Abstract
Replication-independent incorporation of variant histone H3.3 has a profound impact on chromatin function and numerous cellular processes, including the differentiation of muscle cells. The histone chaperone HIRA and H3.3 have essential roles in MyoD regulation during myoblast differentiation. However, the precise mechanism that determines the onset of H3.3 deposition in response to differentiation signals is unclear. Here we show that HIRA is phosphorylated by Akt kinase, an important signaling modulator in muscle cells. By generating a phosphospecific antibody, we found that a significant amount of HIRA was phosphorylated in myoblasts. The phosphorylation level of HIRA and the occupancy of phosphorylated protein on muscle genes gradually decreased during cellular differentiation. Remarkably, the forced expression of the phosphomimic form of HIRA resulted in reduced H3.3 deposition and suppressed the activation of muscle genes in myotubes. Our data show that HIRA phosphorylation limits the expression of myogenic genes, while the dephosphorylation of HIRA is required for proficient H3.3 deposition and gene activation, demonstrating that the phosphorylation switch is exploited to modulate HIRA/H3.3-mediated muscle gene regulation during myogenesis.
Introduction
Histone incorporation by histone chaperones specifies chromatin structure by organizing nucleosomes. One variant form of histone H3, H3.3, is deposited onto chromatin in a replication-independent manner1, 2 and preferentially enriched at the promoters of actively transcribed genes and the upstream regulatory regions of genes in myoblasts and mouse embryonic stem (ES) cells for chromatin decondensation and is a marker of current and anticipated target gene activation.3, 4, 5 H3.3 often co-ordinates with other variant histones, such as H2AZ, to maintain DNA elements for transcription factor binding through efficient nucleosome disruption,6 suggesting that H3.3 serves as an euchromatic modulator.7 Indeed, human and mouse cell lines require H3.3 to induce a series of stress-responsive genes upon exposure to interferons or heat shock,8, 9 and to bookmark damaged chromatin for transcription recovery after genotoxic stress.10 In addition, recent studies have revealed that H3.3 also accumulates in pericentromeric or telomeric regions, which are typical constitutive heterochromatin.11 Because of the opposite roles of H3.3 in chromatin regulation and physiological functions, the detailed regulatory mechanism of the replication-independent pathway, which spatially and timely incorporates H3.3 according to cellular requirements, should be investigated to determine the functional profile of H3.3.
The histone chaperone HIRA complex, which consists of HIRA, Cabin1 and UBN1, is associated with euchromatic and heterochromatic loci, and is essential for the activation of mesodermal and angiogenic genes, as well as a subset of cell cycle-regulating genes, by mediating H3.3 incorporation.12, 13 In addition, upon the senescence of human fibroblasts, HIRA draws senescence-associated heterochromatin foci (SAHF) in concert with another histone chaperone, Asf1a.14 Such strong functional relevance between HIRA and H3.3 suggests that HIRA-mediated H3.3 incorporation could be tightly regulated in a variety of cellular events. One mechanism that has recently been proposed for the repressive function of H3.3 in mouse ES cells is that HIRA-dependent H3.3 incorporation contributes to PRC2 recruitment and to the maintenance of H3 K27 trimethylation at bivalent promoters to prevent mouse ES cells from differentiating toward an extraembryonic trophectoderm lineage.15
Another histone chaperone, DAXX, also has a distinctive role in H3.3 loading.16, 17 DAXX directly binds H3.3 and cooperates with ATRX to mediate the deposition and remodeling of H3.3 nucleosomes at telomeric and pericentromeric heterochromatin. DAXX also mediates gene-specific regulation at transcribing loci during neuronal activation. Interestingly, neuronal depolarization-induced target gene activation and H3.3 deposition require the dephosphorylation of the DAXX protein, indicating that upstream signaling devices operate to target the histone chaperone activity of DAXX.18 However, as for HIRA, its regulatory mechanism upon changes in cellular circumstances have not been described. HIRA is also phosphorylated in the S phase and during senescence of primary human fibroblast cells by cdk2 and GSK3 β, respectively.19, 20 The phosphorylation of HIRA by GSK3β facilitates SAHF formation; however, the functional implications of HIRA modification in relation to H3.3 incorporation are unclear.
We have shown that HIRA/Asf1a-mediated H3.3 incorporation is an essential event for MyoD activation during muscle cell differentiation.3 While HIRA and Asf1a are pre-recruited to the regulatory elements of muscle genes, such as MyoD, in proliferating cells, H3.3 incorporation increases only when the myogenic program is activated, implying that the activity of HIRA rather than protein recruitment is targeted by the differentiation signal. In this study, we investigated a post-translational modification of HIRA that modulates histone chaperone activity. We propose that the phosphorylation of serine (S) 650 in human HIRA (648 in mouse HIRA), which is mediated by Akt1, is an important regulatory switch, and that it guides muscle gene regulation during cellular differentiation.
Materials and methods
Cell culture
C2C12 myoblasts were obtained from the American Type Culture Collection (ATCC) and grown in Dulbecco's modified Eagle's medium with 15% fetal bovine serum (Hyclone), 1% penicillin and streptomycin. C2C12 myoblasts were induced to differentiate into myotubes by replacing the culture medium with differentiation medium composed of Dulbecco's modified Eagle's medium supplemented with 2% horse serum, 1% penicillin, and streptomycin. Cells were cultivated in the differentiation medium for 3 days.
