ABSTRACT
Autolysins, also known as peptidoglycan hydrolases, are enzymes that hydrolyze specific bonds within bacterial cell wall peptidoglycan during cell division and daughter cell separation. Within the genome of Lactobacillus acidophilus NCFM, there are 11 genes encoding proteins with peptidoglycan hydrolase catalytic domains, 9 of which are predicted to be functional. Notably, 5 of the 9 putative autolysins in L. acidophilus NCFM are S-layer-associated proteins (SLAPs) noncovalently colocalized along with the surface (S)-layer at the cell surface. One of these SLAPs, AcmB, a β-N-acetylglucosaminidase encoded by the gene lba0176 (acmB), was selected for functional analysis. In silico analysis revealed that acmB orthologs are found exclusively in S-layer- forming species of Lactobacillus. Chromosomal deletion of acmB resulted in aberrant cell division, autolysis, and autoaggregation. Complementation of acmB in the ΔacmB mutant restored the wild-type phenotype, confirming the role of this SLAP in cell division. The absence of AcmB within the exoproteome had a pleiotropic effect on the extracellular proteins covalently and noncovalently bound to the peptidoglycan, which likely led to the observed decrease in the binding capacity of the ΔacmB strain for mucin and extracellular matrices fibronectin, laminin, and collagen in vitro. These data suggest a functional association between the S-layer and the multiple autolysins noncovalently colocalized at the cell surface of L. acidophilus NCFM and other S-layer-producing Lactobacillus species.
IMPORTANCE Lactobacillus acidophilus is one of the most widely used probiotic microbes incorporated in many dairy foods and dietary supplements. This organism produces a surface (S)-layer, which is a self-assembling crystalline array found as the outermost layer of the cell wall. The S-layer, along with colocalized associated proteins, is an important mediator of probiotic activity through intestinal adhesion and modulation of the mucosal immune system. However, there is still a dearth of information regarding the basic cellular and evolutionary function of S-layers. Here, we demonstrate that multiple autolysins, responsible for breaking down the cell wall during cell division, are associated with the S-layer. Deletion of the gene encoding one of these S-layer-associated autolysins confirmed its autolytic role and resulted in reduced binding capacity to mucin and intestinal extracellular matrices. These data suggest a functional association between the S-layer and autolytic activity through the extracellular presentation of autolysins.
INTRODUCTION
Beneficial microorganisms such as probiotics are defined by the FAO/WHO as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (1). Lactobacillus acidophilus NCFM is a generally recognized as safe, industrially significant lactic acid bacterium which has been sold commercially and consumed in various probiotic food formulations for more than 35 years (2). Predicated by the availability of a fully sequenced and annotated genome (3), L. acidophilus NCFM is one of the most studied and well-characterized probiotic bacteria (2, 4–8). Most notably, the probiotic activity of L. acidophilus is mediated by cell surface-associated components which interact with the host gastrointestinal mucosa and immune system (9–11).
As in other Gram-positive bacteria, the cell envelope of L. acidophilus is characterized by a lipid membrane surrounded by a thick peptidoglycan sacculus with a complex assemblage of macromolecules, including teichoic acids, polysaccharides, and proteins (12, 13). The peptidoglycan is composed of glycan chains consisting of alternating N-acetylglucosamine and N-acetylmuramic acid, linked via β-1,4 bonds and covalently cross-linked with peptide chains. Among the numerous functions of peptidoglycan are the maintenance of cell shape (14), integrity from osmotic pressure (15), and the presentation of proteins (12). Some of these proteins are covalently linked to the peptidoglycan via sortase and LPXTG motif recognition (16), while many others, including proteins which comprise the surface (S)-layer, are noncovalently attached through cell wall binding domains (CWBD) (17–19).
S-layers are semiporous, proteinaceous crystalline arrays consisting of self-assembling (glyco)protein subunits called S-layer proteins (SLPs). While S-layers can be found in all prokaryotes, including Gram-positive and Gram-negative organisms and many species of Archaea, S-layers are not ubiquitous to all microorganisms (17). In L. acidophilus, the S-layer monolayer is composed of a dominant protein constituent, SlpA (46 kDa), with minor constituents SlpB (47 kDa) and SlpX (51 kDa) (20). Because the S-layer is presented as the outermost layer of proteins on the cell wall, it has been the target for functional analysis of probiotic-host interactions. In fact, in vitro studies using intestinal epithelial cell lines suggest that the S-layer is a major factor in intestinal adhesion for L. acidophilus (21, 22). Furthermore, SlpA of L. acidophilus NCFM has been shown to bind dendritic cell C-type lectin receptors (23) and exert regulatory signals which mitigate inflammatory disease states and promote maintenance of healthy intestinal barrier function (24).
Despite the apparent importance of the S-layer for probiotic-host interactions, there is still a great deal that is not known about the composition and evolutionary function of S-layers. Complete functional analysis of the S-layer in L. acidophilus has been limited due to the apparent essentiality of the S-layer for cell survival and the ensuing difficulty of creating a stable deletion mutant of SlpA in L. acidophilus (Y. J. Goh and T. R. Klaenhammer, unpublished data). Exoproteomic analysis of L. acidophilus NCFM and other S-layer-forming lactobacilli has revealed the presence of numerous S-layer-associated proteins (SLAPs), which are colocalized with the S-layer through noncovalent association with the cell wall peptidoglycan (10, 25). In addition to uncharacterized proteins with putative probiotic activity, numerous autolysins were found in these SLAP fractions (10, 25).
