Abstract
Ataxia-telangiectasia (A-T) is a rare multi-system disorder caused by mutations in the ATM gene. Significant heterogeneity exists in the underlying genetic mutations and clinical phenotypes. A number of mouse models have been generated that harbor mutations in the distal region of the gene, and a recent study suggests the presence of residual ATM protein in the brain of one such model. These mice recapitulate many of the characteristics of A-T seen in humans, with the notable exception of neurodegeneration. In order to study how an N-terminal mutation affects the disease phenotype, we generated an inducible Atm mutant mouse model (Atmtm1Mmpl/tm1Mmpl, referred to as A-T [M]) predicted to express only the first 62 amino acids of Atm. Cells derived from A-T [M] mutant mice exhibited reduced cellular proliferation and an altered DNA damage response, but surprisingly, showed no evidence of an oxidative imbalance. Examination of the A-T [M] animals revealed an altered immunophenotype consistent with A-T. In contrast to mice harboring C-terminal Atm mutations that disproportionately develop thymic lymphomas, A-T [M] mice developed lymphoma at a similar rate as human A-T patients. Morphological analyses of A-T [M] cerebella revealed no substantial cellular defects, similar to other models of A-T, although mice display behavioral defects consistent with cerebellar dysfunction. Overall, these results suggest that loss of Atm is not necessarily associated with an oxidized phenotype as has been previously proposed and that loss of ATM protein is not sufficient to induce cerebellar degeneration in mice.
Introduction
Ataxia-telangiectasia (A-T) is a rare hereditary disorder affecting approximately 1/40 000 live births, with symptoms that include radiosensitivity, immunodeficiency, increased incidence of malignancy and progressive ataxia that is believed to be due to cerebellar degeneration (1–7). A-T patients harbor mutations that are dispersed throughout the ATM gene, and most mutations are predicted to generate protein truncations (8–10). The heterogeneity of ATM mutations is reflected in the significant variety observed in the clinical presentation of A-T disease pathologies (11–13).
Several different transgenic models of A-T, with differing mutations, have been created in order to study this disease, but these display a confusing set of phenotypes. In particular, all mouse models of A-T are associated with a high rate of tumor development in the mutant mice, with a predominance of lymphomas. In contrast, although there is a marked elevation in the frequency of cancer in individuals with A-T compared with the rest of the population, malignancies appear to be restricted to ∼10–20% of A-T patients (14–17). Moreover, mouse models of A-T show relatively minor alterations in cerebellar structure, although defects in Purkinje neurons were described (18,19). In addition, it has also been shown that ATM kinase-inactive mutations in mice lead to embryonic lethality in mice (20,21).
Based on the inability to detect the ATM protein by immunoblotting, the truncating and in-frame deletions used in existing mouse models of A-T mutations were believed to destabilize the Atm product and result in a complete loss of the protein (22–29). A recent study, however, found a novel, brain-specific splicing event in the Atmtm1Awb/tm1Awb mouse (here referred to as A-T [A]) that results in nearly full-length Atm mRNA, and a catalytically active protein that effectively responds to DNA damage (30). The potential presence of truncated Atm introduces an additional level of complexity, making it difficult to determine what aspects of A-T can be ascribed to the loss of ATM, or are instead due to altered functions of a residual truncated ATM protein, which may modulate secondary pathways, such as oxidative stress (31–33).
We generated an inducible Atm mutant mouse designed to cause a truncating mutation in the proximal region of the gene by removing exon 4. This mutation results in a severely truncated ATM protein fragment consistent with mutations observed in canonical cases of A-T in humans (10,11,29,34) and lacking all known sites by which ATM interacts with other proteins. We found that while the resulting mutant mice exhibit multiple features present in other murine mutant models of A-T, several marked distinctions with existing models were observed. The most prominent differences include an absence of detectable oxidative imbalance, a decreased incidence of cancers and an altered ability of cells to engage in DNA damage repair. The absence of tumors, which allows for an extended survival period, was not associated with late onset cerebellar defects, contrary to suggestions made in regards to other A-T models where an absence of cerebellar degeneration was often explained by the premature cancer phenotype and death. While we could not identify morphological defects in the cerebellum in our new mouse model, animals displayed robust behavioral defects consistent with cerebellar dysfunction.
Results
Generation of Atmtm1Mmpl mice
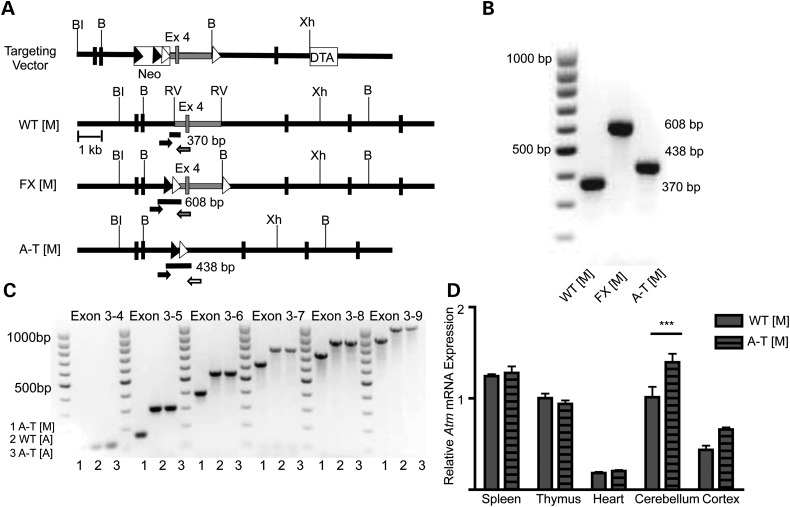
To study the effects of a truncating null Atm mutation, we generated mice containing two loxP sites flanking exon 4, (Atmtm1Mmpl flx/flx referred to as FX [M]). We first inserted an FRT-Neo-FRT-loxP cassette into intron 3 and an AvrII-BamHI-loxP site into intron 4 of the mouse Atm gene (Fig. 1A). A chimeric male generated from ES cell clones was crossed with ROSA26Sortm1(FLP1)Dym/J female mice to remove the neomycin cassette. To induce loss of Atm in all tissues, we crossed the FX [M] mice with CMV-Cre B6.C-Tg (CMV-cre1Cgn/J, Jackson Laboratory) transgenic mice, demonstrating that the loxP flanked exon 4 of the Atm gene could be efficiently deleted in vivo, thereby generating the novel Atm mutant allele (Atmtm1Mmpl/tm1Mmpl, referred to as A-T [M] throughout the manuscript [Fig. 1A]). Cre-mediated excision of Atm exon 4 is comparable to mutations found in humans (10,11,29,34) and is predicted to cause a frameshift mutation through the splicing of exon 3 to exon 5. Using PCR analysis of genomic DNA samples, we were able to identify the wild-type Atm allele (Atmtm1Mmpl+/+, 370 bp, referred to as WT [M]), the floxed, but not excised allele (FX [M], 608 bp), and the knock-out allele (A-T [M], 438 bp) (Fig. 1A and B). In light of recent reports of the embryonic lethality observed in mice carrying Atm kinase-inactive mutations (20,21), we also examined if the offspring were obtained in the expected Mendelian ratios when we performed heterozygous crosses of Atmtm1Mmpl/+ mice. From 66 litters, we obtained 549 live births containing 278 Atmtm1Mmpl/+ (50.6%), 158 Atmtm1Mmpl+/+ (28.8%) and 113 Atmtm1Mmpl/tm1Mmpl (20.6%) mice, which was significantly different from the predicted Mendelian ratios (Chi-square = 7.466; degrees of freedom 2; P = 0.023). This suggests that loss of full-length ATM is associated with decreased embryonic development and/or postnatal viability.
Figure 1.
Generation of the Atmtm1Mmpl allele. (A) Strategy to introduce loxP sites into introns 3 and 4 of the mouse Atm gene by homologous recombination in ES cells. BI = BstEII; B = BamHI; RV = EcoRV; Xh = XhoI; Neo = neomycin positive selection cassette; DTA = diphtheria toxin A negative selection cassette. FRT sites are indicated with black triangles, loxP sites shown as white triangles. Genotyping primers; primer 1 (black arrow), primer 2 (gray arrow), primer 3 (white arrow). (B) PCR genotyping distinguishes between WT (M) (370 bp), FX [M] (608 bp) and A-T [M] (438 bp). (C) PCR analysis of cerebellar Atm transcripts from A-T [M], WT [A] and A-T [A] is consistent with the absence of exon 4 (145 bp) in the mRNA of the A-T [M] mice. The presence of single PCR bands using multiple primer sets demonstrates the absence of splice variants in exons 3–9 of the mouse Atm gene in cerebellar tissue. (D) qPCR analysis of Atm expression in the spleen, thymus, heart, cerebellum and cortex of A-T [M] and WT [M] mice. Atm expression in all tissues normalized to the expression of Atm in the wild thymus. Atm transcripts were significantly higher in the cerebellum of A-T [M] mice but not the spleen, thymus, heart or cortex as compared with WT [M] controls. A-T [M] refers to (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl+/+), FX [M] (Atmtm1Mmpl flx/flx). ***P < 0.001 relative to control treatment; ANOVA followed by Bonferroni's post hoc test. Data plotted as mean ± S.E.M. of (n ≥ 3) independent samples.
Recent reports by Li and colleagues (30) demonstrated the presence of a novel mRNA splice variant in the cerebella of a well-studied A-T mouse model (Atmtm1Awb/tm1Awb (23), referred to as A-T [A] in this manuscript). These mice were found to express a brain-specific splice variant that encoded a truncated, but catalytically active ATM protein (30). In order to ascertain the level of Atm expression, we performed qPCR using a primer/probe set specific for the Atm exon 57–58 junction. We were able to detect Atm mRNA in the spleen, thymus, heart and cerebellum of both WT [M] and A-T [M] mice. The level of Atm mRNA in A-T [M] mice was unchanged in the spleen, thymus and heart, but was significantly higher in the cerebellum (Fig. 1D).
In the A-T [M] mouse, we predicted that the first 62 amino acid codons of the mRNA are synonymous with the wild-type mRNA, but would encode multiple stop codons following the eighty-third amino acid. The absence of transcripts containing exon 3 to exon 7 splicing in the A-T [M] mouse would, however, be especially significant because this transcript would contain an in-frame deletion that would allow for the translation of the majority of the ATM protein. For the analysis of brain-specific Atm splice variants, we first generated cDNA from the cerebella of Atmtm1Awb+/+ (referred to as WT [A]), A-T [A] and A-T [M] animals. Using conventional PCR, we found that Atm mRNA from A-T [M] mice was 145 base pairs smaller than the wild-type transcript, corresponding to the deletion of Atm exon 4 from the genome. The cDNA from these animals also generated a single PCR product using primer pairs designed to probe for the presence of downstream exons 5 through 9. This indicated the absence of splice variants within the Atm regions tested in the cerebellar tissue (Fig. 1C).
Although the conventional PCR data indicated the absence of compensatory splicing in the immediate vicinity of the exon 4 deletion, we wanted to confirm our initial analysis while also testing for alternative splicing in other regions of the message. Total RNA was collected from the cerebella of postnatal day 8 A-T [M] and WT [M] mice, then profiled by RNAseq analysis as described in the methods. Our analysis confirmed the absence exon 4 and the presence of the exon 3/5 splice product in all detectable A-T [M] cerebellar Atm mRNA. No compensatory splicing that would be predicted to generate a functional protein product was detected in any regions of the Atm mRNA (Supplementary Material, Table S1 and Fig. S1).
In contrast to the lack of Atm mRNA expression observed in the thymus and spleen of A-T [A] mice (30), Atm message was found in the thymus, spleen, heart and cerebellum of A-T [M] mice. We therefore also explored the status of Atm mRNA splicing in A-T [M] mice using conventional sequencing analysis. In the A--T [M] spleen, the exon 3/5 splice product was detected in conjunction with the presence of multiple in-frame stop codons <100 nucleotides downstream of the new exon/exon junction (Supplementary Material, Fig. S2).
Loss of ATM impairs cell proliferation and alters the DNA damage response in A-T [M] cells
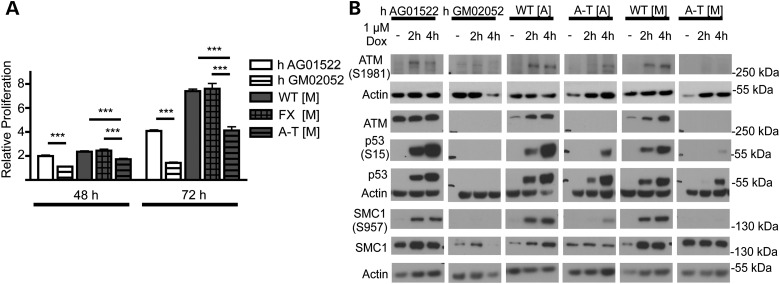
A-T fibroblasts have been characterized as having impaired proliferation and defects in the DNA damage response following treatment with radiomimetic drugs or ionizing radiation (35–38). We examined the growth properties of fibroblasts isolated from the A-T [M] mice. As an additional control we used human A-T cells derived from patient fibroblasts (h A-T GM02052). These cells harbor a 103C>T transition that generates a truncating mutation in exon 5 of ATM (34) and is hence similar to the truncating null mutation at the proximal region of Atm that we introduced in our A-T [M] mice. When grown in the same culture conditions, the mouse and human A-T fibroblasts proliferated at a reduced rate as compared with the appropriate control fibroblasts (Fig. 2A).
