Abstract
Cancer stem cells are a subpopulation of cancer cells characterized by self-renewal ability, tumorigenesis and drug resistance. The aim of this study was to investigate the role of HMGA1, a chromatin remodeling factor abundantly expressed in many different cancers, in the regulation of cancer stem cells in ovarian cancer. Spheroid-forming cancer stem cells were isolated from A2780, SKOV3 and PA1 ovarian cancer cells by three-dimensional spheroid culture. Elevated expression of HMGA1 was observed in spheroid cells along with increased expression of stemness-related genes, such as SOX2, KLF4, ALDH, ABCB1 and ABCG2. Furthermore, spheroid A2780 cells, compared with adherent cells, showed higher resistance to chemotherapeutic agents such as paclitaxel and doxorubicin. HMGA1 knockdown in spheroid cells reduced the proliferative advantage and spheroid-forming efficiency of the cells and the expression of stemness-related genes. HMGA1 overexpression in adherent A2780 cells increased cancer stem cell properties, including proliferation, spheroid-forming efficiency and the expression of stemness-related genes. In addition, HMGA1 regulated ABCG2 promoter activity through HMGA1-binding sites. Knockdown of HMGA1 in spheroid cells reduced resistance to chemotherapeutic agents, whereas the overexpression of HMGA1 in adherent ovarian cancer cells increased resistance to chemotherapeutic agents in vitro. Furthermore, HMGA1-overexpressing A2780 cells showed a significant survival advantage after chemotherapeutic agent treatment in a xenograft tumorigenicity assay. Together, our results provide novel insights regarding the critical role of HMGA1 in the regulation of the cancer stem cell characteristics of ovarian cancer cells, thus suggesting that HMGA1 may be an important target in the development of therapeutics for ovarian cancer patients.
Introduction
Cancer stem cells (CSCs) were first identified in myeloid leukemia with the cell surface marker combination of CD34+ and CD38−.1 Thereafter, CSCs were identified in other solid tumors, including breast,2 brain,3 prostate,4 pancreatic5 and colon cancers.6 In 2008, ovarian CSCs were identified from primary human tumors.7 Many papers have reported on the features of CSCs, including the main characteristics of self-renewal and differentiation into multiple cell types. Spheroid-forming cells have commonly been found in the ascites of ovarian cancer patients,8 and they are able to initiate tumors.9 They show a reduced response to chemotherapeutic drugs10 and have been thought to have an important role in cancer metastasis.11 CSCs share several properties with pluripotent stem cells, as both have the abilities of self-renewal and differentiation.12 Current evidence has shown that key regulators of embryonic stem cells, including Oct4 and Sox2, are overexpressed in poorly differentiated tumors.13 Therefore, we investigated the expression of stemness-related genes in ovarian CSCs, which were isolated through spheroid culture of ovarian cancer cells, in comparison with ovarian cancer cells.
HMGA1 is essential for the cellular reprogramming of somatic cells to induce pluripotent stem cells via four reprogramming factors (OCT4, SOX2, KLF4 and c-MYC).14 HMGA proteins are architectural factors constituting clinical hubs in the chromatin network.15 HMGA proteins, including HMGA1 (with the splice variants HMGA1a and HMGA1b) and the highly related HMGA2, bind AT-rich DNA stretches, forming stereo-specific enhanceosomes on the promoter/enhancer regions of genes involved in regulation of transcription.16 These proteins are expressed in many cancer tissues and during embryogenesis but are expressed at low levels or are absent in normal adult tissues.17 HMGA1 has been shown to regulate tumor progression by reprogramming differentiated cells into poorly differentiated CSCs. Several studies have reported that HMGA1 has important roles in the self-renewal and differentiation of stem cells.18, 19, 20, 21 In recent years, HMGA1 expression has been reported in breast cancer and colon cancer cells, in which HMGA1 has been suggested to be a master regulator of tumor progression and metastasis by regulating the epithelial-to-mesenchymal transition.20, 22, 23, 24
In this study, we found that HMGA1 dramatically influenced the drug resistance of ovarian CSCs, altered the expression of genes related to stem cell characteristics, and increased cell growth and viability. In addition, HMGA1 regulated ABCG2 promoter activity and boosted transcriptional activity. These results suggest that HMGA1 is a critical regulator of the maintenance of CSC characteristics in ovarian cancer and may provide a novel opportunity in developing treatments to cure ovarian cancer.
Materials and methods
Materials
Cell culture plates for adherent cells were purchased from Thermo Fisher Scientific (Waltham, MA, USA). For culture of spheroid cells, culture plates with an ultra-low-attachment surface were purchased from Corning Life Sciences (Tewksbury, MA, USA). Neurobasal medium, RPMI 1640, FBS, B-27 supplement, penicillin, streptomycin and Accutase cell detachment solution were purchased from Life Technologies (Grand Island, NY, USA). Basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) were purchased from R&D Systems (Minneapolis, MN, USA). The human ovarian cancer cell line A2780 was purchased from the American Type Culture Collection (Manassas, VA, USA). Antibodies against OCT4 (sc-8628) and SOX2 (sc-17320) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), and antibodies against KLF4 (ab151733), HMGA1 (ab129153) and ABCG2 (ab3380) were purchased from Abcam (Boston, MA, USA). Anti-aldehyde dehydrogenase 1 (ALDH1) antibody (611194) was purchased from BD Biosciences (San Jose, CA, USA). Antibodies against ABCB1 (12683) was purchased from Cell Signaling Technology (Danvers, MA, USA).
Cell culture
A2780 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and penicillin/streptomycin and maintained at 37 °C in 5% CO2. For isolation of sphere-forming cells, A2780 cells were detached with trypsin/EDTA solution and seeded in CSC culture medium (Neurobasal medium supplemented with 20 ng ml−1 bFGF, 10 ng ml−1 EGF, 2.5 μg ml−1 amphotericin B, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and B-27 supplement) on ultra-low-attachment culture plates. Two weeks after plating, spheroids that could not be dissociated by pipetting were formed and were isolated from non-spheroid-forming cells by centrifugation (800 r.p.m. for 2 min). The spheroids were dissociated by treatment with Accutase cell detachment solution and maintained in CSC culture medium in low-attachment-surface plates.
