ABSTRACT
Aspergillus carbonarius is the main responsible fungus of ochratoxin A (OTA) contamination of grapes and derived products. To date, the biosynthetic mechanism of this mycotoxin has been partially elucidated. Availability of genome sequence of A. carbonarius has allowed the identification of a putative gene cluster involved in OTA biosynthesis. This region hosts the previously characterized AcOTAnrps and AcOTApks genes encoding two key enzymes of the biosynthetic pathway. At about 4,400 nucleotides downstream of these loci, a gene encoding a putative flavin dependent-halogenase came out from the annotation data. Its proximity to OTA biosynthetic genes and its sequence analysis have suggested a role in the biosynthesis of OTA, directed to the introduction of the chlorine atom in the C-5 position of the final molecular structure of this mycotoxin. The deduced protein sequence of the halogenase gene, we designated AcOTAhal, shows a high similarity to a halogenase that is located in the OTA cluster of A. niger. The deletion of the halogenase gene completely eliminated the production of ochratoxin A in A. carbonarius and determined a significant increase of ochratoxin B, as confirmed by mass spectrometry analysis. Moreover, its expression profile was similar to the two biosynthetic genes previously identified, AcOTApks and AcOTAnrps, indicating a strong correlation of the AcOTAhal gene with the kinetics of OTA accumulation in A. carbonarius. Therefore, experimental evidence confirmed that the chlorination step which converts OTB in OTA represents the final stage of the biosynthetic pathway, supporting our earlier hypothesis on the order of enzymatic steps of OTA biosynthesis in A. carbonarius.
IMPORTANCE Ochratoxin A is a potent mycotoxin classified as a possible carcinogen for humans, and Aspergillus carbonarius is the main agent responsible for OTA accumulation in grapes. We demonstrate here that a flavin-halogenase is implicated in the biosynthesis of OTA in A. carbonarius. The encoding gene, AcOTAhal, is contiguous to biosynthetic genes that we have already described (nrps and pks), resulting as part of the biosynthetic cluster. The encoded protein is responsible of the introduction of chlorine atom in the final molecular structure and acts at the last step in the pathway. This study can be considered a continuation of an earlier study wherein we started to clarify the molecular basis of OTA biosynthesis in A. carbonarius, which has not been completely elucidated until now. This research represents an important step forward to a better understanding of the production mechanism, which will contribute to the development of improved control strategies to reduce the risk of OTA contamination in food products.
INTRODUCTION
The biosynthetic genes of most fungal mycotoxins are organized in clusters, and in such a case the identification of one gene frequently facilitates the identification of the other genes involved in the biosynthetic pathway. The biosynthetic clusters of the most important mycotoxins—such as aflatoxins, trichothecenes, fumonisins, and patulin—have been elucidated for some time. This has allowed a better comprehension of the regulatory process behind the mycotoxin biosynthesis, a thorough examination of the producing capacity of fungal populations, and the development of various and efficient molecular diagnostic assays (1–3).
The whole-genome sequencing projects for A. niger and A. carbonarius, two related fungal species responsible for ochratoxin A (OTA) contamination of different food products, have provided the possibility to elucidate some important steps of the biosynthetic pathway of this mycotoxin that, as yet, have not been completely defined.
Ochratoxin A is a polyketide mycotoxin that results in nephrotoxic, teratogenic, immunotoxic, neurotoxic, and hepatotoxic effects (4, 5); moreover, OTA has been classified as a possible human carcinogen (6). Among the OTA producing fungi, Aspergillus carbonarius is the main responsible of OTA contamination of grapes, grape juice, dried vine fruits, must, and wine (7–10).
A polyketide synthase (PKS) and a nonribosomal peptide synthetase (NRPS) have been shown to be involved in two key steps in the biosynthetic pathway of OTA in A. carbonarius. In particular, the PKS encoded by the AcOTApks gene (11) catalyzes the formation of the isocoumarin group during the initial stages of biosynthesis, starting from acetate and malonate, to originate the characteristic pentaketide skeleton of the OTA molecule. The NRPS encoded by the AcOTAnrps gene is involved in the linking of phenylalanine to dihydroisocoumarin ring and was hypothesized to act before the final chlorination step (12). The reaction catalyzed by NRPS results in the production of ochratoxin B (OTB), which is the nonchlorinated analogue of OTA and which subsequently serves as the substrate of chlorination activity to form the final molecular structure of OTA, containing a chlorine atom in the C-5 position of the molecule. This small structural difference, i.e., chlorine in OTA versus hydrogen in OTB, does not seem to be responsible for the different toxicities and instead may be crucial for the differential uptake and binding in cells (13). In fact, whereas OTB appears to be much less toxic than OTA in vivo, recent investigations in vitro have resulted in lower, different, or even equal toxicities of OTB compared to OTA that are likely due to differences in cellular uptake and protein binding (14, 15).
The intervention of a chlorinating enzyme in the OTA biosynthetic pathway has been supposed according to the molecular structure of the mycotoxin (16), and a first evidence of its putative role was reported in Penicillium nordicum (17, 18). Penicillium nordicum is the main OTA contaminant of protein-rich foods such as fermented meats and cheese (7), and it was one of the most investigated ochratoxigenic fungi to elucidate the molecular mechanism of OTA biosynthesis. Two fragments of the chromosomal DNA of P. nordicum were identified to carry genes encoding proteins putatively associated with OTA biosynthesis due to their expression profile and, in addition to a PKS, the proteins found included an NRPS, a transporter protein, and a protein homologous to a bacterial chloroperoxidase (17, 19). Halogenation is a frequent modification of microbial secondary metabolites, and compounds containing carbon-halogen bonds include natural products with a wide range of biological activities (20); moreover, chlorination is the predominant halogenating modification that occurs. In recent years, several halogenating enzymes have been discovered that allowed a better understanding of the biohalogenation process in microorganisms. In particular, flavin-dependent halogenases have been identified as major players in the introduction of halogen into activated organic molecules in natural product biosynthesis (21).
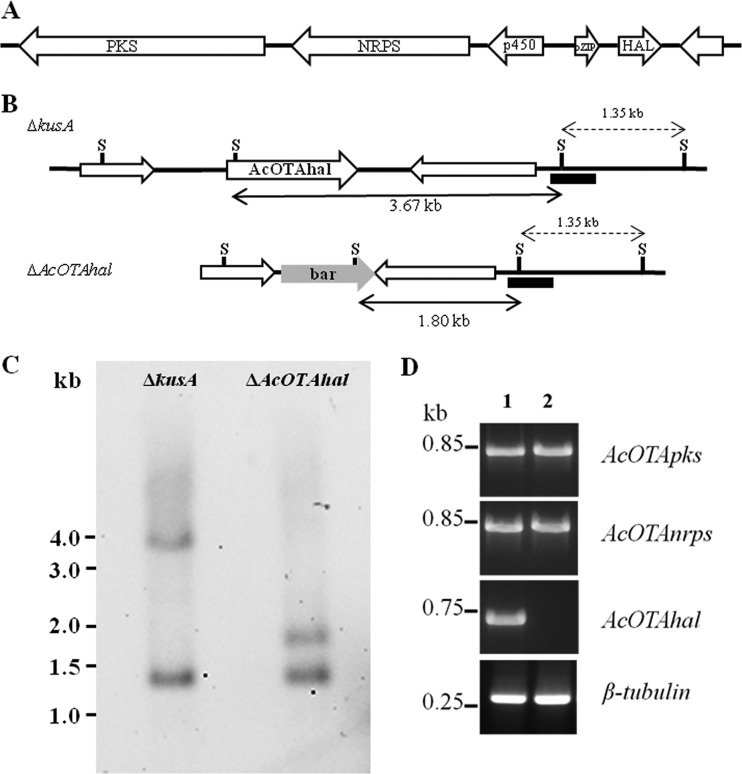
Sequencing of the genomic region of A. carbonarius carrying the pks and nrps genes involved in OTA biosynthesis allowed the identification of a gene encoding a putative flavin-dependent halogenase. Here, we identify and characterize this gene, designated AcOTAhal, by means of a gene-knockout approach. The expression level profiles of OTA pks, nrps, and hal genes were investigated to verify their correlation to the kinetics of OTA accumulation in A. carbonarius. Moreover, the expression profiles of two other genes, which we named AcOTAp450 and AcOTAbZIP, were also analyzed. They encode a cytochrome p450 monooxygenase and a basic leucine zipper (bZIP) transcription factor, respectively, and are located next to the OTA pks, nrps, and hal genes in the assumed biosynthetic cluster (Fig. 1A).
