Version Changes
Revised. Amendments from Version 1
The major changes to the manuscript involve rewriting of the introduction, to highlight the differences between the overall objective of our research program and the objectives of this study. We are interested in studying the genetics of parallel evolution, but this study focuses on revealing differences in expression and genes separating sympatric benthic and limnetic morphs. We added clarifications on several aspects of the work and analyses, for instance providing workflow and sample overview in new Figure 2, adding morphs and descriptions to Figure 1, explaining the sampling and SNP filtering. The reviewers pointed out a mistake in our interpretation of the fate of paralogous genes in salmonids, based on the Rainbow trout genome, which we met by fixing the manuscript and changing the interpretations. The reviewers questioned the choice of transcripts studied with qPCR. We rewrote this section, and added a table that summarizes the qPCR data, and highlights the fact that the data can be used to find candidate genes or paralog groups with expression differences between Arctic charr morphs. The reviewers also highlighted the low efficiency in the qPCR reactions on the natterin-like paralogs. We tried to accommodate those weaknesses by redoing figures, with the appropriate correction factors. Multiple other aspects of the text where rewritten in accordance with the suggestions of the reviewers, which have in our opinion greatly improved the manuscript
Abstract
Species and populations with parallel evolution of specific traits can help illuminate how predictable adaptations and divergence are at the molecular and developmental level. Following the last glacial period, dwarfism and specialized bottom feeding morphology evolved rapidly in several landlocked Arctic charr Salvelinus alpinus populations in Iceland.
To study the genetic divergence between small benthic morphs and limnetic morphs, we conducted RNA-sequencing charr embryos at four stages in early development. We studied two stocks with contrasting morphologies: the small benthic (SB) charr from Lake Thingvallavatn and Holar aquaculture (AC) charr.
The data reveal significant differences in expression of several biological pathways during charr development. There was also an expression difference between SB- and AC-charr in genes involved in energy metabolism and blood coagulation genes. We confirmed differing expression of five genes in whole embryos with qPCR, including lysozyme and natterin-like which was previously identified as a fish-toxin of a lectin family that may be a putative immunopeptide. We also verified differential expression of 7 genes in the developing head that associated consistently with benthic v.s.limnetic morphology (studied in 4 morphs). Comparison of single nucleotide polymorphism (SNP) frequencies reveals extensive genetic differentiation between the SB and AC-charr (~1300 with more than 50% frequency difference). Curiously, three derived alleles in the otherwise conserved 12s and 16s mitochondrial ribosomal RNA genes are found in benthic charr.
The data implicate multiple genes and molecular pathways in divergence of small benthic charr and/or the response of aquaculture charr to domestication. Functional, genetic and population genetic studies on more freshwater and anadromous populations are needed to confirm the specific loci and mutations relating to specific ecological traits in Arctic charr.
Keywords: Salmonids, Aquaculture, ecomorphs, Polymorphism, parallel evolution, immunology, craniofacial divergence, mtDNA
Introduction
Historical contingencies and chance shape organisms during evolution 1, 2, but convergence in phenotype and molecular systems indicates that evolution is to some extent predictable 3, 4. Identification of genes and variants that influence evolved differences is not a trivial task 5. Ideal systems to study the role of chance and necessity in ecological evolution would be related species or populations with readily observable phenotypic variation, living in a tractable ecological setting, and showing parallel evolution of specific traits within/among species/populations. Examples of such species complexes are provided finches of the Galapagos islands 6, while cichlids of the African great lakes also provide an exciting mulit-species system in the same respect 7. The threespine stickleback has also emerged as a model “single species” system 8. The amount of diversity in the feeding specializations of fish provide great opportunities for studying adaptation and divergence at the developmental and genetic level.
One approach to identify pathways related to function or morphological differences between species, populations or ecomorphs is to study gene expression during development 9, 10. For example a microarray study of liver samples from anadromous and resident populations of brown trout ( Salmo trutta), revealed that gene expression in juveniles was more influenced by life history than relatedness 11. Furthermore, Filteau et al. (2013) 12 found a set of coexpressed genes differenting two whitefish morphotypes, implicating Bone morphogenesis protein (BMP) signaling in the development of ecological differences in trophic morphology. Thus we were quite keen to apply RNA-sequencing to analyze ecomorphs in our study system, Arctic charr. Two previous studies have used RNA-seq to study salinity tolerance in adult Arctic charr, and found links between gene expression and quantitative trait loci 13, 14.
Some northern freshwater fish species exhibit frequent parallelism in trophic structures and life history and in several cases are found as distinct resource morphs 8, 15– 19. One of these species, Arctic charr ( Salvelinus alpinus), is well suited for studying the developmental underpinnings of trophic divergence and parallel evolution. Local adaptation has been extensively studied in the salmonid family, to which Arctic charr belongs 20. The family is estimated to be between 88–103 million years old 21, 22. A whole genome duplication event occurred before the radiation of the salmonid family 21– 24 which has provided time for divergence of ohnologous genes (paralogous genes originated by whole genome duplication event). Furthermore, recent estimates from the rainbow trout ( Oncorhynchus mykiss) genome suggest that ohnologous genes are lost at a rate of about 170 genes per million years, and that on the order of 4500 were retained in rainbow trout 22. De novo assembly of genomes and transcriptomes is complicated if many paralogs are present, which is the case in Arctic charr - see 13, 14. In this study we opted for mapping the reads (36 bp) to a related reference genome/transcriptome 25, instead of de novo assembly.
Molecular studies of the highly polymorphic Arctic charr
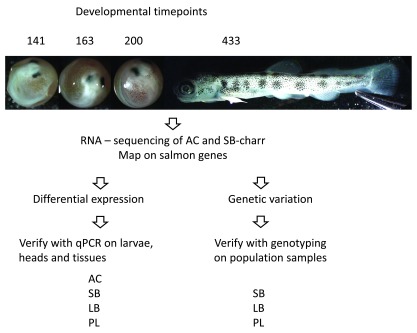
Following the end of the last glacial period, about 10.000 years ago, Arctic charr colonized northern freshwater systems 26. It is found as anadromous or lake/stream residents and exhibits high level of within species polymorphism 17, 26. Charr is also known to harbour substantial phenotypic plasticity, which may promote or reduce divergence 15, 27. Resource polymorphism in charr correlates with ecological attributes 28– 30. For instance small charr with benthic morphology, are found in multiple lavaspring and pond habitats in Iceland 31, and a comparative study of Icelandic lakes 30 found that lakes with greater limnetic habitat, lower nutrients levels, and greater potential for zooplankton consumption appeared to promote resource polymorphism. Some of the larger lakes contain two or more distinct morphs, typically limnetic and benthic forms. Multiple lines of evidence show that these differences stem both from environmental and genetic causes 32– 36. The best studied example of sympatric charr are the four morphs in Lake Thingvallavatn 37; two have a benthic morphotype, a large benthivorous (LB-charr) and a small benthivorous (SB-charr), and two morphs are limnetic, a large piscivorous morph (PI-charr) and small planktivorous morph (PL-charr) 38. Both PL and PI-charr operate in open water and feed on free-swimming prey, PL on planktonic crustaceans and PI on small fish. The PL, LB and SB-charr are presented in Figure 1.
Figure 1. The Arctic charr morphs used in this study.
Adult individuals of the four morphs studied here, from above; the Holar aquaculture charr, the small benthic charr; the planktivorous charr; and the large benthic charr. The latter three all come from Lake Thingvallavatn and were sexually ripe. The morphs differ in size at maturation, body and head shape - mainly lower jaw and length of maxilla and colour pattern in the wild.
Several population genetics studies, using allozymes or mtDNA revealed no differences among charr morphs in Lake Thingvallavatn 39– 41 while other studies using microsatellite markers and nuclear genes, found significant 42– 44 genetic differences among morphs in the lake 45. Importantly Kapralova et al. (2011) 44 concluded that small benthic morphs have evolved repeatedly in Iceland and that gene flow has been reduced between the PL and SB morphs in Lake Thingvallavatn since its formation approximately 10,000 years ago 46. We also discovered genetic separation in immunological genes ( MHCIIα and cath2) between morphs in Iceland and within the lake 45, consistent with ecologically driven evolution of immune functions. Recently qPCR analyses showed that expression of mTOR pathway components in skeletal muscle correlates with the SB-charr form in Iceland 47, but it is unknown whether there is genetic differentiation in those genes or upstream regulators. Because individual genes have distinct histories 48, 49, genome wide methods are needed to identify genes and mutation that associate with divergence. Icelandic aquaculture charr (AC) was founded with fish from the north of Iceland, and has been bred at Holar University College since 1990 50. The Holar AC-charr has responded to artificial selection in growth and performance characteristics, and is now the dominant charr breed in aquaculture in Iceland. While clearly a derived form, it has retained general limnetic craniofacial morphotype ( Figure 1).
In this study we compare SB-charr from Lake Thingvallavatn and AC-charr because i) SB charr represents an extensively studied and derived form of charr, that has been separated from Anadromous fish for approx. 10,000 years, ii) of the availability of abundant AC material and iii) we wanted an extreme contrast, because of budget reasons we could only sequence 8 samples at the time. The AC-charr is included here as a limnetic reference population, in part because we were unable to catch spawning anadromous charr, the ideal outgroup. But by focusing the follow up work on sympatric benthic and limnetic morphs of Lake Thingvallavatn, we can test and verify a subset of the signals found here. The contrast of SB and AC is justified as the data and studies ( 51– 53) building on this data illustrate (see discussion).
The overall objectives of our research program are to investigate the genetics and developmental underpinnings of charr divergence and benthic parallelism. As a step towards this we compare the developmental transcriptome of SB charr and AC charr. The aims of this study are threefold. First, to find genes and pathways related to the development of phenotypic differences between small benthic charr from Lake Thingvallavatn and Icelandic aquaculture charr conforming to a limnetic morphotype. Second, to screen for signals of genetic differentiation between these two charr types. Third, we set out to verify a subset of the expression and genetic signals, in these morphs and two more (benthic and limnetic) morphs from Lake Thingvallavatn. We conduct RNA-sequencing of developing offspring of these two contrasting Arctic charr morphs, reared in common lab environment to minimize the effects of environmentally induced phenotypic plasticity on developmental phenotypes and gene expression. The data reveal genetic changes in nuclear and mitochondrial genes and differential expression of genes that may affect craniofacial and phenotypic traits which separate benthic and limnetic morphotypes in charr.
Methods
Sampling, rearing and developmental series
Overview of the experimental design, RNA sequencing, analyses and follow work is outlined in Figure 2. We set up crosses and reared embryos in the laboratory as described in 51. Embryos from four charr morphs were studied: an aquaculture charr (AC-charr) from the Holar breeding program 50 and three natural morphs from Lake Thingvallavatn; SB, LB and PL-charr 54. Samples of the first two, AC and SB-charr, with contrasting adult size and morphology ( Figure 1), were collected in 2009 and material sent for RNA sequencing. The latter two were sampled in 2010 and were used for qPCR and SNP studies of selected genes. Briefly, in September 2009 we got material from spawning AC-charr from the Holar breeding program, from single parent crosses 50 and spawning SB-charr collected via gill netting in Olafsdrattur in Lake Thingvallavatn. Similarly, in the 2010 spawning season SB-, LB- and PL-charr were collected from Lake Thingvallavatn. For each parent group, eggs from several females (3–10) were pooled and fertilized using milt from several males (3–5) from the same group. Embryos were reared at ~ 5°C under constant water flow and in complete darkness at the Holar University College experimental facilities in Verid, Saudárkrókur. The water temperature was recorded twice daily and the average was used to estimate the relative age of the embryos using tausomite units ( τs) 55. Embryos and juveniles were sampled at designated time points, placed in RNAlater (Ambion) and frozen at −20°C. Post hatching juveniles were reared at the same temperature on standard Aquaculture food. For the investigation of different tissues of adult aquaculture charr (AC) from Hólar (fish size 20–25 cm) were used. Six randomly selected individuals were killed (by cutting through spinal cord) and dissected, and samples were taken from the skin, heart, liver, gills, spleen, intestine and kidney of each fish. The samples were placed in RNAlater (Ambion) and stored at −20°C. We used DNA for population genetic analyses from our previous study 45, eight individuals from each of the three types, PL, LB and SB-charr.
Figure 2. Schematic of RNA sequencing and follow up qPCR and population genetic work.
RNA from embryos of the AC and SB charr at four stages (AC embryos pictured at top) were sequenced with Illumina technology. To verify differentially expressed genes we used RNA from embryos and heads of these four morphs, and tissues from adult AC charr. To verify SNPs we genotyped population samples from three Lake Thingvallavatn morphs (PL, LB and SB).
Fishing in Lake Thingvallavatn was done with permissions obtained both from the owner of the land in Mjóanes and from the Thingvellir National Park commission. Ethics committee approval is not needed for regular or scientific fishing in Iceland (The Icelandic law on Animal protection, Law 15/1994, last updated with Law 157/2012). Sampling was performed by Holar University College Aquaculture Research Station (HUC-ARC) personnel. HUC-ARC has an operational license according to Icelandic law on aquaculture (Law 71/2008), which includes clauses of best practices for animal care and experiments.
RNA extraction and transcriptome sequencing
Embryos of AC- and SB-charr sampled in 2009 were used for transcriptome sequencing. For this we focused on the time covering development of pharyngeal arches and morphogenesis of the head: at 141, 163, 200 and 433 τs (post fertilization). For each combination of morphs and timepoints we pooled RNA from approximately six individuals. RNA extraction and following steps were performed as described earlier 51, 56. Briefly, the embryos were dechorionated and homogenized with a disposable Pellet Pestle Cordless Motor tissue grinder (Kimble Kontes, Vineland, NJ, USA) and RNA was extracted into two size-fractions using the Ambion mirVana kit (Life Technologies, Carlsbad, CA, USA). The high molecular weight fraction was further used for mRNA-seq and RNA quality was analysed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA from samples was pooled - equal contribution of each sample - and first and second strand cDNA synthesis, fragmentation, adapter ligation and amplification were performed using the mRNA-Seq 8-Sample Prep Kit (Illumina, San Diego, CA, USA) according to manufacturer’s instructions. Sequencing was performed at DeCode genetics (Reykjavík, Iceland) using SOLEXA GAII technology (Illumina, San Diego, CA, USA).
The sequencing reads were deposited into the NCBI SRA archive under BioProject identifier PRJNA239766 and with accession numbers: SRX761559, SRX761571, SRX761575, SRX761577, SRX761451, SRX761461, SRX761490 and SRX761501.
The embryos sampled in 2010 were used for qPCR analyses. RNA was extracted from six whole embryos, in two replicates (two repetitions X three fish) (AC and SB sampled at 161 and 200 τs). For the extraction of RNA from heads of AC, SB, LB and PL, 12 embryos (two repetitions X six fish) at 178, 200 and 216 τs were used. Embryos were dechorionated and decapitated in front of the pectoral fin. RNA extraction and cDNA preparation were performed as described previously in 51. Similarly, RNA was extracted from a small piece (approximately 2 mm 2) of skin, heart, liver, gill, spleen, intestine and liver from six adult AC-charr.
Analyses of RNA-seq data and mapping to Salmon EST contigs
As no S. alpinus genome is available and de novo assembly of the 36 bp reads yielded an excessive number of short contigs we chose to assess expression and genetic variation by mapping the reads to 59336 S. salar expressed sequence tag (EST) contigs from the SalmonDB [ 57, downloaded 22. March 2012] and the Arctic charr mitochondrial genome [ 48, NC_000861].
To estimate expression, reads were aligned with RSEM version 1.1.18 with default parameters. RSEM distributes reads that map to multiple locations to the most likely contig, using expectation maximization 58. The read counts for contigs with the same annotation were pooled because some genes were represented by more than one contig, and due to whole genome duplication almost the half of salmonid genes exist as ohnologs 22, 24. Thus the expression tests are done on gene or paralog group level, instead of the contig level. We acknowledge that paralogous genes are not always expressed similarly, but feel its necessary to do this pooling because of the nature of the data. In the remainder of the paper, we will refer to gene or paralog group (the number of underlying contigs is indicated in relevant tables). This brought the number of genes considered down to 16851. Lastly, paralog groups with fewer than 800 mapped reads in the entire dataset were excluded from the analyses, yielding a total of 10496.
