Abstract
POU1F1, a pituitary-specific POU-homeo domain transcription factor, plays an essential role in the specification of the somatotroph, lactotroph and thyrotroph lineages and in the activation of GH1, PRL and TSHβ transcription. Individuals with mutations in POU1F1 present with combined deficiency of GH, PRL and TSH. Here, we identified a heterozygous missense mutation with evidence of pathogenicity, at the POU1F1 locus, in a large family in which an isolated growth hormone deficiency segregates as an autosomal dominant trait. The corresponding p.Pro76Leu mutation maps to a conserved site within the POU1F1 transactivation domain. Bandshift assays revealed that the mutation alters wild-type POU1F1 binding to cognate sites within the hGH-LCR and hGH1 promoter, but not to sites within the PRL promoter, and it selectively increases binding affinity to sites within the hGH-LCR. Co-immunoprecipitation studies reveal that this substitution enhances interactions of POU1F1 with three of its cofactors, PITX1, LHX3a and ELK1, and that residue 76 plays a critical role in these interactions. The insertion of the mutation at the mouse Pou1f1 locus results in a dramatic loss of protein expression despite normal mRNA concentrations. Mice heterozygous for the p.Pro76Leu mutation were phenotypically normal while homozygotes demonstrated a dwarf phenotype. Overall, this study unveils the involvement of POU1F1 in dominantly inherited isolated GH deficiency and demonstrates a significant impact of the Pro76Leu mutation on DNA-binding activities, alterations in transactivating functions and interactions with cofactors. Our data further highlight difficulties in modeling human genetic disorders in the mouse despite apparent conservation of gene expression pathways and physiologic functions.
Introduction
Pituitary development is temporally and spatially regulated by numerous signaling molecules and transcription factors (1). The Pou-homeodomain protein, Pou1f1, initially named Pit1, plays a key role in the development of the anterior pituitary. Pou1f1 autoregulates its own expression (2,3) and the expression of the three signature hormones [growth hormone (Gh), prolactin (Prl) and thyroid-stimulating hormone beta subunit (Tshβ)]. As such, Pou1f1 serves essential functions in the differentiation and proliferation of somatotropes, lactotropes and thyrotropes. The loss of POU1F1 functions results in combined pituitary hormone deficiency (CPHD) syndromes in both mice and humans.
The Pou1f1 protein (291 amino acids) is composed of an N-terminal transactivating domain (TAD) (4) involved in protein–protein interactions, and a homeodomain comprising Pou-specific and Pou-Homeo domains involved in DNA binding and in interactions with transcriptional cofactors (2). Pou1f1 recognizes a weakly conserved A/T-rich consensus sequence (A/T)(A/T)TATNCAT, binds to well-defined sites within the promoters and/or enhancers of multiple target genes (5,6) and stimulates gene transcription in concert with a number of cofactors. Examples of Pou1f1 interactions include the association with Pitx1 via the Pou1f1-TAD to activate the Prl and Gh promoters (7), and association with the LIM domains of Lhx3 through the Pou1f1-homeodomain to activate the Pou1f1, Tshβ and Prl promoters (8,9) as well as the human PRL promoter (9). Importantly, forced co-expression of Pou1f1 along with a member of ETS oncogene family (ELK1, an ubiquitous transcription factor) is capable of activating the endogenous GH1 in the human HEK293 cell line to levels 23-fold greater than measured in the non-transfected cells (10). These studies highlight the central and essential functions of POU1F1 in anterior pituitary development and in corresponding expression of three landmark hormones, GH, PRL and TSHβ.
Naturally occurring mutations in Pou1f1 were initially reported in the Snell and Jackson mice, two dwarf strains with combined deficits in Gh, Tshβ and Prl associated with a hypoplastic anterior pituitary (11). In humans, the first POU1F1 mutations were identified in 1992 (12) and 35 distinct mutations have since been reported worldwide (HGMD: hgmd.cf.ac.uk). The vast majority of these mutations act in a recessive manner with only six demonstrating an autosomal dominant inheritance of hormone deficiency. Although the detailed clinical attributes of patients with the various POU1F1 mutations can vary, these patients consistently display an overall picture of CPHD of GH, PRL and TSH. GH and PRL deficiencies in affected individuals are initially noted early in childhood, whereas the central hypothyroidism tends to appear later in childhood or in adolescence. Radiologic imaging in these individuals often reveals a small anterior pituitary gland with a normal posterior pituitary and infundibulum.
The human growth hormone cluster contains five genes; GH1 is expressed specifically in the pituitary somatotropes while the expression of its four paralogs, GHV, CSA, CSB and CSL (a pseudogene), is specific to the syncytiotrophoblast epithelium lining the placental villi. This multigene locus contrasts with the single-Gh gene locus in the mouse. It was generated by local duplications of the ancestral GH gene at a point subsequent to the divergence of the rodent and primate lineages. The GH1 promoter contains a pair of conserved POU1F1-binding sites within its proximal 200 bp region. In humans, these two sites (hereafter named prox-GH1 and prox-GH2) are not sufficient for high-level expression of GH1 in the pituitary when assayed in mouse transgenic assays (13). Instead, a powerful enhancer, a DNaseI hypersensitive site I (HSI), located 14.5 kb 5′ to the hGH1 promoter, is both necessary and sufficient to drive high levels of GH1 in the somatotrope. This HSI enhancer is a component of the hGH locus control region (LCR) and does not appear to have a correlate in the mouse genome. HSI contains a tightly packed array of three POU1F1-binding sites (HSI-A, HSI-B and HSI-C). These three sites play an essential role in both the activation and the maintenance of hGH1 transcription in the somatotrope (14–17) and in its maintenance in the adult (18). POU1F1 binding at these LCR sites within HSI triggers the formation of an extensive (32 kb) domain of histone acetylation throughout the hGH locus (19) and is essential for bringing the LCR in close proximity (‘looping’) to the hGH1 promoter (20–22). It has been demonstrated that a single-base difference between POU1F1-binding sites at the hGH1 promoter and those at HSI modifies the conformation of the POU1F1/DNA complex, suggesting that these complexes may function through differential cofactor recruitment (23). Thus, POU1F1 appears to have distinct functions in binding to its cognate sites at HSI and within the hGH1 promoter.
Here, we describe a family in which nine members in three generations manifest growth retardation linked to isolated growth hormone deficiency (IGHD). These individuals lack evidence for associated TSH and PRL deficiency. This phenotype was inherited in an autosomal dominant pattern and co-segregated over three generations with a missense mutation within the POU1F1 transcriptional activation domain (TAD). A series of in vitro and in vivo functional assays were carried out to delineate the mechanism(s) underlying this novel dominantly inherited isolated GH deficit.
Results
Identification of a short stature phenotype inherited in an autosomal dominant pattern over three generations of a human kindred
Nine individuals (five females and four males) from the same non-consanguineous Caucasian family originating from the east of France demonstrated findings of severe growth retardation. The short stature phenotype segregated as an autosomal dominant trait over three successive generations. Height standard deviation (SD) scores, at the time of diagnosis, varied from −3 to −5.4 (Fig. 1A) and all patients had a serum GH peak below 5 µg/l (Table 1). The endocrine deficit was limited to a deficiency in growth hormone (i.e. IGHD phenotype) (Table 1). Although basal serum PRL was relatively low in affected individual III.7, the thyrotropin-releasing hormone (TRH) stimulation induced a 5-fold increase suggesting that PRL expression and regulation was not adversely affected. Eight of the affected individuals (II.2, II.4, II.6, II.8, II.10, III.1, III.4 and III.7) benefited from a GH treatment with significant augmentation in linear growth (the ninth individual, I.2, was not treated due to advanced age).
