Abstract
The histone acetyltransferase (HAT) enzymes p300 and CBP are closely related paralogs that serve as transcriptional coactivators and have been found to be dysregulated in cancer and other diseases. p300/CBP is a multidomain protein and possesses a highly conserved bromodomain that has been shown to bind acetylated Lys residues in both proteins and various small molecules, including I-CBP112 and CBP30. Here we show that the ligand I-CBP112 can stimulate nucleosome acetylation up to 3-fold while CBP30 does not. Activation of p300/CBP by I-CBP112 is not observed with the isolated histone H3 substrate but requires a nucleosome substrate. I-CBP112 does not impact nucleosome acetylation by the isolated p300 HAT domain, and the effects of I-CBP112 on p300/CBP can be neutralized by CBP30, suggesting that I-CBP112 likely allosterically activates p300/CBP through bromodomain interactions. Using mass spectrometry and Western blots, we have found that I-CBP112 particularly stimulates acetylation of Lys18 of histone H3 (H3K18) in nucleosomes, an established in vivo site of p300/CBP. In addition, we show that I-CBP112 enhances H3K18 acetylation in acute leukemia and prostate cancer cells in a concentration range commensurate with its antiproliferative effects. Our findings extend the known pharmacology of bromodomain ligands in the regulation of p300/CBP and suggest a novel approach to modulating histone acetylation in cancer.
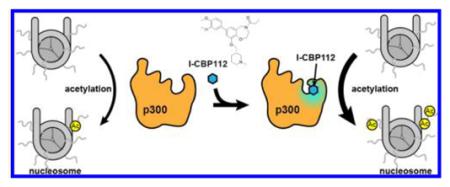
Graphical abstract
p300 and its paralog CBP are multidomain histone acetyltransferases (HATs also known as lysine acetyltrans-ferases or KATs) that catalyze the acetylation of Lys residues in histones and other proteins.1 Serving as transcriptional coactivators, p300 and CBP participate in numerous ways to regulate cell growth, differentiation, and gene expression across many organ systems and physiologic pathways.2–8 Dysregulation of p300/CBP by mutation, altered expression, or other mechanisms has been linked to disease states, including various malignancies such as acute leukemias and prostate cancer.1,5,6,9–14 Furthermore, p300/CBP is critical in development as evidenced by the genetic disorder Rubinstein-Taybi syndrome, which occurs with loss of function mutations in single alleles of either p300 or CBP.15
The p300/CBP HAT domain is centrally located in the protein (Figure 1A);1,16 its three-dimensional structure, catalytic mechanism, regulation by an autoacetylation loop, and substrate specificity have been the subject of a number of biochemical studies.8,17–21 While p300/CBP appears to favor acetylation of Lys residues near other basic amino acids, acetyltransferase activity on histones and other protein substrates is relatively indiscriminate, consistent with its hit- and-run catalytic mechanism.19 Inhibitors of p300/CBP HAT activity have been developed and are under investigation as therapeutics for a number of diseases.5,10,14,22,23 Additional key p300/CBP domains include a well-characterized bromodomain that is just N-terminal to the HAT domain and can bind acetyl-Lys-containing peptides. The bromodomain is one of the most highly conserved domains in p300 and CBP, possessing 96% sequence identity.24 A crystal structure of a section of p300 that includes the bromodomain and the HAT domain flanking a middle RING/PHD domain revealed the potential for allosteric interactions to modulate p300 HAT activity, although this has not been investigated previously.16,25
Figure 1.
Nucleosome acetylation by p300. (A) Full-length p300 and CBP contain multiple functional domains that mediate various other interactions, including the nuclear receptor-interacting domain (NRID), cysteine/histidine-rich domains (C/Hx), an interferon binding homology domain (IHD), a CREB and MYB interaction domain (KIX), a Sin3-interacting domain (SID), an interferon binding domain (IBiD), a glutamine-rich domain, and a proline P×P motif, a bromodomain, a RING/plant homeobox domain (PHD), and a histone acetyltransferase (HAT) domain. Figure not to scale, adapted from refs 1 and 16. (B) Chemical structure of bromodomain ligand I-CBP112. (C) Chemical structure of bromodomain ligand CBP30. (D) Autoradiograph image of the time course of p300 acetylation of recombinant nucleosomes. Reaction mixtures contained 5 μM [14C]acetyl-CoA, 0.5 μM nucleosome, and 10 nM p300. (E) Quantification of duplicates of panel D (average ± span). (F) Acetylation rate (v) vs p300 concentration. Reaction mixtures contained 5 μM [14C]acetyl-CoA and 0.5 μM nucleosome [average ± span (duplicates)].
Several years ago, small molecule ligands highly specific for the BET bromodomain were reported and shown to have a range of pharmacologic effects. 26–29 In addition, potent and selective small molecule ligands for the p300/CBP bromodomain have been described, 3,24,30–34 and we explore two of these here, I-CBP112 and CBP30 (Figure 1B,C), in the context of p300/CBP acetylation of nucleosomes. While binding assays with the isolated p300 and CBP bromodomains have shown that these compounds bind with submicromolar affinities,24,31,32 their effects with purified full-length p300 or CBP have not yet been reported. Here we explore the ability of I-CBP112 and CBP30 to modulate p300/CBP HAT activity and unexpectedly find that I-CBP112, but not CBP30, can stimulate p300/CBP acetylation of the nucleosome substrate. The details of these results and their potential mechanistic implications are described.