Figure 2.
Proliferation and DNA damage response in wild-type and ATM mutant fibroblasts. (A) Proliferation of ATM mutant (GM02052 and A-T [M]) fibroblasts is reduced relative to wild-type controls (AG01522 and WT [M]) as determined by counting Hoechst(+)/PI(−) cells at 24, 48 and 72 h. (B) Representative chemiluminescent protein immunoblot images of ATM mutant and wild-type fibroblasts. Full-length ATM protein was not detected in ATM mutant fibroblasts. Following 2 and 4 h of doxorubicin treatment, ATM mutant fibroblasts show decreased levels of p53, p53(Ser15) and SMC1(Ser957) protein relative to wild-type controls. ***P < 0.001 relative to control treatment; ANOVA followed by Bonferroni's post hoc test. Data plotted as mean ± S.E.M. of (n ≥ 3) independent experiments. A-T [A] refers to (Atmtm1Awb/tm1Awb), WT [A] (Atmtm1Awb+/+), A-T [M] (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl +/+).
As expected from the absence of a splice product, we could not detect full-length ATM protein in the lysates from the A-T [M] mouse fibroblasts (Fig. 2B). When the fibroblasts were treated with doxorubicin, a radiomimetic drug (39,40), for 2 and 4 h, we observed an accumulation of ATM(Ser1981) in both the wild-type and A-T [A] samples, confirming the presence of a residual ATM protein in the A-T [A] cells (30). We did not see such an accumulation in A-T [M] fibroblasts, consistent with an absence of a functional protein. Due to the role of ATM in the stabilization and activation of p53 in response to genotoxic stress (41–43), we next measured the levels of p53 and phosphorylated p53(Ser15) in whole-cell extracts obtained from fibroblasts treated with doxorubicin (Fig. 2B). At 2 and 4 h after doxorubicin treatment, a substantial increase in the levels of total p53 and phosphorylated p53(Ser15) was seen in the wild-type h AG01522, WT [A] and WT [M] fibroblasts. In comparison, p53 and phospho-p53 did not accumulate in h A-T GM02052 fibroblasts, but was seen in mouse A-T [A] cells after 2 h of treatment. Stabilization of p53 was considerably more delayed in mutant fibroblasts isolated from our A-T [M] animals, and was only detected after 4 h of doxorubicin treatment. As phosphorylation of p53(Ser15) can occur independently of ATM activity through other kinases such as ATR (44), we also measured SMC1(Ser957), which is phosphorylated in an ATM-dependent manner in response to ionizing radiation (45). Consistent with the lack of ATM, ATM(Ser1981) and delayed p53(Ser15) induction in our A-T [M] cells, we observed no SMC1(Ser957) at 2 and 4 h after doxorubicin treatment in A-T [M] fibroblasts. In contrast, A-T [A] fibroblasts, showed a small, but detectable level of SMC1(Ser957) at 4 h after doxorubicin treatment (Fig. 2B), suggesting the presence of a partially functional ATM protein in A-T [A] mice.
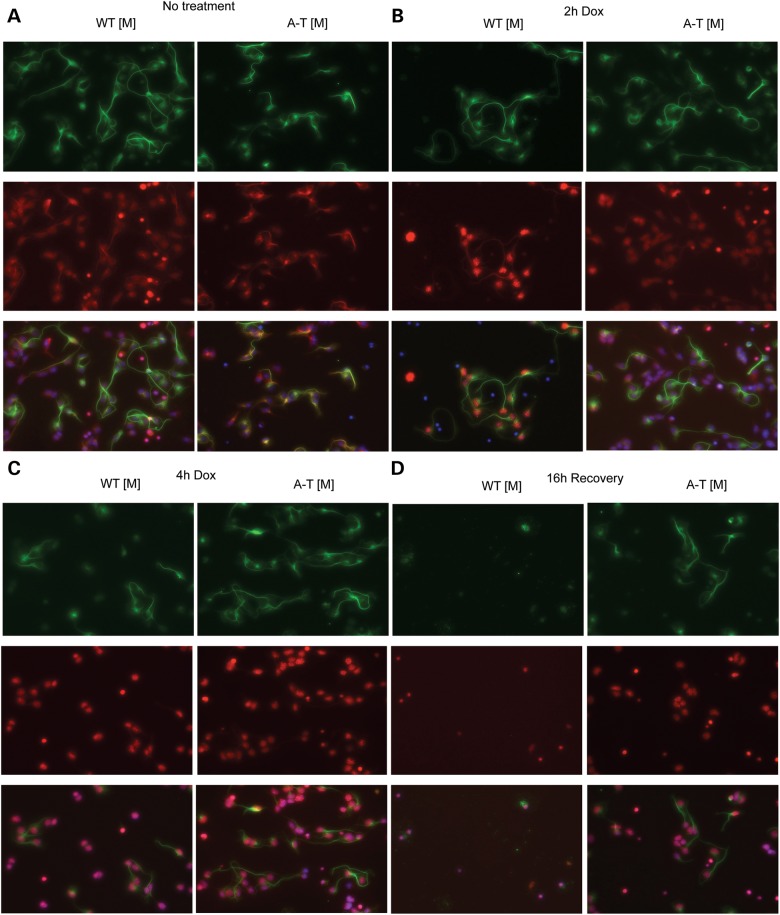
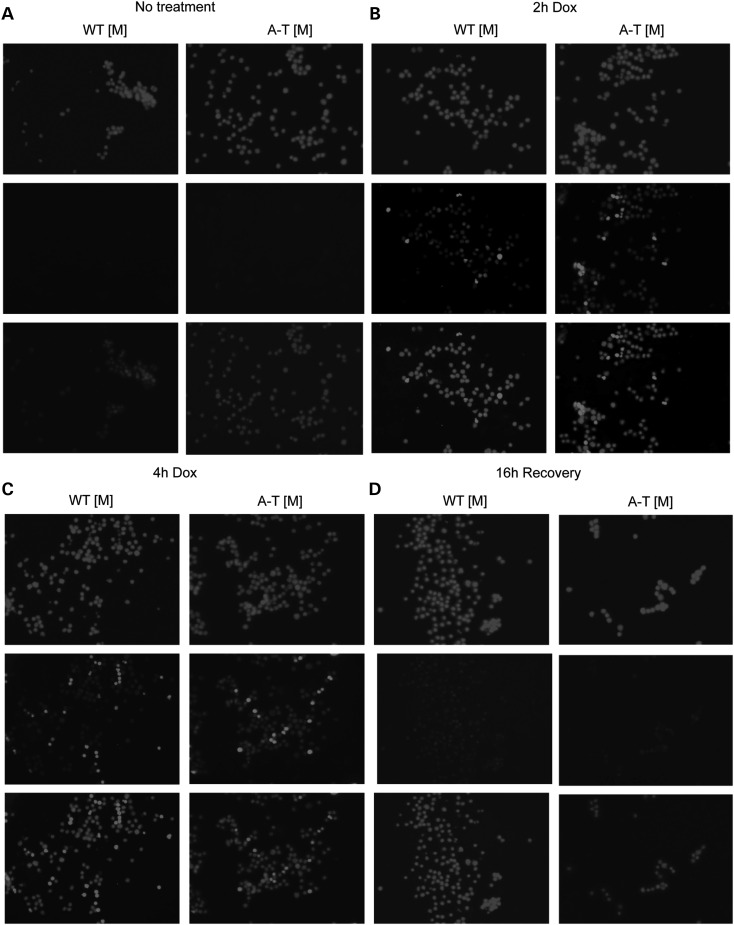
A prominent feature of A-T is the degeneration of cerebellar neurons that is thought, in part, to result from decreased survival signals following DNA damage (46–48). In addition, induction of cell death by ionizing radiation is decreased in several regions of the brain, including the cerebellum of A-T mice when compared with wild-type controls (28). As p53 activation is required for the induction of ionizing-radiation-induced apoptosis (28) and we observed a decrease in p53(Ser15) following doxorubicin treatment of A-T [M] cells, we next examined whether radiomimetic treatment of A-T [M] cerebellar granule neurons (CGN) with doxorubicin resulted in altered cell survival. Consistent with our observation of a delayed DNA damage response in A-T [M] fibroblasts, robust γH2AX phosphorylation and nuclear accumulation was observed in WT [M], but not A-T [M] CGNs 2 h after doxorubicin treatment (Fig. 3B). Both W-T [M] and A-T [M] CGNs showed uniform γH2AX phosphorylation and nuclear accumulation at 4 h after treatment (Fig. 3C). When the doxorubicin was removed and the CGNs were allowed to recover for an additional 16 h, there was widespread cell death in both the WT [M] and A-T [M] cultures. Surprisingly, the A-T [M] CGNs, showed a decreased susceptibility to radiomimetic-induced cell death, as there were substantially more surviving CGNs in the mutant cultures 20 h after the addition of doxorubicin (Fig. 3D).
Figure 3.
Doxorubicin induced γH2AX nuclear accumulation in cerebellar neurons. Mixed cerebellar cultures isolated from P8 mice were left untreated (A) or dosed with 1 µm doxorubicin (B and C) for 2 and 4 h, respectively. (D) Following a 4 h treatment, the 1 µm doxorubicin was removed and the cerebellar cultures were incubated another 24 h before fixation. (B and C) Doxorubicin treated WT [M] neurons (Tubulin β-III, green) show robust nuclear γH2AX (red) accumulation and foci formation at 2 and 4 h. Treatment of A-T [M] neurons (Tubulin β-III, green) with doxorubicin resulted in impaired nuclear accumulation of γH2AX (red) at 2 and 4 h. (D) 24 h after treatment, extensive nuclear γH2AX accumulation was observed in A-T [M] neurons. In contrast, doxorubicin resulted in the death of WT [M] neurons 24 h after treatment. (n = 3). A-T [M] refers to (Atmtm1Mmp1/tm1Mmp1), WT [M] (Atmtm1Mmpl+/+).
Loss of ATM alters the immunophenotype but does not increase the incidence of T-cell lymphoma
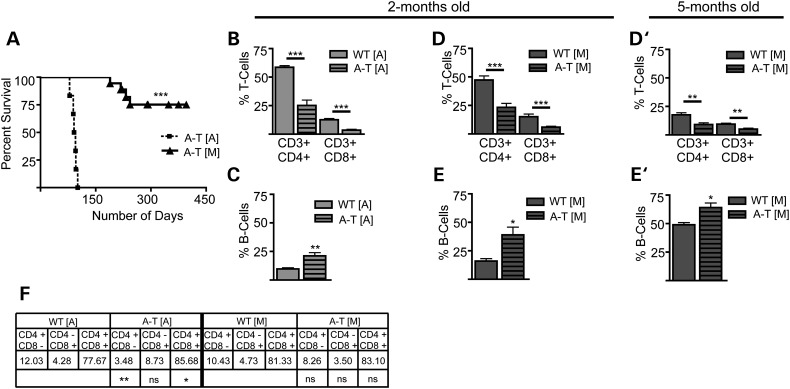
ATM plays a central role in the resolution of DNA double strand breaks required for the maturation of the T-cell receptor, and the impairment of this process has been implicated in the development of the thymic T-cell lymphomas seen in multiple mouse models of A-T and in patients (49–53). Previously generated Atm mutant mice are highly prone to the development of thymic lymphomas and although some variability exists in the time required for these animals to succumb to the disease, tumors may develop as early as 9 weeks of age. Even in strains with reduced lymphoma incidence, these tumors result in the death of mice by 9 months with various penetrance (22,54). Consistent with previous findings, in our hands A-T [A] animals developed aggressive thymic lymphomas with 100% of our A-T [A] mice expiring by 120 days of age due to complications arising from the tumors (Fig. 4A).
Figure 4.
Determination of lifespan and characterization of immunophenotype in Atm mutant mice. (A) Kaplan–Meier survival curve illustrates a significant increase in the survival of A-T [M] (n = 25) mice as compared with the A-T [A] (n = 6) animals. (B) Flow cytometry Analysis of circulating T-cells in A-T [A] animals (light gray bar) shows a significant decrease in CD3+/CD4+ and Cd3+/CD8+ at 2 months of age. (C) The percentage of circulating B-cells (B220+) in A-T [A] mice is significantly increased at 2 months of age. (D and D′) A-T [M] mice show a significant decrease in the percentage of circulating T-cells (CD3+/CD4+ and Cd3+/CD8+) at 2 months and 5 months of age, respectively. (E and E′) A-T [M] mice also had and a significant increase in the percentage of circulating B-cells (B220+) at 2 months and 5 months of age, respectively. (F) Flow cytometry analysis found a significant decrease in the percentage of CD4+/CD8− cells and a significant increase in the CD4+/CD8+ double positive T-cells residing in the thymus of 3-month-old A-T [A] (light gray striped bar) mice as compared with thymocytes harvested from WT [A] mice (light gray bar). *P < 0.05, **P < 0.01, ***P < 0.001 relative to wild-type control; Student's t-test. Data plotted as mean ± S.E.M. of (n ≥ 3) samples isolated from independent animals. A-T [A] refers to (Atmtm1Awb/tm1Awb), WT [A] (Atmtm1Awb+/+), A-T [M] (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl+/+).