A generalized linear model (GLM) with morph and developmental time as explanatory variables was used to find genes with different expression levels between the two charr morphotypes (groups) using the edgeR-package in R 59.
To obtain further insight into the expression profiles of differently expressed genes, we performed clustering analyses on log-transformed cpm-values (counts per million; cpm-function in edgeR). The values for each gene were scaled by mean and standard deviation, and the euclidean distance used for the hclust-function in R 60 with the default settings. We used the hypergeometric-test in goseq 61 to test for gene ontology enrichment. Since we pooled the read-count from different contigs we could unfortunately not take gene length into account in those tests.
Tests of differential expression with qPCR
We previously identified suitable reference genes to study Arctic charr development 51. Here we examined the expression of several genes in whole charr embryos, embryonic heads and adult tissues. Primers were designed using the Primer3 tool 62 and checked for self-annealing and heterodimers according to the MIQE guidelines 63 ( S1 Table). Primers for genes with several paralogs were designed for regions conserved among paralogs, except for natterin-like, where primers were designed to match regions differing in sequence between paralogs. Relative expression was calculated using the 2 −ΔΔ Ct method 64. For the calculation of relative expression of genes in whole embryos, the geometric mean expression of three reference genes, β- Actin (Actb), elongation factor 1 α and Ubiquitin-conjugating enzyme E2 L3, was used for normalization. For visual comparisons among samples, the normalized expression was presented as relative to the expression in AC at 161 τs (calibration sample). For the embryonic head samples Eukaryotic Translation Initiation Factor 5A (If5a1) and Actb were used as reference genes and a biological replicate of AC at 178 ( τs) as the calibrator sample, see 51, 52. Standard errors of relative expression were calculated from the standard errors (SE) of the Δ C T-values with the formula 2 −(ΔΔ Ct+ SE) = minimum fold expression and 2 −(ΔΔ Ct− SE) = maximum fold expression. The statistical analysis was performed using the Δ C T-values with a two-way ANOVA with GLM function in R.
Normal distribution of residuals was confirmed for all data. For the study of expression in the embryonic head we followed a significant morph effect in the ANOVA with Tukey’s post-hoc honest significant difference test, on relative expression ratios (Δ C Ts). Three genes had lower efficiency (as low as 1.72). We acknowledge that the data on those genes may be weak.
Polymorphisms in the Arctic charr transcriptome
For analysis of genetic variation we mapped the reads to the salmon contigs, this time using the Burrow-Wheeler Aligner (BWA) 65 with a seed length of 25, allowing two mismatches. We re-mapped the reads, since BWA allows short indels (RSEM does not) but disregarding them leads to many false SNPs close to indels. To extract candidate polymorphic sites from the Arctic charr transcriptome we ran VarScan2 66 with minimum coverage of 50 reads and minimum minor allele frequency of 0.1 on reads mapped to each S. salar contig for all of the 8 timepoints and morph combinations. This was done separately for reads that mapped uniquely to one contig only (UNI) and reads that mapped to two or more contigs (REP). These SNP-candidates were further processed in R 60, following established principles for variant calling 67. SNP-candidates at 90% frequency or higher in all samples were disregarded, as they reflect differences between Arctic charr and S. salar and are not the focus of this study. SNP-candidates with poor coverage in specific samples - i.e. coverage of five or fewer reads in three or four samples of each morph - were removed. As the SNP analysis was done on individual contigs, differences among paralogs appear in the data. To address this we use the fact that each sample is a pool of few individuals, thus true SNPs are unlikely to have the same frequency in all samples. However, variants reflecting differences between paralogs will have similar frequency all samples (assuming steady difference in their expression in all samples). We evaluated differences between samples with Fisher exact tests, and only SNPs significantly different between samples with a p < 0.05 (with no multiple testing correction) were retained. To compare morphs, read numbers were summed over the four samples from each morph. A conservative approach was taken by focusing on SNP-candidates that showed the largest differences in frequency between morphs (delta), without adjusting for multiple testing (Fisher exact test, p > 5%). SNP-candidates with the highest frequency difference (delta > 95%) were manually processed and redundant candidates removed. A similar approach was used to mine for polymorphisms in Arctic charr mtDNA (NC_000861), using S. salar mtDNA as the outgroup (NC_001960.1).
We wrote a python script to predict the impact of SNPs within the mRNA sequences. Polymorphisms were categorized according to their location (3’UTR, coding, 5’UTR), and those within the coding region into synonymous or non-synonymous.
Verification of candidate SNPs
We chose 12 candidate SNPs for verification (see below). As the AC-charr is not a random breeding population, and because our interest is on differences between wild morphs, we took random samples of spawning SB, LB and PL-charr from Lake Thingvallavatn (8 per morph) from our earlier study 45. Using the same PCR and DNA sequencing approach we genotyped 12 candidate SNPs ( S2 Table). Briefly, we first compared the Salmon genome and ESTs [ 57, downloaded 22. March 2012] and short contigs from our preliminary assembly of the Arctic charr transcriptome. This allowed us to infer the placement of the putative polymorphism in the locus, and design paralog specific primers for PCR (less than 1 kb amplicons). MJ tetrad machine was used for PCR and the program was 5 min. at 95°C, followed by 35 cycles of 30 sec. at 52°C, 1 min. at 72°C, 30 sec. at 95°C, ending with 12°C while waiting on the human. Each individual was genotyped by first amplifying the region of interest using PCR, followed by ExoSAP (Affymetrix), direct sequencing (BigDye) and finally run on an Applied Biosystems 3500xL Genetic Analyzer (Hitachi). Raw data was base-called using the Sequencing Analysis Software v5.4 with KBTMBasecaller v1.41 (Applied Biosystems). Ab1 files were run through Phred and Phrap and imported to Consed for visual editing of ambiguous bases and putative polymorphisms, and for trimming primers. The FASTA files were aligned with ClustalW online [ 68, http://www.ebi.ac.uk/Tools/msa/clustalw2/] and manually inspected in Genedoc 69. All sequences where deposited to Genbank as popsets under the accession numbers KP019972-KP020026.
Comparative genomic analyses of sequence polymorphisms
Two approaches were used for genomic comparisons of verified SNPs in the mitochondrial genome. Using the charr mtDNA sequence we performed both a BLAST search on salmon ESTs (May 2013) and retrieved multiZ alignments of vertebrates from the UCSC genome browser (in September 2013). This yielded several hundred sequences from related fish and other vertebrates. The list was reduced to 20 sequences for visualization, by keeping members of the major taxa but removing more closely related sequences, aligned with ClustalW and manually adjusted in Genedoc. The species and genome versions used are; Human ( Homo sapiens, hg19), Lamprey ( Petromyzon marinus, petMar1), Fugu ( Takifugu rubripes, fr2), Medaka ( Oryzias latipes, oryLat2), Stickleback ( Gasterosteus aculeatus, gasAcu1), Tetraodon ( Tetraodon nigroviridis, tetNig2), Zebrafish ( Danio rerio, danRer6). We also downloaded from NCBI the sequence of whole or partial mtDNA from several fish species; Brown trout ( Salmo trutta, JQ390057 and AF148843), Broad whitefish ( Coregonus nasus, JQ390058), Legless searsid ( Platytroctes apus, AP004107), Pacific menhaden ( Ethmidium maculatum, AP011602), Icefish ( Salanx ariakensis, AP006231 and HM151535), Chain pickerel ( Esox niger, AP013046) and Western Pacific roughy ( Hoplostethus japonicus, AP002938). The three mitochondrial variants (numbered by the S. alpinus mtDNA - NC_000861) are; m1829G>A (CCACGTTGTGAAACCAAC[G/A]TCCGAAGGTGGATTTAGCAGT), m3211T>C (CGTGCAGAAGCGGGCATAAG[T/C]ACATAAGACGAGAAGACCCT) and m3411C>T (CTCTAAGCACCAGAATTT[C/T]TGACCAAAAATGATCCGGC).
Results
RNA sequencing characteristics
Each sample yielded good quality data, with sequencing depth from 49 to 58 million (average: 55 million) reads. To quantify the expression levels, the reads were aligned to a salmon EST-assembly 57. Around 20% of the reads mapped uniquely to the EST data ( S3 Table). A further 30% mapped to two or more contigs, probably representing paralogous genes, recent duplications or repeat-like elements within transcribed regions. A substantial fraction of the RNA-sequencing reads did not map to the contigs from S. salar. Analyses of those reads require an Arctic charr genome sequence or transcriptome assembly from longer and paired end reads, currently underway in our laboratory.
Differential expression during Arctic charr development
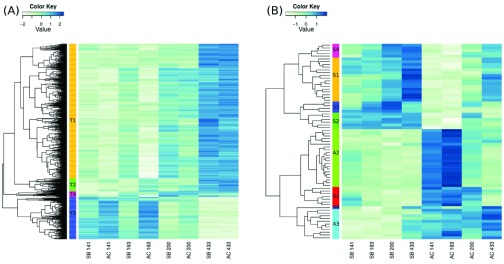
We detected considerable changes in the transcriptome during Arctic charr development ( Figure 3a). The expression of 1603 and 2459 paralog groups differed significantly between developmental timepoints at the 1% and 5% levels of false discovery rate (FDR), respectively ( Dataset 1). The difference was most pronounced between prehatching (timepoints: 141, 163, 200 τs) and post hatching embryos (timepoint 433 τs), as more than 70% of the paralog groups with FDR below 1% had higher expression in the latter ( Figure 3b). Gene Ontology analyses reveal six enriched GO categories (below 10%FDR). The most drastic changes were seen in processes related to glycolysis (GO:0006096, FDR = 0.0009), where the expression of 19 out of 25 paralog groups changed during this developmental period. The other five classes that were differentially expressed during charr development are: ion transport (GO:0006811, FDR = 0.027), blood coagulation (GO:0007596, FDR = 0.03), DNA repair (GO:0006281, FDR = 0.08) and two immune related categories (GO:0019882, FDR = 0.08, GO:0006955, FDR = 0.09). Those results probably reflect developmental changes and/or differences in the environment of embryos before and after hatching.
Figure 3. Heatmap of differentially expressed genes in the Arctic charr developmental transcriptome.
Two morphs (SB and AC) are represented, at four timepoints. ( A) The 1603 genes with expression difference among time points, here clustered into four groups. ( B) The 71 genes differentially expressed between morphs are clustered into 4 groups for each morph. High expression is indicated by blue and low expression by beige.
Differential expression between Arctic charr morphs
We were especially interested in genes showing expression differences between the two morphs as they might implicate pathways involved in the ecological divergence among charr populations. In the data 296 paralog groups were differentially expressed (FDR < 5%) between the morphs (141 higher in SB and 152 higher in AC-charr, Dataset 1). Among genes with higher expression in SB-charr two biological GO categories were enriched: blood coagulation (GO:0007596, p = 0.001) and proteolysis (GO:0006508, p = 0.002). Recall, expression of blood coagulation factors also differed between developmental stages (see above). In AC-charr, genes in three categories: respiratory electron transport chain (GO:0022904, p = 0.0006), ATP synthesis coupled electron transport (GO:0042773, p = 0.002) and neurotransmitter transport (GO:0006836, p = 0.009) have higher expression. The first two GO categories both relate to energy generation in mitochondria and could reflect higher expression of genes with mitochondrial functions in AC-charr. At more stringent FDR (1%), 31 paralog groups, with diverse functional annotations, were higher expressed in SB and 40 genes higher in AC-charr ( Figure 3b, Table 1 and Table 2). The higher expressed ESTs were clustered into 4 groups for each morph, reflecting in some cases functional similarity. For instance SB cluster 3 has three immune related paralog groups: Complement factor D (9), H-2 class I histocompatibility antigen L-D alpha chain (2) and Sushi domain-containing protein 2 (4) ( Table 1). Note, however, that immune genes were not significantly enriched in the GO comparison of morphs. The results suggest genes with mitochondrial function, blood coagulation and other functions are differentially expressed between the morphs, but as few samples were sequenced, qPCR verification was needed.
Validation of gene expression differences in whole embryos and paralog specific expression of natterin genes
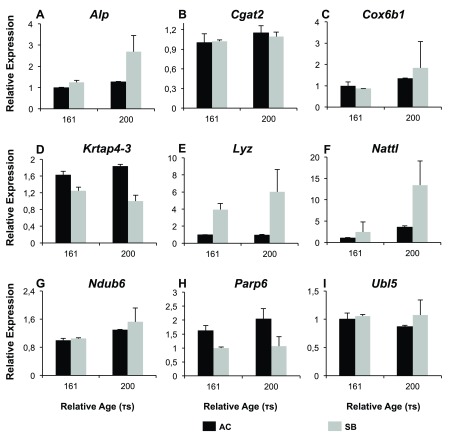
For validation we opted for qPCR analyses of 9 genes/paralog groups in whole embryos and 8 in embryonic heads, which showed differential expression between AC and SB-charr, with statistical support ranging from <1% to about 10% FDR. We studied paralog groups with less FDR support, in part to be able to cast a wider net (see below). Of the nine paralog groups studied in whole embryos, five were confirmed to be differentially expressed between AC and SB-charr at 161 or 200 τs ( Figure 4, S4 Table and Dataset 2). Three genes, Nattl, Alkaline phosphatase (Alp) and Lysozyme C II (Lyz2), had significantly higher expression in SB. The other two, Keratin-associated protein 4-3 (Krtap4-3) and Poly polymerase 6 (Parp6) had higher expression in AC embryos ( Figure 4, S4 Table). No morph and time interaction was detected for any of the genes.
Figure 4. qPCR validation of candidates from transcriptome in whole embryos of Arctic charr.
Relative expression of 9 genes ( A– I) analysed by qPCR in the small benthic (SB) charr from Lake Thingvallavatn and aquaculture (AC) charr at two different developmental timepoints (161 and 200 τs). 5 genes were differentially expressed between the two morphs ( Alp, Krtap4-3, Lyz, Nattl, Parp6), while 4 further genes did not show significant expression differences between morphs ( Cgat2, Cox6B1, Ndub6, Ubl5), see S4 Table. Error bars represent standard deviation calculated from two biological replicates.
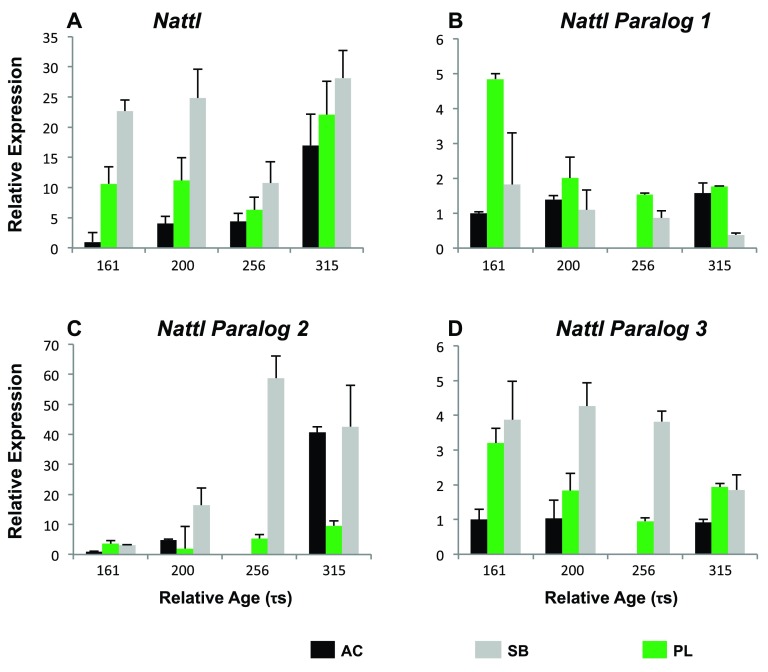
As some genes are represented by different contigs or even paralogs, we set out to disentangle the expression of one paralog group ( Nattl) in detail. We measured the expression of three natterin paralogs ( nattl1, nattl2 and nattl3), by designing qPCR primers that matched divergent regions. These genes caught our interest because the only prior work implicated Natterin as a toxin produced by a tropical fish 70, 71. We studied nattl expression in several developmental stages in AC-, SB- and PL-charr as well as in selected tissues of adult AC-charr. The expression level of the three paralogs differed between morphs and timepoints ( Figure 5 and S5 Table). Overall nattl2 had the highest expression in all morphs. The nattl1 had higher expression in embryos of PL-charr than in AC- and SB-charr, while nattl2 and nattl3 were more expressed in SB-embryos. Note however, the efficiency of the primers for the nattl genes ranged from 1.72 to 1.77, which suggests this data should be interpreted with caution.