EXPERIMENTAL METHODS
Protein Purification
His-p300-Strep2-Flag and His-CBP-Strep2-Flag encoded by the pVL1393 baculovirus expression vector were expressed in Sf9 insect cells and purified as described previously.17 Protein purity and quantification were assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels stained with Coomassie blue alongside a BSA standard. Semisynthetic p300-HAT was prepared using expressed protein ligation as described previously.21,35 Nucleosomes were prepared with Xenopus laevis histones [wild type except for an H3 (C110A) substitution] and a FAM-labeled 225 bp DNA fragment DNA containing the 601 Widom positioning sequence as previously described.36,37
Radioactive Histone Acetyltransferase Assays
Global nucleosome acetylation was assessed by acetyltransferase assays in which nucleosome acetylation was quantified on the basis of the enzyme-dependent incorporation of 14C from the substrate [14C]acetyl-CoA (60 mCi/mmol) into the histone proteins as visualized by densitometry. In vitro reactions were conducted in a buffer containing 50 mM HEPES (pH 7.9), 50 mM NaCl, 1 mM TCEP, and 25 μg/mL BSA at 30 °C and initiated by the addition of acetyl-CoA. Reaction mixtures typically contained 5 μM acetyl-CoA, 0.5–1 μM nucleosome, and 10 nM p300 or 20 nM CBP. When small molecule ligands were added, the DMSO concentration was kept at or below 2.5%, a concentration that did not significantly affect measured rates. On the basis of the linear range of activity from the respective time courses, p300 reactions were generally performed for 5 min and CBP reactions for 10 min at 30 °C (Figure 1D,E). Similarly, enzyme titrations were performed to optimize the concentration at which the extent of substrate turnover was <10% (Figure 1F). Reactions were quenched by the addition of 5× sodium dodecyl sulfate-tris-tricine [167 mM Tris-HCl (pH 6.8), 47% glycerol, 11.7% SDS, 250 mM DTT, and 0.05% Coomassie blue] loading dye and separated on 16% tris-tricine gels also including [14C]BSA.22 Samples of the total reaction mixture were quantified by scintillation counting to determine 14C content. Acetyl transfer was assessed by autoradiography on dried tris-tricine gels to determine 14C in the nucleosome bands calibrated versus [14C]BSA. All assays were performed at least twice. Duplicates typically agreed within 20%.
Immunoblot Histone Acetyltransferase Assays
Acetylation of H3K18 (H3K18ac) was monitored under conditions similar to those of the radioactive assays, although only nonradioactive acetyl-CoA was employed. Reactions were quenched with 4× SDS loading dye [200 mM Tris-HCl (pH 6.8), 40% glycerol, 8% SDS, 50 mM EDTA, 4% β-mercaptoethanol, and 0.08% bromophenol blue] and mixtures heated at 95 °C for 5 min before separation on 4 to 12% bis-tris gels. Proteins were transferred by iBLOT to nitrocellulose, blocked for 1 h with 5% BSA-TBST, and probed overnight at 4 °C with 1:1000 H3K18ac (Millipore 07-354) in 2.5% BSA-TBST followed by anti-rabbit HRP at 1:5000 for 1 h at room temperature in 2.5% BSA-TBST.
Bromodomain Small Molecule Reagents
I-CBP112 (1-[7-(3,4-dimethoxyphenyl)-9-{[(3S)-1-methylpiperidin-3-yl]-methoxy}-2,3,4,5-tetrahydro-1,4-benzoxazepin-4-yl]propan-1-one) and SGC-CBP30 {8-(3-chloro-4-methoxyphenethyl)-4-(3,5-dimethylisoxazol-4-yl)-9-[2-(morpholin-4-yl)propyl]-7,9-diazabicyclo[4.3.0]nona-1(6),2,4,7-tetraene} (hereafter termed I-CBP112 and CBP30, respectively) were gifts from the Structural Genomics Consortium or were obtained from Cayman Chemicals. (±)-JQ1 was obtained from Santa Cruz Biotechnology. Ligands were dissolved in 100% DMSO, which was included under control conditions and kept constant within each experiment.
Mass Spectrometry
Acetylation reactions for mass spectrometry analysis were performed like those for the aforementioned acetyltransferase assays. However, to obtain sufficient signal for detection after subsequent processing, 75 nM p300 or 200 nM CBP enzyme was used as well as 50 μM nonradioactive acetyl-CoA and 1 μM nucleosome. Reactions were quenched with 4 volumes of trichloroacetic acid, mixtures washed with acetone and dried, and free lysines blocked by treatment with propionic anhydride followed by pH adjustment with ammonium hydroxide and incubation for 1 h at 51 °C. The modified proteins were digested in trypsin overnight at 37 °C before separation of peptides on a Waters Acquity UPLC system with a BEH C18 column and identification on a Thermo TSQ Quantum Access triple-quadrupole mass spectrometer. Peptide elution was monitored by selected reaction monitoring for H3 and H4 acetylated and propionylated peptides.17 Six time points were taken for each reaction, and the data were plotted with respect to time to determine rates and standard deviations. Statistical significance was determined using unpaired two-tailed t tests and the Holm–Sidak method to correct for multiple comparisons (α = 5.000%).
Cell Culture
LNCaP and KG1a cells were grown in RPMI 1640 with GlutaMAX (Life Technologies 61870) supplemented with 10 and 20% fetal bovine serum (Sigma 6178), respectively, as well as 1 unit/mL penicillin and streptomycin. Cells were maintained at 37 °C in a 5%/95% CO2/air atmosphere. Drug treatments were performed in RPMI 1640 with GlutaMAX with 2.5% serum to minimize plasma protein binding.