In contrast, A-T [M] mice very rarely developed thymic lymphomas or any other cancers. In a cohort of 25 A-T [M] animals, all animals were alive at 120 days before cohorts were used experimentally or euthanized for tissue analysis. Eight animals were kept alive beyond 120 days: two of these animals (8% of the cohort) succumbed to thymic lymphomas at 124 and 218 days of age, two animals were euthanized due to infections (232 and 243 days of age), three were lost to unknown causes at 146, 156 and 189 days of age and one animal remained without signs of disease well over a year and was finally euthanized (Fig. 4A). The rate of lymphoma development in this small A-T [M] cohort reflects the rates of cancer incidence in human A-T patients, where 10–20% of patients develop malignancies with a majority involving non-Hodgkin lymphoma and leukemia (14–17,55).
To determine whether the low incidence of thymic lymphomas in A-T [M] mice might be due to an absence of the developmental immunologic abnormalities described in other Atm mutant mouse strains, we performed an analysis of peripheral blood (PB) lymphocytes from A-T [A] and A-T [M] mice using flow cytometry. Surface marker staining for CD3, CD4 and CD8 antigens showed a comparable reduction in the percentage of T-cell subsets within the circulating lymphocyte population of A-T [A] and A-T [M] mutant mice at 2 months of age (Fig. 4B and D). When PB was analyzed with the surface marker B220, we also observed a comparable increase in the percentage of B cells within the circulating lymphocyte populations in the A-T [A] and A-T [M] mutant mice at 2 months of age (Fig. 4C and E). The percentage of circulating T and B-cells was also analyzed using the PB of 5-month-old A-T [M] mice, a time point at which all A-T [A] have already succumbed to lymphomas. The decrease in the percentage of circulating T-cells and increase in the percentage of circulating B-cells were both maintained in the PB of the A-T [M] mice (Fig. 4D′ and E′). Thus, a similar alteration in the T and B-cell profiles in PB was observed in both mouse models.
As the development of lymphomas in A-T [A] might be a result of impaired maturation of thymocytes (5,6,25,27,56–61), we next determined whether Atm mutations in A-T [A] or A-T [M] mice altered the development of T-cells within the thymus. Cells isolated from the thymuses of 3-month-old (lymphomic) A-T [A] and 7-month-old (healthy) A-T [M] mice were analyzed by flow cytometry. Thymocytes isolated from the A-T [A] mice showed a significant increase in the levels of immature CD4/CD8 double positive T-cells and a significant decrease in the levels of more mature CD4 and CD8 single positive cells (Fig. 4F). The expansion of the immature CD4+/CD8+ thymocytes is consistent with the phenotype of the T-cell lymphoma that develops in the A-T [A] mice (23), although it is not clear whether such cells are indeed the tumor initiating population. In contrast with observations in A-T [A] mice, T-cell populations isolated from the 7-month-old A-T [M] thymuses showed no change in the distribution of immature CD4/CD8 double positive T-cells and mature T-cells compared with wild-type (Fig. 4F). This observation is consistent with the absence of tumor formation, even at an age at which all of the A-T [A] mice have succumbed to thymic lymphomas.
Although ATM is important for the maturation of thymocytes, its loss does not render these cells more sensitive to radiation-induced apoptosis (28,35,62,63) and likely involves the redundant functions of DNA-PKcs in phosphorylating DNA repair proteins and initiating p53 dependent apoptosis (64). Consistent with these observations, A-T [M] thymocytes treated with doxorubicin exhibited similar levels of γH2AX phosphorylation at 2, 4 and 20 h (Fig. 5) and no overt changes in the relative number of live cells as compared with WT [M] thymocytes.
Figure 5.
Doxorubicin induced γH2AX nuclear accumulation in thymocytes. Mixed thymocyte cultures isolated from 3-month-old mice were left untreated (A) or dosed with 1 µm doxorubicin (B and C) for 2 and 4 h, respectively. (D) Following a 4 h treatment, the 1 µm doxorubicin was removed and the thymocyte cultures were incubated another 24 h before fixation. (B and C) Doxorubicin treated WT [M] and mutant A-T [M] thymocytes show robust nuclear γH2AX (lower panel) accumulation and foci formation at 2 and 4 h, and are resolved 24 h after insult (D). (n = 3). A-T [M] refers to (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl+/+).
Loss of ATM function is not sufficient to increase oxidative stress
It has been suggested that DNA damage is associated with oxidative stress (65), which is one of the driving forces for tumor formation (and other pathologies) and is thought to contribute significantly to the pathogenesis of the disease in a subset of A-T patients and A-T mutant mouse models (32,66–72). It also has been suggested that ATM maintains the redox state of the cell by controlling reactive oxygen species (ROS) formation, regulating mitochondria biogenesis and NADPH synthesis and altering cellular autophagy (73–77). We isolated cells and tissue from our new mouse line and determined their redox status by measuring the levels of intracellular ROS using CM-H2DCFDA fluorescence.
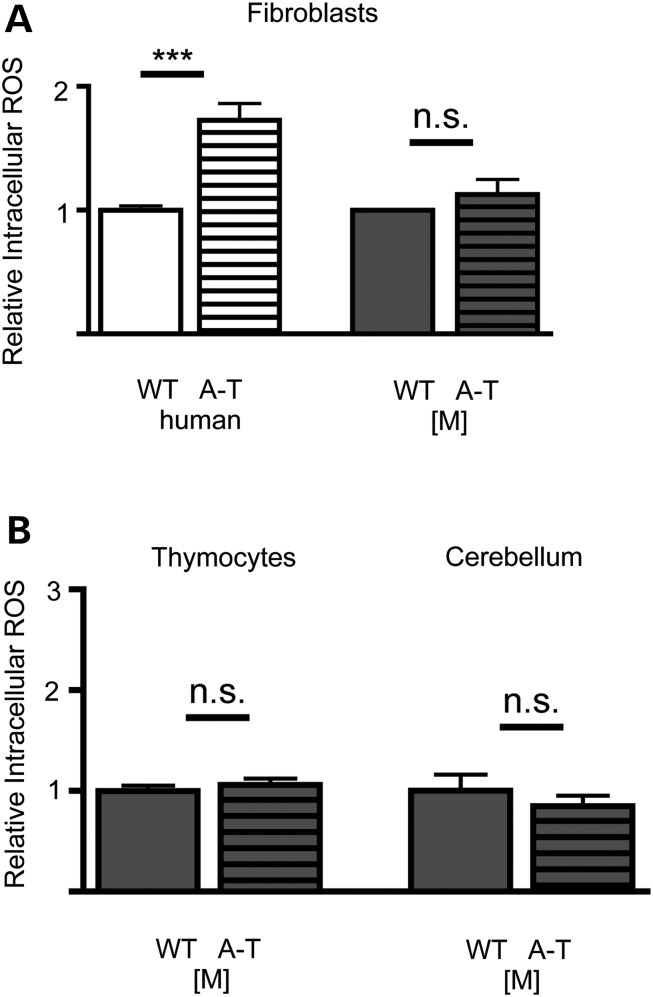
Surprisingly, while we found that the human A-T fibroblasts were more oxidized when compared with their respective controls, fibroblasts isolated from A-T [M] mice showed no significant difference in the levels of intracellular ROS compared with their wild-type controls (Fig. 6A, dark gray bars). As we are particularly interested in the cancer and neural pathology, and as it has been suggested that oxidative stress modulates or contributes to the pathologies observed in A-T [A] animals (32,70,71), we also measured the ROS levels of freshly dissected thymocytes and cerebellar tissue from P7 animals. Utilizing flow cytometry, we found again no significant change in the ROS levels in cells isolated from the A-T [M] mice compared with wild-type controls (Fig. 6B).
Figure 6.
Analysis of the oxidative state in ATM mutants. (A) Flow cytometry analysis revealed that Human ATM mutant (GM02052) are significantly more oxidized than control fibroblasts as determined by treatment with the redox sensitive dye CM-H2DCFDA. No difference in the oxidative state of A-T [M] and WT [M] fibroblasts was detected. (B) No significant differences in the oxidative state of A-T [M] thymocytes and cerebellar tissue were detected as compared with WT [M] controls. Not significant (n.s), ***P < 0.001 relative to prospective wild-type controls; Student's t-test. Data plotted as mean ± S.E.M. of (n ≥ 3) independent experiments and/or tissue samples. A-T [M] (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl+/+).
Loss of ATM function impairs motor behavior
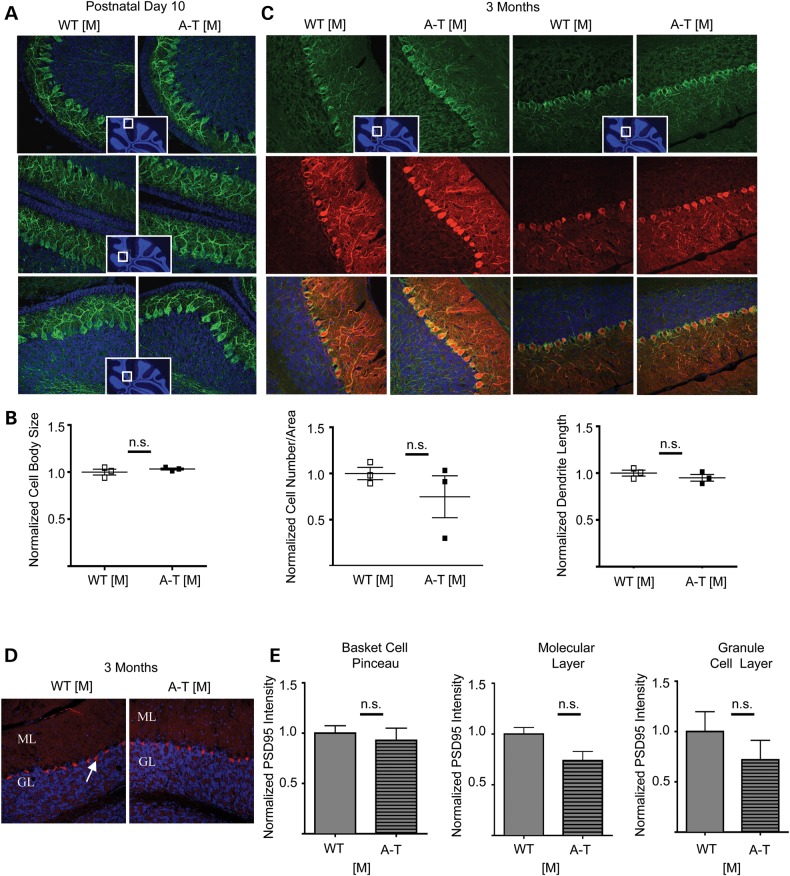
A-T animal models only mildly recapitulate the cerebellar degeneration seen in humans. As the cancer prone A-T [A] is the most widely studied model it has been suggested that lack of a cerebellar degeneration might simply be a consequence of the premature death that does not allow for the full neurological pathology to develop (78). As our A-T [M] animals did not develop tumors, we were able to characterize the neurological sequalae and motor behavior in the absence of a tumor burden. Labeling of sagittal cerebellar sections from 10-day-old and 3-month-old mice with the neurofilament marker SMI32 and Calbindin, a marker for Purkinje neurons, revealed no obvious abnormalities (Fig. 7A–C). While we did see occasional changes in the number and size of the Purkinje neuron cell bodies and changes in dendritic length in some regions in the A-T [M] cerebella compared with WT [M], such changes were not consistent and were not statistically significant (Fig. 7B).
Figure 7.
Immunohistochemical analysis of cerebellar sections. (A) Representative images of cerebellar sections from postnatal day 10 WT [M] and A-T [M] mice using the marker SMI-32 (Green). (B). Multiple parameters were measured for the Purkinje cells of the postnatal day 10, WT [M] and A-T [M] mice including cell body size, cell number per area and dendritic length. No significant (n.s) differences were observed when analyzed using the Student's t-test. Inserts show the location in the cerebellum of the prospective sections. (C) Immunohistochemical analysis of 3-month-old cerebellar sections. Representative images of cerebellar sections from WT [M] and A-T [M] mice using the markers SMI-32 (Green) and Calbindin (Red). The Purkinje cells co-labeled with these two markers and no gross abnormalities were observed in either genotype. Inserts show the location in the cerebellum of the prospective sections. Three mice were analyzed from each genotype. A-T [M] refers to (Atmtm1Mmpl/tm1Mmpl), WT [M] (Atmtm1Mmpl+/+). N = 3 mice per genotype, with at least three sections quantified per genotype. (D) Representative image of cerebella from 3-month-old WT [M] and AT [M] mice labeled with an anti-PSD95 antibody. Arrow indicates labeling in Basket cell pinceau. ML: molecular layer, GL granule layer. (E) No significant differences (n.s) were observed when the fluorescent intensity was quantified within the basket cell pinceau, the molecular layer and the granule layer. N = 3 per genotype. P > 0.05 Students T-test.
Even though there were no obvious abnormalities in gross cerebellar morphology or Purkinje cell number and appearance, we could not discount any alterations occurring on the level of synaptic function and/or synapse maturation. A recent report by Carlessi et al. (79) identified a number of deficits in hiPSC-derived neurons from A-T patients, including decreased expression of PSD95, a synaptic density protein shown to orchestrate synaptic development by driving maturation of glutamatergic synapses and enhancing maturation of the presynaptic terminal in cultured hippocampal neurons (80). To address whether there were changes in expression in our new A-T model in vivo, we labeled cerebellar sections from 3-month-old W-T and A-T [M] animals with PSD95 (Fig. 7D) and quantified the fluorescent intensity in multiple regions: the basket cell pinceau, the molecular layer and the granule layer. As shown in Figure 7E, we did not find any significant differences in the expression of PSD95 in any of these areas.