Autolysins, also known as peptidoglycan hydrolases (PGH), are a class of enzymes responsible for peptidoglycan turnover during cell division and daughter cell separation (13, 26, 27). PGH have numerous catalytic domains and are normally bound to the cell wall through LysM- or SH3-anchoring domains (28, 29). Notably, the PGH identified in the SLAP fractions of L. acidophilus NCFM and other S-layer-forming Lactobacillus species are anchored to the cell wall with noncovalent attachment domains (NCAD) (pfam03217), the same domains found in SLPs (10, 25, 30). In this study, an S-layer-associated β-N-acetylglucosaminidase autolysin, designated AcmB, was selected for functional analysis in L. acidophilus NCFM. Chromosomal deletion of acmB resulted in aberrant cell division, autolysis, and autoaggregation, confirming the role of this SLAP in PGH activity. Further, the absence of AcmB within the exoproteome had a pleiotropic effect on cell surface proteins associated with the peptidoglycan, as measured through the reduced ability of the ΔacmB strain to bind to mucin and extracellular matrices in vitro. Here we present the S-layer as a scaffold for numerous proteins, including autolysins. Analysis of these S-layer-associated autolysins will undoubtedly lead to a more comprehensive understanding of the evolutionary function of the S-layer in S-layer-forming species of Lactobacillus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. L. acidophilus strains were propagated in de Man Rogosa Sharpe (MRS) broth (Difco) under aerobic conditions, statically or on MRS solid medium containing 1.5% (wt/vol) agar (Difco) under anaerobic conditions at 37°C and at 42°C where indicated. Recombinant strains were selected in the presence of 2 μg/ml erythromycin (Sigma-Aldrich, St. Louis, MO) and/or 2 to 5 μg/ml chloramphenicol (Sigma). Escherichia coli strains were grown in brain heart infusion (Difco) medium at 37°C with aeration. E. coli EC101 was grown in the presence of 40 μg/ml kanamycin (Sigma-Aldrich), while NCK1911 and transformants were grown with 40 mg kanamycin and 150 μg/ml erythromycin. Counterselection of L. acidophilus plasmid-free excision recombinants was performed using 5-fluorouracil-supplemented glucose semidefined medium, as previously described (20).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype or characteristic(s)a | Reference or source |
|---|---|---|
| L. acidophilus strains | ||
| NCFM | Human intestinal isolate | 2 |
| NCK1909 | NCFM carrying a 315-bp deletion within the upp gene | 20 |
| NCK1910 | NCK1909 harboring pTRK669; host for pORI-based counterselective integration vector | 20 |
| NCK2395 | NCK1909 carrying a 1,103-bp deletion within the lba0176 gene | This study |
| NCK2397 | NCK2395 harboring pTRK1098 for complementation of lba0176 | This study |
| E. coli strains | ||
| NCK1831 | EC101 host for pORI-based plasmids | 58 |
| NCK1911 | Host harboring pTRK935, Knr, Emr | 20 |
| NCK2394 | Host harboring pTRK1097, Knr, Emr | This study |
| NCK2396 | Host harboring pTRK1098, Emr | This study |
| Plasmids | ||
| pTRK669 | Ori (pWV01), Cmr, RepA+ thermosensitive | 59 |
| pTRK935 | pORI upp-based counterselective integration vector | 20 |
| pTRK882 | Δcat derivative of pGK12 with MCS from pORI28 and cloned Ppgm, Emr | 40 |
| pTRK1097 | pTRK935 with flanking regions of lba0176 cloned into BamHI/SacI site | This study |
| pTRK1098 | pTRK882 with lba0176 cloned into EcoRI/BamHI site | This study |
| Primersb | ||
| Construction of ΔacmB mutant | ||
| 0176BamHIF | GTAATAGGATCCATCTGAGTTGTTTGGTAATG | This study |
| 0176R | CATTATTCACTACTGGGGTA | This study |
| 0176Soe | TACCCCAGTAGTGAATAATGTATTACAGAATCGGTATTCG | This study |
| 0176SacIR | TAAAGTAGAGCTCGCATCATTGTTAATTGATTT | This study |
| Screening of ΔacmB locus | ||
| 0176up | AACCAAAGTTAAATGAAACA | This study |
| 0176dw | CTTAGCTTGCAAATCATAGT | This study |
| Complementation of acmB | ||
| C0176EcoRIF | GATCGAATTCAAGGAGAACGTATATGAAGAAGAGACTTTTGACCCAGC | This study |
| C0176BamHIR | GATCGGATCCTACATGAAGTTAGCTTTTTTAATG | This study |
Km, kanamycin; Em, erythromycin; Cm, chloramphenicol.
Restriction sites are underlined. Ribosome binding sites are in italics. Start codons are in bold.
DNA manipulation and transformation.
Genomic DNA from L. acidophilus strains was isolated using a Fungal/Bacterial DNA MiniPrep kit (Zymo Research). Plasmid DNA from E. coli was isolated using a QIAprep Spin Miniprep kit (Qiagen). Restriction enzyme digestions and ligations were performed using Roche restriction enzymes (Roche Diagnostics) and T4 DNA ligase (New England BioLabs), respectively. PCR primers were designed based on the genomic sequence data and synthesized by Integrated DNA Technologies. PCRs were carried out in Bio-Rad MyCycler thermocyclers (Bio-Rad Laboratories) using Choice-Taq Blue DNA polymerase (Denville Scientific) for screening of recombinants and PfuUltra II fusion HS DNA polymerase (Agilent Technologies) for cloning purposes. PCR amplicons were analyzed on 0.8% agarose gels and purified using QIAquick gel extraction kits (Qiagen).
E. coli EC101 cells were made competent using a rubidium chloride competent cell protocol (31). L. acidophilus cells were prepared for electrotransformation using a modified penicillin treatment protocol (20, 32, 33).
In silico and RNA sequencing analyses.
Predicted peptidoglycan hydrolases were identified from the L. acidophilus NCFM genome (3) (NCBI accession number NC_006814). Homologous sequences were identified and compared using the BLASTn and BLASTp features of NCBI, as well as SANSparallel (34). Signal peptidase cleavage sites for protein sequences were predicted using SignalP 4.1 (35). Protein domains were identified using UniProt and the Pfam protein family database (36, 37). RNA sequencing analysis from a previous study (10) was utilized to examine mRNA expression of the predicted peptidoglycan hydrolases. Gene expression was measured by the normalized transcripts per million (TPM) calculator within Geneious 8.0.5 (38).
Deletion of acmB in L. acidophilus NCFM.
The upp-based counterselection gene replacement method (20) was used to create an internal deletion of 1,103 bp in acmB (lba0176) of NCK1909, a upp-deficient background strain of L. acidophilus NCFM. With splicing by overlap extension PCR (39), the 1-kb regions flanking the deletion target were spliced with a BamHI restricted site added at the upstream end and SacI at the downstream end. This construct was digested with BamHI and SacI and then ligated into the polylinker of the similarly digested integration plasmid pTRK935 and transformed into competent E. coli EC101. The resulting recombinant plasmid, pTRK1097, was transformed into L. acidophilus NCK1909 harboring the helper plasmid pTRK669 (NCK1910). Single crossover integrants were screened as described previously (20). Colonies with the ΔacmB genotype were screened by PCR among the double recombinants recovered on glucose semidefined medium agar plates containing 5-fluorouracil. Deletion of acmB was confirmed by PCR and sequencing, and the resulting ΔacmB mutant was designated NCK2395.
Complementation of acmB in ΔacmB strain of L. acidophilus NCFM.
The ΔacmB strain of L. acidophilus NCFM was complemented using pTRK882, an expression plasmid with the promoter for pgm (lba0185), encoding a phosphoglyceromutase (40). The acmB gene, along with its native ribosome binding site, was amplified with EcoRI and BamHI restriction sites added to the 5′ and 3′ ends of the amplicon and subsequently cloned into the polylinker of pTRK882. The integrity of the insert was confirmed by DNA sequencing. The resulting recombinant plasmid, pTRK1098, was electroporated into the ΔacmB mutant of L. acidophilus NCFM. Transformants were selected by erythromycin resistance, generating NCK2397 for phenotypic comparison.
LiCl extraction of SLAPs.
Noncovalently bound cell surface proteins, including S-layer proteins and S-layer-associated proteins (SLAPs), were extracted from NCK1909 and ΔacmB L. acidophilus NCFM strains using LiCl denaturing salt, as described previously (25). Proteins were quantified via a bicinchoninic acid assay kit (Thermo Scientific) and visualized via SDS-PAGE using precast 4 to 20% Precise Tris-HEPES protein gels (Thermo Scientific). The gels were stained using AcquaStain (Bulldog Bio) according to the instructions of the manufacturer.
Microscopic and morphological assessments.