Cell Proliferation Assays
Cells (6000 KG1a and 13000 LNCaP cells/well) were plated in 96-well flat-bottom plates approximately 24 h prior to drug treatment. After 24 h, 10–20% fetal bovine serum-containing medium was replaced with 2.5% serum medium, and cells were treated with I-CBP112 in 0.18% DMSO; 0.18% DMSO was shown to have negligible cell growth effects under the conditions used in our experiments. After being exposed to I-CBP112 for 66 h, cells were subjected to a final concentration of 0.476% [3H]thymidine per well and allowed to proliferate for an additional 6 h (exposure to I-CBP112 for a total of 72 h). Cells were harvested, and the counts of 3H in each well were taken relative to those treated with vehicle alone to quantify the effect of the ligand on proliferation. Experiments were conducted in triplicate or greater, each on a separate occasion. IC50 values were calculated using a standard dose–response equation
where h represents the Hill coefficient.
RESULTS
Effects of I-CBP112 on p300-Catalyzed Nucleosome Acetylation
We explored the effects of the bromodomain ligand I-CBP112 (Figure 1B) on nucleosome acetylation by full-length p300 (Figure 1A) using a radioactive assay with [14C]acetyl-CoA as the substrate. Recombinant full-length purified p300 and reconstituted nucleosomes composed of Xenopus histones and the 601 Widom DNA sequence were employed. In these assays, the histones were imaged using tris-tricine gels and the degree of acetylation was quantified by phosphorimage analysis, referenced to the [14C]BSA standard. We identified conditions under which p300 acetylation of the nucleosomes showed linearity with respect to time and enzyme concentration (Figure 1D–F). In the presence of I-CBP112, we consistently observed an approximately 2–3-fold global activation of nucleosome acetylation (Figure 2A,B). This was seen across the H2A, H2B, H3, and H4 bands in the octamer and showed an EC50 of 11 ± 3 μM upon global activation of nucleosome acetylation. A time course with I-CBP112 showed that activation of p300 did not alter the linearity of product formation versus time (Figure 2C). We also investigated the dependence of I-CBP112 activation of p300 on nucleosome concentration. In this experiment, HAT activity was not saturable at the highest nucleosome concentration tested, so it was not possible to readily discriminate between a kcat and KM effect on I-CBP112 stimulation (Figure 2D,E).
Figure 2.
Effects of I-CBP112 on full-length p300-mediated nucleosome acetylation. (A) Autoradiograph image of reaction mixtures containing 10 nM p300, 0.5 μM nucleosome, 5 μM [14C]acetyl-CoA and varying concentrations of I-CBP112. (B) Quantification of duplicate reactions in panel A. EC50 obtained from a standard binding isotherm (reported average ± span of duplicates). (C) p300-catalyzed nucleosome acetylation time course with or without I-CBP112 (40 μM) [average ± span (duplicates)]. Comparison of fits with or without I-CBP112 shows p = 0.005 for the linear initial 5 min. (D) p300-catalyzed nucleosome acetylation with variable nucleosome concentration with or without I-CBP112 (40 μM). (E) Quantification of duplicate reactions as seen in panel D [points = average ± SEM (n = 2–3)]. Comparison of fits showed the curves were significantly different (p < 0.0001).
Differential Effects of CBP30 and I-CBP112 on p300-Catalyzed Nucleosome Acetylation
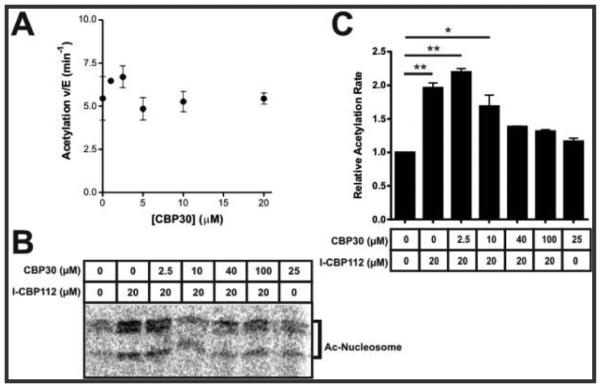
We next tested the effects of CBP30 (Figure 1C), a chemically distinct bromodomain ligand with an approximately 5-fold greater affinity toward the p300/CBP bromodomain than I-CBP112.24,31,32 Interestingly, unlike I-CBP112, CBP30 did not show an impact on p300 catalytic activity toward nucleosomes (Figure 3A). This raised the possibility that the effects of I-CBP112 on p300 catalytic activity may have occurred through a cryptic binding site of I-CBP112 within p300’s HAT domain or perhaps within nucleosomes. We thus investigated this possibility by assessing the effects of I-CBP112 on the isolated p300 HAT domain and its acetylation of nucleosomes. In contrast to full-length p300, the isolated p300 HAT domain was not significantly stimulated by I-CBP112 (Figure 4A,B), suggesting that direct HAT domain or nucleosome interaction was not the molecular basis for I-CBP112 effects. Furthermore, we saw no effect of I-CBP112 on the acetylation of free histone H3 (Figure 4C). Therefore, it is most likely that nucleosomal structure is necessary for I-CBP112 modulation of p300 activity.
Figure 3.