Plasmids
Double-strand oligonucleotides encoding HIRA (640–655) wild type (WT), S650A and S650D were synthesized and cloned into the EcoRl and Xhol sites of the pGEX4T-1 vector. pcDNA3 HA-HIRA was subjected to site-directed mutagenesis to generate pcDNA HA-HIRA S650A or S650D. HA-HIRA WT was PCR-amplified using pcDNA3 HA-HIRA and cloned into the EcoRI-cleaved pBabe hygro vector. pBabe HA-HIRA S650A and S650D were generated by site-directed mutagenesis. All mutations were confirmed by sequencing. For the baculovirus-mediated expression of HIRA, HIRA cDNA was amplified by PCR and subcloned into the pFATBAC1 vector, which provides a Flag tag.
Immunoprecipitation
Cells (293T cells)grown in 6-cm-diameter dishes were transfected using Lipofectamine 2000 (Invitrogen). At 24–48 h after transfection, whole-cell extracts were prepared with NETN Buffer (25 mM Tris-HCl (pH8.0), 1 mM EDTA, 0.15 M NaCl, 0.5% NP-40) containing protease and phosphatase inhibitors. The extracts were subjected to immunoprecipitation using antibodies and A or G beads (GE Healthcare, Pittsburgh, PA, USA).
Chromatin immunoprecipitation
Cells were cross-linked with 1% formaldehyde for 15 min and quenched by adding glycine to a final concentration of 0.125 M. Nuclei were prepared with lysis buffer A (10 mM Tris HCl (pH7.5), 10 mM KCl, 5 mM MgCl2, 0.5% NP40), and resuspended in SDS lysis buffer (50 mM Tris HCl (pH7.9), 10 mM EDTA, 0.5% SDS). For DNA fragmentation, the chromatin solution was sonicated using a Bioruptor (Cosmo Bio, Tokyo, Japan). Antibodies and beads were added and incubated overnight at 4 °C with rotation. DNA was purified using a PCR purification kit (Qiagen, Hilden, Germany). The percentage input (Immunoprecipitation DNA/input DNA) represents the relative amount of immunoprecipitated DNA.3
RNAi and quantitative RT-PCR (qRT-PCR)
Transfection of small interfering RNA (siRNA) was performed using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer's instructions. siRNAs were purchased from Samchully Pharm (Seoul, Korea). The siRNA sequences were as follows: mAkt1 siRNA, 5′-AAGAAGAGACGAUGGACUUTT-3′ mAkt2 siRNA, 5′-GUAUGCCUUCCAGACCCAUTT-3′. Total RNA was purified using NucleoSpin (Machery Nagel, Düren, Germany 740955.250) according to the manufacturer's instructions. Two micrograms of RNA were reverse transcribed using a cDNA synthesis kit (Thermo, Waltham, MA, USA, K1632). Real-time PCR was performed using the CFX96 System (Bio-Rad, Hercules, CA, USA). PCR amplification was carried out using KAPA SYBR FAST Master Mix (KAPA Biosystem, Wilmington, MA, USA), and relative quantification was conducted using the Livak method.21 The sequences of primers used for RT-PCR were as follows: mAkt1 forward, 5′-CCTCAGGATGTGGATCAGCGA-3′ mAkt1 reverse, 5′-CCACAGTCTGAATGGCGGT 3′ mAkt2 forward, 5′-TTCGGCAAGGTCATTCTGGT-3′ mAkt2 reverse, 5′-GGAAGGGGTGCCTGGTATTC-3′.
Protein purification
Escherichia coli DH5α was transformed with pGEX HIRA 640–655 and grown in LB ampicillin medium supplemented with 1 mM IPTG at 25 °C overnight. Cells were collected by centrifugation and resuspended in PBS with 1% Triton X-100, 1 mM DTT and protease inhibitors. Cells were sonicated until the solution was clear. Cell extracts were mixed with glutathione beads (Sigma-Aldrich, St Louis, MO, USA) and rotated at room temperature for 30 min. GST-HIRA/glutathione beads were washed three times with a lysis buffer and then eluted with an elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl (pH8.0)). For the baculovirus expression of Flag-tagged HIRA WT, S650A and S650D, baculoviruses were generated according to the manufacturer's protocol (Gibco-Invitrogen, Waltham, MA, USA) and used to infect Sf9 cells. Expressed proteins were purified using M2 agarose (Sigma-Aldrich) as described previously.22
In vitro kinase assay
Purified HIRA (3 μg) and active Akt1/PKBα (80 ng; Millipore, 14–276) were mixed in a kinase assay buffer (25 mM HEPES (pH8.0), 25 mM MgCl2, 1 mM DTT, 10 μCi 32P-ATP, phosphatase inhibitor cocktail). Reaction mixtures were incubated at 30 °C for 2 h and resolved by SDS-polyacrylamide gel electrophoresis. The gel was stained with Coomassie brilliant blue, destained, and dried. The dried gel was exposed to X-ray film for 1 h. For the non-radioactive kinase assay, the assay mixture was supplemented with 200 μM cold ATP instead of 32P-ATP. SDS-polyacrylamide gel electrophoresis gels were transferred to nitrocellulose membranes and probed with an anti-p-HIRA antibody.