Morphological assessment of L. acidophilus NCFM strain was performed using a phase-contrast light microscope at ×40 magnification (Nikon Eclipse E600). Cells were observed over a growth period of 24 h in MRS broth at 37°C or MRS broth with 5 μg/ml erythromycin for the complemented strain NCK2397. Pictures were taken using a QImaging MicroPublisher 5.0 RTV camera attachment at 1-, 4-, 7-, 14-, and 24-h time points. Cell chain length was measured using Image-Pro Insight software (Media Cybernetics).
Autoaggregation and autolysis assays.
Autoaggregation assays were performed as described previously (41). Bacteria were grown in MRS broth with 5 μg/ml erythromycin where necessary for 16 h, harvested by centrifugation at 1,771 × g for 10 min, and washed twice with phosphate-buffered saline (PBS) (pH 7.4). Washed cells were resuspended in PBS to an adjusted optical density at 600 nm (OD600) of 1. Cell suspensions were mixed by vortexing for 10 s, and autoaggregation was determined over 5 h at room temperature. Every hour, 100 μl of the upper suspension was transferred to a cuvette with 900 μl PBS, and the absorbance (OD600) was measured. Autoaggregation percentages were calculated as follows: 1 − (At/A0) × 100, where At is the OD600 at time (t) = 1, 2, 3, 4, and 5 h and A0 is the OD600 at time zero.
Autolysis was performed as described previously (29) with some modifications. L. acidophilus NCFM strains were grown to late-exponential phase (OD600 of ∼1.0) and were harvested via centrifugation at 1,771 × g for 10 min. Cells were washed once with PBS (pH 7.4) and resuspended in PBS (pH 7.4) supplemented with 0.05% Triton X-100. Suspensions were transferred to sterile 96-well plates with transparent bottoms. OD600 values was assessed every 20 min for 24 h using a FLUOStar Optima microtiter plate reader (BMG Technologies) at 37°C. Autolysis was calculated as the decrease in OD600 relative to the initial OD600 at time zero.
Mucin and ECM adherence assays.
Extracellular matrix (ECM) binding assays were performed as described previously (42). Mucin (type III from porcine stomach; Sigma) was dissolved in PBS to a final concentration of 10 mg/ml. Fibronectin (from human plasma; Sigma), collagen (type IV from human cell culture; Sigma), and laminin (from Engelbreth-Holm-Swarm murine sarcoma/basement membrane; Sigma) were diluted in 50 mM carbonate-bicarbonate buffer (pH 9.6) (Sigma) to a final concentration of 10 μg/ml. For each assay, a Nunc MaxiSorp 96-well microplate (Sigma) was coated with 100 μl/well substrate and incubated at 4°C overnight. The wells were washed twice with PBS to remove excess substrate before blocking with 150 μl/well 2% bovine serum albumin (BSA) solution (Sigma) for 2 h at 37°C. Excess BSA was removed by two washes with PBS (pH 7.4).
Bacterial cells were grown in MRS broth to stationary phase (16 h) in preparation for the assay. Cultures were centrifuged (1,771 × g, 15 min, room temperature), washed once, and resuspended in PBS (pH 4.75). Cell density was adjusted to ∼1 × 108 CFU/ml based on previously calculated OD600/CFU ratios. Cell suspensions (100 μl) were added to each mucin or ECM-coated well. Initial cell counts were enumerated on MRS agar plates. After incubation for 1 h at 37°C, the wells were gently washed five times with 200 μl/well PBS. Adhered cells were recovered by adding 100 μl of 0.05% Triton X-100 solution (FisherBiotech) (prepared in PBS) to each well and agitating on an orbital shaker (200 rpm) for 15 min. Cell suspensions were transferred into 900 μl of 0.1× MRS before being further diluted and plated in duplicate on MRS plates. Colonies were enumerated and expressed as a percentage of relative adherence (mutant CFU/parent CFU), where parent (NCK1909) CFU were defined as 100%.
RESULTS
In silico analysis of autolysins within L. acidophilus NCFM.
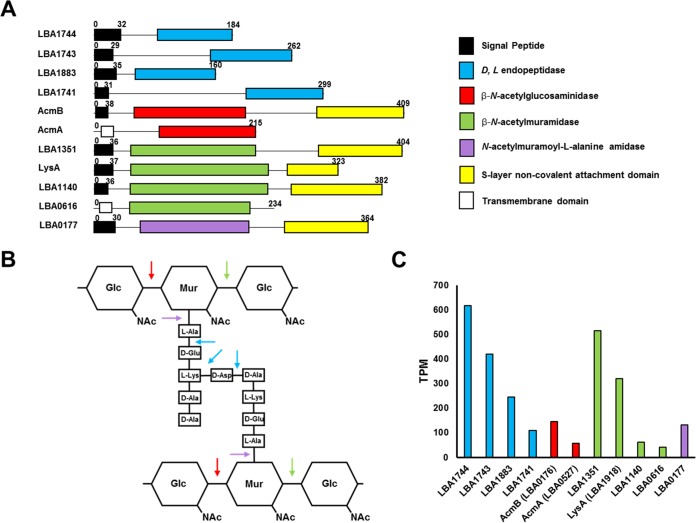
Within the genome of L. acidophilus NCFM, there are 11 genes which encode putative autolysins with predicted PGH activity (Table 2; Fig. 1). These PGH activities can be subdivided into four classes: d- and l-endopeptidases (pfam00877) (Fig. 1A, blue), β-N-acetylglucosaminidases (pfam01832) (Fig. 1A, red), β-N-acetylmuramidases (pfam01183) (Fig. 1A, green), and N-acetylmuramoyl-l-alanine amidases (pfam01510) (Fig. 1A, purple). The four classes of autolysins predicted in L. acidophilus have the required specificities to hydrolyze all components of the peptidoglycan (Fig. 1B). Further, there is apparent redundancy in three of the PGH classes, with four endopeptidases, two β-N-acetylglucosaminidases, and four β-N-acetylmuramidases encoded within the genome (Fig. 1A). With transcriptome sequencing (RNA-seq) data from a previous study (10), mid-logarithmic phase transcriptional profiles of the predicted autolysins were compared to analyze which may be the primary autolysin of each class (Fig. 1C). Based on these data, LBA1744, AcmB, and LBA1351 were found to be the most highly expressed endopeptidase, β-N-acetylglucosaminidase, and β-N-acetylmuramidase, respectively (Fig. 1C). LBA0177 appears to be the sole N-acetylmuramoyl-l-alanine amidase of L. acidophilus NCFM. Two genes with very low expression, AcmA and LBA0616, encoding a β-N-acetylglucosaminidase and a β-N-acetylmuramidase, do not have predicted signal peptide sequences and thus are likely not functional autolysins (Fig. 1A and C).
TABLE 2.