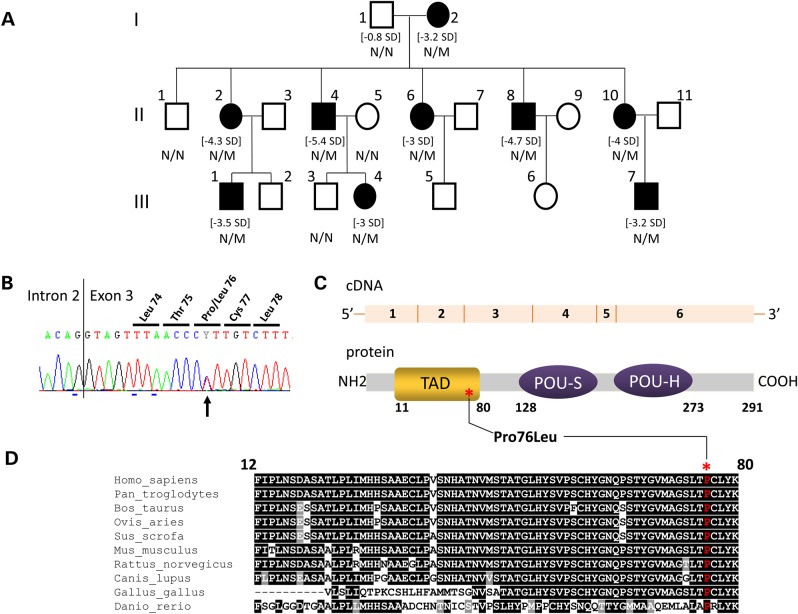
Figure 1.
Identification of a POU1F1 mutation segregating with a short stature phenotype and low serum GH levels in a three generation kindred. (A) Genealogical tree of the IGHD family. Squares: males; circles: females; filled black symbols: IGHD patients. For subjects with growth retardation, their height SD is indicated between brackets. The genotype (N/N: normal, N/M: heterozygous) is indicated under each tested individual. (B) Electrophoregram of the portion of Exon 3 showing (black arrow) the heterozygous c.227C > T transition leading to p.Pro76Leu mutation. The vertical line represents the intron–exon junction. (C) A schematic representation of the POU1F1 cDNA and POU1F1 protein: six exons encoding the 291 amino acids protein consisting of two main domains, the TAD (orange) in which the variation has been identified (noted with an asterisk) and the POU-S and POU-H domains (homeodomain, purple). (D) Evolutionary conservation of proline 76 (noted above with an asterisk): interspecies similarity (shown in one-letter code) of the TAD domain of POU1F1 aligned with sequences of the TAD domain found in nine other vertebrates species: black underlined, total conservation; gray underlined, conservative amino acid substitutions; not underlined, amino acid not conserved and indexed in a different group.
Table 1.
Clinical and endocrinological data of the nine IGHD patients
| Patient | Height SD | GH | T4 | PRL | GH treatment | MRI |
|---|---|---|---|---|---|---|
| I.2 | −3.2 | Not eval | Not eval | Not eval | − | − |
| II.2 | −4.3 | 4 | 70 | Not eval | + | − |
| II.4 | −5.4 | 2.2/2.2 | 78 | Not eval | + | − |
| II.6 | −3 | 1.5/0.5 | 58 | Not eval, normal breast feeding | + | − |
| II.8 | −4.7 | 2 | 94 | Not eval | + | − |
| II.10 | −4 | 3/0.5 | 10.7 | Not eval, normal breast feeding | + | − |
| III.1 | −3.5 | 4.3/3.1 | 10.4 | 3.1 (12.5/TRH) | + | AP hypo |
| III.4 | −3 | 2.4/2.3 | 15.2 | 5.1 (16.7/TRH) | + | AP hypo |
| III.7 | −3.2 | 2/3.1 | 11.9 | 0.9 (4.6/TRH) | + | N |
Height SD, height standard deviation; GH µg/l: arginine/ornithine tests, normal value >10 µg/l; total T4 ng/ml: normal value 40–120 for II.2, II.4, II.6 and II.8 and free T4 pmol/l (in italic) normal value 10–21 for II.10, III.1, III.4 and III.7; PRL ng/ml, PRL before and after TRH stimulation (between brackets): normal value 2.5–20; Not eval: not evaluated; AP hypo: hypoplasia of the anterior pituitary; N: normal.
Magnetic resonance imaging (MRI) of the pituitary region performed in three affected members of the family was found to be normal in one case (III.7) and showed anterior pituitary hypoplasia for two individuals (III.1 and III.4); no abnormalities at the level of posterior pituitary, pituitary stalk, septum pellucidum, corpus callosum or optic nerves were detected. These data allowed us to conclude that a mutation resident in this kindred resulted in an isolated GH deficiency segregating as an autosomal dominant trait.
Affected individuals in the kindred carry a novel mutation at the POU1F1 locus
The low levels of hGH1 expression in the nine short stature individuals and the hypoplasia of the anterior pituitary in two of three studies prompted us to screen for mutations in a defined set of genes critical to GH synthesis and/or pituitary development; GH1, LCR-GH1 (HSI fragment), GHRHR, GHRH, GHSR, GHRL and HESX1 genes (see ‘Materials and Methods’ section for details). The sequence of each of these target genes was normal. Causes of developmental defects of the pituitary were next assayed by the analysis of the PROP1 and POU1F1 genes in individual III.4. While PROP1 analysis revealed a normal sequence, the analysis of POU1F1 revealed heterozygosity for a sequence variant in Exon 3, c.227C>T (Fig. 1B). This base transition, not previously described in ExAC (exac.broadinstitute.org) or in Ensembl (ensembl.org), results in a non-conservative substitution that replaces Proline (Pro) by Leucine (Leu) at codon 76. This Pro76Leu (P76L) substitution maps within the highly conserved TAD and involves a proline residue that is invariant in vertebrates spanning evolution from zebrafish to primates (Fig. 1C and D). To establish its intra-familial segregation, the region of the POU1F1 gene encompassing the mutation (Exon 3) was subsequently sequenced in all available family members. This analysis revealed a perfect segregation of the defined mutation with the short stature phenotype (Fig. 1A). These data lead us to conclude that the dominantly inherited IGHD in this family was due to the defined c.227C>T at the POU1F1 locus.
Nuclear localization of the POU1F1 transcription factor is unaffected by the P76L mutation
To assess the functional consequences of the P76L mutation at the protein level, we first assessed the subcellular distribution of the mutant POU1F1 protein. Expression plasmids encoding human influenza hemagglutinin (HA)-tagged versions of the wild type (WT) and the P76L POU1F1 proteins (pcDNA4-POU1F1_WT-HA and pcDNA4-POU1F1_P76L-HA, respectively) were individually transfected into the human embryonic kidney 293T (HEK293T) cell line. Protein accumulation was assessed in individual cells by immunofluorescence microscopy with an anti-HA antibody (Fig. 2A). The analyses of both the WT and the mutant POU1F1 proteins revealed intense nuclear staining. These data suggest that the mutation fails to alter the nuclear import and retention of POU1F1.
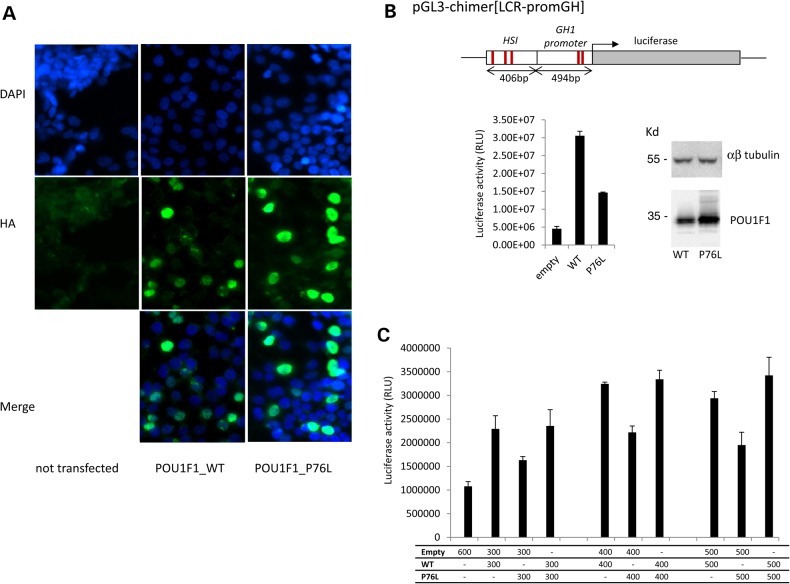
Figure 2.