FIG 1.
(A) Structure of the ochratoxin A cluster in Aspergillus carbonarius. (B) Map of the AcOTAhal locus in ΔkusA and ΔAcOTAhal strains of A. carbonarius. (C) Southern blot hybridizations from genomic DNA digested with SalI. The restriction sites SalI are indicated by “S.” (D) RT-PCR profile of expression of AcOTApks, AcOTAnrps, and AcOTAhal genes in the ΔkusA (lane 1) and ΔAcOTAhal (lane 2) strains grown on a medium permissive for OTA. β-tubulin was used as a control.
The present study, together with our previous studies on the key enzymes PKS and NRPS, represents a step forward in the knowledge of molecular basis of OTA biosynthesis in A. carbonarius and offers new insights for a full understanding of mycotoxin production mechanism and ultimately for improved control of the contamination risk.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The fungal strains used in this study were the wild-type A. carbonarius ITEM 5010 from the Agro-Food Microbial Culture Collection of the Institute of Sciences of Food Production, CNR, Bari, Italy (www.ispa.cnr.it/Collection) and the mutant strains generated from ITEM 5010, KB1039 (ΔkusA), lacking the kusA gene to facilitate homologous integration (22), and AC1501 (ΔkusA ΔAcOTAhal), lacking both kusA and AcOTAhal, the gene encoding the flavin-dependent halogenase. Fungal strains were grown on minimal medium (MM) agar plates (6 g/liter NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4·7H2O, 0.82 g/liter KH2PO4, 1.05 g/liter K2HPO4 [pH 6.5], 10 g/liter glucose, 1 ml/liter Hutner's trace elements, 15 g/liter agar).
Mycelia for protoplasting were obtained from a 100 ml of YEPD (1% yeast extract, 2% peptone, 2% glucose) culture inoculated with 500 μl of conidial suspension (106 conidia/ml), followed by incubation overnight at 30°C at 150 rpm. The mycelia were collected by filtration and resuspended in digestion buffer (1.2 M MgSO4, 50 mM phosphate buffer [pH 5.0]) containing 60 mg/ml VinoTaste Pro (Novozymes), followed by incubation at 30°C for 3 to 5 h with gentle shaking. After digestion, the protoplasts were separated from undigested mycelia by filtration through sterile Miracloth, and the filtrate was then overlaid with sterile 0.4 M ST (0.4 M sorbitol, 100 mM Tris-HCl [pH 8.0]) at a ST/protoplast suspension ratio of 1:5 (vol/vol). After centrifugation at 800 g for 15 min, the interface layer was collected and resuspended in sterile 1 M ST (1 M sorbitol, 100 mM Tris-HCl [pH 8.0]). Protoplasts were collected by centrifugation at 800 × g for 15 min, and the pellet was washed twice with 1 M ST. The final pellet was resuspended in sterile STC (1 M sorbitol, 50 mM Tris-HCl [pH 8.0], 50 mM CaCl2) to a final concentration of 1.25 × 107/ml. Transformations were performed by mixing about 10 μg of transformation cassette with 20% of PEG solution (40% PEG 4000 in STC) and about 1.25 × 106 protoplasts, followed by incubation on ice for 15 min. Then, 1 ml of PEG solution was added to each sample, followed by incubation at room temperature for 15 min. After incubation, each transformation reaction was transferred into 10 ml of recovery media (MM with 1 M sorbitol) and incubated at 30°C for 1 h shaking gently. The protoplasts were plated by inclusion in selective soft agar recovery media supplemented with 100 μg/ml hygromycin B, used as a selection marker for kusA deletion, and 1 mg/ml glufosinate, used as a selection marker for deletion of AcOTAhal. Resistant colonies were selected after 3 to 5 days of incubation at 30°C.
The production of OTA and related metabolites by strains AC1501 and KB1039 was checked on yeast extract sucrose (YES) agar plates (yeast extract, 20 g/liter; sucrose, 150 g/liter; agar, 20 g/liter) inoculated with 100 μl of a conidial suspension (106 conidia/ml) of the strains, followed by incubation in the dark at 25°C. After 7 days of growth, five agar plugs with mycelium (5 mm in diameter) were collected from each plate and stored at −20°C until analysis for OTA and related metabolites. Triplicate cultures were prepared and analyzed for each experiment.
For gene expression analyses, 100 μl of a conidial suspension (106 conidia/ml) from wild-type strain ITEM 5010 of A. carbonarius was inoculated onto MM agar plates overlaid with sterile cellophane membranes. Incubation was carried out in the dark at 25°C. Fungal mycelium was harvested after 2, 3, 4, 5, and 7 days postinoculation and then frozen in liquid nitrogen and stored at −80°C for RNA extraction and OTA content analysis. Triplicate cultures were prepared and analyzed for each experiment.
Nucleic acid extraction, cDNA synthesis, RT-PCR, and quantitative RT-PCR.