Autolysins of L. acidophilus NCFM
| Open reading frame | Identification | UniProt code | PGH domaina | Amino acid length | SignalPb | SLAPc |
|---|---|---|---|---|---|---|
| Endopeptidase | ||||||
| lba1744 | Putative glycosidase | Q5FIB9 | pfam00877 | 184 | + | − |
| lba1741 | Cell wall-associated hydrolase | Q5FIC1 | pfam00877 | 299 | + | − |
| lba1743 | Cell wall-associated hydrolase | Q5FIC0 | pfam00877 | 262 | + | − |
| lba1883 | NLP-P60 secreted protein | Q5FHZ1 | pfam00877 | 160 | + | − |
| β-N-Acetylglucosaminidase | ||||||
| acmA (lba0527) | N-Acetylglucosaminidase | Q5FLL6 | pfam01832 | 215 | − | − |
| acmB (lba0176) | N-Acetylglucosaminidase | Q5FMJ9 | pfam01832 | 409 | + | + |
| β-N-Acetylmuramidase | ||||||
| lba0616 | Putative uncharacterized protein | Q5FLC9 | pfam01183 | 234 | − | − |
| lysA (lba1918) | Lysin | Q5FHV9 | pfam01183 | 323 | + | + |
| lba1140 | Lysin | Q5FJZ4 | pfam01183 | 382 | + | + |
| lba1351 | Lysin | Q5FJE6 | pfam01183 | 404 | + | + |
| N-Acetylmuramoyl-l-alanine amidase | ||||||
| lba0177 | Autolysin/amidase | Q5FMJ8 | pfam01510 | 364 | + | + |
FIG 1.
(A) The modular domain organization of the 11 predicted autolysins of L. acidophilus NCFM. The numbers indicate the corresponding amino acid length. (B) The corresponding specific activity of each catalytic domain on the peptidoglycan structure. (C) RNA-seq transcriptional analysis of the gene encoding each autolysin, with colors corresponding to mRNA transcripts from genes with the following catalytic activities: blue, endopeptidase; red, β-N-acetylglucosaminidase; green, β-N-acetylmuramidase; and purple, N-acetylmuramoyl-l-alanine amidase.
Notably, five of the nine functional autolysins have been identified as SLAP constituents (Table 2) of the noncovalently bound exoproteome in L. acidophilus NCFM and other S-layer-forming species of Lactobacillus acidophilus homology group (10, 25). In fact, these proteins have the noncovalent attachment domain (NCAD) (pfam03217) found in other SLP, suggesting their colocalization at the cell surface along with the S-layer (Fig. 1A, yellow). These SLAPs encompass all of the predicted functional β-N-acetylmuramidases, as well as the sole N-acetylmuramoyl-l-alanine amidase, LBA0177, and the β-N-acetylglucosaminidase, AcmB. The four endopeptidases were not found in the SLAP fractions of the previous studies and do not have the NCAD domains (Fig. 1A). AcmB was selected for functional analysis based on its prevalence in the noncovalently bound exoproteome and conservation as a SLAP in L. acidophilus NCFM and other S-layer-forming species of the homology group (see below).
In silico genomic analysis of AcmB.
Although the gene encoding AcmB (lba0176) was previously annotated as a β-N-acetylmuramidase (3), the protein has a mannosyl-glycoprotein endo-β-N-acetylglucosaminidase catalytic domain (pfam01832), suggesting that AcmB is a β-N-acetylglucosaminidase. Within the chromosome of L. acidophilus NCFM, acmA (lba0527) is the only other gene which also contains the β-N-acetylglucosaminidase domain (Fig. 1A). However, acmA may be truncated, does not have a signal peptide sequence (Fig. 1A), and demonstrates reduced transcriptional expression compared to that of acmB, which does contain a signal peptide sequence (Fig. 1C). For these reasons, AcmB appears to be the primary β-N-acetylglucosaminidase for L. acidophilus NCFM.
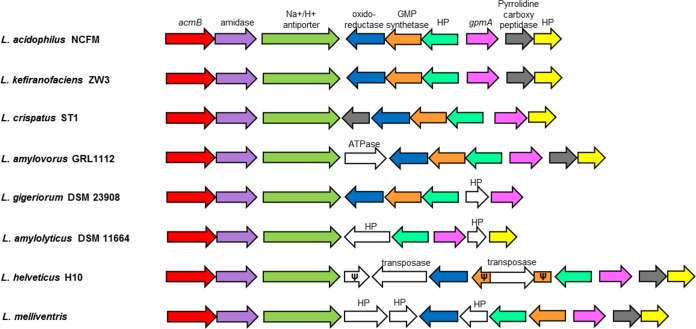
AcmB orthologs are found in numerous Lactobacillus species, including L. amylovorus GRL1112 (81.7% amino acid identity), L. helveticus H10 (78.5%), L. kefiranofaciens ZW3 (75.8%), L. crispatus ST1 (73.3%), L. melliventris (61%), L. amylolyticus DSM 11664 (61.3%), and L. gigeriorum DSM 23908 (59.3%). Most notably, the AcmB orthologs are found exclusively in S-layer-forming species of Lactobacillus, providing further evidence that AcmB is an S-layer-associated protein. There are additional orthologs found in the genomic region surrounding AcmB in these S-layer-forming species, compared to those in L. acidophilus NCFM (Fig. 2). In fact, the genomic region directly downstream of acmB appears to be syntenic and conserved among the seven S-layer-forming species listed above (Fig. 2). Among the orthologs with synteny in this region are genes encoding an S-layer-associated N-acetylmuramoyl-l-alanine amidase, an uncharacterized Na+/H+ ion transporter, an oxidoreductase, a GMP synthetase, a phosphoglyceromutase, a pyrrolidine carboxypeptidase, and two hypothetical proteins (Fig. 2). These genes were all found to be in the genomic region surrounding AcmB in the S-layer-forming strains, with the exception of the pyrrolidine carboxypeptidase, which is absent in L. amylolyticus DSM 11664 and L. gigeriorum DSM 23908 (Fig. 2). In L. kefiranofaciens ZW3, the position of the genes within the region is identical to that in L. acidophilus NCFM and is likewise conserved in L. crispatus ST1, with the exception of the translocation of a pyrrolidine carboxypeptidase gene from the positive strand to the negative strand of the genome (Fig. 2).
FIG 2.
acmB orthologs were found in various S-layer-forming strains of Lactobacillus, including L. amylovorus GRL1112, L. helveticus H10, L. kefiranofaciens ZW3, L. crispatus ST1, L. melliventris, L. amylolyticus DSM 11664, and L. gigeriorum DSM 23908. The genetic region surrounding acmB was highly syntenic in all species examined. Arrows represent genes, while the colors represent specific genes, as follows: red, acmB; purple, amidase; green, Na+/H+ ion transporter; blue, oxidoreductase; orange, GMP synthetase; teal, conserved hypothetical protein (HP); pink, gpmA; dark gray, pyrrolidine carboxypeptidase; and yellow, conserved HP. Genes in white are divergent genes unique to each species where indicated. Ψ indicates a truncated pseudogene.
Deletion of acmB from the chromosome of L. acidophilus NCFM and corresponding complementation of acmB in the ΔacmB mutant.