To determine whether loss of ATM might impair cellular function in the absence of gross changes in anatomical morphology, we conducted behavior analyses assessing cerebellar function. When we tested our animals in the commonly used rotorod paradigm, we did not see a statistically significant difference in the gross motor coordination of 4-, 7- or 9-month-old A-T [M] animals compared with WT controls (Supplementary Material, Fig. S3). In fact, the rotarod staying time was slightly improved in the A-T [M] animals at 7 and 9 months relative to the wild-type controls, and may reflect motivational changes, which can alter performance in this test.
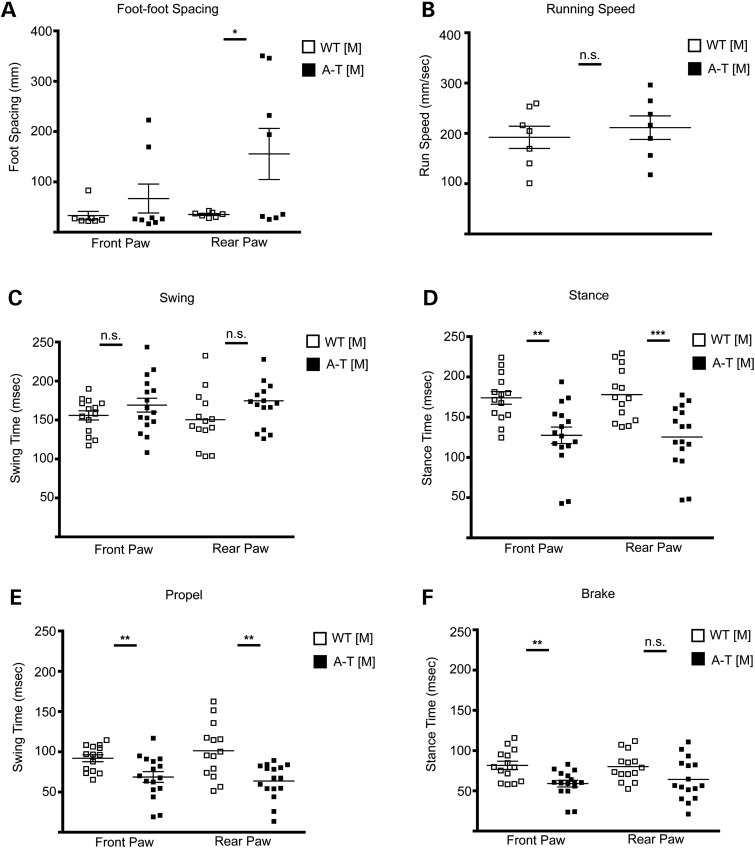
When we examined mice using the GaitScan system (Cleversys Inc.), which enables a more detailed analysis of motor skills linked to cerebellar function and records voluntary movement across an illuminated catwalk, we found evidence of behavioral alterations thought to be related to cerebellar function. We observed an overall significant increase in the foot spacing of the A-T [M] mice (Fig. 8A), which is consistent with a wide-spaced ataxic gait observed in other A-T mouse models (23). During voluntary locomotion, A-T [M] and WT [M] mice did not vary significantly in their running speed (Fig. 8B), which allowed us to compare speed-dependent measures such as stance (time during which the paws are in contact with the runway surface) and swing (time during which the paws are not in contact with the runway surface). A-T [M] animals did show a trend toward increased front and rear paw swing time, and a significant decrease in the stance time of all four limbs of locomotion as compared with WT [M] controls at 3 months of age. (Fig. 8C and D).
Figure 8.
A-T [M] mice show motor function defects. Mouse gait was filmed on a static runway and analyzed for (A) foot spacing, (B) running speed, (C) swing (time in which paw is not in contact with runway surface during each step), (D) stance (time in which paw is in contact with the runway surface during each step), (E) propel and (F) brake time in 3-month-old WT [M] and A-T [M] mice. Not significant (n.s), *P < 0.05, **P < 0.01, ***P < 0.001 relative to wild-type control animals; Student's t-test. Data plotted as mean ± S.E.M. of (n = 7) WT [M] and (n = 8) A-T [M] animals. A-T [M] refers to (Atmtm1Mmpl/tm1Mmp1), WT [M] (Atmtm1Mmpl+/+).
Discussion
Here we present the first example of an Atm amino-terminal targeted mutant mouse model of A-T. Unlike previous mouse models of A-T (23–25,28,30,35,48,81,82), which assume that any protein fragments that are generated by the distal Atm mutations are not functional or are degraded, the A-T [M] mouse was designed to translate a severely truncated amino-terminal Atm fragment. This mutation correlates with a well-characterized 103C-T transition mutation found in regions of North Africa that results in a stop codon at position 35 of the ATM protein (29), and closely resembles the truncating mutations also found in other patients (10,11). The mRNA transcribed by the A-T [M] mouse is predicted to allow for translation of the first 62 amino acid residues of ATM before the frameshift mutation introduced by the deletion of Atm exon 4 causes a non-synonymous substitution of 21 amino acids and multiple premature stop codons. As the deletion of Atm exon 4 leads to the loss of all known putative substrate binding sites (including the p53 and BRCA1 binding sites in the N-terminal region of the protein) (83,84), any residual protein that might be expressed in the cells would not be expected to interact with other cellular proteins that normally interact with Atm.
Alternative mRNA splicing is of particular concern, however, in the generation and analysis of A-T mouse models, as mutations that allow for the production of residual, functional protein product are associated with milder phenotypes, variant clinical presentation and correction of the cellular phenotype (85–88). As much as 48% of ATM mutations may be associated with defects in splicing (89), and alternative splicing has previously been described in the cerebellum of a well-characterized Atm mutant mouse (30). In the current study, we used an NGS strategy to sequence the cerebellar RNA from our WT [M] and A-T [M] mice. As predicted, the deletion of exon 4 in A-T [M] mice resulted in an exon 3/5-frameshift splice mutation. Expression of this mutant mRNA transcript was also detected in the spleen of A-T [M] animals, indicating that the exon 3/5 splice is not a brain-specific variant. This mutation did not generate new cryptic 5′ or 3′ splice sites that would be predicted to generate functional ATM product. While we cannot eliminate the possibility that alternative ATG initiation codons may be used to translate truncated ATM protein, our in vitro analysis of ATM expression and DNA damage response in fibroblasts, CGN and thymocytes strongly suggests that any protein generated from the A-T [M] transcripts serves no currently described function associated with ATM.
In addition to the data regarding the 5′ region of the Atm transcript, our RNAseq analysis also provided new information regarding the splicing of exons 44, 45 and 46 in the WT [M] and A-T [M] mice. Curiously, we found that all of the paired-end 125 nt reads overlapping the 3′ junction in exon 44 from the WT [M] and 90.6% of the same reads from the A-T [M] cerebellar mRNA indicated exon 44/46 splicing (Supplementary Material, Table S1 and Fig. S2). Deletion of exon 45 from the transcript is predicted to result in an in-frame deletion (9 base pairs) and the loss of Ser2157, Leu2158 and Phe2159 that reside in the FAT domain of ATM. The FAT domain is considered important for the structural integrity of ATM (90), but the deletion of these amino acids did not appear to affect the stability and function of ATM in the WT [M] cells as determined by the analysis of the DNA damage response in CGN (Fig. 3). It also remains undetermined if this splice variant is specific to the cerebellum, or is seen in human tissue.
A-T [M] mice closely resembled A-T patients in respect to the development of cancer. While the frequency of cancer is greatly elevated in patients in A-T, it still only occurs in about 10–20% of patients, with the majority of malignancies being of lymphoid origin (14–17,55). A recent study by Genik (54) suggests that the cancer incidence among established A-T mouse models depends on the background strain. While the specific contribution of the background strain to the cancer phenotype is not clear, we suggest that at least one component may be the relative oxidative status of the animal, rather than differences in DNA repair. Comparisons of the DNA repair ability between cells isolated from A-T [A] and A-T [M] mice shows that phosphorylation of p53 at serine15 in response to the radiomimetic drug doxorubicin was delayed in both lines (although somewhat more so in A-T [M] mice), while in contrast, the oxidative burden of A-T [A] and A-T [M] cells was significantly different.
Indeed, oxidative stress has been implicated in the pathogenesis of a variety of cancers including lymphomas [for review see (91–95)]. Consistent with this notion is the observation that treatment of the oxidized A-T [A] mutant mice with antioxidants, or the modulation of mitochondrial function and ROS production by other means, delays the onset of tumor formation in these mice (96). This suggests a link between oxidative stress and an increased susceptibility to the development of thymic lymphomas (66,97). Our new mouse supports such a link. We did not see a high tumor burden in our A-T [M] mice, nor did we find an elevated oxidative status in fibroblasts, lymphocytes or cerebellar cells. These observations from our A-T [M] mice are consistent with the human situation where some A-T patients do not show increased markers of oxidative stress in the PB (91,98). Taken together, these data demonstrate that loss of ATM function alone is not sufficient to cause an increase in oxidative burden. Whether oxidation in other cases is due to residual activity or some other reason is currently unclear.
Loss of ATM function, and its associated high cancer rate in other A-T models, had been shown to affect the maturation of cells of the hematopoietic system. We thus asked whether A-T [M] mice show defects in this cell compartment, despite the absence of oxidative burden and the low tumor burden. A comparison of PB from A-T [M] mice to its wild-type counterpart showed a reduction in the percentage of T cell subsets within the circulating lymphocyte populations at 2 months of age, as well as a comparable increase in the percentage of B cells within the circulating lymphocyte populations in mutant mice. This result confirms that a loss of ATM did indeed perturb some aspect of hematopoietic cell maturation, despite the lack of oxidation. Notably, we did not see some of the changes that were reported in the A-T [A] mouse model, where thymocytes isolated from the mice showed a significant increase in the levels of immature CD4/CD8 double positive T cells and a significant decrease in the levels of more mature CD4 and CD8 single positive cells. As it is likely that this profile represents early consequences of pre-neoplastic changes occurring in the thymus of A-T [A] mice, it might not be surprising that our A-T [M] model lacks this specific defect.
We were surprised by the finding of a low rate of embryonic lethality or reduced postnatal viability in the A-T [M] mice. This phenotype has been described before in mutations that generate a complete ATM protein with an inactive, or ‘dead’ kinase domain (20,21). While it is tempting to speculate that loss if kinase activity in the context of complete ATM protein is responsible for this phenotype, the results from Spring et al. (26) do not support this notion. These authors also generated an in-frame deletion that disrupted the kinase function in the context of expressed ATM protein, but did not report embryonic lethality. Given the contribution of background strain variation to the phenotypic presentation of pathologies in AT mouse models, the embryonic lethality might, like the cancer phenotype, be modulated by the genetic background. A detailed analysis of the penetrance of embryonic lethality in the various A-T animal models might be warranted.
While the availability of mouse strains with a fuller range of mutations may help in investigating the diversity of manifestations seen in A-T, there are also clear discrepancies between mouse models and human disease that are of great importance to understand. Of particular interest is the failure to find gross cerebellar changes, despite evidence of changes in motor control coordination seen in the A-T [M] mice. An analysis of Purkinje cell morphology, which had been previously reported to be possible targets of loss of ATM, did not reveal significant changes in A-T [M] compared with WT [M] littermates. We also could not reproduce findings from Carlessi et al. (79), who reported a significant decrease in the postsynaptic density protein PSD95 in hiPSC-derived neurons from A-T patients. This could mainly be due to the unique expression of PSD95 in the cerebellum compared with other cells. Our data confirm the finding demonstrating that PSD95 is highly expressed in the basket cell terminal plexus (pinceau) that surrounds the base of the Purkinje cell axons between the soma and the myelin sheath (99,100). PSD95 is also expressed in the molecular and granular layer, although its function in these regions remains unclear (100).
In contrast to our in vivo analysis, Carlessi's analysis of PSD95 expression was limited to cells grown in vitro that expressed GFAP, and are thus likely to represent neural stem cells, rather than mature neurons. Furthermore, the authors do not provide any data that would allow for the conclusion that the in vitro generated neurons are cerebellar neurons and are thus comparable to our in vivo labeling. It might be valid in future work to define PSD95 levels outside the cerebellum (i.e. hippocampus), where changes in expression levels have been associated with excitatory synapse maturation and activity (80).
Despite the absence of obvious gross morphological and cellular impairments, A-T [M] animals showed motor impairments. These were, however, only apparent when we used GaitScan analysis. The commonly used rotarod test did not yield reliable results in our hands in both the A-T [M], as well as the A-T [A] animals, where a behavioral defect has been reported before using this test (23). The discrepancy may be due in part to uncooperative mice or/and the insensitivity of this test (101). The GaitScan analysis we used here provides a highly sensitive and noninvasive detection of various characteristic parameters that are related to pathophysiological conditions. Unlike the rotarod test, this is not dependent on motivation and does not lead to exhaustion, fatigue and the potential adverse effect of repeated falls from the rod. The test is also highly automated, provides a vast amount of independent data points, and is far less prone to observer bias. Using these analyses, we found clear differences between WT [M] and A-T [M] mice in parameters thought to be related to cerebellar function. We also noticed a potential segregation of our A-T [M] cohort into animals that are highly affected and some that are relatively normal. This was particularly apparent in the foot-spacing data and might indicate an underlying difference in high and low responders, which warrants further examination.