Conserved nuclear localization and diminished transcriptional activity of the mutant POU1F1 protein. (A) Subcellular localization of POU1F1_WT and POU1F1_P76L in HEK293T cells transfected with the corresponding HA-tagged expression plasmids. Forty-eight hours after transfection, cells were immunostained with mouse anti-HA antibody (1/1000) then Alexa488 (goat anti-mouse 1/2000). Nuclei are stained in blue by DAPI. The two proteins (WT and mutated) were both localized in the nucleus. A control with no transfected cells is also shown. (B) Impact of the Pro76Leu mutation on the transcriptional capability of POU1F1. HEK293T cells were co-transfected with pcDNA3-POU1F1_WT-HA or pcDNA3-POU1F1_P76L-HA in combination with a luciferase reporter ORF under control of the HSI enhancer segment of the hGH LCR linked directly to the intact hGH promoter pGL3-chimer[LCR-promGH]. The previously defined POU1F1-binding sites in HSI and in the hGH promoter are indicated by the red lines. POU1F1 protein expression from the expression vectors was monitored by western blot with anti-POU1F1 polyclonal antibody relative to αβ-tubulin level (top). Luciferase activity represents the means ± SD of triplicate assays; a representative experiment of three experiments. (C) The assessment of a potential dominant-negative effect of the P76L mutation over the WT protein. Co-transfection of the HSI/hGH/Luc reporter with the POU1F1_WT plasmid, POU1F1_P76L plasmid in increasing amounts up to a saturation response or with a 1:1 mixture of the two plasmids.
The P76L POU1F1 mutation has a negative impact on transcriptional activation of the GH gene
The functional impact of the P76L mutation on POU1F1 transcriptional activity was assessed in a luciferase reporter assay. The luciferase open reading frame (ORF) was placed under the transcriptional control of the hGH1 promoter (containing two well-described POU1F1-binding sites) linked to the 404 bp HSI fragment of the hGH LCR encompassing an array of three critical POU1F1-binding sites (Fig. 2B, top). The reporter plasmid was co-transfected into HEK293T cells along with a plasmid expressing the WT or the P76L POU1F1 protein. Western analysis of three independent studies revealed that the mutant protein was expressed at levels equal to or greater than the WT POU1F1 (Fig. 2B, data not shown). The co-transfection with the WT POU1F1 protein increased luciferase reporter activity by 5-fold over that of an empty vector. The transcriptional enhancement of the luciferase reporter by the P76L POU1F1 was 50% relative to the WT (Fig. 2B). It was additionally noted that co-transfection of equal amounts of the plasmid encoding the POU1F1 proteins did not inhibit the transcriptional activity associated with the WT POU1F1 protein (Fig. 2C). These data lead us to conclude that the P76L POU1F1 mutation results in a decrease in the transcriptional activity of the GH1 gene, but argue against a dominant effect of the mutant protein, at least in the context of defined reporter assay.
POU1F1_P76L has a differential impact on POU1F1 binding to cognate sites at HSI of the hGH LCR and at the GH1 promoter
The decreased transcriptional capability of POU1F1_P76L prompted us to compare the binding affinity of the WT and mutant POU1F1 protein toward a subset of cognate-binding targets. This was done by subjecting bacterially generated WT and P76L POU1F1 proteins to surface plasmon resonance (SPR) analysis. For purposes of the analysis, the WT and mutant proteins were generated in parallel with all culture and purification steps held constant. Of note, we observed a 40-fold higher recombinant protein yield from the bacterial cultures expressing the WT versus the mutant protein. This difference appeared to reflect the formation of inclusion bodies in cultures of the mutant protein because the yield of the mutant protein was increased by culturing at lower temperature. This difference in solubility is consistent with an altered conformation of the POU1F1_P76L protein. Biotinylated DNA targets representing the defined POUF1-binding sites of the hGH1 promoter and of HSI were immobilized on a streptavidin (SA) sensor chip and used for the analysis.
For a first qualitative test, equal amounts of the purified recombinant POU1F1_WT and POU1F1_P76L proteins (quantified by Experion; see ‘Materials and Methods’ section) were then loaded as analytes. The recorded resonance units (RUs) showed that the WT and P76L POU1F1 proteins bind the promoter and the HSI DNA targets; the difference of 11RU on HSI target is significant. This result allows us to conclude that POU1F1_P76L binds better than POU1F1_WT at the same concentration under our assay conditions (Fig. 3A). As a control, we performed similar experiments with a recombinant form of POU1F1 carrying a previously identified CPHD missense mutation (24). This mutation in a conserved residue of the homeodomain abolishes the binding of POU1F1 to its DNA targets demonstrated by a dramatic loss of binding in our SPR analysis (Fig. 3A).
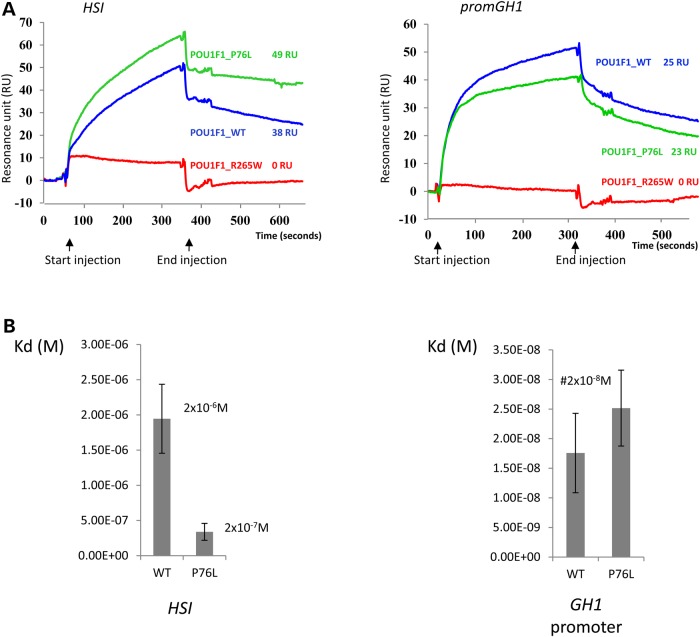
Figure 3.
POU1F1 binding to DNA studied by SPR. (A) Sensorgrams. Biotinylated DNAs (HSI and promGH1) were loaded on a streptavidin chip and binding of 50 nm of purified WT (blue) or P76L (green) or R265W (red) POU1F1 proteins to the two DNA targets was assessed. The recorded RUs for each protein were noted. (B) Kinetics of POU1F1 binding the DNA sites: 0, 10, 20, 30, 40 and 50 nm of each purified protein were successively loaded on the streptavidin chips on which biotinylated DNA POU1F1 targets have been anchored. After data treatment using the BIAevaluation software 4.1, the association and dissociation constants corresponding to a Kd value in molar (in m) were determined for WT and P76L-POU1F1 proteins on each target sequence and represented by histograms: on the HSI (left graph) and on the hGH1 promoter (right graph). Each study was performed in triplicate.