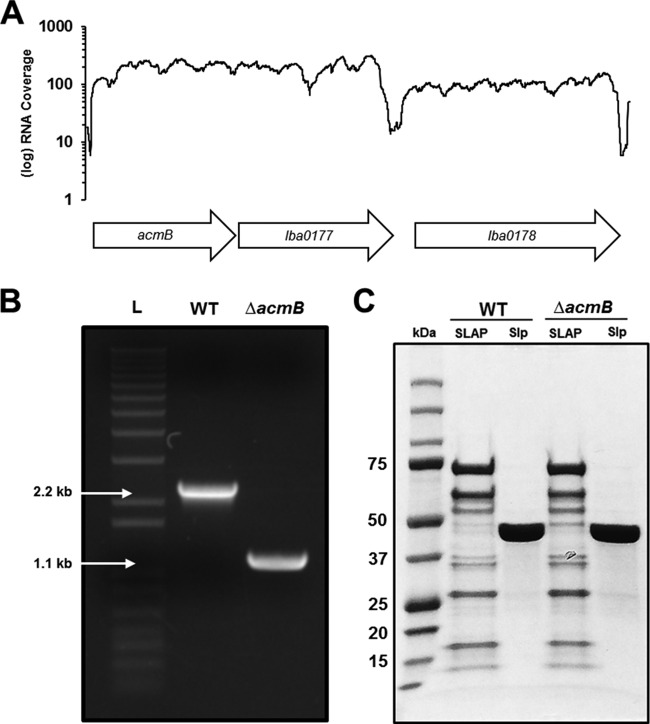
With a upp-based counterselective gene replacement method (20), a markerless chromosomal deletion of acmB was made in a upp-deficient background host of L. acidophilus NCFM. The ΔacmB genotype was confirmed using PCR and DNA sequencing with primers flanking the 1,227-bp acmB gene (Fig. 3A and B). The absence of AcmB from the SLAP fraction of the ΔacmB strain was not observed visually by SDS-PAGE, due to multiple 45-kDa proteins in the fraction (Fig. 3C). Furthermore, the ΔacmB mutant did not appear to have an altered SLAP profile compared to that of the wild type (WT) (Fig. 3C). For phenotypic analysis, acmB was also complemented in the ΔacmB strain using pTRK1098, an expression plasmid with the constitutive pgm promoter.
FIG 3.
The gene encoding AcmB was deleted from the chromosome of L. acidophilus NCFM. (A) RNA-seq analysis demonstrates that acmB is polycistronically expressed with lba0177, which encodes an S-layer-associated N-acetylmuramoyl-l-alanine amidase. (B) Gel electrophoresis of PCR products using the primers indicated in panel A for the parent strain (WT) compared to the ΔacmB mutant. The deletion was confirmed by sequencing. L, DNA ladder. (C) SDS-PAGE of the noncovalently bound extracellular S-layer proteins (SLP) and S-layer-associated proteins (SLAPs) isolated from both the WT and ΔacmB strains.
Phenotypic analyses of the ΔacmB mutant.
Following deletion confirmation, the ΔacmB strain was phenotypically assessed for (i) cell morphology, (ii) autoaggregation and autolytic capacity, and (iii) the ability to bind to mucin and extracellular matrices (ECM) collagen, fibronectin, and laminin.
Cellular morphology of the ΔacmB mutant.
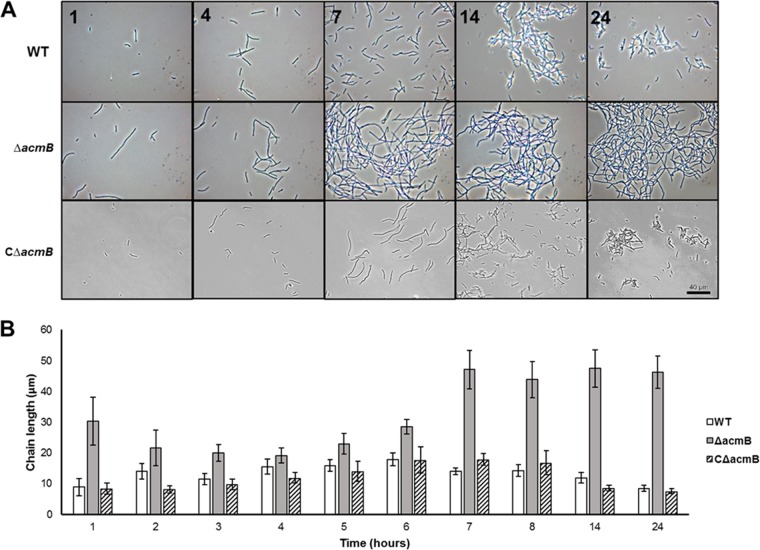
Cellular morphology of the ΔacmB strain was assessed by light microscopy over a 24-h growth time course in MRS culture (Fig. 4). Compared to the WT strain, the ΔacmB mutant had a distinctive morphological phenotype consisting of increased chain lengths and autoaggregation (Fig. 4A). Complementation of acmB in the ΔacmB strain (CΔacmB) resulted in a return to WT morphology (Fig. 4A). This cell division-related morphology was quantified by measuring the chain lengths of dividing cells throughout the 24-h time course (Fig. 4B). Both the WT and CΔacmB strains demonstrated prototypical chain lengths which followed a standard bacterial growth curve (Fig. 4B, white and striped bars). Specifically, chain lengths increased from lag phase to logarithmic phase (hours 1 to 6) and decreased as a result of dechaining during the transition to stationary phase (hours 7 to 24) (Fig. 4B). On the contrary, the ΔacmB mutant had a statistically significant increase in chain length across all measurements (P < 0.001) (Fig. 4B, gray bars). At the start of lag phase (hour 1), the ΔacmB strain had a pronounced increase in chain length (mean [M], 30.32 μm; confidence interval [CI] = 7.78) compared to those for the WT (M, 8.83 μm; CI, 2.77) and CΔacmB (M, 8.34 μm; CI, 1.79) strains, likely due to residual cells from inoculation transfer of the previous stationary-phase culture (Fig. 4B). By early-logarithmic phase, the differences in chain lengths were less evident between the ΔacmB mutant and the WT and CΔacmB strains. However, by mid-logarithmic phase (hour 7) the chain lengths in the ΔacmB culture were considerably longer (M, 47.0423 μm; CI, 6.17) than those for the WT (M, 13.94 μm; CI, 0.58) and CΔacmB (M, 17.85 μm; CI, 1.91) strains (Fig. 4B), suggesting aberrant dechaining and daughter cell separation. For the remainder of the time course, the ΔacmB strain maintained increased chain lengths while the CΔacmB and WT strains underwent normal cell division, as indicated by a concomitant decrease in cell chain lengths (Fig. 4B).
FIG 4.
(A) The cellular morphologies of the wild-type (WT), mutant (ΔacmB), and acmB complemented (CΔacmB) strains were assessed using phase-contrast light microscopy over a 24-h growth period. (B) Chain length measurements were taken for the WT, ΔacmB, and CΔacmB strains. The chain length for the ΔacmB mutant (n = 611 cells) was significantly higher than those for the WT (n = 661 cells) and CΔacmB (n = 316 cells) strains across all time points measured (P < 0.001). Error bars represent confidence intervals.
Autoaggregation and autolysis of the ΔacmB mutant.