Genomic DNA was isolated using the Wizard Magnetic DNA Purification System for Food kit (Promega, Madison, WI) according to the manufacturer's protocol. Total RNA was extracted from frozen mycelium ground in liquid nitrogen using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. RNA samples were treated with RNase-free DNase I (Qiagen) to eliminate any possible DNA contamination. RNA aliquots were stored at −80°C. First-strand cDNA was synthesized using 3 μg of total RNA, oligo(dT)18 and random hexamer primers at a 1:1 ratio, and SuperScript III reverse transcriptase (Invitrogen, San Diego, CA) according to the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) was used to analyze the presence of transcripts of the AcOTApks, AcOTAnrps, and AcOTAhal genes in A. carbonarius strains KB1039 and AC1501. The primer pairs listed in Table 1 were used under the following conditions: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 1 min, and then a final extension step at 68°C for 5 min. Primers Bt2a/Bt2b (23) were used to monitor the β-tubulin gene expression as an endogenous control. The transcription profiles of the above-mentioned three genes and of two other genes placed next to them in the cluster, named AcOTAp450 and AcOTAbZIP, were analyzed at different times during the growth of wild-type A. carbonarius ITEM 5010 on MM agar medium by using real-time quantitative reverse transcription-PCR (qRT-PCR). For the amplification reaction, a SYBR green I assay was performed in the 7500 Fast real-time PCR system (Applied Biosystems, Warrington, United Kingdom) with 2.5× Real master mix SYBR-ROX (5 PRIME GmbH, Hamburg, Germany) in a reaction volume of 25 μl with 200 nM concentrations of each primer for the target and reference genes, except for the primer pair RT_AcOTAbZIP used at 250 nM. The constitutively expressed β-tubulin gene served as an internal reference to normalize target gene expression. The primers used for the qRT-PCR are listed in Table 1. The following amplification conditions were used: an initial denaturation step at 95°C for 2 min, followed by 40 cycles of 10 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The specificity of the PCR amplifications was confirmed by dissociation curve analyses. The relative quantification of gene expression was established using the comparative 2−ΔΔCT method. The PCR efficiency of each oligonucleotide pair was calculated from each linear regression of standard curves. Real-time PCR derived data were quantified relatively by using the Relative Expression Software Tool (REST) (24), taking the divergent efficiencies into account. Three biological replicates were performed for each experiment, and each was tested in triplicate, in addition to a no-template control included for each primer pair.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Primers for the AcOTAhal deletion cassette | |
| OThal-F1 | ATCCATCCGACAAGAAGCAC |
| OThal-F2 | GAGCCTTTGAAGACGGACTG |
| OThal-R3 | TGACCTCCACTAGCTCCAGCGCTCATGCCCATCCCTAGTA |
| OThal-F4 | TCACCGAGATTTAGGTCGACGGCCATTCCAGACCGTATAA |
| OThal-R5 | GCACCAACGAACATCTCTGA |
| OThal-R6 | CCAGGTGAGTCCTGCACATA |
| OThal-BARf | TACTAGGGATGGGCATGAGCGCTGGAGCTAGTGGAGGTCA |
| OThal-BARr | TTATACGGTCTGGAATGGCCGTCGACCTAAATCTCGGTGA |
| Primers for RT-PCR and qRT-PCR | |
| AcOTApks_for | GAACCATTTCCGACCTTCT |
| AcOTApks_rev | TTCGAGGATGGCAAGTAGA |
| AcOTAnrps_for | TCATCTCCGACGAGGAAC |
| AcOTAnrps_rev | CAAAGGAATCCTCGTCACT |
| AcOTAhal_for | GTTGTGCTAGAAGCGGATG |
| AcOTAhal_rev | AAGTACGGGTCGATGAAGG |
| RT_AcOTApks_F | CGTGTCCGATACTGTCTGTGA |
| RT_AcOTApks_R | GCATGGAGTCCTCAAGAACC |
| RT_AcOTAnrps_F | ACGGGTCGCTGCTCTATATC |
| RT_AcOTAnrps_R | ACTCACCACATCAACCACGA |
| RT_AcOTAhal_F | GAACGCCAGTAGAGGGACAG |
| RT_AcOTAhal_R | ATGGAGGTGGTGTTGTTGTG |
| RT_AcOTAp450_F | GTGGTTATCCCGCCCAATAC |
| RT_AcOTAp450_R | TGCCAGATTCATCCCGATAC |
| RT_AcOTAbZip_F | AATGGAACCAGCATTGATCTC |
| RT_AcOTAbZip_R | GACCCAAGCATTCGCTCTA |
| RT3 BT Ac_F | CAAACCGGCCAGTGTGGTA |
| RT3 BT Ac_R | CGGAGGTGCCATTGTAAACA |
| Primers used for Southern hybridization probe | |
| Probe_HalF | CGACAACACAGGCATACAG |
| Probe_HalR | CTTAGGGCTGTTGATCTGC |
Deletion of the AcOTAhal gene in Aspergillus carbonarius.
The deletion cassette for the AcOTAhal gene (Fig. 1B) was generated by a commonly used PCR fusion approach (25), consisting of a fusion of the sequences flanking the coding region of the AcOTAhal gene with the bar gene encoding resistance to glufosinate. The primers used to create the deletion cassette are listed in Table 1. In particular, OTAhalF1/OTAhalR3 and OTAhalF4/OTAhalR6 were used to amplify two ∼1,800-bp fragments, one upstream and one downstream of the targeted gene, which subsequently were fused to the bar resistance marker amplified from the plasmid pCB1530 (Fungal Genetics Stock Center, Kansas City, MO) by using the primer pair uBarR/uBarF, including extension sequences which overlap extension sequences of the primers OTAhalR3 and OTAhalF4. The joined product was amplified with the nested primers OTAhalF2 and OTAhalR5 and used to transform protoplasts prepared from strain KB1039 (ΔkusA).
Selection of ΔAcOTAhal transformants was performed on MM supplemented with 100 μg/ml hygromycin B and 1 mg/ml glufosinate (bioWorld, Dublin, OH). Confirmation of homologous recombination in putative transformants was made by Southern blotting according to the standard procedures (26). Genomic DNA of strain KB1039 and ΔAcOTAhal transformants was digested with SalI restriction enzyme and subsequently hybridized with a 491-bp digoxigenin-labeled probe (PCR DIG probe synthesis kit; Roche, Mannheim, Germany), synthesized with the primers Probe_HalF/Probe_HalR and sharing homology with the AcOTAhal flanking region.
Chemical analyses of fungal extract by HPLC-FLD and HPLC-HRMS.
Standard solutions of OTA (100 μg/ml), OTB (10 μg/ml), and ochratoxin α (OTα) (10 μg/ml) were purchased from Romer Labs Diagnostic GmbH (Tulln, Austria). A mixed standard solution was prepared by mixing the three standard solutions and used for the preparation of calibration solutions that were prepared in methanol containing 0.6% formic acid, 0.02% hydrochloric acid, and 2.5% water. Extracts from agar plug cultures and mycelium were prepared according to the microextraction method of Smedsgaard (27) with some modifications. In particular, 5 plugs of 5 mm diameter or 150 to 620 mg of mycelium collected from cellophane membranes were transferred to a 12 ml test tube and 3 ml of the solvent mixture methanol-dichloromethane-ethyl acetate (1:2:3) containing 1% (vol/vol) formic acid were added. The extraction was performed ultrasonically for 60 min and the supernatant was transferred to a clean vial, evaporated to dryness at 50°C under a gentle stream of nitrogen and redissolved in 1 ml of methanol containing 0.6% formic acid, 0.02% hydrochloric acid and 2.5% water. The reconstituted sample extracts were filtered through a 0.22 μm pore filter and analyzed by high-pressure liquid chromatography with fluorescence detection (HPLC-FLD) and high-pressure liquid chromatography/high-resolution mass spectrometry (HPLC-HRMS).
HPLC-FLD analyses were performed with an Agilent 1260 Infinity system comprising of a binary pump (G1312B), an auto sampler (G1367E) with a 100 μl loop, a fluorescence detector (G1321B) (excitation wavelength, 333 nm; emission wavelength, 460 nm), a thermostatic oven set at 30°C (G1316C), and a software for Microsoft Windows 7 (OpenLAB, CSD, ChemStation Edition). The column used was a Zorbax C18 (150 mm by 4.6 mm, 5-μm particles; Phenomenex, Torrance, CA) with a 3-mm, 0.45-μm-pore-size guard filter (Rheodyne, Cotati, CA). A linear gradient starting from 85% water (A) and 15% acetonitrile (B) going to 100% acetonitrile in 40 min, then maintaining 100% acetonitrile for 3 min, was used. The flow rate of the mobile phase was 1 ml/min. Both eluents contained 0.005% (vol/vol) trifluoroacetic acid. A volume of 10 μl of filtered fungal extract was injected into the HPLC-FLD apparatus.
