Abstract
Defects in organelle dynamics underlie a number of human degenerative disorders, and whole exome sequencing (WES) is a powerful tool for studying genetic changes that affect the cellular machinery. WES may uncover variants of unknown significance (VUS) that require functional validation. Previously, a pathogenic de novo variant in the middle domain of DNM1L (p.A395D) was identified in a single patient with a lethal defect of mitochondrial and peroxisomal fission. We identified two additional patients with infantile encephalopathy and partially overlapping clinical features, each with a novel VUS in the middle domain of DNM1L (p.G350R and p.E379K). To evaluate pathogenicity, we generated transgenic Drosophila expressing wild-type or variant DNM1L. We find that human wild-type DNM1L rescues the lethality as well as specific phenotypes associated with the loss of Drp1 in Drosophila. Neither the p.A395D variant nor the novel variant p.G350R rescue lethality or other phenotypes. Moreover, overexpression of p.A395D and p.G350R in Drosophila neurons, salivary gland and muscle strikingly altered peroxisomal and mitochondrial morphology. In contrast, the other novel variant (p.E379K) rescued lethality and did not affect organelle morphology, although it was associated with a subtle mitochondrial trafficking defect in an in vivo assay. Interestingly, the patient with the p.E379K variant also has a de novo VUS in pyruvate dehydrogenase 1 (PDHA1) affecting the same amino acid (G150) as another case of PDHA1 deficiency suggesting the PDHA1 variant may be pathogenic. In summary, detailed clinical evaluation and WES with functional studies in Drosophila can distinguish different functional consequences of newly-described DNM1L alleles.
Introduction
Mitochondrial diseases are a clinically and genetically heterogeneous group of disorders characterized by defects in mitochondrial function. Diagnosis of mitochondrial disease can be difficult because of phenotypic variability including lactic acidosis, epilepsy, muscle weakness, deafness, optic atrophy and encephalopathy, each of which may be variably present (1–5). These disorders may be inherited in a mitochondrial, X-linked, recessive or dominant manner, or arise de novo. Because over 100 genes in both mitochondrial and nuclear DNA have been associated with mitochondrial disease, the identification of the responsible gene can be challenging (6,7). In addition, the interplay between mitochondria and other organelles makes analysis of cases of possible mitochondrial diseases even more difficult. For example, a number of mitochondrial phenotypes can be observed in patients with peroxisomal disorders such as peroxisomal biogenesis disorders (8).
Given the phenotypic and genetic heterogeneity of mitochondrial and peroxisomal diseases, whole exome sequencing (WES) has emerged as a diagnostic modality for the diagnosis of mitochondrial disorders (7,9). WES is a powerful tool for the diagnosis of genetic disease (10–14). However, WES may uncover novel sequence alterations and variants of unknown significance (VUS) (15) which cannot be categorized as benign or pathogenic due to lack of functional evidence. Even with careful phenotyping, identification of the responsible gene can be even more challenging in patients with two or more potential causative genes. In one series, approximately 5% of individuals referred for WES had two molecular diagnoses (14). In a series of 53 patients with clinically diagnosed mitochondrial disease, several patients had two or more VUS that could not be categorized based on lack of functional data (7).
One method to determine whether a variant is pathogenic is through functional validation in model organisms (16) such as Drosophila (17). Drosophila offer the advantage of diversity and availability of many reagents for genetic manipulation, short generation time compared with mammalian models, and conservation of many human genes (18). Drosophila functional studies of human VUS can shed light on the functional significance of a single VUS or more than one VUS giving rise to a blended phenotype (19).
DNM1L (Dynamin 1-like, synonyms Drp1) is a member of the dynamin superfamily of GTPases and mediates mitochondrial and peroxisomal fission (20–22). One patient has been previously described with a lethal encephalopathy due to defective mitochondrial and peroxisomal fission (MIM #614388). This individual had poor feeding, poor growth, lactic acidosis, seizures, hypotonia, nystagmus, and an abnormal gyral pattern on magnetic resonance imaging (MRI), and passed away at 37 days of life (23). Analysis of very long chain fatty acids revealed elevated cerotic acid, suggesting peroxisomal dysfunction. The patient's fibroblasts exhibited a decreased number of peroxisomes and dysmorphic mitochondria. Sequencing of DNM1L revealed a c.1184C>A, p. A395D variant.
Mice which lack Dlp1, the homologue of DNM1L, die at embryonic day E12.5, indicating a crucial role for this gene in mammalian development and these mice display abnormal mitochondrial and peroxisomal morphology (24). Drosophila Drp1 mutants were identified in a screen for synaptic transmission mutants (25). Fly Drp1 mutants display altered cellular distribution of mitochondria in the nervous system leading to a near absence of mitochondria from synapses and exhibit defects in mitochondrial morphology and synaptic transmission (26,27). Given the evidence across species, DNM1L is clearly a candidate gene for encephalopathy, but thus far, only one case has been reported (23). Here, we report two additional heterozygous missense variants in DNM1L and we use Drosophila to understand the function of these alleles.