Effects of CBP30 on full-length p300-mediated nucleosome acetylation. (A) Quantification of acetyltransferase reaction mixtures containing 10 nM p300, 0.5 μM nucleosomes, 5 μM [14C]acetyl-CoA, and varying concentrations of CBP30 [average ± span (duplicates)]. (B) Autoradiograph image of reaction mixtures containing 10 nM p300, 0.5 μM nucleosome, 5 μM [14C]acetyl-CoA, and designated I-CBP112 and CBP30 concentrations. (C) Quantification of the gel in panel B. Error bars represent the span of data (n = 2). (**p < 0.01; *p < 0.05 by one-way ANOVA with Dunnet correction for a normalized control; for all others, p > 0.35).
Figure 4.
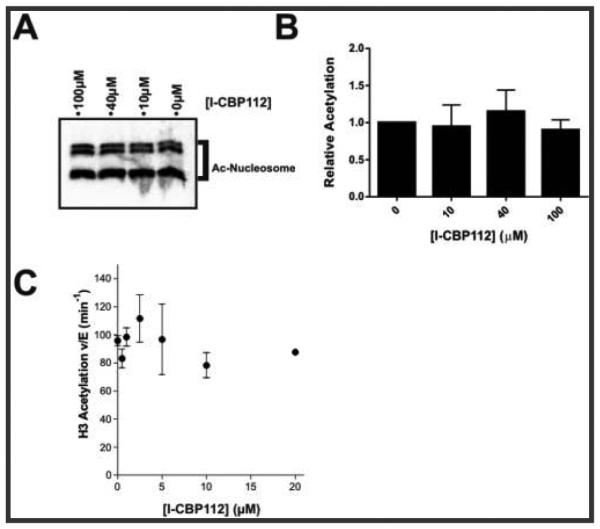
Effects of I-CBP112 on p300 HAT domain-mediated acetylation of nucleosomes and full-length p300-mediated acetylation of H3. (A) Autoradiograph image of reaction mixtures containing semisynthetic p300 HAT, 1.0 μM nucleosome, 5 μM [14C]acetyl-CoA, and varying concentrations of I-CBP112. (B) Quantification of reactions as in panel A [average ± standard error of the mean (n = 3)], (p = 0.8535 by one-way ANOVA). (C) Quantification of acetyltransferase reaction mixtures containing 1.5 nM full-length p300, 20 μM histone H3, 10 μM [14C]acetyl-CoA, and varying concentrations of I-CBP112 [average ± span (duplicates)].
We next performed a competition assay between CBP30 and I-CBP112 on full-length p300 nucleosome acetylation. Using fixed I-CBP112 (20 μM) and variable CBP30 concentrations, we observed that CBP30 could block the I-CBP112-induced activity of p300 on nucleosomes in a concentration-dependent fashion (Figure 3B,C). These results suggest that I-CBP112 stimulates p300-catalyzed nucleosome acetylation by binding to p300’s bromodomain, whereas CBP30’s distinct mode of p300 bromodomain interaction does not manifest in this allosteric activation.24,31,32
Effects of I-CBP112 on Lys Selectivity of p300- and CBP-Catalyzed Nucleosome Acetylation
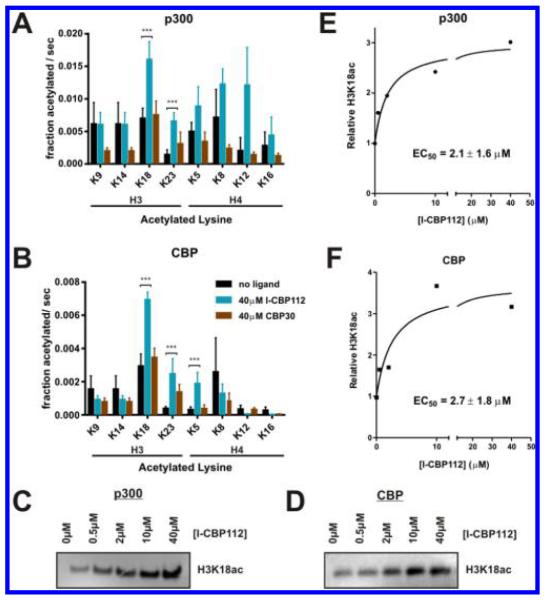
To address the possibility that I-CBP112 influences specific Lys targeting by p300 and CBP, we performed quantitative mass spectrometry in a label-free fashion17,18 to analyze the degree of Lys acetylation catalyzed by these two enzymes with nucleosome substrates. Under initial conditions (<10% total nucleosome acetylation), we found that I-CBP112 significantly enhanced acetylation by p300 at the histone H3K18 and H3K23 sites (Figure 5A). I-CBP112 induced enhanced acetylation of these same sites by CBP as well as at H4K5 (Figure 5B). We were most interested in the effects on H3K18 acetylation because this position is one of the best established sites in cellular studies of p300.5,8,12,38 We therefore performed Western blots to measure H3K18ac levels in nucleosome acetylation reactions with p300 and CBP enzymes (Figure 5C,D). We observed that I-CBP112 stimulated H3K18ac by ~3-fold, consistent with the mass spectrometry results. The EC50’s of activation of I-CBP112 on p300- and CBP-mediated H3K18 acetylation were ~2 μM (Figure 5E,F), modestly lower than I-CBP112’s EC50 for bulk nucleosome acetylation by p300 found in the radioactive assay (Figure 2B).
Figure 5.