HPLC-HRMS analyses were performed with a benchtop single-stage mass spectrometer (Exactive) equipped with a heated electrospray ion source (HESI II) (Thermo Fisher Scientific, Bremen, Germany), coupled to an Accela HPLC system (Thermo Fisher Scientific, San Jose, CA). The HESI II interface was used in the positive-ion mode for 7-methylmellein (7-MM), ethyl ester of phenylalanine (Phe), ethyl ester of OTB, and OTA and in the negative-ion mode for ochratoxin β (OTβ), OTα, Phe, OTB, and ochratoxin C (OTC). The HPLC-HRMS retention times and HR mass data of the metabolites are presented in Table 2. The scan range was 50.2 to 1,003.0 m/z, with a resolution power of 10,000 FWHM (full width at half maximum). Other settings were as follows: sheath gas flow rates 15 and 30 arbitrary units and auxiliary gas flow rates 15 and 10 arbitrary units for positive and negative ions, respectively; sweep gas, 0 arbitrary units; capillary temperature, 300°C; capillary voltage, 4 and 4.5 kV for positive and negative ions, respectively. Xcalibur software (v2.1.0; Thermo Fisher Scientific) was used for data acquisition and processing.
TABLE 2.
Summary of HPLC-HRMS retention times and HR mass data of OTA and other metabolites identified in cultures of KB1039 (ΔkusA) and AC1501 (ΔkusA, ΔAcOTAhal) strainsa
| Compound | RT (min) | Measured mass (Da) | Monoisotopic mass (Da) | Error (mDa) | Formula |
|---|---|---|---|---|---|
| PHE | 3.50 | 165.0787 | 165.0789 | −0.2 | C9H11NO2 |
| OTB ethyl ester | –* | 399.1682 | C22H25NO6 | ||
| OTβ | 13.29 | 222.0524 | 222.0528 | −0.4 | C11H10O5 |
| PHE ethyl ester | 17.00 | 193.1106 | 193.1103 | 0.3 | C11H15NO2 |
| 7-MM | 17.95 | 192.0788 | 192.0786 | 0.2 | C11H12O3 |
| OTα | 28.73 | 256.0132 | 256.0138 | −0.6 | C11H9ClO5 |
| OTC | –* | 431.1136 | C22H22ClNO6 | ||
| OTB | 31.26 | 369.1200 | 369.1212 | −1.2 | C20H19NO6 |
| OTA | 41.87 | 403.0848 | 403.0823 | 2.5 | C20H18ClNO6 |
RT, retention time. *, the OTB ethyl ester and OTC were not detected in cultures of both ΔkusA and ΔAcOTAhal strains.
A volume of 20 μl of filtered fungal extracts were analyzed by HPLC-HRMS for the identification of 7-MM, OTβ, OTα, Phe, ethyl ester of Phe, OTB, ethyl ester of OTB, OTC, and OTA. The column was a Gemini C18 (150 mm by 2 mm, 5-μm particles). The mobile phase was a gradient of water (A) and methanol (B), both containing 0.5% acetic acid and 1 mM ammonium acetate. For the separation of analytes, a multiple linear binary gradient was used: from 20 to 40% B in 3 min, then to 63% B in 35 min, and then maintained at 63% B for 11 min. The column was reequilibrated with 20% B for 10 min prior to the successive injection.
Statistical analysis.
Statistical analysis was performed by using the GraphPad Instat software (Instat, San Diego, CA). Data were subjected to the paired t test (two-tailed P value) to determine the statistical differences when two groups of data were compared. One-way analysis of variance (ANOVA) with posttest (standard parametric methods) was used when three groups of data were compared. Significance was defined as P < 0.05.
Accession number(s).
The genomic and transcript sequences of the AcOTAhal gene were obtained by Sanger sequencing and RNA-seq analysis, respectively. Nucleotide sequences were assembled using CLC Genomics Workbench 7.5 (CLC, Inc., Aarhus, Denmark) and deposited in GenBank under accession no. KU960948. The deduced amino acid sequence was determined by using the ExPASy translation tool (http://web.expasy.org/translate/). The protein sequence was aligned by using the blastp algorithm against nonredundant protein collection at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
RESULTS
Identification and inactivation of AcOTAhal in A. carbonarius.
The analysis of the genomic sequence of A. carbonarius ITEM 5010 corresponding to the OTA biosynthetic cluster allowed the identification of a putative halogenase gene at about 4,400 nucleotides downstream of the region hosting AcOTAnrps and AcOTApks genes (Fig. 1A), previously described as two key genes for OTA biosynthesis (11, 12). According to the annotation data available from JGI (http://jgi.doe.gov/carbonarius/), this hypothetical gene encoded a protein (ID 209543) showing a high similarity with a RadH flavin-dependent halogenase protein of A. niger (accession number XP_001397309). This evidence, as well as its proximity to the other OTA biosynthetic genes, led us to suppose its involvement in the predictable chlorination step of OTA biosynthetic pathway. The functional role of this gene was demonstrated by a gene deletion approach. In particular, a deletion cassette for AcOTAhal was assembled and transformed into strain KB1039 (ΔkusA) of A. carbonarius ITEM 5010. Three transformants determined to be positive by PCR screening for the integration of the bar resistance marker at the AcOTAhal locus were obtained (data not shown). One of them, designated AC1501 (ΔkusA ΔAcOTAhal), was used for subsequent analyses. Southern blot hybridization, performed on genomic DNA of the KB1039 and AC1501 strains digested with SalI, confirmed the complete deletion of AcOTAhal gene in AC1501. In details, the hybridizing bands of 3.67 and 1.35 kb indicated the presence of endogenous gene in KB1039, the ΔkusA strain, whereas the homologous integration of the deletion cassette resulted in a band of 1.80 and 1.35 kb in the mutant strain AC1501, as shown in Fig. 1B and C. An RT-PCR analysis performed on cDNA from AC1501 grown on an OTA permissive medium showed the presence of AcOTApks and AcOTAnrps transcripts, whereas no AcOTAhal transcript was observed, thus confirming the deletion of the gene in the mutant strain (Fig. 1D).
Sequencing and characterization of the AcOTAhal gene.
The sequencing of the AcOTAhal gene from genomic DNA and of the relative transcript from cDNA allowed to verify that the gene is 1,548 bp long and contains four introns. The deduced amino acid sequence encodes a protein of 435 residues with a predicted molecular mass of 47,337 Da. Analysis of conserved domains revealed the presence of a flavin adenine dinucleotide (FAD)-binding domain and a tryptophan halogenase domain. From BLAST analysis, it was determined that the AcOTAhal protein shares 84% similarity with a RadH flavin-dependent halogenase protein of A. niger CBS 513.88 (GenBank accession number XP_001397309) and 83% similarity with a hypothetical protein of P. nordicum DAOMC 185683 (GenBank accession number KOS43943). The AcOTAhal sequence was deposited at NCBI under accession number KU960948.
Expression analysis of OTA genes in A. carbonarius.