Antibodies
The following antibodies were used in this study: anti-HIRA (Active Motif, Carlsbad, CA, USA, WC119 and WC15), anti-Asf1a (Cell Signaling, Danvers, MA, USA, 2990), anti-Cabin1, anti-UBN1 (Abcam, Cambridge, MA, USA), anti-H3 (Abcam, ab1791), anti-H4 (Millipore, Billerica, MA, USA, 07–108), anti- H4S47p (Abcam), anti-H3.3 (Millipore, 09–838), anti-H3.1 and anti-H3.3 (provided by Dr Y Ohkawa), anti-pan-Akt, anti-Akt1, anti-Akt2, anti-p-Akt (S473) (Cell Signaling, 9271), anti-phospho-serine (Abcam, ab9332), anti-phospho-threonine (Abcam, ab9337), anti-myogenin (Santa Cruz, Santa Cruz, CA, USA, sc-576), anti-MHC (Upstate, Billerica, MA, USA, 05–716), 8WG16 (Covance, Princeton, NJ, USA, MMS-126R), anti-HA (Roche, Basel, Switzerland, 11 666 606 001), anti-Flag (Sigma-Aldrich, F3165), anti-Myc (Roche, 11 667 203 001), anti-β-catenin (Santa Cruz, sc-7963), and anti-α-tubulin (AbFrontier, Seoul, Korea, LF-PA0146). The anti-p-HIRA (Abmart, Shanghai, China) antibody was generated using the phospho-peptides RPRKD(pS)RLMPV and was affinity-purified from rabbit serum using a specific column. Antibody specificity was confirmed by checking the absence of cross-reactivity with the control peptide, RPRKDSRLMPV.
Results
HIRA interacts with the Akt kinase
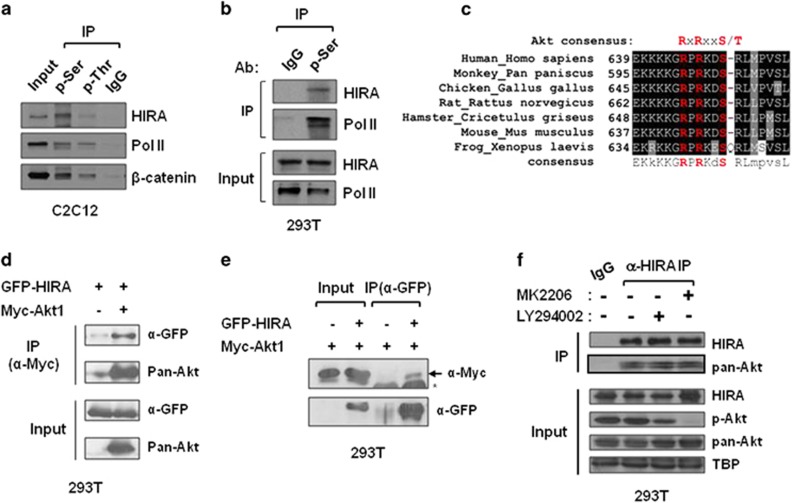
As HIRA regulates MyoD and muscle gene expression through H3.3 incorporation without significant changes in its abundance or occupancy at the target genes during muscle cell differentiation, we hypothesized that histone chaperone activity might be changed by differentiation programs. To investigate the regulatory mode of HIRA, we set out to determine its phosphorylation states. HIRA is known to be phosphorylated in cells, yet the roles of phosphorylation in chromatin organization and chaperone function have not been defined. Phosphorylated proteins were enriched by immunoprecipitation (IP) with anti-p-Ser or p-Thr antibodies, and the presence of the HIRA protein in the pellet was analyzed by immunoblotting, along with that of RNA polymerase II or β-catenin, two well-known phosphorylated proteins, as positive controls. Phosphorylation of HIRA was detected in C2C12 mouse myoblasts and HEK293T cells (Figures 1a and b). To identify a potential kinase that phosphorylates HIRA, we searched putative phosphorylation sequences in HIRA using a kinase-specific phosphorylation prediction tool (GPS2.1, http://gps.biocuckoo.org). The most conspicuous hit was S650 of human HIRA, which coincides with the consensus sequence of Akt substrates (RxRxxS/T) (Supplementary Figure S1). The sequence motif including S650 is conserved throughout vertebrates (Figure 1c). Akt, also known as protein kinase B, is reported to have a role in muscle gene transcription, cell cycle exit, and muscle regeneration, although the downstream targets responsible for these diverse biological effects have not been fully identified.23, 24, 25 To directly determine the functional relevance of HIRA phosphorylation and Akt kinase, we tested the protein–protein interaction between HIRA and Akt1(PKBα). Using extracts of 293T cells expressing green fluorescent protein (GFP)-HIRA and Myc-Akt1, GFP-HIRA was co-precipitated with an anti-Myc antibody (Figure 1d), and the reciprocal IP with an anti-GFP antibody showed the presence of Myc-Akt1 in the pellet (Figure 1e). Mammals encode several isoforms of Akts known as Akt1, Akt2 and Akt3, which have overlapping or distinct functions. To determine whether the HIRA–Akt interaction is isoform specific, 293T cells transfected with GFP-HIRA and HA-Akt2 were assayed by IP, which indicated that HIRA also interacted with Akt2 (Supplementary Figure S2). Furthermore, IP using a monoclonal antibody against HIRA showed that the two proteins interacted at the endogenous protein level (Figure 1f and Supplementary Figure S3). Interestingly, their interaction was not affected by the kinase inhibitors, MK2206 or LY294002, indicating that the proteins associate with each other regardless of the kinase activity of Akt, shown by its phosphorylation status (p-Akt).
Figure 1.
HIRA interacts with Akt1. (a, b) Whole-cell extracts prepared from C2C12 (a) or HEK293T (b) cells were subjected to IP using anti-p-Ser, anti-p-Thr, and control IgG antibodies, they were then probed with anti-HIRA (WC119) to detect phospho-HIRA. Two phospho-proteins, RNA polymerase II and β-catenin, were used as positive controls. (c) Sequences potentially targeted by Akt (RxRxxS/T) in vertebrate homologues were aligned. (d, e) GFP-HIRA was co-expressed with Myc-Akt1 in 293T cells. Whole-cell extracts were subjected to IP and immunoblotted using the indicated antibodies. Five percent of the input sample was loaded in parallel. *Non-specific background. (f) 293T cells were treated with MK2206 (10 μM) or LY294002 (10 μM) and the whole-cell extracts were subjected to IP using anti-HIRA(WC15) or IgG. Anti-pan-Akt, p-Akt(S473), or HIRA(WC119) antibodies were used for immunoblotting. TBP (TATA-binding protein) was used as the loading control. GFP, green fluorescent protein.