Western blotting
Cells were washed twice with HBSS and then lysed in lysis buffer (20 mM Tris-HCl, 1 mM EGTA, 1 mM EDTA, 10 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 30 mM sodium pyrophosphate, 25 mM β-glycerol phosphate, 1% Triton X-100, pH 7.4). The cell lysates were centrifuged for 15 min at 4 °C, and the supernatants were used for western blotting. The lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and then stained with 0.1% Ponceau S solution (Sigma-Aldrich, St Louis, MO, USA) to ensure equal loading of the samples. After being blocked with 5% non-fat milk for 30 min, the membranes were incubated with primary antibodies overnight, and the bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies using the enhanced chemiluminescence western blotting system (Amersham Biosciences, Piscataway, NJ, USA).
Reverse transcription-polymerase chain reaction
All RNA samples were prepared using TRI reagent (Sigma-Aldrich). RNA samples (2 μg) were reverse-transcribed to complementary DNA using M-MLV Reverse transcriptase (Promega, Madison, WI, USA) and 0.5 μg of oligo (dT) 15 primer (Promega). cDNA in 1 μl of the reaction mixture was amplified with [Ready] 2 × GO (Nanohelix) and 10 pmol each of the sense and antisense primers. The thermal cycle profile was as follows: denaturation at 95 °C for 30 s, annealing at 51–55 °C for 30 s depending on the primers used and extension at 72 °C for 90 s. Each PCR reaction was carried out for 18–30 cycles, and the PCR products were size fractionated on 1% gel-RED/agarose gel and visualized under UV transillumination. The primer pairs were as follows: GAPDH, 5′-TCC ATG ACA ACT TTG GTA TCG-3′, 5′-TGT AGC CAA ATT CGT TGT CA-3′ HMGA1, 5′-ATG AGT GAG TCG AGC AA-3′, 5′-TCA CTG CTC CTC CTC CGA-3′.
Cell proliferation assay
Proliferation was determined using two methods: a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and cell counting. To measure cell proliferation, A2780 cells were seeded in a 24-well culture plate at a density of 1 × 104 cells per well. After being cultured under various experimental conditions, the cells were washed twice with HBSS and incubated with 200 μl of MTT solution (0.5 mg ml−1) for 2 h at 37 °C. After the incubation, the MTT solution was removed, the cells were treated with dimethyl sulfoxide (200 μl per well), and the absorbance of the solution at 570 nm was determined using a micro-plate spectrophotometer (TECAN) after dilution to a linear range. Cell counts were determined using Trypan blue solution (Sigma-Aldrich).
Short-hairpin RNA-mediated knockdown
Short-hairpin RNA (shRNA) targeting HMGA1 was purchased from Sigma. For the generation of lentiviral particles, 6.67 μg of targeted viral plasmid, 5 μg of VSVG and 3.33 μg of Δ8.9 were used to transfect 293FT cells using 15 μl of the plasmid mixture with Lipofectamine/Lipofectamine Plus reagent (Life Technologies). At 48 h after transfection, the supernatants containing virus particles were filtered using 0.45 μm filters and concentrated using a Lenti-X Concentrator (Clontech Laboratories, Mountain View, CA, USA) for 24 h at 4 °C. A2780 ovarian cancer cells were infected with concentrated viral supernatants with 10 μg ml−1 Polybrene (Sigma-Aldrich) for 24 h and the medium was changed to RPMI 1640 medium with 10% FBS. A2780 cells expressing shRNA were selected for 1 day with 1 μg ml−1 puromycin and then were maintained in RPMI 1640 supplemented with 10% FBS and 1 μg ml−1 puromycin.
Generation of HMGA1-overexpressing cells
pIRES-puro3 vectors overexpressing HMGA1 or non-target controls were purchased from Addgene (Plasmid #13466). A2780 cells were co-transfected with the pLKO.1-puro plasmid using Lipofectamine 2000 (Invitrogen). The cells were transfected with a control (pLKO.1-puro) or pIRES-puro3-HMGA1, and this was followed by selection with puromycin (1 μg ml−1) for 1 week. To ensure the overexpression of HMGA1, the mRNA or protein levels of HMGA1 were determined by RT-PCR analysis or western blotting.
Cytotoxicity assay
In all, 10 000 cells were seeded into each well of a 24-well plate in 500 μl RPMI 1640 medium with 5% FBS. After 24 h or 48 h of incubation with paclitaxel (0.1 μM) or doxorubicin (1 μM), the cells were harvested and subjected to MTT assays. The percentage of cell survival was expressed relative to that of the untreated control.
Flow cytometry
ALDH (aldehyde dehydrogenase) activity was detected using an Aldefluor assay kit (STEMCELL Technologies, Vancouver, BC, Canada) as described by the manufacturer. Analysis of the fluorescence intensity of the stained cells was performed using a CANTO II (BD Biosciences). The ALDH activity of the sample was determined based on the fluorescence intensity beyond the threshold defined by the reaction with diethylaminobenzaldehyde.
Spheroid-formation assay
Cells were plated in Corning ultra-low-attachment 24-well plates (Corning, NY, USA) at a density of 100 viable cells ml−1 and grown in serum-free Neurobasal medium supplemented with 20 ng ml−1 bFGF, 10 ng ml−1 EGF, 2.5 μg ml−1 amphotericin B, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and B-27 supplement. The cells were allowed to continue to grow for 7 days, and the numbers of spheroids were counted under a microscope.
Computational analysis of HMGA1-binding elements
The HMGA1-binding element was searched for using the MatInspector application, a part of GenomatixSuite software (Genomatix Software GmbH, Munich, Germany).25 The matrices of the HMGA family of architectural transcription factors (Genomatix Matrix Library 8.2) were used with a core similarity threshold of 0.75 and a matrix similarity threshold of Optimal −0.02. Sequences bearing a match to any of the four matrices were termed HMGA1 sequences. The remaining sequences were classified as HMGA1 and scanned for other TF binding site motifs contained in the Genomatix Matrix Library using the standard parameters, as described previously.
Deletion of HMGA1-binding element in ABCG2-promoter reporter constructs
The human ABCG2 promoter construct was kindly provided by Dr Douglas D. Ross.26 The human ABCG2 promoter contains three HMGA1-binding elements: 5′-AACTGACAGAAATTAATTCAAAAGA-3′, 5′-TTGGGTGAACCATTAATAGTTATTC-3′ and 5′-TTCTCGTAGTTAATCACTCTGGTTC-3′. Mutant ABCG2 promoter constructs carrying deletions of HMGA1-binding elements were generated using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer's instructions; the intended mutations were confirmed by sequencing.
