a protein family, as was shown previously for the neuroligin family of adhesion molecules (Shipman et al., 2011). This approach not only enables the study of protein families or mechanistically related proteins, but can also be used to reveal isoform-specific functions by comparing the effects of re-expressing individual members. Furthermore, as splice variants may give rise to further diversification of the Shank family (Lim et al., 1999; Jiang & Ehlers, 2013; Wang et al., 2014), it would be of interest to test how specific isoforms determine the organization and function of the synapse. For instance, short Shank3 isoforms have been described that lack the N-terminal domains ANK and SH3 domains, and such isoforms might provide a means to titrate the number of binding sites for specific factors at the synapse. Moreover, the growing evidence that de novo genetic mutations in synaptic proteins (De Rubeis et al., 2014; Fromer et al., 2014; Iossifov et al., 2014), most notably the Shank family (Jiang & Ehlers, 2013; Leblond et al., 2014), might underlie the development of psychiatric disorders such as intellectual disability and autism urge for a more thorough understanding of the family of Shank proteins at excitatory synapses.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Blanpied Lab, Casper Hoogenraad and Amélie Fréal for helpful discussions and critical evaluation of the manuscript, and Tamar Davis and Minerva Contreras for excellent technical support. This work was supported by the National Institutes of Health (NIH: MH080046 and MH096376), the Dana Foundation, the Broad Foundation, and the Katherine D. and Theodore J. Carski Fund to T.A.B.; and the Netherlands Organization for Scientific Research (NWO-ALW-VENI) and the Federation of European Biochemical Societies (FEBS Return-to-Europe Fellowship) to H.D.M.
Abbreviations
- Abp1
actin-binding protein-1
- AMPAR
amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- Arp2/3
actin-related protein-2/3
- CV
coefficient of variance
- DIV
days in vitro
- DMSO
dimethylsulphoxide
- EB
extracellular buffer
- EF
elliptical buffer
- GFP
green fluorescent protein
- Gly
glycine
- LatA
Latrunculin A
- LTP
long-term potentiation
- MSD
mean squared displacement
- NGS
normal goat serum
- NMDAR
N-methyl-d-aspartate receptor
- N-WASP
neural WASP
- PALM
photoactivated localization microscopy
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PSD
postsynaptic density
- RT
room temperature
- WASP
Wiskott–Aldrich syndrome protein
- WAVE
WASP family verprolin homologous protein
Footnotes
Supporting Information
Additional supporting information can be found in the online version of this article:
Fig. S1. COS7 cells were transfected with rat Shank1-GFP (left panels), rat Shank2-GFP (middle panels) or rat Shank3-GFP (right panels), fixed and stained with anti-Shank1 (top row), anti-Shank2 (middle row) or anti-Shank3 (bottom row). Note that each of the antibodies only recognizes the correct Shank isoform with no observable cross-reactivity.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- Allison DW, Chervin AS, Gelfand VI, Craig AM. Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: maintenance of core components independent of actin filaments and microtubules. J. Neurosci. 2000;20:4545–4554. doi: 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, Scherer SW, Sprengel R, Schratt G, Rappold GA. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum. Mol. Genet. 2012;21:344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied T, Kerr J, Ehlers M. Structural plasticity with preserved topology in the postsynaptic protein network. Proc. Natl. Acad. Sci. USA. 2008a;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Kerr JM, Ehlers MD. Structural plasticity with preserved topology in the postsynaptic protein network. Proc. Natl. Acad. Sci. USA. 2008b;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F, Richter D, Gundelfinger ED, Kreienkamp HJ. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alphafodrin. J. Biol. Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Liedtke T, Spilker C, Dresbach T, Bockmann J, Kreutz MR, Gundelfinger ED. C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J. Neurochem. 2005;92:519–524. doi: 10.1111/j.1471-4159.2004.02910.x. [DOI] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr. Opin. Neurobiol. 2012;22:383–388. doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, Choquet D, Gautreau A, Sibarita JB, Giannone G. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 2014;33:2745–2764. doi: 10.15252/embj.201488837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YK, Hsueh YP. Cortactin-binding protein 2 modulates the mobility of cortactin and regulates dendritic spine formation and maintenance. J. Neurosci. 