We next evaluated the affinity of the WT and the P76L proteins for their DNA targets in a set of kinetic binding studies (see ‘Materials and Methods’ section). The dissociation constants obtained on the GH1 promoter measured using various concentrations of those proteins were similar for the POU1F1_WT and the POU1F1_P76L proteins (Kd of 1.7 × 10−8 M and 2.4 × 10−8 M, respectively) (Fig. 3B, right panel). In contrast, the Kd of the POU1F1_P76L protein was significantly lower than that of the WT protein (i.e. 2.0 × 10−7 M versus 2.0 × 10−6 M) when tested for interaction with HSI (Fig. 3B, left panel). This increased affinity of the POU1F1_P76L protein for the HSI sites was linked to an increased association rate: ka = 5.9 × 102 mol−1 s−1 for the POU1F1_WT and ka = 3.0 × 103 mol−1 s−1 for the POU1F1_P76L protein. These kinetic studies show that the WT POU1F1 protein has higher affinity on the promoter sites than on the HSI sites. These data lead us to conclude that the P76L mutation has a differential impact on the interaction of the POU1F1 protein with different sets of cognate-binding sites.
POU1F1_P76L alters the binding of the POU1F1 WT protein at cognate sites in the GH1 but not PRL promoter
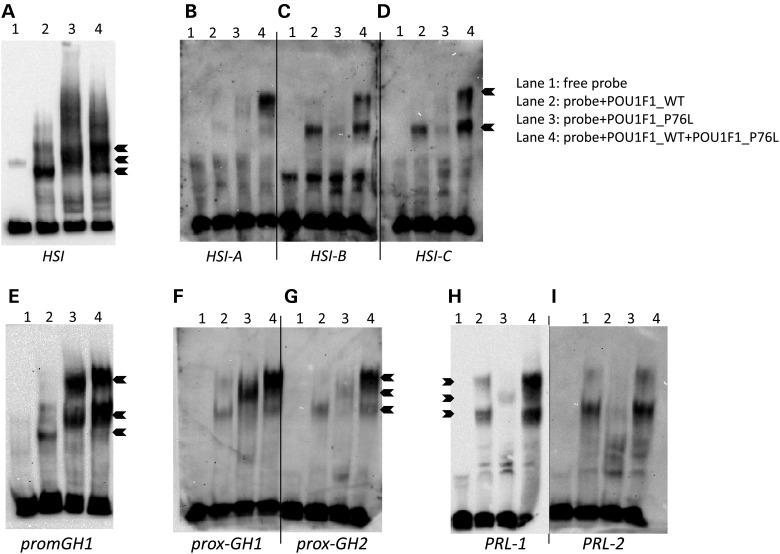
We next compared by electrophoretic mobility shift assay (EMSA) the binding of the WT alone, the POU1F1_P76L alone and a mix of the two proteins, as would occur in individuals heterozygous for the P76L mutation. Binding was assessed for DNA fragments containing the full set of POU1F1-binding sites (HSI and GH1 promoter) or each of the corresponding individual POU1F1-binding sites (HSI-A, HSI-B, HSI-C, prox-GH1 and prox-GH2). Remarkably, the migration patterns were different for the DNA incubated with POU1F1_WT (Lane 2), POU1F1_P76L (Lane 3) and the mix of these two proteins (Lane 4) for the HSI (Fig. 4A–D) and for the hGH1 promoter sites (Fig. 4E–G). These data suggest a modified binding conformation of the WT/mutant dimer (POU1F1_WT/POU1F1_P76L) complex on all POU1F1-binding sites of the GH1 promoter and HSI. Of note, the POUF1_WT/POU1F1_P76L mix showed the same migration pattern as POU1F1_WT when assayed for binding to the prolactin promoter (i.e. PRL-1 and PRL-2) (Fig. 4H and I). These data are consistent with the disease phenotype characterized by a deficit in GH with no PRL deficiency.
Figure 4.
Analysis of POU1F1 binding to DNA by electrophoretic mobility shift assays. (A) Twenty fmoles of biotinylated DNA containing a 212 bp segment of HSI encompassing all three POU1F1-binding sites (A, B and C) were incubated with 200 ng of purified WT (Lane 2) or mutant P76L POU1F1 (Lane 3) protein or with a 1:1 mixture (100 ng of each) of the two proteins (Lane 4). Lane 1 contains the biotinylated DNA target in the absence of added protein. (B–D) Binding to each individual POU1F1-binding site within HSI: 37 bp including one site HSI-A, B, 37 bp HSI-B, C, 38 bp HSI-C, D. (E) Binding to a 70 bp segment of the GH1 promoter encompassing the two POU1F1-binding sites. (F–G) Binding to each of the two POU1F1-binding sites in the hGH1 promoter: 32 bp fragment encompassing prox-GH1 (F) or the 30 bp fragment encompassing prox-GH2 (G). (H and I) Binding to each of the two POU1F1-binding sites in the human PRL gene promoter: 29 bp fragments containing either PRL-1 (H) or PRL-2 (I).
The P76L mutation increases the interaction of POU1F1 with three different POU1F1 transcriptional cofactors
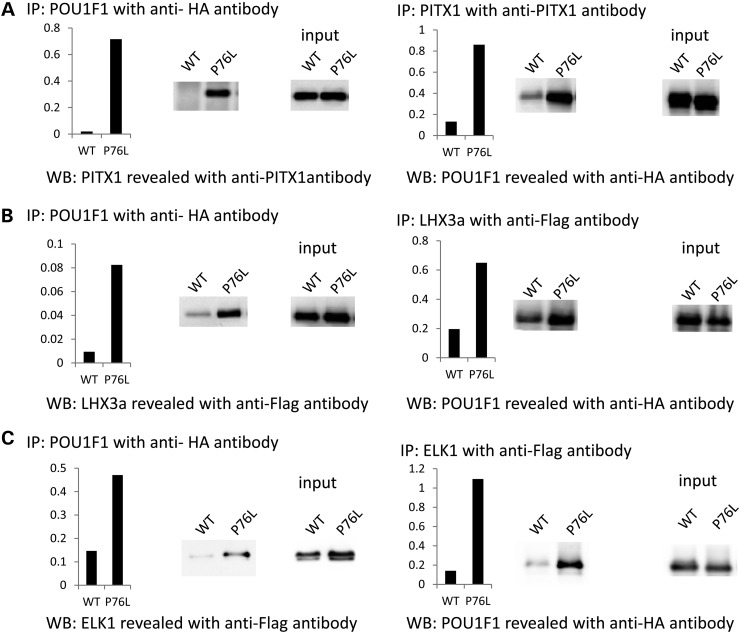
The impact of the POU1F1 mutation on complex formation with two pituitary-specific (PITX1 and LHX3a) transcriptional cofactors and one ubiquitous transcription factor (ELK1) was assessed by co-immunoprecipitations after cotransfection in HEK293T cells of an expression plasmid encoding the HA-tagged POU1F1_WT or POU1F1_P76L with a plasmid encoding PITX1, LHX3a or ELK1. Nuclear proteins were immunoprecipitated with an anti-HA antibody and the pellets were subsequently assayed for each cofactor by western blot (Fig. 5, left panel). In parallel, the nuclear proteins were also immunoprecipitated with antibodies directed against each cofactor and the western blots were revealed with an anti-HA-antibody (Fig. 5, right panel). The data revealed that the P76L mutation enhances complex formation with each partner by 5–10-folds when compared with the WT POU1F1 protein (Fig. 5A–C). A formal possibility is that the difference in amount of complex observed is due to the POU1F1_P76L conformational difference, which would make the HA-tag in the protein complex more accessible to the antibody. Co-immunoprecipitations performed with antibodies directed against each of the interacting proteins allowed us to rule out this possibility.
Figure 5.