Luciferase assay
A2780 cells were plated in 12-well culture plates at 3 × 105 cells per well. Transfection experiments were performed 16 h after cell seeding using Lipofectamine/Lipofectamine Plus (Life Technologies) according to the manufacturer's instructions. Briefly, the transfection mixtures contained 400 ng of reporter construct (ABCG2 reporter constructs, pGL3-basic containing ABCG2 promoter (−1285), deletion of HMGA1-binding element ABCG2 promoter) or 400 ng of the puro3-HMGA1 (Addgene, Cambridge, MA, USA) expression plasmid and 10 ng of an internal control plasmid (pCMV-RL vector containing Renilla luciferase, Promega). The ABCG2 reporter construct was produced as described previously.26 Twenty-four hours later, the cells were harvested, lysed and centrifuged at 2000 g for 3 min at 4 °C, and the luciferase activity was determined according to the manufacturer's instructions (Luciferase Assay System, Promega). All experimental values were averaged from triplicate determinations for each experimental condition, and the experiments were performed in triplicate. Subsequently, the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) using VICTOR3 (Perkin Elmer, Waltham, MA, USA).
Drug resistance assay in a xenograft tumor model
All animal studies adhered to protocols approved by the Pusan National University Institutional Animal Care and Use Committee. HMGA1-overexpresing A2780 cells and parental cells (1 × 105 cells) were resuspended in 50 μl Matrigel solution (1:1 dilution with RPMI) and injected subcutaneously into the right and left flanks of 6- to 8-week-old female BALB/c-nu/nu mice. Mice transplanted with tumor cells were then inspected biweekly for tumor appearance on the basis of visual observation and palpation. Measurement of the length (mm), width (mm) and height (mm) of the tumor masses was performed twice weekly using electronic Vernier calipers, and the tumor volumes (mm3) were calculated as (length × width × height)/2. To confirm drug resistance in vivo, 14 days after A2780 cell injection, paclitaxel (5 mg kg−1) was intraperitoneally injected into nude mice every 3 and 4 days. The tumor sizes were measured every 3–4 days. All of the mice were killed by anesthetic overdose on day 45. Tumorigenicity was measured every 3–4 days beginning at 14 days after A2780 cell injection. All mice were killed by anesthetic overdose on day 42.
Immunohistochemistry analysis
For immunostaining, tumors were removed, formalin-fixed and paraffin-embedded. Sections measuring 6 μm in thickness were taken from the paraffin-embedded specimens at 150 μm intervals and stained with the indicated antibodies; this was followed by washing and mounting in Vectashield medium (Vector Laboratories, Burlingame, CA, USA) with 4',6-diamidino-2-phenylindole for the visualization of nuclei. The stained sections were visualized by laser scanning confocal microscopy (Olympus FluoView FV1000, Center Valley, PA, USA).
Statistical analysis
The data are presented as the mean±s.d. Statistical significance (P<0.05) was determined using two-tailed unpaired Student's t-tests. n⩾3 unless stated otherwise.
Results
Spheroid cells isolated from A2780 ovarian cancer cells exhibit stem cell-like properties
For the generation of spheroids, which exhibit enriched characteristics of CSCs, A2780 ovarian cancer cells were seeded on ultra-low-attachment culture plates in Neurobasal media with EGF, bFGF and B-27 supplement. In these culture conditions, in which cell death was observed due to serum starvation, aggregates of cells were formed and continued to grow in suspension (Figure 1a). Two weeks after plating, certain aggregates compacted into spheroids, which could not be dissociated by pipetting. The spheroids were isolated by brief centrifugation at 800 r.p.m. for 2 min; the spheroids sedimented, whereas single cells stayed in suspension due to weight differences. Floating spheroids and clusters were dissociated by Accutase treatment and re-plated on day 14, which generated secondary spheroids with the appearance of prototypical spheroids (Figure 1a). Using this approach, sustainable spheroids were obtained from A2780 ovarian cancer cells.
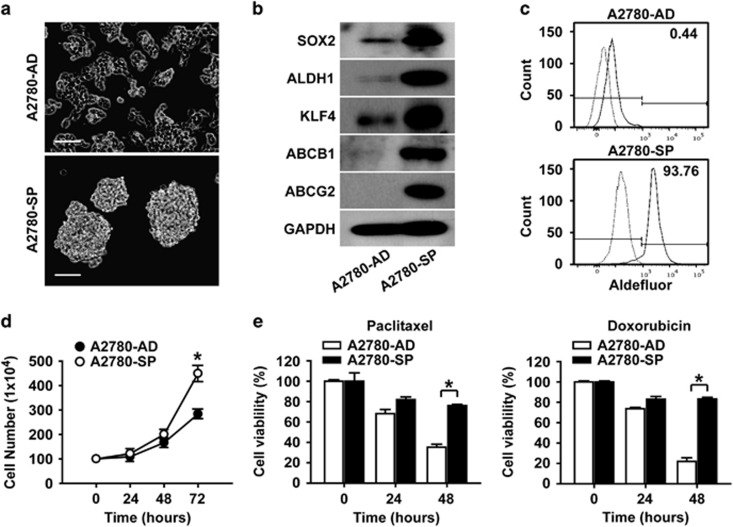
Figure 1.
Spheroid cells from A2780 ovarian cancer cells have stem cell-like characteristics. (a) Bright-field images of spheroids (SP) generated from adherent A2780 ovarian cancer cells (AD) cultured in spheroid-forming conditions (Neurobasal media) are shown. Images of spheroids on day 10 after plating into spheroid culture condition are shown. Scale bar, 100 μm. (b) Western blotting analysis of adherent A2780 cells and spheroids with the indicated markers is shown. (c) Flow cytometric analysis of adherent A2780 cells and spheroid cells after incubation with ALDH substrate is shown. (d) Time course analysis of cell counts after plating 100 cells from A2780 adherent cells or spheroids per well is shown. *P<0.05. (e) Time course results of an MTT assay with A2780 adherent cells and spheroid cells in the absence or presence of paclitaxel (left panel, 0.1 μM) or doxorubicin (right panel, 1 μM) are shown. *P<0.05.