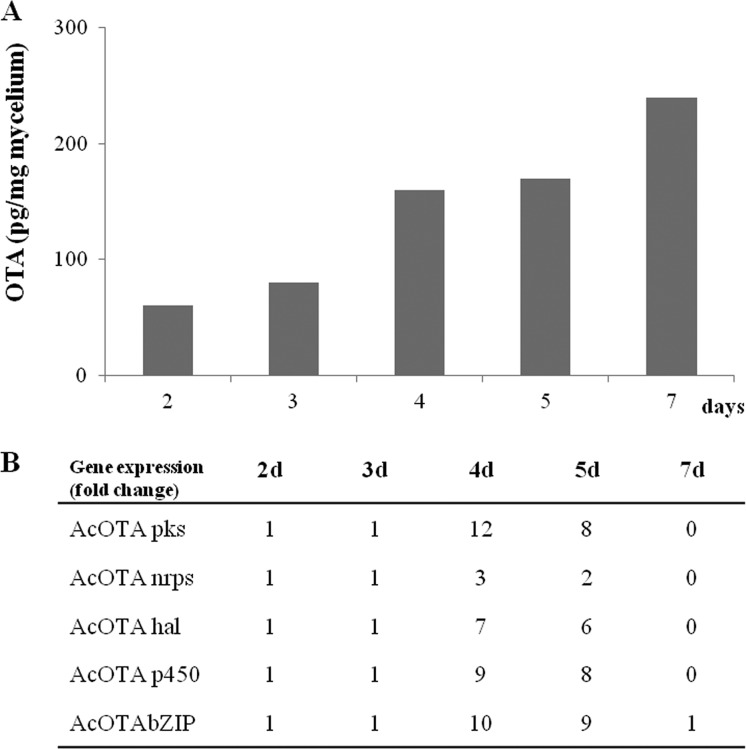
We monitored the transcription profiles of the three biosynthetic genes we have characterized thus far—AcOTApks, AcOTAnrps, and AcOTAhal—and of two other genes—AcOTAp450 and AcOTAbZIP—located next to them and putatively involved in OTA biosynthesis. These latter genes encode a cytochrome P450 oxidase (ID 517149) and a bZIP transcription factor (ID 7821) of A. carbonarius. The expression of the genes was monitored for 7 days in the wild-type strain ITEM 5010 of A. carbonarius during growth on MM, an OTA permissive medium. The observed expression levels were consistent with OTA production. In particular, OTA was detected since the second day of growth and progressively increased up to 240 pg/mg mycelium after 7 days (Fig. 2A). From the relative gene expression analysis, all the transcripts, which were detected since the beginning as well, turned out to be up-expressed at days 4 and 5, with a peak of expression at day 4. In particular, at day 4 upregulations of about 12-fold for AcOTApks, 3-fold for AcOTAnrps, 7-fold for AcOTAhal, 9-fold for AcOTAp450, and 10-fold for AcOTAbZIP were observed compared to day 2. A sharp decrease in expression levels was observed at day 7 for all the genes (Fig. 2B).
FIG 2.
Time course analysis of OTA production (A) and relative expression (x-fold) analysis (B) by qRT-PCR of AcOTApks, AcOTAnrps, AcOTAhal, AcOTAp450, and AcOTAbZIP of the wild-type A. carbonarius ITEM 5010 during growth on MM medium.
Production of OTA, OTα, OTB, and other secondary metabolites in mutant strains of A. carbonarius.
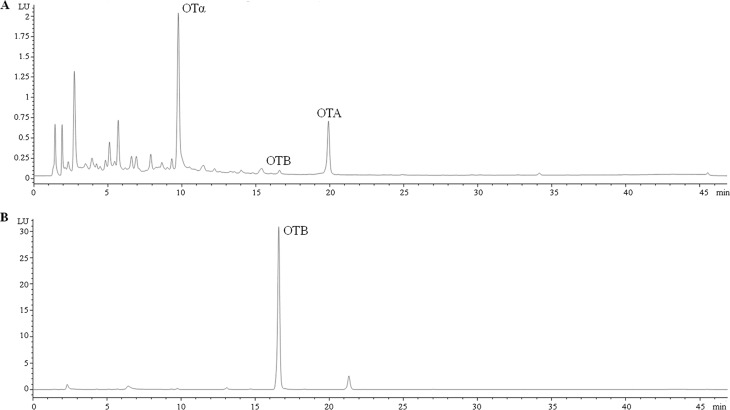
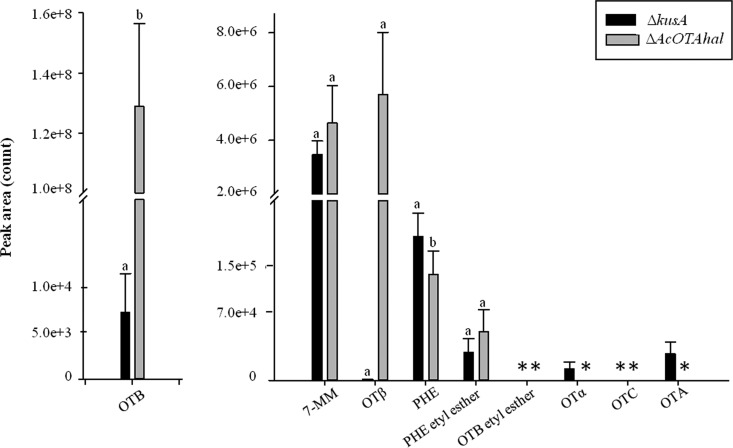
Cultures of A. carbonarius strain KB1039 and AC1501 were analyzed for the presence of OTA, OTα, and OTB by HPLC-FLD. Chemical analyses showed that OTA, OTα, and OTB were detected in the cultures of KB1039, whereas only OTB was detected in the cultures of AC1501 (Fig. 3). In particular, in KB1039, after 7 days, the mean level ± the standard error of the mean of OTA was 0.1 ± 0.04 nmol/cm2 of agar plate, and the presence of the commonly associated metabolites OTα and OTB at mean levels of 1.47 ± 0.31 and 0.04 ± 0.01 nmol/cm2, respectively, was also evident. In cultures of strain AC1501 lacking the AcOTAhal gene, after 7 days, neither OTA nor OTα was detected with limits of detection of 0.001 and 0.003 nmol/cm2, respectively. Interestingly, high levels of OTB (598.6 ± 59.2 nmol/cm2) were measured in the cultures of the mutant strain AC1501 (Fig. 3B). Cultures of A. carbonarius KB1039 and AC1501 strains were also analyzed by HPLC-HRMS to confirm the results of OTA, OTB, and OTα obtained by HPLC-FLD and for the identification of other secondary metabolites presumably involved in the biosynthesis pathway of OTA such as 7-MM, OTβ, Phe, Phe ethyl ester, OTB ethyl ester, and OTC. The simultaneous identification of these metabolites was possible through the use of the Orbitrap mass analyzer, which enables high mass resolution, high mass accuracy, a large dynamic range, and a high mass/charge range (28). OTA, OTB, OTα, OTβ, 7-MM, Phe, and Phe ethyl ester—but not OTB ethyl ester and OTC—were identified in culture extracts of A. carbonarius strain KB1039. The metabolic profile of the mutant strain A. carbonarius AC1501 was different because in addition to the absence of OTB ethyl ester and OTC, OTA and OTα were also not detected, in accordance with the knockout of the AcOTAhal gene in this strain. Specifically, Fig. 4 presents the mean peak area of each metabolite measured by HPLC-HRMS in culture extracts of KB1039 (ΔkusA) and AC1501 (ΔAcOTAhal) prepared and analyzed in triplicate. In AC1501, in addition to the absence of OTα, OTA, OTB ethyl ester, and OTC, a large increase in OTB and OTβ was also evident compared to the KB1039 strain. The increase in OTB was statistically significant (P = 0.0292), whereas the large increase of OTβ in culture extracts of AC1501 was not significant (P = 0.0687) due to the large variability of OTβ production in the three replicates. Comparable mean levels of 7-MM, Phe, and Phe ethyl ester were measured in the cultures of the two strains.
FIG 3.