Based on the abnormal cellular morphology and aberrant dechaining phenotype of the ΔacmB mutant, the autoaggregation and autolytic capacity of this strain were evaluated (Fig. 5). The sedimentation rates of the ΔacmB, WT, and complemented CΔacmB strains were measured over 5 h in PBS (Fig. 5A). For the first 2 h, the three strains had comparable autoaggregation rates. By 3 h, the autoaggregation of the ΔacmB mutant (M, 49.24%; CI, 7.16%) was significantly higher (P < 0.001) than those of the WT (M, 28.69%; CI, 2.68%) and CΔacmB (M, 33.94%; CI, 3.19%) strains (Fig. 5A). The differences were most pronounced at 5 h, at which the ΔacmB mutant had an autoaggregation percentage of 68.05% (CI, 7.73%), compared to 45.42% (CI, 10.35%) and 51.61% (CI, 3.31%) for the WT and CΔacmB strains, respectively (P < 0.01) (Fig. 5A). To assess the autolytic behavior of the ΔacmB, WT, and CΔacmB strains, Triton X-100-induced autolysis assays were performed (Fig. 5B). The rate of autolysis in the ΔacmB mutant was significantly lower than in the WT and complemented strains (P < 0.05). Specifically, Triton X-100-induced cells resulted in 40% autolysis of the ΔacmB population, compared to 50% in the WT and 52% in the CΔacmB strain (Fig. 5B). These data demonstrate that the absence of AcmB in the ΔacmB strain causes an increase in autoaggregation, along with a decrease in stress-induced autolysis.
FIG 5.
(A) Autoaggregation of WT, mutant (ΔacmB), and complemented (CΔacmB) cells. Asterisks indicate statistical significance (P < 0.001). (B) Triton X-100-induced autolysis assays for WT, ΔacmB, and complemented strains. The differences between the ΔacmB mutant and the WT and complemented strains are statistically significant (P < 0.001). Each assay was performed in triplicate; all error bars represent confidence intervals.
Adherence capacity of the ΔacmB mutant.
Extracellular proteins localized to the cell surface are important mediators of probiotic activity, including adhesion to host intestinal epithelial mucus layer and ECM. Because of the irregular morphology of the ΔacmB strain, the ability of this mutant to bind mucin and ECM, including collagen, fibronectin, and laminin, was examined. The binding capacity of the ΔacmB mutant was significantly reduced relative to that of the WT for mucin and all ECM tested (Fig. 6). Specifically, there was a 50% reduction of cells bound to type III porcine mucin (P < 0.002), a 55% reduction of cells bound to type IV human collagen (P < 0.001), a 63% reduction of cells bound to human plasma fibronectin (P < 0.001), and a 65% reduction in adherence to murine laminin (P < 0.001), relative to those of the WT. These data suggest that the absence of AcmB has a pleiotropic effect on the cell surface, which results in decreased binding to various ECM.
FIG 6.
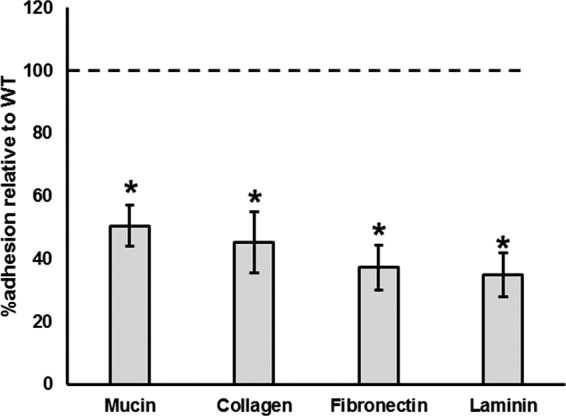
The ability of the ΔacmB mutant to bind to mucin and extracellular matrices (ECM) was assessed. Compared the wild-type (WT) reference (dotted line), the ΔacmB strain showed a significant reduction in binding to mucin, collagen, fibronectin, and laminin. Asterisks indicate statistical significance (P < 0.001). Adherence assays were performed in triplicate; all error bars represent confidence intervals.
DISCUSSION
The Gram-positive cell wall is composed of a thick peptidoglycan sacculus responsible for sustaining cell shape, resistance against environmental and osmotic stresses, and the covalent and noncovalent presentation of proteins (26). Extracellular proteins responsible for the turnover of peptidoglycan during cell division and daughter cell separation are known as peptidoglycan hydrolases (PGH), or autolysins (27). These PGH are divided into four classes: (i) β-N-acetylmuramidases, (ii) β-N-acetylglucosamidases, (iii) N-acetylmuramoyl-l-amidases, and (iv) peptidases (28).
In this study, the PGH complement of L. acidophilus NCFM was identified and AcmB, an S-layer-associated β-N-acetylglucosaminidase, was functionally characterized. Within the genome of L. acidophilus NCFM, 11 genes encoding putative PGH were identified, including four β-N-acetylmuramidases, two β-N-acetylglucosaminidases, one N-acetylmuramoyl-l-amidase, and four peptidases. Nine of these PGH are predicted to be functional based on the presence of signal peptide sequences and RNA transcription analyses. The redundancy of encoded autolysins is consistent with the findings of previous studies identifying the PGH complement in other lactobacilli. L. casei BL23 encodes 13 PGH (43), while L. plantarum WCFS1 encodes 12 (29) and silage-fermenting L. buchneri CD034 encodes 24 (44). Notably, the closely related and S-layer-forming cheese-ripening bacterium L. helveticus DPC 4571 encodes 9 autolysins, including 5 orthologs from the PGH identified in this study (45). Redundancy of PGH within bacterial genomes is widespread (27) due to the essentiality of autolysin activity for cell survival (46). Further, this redundancy may be due to the fact that many of these hydrolases have more than one function. PGH of Bacillus subtilis have numerous described functions beyond autolytic hydrolase activity, including roles in protein turnover and secretion, motility, and competence (47). However, the characterization of autolysin activity in Lactobacillus species, to date, has been primarily focused on the hydrolysis of peptidoglycan sacculi (13).
Autolysins are associated with the cell wall via numerous cell wall binding domains (CWBD) including CHAP domains, GW domains, SH3 domains, and LysM domains (13, 27). Notably, in L. acidophilus, the primary CWBD is the S-layer noncovalent attachment domain (NCAD) (pfam03217), which is responsible for the noncovalent attachment of the S-layer and S-layer-associated proteins in Lactobacillus (10, 30). Five of the nine functional autolysins in L. acidophilus NCFM have the S-layer NCAD domain, suggesting extracellular colocalization with the S-layer. In fact, these five proteins were previously identified in the LiCl-purified SLAP fraction of the L. acidophilus NCFM noncovalently bound exoproteome (25). Based on the in silico prediction of the PGH complement identified in this study and the previously identified SLAPs (25), all encoded β-N-acetylmuramidases, β-N-acetylglucosaminidases, and N-acetylmuramoyl-l-alanine amidases with a signal peptide sequence in L. acidophilus NCFM appear to be SLAPs. These findings are supported by previously published studies on autolysin activity in the S-layer-forming strains L. helveticus ATCC 12046 and L. helveticus ISLC5, in which two autolysins were copurified with the S-layer using LiCl (48, 49). It is also notable that there are two autolysins described in the PGH complement of L. helveticus DPC 4571 which contain the S-layer NCAD domain (45).