Results
Clinical phenotypes
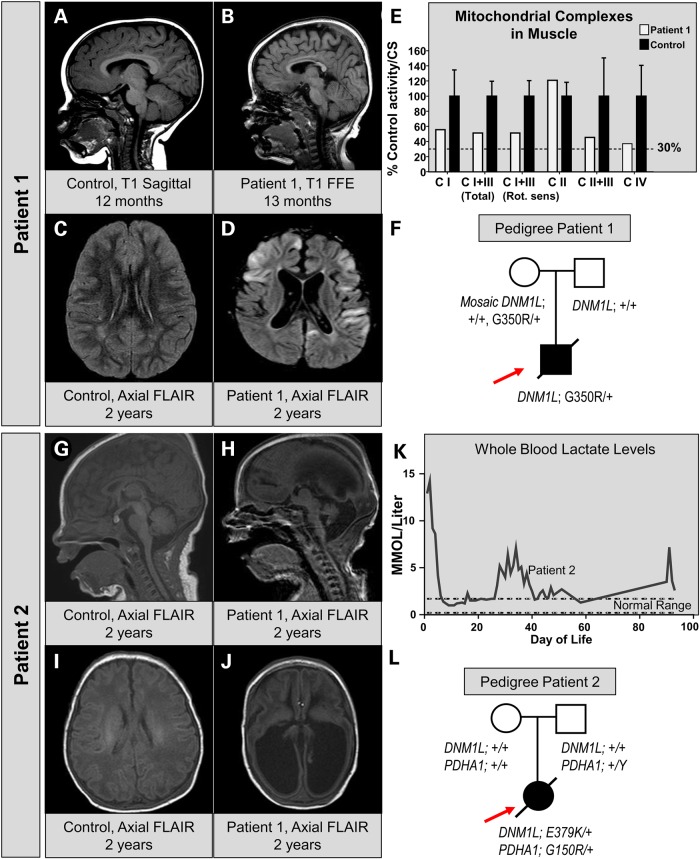
We clinically identified two patients with lactic acidosis, poor feeding, poor growth, developmental delay, and hypotonia. We initially identified Patient 1, a 14 month-old male with global developmental delay (GDD), hypotonia and status epilepticus. He was born at term and had normal development until 5 months of age, when he developed seizures and developmental regression. MRI of the brain revealed a progressive volume loss and demyelination (Fig. 1). At 13 months, MRI showed cerebral volume loss and thinning of the corpus callosum (Fig. 1B versus control in Fig. 1A). By 2 years of age, he had evidence of T2 hyperintense regions in the cortex as well as progressive volume loss (Fig. 1D versus control in Fig. 1C). Though serum lactate levels were initially normal, they were elevated at 4 years of life. A muscle biopsy was performed and respiratory chain enzyme activities were nominally reduced for mitochondrial complexes I, III and IV but did not meet modified Walker criteria (Fig. 1E) (28). Electron microscopy of the muscle revealed mitochondrial pleomorphism as well as some lipid accumulation in the muscle (Supplemental Fig. S1A–C). Plasma very long chain fatty acid levels were normal (Supplemental Fig. S1D and E). The patient passed away at age 5 due to severe status epilepticus with respiratory failure. WES revealed a VUS in the DNM1L gene: c.1048G>A, p.G350R. His father did not exhibit this change and variant analysis of Sanger reads from the maternal blood suggested a low-level (6–8%) maternal mosaicism (Fig. 1F). In addition, this patient had a variant in ALG13 inherited from the mother, but this variant seemed unlikely to be pathogenic given the phenotype and an N-glycan analysis was normal. A full list of candidate genes is shown in Supplementary Material, Table S1.
Figure 1.
Clinical, neuroradiographic and molecular features of two patients with infantile encephalopathy and DNM1L variants. (A) MRI of a control patient at 12 months of age showing normal sagittal T1-weighted images. (B) MRI of Patient 1 at 13 months of age reveals hypoplasia of the corpus callosum and a simplified gyral pattern on T1 sagittal images. (C) MRI of control patient for suspected seizure showing a normal axial FLAIR sequence. (D) Axial FLAIR of Patient 1 at 2 years of age showing multi-focal areas of cortical signal abnormality with swelling and diffusion restriction. Diffuse cortical volume loss is also present. (E) Respiratory chain enzyme activities in muscle. The % control activity for Patient 1 (white bars) is shown for NADH:Ferricyanide dehydrogenase (CI), NADH:cytochrome C reductase total (CI+III), NADH:cytochrome C reductase Rotenone sensitive (CI+III), Succinate dehydrogenase (CII) and Cytochrome C oxidase (CIV). Values were normalized to citrate synthase activity. (F) Pedigree of the patient showing the WES data. Sanger verification was performed and low-level (6–8%) mosaicism was detected in the maternal sample. (G and I) A sagittal MRI in a newborn showing a normal T1 and FLAIR appearance. (H and J) MRI of Patient 2 at 4 days of age reveals ventriculomegaly, absence of the corpus callosum, volume loss and gyral simplification on T1 and FLAIR. (K) Whole blood lactate levels measured in a clinical lab over the time course of Patient 2's medical care at our institution. (L) Pedigree of Patient 2 showing the presence of two independent de novo events, one in DNM1L and one in PDHA1.
Subsequently, Patient 2 was identified at 4 days of life for a persistent lactic acidosis. She was born at 37 3/7 weeks and pregnancy was complicated by intrauterine growth restriction and hydrocephalus. At delivery, she had poor tone and apnea. She was intubated and found to have metabolic acidosis with an elevated lactate. She was placed on thiamine and a ketogenic diet. MRI of the brain showed microcephaly, absence of the corpus callosum and diffuse volume loss with enlarged ventricles (Fig. 1G–J). At 3 months of age, a ventriculoperitoneal shunt was placed for hydrocephalus. She had diffuse hypotonia, global developmental delay, and poor growth. Her parents declined muscle biopsy to evaluate electron transport chain activity. There was persistent elevation of lactate in whole blood (Fig. 1K). The patient passed away at 10 months of age due to pneumonia. WES revealed two de novo changes in mitochondria-related genes, namely a VUS in the PDHA1 gene (c.448G>A, p.G150R), known to be associated with pyruvate dehydrogenase E1 deficiency as well as a VUS in the DNM1L gene (c.1135G>A, p.E379K) (Fig. 1L). This raised the possibility that one or both of these variants were contributing to the patient's phenotype. A full list of candidates is shown in Supplementary Material, Table S1.