To determine whether the spheroids generated from A2780 ovarian cancer cells exhibit CSC-like characteristics, we compared the expression levels of stemness-related genes in the spheroids and in adherent A2780 cells. Ovarian CSCs are known to express stem cell-related genes such as SOX2, KLF4 and ABCG2.27 Therefore, the expression levels of stemness-related genes were analyzed by western blotting. The results showed that the expression levels of SOX2, ALDH1, KFL4, ABCB1 and ABCG2 were higher in spheroids compared with adherent A2780 ovarian cancer cells (Figure 1b). The elevated expression of HMGA1 in spheroids along with SOX2, ALDH1, KLF4 and ABCG2 was observed in SKOV3 and PA1 ovarian cancer cells (Supplementary Figure S1a and b). In addition to the stemness-related genes, the expression of ALDH1, which has been reported to be a CSC marker,28 was increased according to western blotting analysis. Therefore, the percentages of ALDH-expressing cells in adherent cells and spheroids of A2780 cells were examined by flow cytometry analysis. As shown in Figure 1c, the percentages of ALDH+ cells in adherent cells and spheroids of A2780 cells were 0.44 and 93.76%, respectively, correlating with the results from RT-PCR and western blotting. When comparing other CSC characteristics, including proliferative advantage and drug resistance, the spheroid cells showed higher proliferation and higher resistance to paclitaxel or doxorubicin compared with the adherent cells (Figure 1d and e). These results suggest that the spheroid culture of A2780 ovarian cancer cells enriched for cells with CSC characteristics.
HMGA1 is highly expressed in the ovarian CSC population
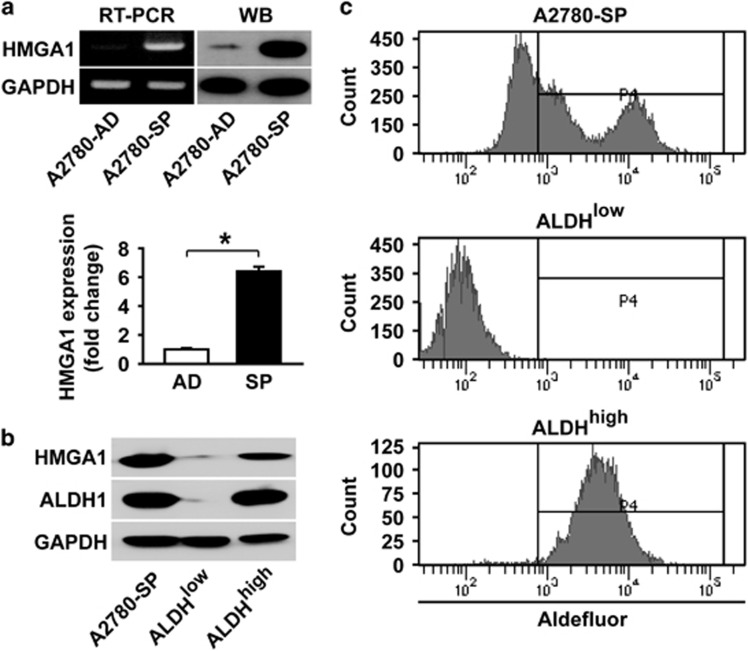
Recent reports have suggested that HMGA1 has an important role in the self-renewal, proliferation and differentiation of embryonic stem cells and CSCs.19, 29 A recent paper has reported that HMGA1 is essential for the reprogramming of somatic cells to induce pluripotent stem cells by introducing reprogramming factors such as OCT4, SOX2, KLF4 and c-MYC.14 When examining HMGA1 expression in adherent A2780 cells and spheroids generated from adherent A2780 cells by RT-PCR and western blotting, high levels of HMGA1 were observed in spheroids compared with adherent A2780 cells (Figure 2a). In further analysis of HMGA1 expression in ALDH+ cells and ALDH− cells isolated from A2780 spheroid cells, significantly higher expression of HMGA1 was observed in ALDH+ cells compared with ALDH− cells (Figure 2b). These results suggest that HMGA1 may have an important role in ovarian CSCs.
Figure 2.
HMGA1 is highly expressed in spheroids from A2780 ovarian cancer cells. (a) RT-PCR analysis and western blotting analysis of A2780 adherent cells and spheroid cells are shown in the upper panels with HMGA1 probes along with GAPDH probes. Quantification of HMGA expression by western blotting after normalization using GAPDH expression is shown in the lower panel. *P<0.05. (b, c) Flow cytometric analysis and sorting of A2780 spheroid cells after incubation with an ALDH substrate is shown. Western blotting analysis of whole spheroid cells, ALDH− cells and ALDH+ cells after sorting is shown with the indicated antibodies.
HMGA1 regulates the expression of stemness-related genes in ovarian cancer cells
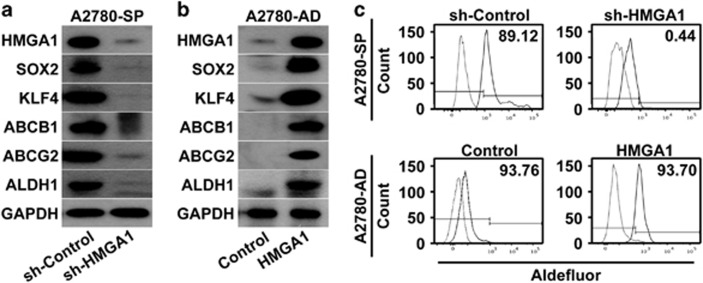
To determine the role of HMGA1 in ovarian CSCs, HMGA1 expression was knocked down in spheroid cells by using an shRNA construct, which successfully reduced the expression of HMGA1 at the protein level (Figure 3a). Western blotting analysis of the expression of stemness-related genes after HMGA1 knockdown, compared with knockdown control, revealed that the expression of SOX2, KLF4, ABCB1, ABCG2 and ALDH1 decreased significantly (Figure 3a). Knockdown of HMGA1 expression in spheroids generated from SKOV3 or PA1 ovarian cancer cells also resulted in decreased expression of SOX2, ALDH1, KLF4 and ABCG2 (Supplementary Figure S1c). To examine whether HMAG1 could induce the expression of stemness-related genes, HMGA1-overexpressing adherent A2780 cells were generated (Figure 3b). Analysis of the expression of stemness-related genes by western blotting revealed that the overexpression of HMGA1 resulted in increased expression of SOX2, KLF4, ABCB1, ABCG2 and ALDH1 in adherent A2780 cells compared with control cells (Figure 3b). Analysis of the expression of ALDH by flow cytometry demonstrated that knockdown of HMGA1 in A2780 spheroid cells resulted in a significant decrease in ALDH+ cells, from 89.12% to 6.01%, whereas the overexpression of HMGA1 in A2780 adherent cells increased the number of ALDH+ cells from 0.44 to 93.7% (Figure 3c). These results strongly suggest that HMGA1 has an important role in regulating the expression of stemness-related genes in ovarian cancer cells.
Figure 3.
