Abstract
Antimony (Sb) is a toxic metalloid that occurs widely at trace concentrations in soil, aquatic systems, and the atmosphere. Nowadays, with the development of its new industrial applications and the corresponding expansion of antimony mining activities, the phenomenon of antimony pollution has become an increasingly serious concern. In recent years, research interest in Sb has been growing and reflects a fundamental scientific concern regarding Sb in the environment. In this review, we summarize the recent research on bacterial antimony transformations, especially those regarding antimony uptake, efflux, antimonite oxidation, and antimonate reduction. We conclude that our current understanding of antimony biochemistry and biogeochemistry is roughly equivalent to where that of arsenic was some 20 years ago. This portends the possibility of future discoveries with regard to the ability of microorganisms to conserve energy for their growth from antimony redox reactions and the isolation of new species of “antimonotrophs.”
INTRODUCTION
Antimony (Sb) occurs widely in soil and aquatic systems. It is a group 15 element in the periodic table, positioned directly below arsenic (As). It exists in four oxidation states (+V, +III, 0, and −III), of which pentavalent antimonate [Sb(V)] and trivalent antimonite [Sb(III)] are the prevalent forms in the environment (1). Being a strong chalcophilic element, Sb frequently cooccurs in sulfidic mineral phases, such as Sb2S3 (stibnite) (1, 2). In the aqueous environments at neutral pH, Sb(V) dominates as Sb(OH)6− under oxic conditions, while Sb(III) is more prevalent as Sb(OH)3 in anoxic environments (3). Furthermore, Sb shares some chemical and toxicological properties with As (4). Antimony and its compounds are considered to be hazardous pollutants by both the U.S. Environmental Protection Agency (5) and the Council of the European Communities (6). In fact, the EPA drinking water standard for Sb is lower than that for As, reflecting its greater overall toxicity. The maximum contaminant level of Sb in drinking water is 6 μg/liter, according to USEPA (7), and the level established by the Council of the European Communities (CEC) is 5 μg/liter (8). Similar to most trace metals, Sb toxicity strongly depends upon its chemical speciation (9). The general order of toxicity for Sb species is greatest in Sb(III), followed by Sb(V) and then organoantimonials (10). Due to its affinity for the thiol groups of glutathione and proteins, exposure to antimony species can cause injury in many organ systems, such as the lungs, heart, liver, and kidney (11, 12).
Antimony contamination in the environment is caused by both natural and anthropogenic activities (1, 13). Natural sources of Sb to the environment include volcanism and the weathering of Sb-bearing crustal rocks and minerals (14, 15). Antimony is widely used in the manufacture of flame retardants, small-arms ammunition, semiconductors, batteries, alloys, pigments, and catalysts (1). For many years, Sb compounds have been used in the treatment of several tropical protozoan diseases, such as leishmaniasis (16). In addition, human activities, especially increased mining and industrial emissions, have significantly accelerated the release of Sb into the environment and the associated exposure of biota to Sb (17, 18).
The present world production and reserves of Sb are estimated at almost 160,000 and 1,800,000 tons, respectively, most of which is from deposits in China, Bolivia, Mexico, Russia, South Africa, and Tajikistan (19). However, the exploitation and utilization of Sb result in increasing Sb contamination in many countries (20–25). Currently, China is the largest producer of Sb, with more than 80% of the world's supply of Sb coming from the mines of Southwest China (20). In China, the Sb concentrations in water (up to 29.4 mg liter−1), sediment (up to 1,163 mg kg−1), and soil (>2 mg kg−1) reported from the mining and smelting areas are extremely elevated compared to typical background concentrations (1 μg liters−1, 0.800 to 3.00 mg kg−1, and 0.57 mg kg−1, respectively) (20). Antimony can be taken up by plants and photosynthetic biofilms and thereby enter the food chains of contaminated environments (2, 26). This can ultimately cause a series of human health risks (27); hence, the problem of Sb pollution demands global attention.
As was the case with As, microorganisms now appear to play an important role in Sb speciation, mobility, and bioavailability in nature (28, 29). For instance, microbial Sb(III) oxidation, which transforms Sb(III) to Sb(V), could be considered a means of environmental Sb bioremediation because Sb(V) could be stably immobilized (e.g., adsorbed) and safely disposed under an oxic environment (30). Some bacteria can utilize the energy generated from the microbial Sb redox reactions to support their growth (31, 32). Therefore, a better comprehension of the mechanisms driving microbe-Sb interactions is important to elucidate the Sb biogeochemical cycle and to further develop strategies for the bioremediation of Sb-contaminated environments. A search in the NCBI PubMed central database using the word “antimony” shows an exponentially growing number of publications over the past few years (Fig. 1A), suggesting an increased interest in this toxic metalloid. However, most of the reviews that have previously been published over the last decade focused on the behavior, bioavailability, and contamination of Sb in the environment. A detailed knowledge of the molecular mechanisms underpinning the interactions of microorganisms with antimony is still very limited compared with that for other toxic metalloids, like arsenic or selenium. Thus, this review covers the latest findings on microbial Sb transformations and describes our current understanding of the enzymes, regulatory mechanisms, and metabolic pathways involved in biogeochemical cycling of Sb.
FIG 1.
The exponential growth (red line) of the number of publications about Sb in NCBI PubMed (A) and the biotransformation pathways of different Sb species (B).
MICROBIAL ANTIMONY CYCLE
Microbial transformations of Sb influence the environmental fate and toxicity of this metalloid. Microbes have coped with the toxicity of Sb using various strategies to thrive in Sb-rich environments, such as Sb(III) efflux, Sb(V) reduction, Sb(III) methylation, and Sb(III) oxidation (29). These microbial Sb transformations mediate the conversion of Sb compounds among Sb(III), Sb(V), and organoantimonials (Fig. 1B).
Antimonite resistance.
Different strategies are employed by microbes to reduce the accumulation of toxic intracellular Sb(III), such as first inhibiting its entrance into the cell, promoting its active extrusion from the cell if it gains entry, or achieving its sequestration in a nontoxic form within the cell (33). Efflux of antimony is one of the most important mechanisms adopted by microorganisms to protect them from the toxicity of Sb. No specialized channel for antimony uptake has been identified, and possibly no such channel has evolved, because antimony is not an essential trace nutrient (11). At physiological pH, Sb is present as noncharged Sb(OH)3 in solution, and because of its structural similarities to glycerol, the uptake of Sb(III) into prokaryotic and eukaryotic cells is often achieved by aquaglyceroporins (34). The glycerol facilitator GlpF in Escherichia coli was the first aquaglyceroporin identified to transport Sb(III) into bacterial cells (35, 36). Later, Fps1p, the yeast homologue of GlpF, was also found to mediate the uptake of Sb(III) into Saccharomyces cerevisiae (33). The deletion of fps1 improved the tolerance level of S. cerevisiae to Sb(III), while constitutive expression of this gene resulted in hypersensitivity. Interestingly, the expression of fps1 was repressed when cells were exposed to Sb(III), indicating a coordinated regulatory network to protect cells from the toxic effects of Sb(III) (33). In Leishmania species, Sb(III) entered cells primarily through an aquaglyceroporin named AQP1 (37). In general, the transcription level of AQP1 correlated well with the accumulation of, and the sensitivity level for, Sb(III) in Leishmania cells (38). It was suggested that the route of Sb(V) influx is different from Sb(III) (39). However, the uptake mechanism of Sb(V) remains unknown. Whether it enters the cells through the phosphate transport systems used by As(V), such as Pit or Pst, remains unresolved.
At present, two different transporter families have been shown to be responsible for prokaryotic Sb(III) efflux: the ArsB protein, which belongs to the ion transporter superfamily, and Acr3p, belonging to the arsenite carrier family. The ars operon that confers both arsenic and antimony resistance has been found on both plasmids and the chromosome (40, 41). The three-gene operon arsRBC was present in E. coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus (42–44). The extended five-gene operon arsRDABC was first found in E. coli plasmids R773 and R46 and on Acidiphilium multivorum plasmid pKW301 (45, 46). Expression of the ars operons was induced in response to the presence of both As(III) and Sb(III) (43). The ArsR acts as a transcriptional repressor regulating the expression of itself and downstream genes of the ars operon. The arsD gene encodes an As chaperone that transfers As(III) and Sb(III) to ArsA, and ArsA acts as an ATPase, which binds to the As(III)/Sb(III) carrier protein ArsB to form an ATP-coupled efflux pump (47). Meng et al. (36) demonstrated that ArsB is a trivalent metalloid/H+ antiporter. In the presence of ArsA, ArsB catalyzes the extrusion of As(III)/Sb(III) by the hydrolysis of ATP, while it can extrude As(III) and Sb(III) by itself using the electrochemical proton gradient. ArsC was shown to be a cytoplasmic As(V) reductase, reducing As(V) to As(III), thereby enabling its efflux from the cell through the ArsAB pump. ArsC might be involved in the resistance to Sb(III) (47, 48), but it is not known whether it is directly related to the intracellular reduction of Sb(V).
Another trivalent metalloid/H+ antiporter, Acr3p, and its homolog, YqcL, which are mainly present in Actinobacteria and Alphaproteobacteria, can substitute for ArsB, also functioning as an Sb(III) efflux pump (28, 49). Acr3p is also found in archaea and eukaryotes (50), in which a three-gene cluster (acr1, acr2, and acr3) is responsible for Sb(III) resistance. Kang et al. (51) showed that deletion of acr3 in Agrobacterium tumefaciens 5A resulted in more sensitivity to Sb(III). In addition, it has been shown that the expression of acr3 is stimulated by Sb(III), and its gene product conferred Sb(III) tolerance in yeast (33, 52, 53). In S. cerevisiae, the cadmium factor protein Ycf1, which belongs to ABC transporter superfamily, is another system conferring Sb(III) tolerance through vacuolar sequestration (54). In Leishmania, the ABC transporter PGPR and ABCI4 were reported to be involved in Sb(III)-thiol extrusion (55).
Antimonate reduction.
Antimonate reduction appears to be widespread in the environment, and it is prone to occur under anaerobic conditions (3). It is known that Sb(V) can be abiotically reduced to Sb(III) by Fe(II)-containing minerals (56–58). A marine macroalga, Sargassum sp., was the first reported organism able to reduce Sb(V) in seawater (59). In the treatment of leishmaniasis, several studies suggested that Sb(V), when used as a prodrug medicine, might be reduced in both the vertebrate host and the parasites (60–63). However, the knowledge of bacterial Sb(V) reduction is limited.
Kulp et al. (64) reported anaerobic bacterial reduction of Sb(V) in anoxic sediments. Sb(V) reduction was coupled to a dissimilatory respiratory pathway, which utilized acetate or lactate as the electron donor. That same year, an Sb(V)-reducing bacterium, Bacillus sp. MLFW-2, was isolated and found to generate energy from anaerobic Sb(V) reduction (32), and another Sb(V)-respiring isolate was isolated from Sb-contaminated industrial sediments (65). The molecular mechanism of bacterial Sb(V) reduction remains unknown, and the enzymes involved in this reaction have not yet been identified.