2012;32:1043–1055. doi: 10.1523/JNEUROSCI.4405-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Fu SC, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Crooks L, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Sean Hill R, Ionita-Laza I, Jimenez Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei J, Lehtimaki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Wang LS, Weiss LA, Jeremy Willsey A, Yu TW, Yuen RK, Study DDD, Homozygosity Mapping Collaborative for, A. Consortium UK, Autism Sequencing C, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barrett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol. Cell. Biol. 1998;18:5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Yonekubo J, Zeng Y, Osisami M, Frohman MA. Design of expression vectors for RNA interference based on miRNAs and RNA splicing. FEBS J. 2006;273:5421–5427. doi: 10.1111/j.1742-4658.2006.05534.x. [DOI] [PubMed] [Google Scholar]
- Duffney LJ, Wei J, Cheng J, Liu W, Smith KR, Kittler JT, Yan Z. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. J. Neurosci. 2013;33:15767–15778. doi: 10.1523/JNEUROSCI.1175-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, Ma K, Dietz DM, Kajiwara Y, Buxbaum JD, Yan Z. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–1413. doi: 10.1016/j.celrep.2015.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol. Psychiatr. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifkova E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J. Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freche D, Pannasch U, Rouach N, Holcman D. Synapse geometry and receptor dynamics modulate synaptic strength. PLoS One. 2011;6:e25122. doi: 10.1371/journal.pone.0025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O’Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Kerr JM, Lu HE, Blanpied TA. A network of networks: cytoskeletal control of compartmentalized function within dendritic spines. Curr. Opin. Neurobiol. 2010a;20:578–587. doi: 10.1016/j.conb.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single- molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010b;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Lu HE, Blanpied TA. Optimization of cell morphology measurement via single-molecule tracking PALM. PLoS One. 2012;7:e36751. doi: 10.1371/journal.pone.0036751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, MacGillavry HD, Lu HE, Blanpied TA. Live-cell PALM of intracellular proteins in neurons. Nanoscale Imaging of Synapses. 2013:93–123. [Google Scholar]
- Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Knight MJ, Proepper C, Bockmann J, Joubert M, Rowan M, Nienhaus GU, Garner CC, Bowie JU, Kreutz MR, Gundelfinger ED, Boeckers TM. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011;30:569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel A, Ahuja R, Gundelfinger ED, Qualmann B, Kessels MM. The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J. Neurosci. 2008;28:10031–10044. doi: 10.1523/JNEUROSCI.0336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Holder JL, Jr, Schaaf CP, Lu H, Chen H, Kang H, Tang J, Wu Z, Hao S, Cheung SW, Yu P, Sun H, Breman AM, Patel A, Lu HC, Zoghbi HY. SHANK3 overexpression causes maniclike behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SP, Gambin Y, Gomez GA, Verma S, Giles N, Michael M, Wu SK, Guo Z, Johnston W, Sierecki E, Parton RG, Alexandrov K, Yap AS. Cortactin scaffolds Arp2/3 and WAVE2 at the epithelial zonula adherens. J. Biol. Chem. 2014;289:7764–7775. doi: 10.1074/jbc.M113.544478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Nolen BJ. Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. Elife. 2013;2:e00884. doi: 10.7554/eLife.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J. Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Iki J, Inoue A, Bito H, Okabe S. Bi-directional regulation of postsynaptic cortactin distribution by BDNF and NMDA receptor activity. Eur. J. Neurosci. 2005;22:2985–2994. doi: 10.1111/j.1460-9568.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JM, Blanpied TA. Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J. Neurosci. 2012;32:658–673. doi: 10.1523/JNEUROSCI.2927-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Engqvist-Goldstein AE, Drubin DG. Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell. 2000;11:393–412. doi: 10.1091/mbc.11.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Kuriu T, Inoue A, Bito H, Sobue K, Okabe S. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J. Neurosci. 