HIRA S650 is phosphorylated by Akt
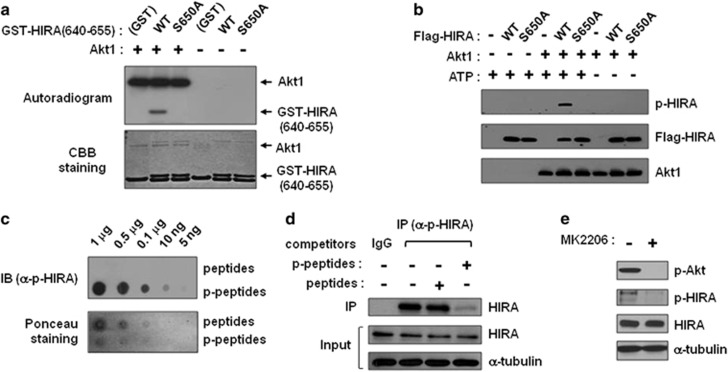
On the basis of their association, we performed an in vitro kinase assay to determine whether Akt1 phosphorylates HIRA. We constructed GST-HIRA(640–655) and purified the proteins using the GST tag. Consistent with their binding, GST-HIRA(640–655) served as a substrate and was phosphorylated by purified Akt1(PKBα), while the GST control or GST-HIRA(S650A) was not (Figure 2a). To examine the functional role of HIRA phosphorylation, a specific antibody recognizing phosphorylated S650 was generated (anti-p-HIRA). The specificity of this antibody was confirmed by a dot blot assay employing unmodified and S650-phosphorylated peptides (645–655; Figure 2c). We then produced full-length wild type and S650A HIRA with a Flag tag from insect cells and performed an in vitro kinase assay with cold ATP, followed by immunoblotting using an anti-p-HIRA antibody. As shown in Figure 2b, HIRA WT but not S650A was strongly recognized by the antibody upon phosphorylation by Akt1 in an ATP-dependent manner. To verify S650 phosphorylation in cells and the specificity of this antibody, we performed IP using a p-HIRA antibody in the presence of S650-phosphorylated or non-phosphorylated peptides acting as competitors. HIRA was efficiently precipitated by the p-HIRA antibody and detected in the pellets, while it was completely out-competed by excess amounts of the p-HIRA peptide, but not by the control peptide (Figure 2d). Furthermore, when Akt activity was inhibited by the allosteric inhibitor, MK2206, the S650 phosphorylation of HIRA decreased with the loss of Akt phosphorylation, indicating that a substantial amount of endogenous HIRA is phosphorylated at S650 and that Akt is the responsible kinase (Figure 2e).
Figure 2.
HIRA is phosphorylated by Akt1 at serine 650. (a) An in vitro kinase assay was performed with purified GST or GST-HIRA(640–655; WT or S650A) and 80 ng of Akt1. Reaction mixtures were subjected to SDS-PAGE followed by autoradiography to detect P32-labeled HIRA. Akt1 shows autophosphorylation. *GST and partial degradation products. (b) Full-length HIRA WT or S650A (3 μg each) purified from insect cells was phosphorylated in the presence and absence of 80 ng of Akt1 and cold ATP. Phosphorylation of HIRA was detected using an anti-phospho-HIRA antibody (p-HIRA). (c) Verification of anti-phospho-HIRA antibody. The specificity of the purified p-HIRA antibody was tested by dot blot assay. The indicated amounts of peptides with phosphorylated Ser 650 (RPRKD(pS)RLMPV) or their unmodified counterparts (RPRKDSRLMPV) were spotted on the membrane, and immunoblotting was performed using an anti p-HIRA antibody. The antibody specifically recognized the p-peptides phosphorylated at Ser650. (d) Whole-cell extracts from 293T cells were subjected to IP using an anti-p-HIRA antibody in the presence or absence of phospho- or unmodified peptides acting as competitors. Immunoprecipitates were probed with WC119 to detect HIRA. Rabbit IgG was used as the negative control. (e) 293T cells were treated with MK2206 (10 μM) for 24 h and whole-cell extracts were used for immunoblotting with the indicated antibodies. Tubulin was used as the loading control. IP, immunoprecipitation; WT, wild type.