In respect to modeling behavioral defects in mice that are very apparent in A-T in humans, it is important to keep in mind that murine models that show severe morphological cerebellar dysgenesis (102), as well as human patients that present with more severe cerebellar dysgenesis and neurodegeneration, such as multiple X-linked disorders (103) do not always develop an obvious ataxia. Thus, it might be critical to consider whether development of pronounced ataxia requires changes in other parts of the CNS, such as the brainstem or spinal cord, or input from other aspects of cerebellar function that may not be present in the mouse compared with humans.
The question also remains for how much of the variability in the A-T phenotype is due to genetic or environmental factors (12). It seems increasingly likely that it is necessary to consider a ‘multiple insult model’ for the development of A-T. The long-lived, non-cancer prone mouse model we have generated will allow for testing whether secondary insults, in combination with the N-terminal mutations, will more fully recapitulate the human neural degeneration pathology. Although oxidative stress appears to be a candidate, it is unlikely that it is a primary factor in the degeneration of cerebellar neurons, at least in connection with the A-T [A] mutation. In the already highly oxidized A-T [A] model, additional oxidative stress induced by LPS or hypoxia was shown to increase the expression of neuronal cell-cycle markers that have been associated with degeneration (104), but no drastic cellular degeneration was detected during the relatively short time period these animals live. As these animals, however, still die prematurely, the question remains of whether an oxidative stressed brain would lead to neuronal degeneration during aging. It appears that whenever oxidative stress is present in the context of A-T, mice develop cancer and die prematurely. On the other hand, decreasing oxidative burden (for example through strain variations) leads to longer-lived animals, but the absence of a high oxidative burden might prevent the full neurological degeneration pathology. Such a scenario seems possible as our A-T [M] mice present with subtle, but robust changes in their behavior, but no gross changes in cerebellar pathology or increased oxidative status. This supports the idea that oxidative stress may not be an initiating event in the disease pathology, but may represent a consequence of other cellular and morphological changes triggered by the mutation of Atm. As our A-T [M] model was constructed to enable inducible loss of ATM in a promoter-specific manner, it will be possible to target loss of ATM to the brain in an oxidized, cancer prone genetic background strain to generate long-lived animals with an inherent oxidative burden in the CNS to shed light on the role of oxidative stress, age and specific Atm mutation in the neurological sequalae of A-T.
Materials and Methods
Animals
All animals were maintained according to University of Rochester Medical Center animal care standards in a non-SPF facility. Heterozygous 129S6/SvEvTac-Atmtm1Awb/J mice were purchased from The Jackson Laboratory and crossed to obtain Atmtm1Awb+/+ (wild-type WT [A]) and Atmtm1Awb/tm1Awb (A-T [A]) mice. Animals of both sexes were used for experimentation and analysis.
Targeting construct, Atmtm1Mmpl flx/flx, and Atmtm1Mmpl/tm1Mmpl mice
A mouse Atm cDNA fragment was used to screen the mouse RPCI-22 (129S6/SvEvTac) mouse library. A 10.75 kb fragment from RP22-399L20 containing introns 1–5 was cloned into pBluescriptKS using XbaI and XhoI. The FRT-Neo-FRT-loxP cassette from Addgene plasmid 13445 (105) was inserted into Atm intron 3 in the sense orientation at the EcoRV site. The orphan AvrII-BamHI-loxP site was inserted into intron 4 in parallel at the EcoRV site. The DTA cassette from Addgene plasmid 13445 was cloned into the XhoI site at the junction of Atm intron five and the pBluescriptKS plasmid for negative selection.
The targeting construct was linearized using BsteII to allow homologous recombination of the plasmid with W4 ES cells (129S6/SvEvTac). Positive selection of clones was performed by the addition of G418. The presence of the orphan AvrII-BamHI-loxP site was confirmed by digesting ES cell DNA with BamHI and performing Southern blotting using a 905 bp fragment generated from mouse Atm DNA with primers (5′-ATC TCG AGA CCA TCC CTC TTC ATG AT) and (5′-ATT CTA GAA GCA GAG GTC AAG ATG CA). Cycle conditions were 2 min at 95°C, 35× (20 s at 95°C, 20 s at 59°C, 30 s at 72°C), 1.5 min at 72°C using PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies 600670). The PCR product was then cloned into pBluescriptKS using XbaI and XhoI and amplified to generate sufficient quantities of probe for Southern blot hybridization. ES cells positive for homologous recombination were injected into C57Bl/6J blastocytes and implanted into pseudopregnant C57Bl/6J females. ES cell electroporation and colony selection, injection of blastocytes and generation of chimeric mice provided as a service by the Gene Targeting and Transgenic Core at the University of Rochester Medical Center.
A chimeric male generated from the ES cell clones was crossed with 129S4/SvJaeSor-GtROSA26Sortm1(FLP1)Dym/J female mice in order to remove the neomycin cassette (generating the Atmtm1Mmpl flx allele). The resulting animals were mated with C57BL/6J mice, producing heterozygous Atmtm1Mmpl flx/+ offspring. Atmtm1Mmpl flx/+ animals were intercrossed to generate Atmtm1Mmpl flx/flx homozygotes (also referred to as FX [M]). To convert the Atmtm1Mmpl flx allele to a null allele, the Atmtm1Mmpl flx/flx homozygotes were bred to CMV-Cre mice (B6.C-Tg(CMV-Cre)1Cgn/J, Jackson Laboratory). We then performed heterozygous crosses of Atmtm1Mmpl/+ animals to generate Atmtm1Mmpl/tm1Mmpl mice (referred to as A-T [M]).
Genotyping
Genomic DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen 69506) as per manufacturer’s protocol. A total of 5 μl of elution buffer containing DNA was transferred to 25 μl PCR reactions (200 μM dNTP, 1.5 mm MgCl2, 1 unit Platinum Taq DNA Polymerase [Life Technologies 10966-026], 2 μM Primer 1 [5′-TTG TGG CAG ACT TGA TAC GTG TCT A]), 1 μM Primer 2 [5′-GGG CAT TAT CTT CCT CTA TCA CCT T] and 1 μM Primer 3 [5′-TGT CAA TGA GCT GAA GGA ATT AGG T]. Cycle conditions were 2 min at 94°C, 30× (20 s at 94°C, 30 s at 55°C, 1 min at 72°C), 1.5 min at 72°C.
Analysis of cerebellar Atm mRNA transcripts
RNA was isolated from the cerebellum of WT [A], A-T [A] and A-T [M] mice using a Nucleospin RNA kit (Macherey-Nagel 740955) as per manufacturer’s protocol. A total of 500 ng of RNA from each sample was converted to cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems 4368814) utilizing random primers with the addition of RNasin Plus RNase inhibitor (Promega N261B). For PCR analysis of the cDNA, samples were diluted 1:10 in nuclease-free water (Life Technologies AM9937). A total of 5 μl of diluted cDNA was transferred to 50 μl PCR reactions (200 μM dNTP, 2.5 units JumpStart Taq DNA Polymerase (Sigma-Aldrich D9307), 0.5 μM of each primer). Primer pairs as follows; exon 3–4 junction (For [5′-TGG GAT GCT GTT TTC AGG TTT TTA C], Rev [5′-TCT GTC TGG AGC TCT GTG TGG T]), exon 3–5 (For [5′-AGG CAT TCT GAT TCC AAA CAA GGA], Rev [5′-TGC TAC AGT CAG CTC CAT ACG]), exon 3–6 (For [5′ GGC ATT CTG ATT CCA AAC AAG GAA], Rev [5′-TGG CAT ACT GAA TAG CCT TGG AAA]), exon 3–7 (For [5′-AGG CAT TCT GAT TCC AAA CAA GGA], Rev [5′ TCT GGC TCC TTG TGG ATG ATG G]), exon 3–8 (For [5′ GGC ATT CTG ATT CCA AAC AAG GAA] Rev [5′-TCC TGA GGA ATA TTT CCC TCG GCT T]), exon 3–9 (For [5′-AGG CAT TCT GAT TCC AAA CAA GGA] Rev [5′ TCA CTT CCC AGC CTA CGT CTA T]). Cycle conditions were 1 min at 94°C, 30× (30 s at 94°C, 30 s at 55°C, 1.5 min at 72°C), 3 min at 72°C.
Quantitative PCR of Atm mRNA transcripts
RNA was isolated from the spleen, thymus, heart, cerebellum and cortex of WT [M] and A-T [M] mice and converted to cDNA and diluted in nuclease-free water as above. A total of 1 μl of diluted cDNA was transferred to 5 μl PCR reactions containing LightCycler 480 Probes Master Mix (Roche 04707494001) and Taqman Gene Expression assays. The Atm (Mm00431914) probe (exon boundary 57–58) was multiplexed with GAPDH (Life Technologies 4352339E) and expression levels were calculated relative to the wild-type thymus using the ΔΔCt method as per the manufacturer’s recommendation when using target assays with amplification efficiencies very close to one (Applied Biosystems Taqman Gene Expression Assays Technical note 127AP05-03).
RNAseq analysis
RNA was isolated from the cerebellum of three WT [M] and three A-T [M] mice using a Nucleospin RNA kit (Macherey-Nagel 740955) as per manufacturer protocol. The University of Rochester Genomics Resource Center then performed RNASeq analysis. RNA concentration was determined using the NanopDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and RNA quality assessed with the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). A total of 1 ng of total RNA was preamplified with the SMARTer Ultra Low Input kit v2 (Clontech, Mountain View, CA, USA) per manufacturer's recommendations. The quantity and quality of the subsequent cDNA was determined using the Qubit Flourometer (Life Technologies, Carlsbad, CA, USA) and the Agilent 2200 TapeStation. A total of 5 ng of cDNA was used to generate Illumina compatible sequencing libraries with the NexteraXT library preparation kit (Illumina, San Diego, CA, USA) per manufacturer's protocols. The amplified libraries were hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina, San Diego, CA, USA) at a concentration of 8 pM per lane. Paired-end reads of 125 nt were generated for each sample and aligned to the mouse genome (GRCm38.p4). Raw reads generated by the Illumina HiSeq2500 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and adapter removal was performed using Trimmomatic version 0.32 with the following parameters: ‘SLIDINGWINDOW:4:20 TRAILING:13 LEADING:13 ILLUMINACLIP:adapters.fasta:2:30:10 MINLEN:15’. Sequence reads were mapped to the mouse genome (GRCm38.p4) with STAR-2.4.2a. Bedtools v2.22.0 and the ATM exon annotations for ATM-001 (isoform1) were then used for the exon level analysis. Atm gene was then visualized using the Integrative Genomics Viewer (106,107).
Atm cDNA sequencing
RNA was isolated from the spleen of A-T [M] mice and converted to cDNA as described above. Exons 2–10 of the Atm gene were then amplified from 1 μl of the RT-PCR reaction (∼500 ng starting RNA template) in 50 μl PCR reactions (250 μM dNTP, 1 μl PfuUltra II Fusion HS DNA Polymerase [Agilent Technologies 600670], 0.2 μM of each primer [5′-TGC TAG CAT GAG TCT AGC ACT CAA TGA TCT], [5′-TGC GGC CGC ACT AGA AGG TTT ACA]). Cycle conditions were 1 min at 95°C, 40× (20 s at 95°C, 20 s at 55°C, 1 min at 72°C), 3 min at 72°C. Amplified cDNA and pCDH vector (System Biosciences CD510B-1) were digested with NheI and NotI (Thermo Scientific ER0975, ER0595) as per the manufacturer’s recommended protocol. Amplified cDNA and vector were subjected to electrophoresis on 1% agarose gels and extracted using a QIAquick Gel Extraction kit (Qiagen 28706). Vector and insert were ligated using the Rapid DNA Ligation kit (Roche Applied Science 11635379001) as per the manufacturer’s recommended protocol. NEB Turbo Competent E. coli (New England BioLabs C2984H) were transformed with 1 μl of ligation reaction. Ampicillin resistant colonies were screened by PCR for the presence of the Atm cDNA fragment in 25 μl reactions (200 μM dNTP, 1.25 units JumpStart Taq DNA Polymerase [Sigma-Aldrich D9307], 0.5 μM of each primer). Primer pairs as follows; For [5′-CGT GTA CGG TGG GAG GTC TAT], Rev [5′-CTA GGC ACC CGT TCA ATT GCC]. Cycle conditions were 1 min at 94°C, 35× (30 s at 94°C, 30 s at 55°C, 3 min at 72°C), 3 min at 72°C. Plasmid DNA was isolated from several positive colonies using a QIAprep Spin Miniprep kit (Qiagen 27106) as per manufacturer’s protocol. A portion of the isolated plasmid DNA was then sequenced using the PCR screening primers listed above by the Genewiz Corporation.
Fibroblast isolation
Human A-T (GM02052) and control (AG01522) fibroblast lines were obtained from the Coriell Cell Repository. Mouse fibroblasts were isolated from ear punches of 5-month-old (WT [M], FX [M] and A-T [M]) mice. Samples were minced with a scalpel and treated with 2.5 mg/ml collagenase type 4 (Worthington LS004189) and 100 Ku/ml DNase (Calbiochem 260913) in Leibovitz L-15 media (Mediatech 10-045-CV) for 1 h at 37°C. Following centrifugation, samples were incubated with 50 U/ml papain (Worthington LS003127) and 75 Ku/ml DNase in Leibovitz L-15 media for 1 h at 37°C. Samples were triturated with 20G needles and then plated in fibroblast growth media [DMEM (Life Technologies 11995-065) supplemented with non-essential amino acids (Life Technologies 11140-050) and 10% FBS] at 5% O2. After two passages, cells were harvested and frozen at −80°C until experimental use.