HPLC-FLD chromatograms of culture extracts of A. carbonarius strain KB1039 (ΔkusA) (A) and strain AC1501 (ΔAcOTAhal) (B) diluted 10-fold. Retention times: OTα, 8.41 min; OTB, 16.5 min; and OTA, 19.9 min.
FIG 4.
Metabolites identified by HPLC-HRMS in cultures of KB1039 (ΔkusA) and AC1501 (ΔAcOTAhal) strains incubated in triplicate for 7 days. Values are means ± the standard errors. Different letters indicate statistical differences within pairs of data (P < 0.05). *, not detected.
DISCUSSION
In contrast to other important mycotoxins, until recently the biosynthesis of OTA has not been completely elucidated. The sequencing of the genome of A. niger and soon after that of A. carbonarius has clarified some aspects of OTA biosynthesis molecular basis and could lead to the definition of the entire biosynthesis pathway.
Specifically, the availability of the genome sequence of A. carbonarius allowed the characterization of the AcOTApks and AcOTAnrps genes involved in OTA biosynthesis, which have been considered the core genes of a supposed OTA biosynthetic cluster, as for most fungal secondary metabolite clusters containing genes coding for these multimodular enzymes (29, 30). The inspection of upstream and downstream regions of OTA cluster and the related annotated genes has resulted in the identification of several genes encoding putative proteins which could have a role in the OTA biosynthetic pathway according to the molecular structure of this mycotoxin. Many of these proteins deserve to be more thoroughly examined, such as a cytochrome P450 oxidase, a predicted transporter protein, one or two fungal specific transcription factors, and the halogenase that is the object of the present study.
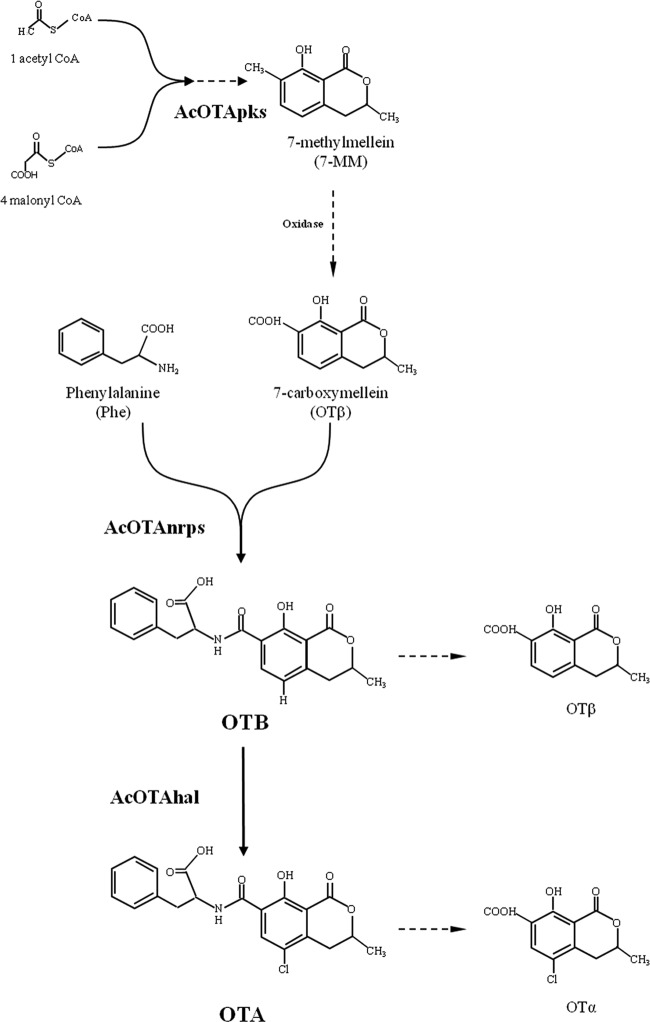
The halogenase encoding gene, which we designated AcOTAhal, was deleted by homologous recombination, and the absence of OTA and OTα in the culture of the mutant strain confirmed its biosynthetic function, since the deletion of this gene interrupted the pathway that would have led to the final production of OTA. In previous work, OTα was recognized as a degradation product of OTA (12), so its absence could be linked to the loss of production of OTA. Further, the HPLC-HRMS analysis supported the order of the reactions in the biosynthetic pathway as proposed by Gallo et al. (12) and outlined in Fig. 5, confirming that the inactivation of the AcOTAhal gene did not compromise the biosynthetic reactions upstream the chlorination step. In fact, in addition to the absence of OTA and OTα, a significant increase in OTB and, to a lesser extent, in OTβ was observed in the cultures of mutant strain AC1501. OTB is the metabolite formed by the condensation of Phe and OTβ by a peptide synthetase (AcOTAnrps), and its accumulation, together with the accumulation of OTβ as unused precursors, is the result of the suppression of the chlorination step, exclusively performed by this halogenating enzyme in the final step of the OTA biosynthetic pathway (Fig. 5). The massive accumulation of OTβ could also be due to the degradation of OTB largely increased in fungal cultures, which could be a possible target of the same proteolytic enzyme that catalyzes the degradation of OTA into OTα (31, 32). Within the panel of metabolites analyzed by HPLC-HRMS, OTC and OTB ethyl ester were not detected in the cultures of both ΔkusA and ΔkusA ΔAcOTAhal mutant strains, confirming that these metabolites should not be involved in the OTA biosynthesis pathway as already observed (12). From the analysis of the deduced protein sequence, the halogenase encoded by AcOTAhal gene contains an FAD-binding domain and a tryptophan halogenase domain.
FIG 5.
Schematic representation of the order of OTA biosynthetic pathway and the proteins demonstrated involved. Dashed arrows represent hypothesized enzymatic steps.
Generally, halogenating enzymes can be grouped in two classes: (i) a less specific haloperoxidase utilizing hydrogen peroxide and a metal cofactor-like heme iron to catalyze electrophilic halogenation reactions and a (ii) highly substrate specific halogenase requiring dioxygen (O2) to activate the halide residue. Another group of halogenating enzymes is constituted by SAM-dependent halogenases which catalyze nucleophilic halogenation reactions. The dioxygen-dependent halogenases are two-component systems which requires α-ketoglutarate or flavin (FADH2-dependent halogenase) as a cosubstrate to keep the halogenase reduced for catalytic turnover (33). FADH2-dependent halogenases activate O2 by means of the formation of flavin-bound hydroperoxide, which then oxidizes the halide. They were discovered when the biosynthesis of the antifungal agent pyrrolnitrin was investigated (34). One of the enzyme involved (PnrA) is a tryptophan halogenase able to halogenate in a site-specific manner l-tryptophan to obtain 7-chlorotryptophan, and in this case the exact positioning of the biosynthetic intermediate at the active site of the enzyme seemed to be of major importance (35). Further investigations also demonstrated that FAD and the chloride residue were bound at the same site (36). Later, the tryptophan 2 halogenase CmdE involved in the biosynthesis of chondramines, mixed PKS/NRPS natural products with cytotoxic activity, was reported to be the first example of tryptophan halogenase integrated in such a modular biosynthetic pathway (37). However, the exact mechanism by which flavin-dependent halogenases operate is still under debate, and mechanistic details, such as which active-site residues participate, may actually vary between members of the family (38). Genes for flavin-dependent halogenase have been found in a number of prokaryotic and fungal biosynthetic gene clusters for secondary metabolites (21). Halogenated natural products are widely distributed in nature, and many of them show biological activity as antibacterials, antifungals, and anticancer agents (39–41). Investigation of biosynthesis of halogenated compounds is interesting also for the biotechnological potential of these enzymes, in particular of FADH2-dependent halogenases. Usually, incorporation of halogen atoms is more advantageous than direct chemical halogenation, which could result in unwanted by-products due to the inaccessibility to specific sites of substrate (42).