One of the five S-layer-associated autolysins is AcmB, a predicted 45-kDa protein with a β-N-acetylglucosaminidase catalytic domain. Further evidence of the association between this autolysin and the S-layer can be seen through examination of the AcmB orthologs in Lactobacillus species. All known AcmB orthologs are found exclusively in S-layer-forming species of the L. acidophilus homology group. Notably, AcmB orthologs are not found in the closely related, but non-S-layer-forming members of the homology group, including Lactobacillus gasseri, Lactobacillus johnsonii, and the progenitor, Lactobacillus delbrueckii subsp. bulgaricus. These data are supported by our recent exoproteomic survey of S-layer- and non-S-layer-forming species of the L. acidophilus homology group (10). Autolysins, including AcmB, were found in the noncovalently bound SLAP fractions of the S-layer-forming L. crispatus ST1, L. amylovorus GRL1112, and L. helveticus CNRZ32, but were not found in the non-S-layer-forming species tested (10).
There are two predicted β-N-acetylglucosaminidases in L. acidophilus NCFM, AcmA and AcmB. Because AcmA does not have a signal peptide sequence and has lower transcriptional expression than the other autolysins, AcmB appears to be the principal β-N-acetylglucosaminidase for L. acidophilus NCFM. To elucidate the role of AcmB in cell wall physiology and autolytic function, a ΔacmB isogenic mutant was created and complemented with an AcmB expression vector. Results indicate that the ΔacmB strain presents an altered cellular morphology consisting of increased chain lengths and autoaggregation due to its altered dechaining phenotype, as well as decreased stress-induced autolysis. The phenotypes of the L. acidophilus NCFM ΔacmB strain are consistent with the characterization of a β-N-acetylglucosaminidase, Acm2, in L. plantarum WCFS1 which similarly presented an altered dechaining and autolysis phenotype (29). Collectively, these data suggest that AcmB is a functioning autolysin involved in peptidoglycan turnover and daughter cell separation during cell division. Further work is necessary to characterize the specific hydrolytic activity of AcmB in L. acidophilus NCFM.
The ΔacmB deletion appeared to have a pleiotropic effect on the cell wall and subsequent presentation of cell surface proteins. The AcmB mutant presented a diminished capacity for binding to mucin and the extracellular matrices (ECM) collagen, fibronectin, and laminin in vitro. The mechanism for this adhesion phenotype remains unclear. It is possible that AcmB may directly interact with ECM in addition to its autocatalytic activity, not unlike the moonlighting proteins enolase, GAPDH, and GroEL, which have demonstrated secondary functions in adhesion to ECM in various Lactobacillus species (50–52). It is also possible that the impaired peptidoglycan hydrolysis in the ΔacmB mutant results in a concomitant impairment of the presentation of extracellular proteins which are covalently or noncovalently bound to the peptidoglycan. It seems most likely, however, that the reduced adhesion phenotype is due to the increased autoaggregation that resulted from the aberrant dechaining phenotype of the ΔacmB strain. Relevant cell surface proteins, including aggregation-promoting factors (42), fibronectin-binding proteins (21, 53), and other adhesins (54–56), may not be as exposed for contact with ECM. This interpretation of the data is consistent with previous observations of an acmA-deficient mutant of Lactococcus lactis MG1363, in which aberrant dechaining resulted in reduced adhesion to glass, polystyrene, and stainless steel (57).
In conclusion, we have shown that the SLAP AcmB is an autolysin involved in cell division and daughter cell separation in L. acidophilus NCFM. AcmB has an effect, directly or indirectly, on the adhesion of L. acidophilus NCFM to mucin and ECM, an important attribute for probiotic bacteria. Orthologs of AcmB are found exclusively in S-layer-forming species of the L. acidophilus homology group. Further, many of these autolysins, including AcmB, have been identified as SLAPs in the noncovalent exoproteomes of the S-layer-forming species of the said homology group. There is a dearth of information regarding the evolutionary function of S-layers in bacteria, especially those in Lactobacillus. Here, we propose that the S-layer may function as a scaffold for multiple proteins, including autolysins. Understanding the biological roles of these autolysins offers important evolutionary insights regarding the essentiality of the S-layer, as well as physiological insights regarding cell division and peptidoglycan hydrolysis in Lactobacillus species that produce S-layers.
ACKNOWLEDGMENTS
This work was supported through funding from the North Carolina Agriculture Foundation (Raleigh, NC) and DuPont Nutrition and Health (Madison, WI).
We acknowledge Yong Jun Goh for her tireless efforts in characterizing the S-layer proteins of Lactobacillus acidophilus NCFM, which has helped shape our overall understanding of the L. acidophilus S-layer, and Yong Jun Goh, Sarah O'Flaherty, and Rosemary Sanozky-Dawes for insightful discussion and critical reading of the manuscript.
Funding Statement
North Carolina Agricultural Foundation and DuPont Nutrition and Health provided funding for this work.
REFERENCES
- 1.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 2.Sanders ME, Klaenhammer TR. 2001. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci 84:319–331. doi: 10.3168/jds.S0022-0302(01)74481-5. [DOI] [PubMed] [Google Scholar]
- 3.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klaenhammer TR, Barrangou R, Buck BL, Azcarate-Peril MA, Altermann E. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol Rev 29:393–409. doi: 10.1016/j.fmrre.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Pfeiler EA, Klaenhammer TR. 2007. The genomics of lactic acid bacteria. Trends Microbiol 15:546–553. doi: 10.1016/j.tim.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Klaenhammer TR, Altermann E, Pfeiler EA, Buck BL, Goh YJ, O'Flaherty S, Barrangou R, Duong T. 2008. Functional genomics of probiotic lactobacilli. J Clin Gastroenterol 42:160–162. doi: 10.1097/MCG.0b013e3181574d48. [DOI] [PubMed] [Google Scholar]
- 7.O'Flaherty S, Klaenhammer TR. 2010. The role and potential of probiotic bacteria in the gut, and the communication between gut microflora and gut/host. Int Dairy J 20:262–268. doi: 10.1016/j.idairyj.2009.11.011. [DOI] [Google Scholar]
- 8.Johnson BR, Klaenhammer TR. 2014. Impact of genomics on the field of probiotic research: historical perspectives to modern paradigms. Antonie Van Leeuwenhoek 106:141–156. doi: 10.1007/s10482-014-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bron PA, Tomita S, Mercenier A, Kleerebezem M. 2013. Cell surface-associated compounds of probiotic lactobacilli sustain the strain-specificity dogma. Curr Opin Microbiol 16:262–269. doi: 10.1016/j.mib.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Johnson BR, Hymes J, Sanozky-Dawes R, DeCrescenzo Henriksen E, Barrangou R, Klaenhammer TR. 2016. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl Environ Microbiol 82:134–145. doi: 10.1128/AEM.01968-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Call EK, Goh YJ, Selle K, Klaenhammer TRK, O'Flaherty S. 2015. Sortase-deficient lactobacilli: effect on immunomodulation and gut retention. Microbiology 161:311–321. doi: 10.1099/mic.0.000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, Bron PA. 2010. The extracellular biology of the lactobacilli. FEMS Microbiol Rev 34:199–230. doi: 10.1111/j.1574-6976.2009.00208.x. [DOI] [PubMed] [Google Scholar]