Bacterial Sb(V) reduction is not only a respiratory pathway but also a promising bioremediation strategy, since Sb(III) can readily precipitate with sulfide or be strongly absorbed by Fe phases in a reducing environment (64, 66, 67). A study by Hockmann et al. (18) indicated that Sb(V) could be rapidly reduced to Sb(III) in anaerobic calcareous soil by the indigenous microorganisms. The generated Sb(III) subsequently bound to the surface of iron (hydr)oxides, which led to the immobilization of Sb. In addition, sulfate-reducing bacteria (SRB) were employed to remove Sb(V) from Sb mine drainage (68). The SRB converted sulfate ions into sulfide that reduced Sb(V) to Sb(III) and resulted in the precipitation of stibnite (Sb2S3). Moreover, a chemoautotrophic microorganism belonging to the Rhizobium genus was found to be able to use H2 as the sole electron donor for the reduction of Sb(V), producing an Sb(III) precipitate in the form of Sb2O3 (69). Therefore, bacterial Sb(V) reduction holds promise for the anaerobic biotreatment of wastewater containing toxic Sb(V).
Antimonite methylation.
Methylation of inorganic Sb can influence the environmental mobility, toxicity, and bioaccumulation of Sb (Fig. 1B) (70). The presence of stibine (STB; SbH3), monomethylstibine (MMSb), and dimethylstibine (DMSb) was first reported in natural waters by Andreae et al. (71). Subsequently, the presence of Sb volatile and methylated species was further observed in freshwater, seawater, geothermal waters, sewage, soils, sediments, and landfill gas (71–74). In addition, methylantimony species have been found in plants, such as pondweed (Potamogeton pectinatus), moss (Drepanocladus sp.), and liverwort (75–77).
In contrast to arsenic biomethylation, which has been known for several decades (78), the biomethylation of Sb is of relatively recent interest. So far, Sb biomethylation has been detected in strains of fungi, methanogenic archaea, and bacteria. The filamentous fungi Scopulariopsis brevicaulis and Phaeolus schweinitzii have been found to generate STB, DMSb, TMSb, and some nonvolatile methylantimony species during aerobic growth (79–82). In addition, the biovolatilization and bioaccumulation of antimony by S. brevicaulis were recently quantified (83). The aerobic yeast Cryptococcus humicolus was also reported to biomethylate both inorganic Sb(III) and Sb(V) (84). It has been suggested that the toxic gases generated from Sb methylation by the fungus S. brevicaulis in crib mattresses might be a cause for sudden infant death syndrome (SIDS) (85). However, further studies indicated that a mix of common environmental Bacillus species strains in crib mattress contributed to the formation of TMSb and some nonvolatile methylantimony species, but no causal relation to SIDS was proven (86, 87).
In the process of the anaerobic digestion of sewage sludge, three methanogenic archaea (Methanobacterium formicicum, Methanosarcina barkeri, and Methanobacterium thermoautotrophicum), a sulfate-reducing bacterium (SRB) (Desulfovibrio vulgaris), and a peptolytic bacterium (Clostridium collagenovorans) were shown to produce TMSb in their culture headspaces; among these, M. formicicum displayed strong methylating activity and could also produce STB, MMSb, and DMSb (88). Another study demonstrated that biomethylation of Sb was stimulated by strains of methanogenic archaea and SRB (89). In addition, a Gram-positive strain, Clostridium glycolicum ASI-1, could convert inorganic Sb into the volatile derivatives STB, DMSb, and TMSb (90). Despite using organic Sb as a substrate, the production of TMSb could only be accomplished by the transformation of trimethyldibromoantimony in pure culture of strain Pseudomonas fluorescens K27 (91). Low yields of MMSb, DMSb, and TMSb by an aerobic Flavobacterium sp. strain suggested that Sb methylation may be a fortuitous process rather than a primary resistance mechanism (70). This is consistent with the results of studies with S. brevicaulis and C. humicolus (80, 81, 92). However, the molecular mechanisms of Sb methylation have not been clarified, and it appears that Sb is methylated much more slowly than is arsenic (29, 93).
Because of the similarity in physicochemical properties and the cooccurrence of arsenic and Sb in the natural environment, it is important to understand the effect of arsenic on Sb biomethylation. It was found that Sb biomethylation by S. brevicaulis, Flavobacterium sp., and Cryptococcus humicolus was enhanced in the presence of arsenic (70, 92, 94), while conversely, arsenic biomethylation was significantly inhibited by the presence of Sb (94). In contrast, Hartmann et al. (92) reported that the addition of arsenic not only enhanced the biomethylation of Sb by C. humicolus but also influenced the speciation of Sb. In addition, Sb biomethylation has been shown to utilize the methylation pathway proposed by Challenger in a study that utilized isotopically labeled antimonite (Sb123) (89), whereby antimony methylation demonstrates a stepwise reduction process of monomethyl-, dimethyl-, and trimethylantimony (78). Therefore, Sb biomethylation probably occurs via similar or identical mechanisms to arsenic and is catalyzed, at least in part, by arsenic methyltransferase (78, 89, 95). However, no studies have directly identified genes and enzymes that are involved in arsenic methylation that are also responsible for Sb biomethylation.
Antimonite oxidation.
Although little is known about the geochemical properties of Sb, some studies have indicated that Sb(III) adsorbs more strongly to surfaces and over a wider range of pH than does Sb(V) (67, 96), a situation that is reversed for arsenic oxyanions (e.g., see reference 97). Thus, Sb(III) oxidation may critically affect the hydrologic mobility of Sb in the environment (Fig. 1B) (67). In nature, Sb(III) is thermodynamically predicted to be dominant in anoxic environments, while Sb(V) is dominant in oxic environments (72, 98). Thus, any Sb(III) molecules that enter oxic environments tend to be oxidized to Sb(V), a situation that is similar to As(III).
Abiotic dark oxidation of Sb(III) with O2 is extremely slow, with a half-life of 170 years at pH 8.5 in homogeneous solutions (99). In contrast, H2O2-linked oxidation is much faster, with a half-life of 118 days for 1 μM H2O2 held at pH 8.0 (99, 100). Several other oxidants also have the potential to oxidize Sb(III) to Sb(V), including natural and synthetic Fe and Mn oxyhydroxides (101, 102), humic acids (103), and iodate (104). In addition, the oxidizing capacities of Fe and Mn oxyhydroxides can transform As species (105, 106), and the amorphous Fe and Mn oxyhydroxides present in natural water and sediment also play a detoxifying role by adsorbing and oxidizing Sb(III). It was found that amorphous Fe and Mn oxyhydroxides, either natural or synthetic, could effectively oxidize Sb(III) under different pH conditions (101).
Antimony(III) can also be oxidized via photo-induced oxidation in natural surface waters, especially when adsorbed to goethite (107). In experiments with seawater, the Sb(III) photo-oxidation rate was increased in the presence of various live phytoplankton species (e.g., Chlorella autotrophica, Dunaliella salina, Nannochloropsis sp., and Tetraselmis subcordiformis), and the oxidation rate increased with higher cell densities (108), although it was not clear if this was a direct metabolic effect or that of an interaction with cellular exudates. In the case of humic acids, the Sb(III) oxidation rate was 9,000 times faster in the light than in the dark (103). It was reported that Sb(III) bound to natural organic matter and mineral particles is oxidized by photo-oxidants more readily because of the change in electron density that results from adsorption (67, 103). Moreover, in the case of Sb(III) adsorbed onto goethite, oxidation only occurs in the light and was pH dependent, increasing at pH values of >5 (107). Since the Sb(III)-oxidizing bacteria reported so far have all been isolated from neutral-pH-range environments (31, 109–115), it appears that microbes may play a major role of Sb(III) oxidation at circumneutral pH.
The first report that bacteria in ore deposits could oxidize Sb(III) to Sb(V) came from a Russian scientist (31), yet beyond that benchmark work, the field remained dormant for nearly 40 years. Although the amount of research currently being conducted on Sb is growing, our understanding of the role that microbial Sb(III) oxidation plays in the biogeochemical cycle of Sb remains far from complete. Nonetheless, the results reported by several research groups in recent years suggest that microbial processes play an important role in the Sb cycle.
Diversity of antimonite-oxidizing bacteria.
An Sb(III)-oxidizing bacterium, Stibiobacter senarmontii, was found to be able to use the energy produced by Sb(III) oxidation with O2 to support chemoautotrophic growth (31). The Sb(III)-oxidizing strains that have been described since that early study generally oxidize Sb(III) during heterotrophic growth, suggesting that this process may serve as a cellular detoxification mechanism rather than one whereby energy is conserved from the oxidation to support the biochemical incorporation of CO2 into the cell's organic matrix. Due to an increased focus on bacterial Sb(III) oxidation, >60 Sb(III)-oxidizing strains were isolated from mining soil (112–114) and contaminated sediments (115–117). Chemoautotrophy, as defined by Sb(III)-dependent growth and inorganic carbon fixation, is more difficult to achieve experimentally. Nonetheless, Terry et al. (115) conducted growth experiments with Variovorax paradoxus IDSBO-4 using radiolabeled [14C]bicarbonate and demonstrated that aerobic Sb(III) oxidation in that strain was coupled to the fixation of CO2 in an apparent chemoautotrophic process, thereby reinforcing the earlier observations made with S. senarmontii and opening the possibility of its wider occurrence in nature.