Co-immunoprecipitation of WT and P76L POU1F1 proteins with three cofactors. HEK293T cells were co-transfected with plasmids pcDNA4-POU1F1-HA expressing POU1F1_WT or P76L (lanes labeled WT and P76L, respectively) and with each of the following three expression vectors: pcDNA3-PITX1 (A), pcDNA3-LHX3a-Flag (B) and pcDNA3-ELK1-Flag (C). Nuclear extracts were co-immunoprecipitated with an anti-HA antibody, and the western blots were revealed with an anti-PITX1 antibody for PITX1 and an anti-Flag antibody for LHX3 and ELK1 (left panels). The same nuclear extracts were co-immunoprecipitated with an anti-PITX1 antibody for PITX1 and an anti-Flag antibody for LHX3 and ELK1, and westerns blots revealed with the anti-HA antibody (right panels); IP: immunoprecipitation, WB: western blots. Quantification of the complex formed, represented by histograms, was calculated as the ratio of protein immunoprecipitated to protein expressed (input).
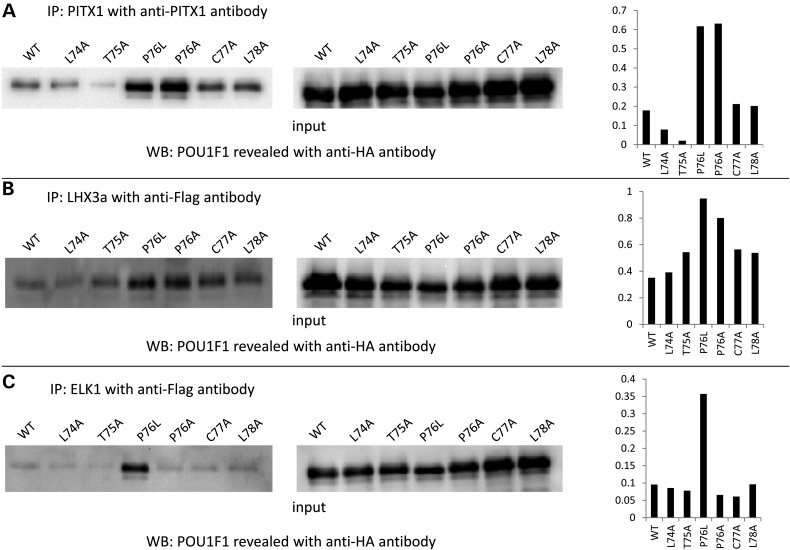
To further evaluate the importance of the peptide surrounding Pro76 in these interactions, we performed similar co-immunoprecipitation experiments with different POU1F1 mutants in which Pro76 and the neighboring (Leu74, Thr75, Cys77 and Leu78) were each individually replaced by alanine residues. The amount of complex formed with PITX1 and LHX3a (Fig. 6A and B, respectively) was increased most prominently by the P76A and P76L substitutions. Moreover, with ELK1 cofactor (Fig. 6C), the leucine substitution seemed to have a critical impact since an alanine did not modify the amount of complex formed. These data demonstrate a major role of the Pro76 residue in the interaction of the POU1F1 transactivating domain with at least three partners.
Figure 6.
Co-immunoprecipitation POU1F1 with three cofactors. Interactions are compared among the WT POU1F1 and a series of derived alanine substitutions surrounding proline 76 site. HEK293T cells were co-transfected with plasmids pcDNA4-POU1F1-HA expressing POU1F1_WT or L74A, T75A, P76L, P76A, C77A, L78A and pcDNA3-PITX1 or pcDNA3-LHX3a-Flag or pcDNA3-ELK1-Flag. Co-immunoprecipitations were performed with an anti-PITX1 antibody (A) or Flag antibodies for LHX3a (B) and ELK1 (C) on nuclear extracts samples. Western blots were generated using an anti-HA antibody. Quantification of the complex formed, represented by histograms (right side), was evaluated as the ratio of protein immunoprecipitated to protein expressed (input).
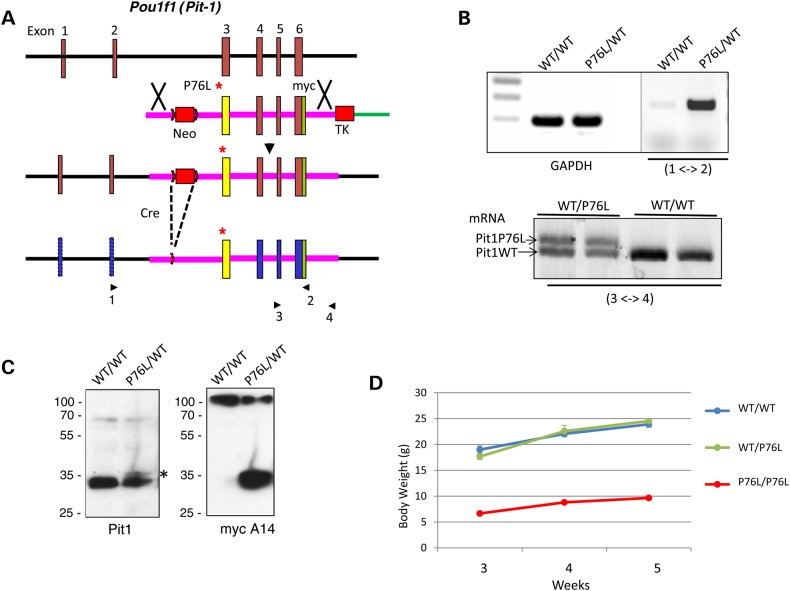
A mouse model of the P76L mutation confirms the adverse impact on gene expression but fails to recapitulate the dominant inheritance pattern
To study the functional consequences of the P76L mutation in an in vivo context, we introduced the P76L mutation into the mouse Pou1f1 locus (see ‘Materials and Methods’ section and Fig. 7A). The expression of P76L encoding Pou1f1 mRNA was confirmed in the pituitaries of P76L/wt mice and the mRNA from the mutant allele was shown to be expressed at equivalent levels to the WT allele by a comparative reverse transcription (RT)/polymerase chain reaction (RT/PCR) (Fig. 7B). Remarkably, however, the expression of the mutant Pou1f1_P76L protein was markedly suppressed, with steady-state levels in the mouse pituitary <10% of output from the endogenous WT locus (Fig. 7C, asterisk). The expression of mRNA from each of three endogenous Pou1f1-dependent genes (mGh, mPrl and mTsh) was assessed and found to be unaltered in mice heterozygous for the P76L mutation (data not shown). Similarly, the expression of the hGH1 gene from the hGH/BAC transgene in the P76L heterozygous mouse was expressed at normal levels (data not shown). Consistent with the low-level expression of Pou1f1 from the mutant locus, mice homozygous for the P76L mutation displayed a dwarf phenotype (Fig. 7D). Overall, these data point to a defect in steady-state expression of the P76L mutant Pou1f1 protein. This deficiency in protein expression is consistent with the poor yield of recombinant protein in the bacterial cultures and may reflect a major alteration in protein solubility or stability in vivo.
Figure 7.