Figure 5. Relative expression of Nattl and its three paralogs during charr development in different morphs.
The expression is graphed for different morphs (SB, AC and PL) at four developmental timepoints (161, 200, 256 & 315 τs, relative to AC-charr at timepoint 161. A) General nattl expression along charr development. B–D) Expression of nattl paralogs 1–3. ANOVA showing the variation among morphs is summarized in S5 Table.
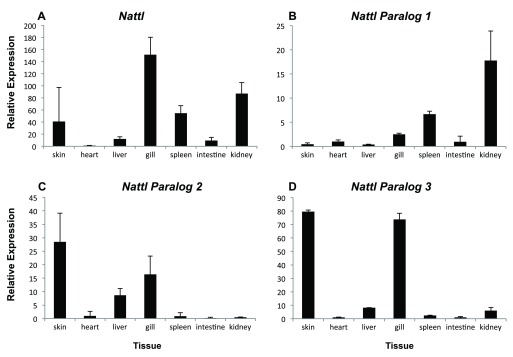
In order to evaluate the hypothesis that nattl genes have immune-related functions we studied expression in adult tissues (in AC-charr). The nattl expression was highest in the gills, followed by expression in kidney, skin and spleen. Low expression levels were detected in liver, intestine and heart ( S1 Figure and S5 Table). The three nattl paralogs followed different patterns, whilst each of them showed significant expression differences among tissues. Nattl1 was mainly expressed in spleen and kidney, while nattl2 showed a significantly higher expression in skin, liver and in gills. Similarly, the relative expression of nattl3 was highest in the gills and skin. This indicates that the three nattl paralogs are expressed in a tissue specific manner, and also differently during the development of the three charr morphs studied here.
Table 1. Differentially expressed genes, with higher expression in the SB morph from Lake Thingvallavatn.
| NR | Name | Abbr | Cont | logFC | logCPM | FDR | Cluster |
|---|---|---|---|---|---|---|---|
| 3766 | Histone H3 embryonic | 1 | 8.71 | 2.74 | 7.80E-035 | S-1 | |
| 5103 | Natterin-like | Nattl | 6 | 2.75 | 7.12 | 7.76E-007 | S-2 |
| 356 | A7J6M9 Putative uncharacterized protein n175R | 1 | 2.33 | 4.66 | 3.30E-006 | S-1 | |
| 6697 | Q1KY05 Main olfactory receptor-like | Sorf | 5 | 3.12 | 6.92 | 9.96E-005 | S-1 |
| 8151 | Sushi domain-containing protein 2 | Susd2 | 4 | 2.20 | 5.55 | 0.0001 | S-3 |
| 1682 | Carcinoembryonic antigen-related cell adhesion
molecule 1 |
Ceacam1 | 3 | 2.55 | 3.83 | 0.0002 | S-1 |
| 6228 | Protein FAM98A | 2 | 1.96 | 4.76 | 0.0003 | S-1 | |
| 7531 | STAM-binding protein-like | Stampbl1 | 2 | 2.07 | 2.62 | 0.0005 | S-1 |
| 6712 | Q1M160 Myc-regulated DEAD box protein | 1 | 1.67 | 3.23 | 0.0009 | S-1 | |
| 2300 | Cytosolic sulfotransferase 3 | Sult3st1 | 3 | 1.73 | 2.13 | 0.0009 | S-1 |
| 2063 | Complement factor D | Cfd | 7 | 1.79 | 6.42 | 0.0016 | S-3 |
| 3326 | Galectin-3-binding protein A | 4 | 1.79 | 3.85 | 0.0017 | S-4 | |
| 3169 | Flocculation protein 11 | Flo11 | 2 | 1.86 | 4.05 | 0.0017 | S-1 |
| 1203 | B5XDY0 H-2 class I histocompatibility antigen
L-D alpha chain |
2 | 1.70 | 2.12 | 0.0028 | S-3 | |
| 9183 | UPI000065D844 related cluster | 2 | 1.97 | 5.55 | 0.0028 | S-1 | |
| 2909 | Epidermis-type lipoxygenase 3 | Loxe3 | 4 | 1.68 | 4.84 | 0.0029 | S-1 |
| 4884 | Myeloperoxidase | Mpo | 4 | 2.20 | 6.78 | 0.0029 | S-1 |
| 10003 | Uridine phosphorylase 1 | Upp1 | 4 | 1.51 | 3.00 | 0.0047 | S-1 |
| 2513 | Desmoglein-1-alpha | Dsg1 | 1 | 1.59 | 2.80 | 0.0054 | S-2 |
| 377 | A7SJA8 Predicted protein (Fragment) | 1 | 1.73 | 2.50 | 0.0055 | S-3 | |
| 9204 | UPI00006A2900 related cluster | 2 | 6.38 | 3.26 | 0.0064 | S-1 | |
| 9642 | UPI00017B1B0F related cluster | 1 | 2.00 | 1.92 | 0.0064 | S-2 | |
| 1965 | Coiled-coil domain-containing protein 136 | Ccdc136 | 2 | 2.15 | 2.32 | 0.0064 | S-2 |
| 9260 | UPI0000F1D4BA PREDICTED | 1 | 1.80 | 2.41 | 0.0065 | S-2 | |
| 738 | Adseverin | Scin | 8 | 1.58 | 5.51 | 0.0073 | S-1 |
| 9678 | UPI00017B4479 related cluster | 1 | 2.18 | 1.97 | 0.0074 | S-4 | |
| 8339 | Testin | Tes | 4 | 1.50 | 4.93 | 0.0080 | S-2 |
| 6840 | Q4SNH3 Chromosome 8 SCAF14543 | 1 | 1.42 | 4.00 | 0.0080 | S-1 | |
| 1668 | Carbohydrate sulfotransferase 6 | Chst7 | 1 | 2.09 | 2.08 | 0.0090 | S-4 |
| 8341 | Testisin | Prss21 | 2 | 2.01 | 2.76 | 0.0090 | S-4 |
| 6373 | Protein asteroid homolog 1 | Aste1 | 6 | 1.29 | 4.24 | 0.0090 | S-4 |
Name: name of unigene or paralog group
Abbr: Abbreviated paralog group or gene name
Cont: Number of contigs
logFC: log Fold Change
logCPM: log Counts Per Million
FDR: False Discovery Rate
The cluster numbering corresponds to Figure 3.
Table 2. Differentially expressed genes, with higher expression in the AC morph.
| NR | Name | Abbr | Cont | logFC | logCPM | FDR | Cluster |
|---|---|---|---|---|---|---|---|
| 3465 | Glutathione S-transferase P 1 | Gstp1 | 1 | -8.35 | 2.45 | 1.12E-019 | A-2 |
| 2475 | Dehydrogenase/reductase SDR family member 7 | Dhrs7 | 2 | -4.88 | 2.15 | 9.67E-014 | A-3 |
| 6945 | Q6NWE8 Sb:cb283 protein | 3 | -6.08 | 3.02 | 2.15E-013 | A-2 | |
| 399 | A8DW32 Predicted protein | 1 | -5.32 | 6.38 | 4.27E-010 | A-1 | |
| 9682 | UPI00017B4B48 related cluster | 2 | -3.70 | 2.81 | 2.61E-008 | A-2 | |
| 9817 | Uncharacterized protein ART2 | 5 | -12.63 | 6.89 | 8.23E-008 | A-2 | |
| 6724 | Q2L0Z2 Putative ATP-dependent RNA helicase | 1 | -3.41 | 1.89 | 1.88E-007 | A-2 | |
| 1197 | B5XD10 Vacuolar proton pump subunit G 1 | Atpv1g1 | 1 | -4.30 | 2.10 | 1.84E-006 | A-2 |
| 5325 | Nucleoside diphosphate kinase B | Nme2 | 1 | -9.85 | 7.63 | 2.51E-006 | A-1 |
| 9205 | UPI0000D5B923: myelin basic protein isoform 1 | Mbpa | 3 | -2.49 | 3.45 | 9.18E-006 | A-3 |
| 6377 | Protein broad-minded | Tbc1d32 | 1 | -2.11 | 2.74 | 4.75E-005 | A-1 |
| 5711 | Pistil-specific extensin-like protein | 1 | -2.16 | 2.60 | 0.0002 | A-3 | |
| 3203 | Formin-like protein 20 | Fmnl2b | 7 | -1.98 | 1.95 | 0.0002 | A-3 |
| 9315 | UPI0000F2EC69: hypothetical protein | 2 | -5.60 | 4.57 | 0.0005 | A-2 | |
| 363 | A7RFV0 Predicted protein (Fragment) | 2 | -1.74 | 4.96 | 0.0010 | A-3 | |
| 6937 | Q6AZT1 MGC81677 protein | 3 | -2.06 | 3.81 | 0.0014 | A-2 | |
| 3756 | Histone H1 | Histh1 | 3 | -2.26 | 4.54 | 0.0017 | A-2 |
| 1133 | B5DGN9 Creatine kinase-1 | Ckm1 | 7 | -4.72 | 5.50 | 0.0017 | A-3 |
| 309 | A1IMH7 CD80-like protein | Cd80 | 12 | -1.94 | 4.29 | 0.0017 | A-2 |
| 7651 | Serine protease ami | 2 | -1.54 | 5.90 | 0.0017 | A-3 | |
| 9935 | Uncharacterized protein C7orf63 homolog | 1 | -1.87 | 1.91 | 0.0025 | A-2 | |
| 5219 | Nostrin | Nostrin | 2 | -2.55 | 3.38 | 0.0029 | A-2 |
| 1855 | Chondroitin sulfate N-acetylgalactosaminyl-
transferase 2 |
Csgalnact2 | 5 | -2.56 | 6.14 | 0.0034 | A-1 |
| 10203 | Xylose isomerase | 6 | -1.55 | 2.43 | 0.0035 | A-3 | |
| 2249 | Cytochrome c oxidase subunit 3 | Cox3 | 11 | -1.78 | 11.15 | 0.0035 | A-3 |
| 180 | 40S ribosomal protein S3-B | Rps3b | 2 | -5.31 | 8.67 | 0.0050 | A-1 |
| 1227 | B6NBL3 Putative uncharacterized protein | 3 | -1.59 | 2.95 | 0.0050 | A-2 | |
| 5055 | NADH-ubiquinone oxidoreductase chain 6 | Nd6 | 2 | -1.48 | 2.65 | 0.0061 | A-3 |
| 4634 | Metallothionein A | Mta | 1 | -3.33 | 5.44 | 0.0064 | A-2 |
| 342 | A5C0J4 Putative uncharacterized protein | 2 | -2.58 | 2.47 | 0.0064 | A-2 | |
| 9698 | UPI00019258B4: similar to epithelial cell
transforming sequence 2 oncogene protein partial |
1 | -2.06 | 2.94 | 0.0064 | A-2 | |
| 5878 | Pro-opiomelanocortin B | Pomcb | 1 | -2.04 | 5.60 | 0.0065 | A-2 |
| 2248 | Cytochrome c oxidase subunit 2 | Cox2 | 9 | -2.21 | 9.83 | 0.0074 | A-2 |
| 1246 | B8JI87 Novel protein similar to vertebrate collagen
type VI alpha 3 (COL6A3) (Fragment) |
1 | -1.69 | 3.16 | 0.0080 | A-3 | |
| 7994 | Sperm-associated antigen 5 | Spag5 | 1 | -2.07 | 3.71 | 0.0080 | A-2 |
| 9515 | UPI000175F90F: similar to pleckstrin homology
domain containing family A member 7 |
1 | -2.00 | 1.87 | 0.0090 | A-2 | |
| 1124 | B5DDZ4 Acta1 protein | Actc1b | 1 | -1.52 | 2.62 | 0.0090 | A-2 |
| 1127 | B5DG94 2-peptidylprolyl isomerase A | Ppia1 | 2 | -2.56 | 5.67 | 0.0090 | A-1 |
| 9175 | UPI000054A3C0 PREDICTED: apolipoprotein B | 3 | -1.32 | 4.09 | 0.0090 | A-4 | |
| 9671 | UPI00017B3C62 related cluster | 1 | -1.51 | 1.92 | 0.0096 | A-1 |
For column header explanation, see footer of Table 1.
Expression differences in the developing heads of benthic and limnetic charr morphs
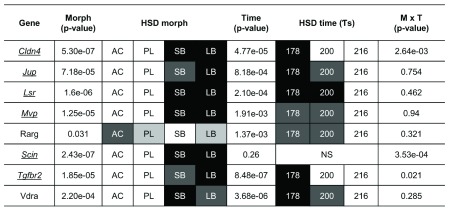
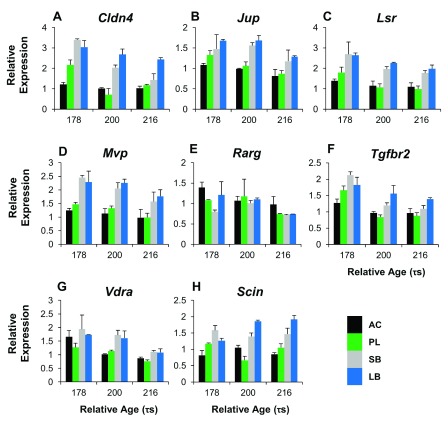
To get a handle on the craniofacial divergence between sympatric Arctic charr morphs we used qPCR to study 8 paralog groups with expression difference in the RNA-seq data (all higher in SB). We focused on those with known craniofacial expression in zebrafish development 72 and compared two benthic (SB, LB) and two limnetic charr (AC, PL). We analyzed heads at three time-points (178, 200 and 218 τs) as this period overlaps with early stages of craniofacial skeletal formation in Arctic charr 73, 74. The qPCR confirmed the higher expression of seven out of these eight genes in the head of benthic charr compared to limnetic charr ( Figure 6, S2 Figure and Dataset 3). These seven genes are Claudin 4 (Cldn4), adseverin (Scin), Junction plakoglobin (Jup), Lipolysis stimulated lipoprotein receptor (Lsr), Major vault protein (Mvp), Transforming growth factor beta receptor II (Tgfbr2) and Vitamin D receptor a (Vdra). The eighth gene, Retinoic acid receptor gamma-A (Rarg) gave a small but significant response in the head, but the effects were reversed, i.e. the expression was higher in AC. The expression difference of the seven genes was, in almost all cases, consistent over the three timepoints studied (See S2 Figure). In summary the qPCR confirmed the differential expression of 12 of the 17 paralog groups studied ( Table 3), some which had 5–10% FDR support. To us that suggests substantial expression differences between these two charr morphs, and that the data can lead to hypotheses about morph specific activity in particular structures, like the developing head.
Figure 6. Expression differences of craniofacial candidate genes in developing head of Arctic charr morphs.
Relative expression ratios, calculated from the qPCR data, were subjected to an ANOVA to test the expression differences amongst four charr groups and three close time points ( τs). The underlined gene names reflect significant difference between SB and AC-charr. A post hoc Tukey’s test (HSD) was performed to determine the effects of morphs, time and morph-time interaction (M X T). White boxes represent low expression, while black boxes represent high expression. The shading represents significant different expression between the samples (α = 0.05, NS = not significant). The genes studied were, Claudin 4 (Cldn4), adseverin (Scin), Junction plakoglobin (Jup), Lipolysis stimulated lipoprotein receptor (Lsr), Major vault protein (Mvp), Transforming growth factor beta receptor II (Tgfbr2) Vitamin D receptor a (Vdra) and Retinoic acid receptor gamma-A (Rarg).