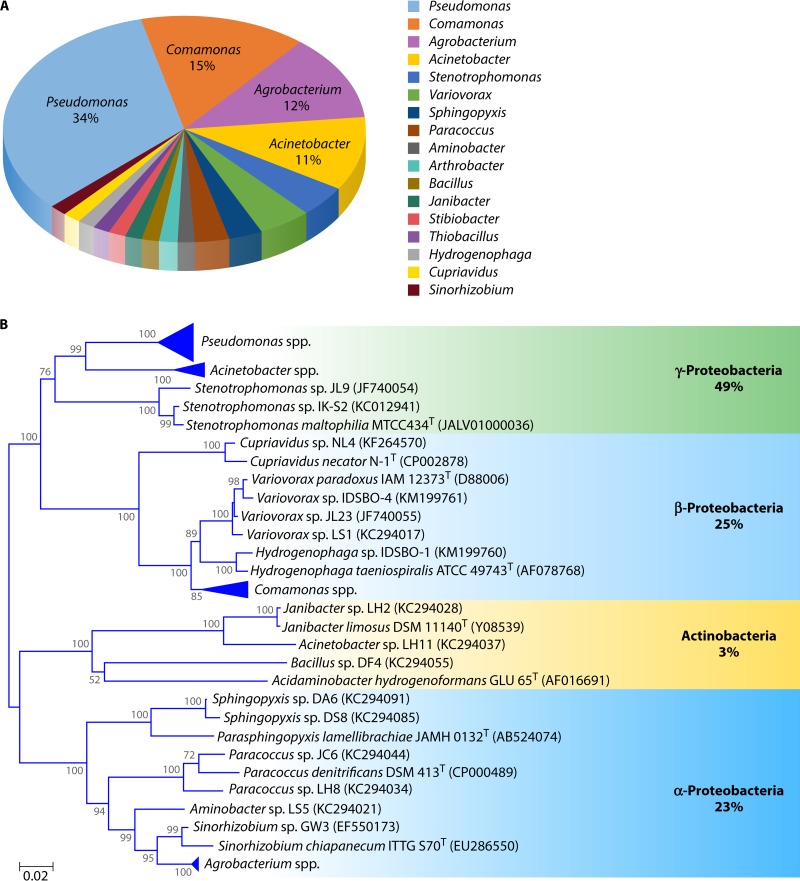
A compilation of the genera found and a phylogenetic tree of the bacterial strains shown to oxidize Sb(III) are given in Fig. 2. The Sb(III)-oxidizing strains identified thus far belong to 17 genera, including Pseudomonas (22 strains), Comamonas (10 strains), Agrobacterium (8 strains), Acinetobacter (7 strains), Stenotrophomonas (3 strains), Variovorax (3 strains), Paracoccus (2 strains), Sphingopyxis (2 strains), Aminobacter (1 strain), Arthrobacter (1 strain), Bacillus (1 strain), Janibacter (1 strain), Stibiobacter (1 strain), Thiobacillus (1 strain), Hydrogenophaga (1 strain), Cupriavidus (1 strain), and Sinorhizobium (1 strain) (31, 109, 111–116). Among all of these Sb(III)-oxidizing strains, Pseudomonas, Comamonas, Agrobacterium, and Acinetobacter are four major genera that make up 34%, 15%, 12%, and 11% of known Sb(III)-oxidizing strains, respectively (Fig. 2A). Of the 65 strains listed in this tally, only two thus far appear to be lithoautotrophs, one of which (S. senarmontii) has been lost. Unlike the case for As(III) (118), to date, there are no examples of anaerobes that can oxidize Sb(III) by using it as an electron donor to support anoxygenic photosynthesis.
FIG 2.
The percentages (A) and a neighbor-joining (NJ) phylogenetic tree (B) based on 16S rRNA gene sequences of the published Sb(III)-oxidizing strains. The Pseudomonas spp. include 22 Sb(III)-oxidizing strains (DA2, DC5, DF12, DF11, DA5, DF3, DF9, DC8, DC7, DS4, DF7, TC13, JC11, DS7, DF8, DF5, DA4, NL6, IK-S1, NL10, NL2, and NL5). The Acinetobacter spp. include seven Sb(III)-oxidizing strains (DC2, LH3, LH4, JL7, DS2, NL1, and NL12). The Comamonas spp. include 10 Sb(III)-oxidizing strains (JL25, JL40, DF1, DS1, DF2, JL13, JL12, JC9, S44, and NL11). The Agrobacterium spp. include eight Sb(III)-oxidizing strains (C58, 5A, GW4, C13, LY4, TS43, TS45, and D14) (see Table S1 in the supplemental material). Among these, the Sb(III) oxidation capabilities of six Agrobacterium (strains C58, C13, LY4, TS43, TS45, and D14) and Sinorhizobium sp. GW3 are unpublished. All of the type strains are used for taxonomic determination without knowing their Sb(III) oxidation abilities. Bootstrap values (>50%) are shown at nodes as percentages of 1,000 replicates. Bar, 0.02 substitutions per nucleotide position.
All of these Sb(III)-oxidizing strains could be classified into Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Actinobacteria (Fig. 2B). Among all of these Sb(III)-oxidizing strains, 49% belong to Gammaproteobacteria, and among these, Pseudomonas and Acinetobacter are two of the most common species (Fig. 2A). Comamonas strains belong to Betaproteobacteria, while Agrobacterium strains are members of the Alphaproteobacteria. The strains belonging to Betaproteobacteria showed the highest Sb(III) oxidation rate; for example, Comamonas testosteroni S44 could completely oxidize 50 μM Sb(III) to Sb(V) within 3 days (114). It is interesting to note that C. testosteroni S44 could not oxidize As(III) (119), indicating that the molecular mechanism of Sb(III) oxidation may at times be different from As(III) oxidation (116). Sb-dependent chemoautotrophic growth of V. paradoxus strain IDSBO-4 was able to oxidize ∼500 μM Sb(III) to Sb(V) over a 10-day incubation period (115). Studies that attempt to show clear Sb-dependent growth are difficult, as they must employ higher concentrations (millimolar range) of this toxic electron donor to elicit significant increases in cell density over the incubation period.
Biotic antimonite oxidation mediated by AioA and AnoA.
Bacterial As(III) oxidation involves the As(III) oxidase AioBA or ArxAB (120, 121). AioBA functions as an aerobic As(III) oxidase (122), while ArxAB catalyzes the anaerobic oxidation of As(III) (121). Based on the similar chemistries between As and Sb, it has been considered that they may share the same resistance and oxidation mechanisms. Indeed, the ars operon conferring arsenic resistance can be induced by Sb(III) and can also transport Sb(III) out of the cell (36). In contrast, Sb(III) does not induce the transcription of aioBA (123). Even though there is a previous study that reported that the oxidation of Sb(III) and As(III) required different biochemical pathways (116), more recent literature has shown that AioBA is able to oxidize Sb(III) both in vivo and in vitro, although the aioBA gene is expressed only by the presence of As(III) and not Sb(III) (123). However, novel Sb(III) oxidation biochemical pathways were also implicated because the disruption of aioA gene only reduced the Sb(III) oxidation rate, but did not eliminate it, thereby implying the cooccurrence of another mechanism (123). Previous studies found that the Comamonas strains could only oxidize Sb(III) but not As(III) (114, 119, 124, 125). Furthermore, Hydrogenophaga taeniospiralis IDSBO-1, isolated by Terry et al. (115), was shown to possess the aioA gene and oxidized As(III), but not Sb(III), under aerobic conditions. That strain also exhibited anaerobic Sb(III) oxidation coupled to the reduction of nitrate via an unknown enzymatic pathway.
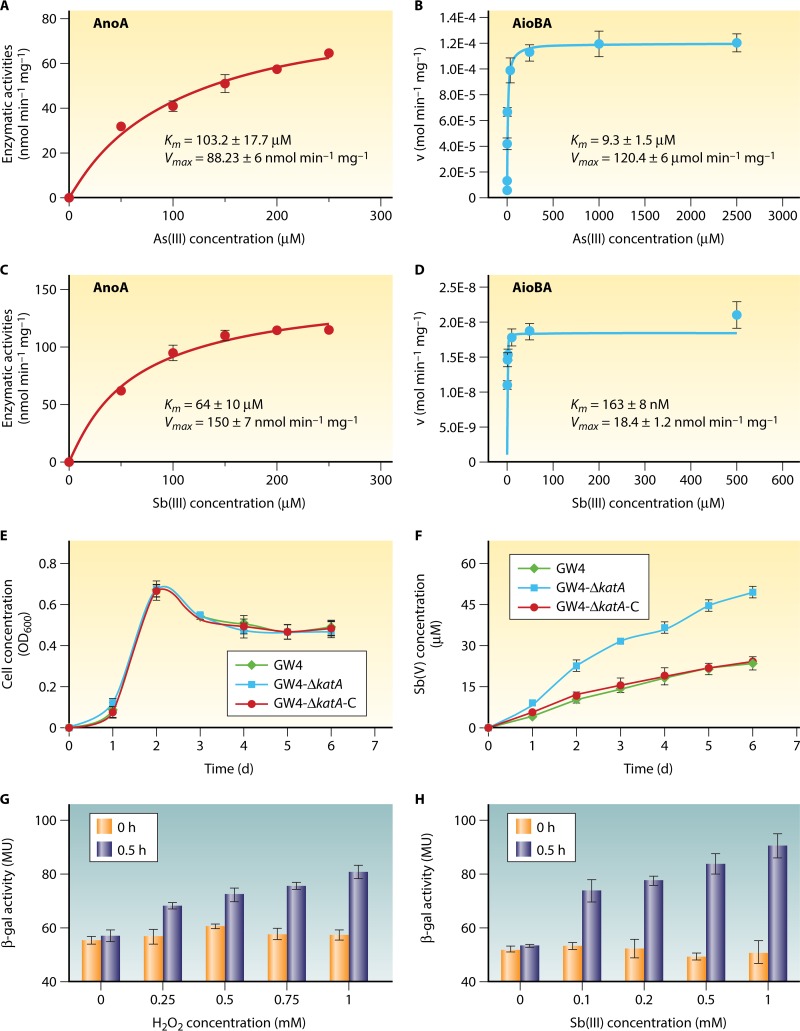
By using a proteomics approach, Li et al. (117) discovered an oxidoreductase (AnoA) in A. tumefaciens GW4 that was induced by the presence of Sb(III). The disruption of anoA reduced resistance to Sb(III) and also decreased the Sb(III) oxidation rate by ∼27% compared with that of the wild-type strain, while the overexpression of anoA increased the Sb(III) oxidation rate by ∼34%. In addition, heterologous expression of AnoA significantly increased the Sb(III) oxidation rate in E. coli (117). Acting as a novel Sb(III) oxidase, AnoA could also oxidize As(III) in vitro, with a Km of 103.2 ± 17.7 μM and a maximum rate of metabolism (Vmax) of 88.23 ± 6 nmol min−1 mg−1 (Fig. 3A). Using Sb(III) as a substrate, AnoA yielded Km and Vmax values of 64 ± 10 μM and 150 ± 7 nmol min−1 mg−1, respectively (Fig. 3C). In contrast, based on published data (123), AioBA yielded Km and Vmax values for As(III) of 9.3 ± 1.5 μM and 120.4 ± 6 μmol min−1 mg−1, respectively (Fig. 3B). The addition of Sb(III) yielded Km and Vmax values of 163 ± 8 nM and 18.4 ± 1.2 nmol min−1 mg−1, respectively (Fig. 3D). These results indicated that AnoA tends to catalyze the Sb(III) oxidation more efficiently than As(III) oxidation, while AioBA is likely to favor oxidation of As(III), although both enzymes could oxidize both As(III) and Sb(III). The existence of the novel Sb(III) oxidase AnoA may explain the occurrence of discernible Sb(III) oxidation in bacteria that lack the As(III) oxidase AioBA (114).
FIG 3.
The comparison of Michaelis-Menten kinetics of AnoA and AioBA for As(III) and Sb(III), and the influence of H2O2 concentration on bacterial Sb(III) oxidation efficiency of A. tumefaciens GW4. (A and B) Kinetic data for As(III). (C and D) Kinetic data for Sb(III). The data from panel A are from our unpublished data, the data from panels B and D are from reference 123, and the data from panel C are from reference 117. (E) The growth curves of strain GW4, the katA mutant strain, and the katA complementary strain. (F) Sb(III) oxidation profiles of the three strains as in panel E (shown with the same symbols in panel E). (G and H) The lacZ reporter assays of katA gene with the addition of H2O2 and Sb(III), respectively (panels G and H have the same symbols). The data are shown as the mean of the results from three replicates, with the error bars representing the standard deviation (SD). v, volume; OD600, optical density at 600 nm; d, days; β-gal, β-galactosidase; MU, Miller units.