The protein encoded by the AcOTAhal gene shows the greatest similarity to a radH flavin-dependent halogenase of A. niger CBS 513.88 and to the hypothetical protein ACN38_g5115 of Penicillium nordicum. The A. niger halogenase is located in the OTA cluster identified in this fungus (43). The genomic regions corresponding to OTA clusters in A. carbonarius and A. niger harbors the putative and already characterized OTA biosynthetic genes organized in the same order and direction of transcription (44). Thus, AcOTAhal is homologous to the A. niger protein above-mentioned and that initially was known as the putative FAD-binding oxidoreductase An15g07880 (43). Recently, Massi et al. (45) reported the results of a PCR screening from which the radH gene was present in the genomes of the OTA-producing isolates of A. niger, whereas the nonproducing isolates did not contain this gene, providing an indirect proof of its involvement in OTA biosynthesis. The P. nordicum protein ACN38_g5115, when subjected to in silico domain analysis, displayed the same Aspergillus halogenase domains as seen in the present study, that is, an FAD-binding domain and a tryptophan halogenase domain. It is not clear whether this halogenating enzyme is involved in OTA biosynthesis. Actually, it is not possible to retrieve the sequence of otachlPN, which was reported to be the putative protein responsible of chlorination step in OTA biosynthesis in P. nordicum (17, 19). However, it is worth mentioning that the rate of similarity among halogenase protein sequences of different origins is quite high, resulting in a AcOTAhal protein similarity higher than 60%, with a great number of halogenating enzymes also from microorganisms that apparently are non-OTA producing. Likely, this is due to the presence of very conserved functional domains. This could also be the reason why the homology between the two halogenases in A. niger and A. carbonarius appears to be higher than the homologies found for the PKSs and NRPSs involved in OTA biosynthesis in the two Aspergillus species (46). Our results are consistent with other studies demonstrating that flavin-dependent halogenases can be found working in parallel with NRPS/PKS enzymes, showing that halogenation takes place alongside synthesis rather than only as a presynthetic or postsynthetic modification (47). In addition to AcOTApks, AcOTAnrps, and AcOTAhal, other genes involved in the biosynthesis of OTA are most likely located in the same genomic region which could be described as the OTA cluster.
In this work, we analyzed the expression profile of AcOTAhal gene and of two other OTA genes we had previously characterized (AcOTApks and AcOTAnrps) during the production of OTA by A. carbonarius ITEM 5010. The results indicated a similar expression profile for the three genes examined and a strong correlation to OTA production kinetics. A similar expression profile was observed for AcOTAp450 and AcOTAbZIP, two genes located between AcOTAnrps and AcOTAhal (Fig. 1A) and that could play a role in the biosynthesis pathway as part of the OTA cluster. It was observed that as long as the genes are upregulated, OTA is accumulated progressively, and the detected upregulation of all the genes preceded the further increase of OTA production occurring at day 7 (Fig. 2). A similar timing of gene activation was also observed in P. nordicum, where OTA biosynthetic genes reached their highest level of transcription before the maximum level of OTA accumulation was detected (19). The AcOTAp450 and AcOTAbZIP genes encode a cytochrome p450 monooxygenase (ID 517149) and a bZIP transcription factor (ID 7821), respectively, which have their homologous genes in the OTA cluster of A. niger. The involvement of a p450 monooxygenase in OTA biosynthesis has already been implied in the OTA-producing A. ochraceus (48) and A. steynii (49) and is likely responsible for an oxidase step for the formation of the precursor metabolite OTβ in the biosynthesis pathway. The bZIP transcription factor in the cluster could play a role in the regulation of OTA gene expression. Fungal secondary metabolic gene clusters often contain one or more transcription factors that are required for expression of the genes for the biosynthetic enzymes (50). In fungi, bZIP (basic leucine zipper) proteins are transcription factors that regulate multiple metabolic processes other than stress response, development, and morphology (3). The transcription profiles of all of the studied genes provide strong evidence for their contribution to the production of OTA.
However, further investigation of A. carbonarius genome sequence, together with analysis of transcriptome under OTA permissive conditions, is needed to define other aspects of OTA biosynthesis and accumulation, such as the regulatory mechanism, in addition to the steps involved in the formation of the molecular structure of OTA. This would contribute to a better comprehension and consequently to the development of improved control strategies aimed to reduce OTA contamination of food products.
ACKNOWLEDGMENTS
The genome sequencing of Aspergillus carbonarius was conducted by the U.S. Department of Energy Joint Genome Institute and supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231.
We appreciate the support of the DOE Biomass Program for the research conducted at Pacific Northwest National Laboratory (PNNL), Richland, WA. We also greatly thank Scott E. Baker and Kenneth S. Bruno from PNNL for providing mutant strain KB1039 and some of the primers used in this study.
REFERENCES
- 1.Russell R, Paterson M. 2006. Identification and quantification of mycotoxigenic fungi by PCR. Process Biochem 41:1467–1474. doi: 10.1016/j.procbio.2006.02.019. [DOI] [Google Scholar]
- 2.Bohnert M, Wackler B, Hoffmeister D. 2010. Spotlights on advances in mycotoxin research. Appl Microbiol Biotechnol 87:1–7. doi: 10.1007/s00253-010-2565-8. [DOI] [PubMed] [Google Scholar]
- 3.Yin W, Keller NP. 2011. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol 49:329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petzinger E, Ziegler K. 2000. Ochratoxin A from a toxicological perspective. J Vet Pharmacol Ther 23:91–98. doi: 10.1046/j.1365-2885.2000.00244.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfohl-Leszkowicz A, Manderville RA. 2007. Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol Nutr Food Res 51:61–99. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- 6.IARC. 1993. Some naturally occurring substances, food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. IARC, Lyon, France. [Google Scholar]
- 7.Lund F, Frisvad JC. 2003. Penicillium verrucosum in cereals indicates production of ochratoxin A. J Appl Microbiol 95:1117–1123. [DOI] [PubMed] [Google Scholar]