Introduction of the P76L mutation into the Pou1f1 locus in the mouse genome. (A) Homologous recombination at the Pou1f1 locus in the mouse genome. The native-Pou1f1 locus is shown on the top and the targeting vector used to insert the P76L mutation into the locus is shown below. The initial recombination product is displayed on the third line and the final locus after deletion of the NeoR cassette via Cre/Lox recombination is displayed at the bottom. The position of the P76L mutation is denoted by the asterisk, the dual selection cassettes, NeoR and thymidine kinase, are represented by the labeled red rectangles. Lox sites are indicated by the arrowhead and the positions of the primers used for selective detection of the wt and mutant Pou1f1 mRNAs are indicated and numbered below the diagram. (B) mRNA expression from the PitP76L locus in the mouse pituitary. Top: Pou1f1 mRNA expression from the P76L locus was specifically detected in a mouse heterozygous for the mutant allele by an RT/PCR assay using a set of primers positioned at Exon 2 (Primer 1) and within the myc epitope segment in Exon 6 (specific to the mutant allele) (Primer 2). Bottom: the relative levels of the mRNA expression from the wt and the mutant locus were directly compared in a mouse heterozygous for the mutant allele (wt/P76L) and a wt mouse (wt/wt) by an RT/PCR analysis using the Exon 5 primer (Primer 3) and a primer within the 3′ untranslated region (Primer 4) in Exon 4, bracketing the myc epitope tag specific to the mutant locus. (C) Protein expression from the P76L locus in the mouse pituitary. Western blot of pituitary extracts from wt/wt and wt/P76L mice were probed with antibodies to mouse Pou1f1 (left blot) and with an antibody to the myc epitope tag (specific to the mutant locus) (right blot). The overall level of Pou1f1_WT protein in the WT/P76L heterozygote pituitary is approximately half of that in the WT/WT mouse. A faint band at the position of the mutant Pit1 protein (asterisk) is migrating at the position predicted from the Pit-1 protein containing the Myc epitope tag. The expression of the P76L Pou1f1 protein was directly verified by re-probing the western with an antibody to the myc epitope. (D) Growth curves of WT/WT, WT/P76L and P76L/P76L mice. The body weights are shown in the Y-axis and the genotypes are indicted for each curve. The analyses of male mice are shown; a parallel analysis of females gave an identical result (not shown).
Discussion
POU1F1 is essential for the formation of the somatotrope, lactotrope and a subset of the thyrotrope lineages. As such, all POU1F1 mutations thus far reported are linked to CPHD, comprising decreases in the expression of GH, PRL and TSH (25). In the current study, we report the first example of a POU1F1 mutation that is linked to an isolated GH deficiency. Functional data reveal that the P76L mutation, located in a highly conserved segment of the transcriptional activation domain (TAD), impacts on the binding patterns of POU1F1 and on the interaction of POU1F1 with protein partners. The predicted conformational change induced by the non-conservative substitution of a proline for a leucine is consistent with the low steady-state levels of the mutant protein in the mouse despite normal levels of mRNA synthesis. Thus the P76L missense mutation results in an unusual situation: a specific disruption of the human GH1 gene expression.
Several lines of evidence demonstrate that the P76L variation identified in POU1F1 is a disease-causing mutation. First, the strict co-segregation of the phenotype with this mutation in nine individuals over three generations. Secondly, the transcriptional activity of the POU1F1_P76L on the chimeric LCR-hGH1 promoter is significantly lower than that of POU1F1_WT. Thirdly, using SPR assays, we show that the P76L mutation leads to an increased affinity of POU1F1 for the LCR sites. Fourthly, bandshift assays reveal that the DNA binding pattern of a mix of POU1F1_WT and POU1F1_P76L is different from that of POU1F1_WT alone on all five cognate-binding sites in the hGH LCR and GH1 promoter, but not on the two cognate sites in the PRL-binding sites. Fifthly, as shown by co-immunoprecipitation studies, the P76L mutation increases the interaction of POU1F1 with three of its known cofactors: PITX1, LHX3a and ELK1.
A predicted impact of the POU1F1_P76L mutation on conformation is consistent with the noted low yield of the mutant protein compared with the WT protein in two distinct settings; in Escherichia coli and in mice carrying the heterozygous Pou1f1 mutation. It should be noted, however, as a formal possibility that the low protein expression in the mouse model may be contributed by the presence of the myc epitope tag, which was present at the mutant locus, but not at the wt Pou1f1 locus.
It is noteworthy that two additional missense mutations have been identified in the human POU1F1 TAD, P14L (26) and P24L (27). Both of these mutations result in a dominantly inherited form of pituitary deficit. However, in both cases, the patients displayed a classical CPHD phenotype rather than the isolated GH loss currently being reported. The mutation site within the TAD, therefore, appears critical in terms of phenotypic consequences. The three-dimensional (3D) structure of the POU1F1 TAD remains undefined and in silico modeling could not be performed to test the relative impacts of the P76L, P14L and P24L mutations on the 3D structure. Of note, however, all three of these constitute a non-conservative substitution of proline for leucine that is predicted to have a major impact on protein secondary structure and tertiary folding.
Our attempt to model the P76L mutation in the mouse was informative in a number of respects. This mutation once introduced into the mouse Pou1f1 locus had no adverse effect on the level of mRNA generated from the locus. Notably, however, the steady level of mutant protein expressed from the P76L locus was markedly suppressed. Mice heterozygous for the mutation had no appreciable decrease in size while mice homozygous were markedly dwarfed. Thus, this mutation results in a recessive dwarf phenotype in the mouse rather than the dominantly inherited phenotype in the human kindred, for which, haploinsufficiency cannot be the mechanism underlying the dominant expression of the disease phenotype since the heterozygous parents of patients with homozygous POU1F1 null mutation have a normal phenotype. Our in vitro data on the coexpression of the mutant and WT POU1F1 proteins argue against a dominant negative effect of the mutant protein over the WT.
The data favor a hypothesis of a toxic gain of function associated with the P76L mutation in humans for two reasons. First, the POU1F1_P76L protein binds the LCR sites with an increased affinity, when compared with POU1F1_WT; this affinity difference might disrupt POU1F1_WT DNA binding at the LCR target sites in heterozygous patients. It is, therefore, tempting to speculate that the mutation could result in a higher occupancy at hGH LCR target sites precluding the binding of POU1F1_WT. Secondly, we observed that the P76L mutation increases the interactions with three different cofactors. The mutant protein might therefore titrate POU1F1-interacting proteins leading to a competition with the WT POU1F1 protein. Moreover, co-immunoprecipitation-alanine-scanning experiments revealed that the amino acid 76 precisely is critical for these interactions; the weak equilibrium of the hGH1 transcriptional complex should be modified through the ability to recruit somatotrope-specific cofactors necessary for high-GH1 transcriptional level. Our data in bandshift assays on the PRL promoter target sites are fully in accordance with the phenotype of the patients with the P76L mutation who had no prolactin deficiency.
Of particular interest is the finding that the POU1F1_WT binds more tightly to promoter sites than those within the LCR. Previous studies have demonstrated that POU1F1 occupies its cognate sites in the hGH1 promoter and at HSI of the hGH LCR with distinct conformations reflecting a single-base pair difference (23). Thus, the missense mutation at position 76 could differentially impact on POU1F1 actions at the hGH promoter via alterations of the HSI function. Such alterations could explain the specific impact of the mutation on the human hGH gene as the mGh gene is lacking the HSI enhancer and PRL, TSHβ and POU1F1 genes are not known to have corresponding LCR control determinants.
In conclusion, the P76L mutation, which segregates perfectly with the severe growth retardation, involves a conformation modification of the POU1F1 protein that affects cofactors and DNA interactions that impact specifically the hGH1 transcriptional level of expression. This constitutes a novel mechanism underlying a dominant form of IGHD in humans, also suggesting that POU1F1 gene should be screened for this phenotype.
Materials and Methods
Patients
All individuals studied in the reported kindred provided their written informed consent to perform genetic studies. All were referred to the pediatric endocrinology outpatient clinic of the Nancy Medical School hospital (CHU of Nancy). Clinical details were assessed using an information sheet established by our laboratory.
Hormonal investigations and MRI
GH plasma values were evaluated after pharmalogical stimulations by arginine and ornithine, prolactin level before and after TRH stimulation, total and free T4 levels, all were measured according to methods at the time of diagnosis. Pituitary MRI was performed on a 0.5 T General Electrics MR max instrument.