2006;26:7693–7706. doi: 10.1523/JNEUROSCI.0522-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, Giuliano F, Stordeur C, Depienne C, Mouzat K, Pinto D, Howe J, Lemiere N, Durand CM, Guibert J, Ey E, Toro R, Peyre H, Mathieu A, Amsellem F, Rastam M, Gillberg IC, Rappold GA, Holt R, Monaco AP, Maestrini E, Galan P, Heron D, Jacquette A, Afenjar A, Rastetter A, Brice A, Devillard F, Assouline B, Laffargue F, Lespinasse J, Chiesa J, Rivier F, Bonneau D, Regnault B, Zelenika D, Delepine M, Lathrop M, Sanlaville D, Schluth-Bolard C, Edery P, Perrin L, Tabet AC, Schmeisser MJ, Boeckers TM, Coleman M, Sato D, Szatmari P, Scherer SW, Rouleau GA, Betancur C, Leboyer M, Gillberg C, Delorme R, Bourgeron T. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl. Acad Sci USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J. Biol. Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Lu HE, MacGillavry HD, Frost NA, Blanpied TA. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 2014;34:7600–7610. doi: 10.1523/JNEUROSCI.4364-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Kerr JM, Blanpied TA. Lateral organization of the postsynaptic density. Mol. Cell Neurosci. 2011;48:321–331. doi: 10.1016/j.mcn.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJ, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Nair D, Hosy E, Petersen JD, Constals A, Giannone G, Choquet D, Sibarita JB. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 2013;33:13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu J, Xiao B, Sala C, Valtschanoff J, Weinberg R, Worley P, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 2003;278:19220–19229. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proepper C, Johannsen S, Liebau S, Dahl J, Vaida B, Bockmann J, Kreutz MR, Gundelfinger ED, Boeckers TM. Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. EMBO J. 2007;26:1397–1409. doi: 10.1038/sj.emboj.7601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J. Neurosci. 2004;24:2481–2495. doi: 10.1523/JNEUROSCI.5479-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. The subcellular organization of cortactin in hippocampus. J. Neurosci. 2004;24:10310–10317. doi: 10.1523/JNEUROSCI.2080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich- Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing P, Real E, Siksou L, Lechaire JP, Boudier T, Boeckers TM, Gertler F, Gundelfinger ED, Triller A, Marty S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur. J. Neurosci. 2006;24:3463–3474. doi: 10.1111/j.1460-9568.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Sala C, Roussignol G, Meldolesi J, Fagni L. Key role of the postsynaptic density scaffold proteins Shank and Homer in the functional architecture of Ca2+ homeostasis at dendritic spines in hippocampal neurons. J. Neurosci. 2005;25:4587–4592. doi: 10.1523/JNEUROSCI.4822-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen AL, Udvardi PT, Shiban E, Spilker C, Balschun D, Skryabin BV, Dieck S, Smalla KH, Montag D, Leblond CS, Faure P, Torquet N, Le Sourd AM, Toro R, Grabrucker AM, Shoichet SA, Schmitz D, Kreutz MR, Bourgeron T, Gundelfinger ED, Boeckers TM. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Seese RR, Babayan AH, Katz AM, Cox CD, Lauterborn JC, Lynch G, Gall CM. LTP induction translocates cortactin at distant synapses in wild-type but not Fmr1 knock-out mice. J. Neurosci. 2012;32:7403–7413. doi: 10.1523/JNEUROSCI.0968-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman SL, Schnell E, Hirai T, Chen BS, Roche KW, Nicoll RA. Functional dependence of neuroligin on a new non-PDZ intracellular domain. Nat. Neurosci. 2011;14:718–726. doi: 10.1038/nn.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P, Higley MJ, Xu W, Czervionke BL, Malenka RC, Sabatini BL. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron. 2008;60:788–802. doi: 10.1016/j.neuron.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R, Larson D, Webb W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- Valtschanoff JG, Weinberg RJ. Laminar organization of the NMDA receptor complex within the postsynaptic density. J. Neurosci. 2001;21:1211–1217. doi: 10.1523/JNEUROSCI.21-04-01211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Han SP, Michael M, Gomez GA, Yang Z, Teasdale RD, Ratheesh A, Kovacs EM, Ali RG, Yap AS. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 2012;23:4601–4610. doi: 10.1091/mbc.E12-08-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Bockers TM, Dityatev7 A, Sala C. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J. Biol. Chem. 2011;286:34839–34850. doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Mol Autism. 2014;5:30. doi: 10.1186/2040-2392-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Xiao M, Nicoll RA. Contribution of cytoskeleton to the internalization of AMPA receptors. Proc. Natl. Acad. Sci. USA. 2001;98:1261–1266. doi: 10.1073/pnas.031573798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.