- 8.Battilani P, Magan N, Logrieco A. 2006. European research on ochratoxin A in grapes and wine. Int J Food Microbiol 111:S2–S4. doi: 10.1016/j.ijfoodmicro.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Perrone G, Susca A, Cozzi G, Ehrlich K, Varga J, Frisvad JC, Meijer M, Noonim P, Mahakarnchanakul W, Samson RA. 2007. Biodiversity of Aspergillus species in some important agricultural products. Stud Mycol 59:53–66. doi: 10.3114/sim.2007.59.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Culebras PV, Crespo-Sempere A, Sánchez-Hervás M, Elizaquivel P, Aznar R, Ramón D. 2009. Molecular characterization of the black Aspergillus isolates responsible for ochratoxin A contamination in grapes and wine in relation to taxonomy of Aspergillus section Nigri. Int J Food Microbiol 132:33–41. doi: 10.1016/j.ijfoodmicro.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Gallo A, Knox BP, Bruno KB, Solfrizzo M, Baker SE, Perrone G. 2014. Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int J Food Microbiol 179:10–11. doi: 10.1016/j.ijfoodmicro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Gallo A, Bruno KS, Solfrizzo M, Perrone G, Mulè G, Visconti A, Baker S. 2012. New insight into the ochratoxin A biosynthetic pathway through deletion of a nonribosomal peptide synthetase gene in Aspergillus carbonarius. Appl Environ Microbiol 78:8208–8218. doi: 10.1128/AEM.02508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heussner AH, Bingle LEH. 2015. Comparative ochratoxin toxicity: a review of the available data. Toxins 7:4253–4282. doi: 10.3390/toxins7104253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knasmüller S, Cavin C, Chakraborty A, Darroudi F, Majer BJ, Huber WW, Ehrlich VA. 2004. Structurally related mycotoxins ochratoxin A, ochratoxin B, and citrinin differ in their genotoxic activities and in their mode of action in human-derived liver (HepG2) cells: implications for risk assessment. Nutr Cancer 50:190–197. doi: 10.1207/s15327914nc5002_9. [DOI] [PubMed] [Google Scholar]
- 15.Mally A, Keim-Heusler H, Amberg A, Kurz M, Zepnik H, Mantle P, Völkel W, Hard GC, Dekant W. 2005. Biotransformation and nephrotoxicity of ochratoxin B in rats. Toxicol Appl Pharmacol 206:43–53. doi: 10.1016/j.taap.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Huff WE, Hamilton PB. 1979. Mycotoxins–their biosynthesis in fungi: ochratoxins–metabolites of combined pathways. J Food Prot 42:815–820. [DOI] [PubMed] [Google Scholar]
- 17.Färber P, Geisen R. 2004. Analysis of differentially expressed ochratoxin A biosynthesis genes of Penicillium nordicum. Eur J Plant Pathol 110:661–669. doi: 10.1023/B:EJPP.0000032405.21833.89. [DOI] [Google Scholar]
- 18.Karolewiez A, Geisen R. 2005. Cloning a part of the ochratoxin A biosynthetic gene cluster of Penicillium nordicum and characterization of the ochratoxin polyketide synthase gene. Syst Appl Microbiol 28:588–595. doi: 10.1016/j.syapm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Geisen R, Schmidt-Heydt M, Karolewiez A. 2006. A gene cluster of the ochratoxin A biosynthetic genes in Penicillium. Mycotoxin Res 22:134–141. doi: 10.1007/BF02956777. [DOI] [PubMed] [Google Scholar]
- 20.Gribble GW. 2004. Natural organohalogens: a new frontier for medicinal agents? J Chem Educ 81:1441–1449. doi: 10.1021/ed081p1441. [DOI] [Google Scholar]
- 21.Neumann CS, Fujimori DG, Walsh CT. 2008. Halogenation strategies in natural product biosynthesis. Chem Biol 15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Kuck U, Hoff B. 2010. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol 86:51–62. doi: 10.1007/s00253-009-2416-7. [DOI] [PubMed] [Google Scholar]
- 23.Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu JH, Hamari Z, Han KH, Seo JA, Reyez-Dominguez Y, Scazzocchio C. 2004. Double joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Smedsgaard J. 1997. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr A 760:264–270. doi: 10.1016/S0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- 28.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. 2005. The Orbitrap: a new mass spectrometer. J Mass Spectrom 40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 29.Walsh CT, Fischbach MA. 2010. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc 132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolouli K, Mossialos D. 2012. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol Lett 34:1393–1403. doi: 10.1007/s10529-012-0919-2. [DOI] [PubMed] [Google Scholar]
- 31.Engelhardt G. 2002. Degradation of ochratoxin A and B by the white rot fungus Pleurotus ostreatus. Mycotoxin Res 18:37–43. doi: 10.1007/BF02946138. [DOI] [PubMed] [Google Scholar]
- 32.Wu Q, Dohnal V, Huang L, Kuča K, Wang X, Chen G, Yuan Z. 2011. Metabolic pathways of ochratoxin A. Curr Drug Metab 12:1–10. [DOI] [PubMed] [Google Scholar]
- 33.Butler A, Sandy M. 2009. Mechanistic considerations of halogenating enzymes. Nature 460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- 34.Hammer PE, Hill DS, Lam ST, Van Pée KH, Ligon JM. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl Environ Microbiol 63:2147–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holzer M, Burd W, Reissig HU, van Pée KH. 2001. Substrate specificity and regioselectivity of tryptophan 7-halogenase from Pseudomonas fluorescens BL915. Adv Synth Catal 343:591–595. doi:. [DOI] [Google Scholar]
- 36.Dong C, Flecks S, Unversucht S, Haupt C, van Pée KH, Naismith JH. 2005. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science 309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rachid S, Krug D, Kunze B, Kochems I, Scharfe M, Zabriskie TM, Blöcker H, Müller R. 2006. Molecular and biochemical studies of chondramide formation—highly cytotoxic natural products from Chondromyces crocatus Cm c5. Chem Biol 13:667–681. doi: 10.1016/j.chembiol.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Smith DRM, Grüschow S, Goss RJM. 2013. Scope and potential of halogenases in biosynthetic applications. Curr Opin Chem Bio 17:276–283. doi: 10.1016/j.cbpa.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M. 1995. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci Biotechnol Biochem 59:1099–1106. doi: 10.1271/bbb.59.1099. [DOI] [PubMed] [Google Scholar]
- 40.Yeh E, Garneau S, Walsh CT. 2005. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc Natl Acad Sci U S A 102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heemstra JR, Walsh CT. 2008. Tandem action of the O2- and FADH2-dependent halogenases KtzQ and KtzR produce 6,7-dichlorotryptophan for kutzneride assembly. J Am Chem Soc 130:14024–14025. doi: 10.1021/ja806467a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner C, El Omari M, König GM. 2009. Biohalogenation: nature's way to synthesize halogenated metabolites. J Nat Prod 72:540–553. doi: 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- 43.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d'Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 44.Ferracin LM, Fier CB, Vieira ML, Monteiro-Vitorello CB, Varani A, Rossi MM, Müller-Santos M, Taniwaki MH, Iamanaka BT, Fungaro MH. 2012. Strain-specific polyketide synthase genes of Aspergillus niger. Int J Food Microbiol 155:137–145. doi: 10.1016/j.ijfoodmicro.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Massi FP, Sartori D, de Souza Ferranti L, Iamanaka BT, Taniwaki MH, Vieira ML, Fungaro MH. 2016. Prospecting for the incidence of genes involved in ochratoxin and fumonisin biosynthesis in Brazilian strains of Aspergillus niger and Aspergillus welwitschiae. Int J Food Microbiol 221:19–28. doi: 10.1016/j.ijfoodmicro.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Gallo A, Ferrara M, Perrone G. 2013. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins 5:717–742. doi: 10.3390/toxins5040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wohlleben W, Stegmann E, Süssmuth RD. 2009. Molecular genetic approaches to analyze glycopeptide biosynthesis. Methods Enzymol 458:459–486. doi: 10.1016/S0076-6879(09)04818-6. [DOI] [PubMed] [Google Scholar]
- 48.O'Callaghan J, Stapleton PC, Dobson AD. 2006. Ochratoxin A biosynthetic genes in Aspergillus ochraceus are differentially regulated by pH and nutritional stimuli. Fungal Genet Biol 43:213–221. doi: 10.1016/j.fgb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Gil-Serna J, Vázquez C, González-Jaén MT, Patiño B. 2015. Clustered array of ochratoxin A biosynthetic genes in Aspergillus steynii and their expression patterns in permissive conditions. Int J Food Microbiol 214:102–108. doi: 10.1016/j.ijfoodmicro.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 50.Hoffmeister D, Keller NP. 2006. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416. doi: 10.1039/B603084J. [DOI] [PubMed] [Google Scholar]