HIRA phosphorylation decreases during myogenic differentiation
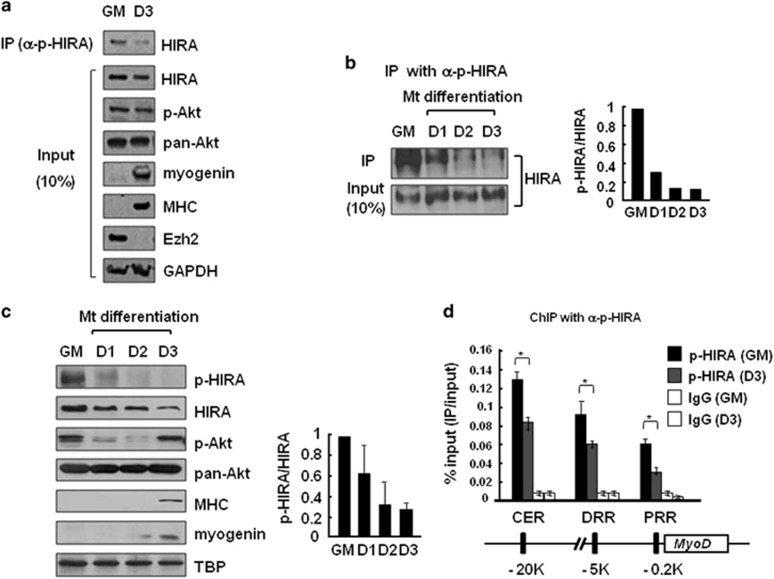
We next tested whether the S650 phosphorylation of HIRA has a role in myoblast differentiation. To delineate the underlying connection, we started by determining whether HIRA phosphorylation is altered during cellular differentiation. S650 in human HIRA corresponds to S648 in mice. We confirmed that the p-HIRA antibody also recognized the phosphorylation of mouse HIRA (Supplementary Figure S4). Proliferating C2C12 myoblasts were differentiated into myotubes by shifting them from a growth (GM) to a differentiation medium for 3 days (D1–D3). Cellular differentiation was monitored by analyzing the markers, Ezh2, myogenin and myosin heavy chain (MHC). IP of whole-cell extracts with p-HIRA antibody followed by probing with an anti-HIRA monoclonal antibody showed that a significant level of HIRA phosphorylation was present in myoblasts of growth medium (GM) (Figure 3a). Interestingly, however, HIRA phosphorylation decreased at differentiation day 3 (D3), while the total HIRA protein level was relatively unchanged (Figures 3a and b). This result was confirmed by the quantitation of p-HIRA signals and their normalization to total HIRA protein levels (Figure 3b, right panel). The reduction in p-HIRA level was also detected by western blotting of whole-cell extracts and the relative ratio of p-HIRA and total HIRA signals (Figure 3c). Interestingly, Akt activity (p-Akt) was also rapidly decreased at D1 and D2 along with p-HIRA, without significant changes in the total level of Akt (pan-Akt), indicating that HIRA phosphorylation and Akt activity are correlated during the early stages of differentiation (D1–D2). However, p-Akt was restored at D3, whereas p-HIRA remained low, suggesting that Akt activity and p-HIRA become uncoupled once differentiation has proceeded past the early stages (Figure 3c).
Figure 3.
HIRA phosphorylation decreases during muscle cell differentiation. The levels of the indicated proteins were monitored in C2C12 cells grown in growth (GM) or differentiating medium for 3 days (D1–3). (a, b) C2C12 cell extracts prepared from myoblasts and myotubes were subjected to IP using an anti-p-HIRA antibody and immunoblotted with an anti-HIRA antibody. Myogenin, MHC and Ezh2 were used as differentiation markers. The p-HIRA/HIRA protein signal ratio in each sample was quantitated and compared through normalization. (c) Whole-cell extracts were analyzed by western blotting using the indicated antibodies. TBP (TATA-binding protein) served as the loading control. The p-HIRA/HIRA protein signal ratio was obtained as above from two independent experiments. (d) ChIP using an anti-p-HIRA antibody was performed to monitor the recruitment of p-HIRA on MyoD regulatory regions in C2C12 myoblasts (GM) and myotubes (D3). Chromatin solutions prepared from each were subjected to IP using an anti-p-HIRA antibody and analyzed in duplicate via quantitative real-time-PCR to measure the relative enrichment of p-HIRA at the indicated loci. The data represent the percentages of ChIP (IP/Input). Error bars represent s.d., n=2. *P<0.05. CER, core enhancer region; ChIP, chromate immunoprecipitation; DRR, distal regulatory region; PRR, proximal regulatory region; siRNA, small interfering RNA.
Next, we investigated whether HIRA was phosphorylated at the chromatin level by chromatin immunoprecipitation. HIRA was preferentially detected in the chromatin fraction throughout differentiation (Supplementary Figure S5). We performed chromatin immunoprecipitation to analyze the phosphorylation states of HIRA bound to the MyoD regulatory regions (CER, DRR and PRR sites). Consistent with the western blotting and IP data (Figures 3a–c), HIRA phosphorylation was significantly reduced in myotubes at D3 compared with phosphorylation in myoblasts (GM) at all three regions, indicating that alterations of the phosphorylation states of HIRA occur on the chromatin (Figure 3d). Taken together, the global and genome-specific analyses show that myoblasts harbor phosphorylated HIRA, whose phosphorylation level decreases during myogenic differentiation.