Based on genome analysis, the antimonite oxidase gene anoA (117) exists in all of the arsenite-oxidizing Agrobacterium and Comamonas strains tested thus far, and the gene exists in other as-yet-untested bacterial strains. To understand the phylogenetic relationship among AnoA in different bacteria, we performed phylogenetic analysis based on amino acid sequences of the putative AnoA from 10 Agrobacterium strains, together with five Rhizobium strains, three Sinorhizobium strains, and two Comamonas strains (see Fig. S1 in the supplemental material). The AnoA can be classified into two main groups. One group contains the AnoA from strains of Agrobacterium, Rhizobium, and Sinorhizobium, and another group contains AnoA of Comamonas strains. This indicates that AnoA in Comamonas has a distant phylogenetic relationship from that in Agrobacterium, Rhizobium, and Sinorhizobium strains. Based on the literature published to date, the deletion of either aioBA or anoA only reduces the Sb(III) oxidation rate but does not completely eliminate Sb(III) oxidation, implying (but not proving) the possible existence of another mechanism(s) of Sb(III) oxidation (117, 123).
Abiotic antimonite oxidation is mediated by H2O2 and possible regulatory mechanisms involved.
In a variety of naturally occurring surface waters, hydrogen peroxide (H2O2) is present at concentrations exceeding 10−7 mol liter−1 (i.e., 10 nM) and is thought to play a key role in the redox chemistry of a number of trace elements in aquatic environments (126–129). Sb(III) oxidation by H2O2 has been studied over a wide range of pH values, ionic strengths, and temperatures, and it may be relevant in surface water with elevated H2O2 with alkaline pH values, such as seawater (100). H2O2 is not only widespread in natural surface water and rainwater but also exists within bacterial cells.
Aberrant electron flow especially under stress conditions from the electron transport chain or cellular redox enzymes to O2 leads to the production of reactive oxygen species (ROS) in bacterial cells (130). The harmful ROS, including superoxides (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (·OH), can cause damage to [Fe-S] clusters, protein carbonylation, membrane lipid peroxidation, and DNA damage (131). Superoxide dismutase (Sod), which catalyzes the dismutation of O2− to H2O2 and O2, is part of a first-line defense against these ROS and commonly occurs in nearly all aerobic bacteria. Catalases and peroxidases represent the second line of defense against ROS by being able to consume H2O2 (131). It has been reported that KatA is a major catalase that can be detected during all phases of growth (132). Therefore, the katA gene in the Sb(III)-oxidizing strain A. tumefaciens GW4 was disrupted, and the mutant strain GW4-ΔkatA and its complemented strain GW4-ΔkatA-C were created. The wild-type strain, GW4-ΔkatA, and GW4-ΔkatA-C showed consistent growth profiles in chemically defined medium (CDM) after the addition of 50 μM Sb(III) (Fig. 3E). The disruption of katA significantly increased the Sb(III) oxidation rate (Fig. 3F), which may have been caused by the increasing cellular H2O2 concentration (130). Consistent with a previous study by Khakimova et al. (133), H2O2 could induce the expression of KatA in strain GW4 (Fig. 3G). Moreover, the addition of Sb(III) was also able to stimulate the expression of katA (Fig. 3H). These results indicated that H2O2 may act as a chemical oxidant in Sb(III) oxidation, along with the aforementioned enzymatic reactions.
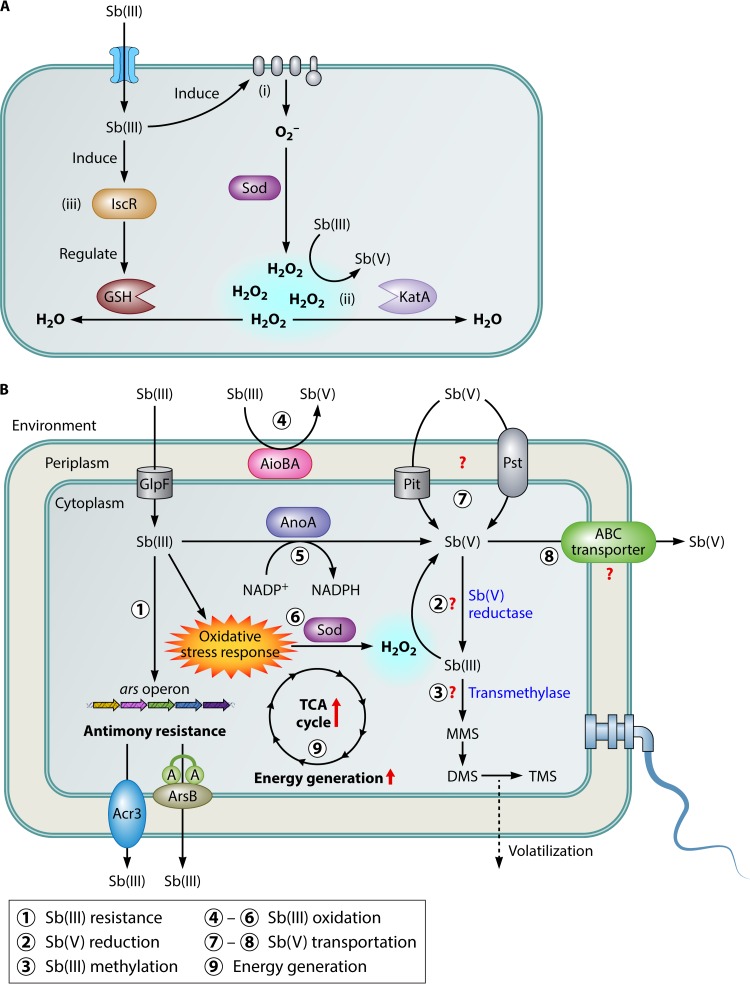
In addition to catalase, glutathione was considered to be a vital component of the bacterial oxidative stress response (134). A previous study with C. testosteroni S44 showed that the [Fe-S] assembly transcription factor IscR could positively contribute to glutathione (GSH) formation, possibly through the regulation of IscS-mediated cysteine desulfurization (134). The expression of iscR was induced by Sb(III), and the deletion of iscR decreased the cellular GSH content. These results suggested that bacterial Sb(III) oxidation was partly under the regulation of IscR (134). The hypothetical model of IscR's regulation of bacterial Sb(III) oxidation may be summarized by the following: (i) Sb(III) can induce the bacterial oxidative stress response, leading to the production of H2O2; (ii) the induced H2O2 oxidizes Sb(III) to Sb(V) under alkaline conditions; and (iii) IscR is involved in the regulation of GSH formation. Then, H2O2 is consumed by KatA and GSH, which might also affect bacterial Sb(III) oxidation (Fig. 4A). Although there are other regulators of the bacterial oxidative response, such as the Mer-like redox sensor SoxR and the LysR regulator OxyR (131), their function(s) with respect to Sb(III) oxidation has not been determined.
FIG 4.
Overview of mechanisms of bacterial antimonite resistance and oxidation. (A) A hypothetical model of IscR's regulation of bacterial Sb(III) oxidation. (i) Sb(III) induced the production of H2O2 via the bacterial oxidative stress response and subsequently H2O2 oxidized Sb(III) to Sb(V). (ii) H2O2 was partially consumed by catalase KatA. (iii) Sb(III) induced the expression of [Fe-S] assembly transcription factor IscR, which could positively contribute to GSH formation. Then, H2O2 was partially consumed by GSH. (B) Cellular events are represented on this model according to the published literature. Sb(III) is taken up through glycerol channel and extruded from the cell by Acr3 and ArsAB, and transportation of Sb(V) remains unknown. Bacteria obtained Sb(III) resistance by the ars operon. In addition, Sb(III) oxidation, Sb(V) reduction, and Sb(III) methylation were also involved in bacterial Sb detoxification. For energy generation, Sb(III) could induce activation of the TCA cycle and produce energy to protect against the toxicity of Sb.
METABOLIC PATHWAYS ASSOCIATED WITH ANTIMONITE RESISTANCE
A proteomics approach was used to study Sb resistance and oxidation in Leishmania spp. (135–138), Miscanthus sinensis (139), and Agrobacterium tumefaciens (117). The proteomics analysis in Leishmania spp. showed that its mechanism of antimony resistance is complex, incorporating aspects of protein folding/chaperones, stress response, antioxidant/detoxification, diverse metabolic processes, RNA/DNA processing, and de novo protein biosynthesis (135–138). The proteomic study of A. tumefaciens GW4 revealed that Sb(III) could influence the Ars resistance, the Sb(III) oxidase AnoA, phosphate metabolism, carbohydrate transport and metabolism, and the metabolism of lipids, purines, and amino acids (Table 1).
TABLE 1.
Proteins induced by the addition of Sb(III) in A. tumefaciens GW4a
| Gene name | Protein name | Accession no. | Upregulated ratio [zero Sb(III):50 μM Sb(III)]b |
|---|---|---|---|
| Antimony resistance | |||
| arsC1 | Arsenate reductase | AFM38847 | 1.0:2.9 |
| arsC2 | Arsenate reductase | AFM38848 | 1.0:4.7 |
| ohr | Organic hydroperoxide resistance protein | KDR90118 | 1.0:3.2 |
| Antimonite oxidation | |||
| anoA | Oxidoreductase | KDR88348 | 1.0:4.1 |
| Phosphate metabolism | |||
| pstS2 | Phosphate-binding protein | KDR86346 | 1.0:2.0 |
| ppa | Putative phosphatase | KDR90647 | 0:11.2 |
| phnM | Metal-dependent hydrolase involved in phosphonate metabolism | KDR86941 | 1.0:2.8 |
| phnI | Putative enzyme of phosphonate metabolism | KDR86951 | 1.0:4.1 |
| afuA | ABC transporter, substrate-binding protein | KDR89957 | 1.0:2.7 |
| Carbohydrate transport and metabolism | |||
| pdhB | Pyruvate dehydrogenase E1 component, beta subunit | KDR89057 | 1.0:2.7 |
| pfp | Pyrophosphate fructose 6-phosphate 1-phosphotransferase | KDR87902 | 1.0:2.8 |
| ugpB1 | Periplasmic glycerol-3-phosphate-binding protein | KDR87393 | 0:5.3 |
| ugpB2 | Periplasmic glycerol-3-phosphate-binding protein | KDR89469 | 0:8.7 |
| acnAb | Aconitate hydratase | KDR88332 | 1.0:2.1 |
| sdhAb | Succinate dehydrogenase | KDR89039 | 1.0:3.3 |
| fumCb | Fumarate hydratase | KDR89425 | 1.0:1.7 |
| Lipid transport and metabolism | |||
| sitA | Manganese ABC transporter, periplasmic-binding protein | KDR90951 | 1.0:2.1 |
| Purine metabolism | |||
| cpdP | 3′,5′-Cyclic-nucleotide phosphodiesterase | KDR86320 | 1.0:25.9 |
| Amino acid metabolism | |||
| trpB | Trp repressor-binding protein | KDR87480 | 1.0:3.9 |
Certain gene/protein and ratio data are from reference 117.