The clinical characteristics of the previously reported patient with the p.A395D variant were compared to the patients reported in this study (Table 1) (23). All three cases share the features of hypotonia, poor feeding, developmental delay, and shortened life span. However, while in the previous case, cerotic acid was elevated, Patient 1 did not exhibit elevations in very long chain fatty acids (VLCFAs), whereas Patient 2 did not have a plasma VLCFA analysis. Both the previous case and Patient 2 showed congenital lactic acidosis, while Patient 1 did not have lactic acidosis until 4 years of life. Given the variability of the phenotypes, it was not clear whether the phenotypes seen in Patient 1 and Patient 2 were due to the pathogenic variants in DNM1L.
Table 1.
Clinical features of the patients in comparison to the previous DNM1L report
| Sequencing findings | Case reported by: Waterham et al. (23) |
Patient 1 | Patient 2 |
|---|---|---|---|
| DNM1L: c.1184C>A (p.A395D) | DNM1L: c.1048G>A (p.G350R) |
DNM1L: c.1135G>A (p.E379K) PDHA 1 c.448G>A (p.G150R) |
|
| Decreased fetal movements | + | − | − |
| Poor feeding | + | + | + |
| Poor growth | + | − | + |
| Developmental delay | + | + | + |
| Developmental regression | N/A | + | − |
| Lactic acidosis | + | + | + |
| Seizures | − | + | − |
| Hypotonia | + | + | + |
| Deep-set eyes | + | − | − |
| Pointed chin | + | − | − |
| Downslanting palpebral fissures | − | − | + |
| Microcephaly | + | − | + |
| Horizontal nystagmus | + | + | − |
| No response to light stimulation | + | − | − |
| Optic discs pale and cupped | + | − | − |
| Optic nerve hypoplasia | − | − | + |
| MRI: abnormal gyral pattern in frontal lobes | + | − | + |
| MRI: dysmyelination | + | − | + |
| Corpus callosum abnormality | − | Thinned | Agenesis |
| Ventriculomegaly | − | + | + |
| Hydrocephalus | − | − | + |
| Very long chain fatty acids | Elevated | Normal | Unknown |
| Age of onset | 4 days | 5 months | Birth |
| Survival | 37 days | 5 years | 11 months |
Missense variants in the middle domain of DNM1L
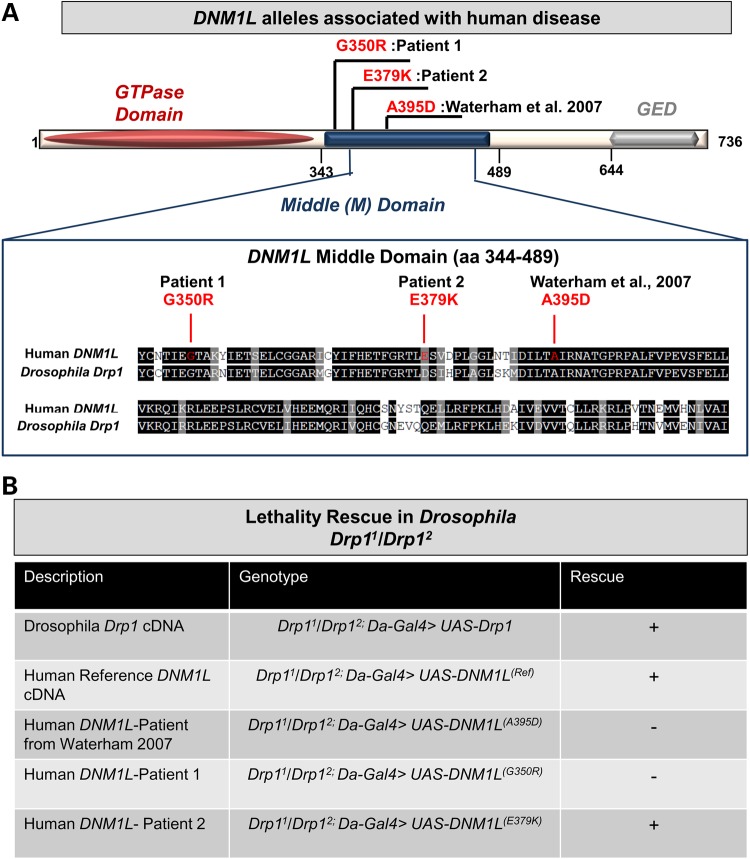
DNM1L encodes a GTPase with an N-terminal GTPase domain, a C-terminal GED domain and a middle (M) domain (Fig. 2A). The middle domain of DNM1L has previously been shown to be important for the tetramerization of DNM1L protein, as missense variations of conserved residues including p.A395 and p.G350, lead to elongated mitochondria in HeLa cells (21). Interestingly, the p.G350D variant was selected for structure-function studies prior to the identification of our patient due to conservation of the amino acid at that position (21). However, Patient 1 had a different missense substitution affecting this amino acid (p.G350R). The glycine at this position is highly conserved and in yeast this residue (G385) is required for the formation of mitochondrial fission complexes, and self-assembly is defective in G385D point mutants (29,30).
Figure 2.
Genetic and domain associations for the DNM1L alleles. (A) DNM1L encodes a protein with an N-terminal GTPase domain, a middle domain and a C-terminal GTPase effector domain (GED). The three variants are missense alleles in the middle domain. Alignment shows homology between human DNM1L and Drosophila Drp1, red lines indicate the three human variants, while the A395D and G350R variants occur in highly conserved residues, the E379K is not conserved. (B) Lethality rescue experiments in Drosophila with human DNM1L alleles. The genotypes are listed as second chromosome; third chromosome. Rescue of lethality is indicated by (+).