Effects of I-CBP112 on Lys site selectivity of full-length p300-mediated nucleosome acetylation. (A) Mass spectrometry quantification of p300-mediated acetylation of residues within the H3 and H4 histone tails shows I-CBP112 specific activation of acetylation of a subset of residues (***p < 0.001). B) Same as panel A but for the CBP enzyme. (C) Representative Western blot for H3K18ac of in vitro reaction mixtures containing 10 nM p300, 1.0 μM nucleosome, 10 μM acetyl-CoA, and varying concentrations of I-CBP112. (D) Same as panel C but with 20 nM CBP enzyme. (E) Quantification of reactions as in panel C [average ± standard error of the fit (n = 3)]. (F) Quantification of reactions as in panel D [average ± standard error of the fit (n = 3)].
Effects of I-CBP112 on Cancer Cell H3K18 Acetylation and Proliferation
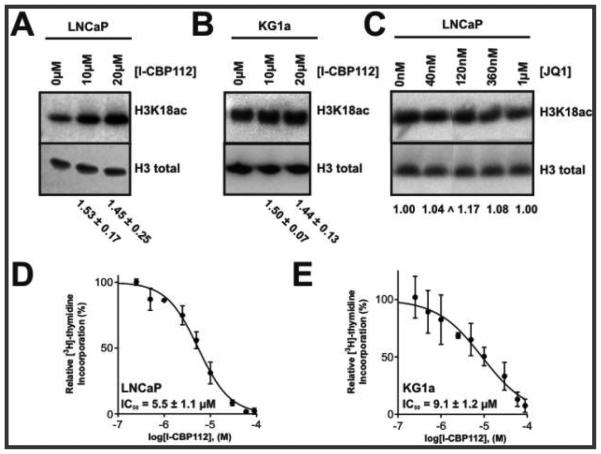
We next explored whether I-CBP112 affected histone H3K18 acetylation in human cancer cell lines. For these studies, we selected an acute myeloid leukemia line, KG1a, and an androgen-dependent prostate cancer cell line, LNCaP, as p300/CBP dysregulation has been implicated in both malignancies.1,5,6,9,11,13 We detected modest but reproducible stimulation of H3K18 acetylation by Western blots with both cell lines at 10 and 20 μM I-CBP112 within 6 h of compound exposure in both the prostate cancer and leukemia cells (Figure 6A,B). This cellular enhancement in H3K18ac induced by I-CBP112 is consistent with the experiments with purified p300/CBP and nucleosomes described above. Because it has been suggested that I-CBP112 might bind weakly to BET bromodomain-containing proteins such as BRD4,32 we tested whether the BET bromodomain inhibitor JQ1 also affected H3K18 acetylation.27 However, JQ1 did not influence H3K18ac levels in its relevant pharmacologic dose range, suggesting that I-CBP112’s effects are not mediated by BET bromodomain binding (Figure 6C). We then examined whether I-CBP112 could influence the proliferation rate of the leukemia and prostate cancer cell lines using a [3H]-thymidine incorporation assay. Indeed, there was a clear inhibition of proliferation of both the prostate cancer and leukemia cells with very similar IC50 values of 5.5 ± 1.1 and 9.1 ± 1.2 μM, respectively. These growth inhibition data nicely correlate with the concentration at which I-CBP112 influences histone acetylation.
Figure 6.
Effects of I-CBP112 on histone H3K18 acetylation and proliferation in cancer cells. (A) Western blots of H3K18 acetylation and total H3 levels upon treatment of LNCaP cell lines with I-CBP112 (4 h). Representative blots of duplicate experiments. Fold change relative to no ligand of the ratios of H3K18ac to total H3 [average ± span (n = 2)]. (B) Same as panel A but in KG1a cells treated for 6 h. (C) Effects of JQ1 on H3K18ac in LNCaP cells (duplicates). All bands from the same gel. (D) I-CBP112 shows an IC50 of 5.5 ± 1.1 μM. The Hill slope = 1.3 ± 0.1 on LNCaP cell proliferation after 72 h ([3H]thymidine incorporation) (n = 3). (E) Same as panel D, but KG1a cells show an IC50 of 9.1 ± 1.2 μM and a Hill slope of 0.82 ± 0.09 (n = 4).
DISCUSSION
As a large and important transcriptional coactivator with established connections to human disease, p300/CBP offers opportunities throughout its numerous domains for targeting by small molecule ligands/inhibitors. Prior efforts have led to compounds that selectively bind the CBP N-terminal region, the KIX domain, the HAT domain, and, as discussed in detail here, the bromodomain (reviewed in refs 1 and 26). We have investigated the bromodomain ligands I-CBP112 and CBP30, and unexpectedly observed that I-CBP112 can stimulate full-length p300/CBP nucleosome acetylation. We have shown that I-CBP112’s p300/CBP activating effect toward nucleosomes is most salient on histone H3K18 acetylation. This is noteworthy because H3K18 is one of p300/CBP’s most preferred sites and is most sharply affected by p300/CBP genetic alteration.8,12,17,38,39 Although other p300/CBP HAT allosteric activators, including TTK21, nemorosone, and CTPB, have been reported previously, their biochemical characterization with in vitro-purified proteins is somewhat limited.2,40–42 In our studies, the degree of p300/CBP activation by I-CBP112 is modest, in the range of 2–3-fold, but it has been observed reproducibly with radioactive assays and mass spectrometry and further confirmed by Western blots. I-CBP112 stimulation of H3K18 acetylation was also detected by Western blotting in cell culture experiments.