Genes tested by qRT-PCR.
The arsenic resistance system (ars) was shown to catalyze the efflux of both As(III) and Sb(III) and was induced by Sb(III) (36). Consistent with these results, the upregulation of the Ars resistance system by Sb(III) could be considered for use as a positive control for the validity of this proteomic analysis in A. tumefaciens GW4. The phosphate system and the proteins involved in phosphate and phosphonate metabolism were both upregulated with the addition of Sb(III) (Table 1). The induction of PstS2 in the presence of Sb(III) (Table 1) implies that Pst2 may have an effect on bacterial Sb(III) oxidation (117).
In comparative proteomic and genomic analyses in the presence or absence of As(III), the enzymes involved in the tricarboxylic acid (TCA) cycle were upregulated after the addition of As(III), indicating the bacteria needed a large amount of energy to resist As(III) toxicity (140–142). In contrast, the TCA cycle in A. tumefaciens GW4 was downregulated by the addition of As(III). Interestingly, this strain could use the energy generated from As(III) oxidation to support growth (Q. Wang, K. Shi, X. Wang, Y. Han, J. Li, L. Wang, J. He, M. Li, and G. Wang, unpublished data), indicating that As(III) is not simply a toxic element to strain GW4 and that it has an energy-linked chemolithotrophic metabolic facet. It is significant that the expression of the enzymes associated with carbohydrate metabolism in A. tumefaciens GW4 was increased by the presence of Sb(III) (Table 1) (117), indicating that strain GW4 possesses totally different resistance mechanisms for As(III) and Sb(III). In addition, reverse transcription-quantitative PCR (qRT-PCR) analysis showed that the genes involved in the TCA cycle were induced by Sb(III), suggesting that strain GW4 may require increased energy to tolerate Sb(III) (Table 1; see also Fig. S2 in the supplemental material).
CONCLUSION AND PERSPECTIVE
This review highlights the recent advances in our understanding of microbial Sb transformations (Fig. 4B). Due to the similar chemical characteristics between As and Sb, the biochemical pathways of Sb(III) oxidation as well as the pathway for dissimilatory Sb(V) reduction were predicted to be shared with As(III) and As(V). However, based on recent literature and our published work, we propose that microbial Sb transformation proceeds by some unique biochemical mechanisms compared with As(III). In the future, particular issues that require attention may include the following points.
(i) It is known that Sb(III) is extruded by the As(III) transporter Acr3 or ArsB. However, the mechanisms of Sb(V) transportation into cells remain unknown. A proteomics study (117) hinted that, as in the case of As(V), a phosphate transport system may be involved in Sb(V) transport; further research to identify the underlying molecular mechanism of Sb(V) importation still needs to be conducted.
(ii) In addition to biologically driven Sb(III) oxidation, some abiotic factors, such as H2O2, are also responsible for bacterial Sb(III) oxidation. The regulation of AnoA's expression and other factors related to abiotic Sb(III) oxidation require further study.
(iii) Dissimilatory microbial Sb(V) reduction using organic substrates as electron donors has been reported. While it is conceivable that the respiratory As(V) reductase ArrAB might be involved in all or part of dissimilatory Sb(V) reduction, this has not been proven, and it remains an open subject for future research scrutiny. Likewise, cytoplasmic Sb(V) reduction, analogous to internal cellular As(V) resistance, reduction, and export, has not been investigated thus far.
(iv) Sb(III) methylation has been described in some microorganisms. To better understand the environmental mobility, toxicity, and biogeochemical cycle of Sb, it is important to clarify mechanisms of Sb(III) methylation and the roles of Sb(III) transmethylase. The volatile methyl and hydride derivatives generated from microbial Sb(III) methylation may represent a significant environmental hazard.
(v) Because of significant and growing Sb environment contamination problems, it is necessary to develop Sb biosensors and bioremediation tools. The expression of AnoA in environmental microbial communities may be a potential biosensor for the monitoring of Sb(III). Anaerobic bacterial Sb(V) reduction holds the possibility of being applied for the remediation of Sb-contaminated environments by producing an immobilized Sb(III) phase. Antimony(III)-oxidizing bacteria may also be applied to enhance phytoremediation effects in combination with plants.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Natural Science Foundation of China (grant 31470226) to G.W., and R.S.O. is supported by the USGS National Research Program.
We are grateful to Birong Yang for technical assistance.
Biographies

Jingxin Li received her B.S. in Biology from Huazhong Agriculture University in 2012, and now she is a Ph.D. student of the College of Life Science and Technology at Huazhong Agriculture University, China. Her major is environmental microbiology, and she does research at the State Key Laboratory of Agricultural Microbiology. In recent years, she mainly studied the molecular mechanism of microbial transformation of toxic metalloids (Sb and As), especially the genes and enzymes responsible for bacterial antimonite oxidation. She has been working in this area for five years.

Qian Wang received her B.S. in Bioengineering from Huazhong Agriculture University in 2007, and got her Ph.D. in Science from Huazhong Agriculture University in 2013. After that, she worked as a postdoc at Huazhong Agriculture University. For up to 10 years, she studied microbial transformation mechanisms of arsenic and antimony and soil arsenic bioremediation using poplar and arsenite-resistant bacteria. Since 2016, she has been a postdoc researcher at Montana State University working on arsenite oxidation regulation and metagenomics of the Yellowstone microbial ecosystem.

Ronald S. Oremland received his B.S. in Biology from Rensselaer Polytechnic Institute in Troy, NY, and his Ph.D. in Marine Sciences from the Rosenstiel School of Marine and Atmospheric Sciences of the University of Miami. He did an NRC postdoc at the NASA Ames Research Center in Moffett Field, CA, after which he joined the U.S. Geological Survey in Menlo Park, CA, where he has been since 1977, currently as a Senior Research Scientist. His research interests lie in the realm of microbial metabolism of toxic metalloids (As, Sb, Se, and Te) and of noxious gases (e.g., methane, methyl bromide, acetylene), as well as microbes and microbial processes occurring in extreme environments, especially soda lakes of the western United States. He is a Fellow of the following societies: ASM, AGU, ISEB, and AAAS.

Thomas R. Kulp received his degrees in Geology from Juniata College in Huntingdon, PA (B.S.), East Carolina University in Greenville, NC (M.S.), and Indiana University in Bloomington, IN (Ph.D.). He was an NRC postdoctoral fellow based at the U.S. Geological Survey in Menlo Park, CA, and later worked there as a research scientist. He joined the faculty at Binghamton University, State University of New York, in 2011 in the Department of Geological Sciences and Environmental Studies. His research investigates the geomicrobiological cycling of toxic metalloids, including As, Sb, and Se. He works with microorganisms from a range of different environmental settings that cycle these metalloids, including contaminated freshwater ecosystems, ground water aquifers, hot springs, and soda lakes. He is a member of AGU, GSA, and ASM.

Christopher Rensing obtained his Ph.D. at the Freie Universität Berlin and later at Martin-Luther Universität Halle-Wittenberg. He was a postdoc working with Barry P. Rosen at Wayne State University Medical School in Detroit, MI. Then, he joined the University of Arizona as an Assistant Professor in 1999 and was promoted to Associate Professor in 2007. In 2011, he joined RTI International as the Director of Research into Food Safety at the Center for Agricultural and Environmental Biotechnology, but shortly after that, he accepted a position as a full professor at the University of Copenhagen in 2012. He holds a rank of full professor at the Institute of Urban Environment, Chinese Academy of Sciences, Xiamen, and became an Adjunct Scientist at the J. Craig Venter Institute in 2016. For his main research, concerning metal-microbe interactions, he is actively investigating mechanisms of such from the cellular to the ecosystem levels.

Gejiao Wang received a B.S. in Plant Physiology and Biochemistry from China Agriculture University in 1984. After that, she worked for five years in the Institute of Microbiology, Chinese Academy of Sciences. She got her Ph.D. in Genetics from the University of Pavia, Italy, in 1995 and was a postdoctoral fellow at Clemson University and the University of Arizona, USA. Since 2004, she has been working as a full professor at Huazhong Agriculture University, China. She devoted herself to the study of microbial transformation of metal(loids) (Sb, As, Cr, and Se) for 15 years. Currently, she is an editorial board member of Applied and Environmental Microbiology and Frontiers in Microbiology.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01375-16.