Altering HMGA1 expression changes the expression of stemness-related gene expression. (a) Western blotting analysis of A2780 spheroid cells with (sh-HMGA1) or without (sh-Control) knockdown of HMGA1 is shown with the indicated probes. (b) Western blotting analysis of HMGA1-overexpressing A2780 cells (HMGA1) or control A2780 cells (Control) is shown with the indicated probes. (c) The results of flow cytometric analysis of A2780 spheroid cells with HMGA1 knockdown in comparison with control A2780 spheroid cells (upper panel) or HMGA1-overexpressing A2780 adherent cells in comparison with control A2780 adherent cells (lower panel) after incubation with ALDH substrate are shown.
HMGA1 regulates CSC-like characteristics in ovarian cancer cells
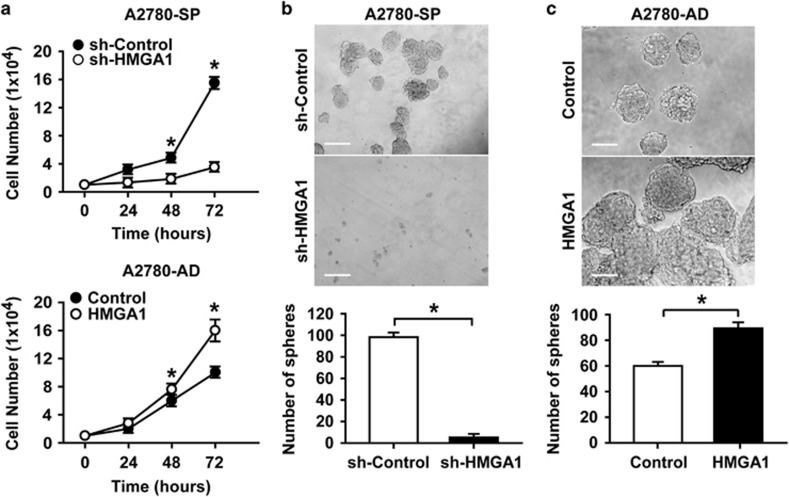
To investigate whether HMGA1 regulates CSC properties in ovarian cancer cells, HMGA1 was knocked down in A2780 spheroid cells, and was overexpressed in A2780 adherent cells. As shown in Figure 4a, the knockdown of HMGA1 in A2780 spheroid cells significantly decreased the proliferative advantage of the spheroid cells (Figure 4a, upper panel). HMGA1-overexpressing A2780 adherent cells showed a significant increase in proliferation in comparison with control adherent cells, with a level of proliferation comparable to that of spheroid cells (Figure 4a, lower panel). Spheroid formation is a characteristic property of ovarian CSCs; therefore, we examined the effect of HMGA1 expression on the formation of spheroids from A2780 cells. When HMGA1 expression was knocked down in A2780 spheroid cells, the sphere-forming ability decreased significantly (Figure 4b). In contrast, when HMGA1 was overexpressed in A2780 adherent cells, the number and size of the spheroids increased significantly (Figure 4c). These results suggest that HMGA1 expression in ovarian cancer cells regulates CSC-like properties.
Figure 4.
HMGA1 expression regulates the CSC characteristics of A2780 ovarian cancer cells. (a) Time course of the cell numbers counted after plating the same number of cells (1 × 104 cells per well). A2780 spheroid cells with HMGA1 knockdown are compared with control A2780 spheroid cells (upper panel), and HMGA1-overexpressing A2780 adherent cells are compared with control A2780 adherent cells (lower panel). *P<0.05 (b) Bright-field images of spheroids generated from A2780 spheroid cells with or without HMGA1 knockdown (upper panels) and counts of the spheroid numbers (lower panel) are shown. Scale bar, 100 μm. *P<0.05 (c) Bright-field images of spheroids generated from HMGA1-overexpressing A2780 adherent cells in comparison with control A2780 adherent cells (upper panels) and counts of the spheroid numbers (lower panel) are shown. Scale bar, 100 μm. *P<0.05.
HMGA1 regulates the drug resistance of ovarian cancer cells
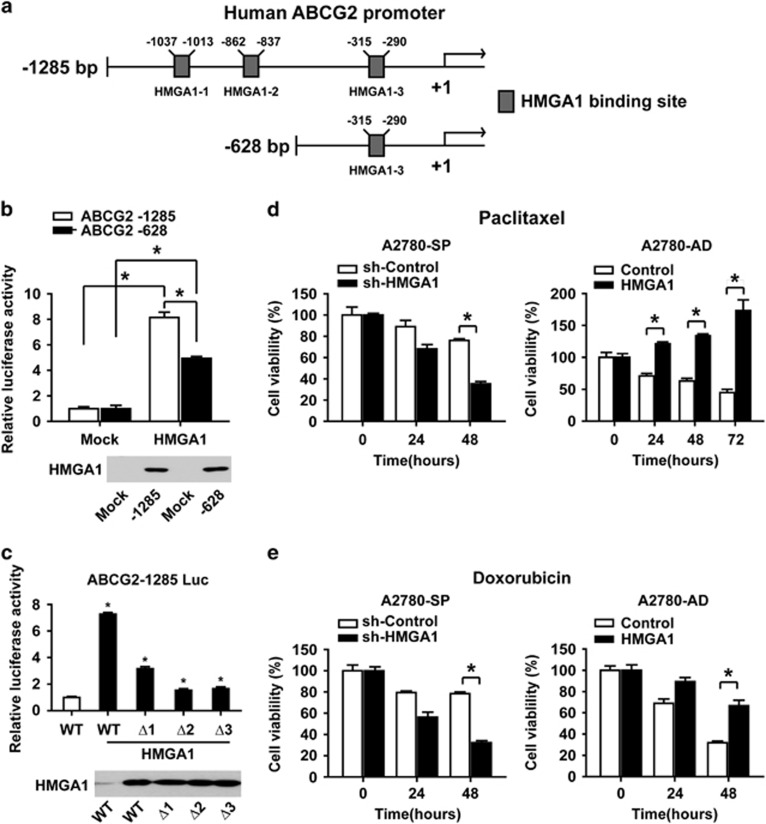
We noted that the expression of ABCG2 was affected by the expression level of HMGA1 (Figure 3a and b). To examine whether the expression of ABCG2 is regulated by HMGA1, we searched for HMGA1-binding sites in the ABCG2 promoter and identified three binding sites for HMGA1 in the 5′ region upstream of the transcription start site in the ABCG2 promoter (Figure 5a).30 To validate the functionality of the putative HMGA1-binding sites, luciferase assays were performed in A2780 cells with two different luciferase constructs containing ABCG2 promoter regions: one with three HMGA1-binding sites (−1285 bp) and another with one HMGA1-binding site (−628 bp). Co-transfection of ABCG2 promoter reporter constructs with the HMGA1-overexpression construct, comparison with the transfection of ABCG2 promoter reporter constructs alone, resulted in significantly increased luciferase activity in (Figure 5b). The ABCG2 promoter −628 bp construct, which lacks two HMGA1-binding sites as compared with the −1285 construct, showed lower activity in comparison with that of the −1285 construct when co-transfected with the HMGA1-overexpression construct. To evaluate the contribution of each binding site to the regulation of ABCG2 expression, deletion constructs were generated for each binding site (ΔHMGA1-1, ΔHMGA1-2, ΔHMGA1-3) in the −1285 construct. The results of the luciferase activity assay after co-transfection of the HMGA1-overexpression construct indicated that the deletion of each binding site resulted in significantly decreased HMGA1-induced luciferase activity (Figure 5c), thus suggesting that HMGA1 binds to more than one site in the 5′ region upstream of the ABCG2 promoter and subsequently regulates the transcriptional activity of ABCG2.