Fibroblast proliferation
Fibroblasts were seeded in fibroblasts growth media at a density of 1000 cells/well in quadruplicate wells of three 96 well plates and incubated 5% O2. After 24, 48 and 72 h, one plate was treated with 10 µg/ml Hoechst (Life Technologies H21492) and 1 µg/ml propidium iodide (PI) (Sigma-Aldrich 81845) for 45 min. Hoechst(+)/PI(−) cells were then counted with a Celigo Cell Cytometer (Brooks). To control for variations in cell survival during plating, the cell counts at 42 and 72 h were normalized to each samples cell count obtained at the 24 h time point.
Doxorubicin treatment and immunoblotting
Fibroblast cultures were grown to confluence on 6-well culture microplates in fibroblast growth media at 5% O2. Cells were treated with 1 µm doxorubicin in fibroblast growth media and harvested in modified radioimmunoprecipitation assay buffer containing protease inhibitors (Roche 11836153001) and phosphatase inhibitors (1 mm Na3VO4 and 5 mm NaF) at 2 and 4 h after treatment. Fibroblasts lysates were subjected to SDS-PAGE on NuPAGE 4–12% Bis-Tris Gels with MES SDS Running Buffer (Life Technologies NP0322 and NP0002). Protein was transferred to PVDF membranes (PerkinElmer NEF1002001PK) using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad) at 100 v for 1.5 h in 1× NuPage Transfer Buffer (Life Technologies NP0006-1). Membranes were blocked with 5% bovine serum albumin (BSA, Sigma-Aldrich A3059) in Tris-buffered saline with 0.05% Tween 20 (Fisher Scientific BP337-100) (TBS-T) for 2 h at room temperature. Following incubation with primary and secondary antibodies, all membranes were washed 3 × 20 min in TBS-T. Membranes were probed with ATM(Ser1981) (1:1000, Cell Signaling 4526S) p53 (1:1000, Santa Cruz sc-1311) and p53Ser15 (1:1000, Cell Signaling 9284S) antibodies in 5% BSA in TBS-T overnight at 4°C, then incubated with Rabbit Anti-Goat IgG (1:1000, Life Technologies 81-1620) or Goat Anti-Rabbit IgG (1:3000 dilution) (Life Technologies G-21234) in 5% BSA in TBS-T for 1 h at room temperature. For ATM expression, membranes were blocked as above, then probed with anti-ATM (1:1000, Cell Signaling 2873) antibody (produced using a synthetic peptide surrounding Val3020 of the human ATM protein) in TBS-T, 5% BSA for 4 days at 4°C. Membranes were then incubated with Goat Anti-Rabbit IgG (1:3000, Life Technologies G-21234). For measurement of actin levels, a 1:10 000 dilution of anti-β-actin (Santa Cruz Biotechnology sc-47778HRP) was used. For signal detection, membranes were incubated with WesternBright ECL Western blotting detection kit (Advansta K-12045) as per manufacturer’s recommendation and imaged with BioBlot BXR film (Laboratory Product Sales BX810).
Cellular DNA damage response
Thymic tissue was harvested from euthanized 3-month-old WT [M], and A-T [M] mice and processed as individual samples. The tissue was processed by grinding the tissue in Hank's Balanced Salt Solution (HBSS) media, then running the suspension through a 100 μM cell strainer. The samples were then pelleted and resuspended in RPMI 1640 (Life Technologies 11875-093) and 10% FBS. The thymocytes were plated at a density of 1 × 10e5 cells/well in 24-well culture plates containing coverslips and previously coated with PLL. After an overnight recovery in a 5% O2 incubator, 1 μM of doxorubicin was added to the wells. Thymocytes were incubated for 2 or 4 h before fixation with 2% paraformaldehyde (PFA). A third set of thymocytes were incubated with doxorubicin for 4 h, then washed and placed in a 5% O2 incubator for 16 h before fixation with 2% PFA. Coverslips were treated with 0.2% Triton followed by anti-γH2AX (1:1000, Novus Biologicals NB100-384) at 4°C overnight. After washing, the coverslips were incubated in secondary antibody and counterstained with Dihydrochloride (DAPI, 1 μg/mL, Life Technologies D1306) before imaging on a Nikon Eclipse TE300 microscope. Images were acquired using a CoolSNAP EZ CCD camera and NIS-Elements imaging software.
Cerebellar cells were isolated from postnatal day 9 WT [M] and A-T [M] mice and processed as individual samples. The tissue was enzymatically digested in a solution of 13.3 mg/mL collagenase type 4 and 80 Ku/mL DNase in Hank's Balanced Salt Solution (HBSS, Life Technologies 14170-112). HBSS containing Sato (putrescine [0.1 mm], selenium [0.224 mm], progesterone [0.2 mm], pathocyte-4 BSA [0.0286% v/v; ICN Biochemicals] approximately according to Bottenstein and Sato [1979]) at 37°C for 25 min. The samples were then pelleted and resuspended in a solution of 20 U/mL papain and 80 Ku/mL DNase in HBSS containing Sato. The tissue was again incubated at 37°C for 25 min, then pelleted and resuspended in a solution of DNase (80 Ku/mL) in DMEM/F12 complete media (Sato, insulin [0.5 mg/mL], and transferrin [10 mg/mL]). The tissue was triturated using 21-, 23- and 26-gauge needles, sequentially. These single cell suspensions were then washed, pelleted and resuspended in Neurobasal media (Life Technologies 21103-049) supplemented with B27 (Invitrogen 10889-038). Mixed cerebellar cells were plated at 4 × 10e4 cells/well in 24-well culture plates containing coverslips and previously coated with PLL. After an overnight recovery in a 5% O2 incubator, 1 μM of doxorubicin was added to the wells and treated as with thymocytes above. Coverslips were treated with 0.2% Triton followed by anti-γH2AX (1:1000, Novus Biologicals NB100-384) and β-III tubulin (1:1000, Sigma-Aldrich T8660) at 4°C overnight. Coverslips were washed, counterstained and imaged as with thymocytes above.
Characterization of PB lymphocytes
The PB was obtained from WT [A], A-T [A], WT [M] and A-T [M] mice by mandibular bleeds and collected into tubes containing 20 units heparin sodium salt (Sigma-Aldrich H3419) per 300 µl blood. Samples were then processed by incubating with 10 ml cold red blood cell lysis buffer (1 mm EDTA, 150 mm NH4Cl, 10 mm NaHCO3) for 15 min at room temperature. The samples were pelleted and resuspended in 100 μl of 1× FACS buffer.
The following antibodies (1:100, BD Biosciences, San Jose, CA, USA) were used: rat anti-CD3-APC-Cy7 (555275), rat anti-CD4-APC (561091), rat anti-CD8a-APC-H7 (560247) and anti-CD45R/B220-PerCP-Cy5.5 (552094). The PB cells were incubated with either the T-cell antibody cocktail or B-cell antibody at 4°C for 15 min, then washed, pelleted and resuspended in 200 μl 1× FACS buffer with DAPI (0.2 μg/ml final). Simply Cellular anti-rat IgG compensation beads (Bangs Laboratory 551) were used for the unstained and single-stained controls, which were treated alongside the PB cells.
The compensation and PB samples were run on a FACSCanto II flow cytometer (BD Biosciences). Analysis of the flow cytometry data was conducted on FlowJo Software (Tree Star, Ashland, OR, USA) by first gating on the live cells (DAPI−), followed by gating of the lymphocyte population based on forward and side scatter, and then exclusion of any doublet populations. All subsequent T-cell analyses were conducted based on the CD3+ peripheral lymphocyte cell population, which was preliminarily used to identify the T-cell lineage. The B-cell analyses were conducted based on the B220+ peripheral lymphocyte cell population, which was used to identify the B-cell lineage.
Characterization of thymocytes
The thymic tissue was harvested from euthanized 3-month-old WT [A], A-T [A], WT [M] and A-T [M] mice and 7-month-old WT [M], and A-T [M] mice. The tissue was processed by grinding the tissue in HBSS media, then running the suspension through a 100 μM cell strainer. The samples were then pelleted and resuspended in 100 μl of 1× FACS buffer. The compensation, staining and analyses were carried out as detailed for the CM-H2DCFDA analysis of the brain tissue and the T-cell analysis of the PB, with the analysis being conducted on the lymphocyte cell population.
Oxidative state of cerebellar tissue
Cerebellar tissues were isolated from postnatal day 7 WT [M] and A-T [M] and processed as individual samples. The tissue was enzymatically digested in a solution of 13.3 mg/mL collagenase type 4 and 80 Ku/mL DNase in Hank's Balanced Salt Solution (HBSS, Life Technologies 14170-112). HBSS containing Sato (putrescine [0.1 mm], selenium [0.224 mm], progesterone [0.2 mm], pathocyte-4 BSA [0.0286% v/v; ICN Biochemicals] approximately according to Bottenstein and Sato [1979]) at 37°C for 25 min. The samples were then pelleted and resuspended in a solution of 20 U/mL papain and 80 Ku/mL DNase in HBSS containing Sato. The tissue was again incubated at 37°C for 25 min, then pelleted and resuspended in a solution of DNase (80 Ku/mL) in DMEM/F12 complete media (Sato, insulin [0.5 mg/mL], and transferrin [10 mg/mL]). The tissue was triturated using 21-, 23- and 26-gauge needles, sequentially. These single cell suspensions were then washed, pelleted and resuspended in 500 μl of 1× FACS buffer (0.5% BSA, 2 mm EDTA in PBS). The cells were incubated with 15 µm CM-H2DCFDA (Life Technologies C6827) at 37°C for 25 min, then washed, pelleted and resuspended in 200 μl 1× FACS buffer with 4′,6-diamidino-2-phenylindole (DAPI, Life Technologies D1306) (0.2 μg/mL final). Cells from both tissue types were used for the unstained, single-stained and positive control, which was a CM-H2DCFDA stained sample that was subsequently treated with 100 μM hydrogen peroxide.
The compensation and brain samples were run on a FACSCanto II flow cytometer (BD Biosciences). Analysis of the flow cytometry data was conducted on FlowJo Software (Tree Star, Ashland, OR, USA) by first gating on the live cells (DAPI−), followed by gating of the relevant cell population based on forward and side scatter, and then exclusion of any doublet populations. From this final population, the DCF+ cell population was determined for each sample.
Fibroblast oxidative state
Fibroblasts were seeded in fibroblasts growth media at a density of 5000 cells/well in triplicate wells of a 96 well plate and incubated at 5% O2. After 24 h, cells were treated with 25 µm CM-H2DCFDA (Life Technologies C6827) for 30 min, followed by 25 µg/ml Hoechst for 30 min. Fluorescent intensity was measured using a Celigo Cell Cytometer (Brooks) and values normalized relative to wild-type controls.
Immunohistochemistry
Postnatal day 10 (P10) and 3-month-old mice were anesthetized with sodium pentobarbital (500 μg/μl) and transcardially perfused using heparin (4 U/l, Sigma-Aldrich, H3393), followed by 4% PFA. The brains were removed and post-fixed in 4% PFA overnight, then transferred to 20% sucrose in 0.1 M Phosphate buffer (PhB)(Na2HPO4 • 7 H2O and NaH2PO4 • 2H2O pH 7.4). The cerebella were embedded in Tissue-Tek O.C.T compound (Sakura Finetek, 4583), snap frozen on dry ice, and 20 μm thick sagittal sections were cut using a cryotome.
The mouse anti-SMI-32 (1:500, Covance SMI-32R-100), rabbit anti-Calbindin (1:200, Cell Signaling 13176S) and rabbit anti-PSD95 (1:200, Cell Signaling 3450S) were detected using either a goat anti-mouse secondary antibody conjugated to Alexa-Fluor 488 (1:2000, Life Technologies A21121), or goat anti-rabbit Alexa-Fluor 568 (1:2000, Life Technologies A11011). The nuclei were visualized using 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, 1 μg/ml, Life Technologies D1306) counterstaining.
All tissue sections were rehydrated in 0.1 M PhB, fixed in 4% PFA, then washed in 0.3% (w/v) Triton X-100 in 0.1 M PhB. After fixation, antigen retrieval was performed on one set of the 3-month-old sections by placing the slides in a solution of sodium citrate (pH 8.5) at 90°C for 12 min. The sections were blocked at room temperature for 1 h in blocking solution (0.1 M PhB, 4% goat serum, 0.3% [w/v] Triton X-100). The primary antibodies were diluted in the blocking solution and incubated on the slides for 24–72 h at 4°C. SMI-32 and Calbindin were applied to the 3-month-old sections that had undergone antigen retrieval. Afterwards, the sections were washed in the Triton X-100 solution, and then incubated with the secondary antibody and DAPI at room temperature for 2 h. The sections were washed in PhB and allowed to dry briefly before cover slipping with Fluoromount G (Southern Biotech 0100-01). The sections were imaged at 40× using a Leica SP1 confocal microscope.