Table 3. Correspondence of transcriptome and qPCR verification on Arctic charr embryos.
| Tissue | Name | Abbr | FDRm | FRDt | Effect | qPCR | Morph |
|---|---|---|---|---|---|---|---|
| Embryo | Alkaline phosphatase | Alp | 0.070 | 0.001 | 0.986 | * | SB |
| Embryo | Chondroitin sulfate
N-acetylgalactosaminyltransferase 2 |
Cgat | 0.004 | 0.331 | -2.556 | ||
| Embryo | Cytochrome c oxidase subunit 6B1 | Cox6b1 | 0.058 | 0.632 | -1.208 | ||
| Embryo | B5X596 Keratin-associated protein 4-3 | Krtap4-3 | 0.012 | 0.278 | -1.986 | * | AC |
| Embryo | Lysozyme C II | Lyz2 | 0.041 | 0.001 | 1.138 | * | SB |
| Embryo | Natterin-like protein | Nattl | 0.000 | 0.000 | 2.755 | * | SB |
| Embryo | NADH dehydrogenase [ubiquinone] 1
beta subcomplex subunit 6 |
Ndub6 | 0.098 | 0.670 | -1.175 | ||
| Embryo | Poly [ADP-ribose] polymerase 6 | Parp6 | 0.108 | 0.379 | -0.986 | * | AC |
| Embryo | Ubiquitin-like protein 5 | Ubl5 | 0.059 | 0.003 | -1.234 | ||
| Head | Claudin-4 | Cldn4 | 0.068 | 0.000 | 1.343 | * | SB/LB |
| Head | Major vault protein | Mvp | 0.065 | 0.528 | 0.958 | * | SB/LB |
| Head | Junction plakoglobin | Jup | 0.051 | 0.006 | 1.147 | * | SB/LB |
| Head | Lipolysis-stimulated lipoprotein receptor | Lsr | 0.013 | 0.043 | 1.369 | * | SB/LB |
| Head | TGF-beta receptor type-2 | Tgfbr2 | 0.065 | 0.013 | 1.728 | * | SB/LB |
| Head | Vitamin D3 receptor A | Vdra | 0.053 | 0.052 | 1.312 | * | SB/LB |
| Head | Retinoic acid receptor gamma-A | Rarg | 0.012 | 0.001 | 1.403 | ||
| Head | Adseverin | Scin | 0.007 | 0.000 | 1.578 | * | SB/LB |
Tissue: which tissue was studied
Abbr: abbreviated paralog group or gene name
FRDm: FDR for comparison of SB and AC-charr in transcriptome
FDRt: FDR for comparison among developmental timepoints in transcriptome
Effect: logarithm of fold change between morphs, positive is higher in SB and negative higher in AC-charr in transcriptome (logFC.morph in supplemental dataset 1)
qPCR: results consistent with transcriptome (*), a blank cell reflects lack of correspondence
Morph: which morph(s) had higher expression in qPCR verification
Analyses of polymorphism in Arctic charr transcriptome
The RNA-seq data also revealed segregating variations with large frequency differences between charr morphs. To uncover candidate SNPs we mapped the reads to all of the S. salar EST-contigs. Filtering on coverage yielded 165,790 candidate SNPs ( Table 4); of those 66.569 came from reads that mapped uniquely and 57.009 candidate SNPs from reads that mapped to more than one contig; with limited overlap between lists. Assuming that the expression of paralogous genes is stable, then differences among paralogs appear as SNPs at similar frequency in all samples. By requiring variant frequency differences (p < 0.05, uncorrected) between samples we reduced the list of candidates by two thirds, yielding over 20.000 candidate SNPs. Note, as cDNA from charr families was sequenced (not a population sample), estimates of SNP frequencies are imprecise. To err on the side of caution, we chose SNP candidates with 50% or higher frequency difference between morphs for further study. The candidate SNPs were also summarized by frequency of the derived allele, in reference to the S. salar sequence. This gave 672 and 872 SNPs at higher frequency, in AC-charr and SB-charr, respectively. The uniquely and multiply mapped reads, revealed approximately similar numbers of candidate SNPs. Gene ontology analysis showed that for derived SNPs in SB, there was an excess of variants in genes related to translation, both as a broad category and specific subgroups ( S6 Table). There was also enrichment of SNPs in genes related to DNA-mediated transposition, DNA integration, DNA replication and oxidation-reduction process. No GO categories were enriched for high frequency derived SNPs in AC. Furthermore, functional effects of the candidate SNPs (UTR, synonymous and non-synonymous) were predicted. The distribution among those categories did not differ between variants detected by uniquely or repeatedly mapped reads, , p = 0.46 ( S7 Table).
Table 4. Candidate SNPs in the Arctic charr transcriptome, filtered by coverage, difference between sample and morphs and frequency difference between morphs.
| SNP-candidates | Morph | Uni | Rep | Total |
|---|---|---|---|---|
| Total | 96231 | 74341 | 165790 | |
| Filter coverage | 66569 | 57009 | 113776 | |
| Diff. Bwn. samples | 21417 | 22252 | 42869 | |
| Diff. Bwn. morphs | 11385 | 12953 | 23974 | |
| Delta > 0.5 | AC | 396 | 285 | 672 |
| Delta > 0.5 | SB | 526 | 353 | 872 |
| Delta > 0.75 | AC | 95 | 68 | 159 |
| Delta > 0.75 | SB | 155 | 95 | 248 |
| Delta > 0.95 a | AC | 17 | 13 | 30 |
| Delta > 0.95 a | SB | 29 | 4 | 33 |
SNP-candidates: found by mapping to S. salar ESTs
Uni/REP: from UNIquely or REPeatedly mapped RNA-reads
Delta: differences in allele frequency between morphs, categorized by which morph had the higher derived allele frequency
aThe number of SNP-candidates before the redundant ones were removed
A total of 60 candidate SNPs are nearly fixed in one morph, with frequency difference between morphs above 95% (after manual inspection of contigs and SNP position three candidates were removed since they represented the same SNP). Of these “fixed” SNPs 46 came from uniquely mapped reads and 14 from reads that mapped more than twice ( Table 5 and Table 6). For the SNPs from uniquely mapped reads, 17 are fixed in AC-charr and 29 in SB-charr. The few genes with two or more polymorphic sites were; Keratin type II cytoskeletal 3 (Krt3), Cysteine sulfinic acid decarboxylase (Csad) and DNA-directed RNA polymerase I subunit RPA12 (Rpa12) with 5, 5 and 2 SNPs respectively. Krt3 and Csad had significant differentiation in both SB and AC. Similarly, 14 SNPs with large differentiation between morphs were predicted from reads that mapped on two or more contigs ( Table 6). Of these, we found two variants in the mitochondrial 60S ribosomal protein L36 (RpL36) and variants in 4 other mitochondrial genes (28S ribosomal protein S18a mitochondrial (MRPS18A), Apoptosis-inducing factor 1 mitochondrial (AIFM1), Isocitrate dehydrogenase [NADP] mitochondrial (acIDH1) and Protein S100-A1 (S100a1)), all at higher frequency in AC-charr. PCR and Sanger sequencing of population samples confirmed SNPs in DNA2-like helicase (Dna2), a gene with nuclear and mitochondrial function, and two other genes Uroporphyrinogen decarboxylase (Urod), and Mid1-interacting protein 1-like (Mid1ip1) ( S2 Table). The candidate variant Eukaryotic translation initiation factor 4 gamma 2 (Eif4g2) was not substantiated by the PCR/sequencing.
Table 5. SNP candidates from uniquely mapped reads.
| (a) Higher frequency in AC morph | |||||||
|---|---|---|---|---|---|---|---|
| Contig | Annotation | Pos | Ref | Var | Freq-SB | Freq-AC | Effect |
| SS2U026955 | Keratin type II cytoskeletal 3 | 300 | A | T | 0.000 | 0.984 | synonymous |
| SS2U026955 | Keratin type II cytoskeletal 3 | 309 | G | A | 0.000 | 0.996 | synonymous |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 192 | C | G | 0.000 | 1.000 | 5prime |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 416 | G | T | 0.000 | 0.961 | G to V |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 945 | C | A | 0.004 | 0.956 | synonymous |
| SS2U043396 | Eukaryotic translation initiation factor 2-alpha
kinase 1 |
134 | A | G | 0.000 | 1.000 | 5prime |
| SS2U043886 | Transcription cofactor HES-6 | 1308 | T | C | 0.000 | 1.000 | 5prime |
| SS2U044339 | Intraflagellar transport protein 52 homolog | 479 | T | C | 0.021 | 1.000 | D to G |
| SS2U045168 | Putative Peptide prediction | 1275 | G | A | 0.000 | 1.000 | 3prime |
| SS2U045328 | E3 ubiquitin-protein ligase DTX3L | 388 | G | A | 0.000 | 0.977 | synonymous |
| SS2U045990 | Low-density lipoprotein receptor-related protein 1 | 135 | T | C | 0.000 | 0.969 | synonymous |
| SS2U048125 a | Transmembrane protein 131-like | 480 | G | A | 0.000 | 1.000 | synonymous |
| SS2U052747 | Uridine 5’-monophosphate synthase | 914 | G | A | 0.000 | 0.951 | synonymous |
| SS2U054542 | Mediator of RNA polymerase II transcription
subunit 20 |
474 | C | T | 0.027 | 0.995 | synonymous |
| SS2U056193 | SUMO-conjugating enzyme UBC9 | 96 | A | T | 0.000 | 1.000 | 3prime |
| SS2U057101 | ETS domain-containing protein Elk-3 | 440 | C | G | 0.000 | 1.000 | 3prime |
| SS2U058860 | Voltage-dependent anion-selective channel
protein 2 |
681 | G | T | 0.000 | 1.000 | 3prime |
| (b) Higher frequency in SB morph | |||||||
| Contig | Annotation | Pos | Ref | Var | Freq-SB | Freq-AC | Effect |
| SS2U000399 | Insulin-like growth factor-binding protein 7 | 598 | C | A | 1.000 | 0.000 | 3prime |
| SS2U004484 | Titin | 387 | G | A | 0.990 | 0.010 | synonymous |
| SS2U026826 | L-asparaginase | 363 | C | T | 1.000 | 0.000 | H to Y |
| SS2U026955 | Keratin type II cytoskeletal 3 | 116 | C | A | 0.996 | 0.031 | T to N |
| SS2U026955 | Keratin type II cytoskeletal 3 | 264 | C | T | 0.970 | 0.008 | synonymous |
| SS2U026955 | Keratin type II cytoskeletal 3 | 317 | C | T | 1.000 | 0.002 | T to M |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 363 | C | T | 1.000 | 0.025 | 5prime |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 387 | C | T | 1.000 | 0.030 | synonymous |
| SS2U033960 | Cysteine sulfinic acid decarboxylase | 657 | T | C | 0.990 | 0.031 | synonymous |
| SS2U034322 | Cyclin-C | 1094 | A | G | 1.000 | 0.000 | 3prime |
| SS2U034431 | Dolichyl-diphosphooligosaccharide–protein
glycosyltransferase subunit 2 |
436 | G | A | 0.992 | 0.000 | G to S |
| SS2U036025 | Nuclear receptor coactivator 4 | 36 | G | A | 1.000 | 0.043 | 5prime |
| SS2U040590 | Glutamyl-tRNA(Gln) amidotransferase subunit A
homolog |
478 | G | A | 0.972 | 0.000 | synonymous |
| SS2U045606 | Superkiller viralicidic activity 2-like 2 | 500 | C | T | 1.000 | 0.000 | synonymous |
| SS2U047816 | Squalene synthase | 1139 | G | A | 1.000 | 0.029 | synonymous |
| SS2U048063 | Lysine-specific demethylase NO66 | 669 | C | T | 1.000 | 0.000 | synonymous |
| SS2U050394 | UPF0542 protein C5orf43 homolog | 596 | G | A | 1.000 | 0.000 | synonymous |
| SS2U050880 a | Transmembrane protein 131-like | 901 | C | T | 1.000 | 0.000 | A to V |
| SS2U052076 | Eukaryotic translation initiation factor 3 subunit A | 824 | C | T | 1.000 | 0.031 | synonymous |
| SS2U053417 | RNA polymerase-associated protein LEO1 | 454 | G | A | 1.000 | 0.049 | synonymous |
| SS2U054333 | Scaffold attachment factor B2 | 382 | G | A | 0.999 | 0.000 | V to M |
| SS2U054705 | Cell division protein kinase 4 | 122 | A | G | 0.971 | 0.000 | 3prime |
| SS2U054965 | DNA-directed RNA polymerase I subunit RPA12 | 106 | G | A | 1.000 | 0.000 | 5prime |
| SS2U054965 | DNA-directed RNA polymerase I subunit RPA12 | 411 | T | G | 1.000 | 0.000 | synonymous |
| SS2U055120 | Chromatin modification-related protein MEAF6 | 350 | A | C | 1.000 | 0.000 | H to P |
| SS2U055153 | Complexin-1 | 1191 | C | A | 1.000 | 0.031 | 3prime |
| SS2U057635 | Mitogen-activated protein kinase 14B | 1370 | A | T | 1.000 | 0.026 | 3prime |
| SS2U058169 | Transmembrane protein 50A | 1214 | C | G | 0.973 | 0.000 | 3prime |
| SS2U058802 | Signal recognition particle 54 kDa protein | 607 | T | A | 0.969 | 0.000 | C to S |
aThose genes are distinct paralogs
Table 6. SNP candidates with significant difference frequency between AC and SB morphs, from reads that mapped to two or more contigs.
| Contig | Annotation | Pos | Ref | Var | Freq-SB | Freq-AC | Effect |
|---|---|---|---|---|---|---|---|
| SS2U004839 | Actin alpha sarcomeric/cardiac | 550 | A | C | 0.015 | 0.999 | 3prime |
| SS2U021298 | 28S ribosomal protein S18a mitochondrial | 462 | A | C | 0.000 | 1.000 | synonymous |
| SS2U041264 | Apoptosis-inducing factor 1 mitochondrial | 341 | C | T | 0.000 | 0.952 | synonymous |
| SS2U054211 a | Cytoplasmic dynein 1 intermediate chain 2 | 136 | T | C | 0.018 | 0.974 | synonymous |
| SS2U054362 a |
Q08CA8 Dynein cytoplasmic 1 intermediate
chain 2 |
945 | A | G | 0.000 | 1.000 | synonymous |
| SS2U055923 | Bystin | 1623 | A | C | 0.000 | 0.983 | 3prime |
| SS2U058758 | Protein S100-A1 | 253 | C | T | 0.000 | 0.984 | synonymous |
| SS2U059000 | Isocitrate dehydrogenase [NADP]
mitochondrial |
1654 | T | C | 0.000 | 0.975 | 3prime |
| SS2U059146 | 60S ribosomal protein L36 | 263 | T | G | 0.009 | 1.000 | synonymous |
| SS2U059146 | 60S ribosomal protein L36 | 470 | A | C | 0.009 | 1.000 | synonymous |
| SS2U036667 | Heterogeneous nuclear ribonucleoprotein K | 813 | C | T | 1.000 | 0.022 | 5prime |
| SS2U042873 | RNA polymerase-associated protein LEO1 | 460 | G | A | 1.000 | 0.000 | synonymous |
| SS2U058455 | Adenylosuccinate lyase | 1616 | C | T | 1.000 | 0.000 | 3prime |
| SS2U058906 | Mid1-interacting protein 1-like | 350 | G | T | 0.985 | 0.000 | E to D |
aThose genes are distinct paralogs
Polymorphism and expression of Arctic charr mtDNA
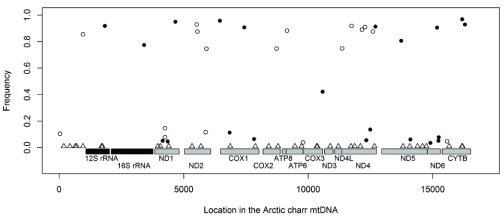
Considering the enrichment of differentially expressed genes related to mitochondrial energy metabolism (above), and high frequency candidate SNPs in several genes with mitochondrial function in AC-charr we decided to study the mitochondrial transcriptome further. The charr studied here reflect metabolic extremes, the aquaculture charr was bred for growth while the small benthic morph is thought to have experienced natural selection for slow metabolism and retarded growth 38, 75. Although mRNA preparation protocols were used for generating cDNA for the RNA-sequencing, a substantial number of reads came from non-polyadenylated sequences. By mapping the reads to mtDNA sequence of Arctic charr we could estimate expression and infer polymorphism both in genes and intergenic regions. There was a clear difference in sequencing coverage, with more than twice as many reads mapped from the AC- compared to SB-charr (mean fold difference 2.27, Wilcoxon test, p < 0.0004). Note, as only two types of fish are compared, the polarity of expression divergence is unknown.