Akt1 is the major kinase responsible for HIRA phosphorylation in myoblasts
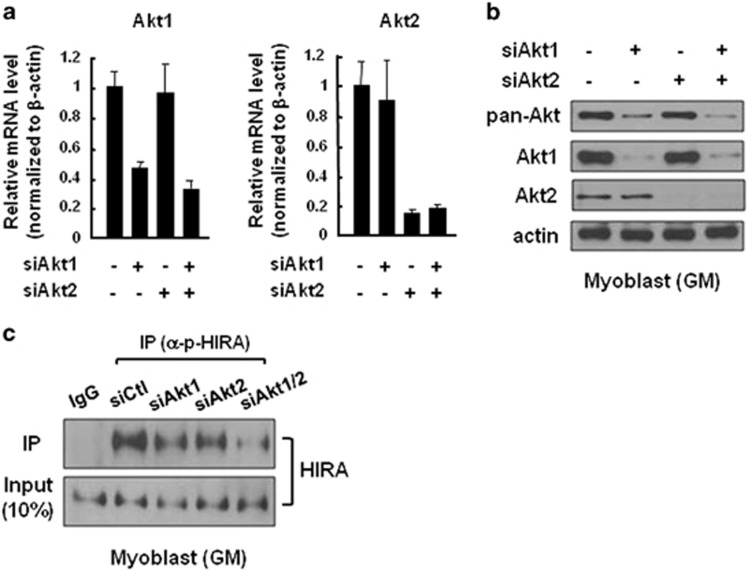
Our data show an intimate correlation between Akt activity and HIRA phosphorylation in myoblasts and during the early stages of differentiation, while the correlation was lost at D3 (Figure 3c). As different Akt isoforms have roles in myogenesis, we examined which Akt isoform was responsible for HIRA phosphorylation in myoblasts. In mammals, Akt1 appears to be critical for the proliferative growth of myoblasts by targeting the cyclin-dependent kinase inhibitor p21 and FoxO1 transcription factor, while Akt2 drives cell cycle exit and cellular differentiation by phosphorylation of GSK3β and p300.26, 27 To identify the Akt isoform required for the phosphorylation of HIRA, we first examined the relative abundances of Akt1 and Akt2 by RNA interference. Knockdown of Akt1 and Akt2 using siRNAs targeting each isoform was specific and efficient (Figure 4a). The depletion of Akt1 in proliferating myoblasts resulted in a significant reduction in the total Akt protein level (pan-Akt), indicating that Akt1 is the predominant isoform that has a role in myoblasts (Figure 4b). Consistently, HIRA phosphorylation was effectively decreased by Akt1 knockdown and was completely eliminated by the depletion of both Akt1 and Akt2, suggesting that HIRA is phosphorylated primarily by Akt1 and that Akt2 seemed to contribute to HIRA phosphorylation as a supplementary kinase in myoblasts (Figure 4c). The expression pattern of Akts during cellular differentiation showed that the Akt1 level decreased gradually during differentiation, whereas that of Akt2 was unchanged or increased at D3 (Supplementary Figure S6). Taken together, these data suggest that HIRA is phosphorylated largely by Akt1 in myoblasts, and the diminished level of p-HIRA might be attributed to the decrease in Akt1 level upon differentiation.
Figure 4.
Akt1 is the major kinase of HIRA in myoblasts. (a) Knock-down of each Akt isoform using specific siRNAs. C2C12 cells were treated with control siRNA or siRNAs specifically targeting Akt1 or Akt2 for 72 h. Total RNA was isolated to determine the steady-state level of each Akt by qRT-PCR. The PCR values were normalized to β-actin and presented as values relative to the control, which was set as 1. (b, c) C2C12 cells were treated with control siRNA or siRNAs for Akt1 or Akt2 for 72 h. Whole-cell extracts were then subjected to western blotting using the indicated antibodies (b) or subjected to IP using an anti-p-HIRA antibody (c), as described in Figure 3. siRNA, small interfering RNA; qRT-PCR, quantitative reverse transcription PCR.
S650 phosphorylation of HIRA does not affect complex formation or histone binding
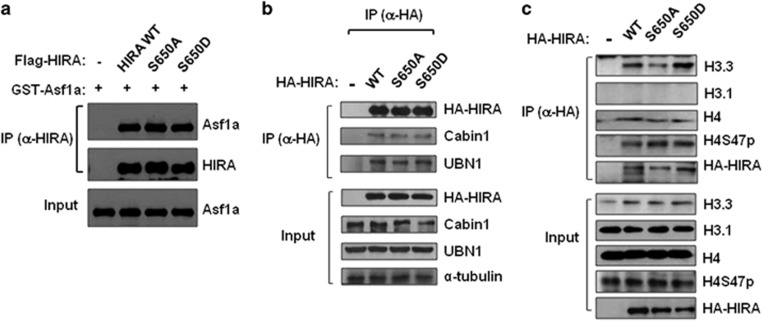
Next, we investigated the functional role of HIRA phosphorylation in H3.3 loading and target gene regulation. To this end, we constructed HA-tagged human HIRA, both the wild type and two mutants (S650A and S650D, phosphodefective and phosphomimicking forms, respectively). Protein phosphorylation often affects interactions with other protein partners. For example, the phosphorylation of Asf1 by Tousled-like kinase (TLK) enhances its binding to histone subunits.28 To test whether HIRA phosphorylation affected its association with the Asf1 histone chaperone, other subunits of the HIRA complex, or histone H3/H4, we partially purified HA-tagged WT or mutants of HIRA by IP. Each HIRA showed efficient binding to Asf1a (Figure 5a) or the other subunits of the HIRA complex, Cabin1 and UBN1 (Figure 5b), indicating that their binding affinity was not affected by the S650 mutation. Furthermore, all HIRAs showed preferential binding toward H3.3 over H3.1, with comparable association efficiencies (Figure 5c). The level of H4 phosphorylation (H4S47p) of the H3.3 nucleosome, reported to be important for HIRA binding,29 was not significantly affected by the S650 mutation (Figure 5c), indicating that S650 phosphorylation of HIRA does not have a direct impact on complex formation or H3.3 histone binding.
Figure 5.
Effect of S650 mutation on protein interactions and H3/H4 histone binding. (a) Purified recombinant Flag-HIRA (WT, S650A and S650D) and GST-Asf1a were incubated in vitro and precipitated using an anti-HIRA antibody (WC15). IP pellets were probed with anti-Asf1a and anti-HIRA (WC119) antibodies. (b, c) HA-HIRA (wild type, S650A, or S650D) was expressed in 293T cells. HA-HIRA was subjected to IP using an anti-HA antibody and the endogenous proteins interacting with HIRA were detected by immunoblotting with the corresponding specific antibodies. WT, wild type.