Figure 5.
HMGA1 regulates ABCG2 promoter activity and drug resistance. (a) Schematic representations of the −1285 and −628 5′ upstream regions of the ABCG2 promoter are shown. HMGA1-binding sites are marked by boxes. (b) The results of a transcriptional activity assay after co-transfecting an ABCG2 promoter construct (−1285 or −628) with an HMGA1-overexpression construct into A2780 cells are shown (upper panel). *P<0.05. The results of western blotting analysis are shown in the lower panel. (c) The results of a transcriptional activity assay after co-transfecting an ABCG2 promoter construct (−1285, ΔHMGA1-1, ΔHMGA1-2 or ΔHMGA1-3) with an HMGA1-overexpression construct into A2780 cells are shown (upper panel). *P<0.05. The results of western blotting analysis are shown in the lower panel. (d) The results from a time course MTT assay with A2780 spheroid cells with HMGA1 knockdown in comparison with control A2780 spheroid cells (left panel), or with HMGA1-overexpressing A280 cells in comparison with control A2780 adherent cells (right panel) in the presence of paclitaxel (0.1 μM), are shown. *P<0.05. (e) The results from a time course MTT assay with A2780 spheroid cells with HMGA1 knockdown in comparison with control A2780 spheroid cells (left panel), or with HMGA1-overexpressing A280 adherent cells in comparison with control A2780 adherent cells (right panel) in the presence of doxorubicin (1 μM), are shown. *P<0.05.
Next, we tested whether HMGA1 expression regulates drug resistance in ovarian cancer cells. HMGA1 expression was knocked down in A2780 spheroid cells or overexpressed in A2780 adherent cells, after incubation with chemotherapeutic agents such as paclitaxel or doxorubicin. Incubation of HMGA1-knockdown A2780 spheroid cells with paclitaxel or doxorubicin resulted in significantly decreased viability compared with that of control spheroid cells (Figure 5d and e, left panels). Incubation of HMGA1-overexpressing A2780 adherent cells with paclitaxel or doxorubicin resulted in a meaningful increase in viability in HMGA1-overexpressing cells compared with control adherent cells (Figure 5d and e, right panels). Together, these results strongly suggest the involvement of HMGA1 in the regulation of drug resistance in ovarian cancer cells.
HMGA1 enhances the drug resistance of ovarian cancer cells in vivo
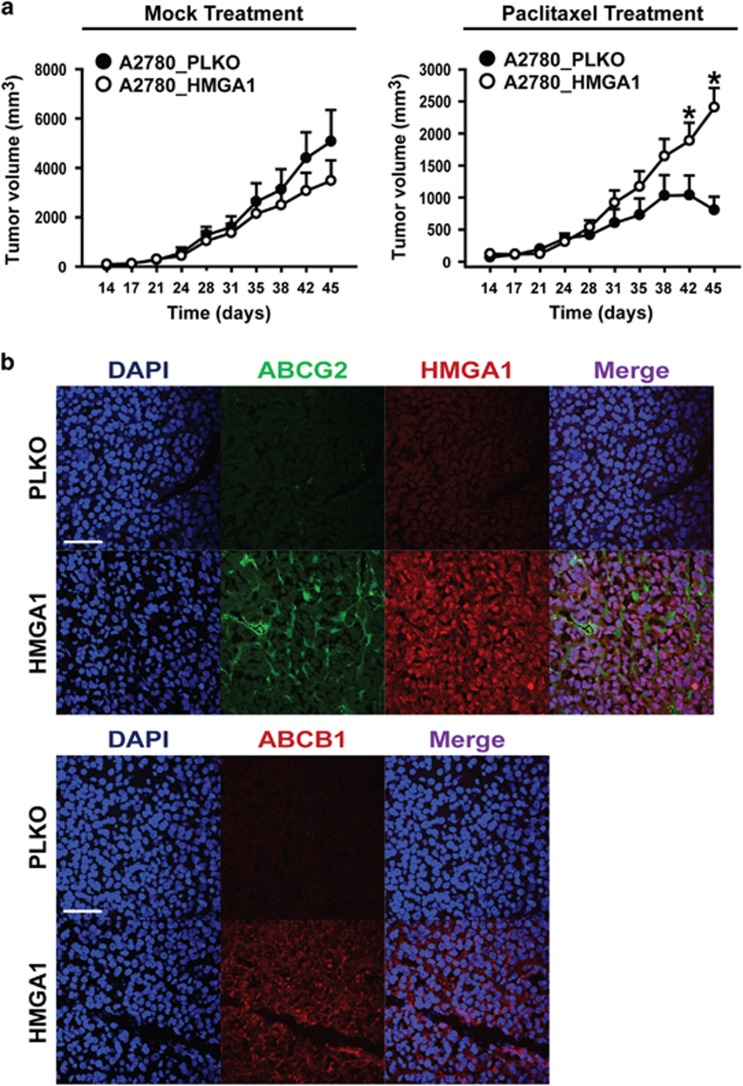
To evaluate the effect of HMGA1 expression in promoting drug resistance in vivo, we performed xenograft tumorigenicity assays with A2780 ovarian cancer cells. When HMGA1-overexpressing A2780 cells and control A2780 cells were subcutaneously injected into nude mice and monitored for tumorigenic activity, HMGA1-overexpressing A2780 cells and control A2780 cells did not show a significant difference in tumor size after HBSS treatment. However, when the mice were treated with paclitaxel, the tumor size after control A2780 cell injection decreased after day 42, whereas the tumor size in HMGA1-overexpressing A2780 cells continued to increase (Figure 6a). Immunohistochemistry of the tumors showed increased expression of HMGA1, ABCG2 and ABCB1 in tumors from HMGA1-overexpressing A2780 cells in comparison with tumors from control A2780 cells (Figure 6b). These results strongly suggest that HMGA1 is an important factor regulating the drug resistance of ovarian cancer cells in vivo.