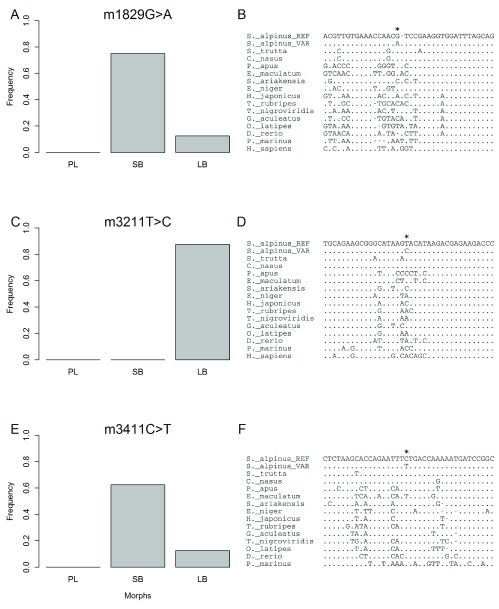
The mapped RNA-reads were used to identify polymorphism and divergence in the entire mitochondrial chromosome. The polymorphisms were found by mapping to mtDNA from a Canadian S. alpinus 48, but ancestral vs. derived status inferred by comparison to S. salar mtDNA. This revealed 82 candidate sites, including 35 that represent divergence between Icelandic and Canadian charr. A total of 20 candidate SNPs had high (more than 50%) frequency difference between SB- and AC-charr ( Figure 7). There was no bias in the distribution of derived SNPs, 11 on the AC branch and 9 in SB. The divergence between Iceland and Canada is particularly little in the 12s and 16s ribosomal RNA genes. Curiously two SNPs in those genes differed strongly in frequency between morphs ( Figure 7). To confirm and better estimate the frequency of variants in the ribosomal genes, we PCR amplified and sequenced two ~550 bp regions in the rRNA genes, comparing three morphs (PL, LB and SB) from Lake Thingvallavatn ( Figure 8A, C & E, S2 Table). The 12s polymorphism (m1829G>A) differed significantly between the morphs ( , p = 0.014), and was at highest frequency in the SB (0% in PL, 12.5% in LB and 75% in SB). Similarly m3411C>T in the 16s was enriched in SB (62.5%) but found at lower frequency in PL (0%) and LB (12.5%) (it differed significantly between morphs, , p = 0.009). The Sanger sequencing also revealed three other polymorphisms in the amplified region, not seen in the transcriptome. Among those m3211T>C in the 16s gene was at 75% frequency in LB, but not found in the other morphs ( , p < 0.0001).
Figure 7. Genetic divergence in the mtDNA between SB- and AC-charr.
The frequency differences between morphs of candidate SNPs, estimated from the RNA-sequencing, graphed along the mtDNA chromosome. The SNPs indicate whether the derived allele is of higher frequency in SB (black dots) or AC (open circles). Sites of divergence between the Icelandic stocks and the Canadian reference sequence are indicated by triangles. The two black boxes represent the rRNA genes and gray boxes the 14 coding sequences (abbreviated names underneath each gene).
Figure 8. Comparative genomics and population genetic differentiation in Arctic charr at 3 mtDNA locations.
Three variants in the 12s and 16s RNA genes are segregating in charr morphs in Lake Thingvallavatn. A, C, E) Frequency of each of those variants in three morphs from Lake Thingvallavatn (PL, LB and SB). A total of 8 individuals were genotyped from each morph, see methods. B, D, F) Aligned are several fish genomes, with Lamprey or humans as outgroups, reflecting a 38 bp window around each of the 3 positions (). Indicated are the two Arctic charr alleles, the reference allele (S._alpinus_REFcharr_WT) and the derived variant (S._alpinus_VARcharr_M). B) Alignment of variant m1829G>A in the 12s rRNA gene in fishes, using humans as an outgroup. D) Similar alignment of a 16s variant, m3211T>C and F) alignment of variant m3411C>T in the 16s rRNA gene.
In order to gauge the potential functionality of those variants we aligned the rRNA genes from nearly hundred fishes and several vertebrates. The position affected by m1829G>A and m3211T>C, in the 12s and 16s rRNAs, are not well conserved in fishes or vertebrates ( Figure 8B & D). However m3411C>T, in the 16s rRNA, alters a position that is nearly invariant in 100 fish genomes ( Figure 8F). The only exception is Pacific menhaden, which curiously also has T in this position. This region could not be aligned properly in other vertebrates. Thus m3411C>T alters a conserved position, but probably not very drastically as the introduced allele is tolerated in another fish species.
The file is tab-delimited and the columns are; “Unigene.Description”: the annotation for that gene/paralog group. “NR.contigs”: number of contigs with this annotation. “logCPM”: count per million, log-scale. "logFC.morph": Mean fold change between the morphs, log-scale. "logFC.T163", "logFC.T200", "logFC.T433": Mean fold change for each timepoints compared to timepoint 141, log-scale. "FDR.morph": P-value for morph difference, multiple testing corrected. "FDR.time": P-value for time differences, multiple testing corrected. "Contigs": SalmonDB id for the contigs with the specific annotation 109.
Copyright: © 2016 Gudbrandsson J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
“Gene Type": Designates the reference and candidate genes. “Gene”: Name of the gene. “Morph”: Which charr type the sample came from. "Relative age": Developmental timepoint, and also indicates the samples from adult fish. "Biological replicate": The two or more biological replicates used. "cDNA No": Marks the cDNA isolation used. "Ct value": Estimate of gene expression. "Sample": Indicates the material used, whole embryos or distinct tissues. "Batch": Demarcates distinct collections of cDNA, applies only to nattl 110.
Copyright: © 2016 Gudbrandsson J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
“Gene Type": Designates the reference and candidate genes. “Gene”: Name of the gene. “Morph”: Which charr type the sample came from. "Relative age": Developmental timepoint. "Biological replicate": The two or more biological replicates used. "cDNA No": Marks the cDNA isolation used. "Ct value": Estimate of gene expression. "Tissue": Indicates the material used 111.
Copyright: © 2016 Gudbrandsson J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Discussion
We are interested in the predictability of evolution at the molecular level, especially whether there exist principles that influence the rewiring of developmental and regulatory systems 4, 76. One way to study this is to identify genetic and developmental effects affecting key traits in species or populations which exhibit parallel evolution. The objective of this study were to get generate hypotheses about the genetic and molecular systems that associate with benthic morphology in charr by mainly focusing on the small benthic morph in Lake Thingvallavatn, Iceland. But as transcriptome were sequenced from embryos of SB-charr and aquaculture charr the data also reflect on the genetics of charr domestication.
Developmental transcriptome of Arctic charr morphs
As no reference genome is available for Arctic charr, we mapped reads to S. salar EST-contigs 57 in order to estimate expression and identify candidate genetic polymorphisms. As many of the contigs are short or have overlapping annotations, we collapsed genes into paralogous genes when appropriate for the expression analysis. The main advantage of this approach was the reduction of the number of statistical tests (and hence an increase in statistical power). The downside is that paralog-specific expression patterns are masked, as our qPCR results of the natterin like gene family show ( Figure 5 and S1 Figure). Recent rainbow trout data shows about 1/4 of paralogs from the latest whole genome duplication event retain the very similar expression patterns 22 indicating that distinct expression patterns of two paralogs is quite common 77. In their analysis of the Arctic charr gill transcriptome, Norman et al. (2014) 13, 14 also used Illumina sequencing technology to evaluate expression. Their reads were longer (2x100 bp) than in this study (36 bp) enabling them to assemble contigs. They did not consider the paralogs in their approach and merged contigs based on sequence identity. Thus the complexity of Arctic charr transcriptome still remains unsolved. Our data reflects differential deployment of several gene classes during Arctic charr development. Studies in salmonids and other fish have demonstrated large changes in expression during early development, including coordinated changes in many cellular and developmental systems 9, 78– 81. Several blood coagulation factors genes showed significant changes during charr development, and were also more highly expressed in the SB-charr. This might reflect differences in the rate of development of blood composition, or tissue composition, in the two morphs. While our main interest is on the derived and repeatedly evolved small benthic charr, the data can also reflect differences due to domestication. In this study we chose to compare SB to AC-charr for several reasons, i) AC-charr has limnetic like head morphology, ii) was available for harvesting of running fish, and iii) because we wanted a strong contrast in this first survey of charr developmental diversity. The AC-charr proved a useful, as the data presented here has already revealed differential expression of several developmental genes and regulators with differential expression between benthic and limnetic charr 51, 52. Previously we found tight correlation of RNA-seq expression and qPCR estimates - using data from this very transcriptome 51. Furthermore, we have actually used the same morphs (AC and SB) and samples in a comparison of the developmental miRNA transcriptome – which reveal that expression of several miRNAs correlates with morph differences 56.
Higher expression of lysozyme II C and natterin-like in SB-charr
Natural selection can shape variation in immunological genes. We decided to study further Lyz2 and the putative immunological genes nattl that had higher expression in SB. The substrate of lysozyme 82 is the bacterial cell wall peptidoglycan and it acts directly on Gram-positive bacteria 83. Lysozyme also promotes the degradation of the outer membrane and therefore indirectly acts also on Gram-negative bacteria 84. Another gene that caught our attention was natterin-like. Natterins were first discovered from the venom gland of the tropical toxic fish species Thalassophryne nattereri 70, 71, and are found by sequence similarity in e.g. zebrafish, Atlantic salmon and here in Arctic charr. The Natterin proteins contain a mannose-binding lectin-like domain (Jacalin-domain). Mannose-binding lectins are pathogen recognition proteins (antibodies) and therefore are important for the acute phase response of fish 85, 86, thus we hypothesized that nattl genes in charr may have immune related functions. The data are consistent with this as the highest expression was found in skin and kidney. This putative immune functions needs to be verified. It is possible that higher expression of those two genes in SB-charr reflect preparation of juveniles for bottom dwelling habitats, which may be rich in bacteria and challenging for immune systems. One can ask whether immunological genes are expected to show similar or less parallelism than others genes shaped by natural selection? The current data does not reflect on this question, but our population genetic work shows genetic variation in immunological genes ( MHCIIα and cath2) does not correlate with the SB-charr ecotype in Iceland 45.
In this study we collapsed contigs into paralog groups for the transcriptome analyses. The disadvantage of this approach is that differential expression of a paralog, can be masked by related genes that do not differ between groups. We looked at this by studying the expression of three paralogs of the natterin like genes in different morphs during Arctic charr development, and among tissues of adult AC-charr. The data suggest that the three nattl genes are expressed differentially between the morphs, thus it is not divergence in the expression of one paralog that explains the general nattl expression disparity in the transcriptome. Certainly, other scenarios could apply to other genes in the transcriptome.
Expression divergence in craniofacial genes in benthic morphs
A study of the skulls of charr post-hatching embryos and juveniles from Lake Thingvallavatn, showed that some elements of the developing head ossified earlier in SB-charr than in PL-charr 87. Morphometric analyses of developing heads (same stages as studied here) demonstrate differences in craniofacial elements between AC- and SB-charr, along a limnetic vs. benthic axis 74. Based on those developmental phenotypes we investigated further genes with roles in craniofacial development that were differentially expressed in the transcriptome. Guided by this transcriptome we had already found two extra-cellular matrix (ECM) remodeling genes, Mmp2 and Sparc and a conserved co-expression module of genes with known roles in craniofacial morphogenesis, to have higher expression in developing heads of benthic Arctic charr morphs than in limnetic morphs 51, 52. Bioinformatic and qPCR analyses suggest the co-expression module may potentially be affected by quantity of the transcription factor ETS2. These studies and the current data confirm the utility of the contrasting developmental transcriptomes for identifying candidate genes with differential expression during head development, as 7 out of 8 candidates were confirmed by qPCR. These genes had consistently higher expression in the developing head of two benthic morphs (SB and LB), and lower in more limnetic fish (AC and PL). The most noteworthy aspect is the fact that three of the morphs (SB, LB and PL) are closely related and live in sympatry in Lake Thingvallavatn 44.
We focused on a few targets of Tgf- β and Ahr signaling pathways because of their role in craniofacial morphogenesis and transcriptional connection 88– 90. Adseverin (Scin) was one of the top differentially expressed genes ( Table 1) and has roles in rearrangements of the actin cytoskeleton, chondrocyte differentiation and skeletal formation 91, 92. Also, in the transcriptome Lsr, Cldn4 and Tgfbr2 had higher expression in SB-charr, and we show that higher expression of those genes associated with the benthic morphotype. Lsr is a molecular component of tri-cellular tight junctions 93 and has been shown to be suppressed upon Tgf- β1 stimulation 94 in a human cell line. Similarly, Cldn4, a tight junction protein with unknown role during embryonic morphogenesis, is a target of the Tgf- β and Ahr signaling pathways 95, 96. Finally, the expression of Tgfbr2, encoding a receptor of Tgf- β was slightly but significantly higher in the head of benthic morphs. Previous studies suggest a crucial role of Tgfbr2 in craniofacial morphogenesis 97.
We also confirmed differential expression of other genes, including two with higher expression in SB-charr. Mvp is the predominant component of cytoplasmic ribonucleoprotein structures called vaults 98, which is highly conserved across eukaryotes. The vaults have been something of an enigma, but are implicated in several processes from signal transmission and immune response 99. The Jup gene also showed higher expression in SB-charr. Finally, higher expression of Vdra, encoding the vitamin D receptor A, was found in the heads of benthic charr. The receptor regulates mineral homeostasis, osteoblast differentiation and bone metabolism 100. A related study from our group, also building on this dataset, mapped in more detail the differential expression of these and other coexpressed genes in limnetic and benthic charr 53.
To summarize, the results show that RNA-sequencing of Aquaculture charr with limnetic craniofacial morphology and small benthic charr can be used to reveal differential expression of genes that associate with limnetic and benthic divergence in craniofacial elements in sympatric charr morphs. It would be interesting if expression of these genes associates with benthic morphology in independently evolved charr populations, as was seen for certain mTOR-pathway genes in muscle of adult SB-charr 47, or even in other species with similar trophic diversity.
Genetics differences between the AC and SB-morphs - possibly in mtDNA function
Previous studies on microsatellite markers documented the population history of charr in Iceland and in particular the parallel evolution of SB-charr 44. Our data confirm genetic differences between SB and AC-charr. By comparing AC and SB-charr, that represents a small benthic resource morph that has evolved repeatedly in Icelandic stream and pond habitats 44, we hoped to implicate genes and pathways involved in adaptation to these special habitats. But the AC-charr is also interesting, as domestication over several decades has led to rapid growth and increased size 50. Morphometrics have not been used to compare the body or craniofacial shape of AC to other charr morphs, but domestication of O. mykiss has affected body shape and fin structure in partiuclar 101. The allele frequency differences and expression divergence observed can reflect neutral population genetic processes and/or selection during domestication or adaptation of SB-charr. By studying expression and allele frequencies in limnetic and benthic morphs from more locations, it may be possible to disentangle these questions. We restricted ourselves to verification of several SNPs, and focused mostly on variants in mtDNA because to us the data suggest interesting divergence in systems related to energy metabolism. First, there is 2X higher expression of respiratory electron transport chain components in AC compared to SB-charr and 100% more mitochondrial derived reads are found in the AC-charr samples. Note that the direction of divergence is unknown, i.e. whether expression was up in AC or down in SB. Second, many derived candidate-SNPs in genes related to mitochondrial function were at high frequency on the AC branch. For instance in S100A1, which has been implicated in mitochondrial regulation in cardiac tissue in humans 102, but its expression is probably not exclusive to this tissue. Third, while the mitochondrial ribosomal genes generally evolve slowly, we do see derived variants at high frequency in the SB and large benthic charr in Lake Thingvallavatn. Specifically, m3411C>T in SB affects a position that is highly conserved among fish, and could affect function of the 16s rRNA. Earlier studies of mitochondrial markers in S. alpinus did not find large signals of divergence within Iceland 40, 42, 45, probably because they studied other genes.