REFERENCES
- 1.Filella M, Belzile N, Chen YW. 2002. Antimony in the environment: a review focused on natural waters. I. Occurrence. Earth Sci Rev 57:125–176. [Google Scholar]
- 2.Dovick MA, Kulp TR, Arkle RS, Pilliod DS. 2016. Bioaccumulation trends of arsenic and antimony in a freshwater ecosystem affected by mine drainage. Environ Chem 13:149–159. doi: 10.1071/EN15046. [DOI] [Google Scholar]
- 3.Filella M, Belzile N, Chen YW. 2002. Antimony in the environment: a review focused on natural waters. II. Relevant solution chemistry. Earth Sci Rev 59:265–285. [Google Scholar]
- 4.Nies DH. 1999. Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 5.US Environmental Protection Agency. 1979. Water related fate of the 129 priority pollutants, vol 1. EP-440/4-79029A. US EPA, Washington, DC. [Google Scholar]
- 6.Council of the European Communities. 1976. Council directive 76/464/EEC of 4 May 1976 on pollution caused by certain dangerous substances discharged into the aquatic environment of the Community. Council of the European Communities, Brussels, Belgium: http://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:31976L0464&from=EN. [Google Scholar]
- 7.US Environmental Protection Agency. 1999. National primary drinking water standards. Document 810F94001. US EPA Office of Water, Washington, DC. [Google Scholar]
- 8.Council of the European Communities. 1998. Council directive 98/83/EC of 3 November 1998 on the quality of water intended for human consumption. Council of the European Communities, Brussels, Belgium: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:31998L0083. [Google Scholar]
- 9.Gebel T. 1997. Arsenic and antimony: comparative approach on mechanistic toxicology. Chem Biol Interact 107:131–144. doi: 10.1016/S0009-2797(97)00087-2. [DOI] [PubMed] [Google Scholar]
- 10.Wilson SC, Lockwood PV, Ashley PM, Tighe M. 2010. The chemistry and behaviour of antimony in the soil environment with comparisons to arsenic: a critical review. Environ Pollut 158:1169–1181. doi: 10.1016/j.envpol.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Fowler BA, Goering PL. 1991. Antimony, p 743–750. In Merian E. (ed), Metals and their compounds in the environment: occurrence, analysis and biological relevance. VCH, Weinheim, Germany. [Google Scholar]
- 12.Murciego AM, Sánchez AG, González MA, Gil EP, Gordillo CT, Fernández JC, Triguero TB. 2007. Antimony distribution and mobility in topsoils and plants (Cytisus striatus, Cistus ladanifer and Dittrichia viscosa) from polluted Sb-mining areas in Extremadura (Spain). Environ Pollut 145:15–21. doi: 10.1016/j.envpol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Filella M, Williams PA, Belzile N. 2009. Antimony in the environment: knowns and unknowns. Environ Chem 6:95–105. doi: 10.1071/EN09007. [DOI] [Google Scholar]
- 14.Hinkley TK, Lamothe PJ, Wilson SA, Finnegan DL, Gerlach TM.. 1999. Metal emissions from Kilauea, and a suggested revision of the estimated worldwide metal output by quiescent degassing of volcanoes, Earth Planet Sci Lett 170:315–325. [Google Scholar]
- 15.Smichowski P. 2008. Antimony in the environment as a global pollutant: a review on analytical methodologies for its determination in atmospheric aerosols. Talanta 75:2–14. doi: 10.1016/j.talanta.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Vásquez L, Scorza Dagert JV, Scorza JV, Vicuña-Fernández N, de Peña YP, López S, Bendezú H, Rojas E, Vásquez L, Pérez B. 2006. Pharmacokinetics of experimental pentavalent antimony after intramuscular administration in adult volunteers. Curr Ther Res Clin Exp 67:193–203. doi: 10.1016/j.curtheres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An YJ, Kim M. 2009. Effect of antimony on the microbial growth and the activities of soil enzymes. Chemosphere 74:654–659. doi: 10.1016/j.chemosphere.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Hockmann K, Lenz M, Tandy S, Nachtegaal M, Janousch M, Schulin R. 2014. Release of antimony from contaminated soil induced by redox changes. J Hazard Mater 275:215–221. doi: 10.1016/j.jhazmat.2014.04.065. [DOI] [PubMed] [Google Scholar]
- 19.United States Geological Survey. 2015. Mineral commodity summaries 2015. US Geological Survey, Reston, VA: http://minerals.usgs.gov/minerals/pubs/mcs/index.html. [Google Scholar]
- 20.He M, Wang X, Wu F, Fu Z. 2012. Antimony pollution in China. Sci Total Environ 421–422:41–50. [DOI] [PubMed] [Google Scholar]
- 21.Ettler V, Mihaljevič M, Šebek O, Nechutný Z. 2007. Antimony availability in highly polluted soils and sediments—a comparison of single extractions. Chemosphere 68:455–463. doi: 10.1016/j.chemosphere.2006.12.085. [DOI] [PubMed] [Google Scholar]
- 22.Flynn HC, Meharg AA, Bowyer PK, Paton GI. 2003. Antimony bioavailability in mine soils. Environ Pollut 124:93–100. doi: 10.1016/S0269-7491(02)00411-6. [DOI] [PubMed] [Google Scholar]
- 23.Maher WA. 2009. Antimony in the environment—the new global puzzle. Environ Chem 6:93–94. doi: 10.1071/EN09036. [DOI] [Google Scholar]
- 24.Wilson NJ, Craw D, Hunter K. 2004. Antimony distribution and environmental mobility at an historic antimony smelter site, New Zealand. Environ Pollut 129:257–266. doi: 10.1016/j.envpol.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Pratas J, Prasad MNV, Freitas H, Conde L. 2005. Plants growing in abandoned mines of Portugal are useful for biogeochemical exploration of arsenic, antimony, tungsten and mine reclamation. J Geochem Explor 85:99–107. doi: 10.1016/j.gexplo.2004.11.003. [DOI] [Google Scholar]
- 26.Telford K, Maher W, Krikowa F, Foster S, Ellwood MJ, Ashley PM, Lockwood PV, Wilson SC. 2009. Bioaccumulation of antimony and arsenic in a highly contaminated stream adjacent to the Hillgrove Mine, NSW, Australia. Environ Chem 6:133–143. doi: 10.1071/EN08097. [DOI] [Google Scholar]
- 27.Fu Z, Wu F, Mo C, Liu B, Zhu J, Deng Q, Liao H, Zhang Y. 2011. Bioaccumulation of antimony, arsenic, and mercury in the vicinities of a large antimony mine, China. Microchem J 97:12–19. doi: 10.1016/j.microc.2010.06.004. [DOI] [Google Scholar]
- 28.Kruger MC, Bertin PN, Heipieper HJ, Arsène-Ploetze F. 2013. Bacterial metabolism of environmental arsenic–mechanisms and biotechnological applications. Appl Microbiol Biotechnol 97:3827–3841. doi: 10.1007/s00253-013-4838-5. [DOI] [PubMed] [Google Scholar]
- 29.Filella M, Belzile N, Lett MC. 2007. Antimony in the environment: a review focused on natural waters. III. Microbiota relevant interactions. Earth-Sci Rev 80:195–217. [Google Scholar]
- 30.Xi J, He M, Lin C. 2011. Adsorption of antimony(III) and antimony(V) on bentonite: kientics, thermodynamics and anion competition. Microchem J 97:85–91. doi: 10.1016/j.microc.2010.05.017. [DOI] [Google Scholar]
- 31.Lialikova NN. 1974. Stibiobacter senarmontii: a new microorganism oxidizing antimony. Mikrobiologiia 43:941–943. (In Russian.) [PubMed] [Google Scholar]
- 32.Abin CA, Hollibaugh JT. 2014. Dissimilatory antimonite reduction and production of antimony trioxide microcrystals by a novel microorganism. Environ Sci Technol 48:681–688. doi: 10.1021/es404098z. [DOI] [PubMed] [Google Scholar]
- 33.Wysocki R, Chéry CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, Tamás MJ. 2001. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol Microbiol 40:1391–1401. doi: 10.1046/j.1365-2958.2001.02485.x. [DOI] [PubMed] [Google Scholar]
- 34.Porquet A, Filella M. 2007. Structural evidence of the similarity of Sb(OH)3 and As(OH)3 with glycerol: implications for their uptake. Chem Res Toxicol 20:1269–1276. doi: 10.1021/tx700110m. [DOI] [PubMed] [Google Scholar]
- 35.Sanders OI, Rensing C, Kuroda M, Mitra B, Rosen BP. 1997. Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J Bacteriol 179:3365–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng YL, Liu Z, Rosen BP. 2004. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J Biol Chem 279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- 37.Gourbal B, Sonuc N, Bhattacharjee H, Legare D, Sundar S, Ouellette M, Rosen BP, Mukhopadhyay R. 2004. Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem 279:31010–31017. doi: 10.1074/jbc.M403959200. [DOI] [PubMed] [Google Scholar]
- 38.Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. 2005. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol 57:1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x. [DOI] [PubMed] [Google Scholar]
- 39.Brochu C, Wang J, Roy G, Messier N, Wang XY, Saravia NG, Ouellette M. 2003. Antimony uptake systems in the protozoan parasite Leishmania and accumulation differences in antimony resistant parasites. Antimicrob Agents Chemother 47:3073–3079. doi: 10.1128/AAC.47.10.3073-3079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butcher BG, Deane SM, Rawlings DE. 2000. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl Environ Microbiol 66:1826–1833. doi: 10.1128/AEM.66.5.1826-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosen BP. 2002. Biochemistry of arsenic detoxification. FEBS Lett 529:86–92. doi: 10.1016/S0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 42.Carlin A, Shi W, Dey S, Rosen BP. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177:981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato T, Kobayashi Y. 1998. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J Bacteriol 180:1655–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver S. 1998. Genes for all metals—a bacterial view of the periodic table. The 1996 Thom Award lecture. J Ind Microbiol Biotechnol 20:1–12. [DOI] [PubMed] [Google Scholar]
- 45.Rosen BP, Borbolla MG. 1984. A plasmid-encoded arsenite pump produces arsenite resistance in Escherichia coli. Biochem Biophys Res Commun 124:760–765. doi: 10.1016/0006-291X(84)91023-4. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki K, Wakao N, Kimura T, Sakka K, Ohmiya K. 1998. Expression and regulation of the arsenic resistance operon of Acidiphilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl Environ Microbiol 64:411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu C, Zhou T, Kuroda M, Rosen BP. 1998. Metalloid resistance mechanisms in prokaryotes. J Biochem 123:16–23. doi: 10.1093/oxfordjournals.jbchem.a021904. [DOI] [PubMed] [Google Scholar]
- 48.Martin P, DeMel S, Shi J, Gladysheva T, Gatti DL, Rosen BP, Edwards BF. 2001. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 9:1071–1081. doi: 10.1016/S0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 49.Achour AR, Bauda P, Billard P. 2007. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res Microbiol 158:128–137. doi: 10.1016/j.resmic.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Rosen BP. 1999. Families of arsenic transporters. Trends Microbiol 7:207–212. doi: 10.1016/S0966-842X(99)01494-8. [DOI] [PubMed] [Google Scholar]