All three variants in the DNM1L gene are located in the middle domain of the protein in a highly conserved region, although the E379K is not a conserved amino acid (Fig. 2A). Missense substitutions in this region have been shown to exhibit dominant-negative effects in vitro due to the middle domain's role in oligomerization (21). This region is also of interest because it shares homology with dynamins (Supplementary Material, Fig. S2A). Though it shares significant homology with other dynamin genes in the genome (DNM1, DNM2 and DNM3), DNM1L is distinct because of its functional role in organelle fission. Variants within the middle domain of dynamin proteins can result in very different phenotypes. For example, the DNM2 gene is associated with both centronuclear myopathy (MIM 160150) and Charcot-Marie Tooth disease type 2M (MIM 606482). Interestingly, missense variants in the middle domain of DNM2 are thought to be associated with centronuclear myopathy rather than CMT. However, we recently reported two cases of CMT rather than centronuclear myopathy with middle domain variants (19) (Supplementary Material, Fig. S2A), one of which had been observed previously (31). In addition, the allele frequencies of heterozygous missense variants in the exome aggregation consortium (ExAC) and EVS databases suggest the possibility of selection against missense variants in the DNM1L middle domain (Supplementary Material, Fig. S2B). Importantly, neither the p.G350R nor the p.E379K variant was observed in ExAC. Based on the observations that missense variants affecting the DNM1L middle domain are rare, and that pathogenic variants in very similar proteins, namely dynamins, underlie a spectrum of neurologic disease, we hypothesized that these variants were pathogenic in our patients.
Human DNM1L rescues Drosophila Drp1 mutants
We therefore undertook a functional study of DNM1L in Drosophila melanogaster. Drosophila Drp1 mutants are lethal with defects in mitochondrial trafficking to synapses, mitochondrial morphology and synaptic transmission (27). The Drp11 and Drp12 alleles are Ethyl-Methane Sulfonate (EMS) induced point mutations, which are lethal. Transheterozygous Drp11/Drp12 are larval lethal with mitochondrial trafficking defects (27,32). We generated transgenic flies carrying the human DNM1L gene with and without the three human variants. Because Drosophila Drp1 is the closest homolog of DNM1L, we first crossed the transgenes into Drp1 backgrounds to determine if the human reference sequence DNM1L(Ref) construct was able to rescue a Drp1 fly mutant. By expressing human DNM1L(Ref) ubiquitously with Da-Gal4, we rescued the lethality of Drp1 (Drp11/Drp12) mutants (Fig. 2B). However, the DNM1L(A395D) from the previously reported case as well as DNM1L(G350R) observed in WES from Patient 1 did not rescue lethality (Fig. 2B). In addition, both of these alleles exhibited some toxicity on a sensitized (Drp11/+) and in a wild-type Drosophila background (Supplementary Material, Fig. S2C). In contrast, the DNM1L(E379K) variant was able to rescue lethality (Fig. 2B) and did not exhibit toxicity with overexpression (Supplementary Material, Fig. S2C).
DNM1L variants and dominant-negative effects on organelle fission
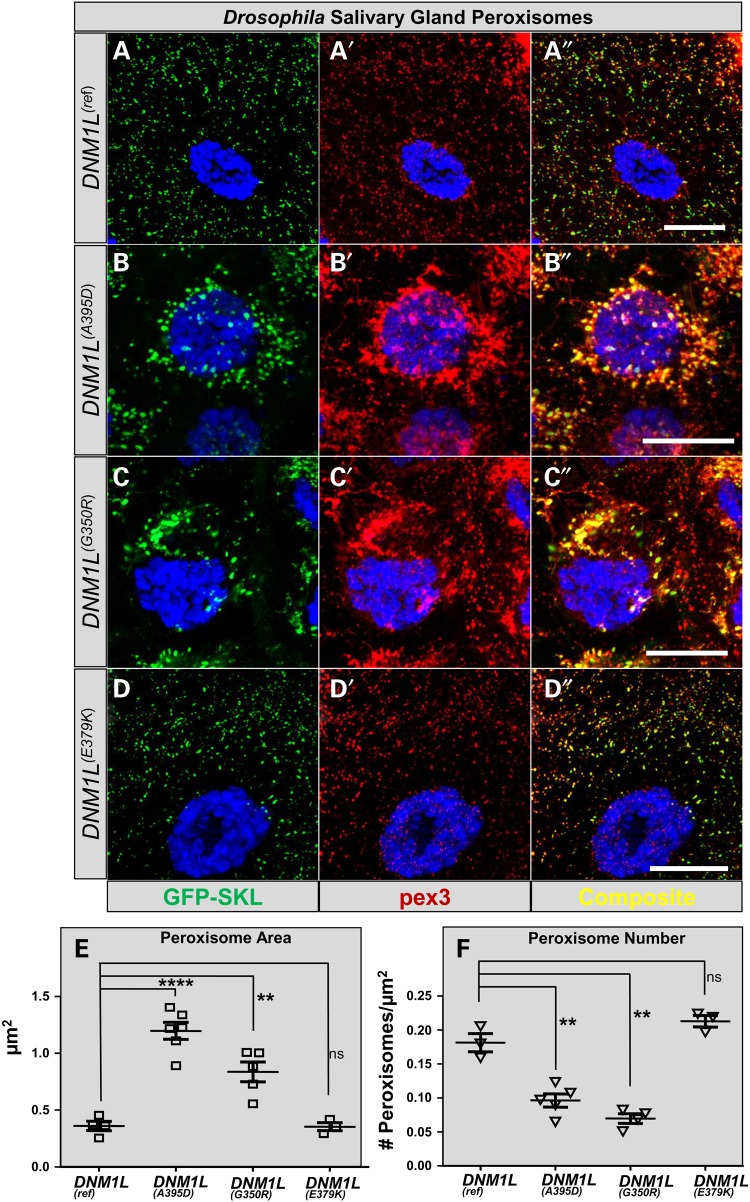
The DNM1L protein is part of the machinery that allows both mitochondria and peroxisomes to undergo fission. We therefore examined peroxisomal morphology in third instar larval salivary glands by over-expressing the DNM1L constructs in the presence of a peroxisomal GFP-SKL marker (ActinGal4>UAS-GFP-SKL) and anti-Pex3 antibody (33) (Fig. 3). Given studies showing p.A395D overexpression can recapitulate peroxisomal phenotypes resulting from DNM1L loss (23), we determined if the VUS in our cases had similar effects. Overexpression of DNM1L(Ref) has no effect on peroxisomal morphology, as the salivary gland peroxisomes are approximately 0.3 µm2 (Fig. 3A–A″ and E). In contrast, expression of the DNM1L(A395D) and DNM1L(G350R) both led to dramatic increase in peroxisomal size and altered cellular distribution (Fig. 3B, C″ and E). However, the DNM1L(E379K) had no effect on peroxisomal size (Fig. 3D-D″ and E). Increased peroxisomal size with DNM1L(A395D) and DNM1L(G350R) was associated with a decreased number of total peroxisomes per cell (Fig. 3F). The results were similar on a Drp1/+ (sensitized) background (data not shown). Therefore, the p.G350R variant in Patient 1 exhibits a strong dominant negative effect on peroxisomal morphology similar to p.A395D. However, the p.E379K variant in Patient 2 did not cause any obvious peroxisomal effects.