The mitochondrion is more than a powerhouse, it integrates metabolism, cell cycle and apoptosis 103. The number of mitochondria and its functions are known to correlate with environmental attributes. For instance in Antarctic fishes under extreme cold, higher numbers of mitochondria are found in muscle and heart cells 104. Our data suggest an expression difference between morphs that could reflect differences in total number of mitochondrion, the number of mtDNA copies per mitochondrion or cell, or difference in RNA expression from the mtDNA, possibly due to evolution of mtDNA related to diet and/or temperature 105. In sum, the results suggest divergence (adaptive or neutral) in mitochondrial function, due to the domestication of aquaculture charr and/or adaptation of the small benthic charr to its habitat in Lake Thingvallavatn. But further work is needed to map out the expression differences of mitochondrial related genes in more SB and anadromous charr morphs (representing the ancestral state). The mtDNA signals could also be investigated in populations along ecological clines (e.g. temperature) or with respect to life history 106.
Conclusions
The data presented here suggest genetic and expression changes in multiple systems associate with divergence among the highly polymorphic and rapidly evolving Arctic charr in Iceland. The data reveal differential expression of two immunological genes between morphs and of several craniofacial developmental genes, that may help sculpture benthic vs. limnetic heads. The genetic data suggest among other things differentiation in the charr mtDNA between the SB and AC-charr morphs. It must be acknowledged that it is not trivial to identify genes affecting variation in ecologically important phenotypes, like shape 107, 108. Our broad interest is in how natural selection tweaks genetic regulatory systems, for instance via genetic changes in regulatory sequences or post transcriptional modifiers relating to adaptations. Genetic changes affecting gene expression can be raw material for adaptation, but could also rise in frequency due to reverberations in regulatory cascades 76. Following this work we plan to study the degree of developmental and population genetics parallelism of the small benthic charr, typically found in cold springs and small pond habitats in Iceland with lava substratum 29, 44. The availability of charr populations at different stages of divergence sets the stage for future genomic studies of the roles of genes, environment and plasticity for shaping this polymorphic species.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Gudbrandsson J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
The sequencing reads were deposited into the NCBI SRA archive under BioProject identifier PRJNA239766 and with accession numbers: SRX761559, SRX761571, SRX761575, SRX761577, SRX761451, SRX761461, SRX761490 and SRX761501.
All DNA sequences where deposited to Genbank as popsets under the accession numbers KP019972-KP020026.
F1000Research: Dataset 1. Parameters and multiple testing corrected p-values for expression analysis, 10.5256/f1000research.6402.d48005 109
F1000Research: Dataset 2. qPCR data for tests of expression in charr developing embryos and adult tissues., 10.5256/f1000research.6402.d48006 110
F1000Research: Dataset 3. qPCR data for tests of expression in charr developing embryo heads., 10.5256/f1000research.6402.d48007 111
Acknowledgements
We would like to thank Baldur Kristjansson for help with genomic alignments. We are very grateful to Droplaug N. Magnusdottir, Gudbjorg Th. Orlygsdottir, Steinunn Snorradottir and Olafur Th. Magnusson at deCODE Genetics for help with the Illumina sequencing. Special thanks to Fredrik Holm for assembling Figure 1.
Funding Statement
This project was supported by The Icelandic Center for Research (grant number: 100204011) to SSS, AP, ZOJ and BKK, The University of Iceland Research/Doctoral Fund to JG and KHK and University of Iceland research fund to AP, SSS and ZOJ.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 1 approved
Supporting Information
S1 Figure. Relative expression of nattl and nattl 1–3 in tissues of adult AC-charr.
Relative expression of Natterin ( A) & Natterin paralogs 1–3 ( B– D) within different tissues (skin, heart, liver, gill, spleen, intestine & kidney) of adult aquaculture charr (RT-qPCR); expression plotted for different tissues, relative to heart tissue (lowest expression levels).
S2 Figure. Relative expression of selected craniofacial candidate genes.
Relative expression of 12 candidate genes with characterized craniofacial expression during zebrafish development (ZFIN website) in the head of SB, LB, PL and AC at three time points in development. In the transcriptome data all of the genes had shown higher expression in SB at 200 τs. The expression is normalized to the geometric means of two craniofacial reference genes ( ACTB and IF5A1). Expression is relative to a replicate of AC morph at 200 (τs), set to one. Error bars represent standard deviation calculated from two biological replicates and each biological replicate contains homogenate of six heads.
Supplemental Table S1 A. qPCR primers used in this study.
| Gene | Description | Primer Sequence (5’- 3’) | Product
Size (bp) |
PCR
Efficiency |
Melting
Temperature (°C) |
Exon
Boundary |
|---|---|---|---|---|---|---|
| Actb | Beta Cytoskeletal Actin | F-GAAGATCAAGATCATCGCCC
R-CAGACTCGTCGTACTCCTGCT |
122 | 1.95 | 80.5 ± 0.7 | Yes |
| Alp | Alkaline phosphatase | F-ACAGCATACCTCTGTGGGG
R-GGTGGCATGGTTCACACG |
177 | 1.90 | 85.12 ± 0.5 | Yes |
| Cldn4 | Claudin-4 | F-GTGCTGTGC CATCCCAAG
R-CACCACACAGGTCATCCACA |
100 | 1.98 | 80.4 ± 0.6 | Yes |
| Cgat2 | Chondroitin beta-1,4-N-
acetylgalactosaminyltransferase 2 |
F-GAGAGCCACTTTACTGAGGGG
R-GAATGGACGGAAAAGAGTAACG |
120 | 1.98 | 81.86 ± 0.3 | Yes |
| Cox6b1 | Cytochrome c oxidase subunit VIb
isoform 1 |
F-GAGGGTCTACAAATCACTGTGC
R-CCTGGAGTCCTACTCATACAAACAT |
147 | 1.93 | 82.22 ± 0.7 | Yes |
| Ef1α | Eukaryotic Translation Elongation
Factor 1 Alpha |
F-GAAGATCGGCTATAACCCTGC
R-ACCTTCCATCCCTTGAACC |
111 | 1.94 | 81.36 ± 0.4 | Yes |
| If5a1 | Eukaryotic Translation Initiation
Factor 5A |
F-GGCTTCGTGGTGCTGAAG
R-CCATGTGGACCTTAGCGTG |
91 | 1.91 | 80.76 ± 0.6 | Yes |
| Jup | Junction plakoglobin | F-CACAGCAGACATACCAGGATG G
R-CTGGCGATCTCTCCCCTGTT |
109 | 1.97 | 81.0 ± 0.3 | Yes |
| Krtap4–3 | Keratin-associated protein 4–3 | F-GCGGGACATCTACACTGCTTA
R-AGAAGGCTAAAGTCTTAGTGACTATC |
151 | 1.89 | 81.88 ± 0.6 | Yes |
| Lsr | Lipolysis-stimulated lipoprotein
receptor |
F-TGCTGTCACTCTGGGCGA
R-CCGTCTGGGCAAGGTTCA G |
80 | 1.91 | 80.77 ± 0.5 | Yes |
| Lyz | Lysozyme | F-TTCCAGATCAACAGCCGCTA
R-GATCGCCACTGTGATGTCAT |
111 | 1.94 | 81.87 ± 0.7 | Yes |
| Mvp | Major vault protein | F-ACCAACTCCCAGGAGGCT
R-CCTCTCCAGACGACCACG |
75 | 1.97 | 78.93 ± 0.3 | Yes |
| Nattl | Natterin-like protein | F-GTGAAAGTCACCTGCATGAATG
R-CATCTCTCCTTTGTGGATACCC |
104 | 1.98 | 78.81 ± 0.8 | No |
| Nattl-1 | Natterin-like protein paralog-1 | F-AATCCGTGTCCTACCACAATGA
R-GGTGTGTCGGTCAAAGCA |
135 | 1.77 | 78.03 ± 0.1 | No |
| Nattl-2 | Natterin-like protein paralog-1 | F-TGAAATVTVTGTCTCATCACAAC
R-GGATCTGGTCGAGGTGGC |
163 | 1.72 | 80.50 ± 0.2 | No |
| Nattl-3 | Natterin-like protein paralog-1 | F-GTGACATCCGTTTCTCACCAG
R-GATGTGTCGGTCAAAGCG |
138 | 1.77 | 79.12 ± 0.2 | No |
| Ndub6 | NADH dehydrogenase 1 beta
subcomplex subunit 6 |
F-TGGTGGAGTGTTCGCCTT
R-CTCTCTGGGAGGTCTGGAA |
171 | 1.89 | 82.40 ± 0.3 | Yes |
| Parp6 | Poly (ADP-Ribose) Polymerase
Family, Member 6 |
F-CCGTATGAATACCGTTCCACAGG
R-CACCCAGATGTTGCCGTGCTT |
147 | 1.93 | 81.87 ± 0.7 | Yes |
Supplemental Table S1 B.
| Gene | Description | Primer Sequence (5’- 3’) | Product
Size (bp) |
PCR
Efficiency |
Melting
Temperature (°C) |
Exon
Boundary |
|---|---|---|---|---|---|---|
| Rarg | Retinoic acid receptor
gamma-A |
F-AAGGCGAGCCCCTTCTTC
R-TGCTCTGGGTCTCCACCG |
82 | 1.92 | 78.62 ± 0.3 | Yes |
| Scin | Scinderin/Adseverin | F-CACCTGATCCCAGACATCCAA
R-CCTCACTCAACAACCTCGC |
136 | 1.90 | 83.24 ± 0.7 | No |
| Tgfbr2 | TGF-beta receptor
type-2 |
F-CTGCTCCGAGGACGAGTG
R-ACCGACACCACCTGGGAG |
72 | 1.93 | 79.02 ± 0.5 | Yes |
| Ubl5 | Ubiquitin-like protein 5 | F-AATAAGGATGATTGAGGTGGTTTG
R-ATGAGCTTCTTCAGGTCTCC |
99 | 1.95 | 78.44 ± 0.3 | Yes |
| Ub2l3 | Ubiquitin-Conjugating
Enzyme E2L 3 |
F-CGAGAAGGGACAGGTGTGTC
R-ACCAACGCAATCAGGGACT |
96 | 1.93 | 79.62 ± 0.3 | Yes |
| Vdra | Vitamin D3 receptor A | F-CGTCACCAAGGCGGGTCA
R-TGGAGCTTG AGTTTCTTCAGGC |
81 | 1.93 | 78.12 ± 0.3 | Yes |
Supplemental Table S2 A. Verification of candidate polymorphisms.
Primer sequences, melting temperatures and primary data.
| Sequence | Position | Forward primer | Reverse primer | Tm
forward |
Tm
reverse |
Paralogs |
|---|---|---|---|---|---|---|
| NC_000861.1 | 1829 | GTGCCTCAGACCCACCTAGA | TCTGTCGCCCGTACTAAGGT | 60.26 | 59.76 | No |
| NC_000861.1 | 3119 | GGCCAGAGTAAACACCGAGA | CCTGGATTACTCCGGTCTGA | 60.25 | 60.07 | No |
| NC_000861.1 | 3411 | GGCCAGAGTAAACACCGAGA | CCTGGATTACTCCGGTCTGA | 60.25 | 60.07 | No |
| NC_000861.1 | 8876 | GACGTCCTTCACTCCTGAGC | GGGCTCATAAACTGGTCGAA | 59.99 | 60.07 | No |
| NC_000861.1 | 15240 | ACCCTAAAACCGAACGATCC | TGGCTAGGAAGAGTCCGGTA | 60.19 | 59.83 | No |
| SS2U034121 | 233 | CTCAACGTGCTTGACCAGTG | CCCTTACCCTCCAGGATCTC | 60.5 | 59.89 | Yes |
| SS2U054644 | 1037 | AAGGACGGCCACTATGGTCT | GGGGCATAGAGTGCACAGG | 60.9 | 61.65 | Yes |
| SS2U054644 | 1188 | TCAGAGATAGTGAAGAAGATGCTG | CGTACTTGATAAGACCTGTCGGTA | 57.92 | 59.62 | No |
| SS2U054644 | 1283 | TCAGAGATAGTGAAGAAGATGCTG | CGTACTTGATAAGACCTGTCGGTA | 57.92 | 59.62 | No |
| SS2U055283 | 1822 | TGTGTGAGGTGGTTGAGGAG | GGGTCATTGCTCCCTACAGA | 59.7 | 60.07 | No |
| SS2U055923 | 615 | GTGGACCCAGAGGATGAGAA | AGAACCTGCTCCCAGTTTGA | 60.05 | 59.84 | No |
| SS2U058906 | 350 | GCCAAAACCTCCACAATGAT | AACTGGCCTTCCAGATCAGA | 59.8 | 59.8 | Yes/No |
Paralogs: indicates whether the PCR and sequencing yielded mixed products, indicative of paralogous genes.
Supplemental Table S2 B.
| Sequence | Genome contig | Gene name | Position | Ref | Var | Freq_AC | Freq_SB | FreqP_PL | FreqP_SB | FreqP_LB |
|---|---|---|---|---|---|---|---|---|---|---|
| NC_000861.1 | n.a. | 12S ribosomal RNA | 1829 | G | A | 0 / 53 | 77 / 81 | 0 / 6 | 3 / 4 | 1 / 8 |
| NC_000861.1 | n.a. | 16S ribosomal RNA | 3119 | A | T | 46 / 87 | 18 / 28 | 0 / 8 | 0 / 8 | 0 / 8 |
| NC_000861.1 | n.a. | 16S ribosomal RNA | 3411 | C | T | 0 / 119 | 26 / 33 | 0 / 8 | 5 / 8 | 1 / 8 |
| NC_000861.1 | n.a. | tRNA-Lys | 8876 | C | A | 73 / 779 | 74 / 352 | 0 / 4 | 0 / 4 | n.a. |
| NC_000861.1 | n.a. | NADH
dehydrogenase 6 |
15240 | G | A | 2 / 3608 | 137 / 2702 | 2 / 4 | 0 / 4 | n.a. |
| SS2U034121 | AGKD01052493.1 | Eukaryotic
translation initiation factor 4 gamma 2 |
233 | C | T | 0 / 95 | 22 / 40 | 2 / 4 | 2 / 2 | n.a. |
| SS2U054644 | AGKD01031893.1 | Uroporphyrinogen
decarboxylase |
1037 | G | A | 28 / 33 | 0 / 56 | n.a. | n.a. | n.a. |
| SS2U054644 | AGKD01031893.1 | Uroporphyrinogen
decarboxylase |
1188 | C | T | 0 / 53 | 19 / 25 | 4 / 4 | 4 / 4 | n.a. |
| SS2U054644 | AGKD01031893.1 | Uroporphyrinogen
decarboxylase |
1283 | G | A | 4 / 60 | 12 / 15 | 4 / 4 | 4 / 4 | n.a. |
| SS2U055283 | AGKD01013777.1 | DNA2-like helicase | 1822 | G | A | 1 / 65 | 25 / 50 | 3 / 4 | n.a. | n.a. |
| SS2U055923 | AGKD01022586.1 | Bystin | 615 | G | A | 106/109 | 7 / 190 | 0 / 4 | n.a. | n.a. |
| SS2U058906 | AGKD01005918.1 | Mid1-interacting
protein 1-like |
350 | G | T | 0 / 49 | 67 / 68 | 4 / 4 | 4 / 4 | n.a. |
Sequence: name of the genebank sequence or EST-contig used as reference for mapped reads.
Genome contig: name of salmon genome (ICSASG_v1) contig with best sequence match to the respective EST-contig.
Ref: Reference variant.
Var: The derived variant.
Freq_AC and Freq_SB: Frequency of variant reads as fraction of total numbers of reads mapped in Aquaculture (AC) or Small benthic (SB).
FreqP: The frequency of variant in genotyping by PCR and direct sequencing, as a fraction of total number of chromosomes sequenced.
Supplemental Table S3. Mapping of Illumina reads to S. salar EST data.