We demonstrate in this study that I-CBP112’s stimulatory effects on p300/CBP are likely to be mediated by p300/CBP’s bromodomain. First, we note that I-CBP112 is established to bind the p300/CBP bromodomain with strong affinity using reliable biophysical measurements and X-ray crystallography.3,32 Second, I-CBP112 does not stimulate a p300/CBP HAT domain-only construct, consistent with a bromodomain requirement. Third, the bromodomain ligand CBP30, which does not stimulate p300/CBP, competes against the I-CBP112 effect. We lack sufficient information to propose a precise mechanism of the I-CBP112 effect, but it may likely be due unique attributes of its footprint on the bromodomain and/or specific induced conformational changes.32 The EC50 of I-CBP112-mediated p300/CBP activation (low micromolar range) is somewhat higher than the measured Kd for the bromodomain binding interaction [~600 nM using isothermal titration calorimetry (http://www.thesgc.org/chemical-probes/I-CBP112)]. This difference in apparent affinities could be due to the effects of studying the isolated bromodomain for the calorimetry studies, rather than the more indirect enzymologic impact using full-length p300/CBP proteins measured here. The structural basis for I-CBP112’s allosteric activation of p300/CBP is not yet clear. The fact that such activation is visible with only a nucleosome substrate and not simply isolated histone H3 protein suggests that complex, multipronged interactions involving p300/CBP and specific aspects of the nucleosome are involved. We cannot exclude interactions of the p300/CBP bromodomain with the autoacetylated loop21 that may be influenced by I-CBP112, although we have no direct evidence of this. Because CBP30 does not demonstrate the p300/CBP stimulatory effect mediated by I-CBP112, bromodomain occupancy per se is insufficient to account for I-CBP112’s enzymatic actions. On the basis of the available information, we hypothesize that I-CBP112 can trigger conformational interactions through the RING/PHD region to the HAT domain that in turn influence nucleosomal interactions.16,25 The fact that CBP30 lacks I-CBP112’s pharmacologic behavior may be related to its distinct mode of binding to the p300/CBP bromodomain observed in X-ray crystal structures.31,32 This activation by I-CBP112 partially parallels the allosteric activation seen upon association of unmethylated H3K4 peptide with the PHD1 domain of lysine demethylase 5A (KDM5A/RBP2/JARID1A), which stimulates the demethylation of both peptide and nucleosomal trimethylated H3 lysine 4 (H3K4me3) by the adjacent catalytic domain.43
As p300/CBP loss of function mutations are well-established in various malignancies,1 activators of p300/CBP could help restore balance to the dynamic equilibrium between acetyltransferase action and histone deacetylases. Indeed, it has been reported previously3,32 and we confirm here that I-CBP112 does have antiproliferative action against cancer. The significant inhibition of proliferation could be a result of stimulation of p300/CBP HAT activity on nucleosomes, as observed in this study, or perhaps though modulation of p300 acetylation of non-histone substrates or other protein–protein interaction functions. Such altered patterns of Lys acetylation or p300/CBP recruitment could tip the balance in gene expression networks that ultimately inhibit cancer cell growth, although further studies will be needed to obtain detailed mechanistic insights into the pharmacology of I-CBP112.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Structural Genomics Consortium for their generous gift of I-CBP112 and CBP30.
Funding
This work was supported by National Institutes of Health (NIH) Grant GM62437 to P.A.C., NIH Grant GM84192 to G.D.B., and NIH Grant GM102503 to A.J.A. B.E.Z. was supported by a Jane Coffin Childs postdoctoral fellowship, and B.L. was supported by the Austrian Science Fund FWF, Grant W1101-B12 to Zoran Culig. This work was also supported by the FAMRI foundation.
ABBREVIATIONS
- acetyl-CoA
acetyl-coenzyme A
- Ac
acetylation
- BEH
ethylene-bridged hybrid
- BET
bromodomain and extra-terminal family
- BRD4
bromodomain-containing 4 protein
- BSA
bovine serum albumin protein
- CBP
CREB binding protein
- I-CBP112
1-[7-(3,4-dimethoxyphenyl)-9-{[(3S)-1-methylpiperidin-3-yl]-methoxy}-2,3,4,5-tetrahydro-1,4-benzoxazepin-4-yl)propan-1-one]
- CBP30 or SGC-CBP30
8-(3-chloro-4-methoxyphenethyl)-4-(3,5-dimethylisoxazol-4-yl)-9-[2-(morpholin-4-yl)-propyl]-7,9-diazabicyclo[4.3.0]nona-1(6),2,4,7-tetraene
- DMSO
dimethyl sulfoxide
- H3K18
histone 3, lysine 18
- HAT
histone acetyltransferase
- HEPES
4-(2-hydroxyethyl)-1-piper-azineethanesulfonic acid
- HRP
horseradish peroxidase
- JQ1 or (±)-SGCBD01
[(R,S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-1-thia-5,7,8,9a-tetraazacyclopenta[e]azulen-6-yl]acetic acid tert-butyl ester
- KAT
lysine acetyltransferase
- TBST
tris-buffered saline tween
- TCEP
tris(2-carboxyethyl)phosphine
- UPLC
ultraperformance liquid chromatography.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00480.
One supplementary figure showing the purification and quantification of full-length p300 and CBP (PDF)
Notes
The authors declare the following competing financial interest(s): P.A.C. is a cofounder and shareholder in Aylin Therapeutics, which is developing p300/CBP inhibitors.