- 51.Kang YS, Shi Z, Bothner B, Wang G, McDermott TR. 2015. Involvement of the Acr3 and DctA anti-porters in arsenite oxidation in Agrobacterium tumefaciens 5A. Environ Microbiol 17:1950–1962. doi: 10.1111/1462-2920.12468. [DOI] [PubMed] [Google Scholar]
- 52.Bobrowicz P, Ułaszewski S. 1998. Arsenical-induced transcriptional activation of the yeast Saccharomyces cerevisiae ACR2 and ACR3 genes requires the presence of the ACR1 gene product. Cell Mol Biol Lett 3:13–20. [Google Scholar]
- 53.Maciaszczyk-Dziubinska E, Migocka M, Wysocki R. 2011. Acr3p is a plasma membrane antiporter that catalyzes As(III)/H+ and Sb(III)/H+ exchange in Saccharomyces cerevisiae. Biochim Biophys Acta 7:1855–1859. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh M, Shen J, Rosen BP. 1999. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manzano JI, García-Hernández R, Castanys S, Gamarro F. 2013. A new ABC half-transporter in Leishmania major is involved in resistance to antimony. Antimicrob Agents Chemother 57:3719–3730. doi: 10.1128/AAC.00211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirsch R, Scheinost AC, Rossberg A, Banerjee D, Charlet L. 2008. Reduction of antimony by nano-particulate magnetite and mackinawite. Mineral Mag 72:185–189. doi: 10.1180/minmag.2008.072.1.185. [DOI] [Google Scholar]
- 57.Leuz AK, Hug S, Moench H, Wehrli B, Johnson CA. 2002. The redox chemistry of antimony in lakes. Geochim Cosmochim Acta 66:450. [Google Scholar]
- 58.Mitsunobu S, Takahashi Y, Sakai Y. 2008. Abiotic reduction of antimony(V) by green rust (Fe4(II)Fe2(III)(OH)12SO4·3H2O). Chemosphere 70:942–947. doi: 10.1016/j.chemosphere.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 59.Kantin R. 1983. Chemical speciation of antimony in marine algae. Limnol Oceanogr 28:165–168. doi: 10.4319/lo.1983.28.1.0165. [DOI] [Google Scholar]
- 60.Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D. 2001. Novel intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J Biol Chem 276:3971–3976. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- 61.Ferreira CS, Martins PS, Demicheli C, Brochu C, Ouellette M, Frézard F. 2003. Thiol-induced reduction of antimony(V) into antimony(III): a comparative study with trypanothione, cysteinylglycine, cysteine and glutathione. Biometals 16:441–446. doi: 10.1023/A:1022823605068. [DOI] [PubMed] [Google Scholar]
- 62.Wyllie S, Fairlamb AH. 2006. Differential toxicity of antimonial compounds and their effects on glutathione homeostasis in a human leukaemia monocyte cell line. Biochem Pharmacol 71:257–267. doi: 10.1016/j.bcp.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 63.Hansen C, Hansen EW, Hansen HR, Gammelgaard B, Stürup S. 2011. Reduction of Sb(V) in a human macrophage cell line measured by HPLC-ICP-MS. Biol Trace Elem Res 144:234–243. doi: 10.1007/s12011-011-9079-9. [DOI] [PubMed] [Google Scholar]
- 64.Kulp TR, Miller LG, Braiotta F, Webb SM, Kocar BD, Blum JS, Oremland RS. 2014. Microbiological reduction of Sb(V) in anoxic freshwater sediments. Environ Sci Technol 48:218–226. doi: 10.1021/es403312j. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen VK, Lee JU. 2014. Isolation and characterization of antimony-reducing bacteria from sediments collected in the vicinity of an antimony factory. Geomicrobiol J 31:855–861. doi: 10.1080/01490451.2014.901440. [DOI] [Google Scholar]
- 66.Polack R, Chen YW, Belzile N. 2009. Behaviour of Sb(V) in the presence of dissolved sulfide under controlled anoxic aqueous conditions. Chem Geol 262:179–185. doi: 10.1016/j.chemgeo.2009.01.008. [DOI] [Google Scholar]
- 67.Leuz AK, Monch H, Johnson CA. 2006. Sorption of Sb(III) and Sb(V) to goethite: influence on Sb(III) oxidation and mobilization. Environ Sci Technol 40:7277–7282. [DOI] [PubMed] [Google Scholar]
- 68.Wang H, Chen F, Mu S, Zhang D, Pan X, Lee DJ, Chang JS. 2013. Removal of antimony (Sb(V)) from Sb mine drainage: biological sulfate reduction and sulfide oxidation-precipitation. Bioresour Technol 146:799–802. doi: 10.1016/j.biortech.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Lai CY, Wen LL, Zhang Y, Luo SS, Wang QY, Luo YH, Chen R, Yang X, Rittmann BE, Zhao HP. 2016. Autotrophic antimonate bio-reduction using hydrogen as the electron donor. Water Res 88:467–474. doi: 10.1016/j.watres.2015.10.042. [DOI] [PubMed] [Google Scholar]
- 70.Jenkins RO, Forster SN, Craig PJ. 2002. Formation of methylantimony species by an aerobic prokaryote: Flavobacterium sp. Arch Microbiol 178:274–278. doi: 10.1007/s00203-002-0456-9. [DOI] [PubMed] [Google Scholar]
- 71.Andreae MO, Asmodé JF, Foster P, Van ‘t Dack L. 1981. Determination of antimony(III), antimony(V), and methylantimony species in natural waters by atomic absorption spectrometry with hydride generation. Anal Chem 53:1766–1771. doi: 10.1021/ac00235a012. [DOI] [Google Scholar]
- 72.Andreae MO, Froelich PN Jr. 1984. Arsenic, antimony, and germanium biogeochemistry in the Baltic Sea. Tellus 36:101–117. [Google Scholar]
- 73.Andrewes P, Cullen WR. 2003. Organoantimony compounds in the environment, p 277–303. In Craig PJ. (ed), Organometallic compounds in the environment. John Wiley & Sons, New York, NY. [Google Scholar]
- 74.Hirner AV, Feldmann J, Krupp E, Gruèmping R, Goguel R, Cullen WR. 1998. Metal(loid)organic compounds in geothermal gases and waters. Org Geochem 29:1765–1778. doi: 10.1016/S0146-6380(98)00153-3. [DOI] [Google Scholar]
- 75.Dodd M, Pergantis SA, Cullen WR, Li H, Eigendorf GK, Reimer KJ. 1996. Antimony speciation in freshwater plant extracts by using hydride generation-gas chromatography-mass spectrometry. Analyst 121:223–228. doi: 10.1039/AN9962100223. [DOI] [Google Scholar]
- 76.Craig PJ, Forster SN, Miller D, Jenkins RO. 1999. An analytical method for the detection of methylantimony species in environmental matrices: methylantimony levels in some UK plant material. Analyst 124:1243–1248. [Google Scholar]
- 77.Koch I, Wang L, Feldmann J, Andrewes P, Reimer KJ, Cullen WR. 2000. Antimony species in environmental samples. Intern J Environ Anal Chem 77:111–131. doi: 10.1080/03067310008032676. [DOI] [Google Scholar]
- 78.Challenger F. 1945. Biological methylation. Chem Rev 36:315–361. doi: 10.1021/cr60115a003. [DOI] [Google Scholar]
- 79.Jenkins RO, Craig PJ, Goessler W, Miller D, Ostah N, Irgolic KJ. 1998. Biomethylation of inorganic antimony compounds by an aerobic fungus: Scopulariopsis brevicaulis. Environ Sci Technol 32:882–885. doi: 10.1021/es970824p. [DOI] [Google Scholar]
- 80.Craig PJ, Jenkins RO, Dewick R, Miller D. 1999. Trimethylantimony generation by Scopulariopsis brevicaulis during aerobic growth. Sci Total Environ 229:83–88. [Google Scholar]
- 81.Andrewes P, Cullen WR, Feldmann J, Koch I, Polishchuk E, Reimer KJ. 1998. The production of methylated organoantimony compounds by Scopulariopsis brevicaulis. Appl Organomet Chem 12:827–842. doi:. [DOI] [Google Scholar]
- 82.Andrewes P, Cullen WR, Polishchuk E, Reimer KJ. 2001. Antimony biomethylation by the wood rotting fungus Phaeolus schweinitzii. Appl Organomet Chem 15:473–480. doi: 10.1002/aoc.131. [DOI] [Google Scholar]
- 83.Boriová K, Čerňanský S, Matúš P, Bujdoš M, Simonovičová A. 2014. Bioaccumulation and biovolatilization of various elements using filamentous fungus Scopulariopsis brevicaulis. Lett Appl Microbiol 59:217–223. doi: 10.1111/lam.12266. [DOI] [PubMed] [Google Scholar]
- 84.Smith LM, Maher WA, Craig PJ, Jenkins RO. 2002. Speciation of antimony compounds in culture headspace gases of Cryptococcus humicolus using solid phase microextraction and gas chromatography-mass spectrometry. Appl Organomet Chem 16:287–293. doi: 10.1002/aoc.303. [DOI] [Google Scholar]
- 85.Richardson BA. 1994. Sudden infant death syndrome: a possible primary cause. J Forensic Sci Soc 34:199–204. doi: 10.1016/S0015-7368(94)72915-7. [DOI] [PubMed] [Google Scholar]
- 86.Jenkins RO, Morris TA, Craig PJ, Goessler W, Ostah N, Wills KM. 2000. Evaluation of cot mattress inner foam as a potential site for microbial generation of toxic gases. Hum Exp Toxicol 19:693–702. doi: 10.1191/096032700670028460. [DOI] [PubMed] [Google Scholar]
- 87.Warnock DW, Delves HT, Campbell CK, Croudace IW, Davey KG, Johnson EM, Sieniawska C. 1995. Toxic gas generation from plastic mattresses and sudden infant death syndrome. Lancet 346:1516–1520. doi: 10.1016/S0140-6736(95)92051-X. [DOI] [PubMed] [Google Scholar]
- 88.Michalke K, Wickenheiser EB, Mehring M, Hirner AV, Hensel R. 2000. Production of volatile derivatives of metal(loid)s by microflora involved in anaerobic digestion of sewage sludge. Appl Environ Microbiol 66:2791–2796. doi: 10.1128/AEM.66.7.2791-2796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wehmeier S, Feldmann J. 2005. Investigation into antimony mobility in sewage sludge fermentation. J Environ Monit 7:1194–1199. doi: 10.1039/b509538g. [DOI] [PubMed] [Google Scholar]
- 90.Meyer J, Annette S, Klaus M, Hensel R. 2007. Volatilisation of metals and metalloids by the microbial population of an alluvial soil. Syst Appl Microbiol 30:229–238. doi: 10.1016/j.syapm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 91.Gürleyük H, Fleet-Stalder VV, Chasteen TG. 1997. Confirmation of the biomethylation of antimony compounds. Appl Organomet Chem 11:471–483. doi:. [DOI] [Google Scholar]
- 92.Hartmann LM, Craig PJ, Jenkins RO. 2003. Influence of arsenic on antimony methylation by the aerobic yeast Cryptococcus humicolus. Arch Microbiol 180:347–352. doi: 10.1007/s00203-003-0600-1. [DOI] [PubMed] [Google Scholar]
- 93.Filella M. 2010. Alkyl derivatives of antimony in the environment. Met Ions Life Sci 7:267–301. [DOI] [PubMed] [Google Scholar]
- 94.Andrewes P, Cullen WR, Polishchuk E. 2000. Arsenic and antimony biomethylation by Scopulariopsis brevicaulis: interaction of arsenic and antimony compounds. Environ Sci Technol 34:2249–2253. [Google Scholar]
- 95.Andrewes P, Cullen WR, Polishchuk E. 2000. Antimony biomethylation by Scopulariopsis brevicaulis: characterization of intermediates and the methyl donor. Chemosphere 41:1717–1725. [DOI] [PubMed] [Google Scholar]
- 96.Thanabalasingam P, Pickering WF. 1990. Specific sorption of antimony (III) by the hydrous oxides of Mn, Fe, and Al. Water Air Soil Pollut 49:175–185. doi: 10.1007/BF00279519. [DOI] [Google Scholar]
- 97.Zobrist J, Dowdle PR, Davis JA, Oremland RS. 2000. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ Sci Technol 34:4747–4753. doi: 10.1021/es001068h. [DOI] [Google Scholar]