HIRA S650 phosphorylation suppresses target gene activation by suppressing H3.3 incorporation
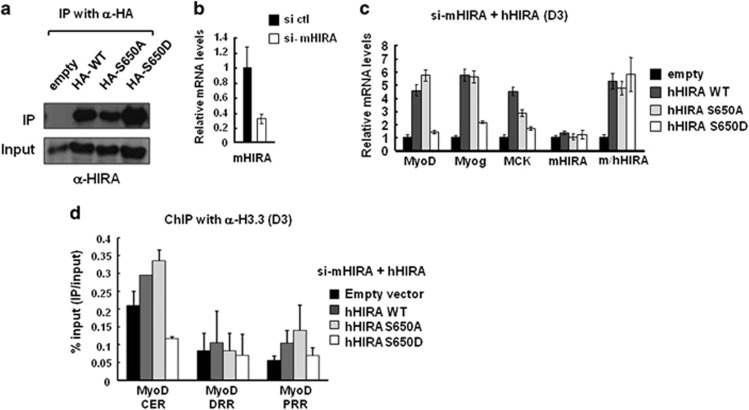
Next, we generated C2C12 myoblasts that stably expressed human HIRA (hHIRA) to examine the effect of phosphorylation on the regulation of endogenous genes. The expression of WT and HIRA mutants was confirmed by IP and western blotting (Figure 6a). Then, cells were treated with siRNAs to deplete endogenous mouse HIRA (mHIRA) to reveal the effects of the mutations, and cells were then differentiated into myotubes. Mouse-specific siRNAs efficiently depleted endogenous mHIRA but had no effect on the expression of hHIRA (Figures 6b and c). Under these conditions, the messenger RNA levels of muscle genes (MyoD, myogenin and MCK) were analyzed by RT-PCR. Interestingly, hHIRA WT and S650A mediated activation of muscle genes in myotubes. However, hHIRA S650D was defective in muscle gene activation, although the HIRA protein levels were similar (Figure 6c), suggesting an inhibitory role of HIRA phosphorylation in muscle gene activation. To directly address the role of HIRA phosphorylation in histone chaperoning function, the H3.3 level was examined by chromatin immunoprecipitation. A substantial amount of H3.3 accumulates in the CER prior to myogenic activation in a HIRA-independent manner, while upon terminal differentiation, all three regions, including the CER, increase the H3.3 occupancy in a HIRA-dependent manner to promote gene activation.3 We also observed that the H3.3 level was low in the PRR and DRR without functional HIRA, with the exception of in CER (Figure 6d). However, deposition of H3.3 was indeed increased in the CER and PRR regions by the expression of WT and S650A, but not of S650D. As the total level of nucleosomal H3 was not changed during differentiation (Supplementary Figure S7), the specific incorporation of H3.3 into chromatin seemed to be affected by S650D, indicating that persistent phosphorylation could result in the inhibition of gene activation. Because H3.3 incorporation through dynamic histone chaperoning is a prerequisite for muscle gene activation,3, 30, 31 our data suggest that the phosphorylation of HIRA is critical for the on-time activation of muscle genes and for muscle cell differentiation.
Figure 6.
Phosphorylation mimicking form of HIRA prevents the incorporation of H3.3 and the activation of muscle genes. (a) C2C12 cells stably expressing empty vectors or each type of HA-hHIRA were generated and confirmed by IP. Cell extracts were subjected to IP using an anti-HA antibody and probed with an anti-WC119 HIRA antibody that recognized both human and mouse HIRA proteins. (b) C2C12 cells were treated with siRNAs specifically targeting endogenous mouse HIRA. The mHIRA knockdown efficiency was monitored by qRT-PCR. (c) C2C12 cell lines were treated with mHIRA-targeting siRNAs, induced to differentiate for 3 days, and subjected to qRT-PCR for analysis of mRNA levels. (d) H3.3 ChIP was performed to monitor H3.3 incorporation in the MyoD regulatory regions in the indicated C2C12 cell lines. Chromatin solutions prepared from myotubes were subjected to IP using an anti-H3.3 antibody. Error bars represent s.d., n=2. IP, immunoprecipitation; mRNA, messenger RNA; qRT-PCR, quantitative reverse transcription PCR; siRNA, small interfering RNA.
Discussion
Myogenesis is a complex and tightly regulated cellular process that is executed by the concerted action of upstream signaling and downstream gene expression pathways. One major signaling component in the myogenic program is Akt, which is activated by insulin/IGF stimulation, and its activity is critical for both myoblast proliferation and differentiation.32 Our data show that HIRA phosphorylation by Akt1 limits HIRA activity and deters it from H3.3 loading and that its phosphorylation rapidly disappears to promote efficient H3.3 transfer and muscle gene activation for a prompt response to myogenic differentiation cues. In this study, HIRA was more efficiently targeted by Akt1 in myoblasts. Given its roles in myogenesis, Akt1 likely maintains myoblast proliferation in part by targeting HIRA to inhibit precocious H3.3 deposition at muscle loci.
It is noteworthy that HIRA phosphorylation was low in myotubes even with the restoration of total Akt activity. In fact, Akt isoforms often differ in their abundance, function and tissue specificity. Akt1 has a role in the proliferative growth of myoblasts, while Akt2 is upregulated during myogenesis and mediates cell cycle exit and cellular differentiation. As different Akts often show different substrate affinities, it is possible that they target HIRA differently during differentiation to control its activity in a cell-type-specific manner.