Figure 3.
Dominant effects of DNM1L expression on Drosophila salivary gland peroxisomes. (A–A″) Drosophila salivary gland cells are shown with expression of the reference DNM1L with Actin-Gal4 alongside UAS-EGFP-SKL (courtesy of Hamed Jefar-Nejad) and co-stained with Drosophila anti-Pex3 antibody produces salivary gland cells that are indistinguishable from GFP-SKL expression alone (data not shown). (B–B″) The p.A395D construct (seen in Waterham et al. (23)) produces enlarged peroxisomes with abnormal distribution. Fewer peroxisomes are apparent in the cell. (C–C″) The p.G350R construct (seen in Patient 1) produces enlarged peroxisomes with abnormal distribution; fewer peroxisomes are apparent in the cell, a phenotype similar to pA395D. (D–D″) The p.E379K construct (Patient 2) does not appear to have a dramatic effect on peroxisomal size compared with DNM1L(Ref). (E) Quantification of the peroxisomal area per peroxisome. In cells expressing the DNM1L(Ref) peroxisomes have an average size of 0.3 µm2. (F) Quantification of peroxisomal number per µm2 of cytoplasm. Error bars = 25 µm2.
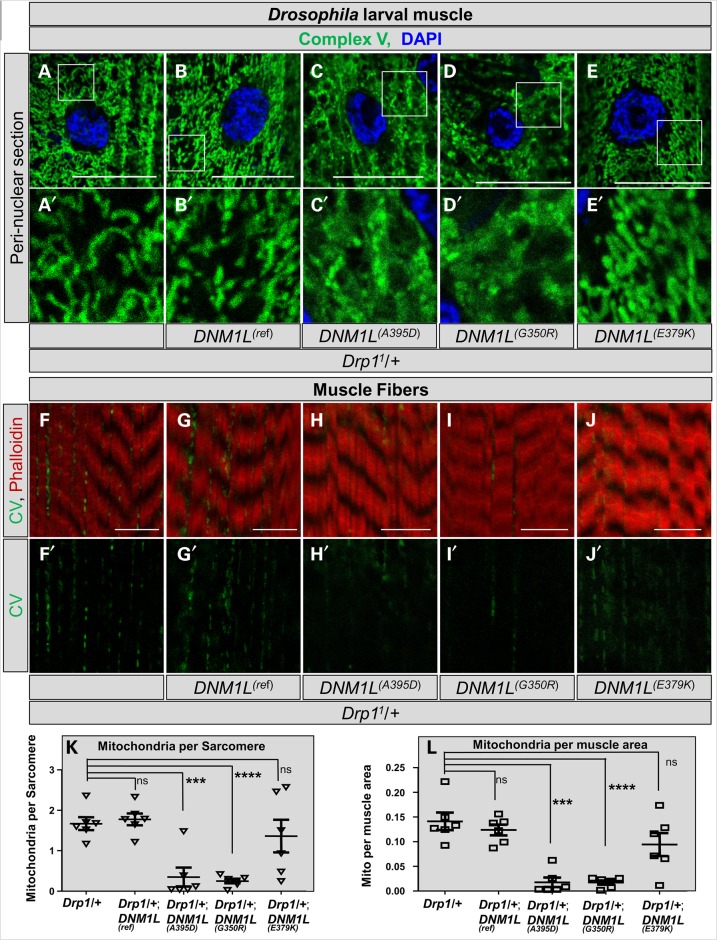
Next, we examined mitochondria in muscle of third-instar larvae in a sensitized genetic background (Drp11/+) by driving expression with MEF2-Gal4. Again, we observed a remarkable alteration in morphology of muscle mitochondria with DNM1L(A395D) and DNM1L(G350R), but not DNM1L(E379K) compared with DNM1L(Ref) (Fig. 4A–E). The mitochondrial distribution in muscle was also altered. Mitochondria are normally seen intercalating between muscle fibers (Fig. 4F–F′). In contrast, there was a paucity of mitochondria between sarcomeres in muscle and reduced mitochondrial numbers and size in both the Drp11/+;DNM1L(A395D) and Drp11/+;DNM1L(G350R) larvae, but not the Drp11/+; DNM1L(E379K) larvae when compared to Drp11/+;DNM1L(Ref) (Fig. 4F–M). We also observed strong effects on mitochondrial morphology with DNM1L(A395D) and DNM1L(G350R) when overexpressed in a wild-type background, suggesting this is a dominant-negative effect (data not shown).
Figure 4.
Drosophila larval muscle mitochondrial size and number. (A–A′) Control third instar larval muscle stained with Complex V and DAPI and imaged at the level of the nucleus. Control mitochondria have clear separation and fibrillar morphology. (B–B′) Mitochondria from DNM1L(Ref) expressed by MEF2-Gal4, a muscle-specific enhancer, showing normal morphology. (C–C′, D–D′) Mitochondria from DNM1L(A395D) and DNM1L(G350R) expressed by MEF2-Gal4 showing mats of inter-connected mitochondria. (E–E′) Mitochondria from DNM1L(E379K) expressed by MEF2-Gal4 showing clear normal morphology. (F–F′) Control third instar larval muscle stained with Complex V and Phalloidin and imaged at the level of the muscle fibers. Control mitochondria are clearly present intercalating between the fibers. (G–G′) Mitochondria from DNM1L(Ref) intercalate between muscle fibers. (H–H′, I–I′) DNM1L(A395D) and DNM1L(G350R) show near absence of mitochondria. (J–J′) DNM1LE379K) shows normal numbers of mitochondria. (K–K′) Quantification of number of mitochondria per sarcomere from experiments shown in (F)–(J). (K–K′) Quantification of mitochondria per muscle area from experiments shown in (F)–(J).