- 13.Chapot-Chartier MP, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabeen MT, Jacobs-Wagner C. 2005. Bacterial cell shape. Nat Rev Microbiol 3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 15.Vollmer W, Seligman SJ. 2010. Architecture of peptidoglycan: more data and more models. Trends Microbiol 18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Call EK, Klaenhammer TR. 2013. Relevance and application of sortase and sortase-dependent proteins in lactic acid bacteria. Front Microbiol 4:73. doi: 10.3389/fmicb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hynönen U, Palva A. 2013. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol 97:5225–5243. doi: 10.1007/s00253-013-4962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smit E, Oling F, Demel R, Martinez B, Pouwels PH. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterization of domains responsible for S-protein assembly and cell wall binding. J Mol Biol 305:245–257. doi: 10.1006/jmbi.2000.4258. [DOI] [PubMed] [Google Scholar]
- 19.Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J 19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh YJ, Azcarate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, Lortal S, Klaenhammer TR. 2009. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 75:3093–3105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck BL, Altermann E, Svingerud T, Klaenhammer TR. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 71:8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frece J, Kos B, Svetec IK, Zgaga Z, Mrsa V, Susković J. 2005. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J Appl Microbiol 98:285–292. doi: 10.1111/j.1365-2672.2004.02473.x. [DOI] [PubMed] [Google Scholar]
- 23.Konstantinov SR, Smidt H, de Vos WM, Bruijns SCM, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, van Kooyk Y. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A 105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, Owen JL, Colliou N, Li E, Johannssen T, Lepenies B, Klaenhammer TR, Mohamadzadeh M. 2015. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J 34:881–895. doi: 10.15252/embj.201490296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson B, Selle K, O'Flaherty S, Goh YJ, Klaenhammer T. 2013. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159:2269–2282. doi: 10.1099/mic.0.070755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 27.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 28.Layec S, Decaris B, Leblond-Bourget N. 2008. Diversity of Firmicutes peptidoglycan hydrolases and specificities of those involved in daughter cell separation. Res Microbiol 159:507–515. doi: 10.1016/j.resmic.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Rolain T, Bernard E, Courtin P, Bron PA, Kleerebezem M, Chapot-Chartier MP, Hols P. 2012. Identification of key peptidoglycan hydrolases for morphogenesis, autolysis, and peptidoglycan composition of Lactobacillus plantarum WCFS1. Microb Cell Fact 11:137. doi: 10.1186/1475-2859-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boot HJ, Kolen CP, Pouwels PH. 1995. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J Bacteriol 177:7222–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Walker DC, Aoyama K, Klaenhammer TR. 1996. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol Lett 138:233–237. doi: 10.1111/j.1574-6968.1996.tb08163.x. [DOI] [PubMed] [Google Scholar]
- 33.Wei MQ, Rush CM, Norman JM, Hafner LM, Epping RJ, Timms P. 1995. An improved method for the transformation of Lactobacillus strains using electroporation. J Microbiol Methods 21:97–109. doi: 10.1016/0167-7012(94)00038-9. [DOI] [Google Scholar]
- 34.Somervuo P, Holm L. 2015. SANSparallel: interactive homology search against Uniprot. Nucleic Acids Res 43:W24–W29. doi: 10.1093/nar/gkv317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 36.UniProt Consortium. 2011. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222−D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 40.Duong T, Miller MJ, Barrangou R, Azcarate-Peril MA, Klaenhammer TR. 2011. Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microbiol Biotechnol 4:357–367. doi: 10.1111/j.1751-7915.2010.00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol 94:981–987. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 42.Goh YJ, Klaenhammer TR. 2010. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 76:5005–5012. doi: 10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regulski K, Courtin P, Meyran M, Claes IJJ, Lebeer S, Vanderleyden J, Hols P, Guillot A, Chapot-Chartier MA. 2012. Analysis of the peptidoglycan hydrolase complement of Lactobacillus casei and characterization of the major γ-d-glutamyl-l-lysyl-endopeptidase. PLoS One 7:e32301. doi: 10.1371/journal.pone.0032301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anzengruber J, Courtin P, Claes IJJ, Debreczeny M, Hofbauer S, Obinger C, Chapot-Chartier MA, Vanderleyden J, Messner P, Schäffer C. 2014. Biochemical characterization of the major N-acetylmuramidase from Lactobacillus buchneri. Microbiology 160:1807–1819. doi: 10.1099/mic.0.078162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jebava I, Plockova M, Lortal S, Valence F. 2011. The nine peptidoglycan hydrolases genes in Lactobacillus helveticus are ubiquitous and early transcribed. Int J Food Microbiol 148:1–7. doi: 10.1016/j.ijfoodmicro.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 48.Lortal S, Rousseau M, Boyaval P, Heijenoort JV. 1991. Cell wall and autolytic system of Lactobacillus helveticus ATCC 12046. Microbiology 137:549–559. [Google Scholar]
- 49.Valence F, Lortal S. 1995. Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl Environ Microbiol 61:3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, Marasco R, Sacco M. 2009. Surface displaced alpha-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb Cell Fact 8:14. doi: 10.1186/1475-2859-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kainulainen V, Loimaranta V, Pekkala A, Edelman S, Antikainen J, Kylvaja R, Laaksonen M, Laakkonen L, Finne J, Korhonen TK. 2012. Glutamine synthetase and glucose-6-phosphate isomerase are adhesive moonlighting proteins of Lactobacillus crispatus released by epithelial cathelicidin LL-37. J Bacteriol 194:2509–2519. doi: 10.1128/JB.06704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kainulainen V, Korhonen TK. 2014. Dancing to another tune—adhesive moonlighting proteins in bacteria. Biology 3:178–204. doi: 10.3390/biology3010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hymes J, Johnson BR, Barrangou R, Klaenhammer TR. 2016. Functional analysis of an S-layer associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 82:2676–2685. doi: 10.1128/AEM.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altermann E, Buck LB, Cano R, Klaenhammer TR. 2004. Identification and phenotypic characterization of the cell-division protein CdpA. Gene 342:189–197. doi: 10.1016/j.gene.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Buck LB, Azcarate-Peril MA, Klaenhammer TR. 2009. Role of autoinducer-2 on the adhesion ability of Lactobacillus acidophilus. J Appl Microbiol 107:269–279. doi: 10.1111/j.1365-2672.2009.04204.x. [DOI] [PubMed] [Google Scholar]
- 56.O'Flaherty S, Klaenhammer TR. 2010. Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology 156:3360–3367. doi: 10.1099/mic.0.043158-0. [DOI] [PubMed] [Google Scholar]
- 57.Mercier C, Durrieu C, Briandet R, Domakova E, Tremblay J, Buist G, Kulakauska S. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol Microbiol 46:235–243. doi: 10.1046/j.1365-2958.2002.03160.x. [DOI] [PubMed] [Google Scholar]
- 58.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell WM, Klaenhammer TR. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl Environ Microbiol 67:4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]