Figure 6.
HMGA1 overexpression enhances the drug resistance of ovarian cancer cells in a xenograft tumor model. (a) Tumor size was measured on the indicated day after the subcutaneous injection of HMGA1-overexpressing A2780 or control A2780 cells into nude mice (1 × 105 per injection) with HBSS or paclitaxel treatment (n=6). Paclitaxel was injected at each data point. *P<0.05. (b) Immunohistochemistry results of tumors harvested on day 45. DAPI and the indicated antibodies are shown. Scale bar, 50 μm.
Discussion
Ovarian cancer has recently been evaluated as a CSC-related disease; thus, understanding the cellular and molecular properties of the CSC population in ovarian cancer is important to understanding and curing this deadly disease. In our study, the CSC population was enriched in ovarian cancer cells through spheroid formation, which is a defining characteristic of epithelial stem cells.29, 31, 32 CSC-enriched spheroids, compared with adherent cells, showed higher expression of stemness-related genes, including SOX2, KLF4 and ADLH1. We found that HMGA1 expression was also significantly higher in spheroids than in adherent cells. Knockdown of HMGA1 in spheroid cells resulted in the decreased expression of stemness-related genes, whereas the overexpression of HMGA1 in adherent cells led to the increased expression of stemness-related genes. HMGA1 is known to contribute to neoplastic transformation, cell proliferation and anchorage-independent growth in numerous human cancers.33, 34 Silencing of HMGA1 expression in ovarian cancer OVCAR cells results in inhibited cellular growth.35 Increasing evidence suggests that HMGA1 is a master regulator of not only normal stem cells but also CSCs.36 In colon cancer cells, HMGA1 silencing results in impaired colon sphere formation and reduced self-renewal efficiency in studies of the stem-like state.20, 24 Moreover, HMGA1 drives metastatic progression in triple-negative breast cancer cells by reprogramming the cancer cells to a stem-like state, and HMGA1 knockdown depletes the CSC population.23 It has been reported that HMGA1 promotes the dedifferentiation and reprogramming of somatic cells to a pluripotent stem cell state by inducing stem cell transcriptional networks.14 Therefore, these results suggest that HMGA1 has an important role in maintaining and inducing CSC-related gene expression and that HMGA1 is a promising target for therapies directed at eradicating CSCs.
We observed that HMGA1 was highly expressed in the ALDH+ population in comparison with the ALDH− population, and HMGA1 knockdown decreased the ALDH+ population, whereas HMGA1 overexpression increased the ALDH+ population in ovarian cancer cells. ALDH catalyzes the dehydrogenation of aldehydes, which can cause inactivation of enzymes, DNA damage and cell death.37 ALDH has drawn attention as a potential prognostic marker of various types of cancer.38 ALDH has an important role in the normal function of stem cells, and recent reports have shown the utility of ALDH in the isolation and identification of CSCs.39 High levels of migratory behavior, clonogenic and tumorigenic activity, and metastatic potential have been reported in high-ALDH populations in prostate cancer, non-small-cell lung cancer, bladder cancer and cervical cancer.40, 41, 42, 43 Recent reports have identified CSCs in ovarian cancer in the high-ALDH population, which overlaps with the side population and maintains platinum resistance.28, 44, 45 These results suggest that HMGA1 expression, which is intimately related to ALDH activity in ovarian CSC-enriched cells, may be a prognostic marker for ovarian cancer patients.
In the present study, we demonstrated that HMGA1 regulates the expression of multidrug resistance proteins, such as ABCB1 and ABCG2. Silencing of HMGA1 expression in ovarian CSCs abrogated resistance to chemotherapeutic reagents, including paclitaxel and doxorubicin, in vitro. Moreover, silencing of HMGA1 expression led to the increased sensitivity of A2780 cells to paclitaxel treatment in an in vivo xenograft tumor model. Consistently with these results, the association of HMGA1 overexpression with resistance to anti-neoplastic drugs in various cancers has been suggested.46 In pancreatic adenocarcinoma, lentivirus-mediated RNA interference of HMGA1 enhances chemosensitivity to gemcitabine, and HMGA1 has been suggested to be a molecular determinant of chemoresistance.47, 48 In colon cancer cells and thyroid cancer cells, silencing HMGA1 expression results in increased sensitivity to anti-neoplastic drugs such as Cetuximab, 5-Fluorouracil or doxorubicin.49 Together with the results from this study, which indicate that HMGA1 upregulates ABCG2 promoter activity in an HMGA1-binding site-dependent manner, these results suggest that HMGA1 is a key regulator of drug resistance in ovarian cancer cells.
HMGA1 forms an enhanceosome with recruited transcription factors and repositions nucleosomes for the expression of different sets of genes.50 In embryonic stem cells, HMGA1 maintains a poorly differentiated, pluripotent state by regulating epigenetic remodeling and transcriptional networks.14 The forced expression of HMGA1 prevents the differentiation of embryonic stem cells by maintaining high expression levels of stem cell genes involved in pluripotency and self-renewal, such as Oct4 and c-Myc. In addition, HMGA1 promotes the reprogramming of somatic cells into induced pluripotent stem cells via reprogramming factors. During the reprogramming process, HMGA1 induces the expression of LIN28, c-MYC and SOX2.14 In the present study, we showed that the silencing of HMGA1 led to the decreased expression of SOX2 and KLF4 in A2780 spheroid cells. These results suggest an essential role of HMGA1 in the transcriptional regulation of stemness-associated genes in CSCs.
Together, our results demonstrate that HMGA1 is a critical regulator for maintaining CSC-like characteristics in ovarian cancer. Therefore, HMGA1 may be a novel therapeutic target for highly metastatic and drug resistant ovarian cancer.