One interesting possibility is that the phosphoserine of HIRA could be targeted by differentiation-responsive phosphatases in myotubes. One of the major modulators of early muscle cell differentiation is the calcineurin/NFAT pathway. Calcineurin is a Ca2+/calmodulin regulated Ser/Thr phosphatase, and once activated, it targets transcription factors such as NFAT to induce the transcription of muscle genes, while calcineurin inhibitors antagonize muscle gene expression and block the formation of multinucleated myotubes.33, 34, 35 Despite its role in the early stages of myogenesis and the later steps of muscle development, a full list of its molecular targets is not yet available. Interestingly, Cabin1, a component of the HIRA complex, was originally identified as a calcineurin-binding protein that inhibits the calcineurin-mediated signaling pathway in T cells.36 In addition, Cabin1 functions as a myogenic inhibitor through the repression of MEF2 by tethering histone deacetylase and/or histone methyltransferase complexes.37 Therefore, it is possible that Cabin1 within the HIRA complex counteracts the activity of phosphatase to maintain the phosphorylation state of the HIRA protein. Analogous to this, neuronal gene activation by H3.3 was reported to be mediated by the calcineurin-mediated dephosphorylation of DAXX upon synaptic stimulation.18
What might be the functional consequences of HIRA phosphorylation? In the case of Asf1, TLK phosphorylation of the C-terminal tail of Asf1 facilitates nucleosome assembly and the interaction with histones.28 The phosphorylation of DAXX is important for the calcium-dependent regulation of its activity without having a significant effect on its association with H3.3 or ATRX.38 In our study, we failed to detect any effect of S650 mutation on H3.3/H4 binding or on the composition of the HIRA complex. However, HIRA S650D showed a marked defect in H3.3 deposition and muscle gene activation compared with the WT and S650A. As multiple transcription factors and ATP-dependent chromatin remodeling factors co-localize with HIRA complexes across the genome, those interacting partners are likely to be potential candidates for future investigations to understand the effect of HIRA phosphorylation on transcriptional regulation.39
In summary, we have demonstrated for the first time that HIRA is targeted by myogenic signaling pathways for the timely control of chromatin and muscle gene expression. HIRA might be pre-recruited to the muscle genes and phosphorylated to inhibit the expression of target genes until necessary. Future exploration of the differential control of phosphorylation and the effect of HIRA modification with regard to chromatin remodeling may reveal the complex signaling network and epigenetic outcomes that underlie muscle development.
Acknowledgments
We thank Y Ohkawa (Kyushu University) for providing the H3.1 and H3.3 antibodies. This work was supported by the Mid-Career Researcher Program (2013R1A2A2A01014702 and 2015R1A2A1A15055713 to E-JC) and the Basic Science Research Program (2010-0028646 to E-JC) through the National Research Foundation of Korea and the Korean government (MSIP) (no. 2012R1A3A2048767 to H-DY).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004; 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 2002; 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW et al. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci USA 2011; 108: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 2013; 27: 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet 2009; 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res 2011; 21: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Smith M, Kanno T, Dasenbrock H, Nishiyama A, Ozato K. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem 2009; 284: 12217–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Heo K, Choi J, Kim K, An W. Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1gamma. Nucleic Acids Res 2011; 39: 8329–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 2013; 155: 94–106. [DOI] [PubMed] [Google Scholar]
- Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev 2010; 20: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N et al. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem 2010; 285: 41567–41577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA et al. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol Cell Biol 2011; 31: 4107–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 2005; 8: 19–30. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 2013; 155: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 2010; 24: 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA 2010; 107: 14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod D, Bartesaghi S, Khelifi A, Bellodi C, Berliocchi L, Nicotera P et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron 2012; 74: 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Nelson DM, Ye X, Baker K, DeCaprio JA, Seeholzer S et al. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol Cell Biol 2001; 21: 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Kennedy A, Banumathy G, Zhang R, Adams PD. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell 2007; 27: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 2010; 140: 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron-Milhavet L, Franckhauser C, Rana V, Berthenet C, Fisher D, Hemmings BA et al. Only Akt1 is required for proliferation, while Akt2 promotes cell cycle exit through p21 binding. Mol Cell Biol 2006; 26: 8267–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotwein P, Wilson EM. Distinct actions of Akt1 and Akt2 in skeletal muscle differentiation. J Cell Physiol 2009; 219: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S, Anguiano M, Rotwein P. Defining Akt actions in muscle differentiation. Am J Physiol Cell Physiol 2012; 303: C1292–C1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Spangenburg EE, Booth FW. Forkhead transcription factor FoxO1 transduces insulin-like growth factor's signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol 2003; 196: 523–531. [DOI] [PubMed] [Google Scholar]
- van der Velden JL, Langen RC, Kelders MC, Wouters EF, Janssen-Heininger YM, Schols AM. Inhibition of glycogen synthase kinase-3beta activity is sufficient to stimulate myogenic differentiation. Am J Physiol Cell Physiol 2006; 290: C453–C462. [DOI] [PubMed] [Google Scholar]
- Klimovskaia IM, Young C, Stromme CB, Menard P, Jasencakova Z, Mejlvang J et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun 2014; 5: 3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Pu M, Hu G, Wen W, Dong Z, Zhao K et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev 2011; 25: 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Maehara K, Sato Y, Konno D, Tachibana T, Kimura H et al. Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle. Nucleic Acids Res 2015; 43: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S et al. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J 2012; 31: 2994–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JD, Kothary R. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet Muscle 2011; 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday BB, Mitchell PO, Kegley KM, Pavlath GK. Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation 2003; 71: 217–227. [DOI] [PubMed] [Google Scholar]
- Friday BB, Horsley V, Pavlath GK. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J Cell Biol 2000; 149: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM et al. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev 1998; 12: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Youn HD, Loh C, Stolow M, He W, Liu JO. Cabin 1, a negative regulator for calcineurin signaling in T lymphocytes. Immunity 1998; 8: 703–711. [DOI] [PubMed] [Google Scholar]
- Jang H, Choi DE, Kim H, Cho EJ, Youn HD. Cabin1 represses MEF2 transcriptional activity by association with a methyltransferase, SUV39H1. J Biol Chem 2007; 282: 11172–11179. [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 2012; 491: 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep 2013; 3: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.