Mutation search
Genomic DNA was isolated from blood samples obtained from each individual using a standard technique. All coding exons and intron–exon boundaries of the GH1, LCR-GH1 (AF_010280), GHRHR, GHRH, GHSR, GHRL, HESX1, PROP1 and POU1F1 (NM_000306) genes were amplified using sets of primers available on request. Sequences were performed according to the thermal cycle sequencing Big dye terminator protocol (ABI Prism 310 Genetic Analyser, PerkinElmer Applied Biosystems).
Plasmid constructs
The full-length POU1F1 and LHX3a cDNAs (9) were subcloned into the pcDNA4 or pcDNA3 expression vectors in which HA or Flag tags were inserted in C-terminal, respectively. The PITX1 cDNA, amplified from pituitary cDNA (Clonetech), was cloned into pcDNA3. The ELK1 cDNA was subcloned from pCGN-ELK1 (Addgene, 27156) into pcDNA3 containing a tag Flag in C-terminal. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to generate plasmids encoding different POU1F1 mutants, pcDNA4-POU1F1-HA-(P76L, L74A, T75A, P76A, C77A and L78A).
The luciferase reporter plasmid was constructed as follows: 406 bp of the LCR (HSI, including the three POU1F1 target sites, AF_010280) and 494 bp of the promoter (including the two POU1F1-binding sites) were linked after amplification using compatible restriction enzyme sites and subcloned into the pGL3-basic-Luciferase plasmid (Promega) to obtain pGL3-chimer[LCR-promGH] plasmid (Fig. 2B).
Cell culture and transfection
HEK293T cells, obtained from the American Type Culture Collection (Manassas, VA, USA), were grown at 37°C in Dulbecco's modified Eagle's medium (Invitrogen), with 10% fetal calf serum. All transfections were performed at 60% confluence using the Fugene method (Promega) according to the manufacturer's protocol.
Luciferase activity assays and western blot
HEK293T cell extracts were prepared and assayed for luciferase activity, using the Promega assay system, 48 h after co-transfection of the pGL3-chimer[LCR-promGH] reporter gene (100 ng) together with either the empty pcDNA3 expression vector and/or the POU1F1-cDNA WT and/or mutant pcDNA4 constructs at different amounts. Each transfection experiment was carried out in triplicate and was independently replicated at least three times. Cell extracts were separated on a 10% polyacrylamide gel, then transferred to a nitrocellulose membrane and probed with an anti-POU1F1 polyclonal antibody (sc-16288, Santa Cruz Biotechnology) and an anti-αβ-tubulin polyclonal antibody (2148, Cell Signaling).
Protein purification
The full-length POU1F1 cDNA was cloned at the BamHI NotI sites into the pGEX-6P-1 vector (GE Healthcare) including a PreScission protease site to remove the Gluthatione S-transferase (GST) tag. GST-tagged fusion proteins were produced in the BL21 DE3 star strain of E. coli (Invitrogen). After induction for 3 h with 0.5 mm isopropyl-β-d-thiogalactopyranoside at 37°C for the GST-POU1F1_WT, at 25°C during 4 h for the GST-POU1F1_P76L, at 15°C for 4 h for the GST-POU1F1_R265W, the bacterial pellet was freezed/thawed three times followed by sonication in PBS1X (200 ml/l of culture), 1% Triton X-100, 10% glycerol and 1 mg/ml lysozyme and finally clarified by centrifugation at 22.000 × g for 20 min. Supernatants were incubated for 30 min at room temperature with Glutathione Sepharose beads (GE Healthcare), then washed 4-folds in PBS1X. The last wash was performed in cleavage buffer [50 mm Tris–HCl pH7.0, 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 1 mm phenylmethylsulfonyl fluoride], then beads were incubated for 4 h at 4°C with 600 µl of PreScission protease (1 U/µl, GE Healthcare) in 40 ml of a cleavage buffer. Centrifugation of beads at 500 × g 15 min at 4°C allowed to collect soluble purified protein, which is concentrated on Corning Spin-X UF 20 ml, 10 000 MWCO. In 10 mM 4-(2-hydroxyethyl)-1piperazineethanesulfonic acid (HEPES) pH7.4, 150 mM NaCl, 3 mM EDTA (HBS) buffer, 2 to 5 ml of protein was dialyzed overnight. All preparations of proteins were checked on Experion (Bio-Rad) instrument for their quality and quantification.
Surface plasmon resonance analysis
Real-time DNA–protein interaction assays were performed using a Biacore 3000 instrument controlled by Biacore 3000 Control Software v4.1 (GE Healthcare). All experiments were done at 25°C. After preincubation in 1 M NaCl, 50 mm NaOH for 1 min (three times), 75 µl of 500 nm biotinylated DNA was covalently coupled to a SA sensor chip (GE Healthcare) (5 µl/min) in HBS-EP running buffer [10 mm HEPES (pH7.4), 150 mm NaCl, 3 mm EDTA, 0.005% surfactant P20]. The chip was then washed 1 min (5 µl/min) with 1 m NaCl, 50 mm NaOH. Real-time monitoring was displayed in a sensorgram as an RU versus time (s). The 70 bp fragment of the GH1 promoter (two POU1F1-binding sites) was generated by annealing a sense (5′ biotinylated) and antisense oligonucleotide and 900RU were obtained. A 212 bp PCR product corresponding to the HSI (three POU1F1-binding sites) was amplified from genomic DNA using a biotinylated primer and 1900RU were anchored. Binding studies were performed during 5 min with 25 µl of 50 nm purified protein in HBS-EP, 1 mm MgCl2. The chip surface was regenerated using 0.1% sodium dodecyl sulphate (SDS) (flow rate: 30 μl/min; contact time: 30 s). Kinetic studies were performed at least in triplicate. Defined concentrations (from 0 to 50 nm) of POU1F1 proteins were injected with a 5 min association phase and 8 min dissociation phase and their dose-dependency response was measured. Data were analyzed with the BIAevaluation software 4.1 and the Kd dissociation constant was determined using the Fit kinetic simultaneous KA/KD (1:1 binding; Langmuir algorithm) and validated when the χ2 was <10 (this value means that the model used for fitting adequately describes the data).
Subcellular localization
HEK293T cells were seeded at 50% confluence on a strip into each chamber of a six-chamber tissue culture plate. After transfection of pcDNA4-POU1F1_WT-HA or pcDNA4-POU1F1_P76L-HA, cells were fixed 24 h in 4% paraformaldehyde and permeabilized in PBS1X-Triton 0.1%. Slides were then blocked with PBS1X-Triton 0.1%-BSA10% and incubated with an anti-HA mouse monoclonal antibody (Sigma) (1/1000) for 1 h. The strips were washed and incubated with the Alexa488-goat anti-mouse secondary antibody (Invitrogen) (1/2000) for 1 h. Nuclear counterstaining was performed with Vectashield containing 4′,6-diamidino-2-phenyllindole dihydrochloride (DAPI) (Vector Laboratories). Immunostaining was then visualized on a Nikon eclipse 80i microscope, and images were captured using a Qimaging (Retiga 2000R) camera and Image Pro Express 6.0 software.
Co-immunoprecipitation
HEK293T cells (3 × 106) were transfected with 1.5 µg of pcDNA4-POU1F1_WT-HA or of pcDNA4-POU1F1_P76L-HA plasmids and 1.5 µg of one of the plasmid encoding the cofactor tested (pcDNA3-PITX1, pcDNA3-LHX3a-Flag or pcDNA3-ELK1-Flag). Nuclear extracts prepared after 30 h of expression were divided into two parts and incubated overnight with each antibody, using Universal magnetic CoIp kit (Active Motif) following the manufacturer's instructions. Purified proteins from magnetic beads were then resolved by SDS-polyacrylamide gel electrophoresis (PAGE), immunoblotted and revealed by chemiluminescence (Supersignal West Dura Chemiluminescent Substrate, Pierce, Thermo Scientific). The Quantity One 1D analysis software (Bio-Rad) was used to visualize and to quantify (Volume Rectangle tool) the complex formed between the two proteins: histograms represent protein immunoprecipitated/input protein expression ratios.