Cell quantification
Purkinje cell analysis: Purkinje cells, labeled with SMI-32, from P10 WT [M] and A-T [M] were analyzed for a variety of parameters: cell body size, cell number per area and dendritic length. The cell body size was determined by measuring the widest part of the soma for each Purkinje cell within a field of view (FOV). The cell number was determined by counting the number of SMI-32+/DAPI+ Purkinje cells per inch2 in a FOV, with the area measurement spanning from the Purkinje cell layer to the external granular layer. The relative dendritic length was quantified by taking a measurement from the top of the Purkinje cell to the base of the dendritic tree. Three mice from each genotype were analyzed, with at least three sections per mouse, and at least three FOV per section. The cerebellar images were analyzed in ImageJ (108).
PSD95 Quantification: Three-month-old cerebella from WT [M] and A-T [M] were stained with PSD95 as described. Using the Leica software (LAS AF v.2.3.6) on a Leica SP1 confocal microscope, the fluorescent intensity either within the individual pinceau, the molecular layer or the granule layer was measured within a FOV. Three mice per genotype were used for this analysis, with four sections, and at least 6 FOV per section.
Behavioral tests
At 4, 7 and 9 months of age WT [M] and A-T [M] mice (n ≥ 7) of mixed sex were given two training trials (interval of 1 h) with the rotarod at a constant speed of 10 rpm for a 5-minute period. After 5 days, mice were tested at a constant speed of 16 rpm, with the average latency to failure (fall or passive rotation) over two trials (1 h interval) being recorded. After 2 days, mice were tested using a rotarod adjusted to accelerate from 10 rpm to 28 rpm over a 5 min period, and the average latency was recorded as above.
At 3 months of age WT [M] (n = 7) and A-T [M] (n = 8) of mixed sex were tested with the GaitScan system (Cleversys). In this experimental setup, the mice voluntarily traverse a glass plate illuminated with low intensity green light that allows for the separation of the darkened body from the reflective paws. A high-speed digital video camera records the ventral view and the software automatically determines the footprint locations. The foot spacing, running speed, stance, swing, propel and break times are then determined from video segments capturing the mouse engaged in continuous forward locomotion. The data acquisition and analysis of the gait for each animal were done using the GaitScan software (Cleversys) to identify each paw with the corresponding foot spacing, running speed, stance, swing, propel and brake time during locomotion. The data points from the front or rear paw for each individual animal were averaged to obtain combined front and rear paw data.
Statistical analyses
All experiments were performed at least three independent times or n ≥ 3 animals. Bar graphs represent mean ± S.E.M. unless otherwise indicated. Statistical tests used were the unpaired, two-tailed Student's t-test and two-way ANOVA with Bonferroni's multiple comparison post hoc test as indicated in the figure legends. P-values <0.05 were considered statistically significant and are marked by ‘*’. Data that did not reach statistical significance are marked by ‘n.s.’ (no significant difference). All statistical analyses were performed using Prism4.0 (GraphPad software, Graphpad, La Jolla, CA, USA)
Supplementary Material
Funding
This work was supported by grants from the National Institutes of Health (HD055550M and NS061339 to M.M.P.); a pilot grant from the University of Rochester, Department of Biomedical Genetics to M.M.P.; in part by the A-T Children's Foundation to M.M.P., in part by a training grant from NYSTEM (New York State Stem Cell Science) to B.K. and in part by “Sparks for Children's Health” to M.N.
Supplementary Material
Acknowledgements
We would like to thank Prof. Lin Gan, Director of the transgenic Core Facility at the University of Rochester, for his invaluable help and insights. We also would like to thank Michelle Lacagnina, Kelly Leuer-Bisciotti and Katherine Hutton for excellent technical assistance on various aspects of the work. RNASeq was performed by the University of Rochester Genomics Resource Center.
Conflict of Interest statement. None declared.
References
- 1.Chun H.H., Gatti R.A. (2004) Ataxia-telangiectasia, an evolving phenotype. DNA Repair (Amst), 3, 1187–1196. [DOI] [PubMed] [Google Scholar]
- 2.Hecht F., McCaw B.K. (1974) Letter: chromosomally marked lymphocyte clones in ataxia-telangiectasia. Lancet, 1, 563–564. [DOI] [PubMed] [Google Scholar]
- 3.Gotoff S.P., Amirmokri E., Liebner E.J. (1967) Ataxia telangiectasia. Neoplasia, untoward response to x-irradiation, and tuberous sclerosis. Am. J. Dis. Child, 114, 617–625. [DOI] [PubMed] [Google Scholar]
- 4.Taylor A.M., Metcalfe J.A., Oxford J.M., Harnden D.G. (1976) Proceedings: The radiosensitivity at the chromosomal level of lymphocytes from patients with ataxia telangiectasia. Br. J. Radiol., 49, 561. [PubMed] [Google Scholar]
- 5.Peterson R.D., Kelly W.D., Good R.A. (1964) Ataxia-telangiectasia. Its association with a defective thymus, immunological-deficiency disease, and malignancy. Lancet, 1, 1189–1193. [DOI] [PubMed] [Google Scholar]
- 6.Eisen A.H., Karpati G., Laszlo T., Andermann F., Robb J.P., Bacal H.L. (1965) Immunologic deficiency in ataxia telangiectasia. N. Engl. J. Med., 272, 18–22. [DOI] [PubMed] [Google Scholar]
- 7.Boder E., Sedgwick R.P. (1958) Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics, 21, 526–554. [PubMed] [Google Scholar]
- 8.Savitsky K., Sfez S., Tagle D.A., Ziv Y., Sartiel A., Collins F.S., Shiloh Y., Rotman G. (1995) The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum. Mol. Genet., 4, 2025–2032. [DOI] [PubMed] [Google Scholar]
- 9.Concannon P., Gatti R.A. (1997) Diversity of ATM gene mutations detected in patients with ataxia-telangiectasia. Hum. Mutat., 10, 100–107. [DOI] [PubMed] [Google Scholar]
- 10.Sandoval N., Platzer M., Rosenthal A., Dork T., Bendix R., Skawran B., Stuhrmann M., Wegner R.D., Sperling K., Banin S. et al. (1999) Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum. Mol. Genet., 8, 69–79. [DOI] [PubMed] [Google Scholar]
- 11.Li A., Swift M. (2000) Mutations at the ataxia-telangiectasia locus and clinical phenotypes of A-T patients. Am. J. Med. Genet., 92, 170–177. [DOI] [PubMed] [Google Scholar]
- 12.Taylor A.M., Byrd P.J. (2005) Molecular pathology of ataxia telangiectasia. J. Clin. Pathol., 58, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhagen M.M., Last J.I., Hogervorst F.B., Smeets D.F., Roeleveld N., Verheijen F., Catsman-Berrevoets C.E., Wulffraat N.M., Cobben J.M., Hiel J. et al. (2012) Presence of ATM protein and residual kinase activity correlates with the phenotype in ataxia-telangiectasia: a genotype-phenotype study. Hum. Mutat., 33, 561–571. [DOI] [PubMed] [Google Scholar]
- 14.Morrell D., Cromartie E., Swift M. (1986) Mortality and cancer incidence in 263 patients with ataxia-telangiectasia. J. Natl. Cancer Inst., 77, 89–92. [PubMed] [Google Scholar]
- 15.Olsen J.H., Hahnemann J.M., Borresen-Dale A.L., Brondum-Nielsen K., Hammarstrom L., Kleinerman R., Kaariainen H., Lonnqvist T., Sankila R., Seersholm N. et al. (2001) Cancer in patients with ataxia-telangiectasia and in their relatives in the nordic countries. J. Natl. Cancer Inst., 93, 121–127. [DOI] [PubMed] [Google Scholar]
- 16.Sandoval C., Swift M. (1998) Treatment of lymphoid malignancies in patients with ataxia-telangiectasia. Med. Pediatr. Oncol., 31, 491–497. [DOI] [PubMed] [Google Scholar]
- 17.Taylor A.M., Metcalfe J.A., Thick J., Mak Y.F. (1996) Leukemia and lymphoma in ataxia telangiectasia. Blood, 87, 423–438. [PubMed] [Google Scholar]
- 18.Li P., Goto H., Kasahara K., Matsuyama M., Wang Z., Yatabe Y., Kiyono T., Inagaki M. (2012) P90 RSK arranges Chk1 in the nucleus for monitoring of genomic integrity during cell proliferation. Mol. Biol. Cell, 23, 1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y., Herrup K. (2005) Loss of neuronal cell cycle control in ataxia-telangiectasia: a unified disease mechanism. J. Neurosci., 25, 2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel J.A., Pellegrini M., Lee B.S., Guo Z., Filsuf D., Belkina N.V., You Z., Paull T.T., Sleckman B.P., Feigenbaum L. et al. (2012) Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol., 198, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto K., Wang Y., Jiang W., Liu X., Dubois R.L., Lin C.S., Ludwig T., Bakkenist C.J., Zha S. (2012) Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J. Cell Biol., 198, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavin M.F. (2013) The appropriateness of the mouse model for ataxia-telangiectasia: neurological defects but no neurodegeneration. DNA Repair (Amst), 12, 612–619. [DOI] [PubMed] [Google Scholar]
- 23.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D. et al. (1996) Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell, 86, 159–171. [DOI] [PubMed] [Google Scholar]
- 24.Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. (1996) Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA, 93, 13084–13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borghesani P.R., Alt F.W., Bottaro A., Davidson L., Aksoy S., Rathbun G.A., Roberts T.M., Swat W., Segal R.A., Gu Y. (2000) Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. USA, 97, 3336–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spring K., Cross S., Li C., Watters D., Ben-Senior L., Waring P., Ahangari F., Lu S.L., Chen P., Misko I. et al. (2001) Atm knock-in mice harboring an in-frame deletion corresponding to the human ATM 7636del9 common mutation exhibit a variant phenotype. Cancer Res., 61, 4561–4568. [PubMed] [Google Scholar]
- 27.Xu Y., Baltimore D. (1996) Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev., 10, 2401–2410. [DOI] [PubMed] [Google Scholar]
- 28.Herzog K.H., Chong M.J., Kapsetaki M., Morgan J.I., McKinnon P.J. (1998) Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science, 280, 1089–1091. [DOI] [PubMed] [Google Scholar]
- 29.Gilad S., Bar-Shira A., Harnik R., Shkedy D., Ziv Y., Khosravi R., Brown K., Vanagaite L., Xu G., Frydman M. et al. (1996) Ataxia-telangiectasia: founder effect among north African Jews. Hum. Mol. Genet., 5, 2033–2037. [DOI] [PubMed] [Google Scholar]
- 30.Li J., Chen J., Vinters H.V., Gatti R.A., Herrup K. (2011) Stable brain ATM message and residual kinase-active ATM protein in ataxia-telangiectasia. J. Neurosci., 31, 7568–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrose M., Goldstine J.V., Gatti R.A. (2007) Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum. Mol. Genet., 16, 2154–2164. [DOI] [PubMed] [Google Scholar]
- 32.Barlow C., Dennery P.A., Shigenaga M.K., Smith M.A., Morrow J.D., Roberts L.J. 2nd, Wynshaw-Boris A., Levine R.L. (1999) Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc. Natl. Acad. Sci. USA, 96, 9915–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barzilai A., Rotman G., Shiloh Y. (2002) ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair (Amst.), 1, 3–25. [DOI] [PubMed] [Google Scholar]
- 34.Gilad S., Khosravi R., Shkedy D., Uziel T., Ziv Y., Savitsky K., Rotman G., Smith S., Chessa L., Jorgensen T.J. et al. (1996) Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet., 5, 433–439. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. (1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev., 10, 2411–2422. [DOI] [PubMed] [Google Scholar]
- 36.Shiloh Y., Becker Y. (1982) Reduced inhibition of replicon initiation and chain elongation by neocarzinostatin in skin fibroblasts from patients with ataxia telangiectasia. Biochim. Biophys. Acta., 721, 485–488. [DOI] [PubMed] [Google Scholar]
- 37.Paterson M.C., Smith B.P., Lohman P.H., Anderson A.K., Fishman L. (1976) Defective excision repair of gamma-ray-damaged DNA in human (ataxia telangiectasia) fibroblasts. Nature, 260, 444–447. [DOI] [PubMed] [Google Scholar]
- 38.Shiloh Y., Tabor E., Becker Y. (1983) Abnormal response of ataxia-telangiectasia cells to agents that break the deoxyribose moiety of DNA via a targeted free radical mechanism. Carcinogenesis, 4, 1317–1322. [DOI] [PubMed] [Google Scholar]
- 39.Ross W.E., Bradley M.O. (1981) DNA double-stranded breaks in mammalian cells after exposure to intercalating agents. Biochim. Biophys. Acta., 654, 129–134. [DOI] [PubMed] [Google Scholar]
- 40.Tewey K.M., Rowe T.C., Yang L., Halligan B.D., Liu L.F. (1984) Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science, 226, 466–468. [DOI] [PubMed] [Google Scholar]
- 41.Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science, 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- 42.Kastan M.B., Zhan Q., el-Deiry W.S., Carrier F., Jacks T., Walsh W.V., Plunkett B.S., Vogelstein B., Fornace A.J. Jr (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 43.Khosravi R., Maya R., Gottlieb T., Oren M., Shiloh Y., Shkedy D. (1999) Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Natl. Acad. Sci. USA, 96, 14973–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das K.C., Dashnamoorthy R. (2004) Hyperoxia activates the ATR-Chk1 pathway and phosphorylates p53 at multiple sites. Am. J. Physiol. Lung Cell Mol. Physiol., 286, L87–L97. [DOI] [PubMed] [Google Scholar]