REFERENCES
- (1).Dancy BM, Cole PA. Protein Lysine Acetylation by p300/CBP. Chem. Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chatterjee S, Mizar P, Cassel R, Neidl R, Selvi BR, Mohankrishna DV, Vedamurthy BM, Schneider A, Bousiges O, Mathis C, Cassel J-C, Eswaramoorthy M, Kundu TK, Boutillier A-L. A Novel Activator of CBP/p300 Acetyl-transferases Promotes Neurogenesis and Extends Memory Duration in Adult Mice. J. Neurosci. 2013;33:10698–10712. doi: 10.1523/JNEUROSCI.5772-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Conery AR, Centore RC, Neiss A, Keller PJ, Joshi S, Spillane KL, Sandy P, Hatton C, Pardo E, Zawadzke L, Bommi-Reddy A, Gascoigne KE, Bryant BM, Mertz JA, Sims RJ. Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma. eLife. 2016;5:e10483. doi: 10.7554/eLife.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fan S, Ma YX, Wang C, Yuan R-Q, Meng Q, Wang J-A, Erdos M, Goldberg ID, Webb P, Kushner PJ, Pestell RG, Rosen EM. p300 Modulates the BRCA1 Inhibition of Estrogen Receptor Activity. Cancer Res. 2002;62:141–151. [PubMed] [Google Scholar]
- (5).Giotopoulos G, Chan W-I, Horton SJ, Ruau D, Gallipoli P, Fowler A, Crawley C, Papaemmanuil E, Campbell PJ, Göttgens B, Van Deursen JM, Cole PA, Huntly BJP. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene. 2016;35:279–289. doi: 10.1038/onc.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ianculescu I, Wu D-Y, Siegmund KD, Stallcup MR. Selective roles for CBP and p300 as coregulators for androgen-regulated gene expression in advanced prostate cancer cells. J. Biol. Chem. 2012;287:4000–4013. doi: 10.1074/jbc.M111.300194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Iyer NG, Chin S-F, Ozdag H, Daigo Y, Hu D-E, Cariati M, Brindle K, Aparicio S, Caldas C. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7386–7391. doi: 10.1073/pnas.0401002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DAL, Tindall DJ. p300 in Prostate Cancer Proliferation and Progression. Cancer Res. 2003;63:7638–7640. [PubMed] [Google Scholar]
- (10).Gao X, Lin J, Ning Q, Gao L, Yao Y, Zhou J, Li Y, Wang L, Yu L. A Histone Acetyltransferase p300 Inhibitor C646 Induces Cell Cycle Arrest and Apoptosis Selectively in AML1-ETO-Positive AML Cells. PLoS One. 2013;8:e55481. doi: 10.1371/journal.pone.0055481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Heery DM, Fischer PM. Pharmacological targeting of lysine acetyltransferases in human disease: a progress report. Drug Discovery Today. 2007;12:88–99. doi: 10.1016/j.drudis.2006.11.012. [DOI] [PubMed] [Google Scholar]
- (12).Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus Small e1a Alters Global Patterns of Histone Modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Isharwal S, Miller MC, Marlow C, Makarov DV, Partin AW, Veltri RW. p300 (histone acetyltransferase) biomarker predicts prostate cancer biochemical recurrence and correlates with changes in epithelia nuclear size and shape. Prostate. 2008;68:1097–1104. doi: 10.1002/pros.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Santer FR, Höschele PPS, Oh SJ, Erb HHH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z. Inhibition of the Acetyltransferases p300 and CBP Reveals a Targetable Function for p300 in the Survival and Invasion Pathways of Prostate Cancer Cell Lines. Mol. Cancer Ther. 2011;10:1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- (15).Roelfsema JH, Peters DJM. Rubinstein–Taybi syndrome: clinical and molecular overview. Expert Rev. Mol. Med. 2007;9:1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- (16).Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- (17).Henry RA, Kuo Y-M, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry. 2013;52:5746–5759. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Henry RA, Kuo Y-M, Bhattacharjee V, Yen TJ, Andrews AJ. Changing the selectivity of p300 by acetyl-CoA modulation of histone acetylation. ACS Chem. Biol. 2015;10:146–156. doi: 10.1021/cb500726b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- (20).Maksimoska J, Segura-Peña D, Cole PA, Marmorstein R. Structure of the p300 Histone Acetyltransferase Bound to Acetyl-Coenzyme A and Its Analogues. Biochemistry. 2014;53:3415–3422. doi: 10.1021/bi500380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- (22).Dancy BM, Crump NT, Peterson DJ, Mukherjee C, Bowers EM, Ahn Y-H, Yoshida M, Zhang J, Mahadevan LC, Meyers DJ, Boeke JD, Cole PA. Live-Cell Studies of p300/CBP Histone Acetyltransferase Activity and Inhibition. Chem-BioChem. 2012;13:2113–2121. doi: 10.1002/cbic.201200381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gajer JM, Furdas SD, Gründer A, Gothwal M, Heinicke U, Keller K, Colland F, Fulda S, Pahl HL, Fichtner I, Sippl W, Jung M. Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis. 2015;4:e137. doi: 10.1038/oncsis.