Numbers of reads aligning to salmon reference for each sample.
| Alignment
per read |
SB 141 | SB 163 | SB 200 | SB 433 | AC 141 | AC 163 | AC 200 | AC 433 |
|---|---|---|---|---|---|---|---|---|
| 0 | 33088778 | 30492314 | 27175901 | 25569628 | 32159386 | 30051365 | 31267710 | 28563169 |
| 1 | 6979368 | 11791558 | 11449549 | 11058555 | 11599602 | 11320997 | 11027195 | 10650748 |
| 2 | 2742358 | 4021683 | 3814418 | 3734404 | 4328402 | 4523686 | 3959198 | 3655786 |
| 3 | 2099068 | 2964994 | 2748108 | 2651522 | 3111277 | 3332577 | 2878729 | 2515303 |
| 4 | 1228292 | 1777846 | 1720902 | 1968251 | 1977738 | 2182392 | 1929818 | 1980420 |
| 5 | 914704 | 1317556 | 1284262 | 1434314 | 1471739 | 1679277 | 1447604 | 1426744 |
| 6 | 645264 | 946579 | 938290 | 1087959 | 1001350 | 1083025 | 1045157 | 1081063 |
| 7 | 425856 | 595785 | 578175 | 726290 | 657220 | 750523 | 690286 | 735351 |
| 8 | 293065 | 428003 | 424426 | 590100 | 530040 | 591332 | 527821 | 579860 |
| 9 | 206205 | 319401 | 334861 | 455838 | 296169 | 334264 | 387901 | 485653 |
| 10+ | 749074 | 1419362 | 1761275 | 3041930 | 1092980 | 1189781 | 1967857 | 3294222 |
| Total reads | 49372032 | 56075081 | 52230167 | 52318791 | 58225903 | 57039219 | 57129276 | 54968319 |
Supplemental Table S4. ANOVAs on qPCR data.
Expression of nine genes was analyzed in whole SB- and AC-charr embryos, at two developmental timepoints (161 and 200 τs).
| Gene | Term | Df | F value | p value | Significance | FDR RNA-seq |
|---|---|---|---|---|---|---|
| Alp | Morph | 1 | 13.4797 | 0.0214 | * | 0.0697 |
| Time | 1 | 14.9526 | 0.0180 | * | 0.0012 | |
| M x T | 1 | 3.9519 | 0.1177 | . | ||
| Cgat2 | Morph | 1 | 0.0257 | 0.8804 | . | 0.0035 |
| Time | 1 | 1.5141 | 0.2859 | . | 0.3312 | |
| M x T | 1 | 0.1866 | 0.6880 | . | ||
| Cox6B1 | Morph | 1 | 0.0898 | 0.7793 | . | 0.0580 |
| Time | 1 | 3.8312 | 0.1219 | . | 0.6320 | |
| M x T | 1 | 0.7359 | 0.4393 | . | ||
| Krtap4–3 | Morph | 1 | 30.0255 | 0.0054 | ** | 0.0121 |
| Time | 1 | 0.3902 | 0.5661 | . | 0.2784 | |
| M x T | 1 | 4.5225 | 0.1006 | . | ||
| Lyz | Morph | 1 | 64.1566 | 0.0013 | ** | 0.0406 |
| Time | 1 | 1.0390 | 0.3657 | . | 0.0005 | |
| M x T | 1 | 1.2026 | 0.3344 | . | ||
| Nattl | Morph | 1 | 8.1148 | 0.0465 | * | 7.718e-07 |
| Time | 1 | 14.6659 | 0.0186 | * | 6.714e-14 | |
| M x T | 1 | 0.2958 | 0.6154 | . | ||
| Ndub6 | Morph | 1 | 0.7447 | 0.4368 | . | 0.0982 |
| Time | 1 | 7.3316 | 0.0537 | . | 0.6698 | |
| M x T | 1 | 0.2269 | 0.6587 | . | ||
| Parp6 | Morph | 1 | 11.2682 | 0.0284 | * | 0.1076 |
| Time | 1 | 0.7393 | 0.4384 | . | 0.3789 | |
| M x T | 1 | 0.2343 | 0.6537 | . | ||
| Ubl5 | Morph | 1 | 1.1420 | 0.3454 | . | 0.0587 |
| Time | 1 | 0.2434 | 0.6476 | . | 0.0025 | |
| M x T | 1 | 0.3974 | 0.5627 | . |
Significance: p > 0.05; * p < 0.05; ** p < 0.01.
FDR RNA-seq: indicates significance of Morph and Time effects in the transcriptome data.
Supplemental Table S5. ANOVAs on Natterin-like qPCR on adults.
Studied were levels of Natterin-like and Natterin-like Paralogs 1–3 in Arctic charr whole embryos (among SB, AC and PL morphs) and tissues from adult AC-charr.
| Gene | Term | Df | F value | p value | Significance |
|---|---|---|---|---|---|
| Nattl | Morph | 2 | 11.5515 | 0.0002 | *** |
| Time | 5 | 8.3202 | 3.99e-05 | *** | |
| M x T | 9 | 4.4758 | 0.0007 | *** | |
| Nattl1 | Morph | 2 | 19.4070 | 0.0001 | *** |
| Time | 3 | 5.9346 | 0.0089 | ** | |
| M x T | 5 | 4.5761 | 0.0126 | * | |
| Nattl2 | Morph | 2 | 14.2921 | 0.0005 | *** |
| Time | 3 | 15.0463 | 0.0001 | *** | |
| M x T | 5 | 3.2462 | 0.0404 | * | |
| Nattl3 | Morph | 2 | 34.4888 | 6.33e-06 | *** |
| Time | 3 | 4.4204 | 0.0238 | * | |
| M x T | 5 | 4.1843 | 0.0174 | * | |
| Nattl | Tissue | 6 | 15.468 | 1.42e-08 | *** |
| Nattl1 | Tissue | 6 | 12.022 | 0.0002 | *** |
| Nattl2 | Tissue | 6 | 7.6811 | 0.0011 | ** |
| Nattl3 | Tissue | 6 | 46.182 | 8.89e-06 | *** |
Significance: p > 0.05; * p < 0.05; ** p < 0.01.
Supplemental Table S6. Gene Ontology analyses of derived SNPs in SB-charr.
| Category | Observed | In category | TERM | FDR adjusted p-value |
|---|---|---|---|---|
| GO:0006412 | 24 | 189 | translation | 4.34E-006 |
| GO:0006396 | 8 | 32 | RNA processing | 0.0016 |
| GO:0006414 | 6 | 19 | translational elongation | 0.0038 |
| GO:0006313 | 5 | 20 | transposition, DNA-mediated | 0.0498 |
| GO:0015074 | 5 | 21 | DNA integration | 0.0510 |
| GO:0006260 | 6 | 35 | DNA replication | 0.0679 |
| GO:0055114 | 20 | 285 | oxidation-reduction process | 0.0679 |
Supplemental Table S7. Predicted effect of SNP-candidates differing in frequency between charr morphs.
| Effect on transcribed region | Uni_SB | Uni_AC | Rep_SB | Rep_AC |
|---|---|---|---|---|
| 5´prime | 32 | 19 | 35 | 24 |
| Synonymous | 232 | 179 | 176 | 113 |
| Non-synonymous | 112 | 72 | 81 | 72 |
| 3´prime | 147 | 123 | 59 | 74 |
From RNA-reads that mapped to one (Uni) or more (Rep) S. salar ESTs.
The candidate SNPs frequencies differ more than 50% between SB and AC-charr, summarized by which morph with higher frequency of the derived allele.
References
- 1. Gould SJ: Ontogeny and Phylogeny. Harvard University Press,1977. Reference Source [Google Scholar]
- 2. Jacob F: Evolution and tinkering. Science. 1977;196(4295):1161–1166. 10.1126/science.860134 [DOI] [PubMed] [Google Scholar]
- 3. Stern DL, Orgogozo V: The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62(9):2155–77. 10.1111/j.1558-5646.2008.00450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stern DL, Orgogozo V: Is genetic evolution predictable? Science. 2009;323(5915):746–751. 10.1126/science.1158997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rockman MV: The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2012;66(1):1–17. 10.1111/j.1558-5646.2011.01486.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abzhanov A, Protas M, Grant BR, et al. : Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305(5689):1462–1465. 10.1126/science.1098095 [DOI] [PubMed] [Google Scholar]
- 7. Albertson RC, Kocher TD: Genetic and developmental basis of cichlid trophic diversity. Heredity (Edinb). 2006;97(3):211–221. 10.1038/sj.hdy.6800864 [DOI] [PubMed] [Google Scholar]
- 8. Cresko WA, McGuigan KL, Phillips PC, et al. : Studies of threespine stickleback developmental evolution: progress and promise. Genetica. 2007;129(1):105–126. 10.1007/s10709-006-0036-z [DOI] [PubMed] [Google Scholar]
- 9. Domazet-Lošo T, Tautz D: A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468(7325):815–8. 10.1038/nature09632 [DOI] [PubMed] [Google Scholar]
- 10. Bozinovic G, Sit TL, Hinton DE, et al. : Gene expression throughout a vertebrate’s embryogenesis. BMC genomics. 2011;12:132. 10.1186/1471-2164-12-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giger T, Excoffier L, Day PJ, et al. : Life history shapes gene expression in salmonids. Curr Biol. 2006;16(8):R281–2. 10.1016/j.cub.2006.03.053 [DOI] [PubMed] [Google Scholar]
- 12. Filteau M, Pavey SA, St-Cyr J, et al. : Gene coexpression networks reveal key drivers of phenotypic divergence in lake whitefish. Mol Biol Evol. 2013;30(6):1384–96. 10.1093/molbev/mst053 [DOI] [PubMed] [Google Scholar]
- 13. Norman JD, Ferguson MM, Danzmann RG: Transcriptomics of salinity tolerance capacity in Arctic charr ( Salvelinus alpinus): a comparison of gene expression profiles between divergent QTL genotypes. Physiol Genomics. 2014;46(4):123–37. 10.1152/physiolgenomics.00105.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Norman JD, Ferguson MM, Danzmann RG: An integrated transcriptomic and comparative genomic analysis of differential gene expression in Arctic charr ( Salvelinus alpinus) following seawater exposure. J Exp Biol. 2014;217(Pt 22):4029–4042. 10.1242/jeb.107441 [DOI] [PubMed] [Google Scholar]
- 15. Skúlason S, Smith TB: Resource polymorphisms in vertebrates. Trends Ecol Evol. 1995;10(9):366–370. 10.1016/S0169-5347(00)89135-1 [DOI] [PubMed] [Google Scholar]
- 16. Snorrason SS, Skúlason S: Adaptive Speciation in Northern Freshwater Fishes. In Ulf Dieckmann, Michael Doebeli, Johan A. J. Metz, and Diethard Tautz, editors, Adaptive speciation.Cambridge University Press, Cambridge, chapter 10,2004;210–228. 10.1017/CBO9781139342179.012 [DOI] [Google Scholar]
- 17. Klemetsen A: The Charr Problem Revisited: Exceptional Phenotypic Plasticity Promotes Ecological Speciation in Postglacial Lakes. Freshwater Rev. 2010;3(1):49–74. 10.1608/FRJ-3.1.3 [DOI] [Google Scholar]
- 18. Bernatchez L, Renaut S, Whiteley AR, et al. : On the origin of species: insights from the ecological genomics of lake whitefish. Philos Trans R Soc Lond B Biol Sci. 2010;365(1547):1783–800. 10.1098/rstb.2009.0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merilä J: Nine-spined stickleback ( Pungitius pungitius): an emerging model for evolutionary biology research. Ann N Y Acad Sci. 2013;1289:18–35. 10.1111/nyas.12089 [DOI] [PubMed] [Google Scholar]
- 20. Fraser DJ, Weir LK, Bernatchez L, et al. : Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity (Edinb). 2011;106(3):404–20. 10.1038/hdy.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macqueen DJ, Johnston IA: A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc Biol Sci. 2014;281(1778): 20132881. 10.1098/rspb.2013.2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berthelot C, Brunet F, Chalopin D, et al. : The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5: 3657. 10.1038/ncomms4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davidson WS, Koop BF, Jones SJ, et al. : Sequencing the genome of the Atlantic salmon ( Salmo salar). Genome Biol. 2010;11(9):403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moghadam HK, Ferguson MM, Danzmann RG: Whole genome duplication: Challenges and considerations associated with sequence orthology assignment in Salmoninae. J Fish Biol. 2011;79(3):561–574. 10.1111/j.1095-8649.2011.03030.x [DOI] [PubMed] [Google Scholar]
- 25. Vijay N, Poelstra JW, Künstner A, et al. : Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol. 2013;22(3):620–34. 10.1111/mec.12014 [DOI] [PubMed] [Google Scholar]
- 26. Noakes DLG: Charr truth: Sympatric differentiation in Salvelinus species. Environ Biol Fishes. 2008;83(1):7–15. 10.1007/s10641-008-9379-x [DOI] [Google Scholar]
- 27. Ghalambor CK, Hoke KL, Ruell EW, et al. : Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525(7569):372–5. 10.1038/nature15256 [DOI] [PubMed] [Google Scholar]
- 28. Adams CE, Fraser D, Wilson AJ, et al. : Patterns of phenotypic and genetic variability show hidden diversity in Scottish Arctic charr. Ecol Freshwater Fish. 2007;16(1):78–86. 10.1111/j.1600-0633.2006.00182.x [DOI] [Google Scholar]
- 29. Kristjánsson BK, Malmquist HJ, Ingimarsson F, et al. : Relationships between lake ecology and morphological characters in Icelandic Arctic charr, Salvelinus alpinus. Biol J Linn Soc. 2011;103(4):761–771. 10.1111/j.1095-8312.2011.01670.x [DOI] [Google Scholar]
- 30. Woods PJ, Skúlason S, Snorrason SS, et al. : Intraspecific diversity in Arctic charr, Salvelinus alpinus, in Iceland: II. Which environmental factors influence resource polymorphism in lakes? Evolutionary Ecol Res. 2012;14(8):993–1013. Reference Source [Google Scholar]
- 31. Kristjánsson BK, Skúlason S, Snorrason SS, et al. : Fine-scale parallel patterns in diversity of small benthic Arctic charr ( Salvelinus alpinus) in relation to the ecology of lava/groundwater habitats. Ecol Evol. 2012;2(6):1099–112. 10.1002/ece3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skúlason S, Noakes DL, Snorrason SS: Ontogeny of trophic morphology in four sympatric morphs of Arctic charr Salvelinus alpinus in Thingvallavatn, Iceland. Biol J Linn Soc. 1989;38(3):281–301. 10.1111/j.1095-8312.1989.tb01579.x [DOI] [Google Scholar]
- 33. Skúlason S, Snorrason SS, Ota D, et al. : Genetically based differences in foraging behaviour among sympatric morphs of Arctic charr (Pisces: Salmonidae). Animal Behaviour. 1993;45(6):1179–1192. 10.1006/anbe.1993.1140 [DOI] [Google Scholar]
- 34. Skúlason S, Snorrason SS, Noakes DLG, et al. : Genetic basis of life history variations among sympatric morphs of Arctic char Salvelinus alpinus. Can J Fish Aquat Sci. 1996;53(8):1807–1813. 10.1139/f96-098 [DOI] [Google Scholar]
- 35. Parsons KJ, Skúlason S, Ferguson M: Morphological variation over ontogeny and environments in resource polymorphic Arctic charr ( Salvelinus alpinus). Evol Dev. 2010;12(3):246–257. 10.1111/j.1525-142X.2010.00410.x [DOI] [PubMed] [Google Scholar]
- 36. Parsons KJ, Sheets HD, Skúlason S, et al. : Phenotypic plasticity, heterochrony and ontogenetic repatterning during juvenile development of divergent Arctic charr ( Salvelinus alpinus). J Evol Biol. 2011;24(8):1640–1652. 10.1111/j.1420-9101.2011.02301.x [DOI] [PubMed] [Google Scholar]
- 37. Sandlund TO, Gunnarsson K, Jónasson PM, et al. : The Arctic charr Salvelinus alpinus in Thingvallavatn. Oikos. 1992;64(1/2):305–351. 10.2307/3545056 [DOI] [Google Scholar]
- 38. Snorrason SS, Skúlason S, Jonsson B, et al. : Trophic specialization in Arctic charr Salvelinus alpinus (Pisces; Salmonidae): morphological divergence and ontogenetic niche shifts. Biol J Linn Soc. 1994;52(1):1–18. 10.1111/j.1095-8312.1994.tb00975.x [DOI] [Google Scholar]
- 39. Magnusson KP, Ferguson MM: Genetic analysis of four sympatric morphs of Arctic charr, Salvelinus alpinus, from Thingvallavatn, Iceland. Environ Biol Fishes. 1987;20(1):67–73. 10.1007/BF00002026 [DOI] [Google Scholar]
- 40. Danzmann RG, Ferguson MM, Skulason S, et al. : Mitochondrial DNA diversity among four sympatric morphs of Arctic charr, Salvelinus alpinus L., from Thingvallavatn, Iceland. J Fish Biol. 1991;39(5):649–659. 10.1111/j.1095-8649.1991.tb04395.x [DOI] [Google Scholar]
- 41. Pálsson S, Árnason E: Sequence variation for cytochrome b genes of three salmonid species from Iceland. Aquaculture. 1994;128(1–2):29–39. 10.1016/0044-8486(94)90099-X [DOI] [Google Scholar]
- 42. Volpe JP, Ferguson MM: Molecular genetic examination of the polymorphic Arctic charr Salvelinus alpinus of Thingvallavatn, Iceland. Mol Ecol. 1996;5(6):763–72. 10.1111/j.1365-294X.1996.tb00372.x [DOI] [PubMed] [Google Scholar]