Electrophoretic mobility shift assay
The LightShift Chemiluminescent EMSA kit (Pierce) was used for the study. Twenty fmoles of different Biotin end-labeled DNA duplexes (promGH1 70 bp, HSI 212 bp, prox-GH1 32 bp, prox-GH2 30 bp, HSI-A 37 bp, HSI-B 37 bp, HSI-C 38 bp, PRL1 (=3P) 29 bp, PRL2 (=1P) 29 bp (23) were incubated for 20 min at room temperature with 200 ng of POU1F1 purified proteins. The DNA–protein complexes were subjected to a 5 or 6% native-PAGE and transferred using the PierceG2 Fast-blotter system (Thermo-Scientific) to a nylon membrane (Biodyne B, Pierce). After transfer, the membrane was immediately cross-linked (UV Stratalinker 2400, Stratagene) using the autocrosslink program. A chemiluminescent method utilizing a luminol/enhancer solution and a stable peroxide solution (Pierce) was used as described by the manufacturer.
Mouse model
The P76L mutation was introduced into the mouse genome via standard homologous recombination in mouse embryonic stem (ES) cells. The recombinant ES cells were validated for the presence of the correspondingly introduced single-nucleotide substitution as well as the presence of the NeoR selection marker and Myc epitope tag by direct sequencing of the Pou1f1 locus of an adult wt/P76L mouse tail DNA. The NeoR cassette, flanked by a unidirectional set of LoxP sites, was deleted by crossing the mouse with an EIIA-Cre mouse. The final mutant gene was validated by multiple-targeted PCR and sequence analyses.
Funding
The work was supported by INSERM.
Acknowledgements
We thank all patients and family members for their cooperation.
Conflict of Interest statement. None declared.
References
- 1.Rizzoti K. (2015) Genetic regulation of murine pituitary development. J. Mol. Endocrinol., 54, R55–R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen B., Rosenfeld M.G. (2001) POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev., 22, 2–35. [DOI] [PubMed] [Google Scholar]
- 3.Ho Y., Cooke N.E., Liebhaber S.A. (2015) An autoregulatory pathway establishes the definitive chromatin conformation at the pit-1 locus. Mol. Cell Biol., 35, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingraham H.A., Flynn S.E., Voss J.W., Albert V.R., Kapiloff M.S., Wilson L., Rosenfeld M.G. (1990) The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell, 61, 1021–1033. [DOI] [PubMed] [Google Scholar]
- 5.Andersen B., Rosenfeld M.G. (1994) Pit-1 determines cell types during development of the anterior pituitary gland. A model for transcriptional regulation of cell phenotypes in mammalian organogenesis. J. Biol. Chem., 269, 29335–29338. [PubMed] [Google Scholar]
- 6.Savage J.J., Yaden B.C., Kiratipranon P., Rhodes S.J. (2003) Transcriptional control during mammalian anterior pituitary development. Gene, 319, 1–19. [DOI] [PubMed] [Google Scholar]
- 7.Szeto D.P., Ryan A.K., O'Connell S.M., Rosenfeld M.G. (1996) P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc. Natl Acad. Sci. USA, 93, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach I., Rhodes S.J., Pearse R.V., Heinzel T., Gloss B., Scully K.M., Sawchenko P.E., Rosenfeld M.G. (1995) P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc. Natl Acad. Sci. USA, 92, 2720–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobrier M.-L., Brachet C., Vié-Luton M.-P., Perez C., Copin B., Legendre M., Heinrichs C., Amselem S. (2012) Symptomatic heterozygotes and prenatal diagnoses in a nonconsanguineous family with syndromic combined pituitary hormone deficiency resulting from two novel LHX3 mutations. J. Clin. Endocrinol. Metab., 97, E503–E509. [DOI] [PubMed] [Google Scholar]
- 10.Yang X., Jin Y., Cattini P.A. (2010) Appearance of the pituitary factor Pit-1 increases chromatin remodeling at hypersensitive site III in the human GH locus. J. Mol. Endocrinol., 45, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Crenshaw E.B.d., Rawson E.J., Simmons D.M., Swanson L.W., Rosenfeld M.G. (1990) Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature, 347, 528–533. [DOI] [PubMed] [Google Scholar]
- 12.Radovick S., Nations M., Du Y., Berg L.A., Weintraub B.D., Wondisford F.E. (1992) A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science, 257, 1115–1118. [DOI] [PubMed] [Google Scholar]
- 13.Jones B.K., Monks B.R., Liebhaber S.A., Cooke N.E. (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell Biol., 15, 7010–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennani-Baïti I.M., Asa S.L., Song D., Iratni R., Liebhaber S.A., Cooke N.E. (1998) DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc. Natl Acad. Sci. USA, 95, 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shewchuk B.M., Asa S.L., Cooke N.E., Liebhaber S.A. (1999) Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J. Biol. Chem., 274, 35725–35733. [DOI] [PubMed] [Google Scholar]
- 16.Su Y., Liebhaber S.A., Cooke N.E. (2000) The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J. Biol. Chem., 275, 7902–7909. [DOI] [PubMed] [Google Scholar]
- 17.Fleetwood M.R., Ho Y., Cooke N.E., Liebhaber S.A. (2012) DNase I hypersensitive site II of the human growth hormone locus control region mediates an essential and distinct long-range enhancer function. J. Biol. Chem., 287, 25454–25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho Y., Shewchuk B.M., Liebhaber S.A., Cooke N.E. (2013) Distinct chromatin configurations regulate the initiation and the maintenance of hGH gene expression. Mol. Cell Biol., 33, 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho Y., Elefant F., Cooke N., Liebhaber S. (2002) A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell, 9, 291–302. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y., Tadevosyan A., Liebhaber S.A., Cooke N.E. (2008) The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep., 9, 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo E.J., Cooke N.E., Liebhaber S.A. (2012) An RNA-independent linkage of noncoding transcription to long-range enhancer function. Mol. Cell Biol., 32, 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo E.J., Brown C.D., Tsai Y.-C., Cooke N.E., Liebhaber S.A. (2015) Autonomous actions of the human growth hormone long-range enhancer. Nucleic Acids Res., 43, 2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shewchuk B.M., Ho Y., Liebhaber S.A., Cooke N.E. (2006) A single base difference between Pit-1 binding sites at the hGH promoter and locus control region specifies distinct Pit-1 conformations and functions. Mol. Cell Biol., 26, 6535–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turton J.P., Strom M., Langham S., Dattani M.T., Le Tissier P. (2012) Two novel mutations in the POU1F1 gene generate null alleles through different mechanisms leading to combined pituitary hormone deficiency. Clin. Endocrinol. (Oxf.), 76, 387–393. [DOI] [PubMed] [Google Scholar]
- 25.Pfäffle R., Klammt J. (2011) Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract. Res. Clin. Endocrinol. Metab., 25, 43–60. [DOI] [PubMed] [Google Scholar]
- 26.Fofanova O.V., Takamura N., Kinoshita E., Yoshimoto M., Tsuji Y., Peterkova V.A., Evgrafov O.V., Dedov I.I., Goncharov N.P., Yamashita S. (1998) Rarity of PIT1 involvement in children from Russia with combined pituitary hormone deficiency. Am. J. Med. Genet., 77, 360–365. [DOI] [PubMed] [Google Scholar]
- 27.Ohta K., Nobukuni Y., Mitsubuchi H., Fujimoto S., Matsuo N., Inagaki H., Endo F., Matsuda I. (1992) Mutations in the Pit-1 gene in children with combined pituitary hormone deficiency. Biochem. Biophys. Res. Commun., 189, 851–855. [DOI] [PubMed] [Google Scholar]