DNM1L variants and mitochondrial trafficking
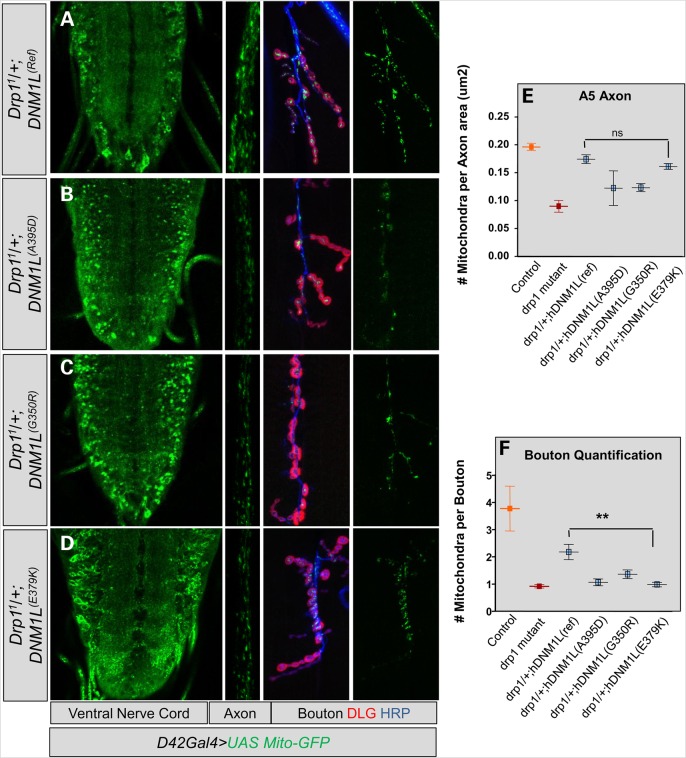
Drosophila Drp1 mutants exhibit altered distribution of mitochondria in the nervous system (27,32). We examined this phenotype for the human VUS on a sensitized background (Drp11/+). The results were similar to overexpression on a wild-type background (data not shown). We again noted altered mitochondrial trafficking in the ventral nerve cord, axons and synaptic boutons of Drp11/+;DNM1L(A395D) and Drp11/+;DNM1L(G350R) larvae (Fig. 5A–C). In addition, while Drp11/+;DNM1L(E379K) larvae appeared to have normal mitochondrial trafficking in the VNC and in the axon, we observed a clear trafficking defect at the level of the bouton in the Drp11/+;DNM1L(E379K) larvae which was statistically significant and consistent with that seen with the other two variants (Fig. 5D and F). Therefore, the trafficking experiments suggest that while the DNM1L(E379K) variant does not exhibit a strong dominant-negative effects on organelle morphology in our assay the DNM1L(E379K) larvae do exhibit trafficking defects.
Figure 5.
Effect of DNM1L variants on mitochondrial brain trafficking. (A) Left, Drosophila larval brain ventral nerve cord on with normal mitochondrial distribution. DNM1L(Ref) was expressed by D42-Gal4 in a sensitized background (Drp11/+). Center, mitochondrial trafficking in the axon at the A5 abdominal segment. Right, mitochondria in the synaptic bouton counterstained with HRP (blue) and Discs Large (DLG, red). (B and C) Expression of DNM1L(A395D) and DNM1L(G350R) dramatically alter distribution of mitochondria with a defect in trafficking mitochondria. (D) A defect in mitochondrial trafficking observed in the synaptic boutons of DNM1L(E379K)-expressing larvae. (E) Quantification of number of mitochondria per axon area for A5 axonal segments for the experiment shown in (A)–(D). (F) Quantification of number of mitochondrial per synaptic bouton for the experiment shown in (A)–(D).
Discussion
Here we report two cases with encephalopathy and missense mutations in DNM1L, the gene underlying the lethal encephalopathy phenotype (MIM 614388) noted in one previous case. Because of the known role for DNM1L in peroxisomal and mitochondrial fission and the Drp1 mutant phenotypes in Drosophila we studied the effect of these variants on organelle fission and trafficking. Our Drosophila studies suggest a dominant negative effect on peroxisomal and mitochondrial morphology for the p.A395D allele reported by Waterham et al. (23) and the p.G350R variant observed in Patient 1. The E379K allele observed in Patient 2 was able to rescue lethality of Drosophila Drp1 mutants and did not cause obvious organelle morphology defects. This finding is relevant to the clinical phenotype of Patient 2 as she had two de novo variants, one in DNM1L and one in PDHA1. She exhibited a severe cortical atrophy, dilated ventricles and an incomplete corpus callosum, similar to those seen in other cases of female PDHA1 deficiency (34). Moreover, the PDHA1 missense allele affects the same amino acid as an allele from that study, glycine at position 150 (34). The patient in that report exhibited an overall similar brain phenotype to Patient 2 but with later onset of lactic acidemia, a lower lactate level and less severe brain abnormalities (34). The difference in severity and phenotype could be explained by differences between the amino acid change (G150R versus G150E), differences in the inherited genetic background of Patient 2, or differences related to the additional de novo event in Patient 2 in DNM1L. The p.E379K allele did not produce the strong-dominant negative effects of the other alleles but it did exhibit an abnormal phenotype in an assay for the trafficking of mitochondria to synaptic boutons. In any case, the fly studies allow us to distinguish this range of possibilities all suggesting a minimal effect of p.E379K, from the strong dominant-negative effects seen in the other alleles.