- 43. Wilson AJ, Gíslason D, Skúlason S, et al. : Population genetic structure of Arctic charr, Salvelinus alpinus from northwest Europe on large and small spatial scales. Mol Ecol. 2004;13(5):1129–42. 10.1111/j.1365-294X.2004.02149.x [DOI] [PubMed] [Google Scholar]
- 44. Kapralova KH, Morrissey MB, Kristjánsson BK, et al. : Evolution of adaptive diversity and genetic connectivity in Arctic charr ( Salvelinus alpinus) in Iceland. Heredity (Edinb). 2011;106(3):472–487. 10.1038/hdy.2010.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kapralova KH, Gudbrandsson J, Reynisdottir S, et al. : Differentiation at the MHCIIα and Cath2 loci in sympatric Salvelinus alpinus resource morphs in Lake Thingvallavatn. PLoS One. 2013;8(7):e69402. 10.1371/journal.pone.0069402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saemundsson K: Geology of the Thingvallavatn Area. Oikos. 1992;64(1/2):40–68. 10.2307/3545042 [DOI] [Google Scholar]
- 47. Macqueen DJ, Kristjánsson BK, Paxton CG, et al. : The parallel evolution of dwarfism in Arctic charr is accompanied by adaptive divergence in mTOR-pathway gene expression. Mol Ecol. 2011;20(15):3167–84. 10.1111/j.1365-294X.2011.05172.x [DOI] [PubMed] [Google Scholar]
- 48. Doiron S, Bernatchez L, Blier PU: A comparative mitogenomic analysis of the potential adaptive value of Arctic charr mtDNA introgression in brook charr populations ( Salvelinus fontinalis Mitchill). Mol Biol Evol. 2002;19(11):1902–9. 10.1093/oxfordjournals.molbev.a004014 [DOI] [PubMed] [Google Scholar]
- 49. Miller W, Schuster SC, Welch AJ, et al. : Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci U S A. 2012;109(36):E2382–90. 10.1073/pnas.1210506109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svavarsson E: Árangur í kynbótum á bleikju og næstu skref [reference in Icelandic]. In Fræðaþing landbúnaðarsins 42007;121–125. Reference Source [Google Scholar]
- 51. Ahi EP, Guðbrandsson J, Kapralova KH, et al. : Validation of reference genes for expression studies during craniofacial development in arctic charr. PLoS One. 2013;8(6):e66389. 10.1371/journal.pone.0066389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahi EP, Kapralova KH, Pálsson A, et al. : Transcriptional dynamics of a conserved gene expression network associated with craniofacial divergence in Arctic charr. Evodevo. 2014;5(1):40. 10.1186/2041-9139-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ahi EP, Steinhäuser SS, Pálsson A, et al. : Differential expression of the aryl hydrocarbon receptor pathway associates with craniofacial polymorphism in sympatric Arctic charr. Evodevo. 2015;6(1):27. 10.1186/s13227-015-0022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snorrason SS, Skúlason S, Sandlund OT, et al. : Shape polymorphism in Arctic charr, Salvelinus alpinus. Physiol Ecol Japan. 1989;1:393–404. Reference Source [Google Scholar]
- 55. Gorodilov YN: Description of the early ontogeny of the Atlantic salmon, Salmo salar, with a novel system of interval (state) identification. Environ Biol Fishes. 1996;47(2):109–127. 10.1007/BF00005034 [DOI] [Google Scholar]
- 56. Kapralova KH, Franzdóttir SR, Jónsson H, et al. : Patterns of miRNA expression in Arctic charr development. PLoS One. 2014;9(8):e106084. 10.1371/journal.pone.0106084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Di Génova A, Aravena A, Zapata L, et al. : SalmonDB: a bioinformatics resource for Salmo salar and Oncorhynchus mykiss. Database (Oxford). 2011;2011: bar050. 10.1093/database/bar050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li B, Dewey CN: RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robinson MD, McCarthy DJ, Smyth GK: edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. R Core Team: R: A Language and Environment for Statistical Computing.2014. [Google Scholar]
- 61. Young MD, Wakefield MJ, Smyth GK, et al. : Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Untergasser A, Cutcutache I, Koressaar T, et al. : Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15),e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bustin SA, Benes V, Garson JA, et al. : The MIQE guidelines: Minimum Information for publication of Quantitative real-time PCR Experiments. Clin Chem. 2009;55(4):611–22. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 64. Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔ CT Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 65. Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koboldt DC, Zhang Q, Larson DE, et al. : VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Piskol R, Ramaswami G, Li JB, et al. : Reliable identification of genomic variants from RNA-seq data. Am J Hum Genet. 2013;93(4):641–651. 10.1016/j.ajhg.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Larkin MA, Blackshields G, Brown NP, et al. : Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 69. Nicholas KB, Nicholas HB, Deerfield DW: GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS. 1997;4(14). Reference Source [Google Scholar]
- 70. Magalhães GS, Lopes-Ferreira M, Junqueira-de Azevedo IL, et al. : Natterins, a new class of proteins with kininogenase activity characterized from Thalassophryne nattereri fish venom. Biochimie. 2005;87(8):687–99. 10.1016/j.biochi.2005.03.016 [DOI] [PubMed] [Google Scholar]
- 71. Magalhães GS, Junqueira-de-Azevedo IL, Lopes-Ferreira M, et al. : Transcriptome analysis of expressed sequence tags from the venom glands of the fish Thalassophryne nattereri. Biochimie. 2006;88(6):693–9. 10.1016/j.biochi.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 72. Sprague J, Bayraktaroglu L, Clements D, et al. : The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34(Database issue):D581–D585. 10.1093/nar/gkj086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eiríksson GM, Skúlason S, Snorrason SS: Heterochrony in skeletal development and body size in progeny of two morphs of Arctic charr from Thingvallavatn, Iceland. J Fish Biol. 1999;55(sa):175–185. 10.1111/j.1095-8649.1999.tb01054.x [DOI] [Google Scholar]
- 74. Kapralova KH, Jónsson ZO, Pálsson A, et al. : Bones in motion: Ontogeny of craniofacial development in sympatric arctic charr morphs. Dev Dyn. 2015;244(9):1168–1178. 10.1002/dvdy.24302 [DOI] [PubMed] [Google Scholar]
- 75. Jonsson B, Skúlason S, Snorrason SS, et al. : Life History Variation of Polymorphic Arctic Charr ( Salvelinus alpinus) in Thingvallavatn, Iceland. Can J Fish Aquat Sci. 1988;45(9):1537–1547. 10.1139/f88-182 [DOI] [Google Scholar]
- 76. Palsson A, Wesolowska N, Reynisdóttir S, et al. : Naturally occurring deletions of hunchback binding sites in the even-skipped stripe 3+7 enhancer. PLoS One. 2014;9(5);e91924. 10.1371/journal.pone.0091924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zou C, Lehti-Shiu MD, Thomashow M, et al. : Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 2009;5(7):e1000581. 10.1371/journal.pgen.1000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jantzen SG, Sanderson DS, von Schalburg KR, et al. : A 44K microarray dataset of the changing transcriptome in developing Atlantic salmon ( Salmo salar L.). BMC Res Notes. 2011;4:88. 10.1186/1756-0500-4-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Drivenes Ø, Taranger GL, Edvardsen RB: Gene expression profiling of Atlantic cod ( Gadus morhua) embryogenesis using microarray. Mar Biotechnol (NY). 2012;14(2):167–76. 10.1007/s10126-011-9399-y [DOI] [PubMed] [Google Scholar]
- 80. Bougas B, Audet C, Bernatchez L: The influence of parental effects on transcriptomic landscape during early development in brook charr ( Salvelinus fontinalis, Mitchill). Heredity (Edinb). 2013;110(5):484–491. 10.1038/hdy.2012.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Piasecka B, Lichocki P, Moretti S, et al. : The hourglass and the early conservation models--co-existing patterns of developmental constraints in vertebrates. PLoS Genet. 2013;9(4):e1003476. 10.1371/journal.pgen.1003476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fleming A: On a Remarkable Bacteriolytic Element Found in Tissues and Secretions. Proc R Soc Lond B. 1922;93(653):306–317. 10.1098/rspb.1922.0023 [DOI] [Google Scholar]
- 83. Chipman DM, Sharon N: Mechanism of lysozyme action. Science. 1969;165(3892):454–465. 10.1126/science.165.3892.454 [DOI] [PubMed] [Google Scholar]
- 84. Subramanian S, MacKinnon SL, Ross NW: A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp Biochem Physiol B Biochem Mol Biol. 2007;148(3):256–63. 10.1016/j.cbpb.2007.06.003 [DOI] [PubMed] [Google Scholar]
- 85. Magnadóttir B: Innate immunity of fish (overview). Fish Shellfish Immunol. 2006;20(2):137–51. 10.1016/j.fsi.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 86. Magnadottir B: Immunological control of fish diseases. Mar Biotechnol (NY). 2010;12(4):361–79. 10.1007/s10126-010-9279-x [DOI] [PubMed] [Google Scholar]
- 87. Eiriksson GM: Heterochrony in bone development and growth in two morphs of Arctic charr ( Salvelinus alpinus) from Thingvallavatn, Iceland . Master’s thesis, University of Iceland,1999. [Google Scholar]
- 88. Chai Y, Ito Y, Han J: TGF-beta signaling and its functional significance in regulating the fate of cranial neural crest cells. Crit Rev Oral Biol Med. 2003;14(2):78–88. 10.1177/154411130301400202 [DOI] [PubMed] [Google Scholar]
- 89. Puga A, Tomlinson CR, Xia Y: Ah receptor signals cross-talk with multiple developmental pathways. Biochem Pharmacol. 2005;69(2):199–207. 10.1016/j.bcp.2004.06.043 [DOI] [PubMed] [Google Scholar]
- 90. Goodale BC, La Du JK, Bisson WH, et al. : AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS One. 2012;7(1):e29346. 10.1371/journal.pone.0029346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nurminsky D, Magee C, Faverman L, et al. : Regulation of chondrocyte differentiation by actin-severing protein adseverin. Dev Biol. 2007;302(2):427–37. 10.1016/j.ydbio.2006.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vieira FA, Thorne MA, Stueber K, et al. : Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation. Gen Comp Endocrinol. 2013;191:45–58. 10.1016/j.ygcen.2013.05.025 [DOI] [PubMed] [Google Scholar]
- 93. Furuse M, Oda Y, Higashi T, et al. : Lipolysis-stimulated lipoprotein receptor: a novel membrane protein of tricellular tight junctions. Ann N Y Acad Sci. 2012;1257:54–8. 10.1111/j.1749-6632.2012.06486.x [DOI] [PubMed] [Google Scholar]
- 94. Jazag A, Ijichi H, Kanai F, et al. : Smad4 silencing in pancreatic cancer cell lines using stable RNA interference and gene expression profiles induced by transforming growth factor-beta. Oncogene. 2005;24(4):662–71. 10.1038/sj.onc.1208102 [DOI] [PubMed] [Google Scholar]
- 95. Planchart A, Mattingly CJ: 2,3,7,8-Tetrachlorodibenzo- p-dioxin upregulates FoxQ1b in zebrafish jaw primordium. Chem Res Toxicol. 2010;23(3):480–7. 10.1021/tx9003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hering NA, Andres S, Fromm A, et al. : Transforming growth factor- β, a whey protein component, strengthens the intestinal barrier by upregulating claudin-4 in HT-29/B6 cells. J Nutr. 2011;141(5):783–9. 10.3945/jn.110.137588 [DOI] [PubMed] [Google Scholar]
- 97. Ito Y, Yeo JY, Chytil A, et al. : Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130(21):5269–80. 10.1242/dev.00708 [DOI] [PubMed] [Google Scholar]
- 98. Kedersha NL, Miquel MC, Bittner D, et al. : Vaults. II. Ribonucleoprotein structures are highly conserved among higher and lower eukaryotes. J Cell Biol. 1990;110(4):895–901. 10.1083/jcb.110.4.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Berger W, Steiner E, Grusch M, et al. : Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66(1):43–61. 10.1007/s00018-008-8364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. van Driel M, Pols HA, van Leeuwen JP: Osteoblast differentiation and control by vitamin D and vitamin D metabolites. Curr Pharm Des. 2004;10(21):2535–55. 10.2174/1381612043383818 [DOI] [PubMed] [Google Scholar]
- 101. Pulcini D, Wheeler PA, Cataudella S, et al. : Domestication shapes morphology in rainbow trout Oncorhynchus mykiss. J Fish Biol. 2013;82(2):390–407. 10.1111/jfb.12002 [DOI] [PubMed] [Google Scholar]
- 102. Völkers M, Rohde D, Goodman C, et al. : S100A1: a regulator of striated muscle sarcoplasmic reticulum Ca 2+ handling, sarcomeric, and mitochondrial function. J Biomed Biotechnol. 2010;2010: 178614. 10.1155/2010/178614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. McBride HM, Neuspiel M, Wasiak S: Mitochondria: more than just a powerhouse. Curr Biol. 2006;16(14):R551–60. 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- 104. O’Brien KM, Mueller IA: The unique mitochondrial form and function of Antarctic channichthyid icefishes. Integr Comp Biol. 2010;50(6):993–1008. 10.1093/icb/icq038 [DOI] [PubMed] [Google Scholar]
- 105. William J, Ballard O, Pichaud N: Mitochondrial DNA: more than an evolutionary bystander. Functional Ecol. 2014;28(1):218–231. 10.1111/1365-2435.12177 [DOI] [Google Scholar]
- 106. Teacher AG, André C, Merilä J, et al. : Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. BMC Evol Biol. 2012;12:248. 10.1186/1471-2148-12-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Palsson A, Gibson G: Association between nucleotide variation in Egfr and wing shape in Drosophila melanogaster. Genetics. 2004;167(3):1187–1198. 10.1534/genetics.103.021766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Palsson A, Dodgson J, Dworkin I, et al. : Tests for the replication of an association between Egfr and natural variation in Drosophila melanogaster wing morphology. BMC Genet. 2005;6:44. 10.1186/1471-2156-6-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gudbrandsson J, Ahi E, Franzdottir S, et al. : Dataset 1 in: The developmental transcriptome of contrasting Arctic charr ( Salvelinus alpinus) morphs. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed]
- 110. Gudbrandsson J, Ahi E, Franzdottir S, et al. : Dataset 2 in: The developmental transcriptome of contrasting Arctic charr ( Salvelinus alpinus) morphs. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed]
- 111. Gudbrandsson J, Ahi E, Franzdottir S, et al. : Dataset 3 in: The developmental transcriptome of contrasting Arctic charr ( Salvelinus alpinus) morphs. F1000Research. 2015. Data Source [DOI] [PMC free article] [PubMed]