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hay DA, Fedorov O, Martin S, Singleton DC, Tallant C, Wells C, Picaud S, Philpott M, Monteiro OP, Rogers CM, Conway SJ, Rooney TPC, Tumber A, Yapp C, Filippakopoulos P, Bunnage ME, Müller S, Knapp S, Schofield CJ, Brennan PE. Discovery and Optimization of Small-Molecule Ligands for the CBP/p300 Bromodomains. J. Am. Chem. Soc. 2014;136:9308–9319. doi: 10.1021/ja412434f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rack JGM, Lutter T, Kjæreng Bjerga GE, Guder C, Ehrhardt C, Värv S, Ziegler M, Aasland R. The PHD finger of p300 Influences Its Ability to Acetylate Histone and Non-Histone Targets. J. Mol. Biol. 2014;426:3960–3972. doi: 10.1016/j.jmb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- (26).Brand M, Measures AM, Wilson BG, Cortopassi WA, Alexander R, Höss M, Hewings DS, Rooney TPC, Paton RS, Conway SJ. Small Molecule Inhibitors of Bromodomain-Acetyl-lysine Interactions. ACS Chem. Biol. 2015;10:22–39. doi: 10.1021/cb500996u. [DOI] [PubMed] [Google Scholar]
- (27).Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Smith SG, Zhou M-M. The Bromodomain: A New Target in Emerging Epigenetic Medicine. ACS Chem. Biol. 2016;11:598–608. doi: 10.1021/acschembio.5b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ghosh S, Taylor A, Chin M, Huang H-R, Conery AR, Mertz JA, Salmeron A, Dakle PJ, Mele D, Cote A, Jayaram H, Setser JW, Poy F, Hatzivassiliou G, DeAlmeida-Nagata D, Sandy P, Hatton C, Romero FA, Chiang E, Reimer T, Crawford T, Pardo E, Watson VG, Tsui V, Cochran AG, Zawadzke L, Harmange J-C, Audia JE, Bryant BM, Cummings RT, Magnuson SR, Grogan JL, Bellon SF, Albrecht BK, Sims RJ, Lora JM. Regulatory T Cell Modulation by CBP/EP300 Bromodomain Inhibition. J. Biol. Chem. 2016;291:13014–13027. doi: 10.1074/jbc.M115.708560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hammitzsch A, Tallant C, Fedorov O, O’Mahony A, Brennan PE, Hay DA, Martinez FO, Al-Mossawi MH, de Wit J, Vecellio M, Wells C, Wordsworth P, Müller S, Knapp S, Bowness P. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10768–10773. doi: 10.1073/pnas.1501956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Picaud S, Fedorov O, Thanasopoulou A, Leonards K, Jones K, Meier J, Olzscha H, Monteiro O, Martin S, Philpott M, Tumber A, Filippakopoulos P, Yapp C, Wells C, Che KH, Bannister A, Robson S, Kumar U, Parr N, Lee K, Lugo D, Jeffrey P, Taylor S, Vecellio ML, Bountra C, Brennan PE, O’Mahony A, Velichko S, Müller S, Hay D, Daniels DL, Urh M, La Thangue NB, Kouzarides T, Prinjha R, Schwaller J, Knapp S. Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res. 2015;75:5106–5119. doi: 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rooney TPC, Filippakopoulos P, Fedorov O, Picaud S, Cortopassi WA, Hay DA, Martin S, Tumber A, Rogers CM, Philpott M, Wang M, Thompson AL, Heightman TD, Pryde DC, Cook A, Paton RS, Müller S, Knapp S, Brennan PE, Conway SJ. A Series of Potent CREBBP Bromodomain Ligands Reveals an Induced-Fit Pocket Stabilized by a Cation–π. Interaction. Angew. Chem., Int. Ed. 2014;53:6126–6130. doi: 10.1002/anie.201402750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gerona-Navarro G, Yoel-Rodríguez, Mujtaba S, Frasca A, Patel J, Zeng L, Plotnikov AN, Osman R, Zhou M-M. Rational Design of Cyclic Peptide Modulators of the Transcriptional Coactivator CBP: A New Class of p53 Inhibitors. J. Am. Chem. Soc. 2011;133:2040–2043. doi: 10.1021/ja107761h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Patel A, Chakravarthy S, Morrone S, Nodelman IM, McKnight JN, Bowman GD. Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res. 2013;41:1637–1648. doi: 10.1093/nar/gks1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning1. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- (38).Haery L, Lugo-Picó JG, Henry RA, Andrews AJ, Gilmore TD. Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth. Mol. Cancer. 2014;13:29. doi: 10.1186/1476-4598-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic Reprogramming by Adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small Molecule Modulators of Histone Acetyltransferase p300. J. Biol. Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- (41).Dal Piaz F, Tosco A, Eletto D, Piccinelli AL, Moltedo O, Franceschelli S, Sbardella G, Remondelli P, Rastrelli L, Vesci L, Pisano C, De Tommasi N. The Identification of a Novel Natural Activator of p300 Histone Acetyltranferase Provides New Insights into the Modulation Mechanism of this Enzyme. Chem-BioChem. 2010;11:818–827. doi: 10.1002/cbic.200900721. [DOI] [PubMed] [Google Scholar]
- (42).Mantelingu K, Kishore AH, Balasubramanyam K, Kumar GVP, Altaf M, Swamy SN, Selvi R, Das C, Narayana C, Rangappa KS, Kundu TK. Activation of p300 Histone Acetyltransferase by Small Molecules Altering Enzyme Structure: Probed by Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. B. 2007;111:4527–4534. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- (43).Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJS, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 2015;6:6204. doi: 10.1038/ncomms7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.