Another interesting feature of the Drosophila functional analysis relates to the similarity between the p.A395D allele and the p.G350R allele in peroxisomal and mitochondrial morphology. However, the p.A395D appeared to have greater toxicity when compared with p.G350R. The slight difference in severity might relate to the observation that Patient 1 exhibited normal plasma VLCFA levels compared with the abnormalities reported for p.A395D. Prior to this study, peroxisomal fission had not been studied in Drosophila, but whether distinct amino acids in the middle domain of DNM1L have distinct roles in mitochondrial versus peroxisomal fission is a hypothesis that can be explored through further studies of human DNM1L middle domain variants.
In conclusion, WES is a powerful diagnostic tool for infantile mitochondrial and peroxisomal phenotypes. In addition, patients like Patient 2 who have two de novo variants present a diagnostic challenge. Functional exploration of human gene variants in Drosophila is informative in these cases. Our data suggest that DNM1L variants may need to be considered in a range of encephalopathies. Our data also suggest that studying organelle dynamics in Drosophila can aid in determining the pathogenicity of variants linked to organelle dysfunction and rare disease phenotypes.
Materials and Methods
Clinical cases and ethics statement
Both patients were enrolled in IRB-approved human studies at University of Texas Houston (Patient 1) and Baylor College of Medicine (Patient 2), as part of the Biochemical and Cell Biology Correlates of Peroxisomal Disorders study. Clinical case histories presented represent standard clinical care including radiologic, biochemical and molecular testing. A plasma sample from Patient 1 was sent for VLCFA levels on a clinical basis as described (35).
Whole-exome capture, sequencing and data analysis
Both patients underwent WES through the Whole Genome Laboratory (https://www.bcm.edu/research/medical-genetics-labs/index.cfm?PMID=21319) using methods described (36). Produced sequence reads were aligned to the GRCh37 (hg19) human genome reference assembly using the HGSC Mercury analysis pipeline (http://www.tinyurl.com/HGSC-Mercury/). Variants were determined and called using the Atlas2 suite to produce a variant call file (37). For the population comparisons we utilized data from the Exome Aggregation Consortium (ExAC), Cambridge, MA, USA (URL: http://exac.broadinstitute.org) [November 2015] and Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA (URL: http://evs.gs.washington.edu/EVS/) [November 2015]. Parental studies for DNM1L and PDHA1 as well as other variants noted in Supplementary Material, Table S1 were performed by Sanger confirmation in proband and sequencing in parental blood DNA samples. The % mosaicism for the maternal sample for Patient 1 was determined by examination of the Sanger traces.
Drosophila transgenics
We generated human DNM1L with and without the three variants of interest in a series of constructs which were codon-optimized for Drosophila expression (GeneArt™). We then subcloned these constructs in the pUAST-attB vector and generated transgenic flies by injecting prepared DNA into embryos (38). We targeted the VK00033 site for site-specific integration (y[1] w[1118]; PBac[y[+]-attP-3B]VK00033) (39,40).
Drosophila genetics
The Drp11 and Drp12 alleles were those reported (27). Transgenic DNM1L constructs were crossed into these genetic backgrounds.
Peroxisomal morphology studies
Two peroxisomal reporters were used in third instar larval salivary gland, a UAS-GFP-SKL construct generated by subcloning a c-terminal SKL tagged GFP into the UAS vector and a transgenic insertion on second chromosome was recombined with Actin-GAL4 (y1 w*; P{Act5C-GAL4}25FO1/CyO, y+). Pex3 staining was performed as described (33). Confocal images were quantified using ImageJ software.
Mitochondrial studies
Mitochondrial trafficking and quantification at the third instar neuromuscular junction was assayed as described (32).
Note Added in Proof
During submission and acceptance of this manuscript additional reports of cases of dominant (41) and recessive (42) cases related to DNM1L have been published.
Supplementary Material
Funding
This work was funded by the National Institutes of Neurological Disorders and Stroke (K08NS076547 to M.F.W.), the Simmons Family Foundation Collaborative Award (to M.F.W. and H.J.B.), the Clayton Murphy Peroxisomal Disorders Research Funds, and the Baylor College of Medicine Medical Genetics Training Grant T32-GM07526-37 (L.A.R.).
Supplementary Material
Acknowledgements
We thank the families for their participation in the research. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. The authors would also like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Conflict of Interest statement: None declared.