Acknowledgments
This research was supported in part by programs of the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2015R1A5A2009656; NRF-2015R1B1A1A01060977) and the Cancer Control Ministry for Health Welfare and Family Affairs of Korea (0920050). Confocal microscopy data were acquired in the Advanced Neural Imaging Center in KBRI, located in Daegu, South Korea.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–737. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003; 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T et al. Identification of human brain tumour initiating cells. Nature 2004; 432: 396–401. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005; 65: 10946–10951. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007; 1: 313–323. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007; 445: 106–110. [DOI] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res 2008; 68: 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleson KM, Boente MP, Pambuccian SE, Skubitz AP. Disaggregation and invasion of ovarian carcinoma ascites spheroids. J Transl Med 2006; 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietarska M, Maugard CM, Filali-Mouhim A, Alam-Fahmy M, Tonin PN, Provencher DM et al. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol Carcinog 2007; 46: 872–885. [DOI] [PubMed] [Google Scholar]
- L'Esperance S, Bachvarova M, Tetu B, Mes-Masson AM, Bachvarov D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics 2008; 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shield K, Riley C, Quinn MA, Rice GE, Ackland ML, Ahmed N. Alpha2beta1 integrin affects metastatic potential of ovarian carcinoma spheroids by supporting disaggregation and proteolysis. J Carcinog 2007; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca E, Cocola C, Sabour D, Pelucchi P, Bertalot G, Palumbo O et al. Overlapping genes may control reprogramming of mouse somatic cells into induced pluripotent stem cells (iPSCs) and breast cancer stem cells. In Silico Biol 2010; 10: 207–221. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS ONE 2012; 7: e48533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Nuclear functions of the HMG proteins. Biochim Biophys Acta 2010; 1799: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarra R, Zammitti S, Lo Sardo A, Maurizio E, Arnoldo L, Pegoraro S et al. HMGA molecular network: From transcriptional regulation to chromatin remodeling. Biochim Biophys Acta 2010; 1799: 37–47. [DOI] [PubMed] [Google Scholar]
- Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A et al. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett 2004; 574: 1–8. [DOI] [PubMed] [Google Scholar]
- Shah SN, Resar LM. High mobility group A1 and cancer: potential biomarker and therapeutic target. Histol Histopathol 2012; 27: 567–579. [DOI] [PubMed] [Google Scholar]
- Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, Croce CM et al. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J 2003; 17: 1496–1498. [DOI] [PubMed] [Google Scholar]
- Belton A, Gabrovsky A, Bae YK, Reeves R, Iacobuzio-Donahue C, Huso DL et al. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS ONE 2012; 7: e30034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldenfrei A, Belton A, Kowalski J, Talbot CC Jr, Di Cello F, Poh W et al. HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genomics 2011; 12: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro S, Ros G, Piazza S, Sommaggio R, Ciani Y, Rosato A et al. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget 2013; 4: 1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SN, Cope L, Poh W, Belton A, Roy S, Talbot CC Jr et al. HMGA1: a master regulator of tumor progression in triple-negative breast cancer cells. PLoS ONE 2013; 8: e63419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca F, Colamaio M, Federico A, Gemei M, Tosti N, Bastos AU et al. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget 2014; 5: 3234–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005; 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- Bailey-Dell KJ, Hassel B, Doyle LA, Ross DD. Promoter characterization and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta 2001; 1520: 234–241. [DOI] [PubMed] [Google Scholar]
- Zhou N, Wu X, Yang B, Yang X, Zhang D, Qing G. Stem cell characteristics of dormant cells and cisplatininduced effects on the stemness of epithelial ovarian cancer cells. Mol Med Rep 2014; 10: 2495–2504. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Suzuki N, Makino H, Furui T, Morii E, Aoki H et al. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecol Oncol 2015; 137: 299–305. [DOI] [PubMed] [Google Scholar]
- Huso TH, Resar LM. The high mobility group A1 molecular switch: turning on cancer - can we turn it off? Expert Opin Ther Targets 2014; 18: 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M et al. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 2003; 278: 14146–14152. [DOI] [PubMed] [Google Scholar]
- Seo EJ, Kwon YW, Jang IH, Kim DK, Lee SI, Choi EJ et al. Autotaxin regulates maintenance of ovarian cancer stem cells through lysophosphatidic acid-mediated autocrine mechanism. Stem Cells 2016; 34: 551–564. [DOI] [PubMed] [Google Scholar]
- Choi EJ, Seo EJ, Kim DK, Lee SI, Kwon YW, Jang IH et al. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget 2016; 7: 3506–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LJ, Maher JF, Bunton TE, Resar LM. The oncogenic properties of the HMG-I gene family. Cancer Res 2000; 60: 4256–4261. [PubMed] [Google Scholar]
- Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, Bunton TE et al. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol 2000; 20: 5490–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Zhang Y, Fu J, Zhang G. Knockdown of HMGA1 expression by short/small hairpin RNA inhibits growth of ovarian carcinoma cells. Biotechnol Appl Biochem 2012; 59: 1–5. [DOI] [PubMed] [Google Scholar]
- Yanagisawa BL, Resar LM. Hitting the bull's eye: targeting HMGA1 in cancer stem cells. Expert Rev Anticancer Ther 2014; 14: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 2008; 4: 697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pors K, Moreb JS. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discov Today 2014; 19: 1953–1963. [DOI] [PubMed] [Google Scholar]
- Xu X, Chai S, Wang P, Zhang C, Yang Y, Yang Y et al. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett 2015; 369: 50–57. [DOI] [PubMed] [Google Scholar]
- van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res 2010; 70: 5163–5173. [DOI] [PubMed] [Google Scholar]
- Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res 2014; 20: 4154–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Zheng PS. High aldehyde dehydrogenase activity identifies cancer stem cells in human cervical cancer. Oncotarget 2013; 4: 2462–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev 2010; 19: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S et al. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS ONE 2014; 9: e107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Torigoe T, Morita R, Kuroda T, Takahashi A, Matsuzaki J et al. Ovarian cancer stem cells are enriched in side population and aldehyde dehydrogenase bright overlapping population. PLoS ONE 2013; 8: e68187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Sepe R, Puca F, Fusco A. High mobility group a proteins as tumor markers. Front Med 2015; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau SS, Ashley SW, Whang EE. Lentivirus-mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J Gastrointest Surg 2006; 10: 1254–1262. [DOI] [PubMed] [Google Scholar]
- Liau SS, Whang E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin Cancer Res 2008; 14: 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo D, Mussnich P, Rosa R, Bianco R, Tortora G, Fusco A. High mobility group A1 protein expression reduces the sensitivity of colon and thyroid cancer cells to antineoplastic drugs. BMC Cancer 2014; 14: 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Wolffe AP. Substrate structure influences binding of the non-histone protein HMG-I(Y) to free nucleosomal DNA. Biochemistry 1996; 35: 5063–5074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.