- 45.Kim W.J., Vo Q.N., Shrivastav M., Lataxes T.A., Brown K.D. (2002) Aberrant methylation of the ATM promoter correlates with increased radiosensitivity in a human colorectal tumor cell line. Oncogene, 21, 3864–3871. [DOI] [PubMed] [Google Scholar]
- 46.Chan S.F., Sances S., Brill L.M., Okamoto S., Zaidi R., McKercher S.R., Akhtar M.W., Nakanishi N., Lipton S.A. (2014) ATM-dependent phosphorylation of MEF2D promotes neuronal survival after DNA damage. J. Neurosci., 34, 4640–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biton S., Dar I., Mittelman L., Pereg Y., Barzilai A., Shiloh Y. (2006) Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J. Biol. Chem., 281, 17482–17491. [DOI] [PubMed] [Google Scholar]
- 48.McKinnon P.J. (2012) ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol., 7, 303–321. [DOI] [PubMed] [Google Scholar]
- 49.Hollis R.J., Kennaugh A.A., Butterworth S.V., Taylor A.M. (1987) Growth of large chromosomally abnormal T cell clones in ataxia telangiectasia patients is associated with translocation at 14q11. A model for other T cell neoplasia. Hum. Genet., 76, 389–395. [DOI] [PubMed] [Google Scholar]
- 50.Heppell A., Butterworth S.V., Hollis R.J., Kennaugh A.A., Beatty D.W., Taylor A.M. (1988) Breakage of the T cell receptor alpha chain locus in non malignant clones from patients with ataxia telangiectasia. Hum. Genet., 79, 360–364. [DOI] [PubMed] [Google Scholar]
- 51.Russo G., Isobe M., Pegoraro L., Finan J., Nowell P.C., Croce C.M. (1988) Molecular analysis of a t(7;14)(q35;q32) chromosome translocation in a T cell leukemia of a patient with ataxia telangiectasia. Cell, 53, 137–144. [DOI] [PubMed] [Google Scholar]
- 52.Lipkowitz S., Stern M.H., Kirsch I.R. (1990) Hybrid T cell receptor genes formed by interlocus recombination in normal and ataxia-telangiectasis lymphocytes. J. Exp. Med., 172, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liyanage M., Weaver Z., Barlow C., Coleman A., Pankratz D.G., Anderson S., Wynshaw-Boris A., Ried T. (2000) Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood, 96, 1940–1946. [PubMed] [Google Scholar]
- 54.Genik P.C., Bielefeldt-Ohmann H., Liu X., Story M.D., Ding L., Bush J.M., Fallgren C.M., Weil M.M. (2014) Strain background determines lymphoma incidence in atm knockout mice. Neoplasia, 16, 129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hecht F., Hecht B.K. (1990) Cancer in ataxia-telangiectasia patients. Cancer Genet. Cytogenet., 46, 9–19. [DOI] [PubMed] [Google Scholar]
- 56.Peterson R.D., Cooper M.D., Good R.A. (1966) Lymphoid tissue abnormalities associated with ataxia-telangiectasia. Am. J. Med., 41, 342–359. [DOI] [PubMed] [Google Scholar]
- 57.Fiorilli M., Businco L., Pandolfi F., Paganelli R., Russo G., Aiuti F. (1983) Heterogeneity of immunological abnormalities in ataxia-telangiectasia. J. Clin. Immunol., 3, 135–141. [DOI] [PubMed] [Google Scholar]
- 58.Hathcock K.S., Bowen S., Livak F., Hodes R.J. (2013) ATM influences the efficiency of TCRbeta rearrangement, subsequent TCRbeta-dependent T cell development, and generation of the pre-selection TCRbeta CDR3 repertoire. PLoS One, 8, e62188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowen S., Wangsa D., Ried T., Livak F., Hodes R.J. (2013) Concurrent V(D)J recombination and DNA end instability increase interchromosomal trans-rearrangements in ATM-deficient thymocytes. Nucleic Acids Res., 41, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isoda T., Takagi M., Piao J., Nakagama S., Sato M., Masuda K., Ikawa T., Azuma M., Morio T., Kawamoto H. et al. (2012) Process for immune defect and chromosomal translocation during early thymocyte development lacking ATM. Blood, 120, 789–799. [DOI] [PubMed] [Google Scholar]
- 61.Matei I.R., Gladdy R.A., Nutter L.M., Canty A., Guidos C.J., Danska J.S. (2007) ATM deficiency disrupts Tcra locus integrity and the maturation of CD4+CD8+ thymocytes. Blood, 109, 1887–1896. [DOI] [PubMed] [Google Scholar]
- 62.Westphal C.H., Rowan S., Schmaltz C., Elson A., Fisher D.E., Leder P. (1997) atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat. Genet., 16, 397–401. [DOI] [PubMed] [Google Scholar]
- 63.Gurley K.E., Kemp C.J. (2007) Ataxia-telangiectasia mutated is not required for p53 induction and apoptosis in irradiated epithelial tissues. Mol. Cancer Res., 5, 1312–1318. [DOI] [PubMed] [Google Scholar]
- 64.Callen E., Jankovic M., Wong N., Zha S., Chen H.T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A. et al. (2009) Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell, 34, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watters D.J. (2003) Oxidative stress in ataxia telangiectasia. Redox Rep., 8, 23–29. [DOI] [PubMed] [Google Scholar]
- 66.Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B. (2012) Mitochondrial dysfunction in ataxia-telangiectasia. Blood, 119, 1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meredith M.J., Dodson M.L. (1987) Impaired glutathione biosynthesis in cultured human ataxia-telangiectasia cells. Cancer Res., 47, 4576–4581. [PubMed] [Google Scholar]
- 68.Reichenbach J., Schubert R., Schindler D., Muller K., Bohles H., Zielen S. (2002) Elevated oxidative stress in patients with ataxia telangiectasia. Antioxid. Redox Signal, 4, 465–469. [DOI] [PubMed] [Google Scholar]
- 69.Reichenbach J., Schubert R., Schwan C., Muller K., Bohles H.J., Zielen S. (1999) Anti-oxidative capacity in patients with ataxia telangiectasia. Clin. Exp. Immunol., 117, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamsler A., Daily D., Hochman A., Stern N., Shiloh Y., Rotman G., Barzilai A. (2001) Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice. Cancer Res., 61, 1849–1854. [PubMed] [Google Scholar]
- 71.Quick K.L., Dugan L.L. (2001) Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann. Neurol., 49, 627–635. [PubMed] [Google Scholar]
- 72.Ludwig L.B., Valiati V.H., Palazzo R.P., Jardim L.B., da Rosa D.P., Bona S., Rodrigues G., Marroni N.P., Pra D., Maluf S.W. (2013) Chromosome instability and oxidative stress markers in patients with ataxia telangiectasia and their parents. Biomed. Res. Int., 2013, 762048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Semlitsch M., Shackelford R.E., Zirkl S., Sattler W., Malle E. (2011) ATM protects against oxidative stress induced by oxidized low-density lipoprotein. DNA Repair (Amst.), 10, 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eaton J.S., Lin Z.P., Sartorelli A.C., Bonawitz N.D., Shadel G.S. (2007) Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J. Clin. Invest., 117, 2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D. et al. (2010) ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA, 107, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cosentino C., Grieco D., Costanzo V. (2011) ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J., 30, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finkel T. (2011) Signal transduction by reactive oxygen species. J. Cell Biol., 194, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weyemi U., Redon C.E., Aziz T., Choudhuri R., Maeda D., Parekh P.R., Bonner M.Y., Arbiser J.L., Bonner W.M. (2015) NADPH oxidase 4 is a critical mediator in Ataxia telangiectasia disease. Proc. Natl. Acad. Sci. USA, 112, 2121–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carlessi L., Fusar Poli E., Bechi G., Mantegazza M., Pascucci B., Narciso L., Dogliotti E., Sala C., Verpelli C., Lecis D. et al. (2014) Functional and molecular defects of hiPSC-derived neurons from patients with ATM deficiency. Cell Death Dis., 5, e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El-Husseini A.E., Schnell E., Chetkovich D.M., Nicoll R.A., Bredt D.S. (2000) PSD-95 involvement in maturation of excitatory synapses. Science, 290, 1364–1368. [PubMed] [Google Scholar]
- 81.Watters D., Kedar P., Spring K., Bjorkman J., Chen P., Gatei M., Birrell G., Garrone B., Srinivasa P., Crane D.I. et al. (1999) Localization of a portion of extranuclear ATM to peroxisomes. J. Biol. Chem., 274, 34277–34282. [DOI] [PubMed] [Google Scholar]
- 82.Zha S., Guo C., Boboila C., Oksenych V., Cheng H.L., Zhang Y., Wesemann D.R., Yuen G., Patel H., Goff P.H. et al. (2011) ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature, 469, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.You Z., Chahwan C., Bailis J., Hunter T., Russell P. (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol., 25, 5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lavin M.F., Scott S., Gueven N., Kozlov S., Peng C., Chen P. (2004) Functional consequences of sequence alterations in the ATM gene. DNA Repair (Amst.), 3, 1197–1205. [DOI] [PubMed] [Google Scholar]
- 85.McConville C.M., Stankovic T., Byrd P.J., McGuire G.M., Yao Q.Y., Lennox G.G., Taylor M.R. (1996) Mutations associated with variant phenotypes in ataxia-telangiectasia. Am. J. Hum. Genet., 59, 320–330. [PMC free article] [PubMed] [Google Scholar]
- 86.Gilad S., Chessa L., Khosravi R., Russell P., Galanty Y., Piane M., Gatti R.A., Jorgensen T.J., Shiloh Y., Bar-Shira A. (1998) Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am. J. Hum. Genet., 62, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilad S., Khosravi R., Harnik R., Ziv Y., Shkedy D., Galanty Y., Frydman M., Levi J., Sanal O., Chessa L. et al. (1998) Identification of ATM mutations using extended RT-PCR and restriction endonuclease fingerprinting, and elucidation of the repertoire of A-T mutations in Israel. Hum. Mutat., 11, 69–75. [DOI] [PubMed] [Google Scholar]
- 88.Du L., Pollard J.M., Gatti R.A. (2007) Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc. Natl. Acad. Sci. USA, 104, 6007–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A. et al. (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet., 64, 1617–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lempiainen H., Halazonetis T.D. (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J., 28, 3067–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. (2010) Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med., 49, 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cremona C.A., Behrens A. (2013) ATM signalling and cancer. Oncogene, 33, 3351–3360. [DOI] [PubMed] [Google Scholar]
- 93.Verschoor M.L., Ungard R., Harbottle A., Jakupciak J.P., Parr R.L., Singh G. (2013) Mitochondria and cancer: past, present, and future. Biomed. Res. Int., 2013, 612369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lightfoot T.J., Skibola C.F., Smith A.G., Forrest M.S., Adamson P.J., Morgan G.J., Bracci P.M., Roman E., Smith M.T., Holly E.A. (2006) Polymorphisms in the oxidative stress genes, superoxide dismutase, glutathione peroxidase and catalase and risk of non-Hodgkin’s lymphoma. Haematologica, 91, 1222–1227. [PubMed] [Google Scholar]
- 95.Kastan M.B. (2007) Our cells get stressed too! Implications for human disease. Blood Cells Mol. Dis., 39, 148–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reliene R., Fleming S.M., Chesselet M.F., Schiestl R.H. (2008) Effects of antioxidants on cancer prevention and neuromotor performance in Atm deficient mice. Food Chem. Toxicol., 46, 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schubert R., Erker L., Barlow C., Yakushiji H., Larson D., Russo A., Mitchell J.B., Wynshaw-Boris A. (2004) Cancer chemoprevention by the antioxidant tempol in Atm-deficient mice. Hum. Mol. Genet., 13, 1793–1802. [DOI] [PubMed] [Google Scholar]
- 98.Degan P., d'Ischia M., Pallardo F.V., Zatterale A., Brusco A., Calzone R., Cavalieri S., Kavakli K., Lloret A., Manini P. et al. (2007) Glutathione levels in blood from ataxia telangiectasia patients suggest in vivo adaptive mechanisms to oxidative stress. Clin. Biochem., 40, 666–670. [DOI] [PubMed] [Google Scholar]
- 99.Kim E., Niethammer M., Rothschild A., Jan Y.N., Sheng M. (1995) Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature, 378, 85–88. [DOI] [PubMed] [Google Scholar]
- 100.Hunt C.A., Schenker L.J., Kennedy M.B. (1996) PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J. Neurosci., 16, 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wahlsten D., Rustay N.R., Metten P., Crabbe J.C. (2003) In search of a better mouse test. Trends Neurosci., 26, 132–136. [DOI] [PubMed] [Google Scholar]
- 102.Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J. (1999) Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell, 97, 689–701. [DOI] [PubMed] [Google Scholar]
- 103.Zanni G., Bertini E.S. (2011) X-linked disorders with cerebellar dysgenesis. Orphanet. J. Rare Dis., 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Y., Hui C.W., Li J., Herrup K. (2014) The interaction of the atm genotype with inflammation and oxidative stress. PLoS One, 9, e85863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoch R.V., Soriano P. (2006) Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development, 133, 663–673. [DOI] [PubMed] [Google Scholar]
- 106.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.