- 98.Chen YW, Deng TL, Filella M, Belzile N. 2003. Distribution and early diagenesis of antimony species in sediments and porewaters of freshwater lakes. Environ Sci Technol 37:1163–1168. doi: 10.1021/es025931k. [DOI] [PubMed] [Google Scholar]
- 99.Leuz AK, Johnson CA. 2005. Oxidation of Sb(III) to Sb(V) by O2 and H2O2 in aqueous solutions. Geochim Cosmochim Acta 69:1165–1172. doi: 10.1016/j.gca.2004.08.019. [DOI] [Google Scholar]
- 100.Quentel F, Filella M, Elleouet C, Madec CL. 2004. Kinetic studies on Sb(III) oxidation by hydrogen peroxide in aqueous solution. Environ Sci Technol 38:2843–2848. doi: 10.1021/es035019r. [DOI] [PubMed] [Google Scholar]
- 101.Belzile N, Chen YW, Wang Z. 2001. Oxidation of antimony(III) by amorphous iron and manganese oxyhydroxides. Chem Geol 174:379–387. doi: 10.1016/S0009-2541(00)00287-4. [DOI] [Google Scholar]
- 102.Leuz AK, Hug SJ, Wehrli B, Johnson CA. 2006. Iron-mediated oxidation of antimony(III) by oxygen and hydrogen peroxide compared to arsenic(III) oxidation. Environ Sci Technol 40:2565–2571. [DOI] [PubMed] [Google Scholar]
- 103.Buschmann JS, Canonica S, Sigg L. 2005. Photoinduced oxidation of antimony(III) in the presence of humic acids. Environ Sci Technol 39:5335–5341. doi: 10.1021/es050269o. [DOI] [PubMed] [Google Scholar]
- 104.Quentel F, Filella M, Elleouet C, Madec CL. 2006. Sb(III) oxidation by iodate in seawater: a cautionary tale. Sci Total Environ 355:259–263. doi: 10.1016/j.scitotenv.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 105.Oscarson DW, Huang PM, Defosse C, Herbillon A. 1981. Oxidative power of Mn(IV) and Fe(III) oxides with respect to As(III) in terrestrial and aquatic environments. Nature 291:50–51. doi: 10.1038/291050a0. [DOI] [Google Scholar]
- 106.Vitre RD, Belzile N, Tessier A. 1991. Speciation and adsorption of arsenic on diagenetic iron oxyhydroxides. Limnol Oceanogr 36:1480–1485. doi: 10.4319/lo.1991.36.7.1480. [DOI] [Google Scholar]
- 107.Fan JX, Wang YJ, Fan TT, Cui XD, Zhou DM. 2014. Photo-induced oxidation of Sb(III) on goethite. Chemosphere 95:295–300. doi: 10.1016/j.chemosphere.2013.08.094. [DOI] [PubMed] [Google Scholar]
- 108.Li SX, Zheng FY, Hong HS, Deng NS, Zhou XY. 2006. Photo-oxidation of Sb(III) in the seawater by marine phytoplankton-transition metals-light system. Chemosphere 65:1432–1439. doi: 10.1016/j.chemosphere.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 109.Torma AE, Gabra GG. 1977. Oxidation of stibnite by Thiobacillus ferrooxidans. Antonie Van Leeuwenhoek 43:1–6. doi: 10.1007/BF02316204. [DOI] [PubMed] [Google Scholar]
- 110.Fan H, Su C, Wang Y, Yao J, Zhao K, Wang Y, Wang G. 2008. Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, northwestern China. J Appl Microbiol 105:529–539. doi: 10.1111/j.1365-2672.2008.03790.x. [DOI] [PubMed] [Google Scholar]
- 111.Luo G, Shi Z, Wang H, Wang G. 2012. Skermanella stibiiresistens sp. nov., a highly antimony-resistant bacterium isolated from coal-mining soil, and emended description of the genus Skermanella. Int J Syst Evol Microbiol 62:1271–1276. doi: 10.1099/ijs.0.033746-0. [DOI] [PubMed] [Google Scholar]
- 112.Hamamura N, Fukushima K, Itai T. 2013. Identification of antimony- and arsenic-oxidizing bacteria associated with antimony mine tailing. Microbes Environ 28:257–263. doi: 10.1264/jsme2.ME12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi Z, Cao Z, Qin D, Zhu W, Wang Q, Li M, Wang G. 2013. Correlation models between environmental factors and bacterial resistance to antimony and copper. PLoS One 8:78533. doi: 10.1371/journal.pone.0078533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li J, Wang Q, Zhang S, Qin D, Wang G. 2013. Phylogenetic and genome analyses of antimony-oxidizing bacteria isolated from antimony mined soil. Int Biodeterior Biodegrad 76:76–80. doi: 10.1016/j.ibiod.2012.06.009. [DOI] [Google Scholar]
- 115.Terry LR, Kulp TR, Wiatrowski H, Miller LG, Oremland RS. 2015. Microbiological oxidation of antimony(III) with oxygen or nitrate by bacteria isolated from contaminated mine sediments. Appl Environ Microbiol 81:8478–8488. doi: 10.1128/AEM.01970-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lehr CR, Kashyap DR, McDermott TR. 2007. New insights into microbial oxidation of antimony and arsenic. Appl Environ Microbiol 73:2386–2389. doi: 10.1128/AEM.02789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Wang Q, Li M, Yang B, Shi M, Guo W, McDermott TR, Rensing C, Wang G. 2015. Proteomics and genetics for identification of a bacterial antimonite oxidase in Agrobacterium tumefaciens. Environ Sci Technol 49:5980–5989. doi: 10.1021/es506318b. [DOI] [PubMed] [Google Scholar]
- 118.Kulp TR, Hoeft SE, Asao M, Madigan MT, Hollibaugh JT, Fisher JC, Stolz JF, Culbertson CW, Miller LG, Oremland RS. 2008. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321:967–970. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- 119.Xiong JB, Li DM, Li H, He M, Miller SJ, Yu L, Rensing C, Wang G. 2011. Genome analysis and characterization of zinc efflux systems of a highly zinc-resistant bacterium, Comamonas testosteroni S44. Res Microbiol 162:671–679. doi: 10.1016/j.resmic.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 120.Stolz JF, Basu P, Santini JM, Oremland RS. 2006. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 121.Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S, Oremland RS, Stolz J, Saltikov CW. 2012. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol 14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- 122.Silver S, Phung LT. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71:599–608. doi: 10.1128/AEM.71.2.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Q, Warelow TP, Kang YS, Romano C, Osborne TH, Lehr CR, Bothner B, McDermott TR, Santini JM, Wang G. 2015. Arsenite oxidase also functions as an antimonite oxidase. Appl Environ Microbiol 81:1959–1965. doi: 10.1128/AEM.02981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu W, Huang J, Li M, Li X, Wang G. 2014. Genomic analysis of Skermanella stibiiresistens type strain SB22T. Stand Genomic Sci 9:1211–1220. doi: 10.4056/sigs.5751047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu L, Zhu WT, Cao Z, Xu B, Wang G, Luo M. 2015. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genomics 16:110. doi: 10.1186/s12864-015-1314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cooper WJ, Zika RG. 1983. Photochemical formation of hydrogen peroxide in surface and ground waters exposed to sunlight. Science 220:711–712. doi: 10.1126/science.220.4598.711. [DOI] [PubMed] [Google Scholar]
- 127.Zika RG, Moffett JW, Petasne RG, Cooper WJ, Saltzman ES. 1985. Spatial and temporal variations of hydrogen peroxide in Gulf of Mexico waters. Geochim Cosmochim Acta 49:1173–1184. doi: 10.1016/0016-7037(85)90008-0. [DOI] [Google Scholar]
- 128.Cooper WJ, Shao C, Lean DRS, Gordon AS, Scully FE. 1994. Factors affecting the distribution of H2O2 in surface waters, p 391–422. In Comstock MJ. (ed), Environmental chemistry of lakes and reservoirs. American Chemical Society, Washington, DC. [Google Scholar]
- 129.Price D, Mantoura RFC, Worsfold PJ. 1998. Shipboard determination of hydrogen peroxide in the western Mediterranean Sea using flow injection with chemiluminescence detection. Anal Chim Acta 377:145–155. doi: 10.1016/S0003-2670(98)00621-7. [DOI] [Google Scholar]
- 130.Ma JF, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey JE, Vasil ML, Monaco JJ, Hassett DJ. 1999. Bacterioferritin a modulate catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol 12:3730–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 132.Hassett DJ, Charniga L, Bean KA, Ohman DE, Cohen MS. 1992. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun 60:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu HL, Zhuang P, Zhang SZ, Rensing C, Huang J, Li J, Wang G. 2015. Global regulator IscR positively contributes to antimonite resistance and oxidation in Comamonas testosteroni S44. Front Mol Biosci 2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.El Fadili K, Drummelsmith J, Roy G, Jardim A, Ouellette M. 2009. Down regulation of KMP-11 in Leishmania infantum axenic antimony resistant amastigotes as revealed by a proteomic screen. Exp Parasitol 123:51–57. doi: 10.1016/j.exppara.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 136.Walker J, Gongora R, Vasquez JJ, Drummelsmith J, Burchmore R, Roy G, Ouellette M, Gomez MA, Saravia NG. 2012. Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol Biochem Parasitol 183:166–176. doi: 10.1016/j.molbiopara.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 137.Matrangolo FSV, Liarte DB, Andrade LC, de Melo MF, Andrade JM, Ferreira RF, Santiago AS, Pirovani CP, Silva-Pereira RA, Murta SM. 2013. Comparative proteomic analysis of antimony-resistant and -susceptible Leishmania braziliensis and Leishmania infantum chagasi lines. Mol Biochem Parasitol 190:63–75. doi: 10.1016/j.molbiopara.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 138.Vincent IM, Racine G, Légaré D, Ouellette M. 2015. Mitochondrial proteomics of antimony and miltefosine resistant Leishmania infantum. Proteomes 3:328–346. doi: 10.3390/proteomes3040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xue L, Ren H, Li S, Gao M, Shi S, Chang E, Wei Y, Yao X, Jiang Z, Liu J. 2015. Comparative proteomic analysis in Miscanthus sinensis exposed to antimony stress. Environ Pollut 201:150–160. doi: 10.1016/j.envpol.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 140.Carapito C, Muller D, Turlin E, Koechler S, Danchin A, Van Dorsselaer A, Leize-Wagner E, Bertin PN, Lett MC. 2006. Identification of genes and proteins involved in the pleiotropic response to arsenic stress in Caenibacter arsenoxydans, a metalloresistant beta-proteobacterium with an unsequenced genome. Biochimie 88:595–606. doi: 10.1016/j.biochi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 141.Weiss S, Carapito C, Cleiss J, Koechler S, Turlin E, Coppee JY, Heymann M, Kugler V, Stauffert M, Cruveiller S, Médigue C, Van Dorsselaer A, Bertin PN, Arsène-Ploetze F. 2009. Enhanced structural and functional genome elucidation of the arsenite-oxidizing strain Herminiimonas arsenicoxydans by proteomics data. Biochimie 91:192–203. doi: 10.1016/j.biochi.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 142.Cleiss-Arnold J, Koechler S, Proux C, Fardeau ML, Dillies MA, Coppee JY, Arsène-Ploetze F, Bertin PN. 2010. Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. doi: 10.1186/1471-2164-11-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.