References
- 1.DiMauro S., Schon E.A., Carelli V., Hirano M. (2013) The clinical maze of mitochondrial neurology. Nat. Rev. Neurol., 9, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leonard J.V., Schapira A.H. (2000) Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet, 355, 299–304. [DOI] [PubMed] [Google Scholar]
- 3.Leonard J.V., Schapira A.H. (2000) Mitochondrial respiratory chain disorders II: neurodegenerative disorders and nuclear gene defects. Lancet, 355, 389–394. [DOI] [PubMed] [Google Scholar]
- 4.Lombes A., Aure K., Bellanne-Chantelot C., Gilleron M., Jardel C. (2014) Unsolved issues related to human mitochondrial diseases. Biochimie, 100, 171–176. [DOI] [PubMed] [Google Scholar]
- 5.Tuppen H.A., Blakely E.L., Turnbull D.M., Taylor R.W. (2010) Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta, 1797, 113–128. [DOI] [PubMed] [Google Scholar]
- 6.Lieber D.S., Hershman S.G., Slate N.G., Calvo S.E., Sims K.B., Schmahmann J.D., Mootha V.K. (2014) Next generation sequencing with copy number variant detection expands the phenotypic spectrum of HSD17B4-deficiency. BMC Med. Genet., 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L., Smertenko T., Alston C.L., Neeve V.C., Best A. et al. (2014) Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA, 312, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrader M., Costello J., Godinho L.F., Islinger M. (2015) Peroxisome-mitochondria interplay and disease. J. Inherit. Metab. Dis., 38, 681–702. [DOI] [PubMed] [Google Scholar]
- 9.Bonnen P.E., Yarham J.W., Besse A., Wu P., Faqeih E.A., Al-Asmari A.M., Saleh M.A., Eyaid W., Hadeel A., He L. et al. (2013) Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet., 93, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamshad M.J., Shendure J.A., Valle D., Hamosh A., Lupski J.R., Gibbs R.A., Boerwinkle E., Lifton R.P., Gerstein M., Gunel M. et al. (2012) The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am. J. Med. Genet. A, 158A, 1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T. et al. (2015) The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet., 97, 199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. et al. (2014) Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA, 312, 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. et al. (2013) Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N. Engl. J. Med., 369, 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. et al. (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E. and Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. (2008) ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med., 10, 294–300. [DOI] [PubMed] [Google Scholar]
- 16.Robinson P.N., Kohler S., Oellrich A., Sanger Mouse Genetics Project, Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D. et al. (2014) Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res., 24, 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wangler M.F., Yamamoto S., Bellen H.J. (2015) Fruit flies in biomedical research. Genetics, 199, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shulman J.M. (2015) Drosophila and experimental neurology in the post-genomic era. Exp. Neurol., 274, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto S., Jaiswal M., Charng W.L., Gambin T., Karaca E., Mirzaa G., Wiszniewski W., Sandoval H., Haelterman N.A., Xiong B. et al. (2014) A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell, 159, 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.R., Blackstone C. (2007) Drp1 phosphorylation and mitochondrial regulation. EMBO Rep., 8, 1088–1089; author reply 1089–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C.R., Manlandro C.M., Arnoult D., Stadler J., Posey A.E., Hill R.B., Blackstone C. (2010) A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J. Biol. Chem., 285, 32494–32503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A., Schrader M. (2003) Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem., 278, 8597–8605. [DOI] [PubMed] [Google Scholar]
- 23.Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., Leonard J.V. (2007) A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med., 356, 1736–1741. [DOI] [PubMed] [Google Scholar]
- 24.Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y. et al. (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol., 11, 958–966. [DOI] [PubMed] [Google Scholar]
- 25.Zhai R.G., Hiesinger P.R., Koh T.W., Verstreken P., Schulze K.L., Cao Y., Jafar-Nejad H., Norga K.K., Pan H., Bayat V. et al. (2003) Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl Acad. Sci. USA, 100, 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rikhy R., Kamat S., Ramagiri S., Sriram V., Krishnan K.S. (2007) Mutations in dynamin-related protein result in gross changes in mitochondrial morphology and affect synaptic vesicle recycling at the Drosophila neuromuscular junction. Genes Brain Behav., 6, 42–53. [DOI] [PubMed] [Google Scholar]
- 27.Verstreken P., Ly C.V., Venken K.J., Koh T.W., Zhou Y., Bellen H.J. (2005) Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron, 47, 365–378. [DOI] [PubMed] [Google Scholar]
- 28.Bernier F.P., Boneh A., Dennett X., Chow C.W., Cleary M.A., Thorburn D.R. (2002) Diagnostic criteria for respiratory chain disorders in adults and children. Neurology, 59, 1406–1411. [DOI] [PubMed] [Google Scholar]
- 29.Bhar D., Karren M.A., Babst M., Shaw J.M. (2006) Dimeric Dnm1-G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem., 281, 17312–17320. [DOI] [PubMed] [Google Scholar]
- 30.Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J.E., Nunnari J. (2006) Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem., 281, 2177–2183. [DOI] [PubMed] [Google Scholar]
- 31.Gallardo E., Claeys K.G., Nelis E., Garcia A., Canga A., Combarros O., Timmerman V., De Jonghe P., Berciano J. (2008) Magnetic resonance imaging findings of leg musculature in Charcot-Marie-Tooth disease type 2 due to dynamin 2 mutation. J. Neurol., 255, 986–992. [DOI] [PubMed] [Google Scholar]
- 32.Sandoval H., Yao C.K., Chen K., Jaiswal M., Donti T., Lin Y.Q., Bayat V., Xiong B., Zhang K., David G. et al. (2014) Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. Elife, 3, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faust J.E., Manisundaram A., Ivanova P.T., Milne S.B., Summerville J.B., Brown H.A., Wangler M., Stern M., McNew J.A. (2014) Peroxisomes are required for lipid metabolism and muscle function in Drosophila melanogaster. PLoS One, 9, e100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ah Mew N., Loewenstein J.B., Kadom N., Lichter-Konecki U., Gropman A.L., Martin J.M., Vanderver A. (2011) MRI features of 4 female patients with pyruvate dehydrogenase E1 alpha deficiency. Pediatr. Neurol., 45, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser A.B., Kreiter N., Bezman L., Lu S., Raymond G.V., Naidu S., Moser H.W. (1999) Plasma very long chain fatty acids in 3,000 peroxisome disease patients and 29,000 controls. Ann. Neurol., 45, 100–110. [DOI] [PubMed] [Google Scholar]
- 36.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. et al. (2013) Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med., 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA, 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venken K.J., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P. et al. (2009) Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods, 6, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venken K.J., He Y., Hoskins R.A., Bellen H.J. (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science, 314, 1747–1751. [DOI] [PubMed] [Google Scholar]
- 41.Vanstone J.R., Smith A.M., McBride S., Naas T., Holcik M., Antoun G., Harper M.E., Michaud J., Sell E., Chakraborty P. et al. (2015) DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur. J. Hum. Genet., doi:10.1038/ejhg.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon G., Malam Z., Paton T., Marshall C.R., Hyatt E., Ivakine Z., Scherer S.W., Lee K.S., Hawkins C., Cohn R.D. et al. (2016) Lethal disorder of mitochondrial fission caused by mutations in DNM1L. J. Pediatr., doi:10.1016/j.jpeds.2015.12.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.