ABSTRACT
Sewage spills can release antibiotic-resistant bacteria into surface waters, contributing to environmental reservoirs and potentially impacting human health. Vancomycin-resistant enterococci (VRE) are nosocomial pathogens that have been detected in environmental habitats, including soil, water, and beach sands, as well as wildlife feces. However, VRE harboring vanA genes that confer high-level resistance have infrequently been found outside clinical settings in the United States. This study found culturable Enterococcus faecium harboring the vanA gene in water and sediment for up to 3 days after a sewage spill, and the quantitative PCR (qPCR) signal for vanA persisted for an additional week. Culturable levels of enterococci in water exceeded recreational water guidelines for 2 weeks following the spill, declining about five orders of magnitude in sediments and two orders of magnitude in the water column over 6 weeks. Analysis of bacterial taxa via 16S rRNA gene sequencing showed changes in community structure through time following the sewage spill in sediment and water. The spread of opportunistic pathogens harboring high-level vancomycin resistance genes beyond hospitals and into the broader community and associated habitats is a potential threat to public health, requiring further studies that examine the persistence, occurrence, and survival of VRE in different environmental matrices.
IMPORTANCE Vancomycin-resistant enterococci (VRE) are harmful bacteria that are resistant to the powerful antibiotic vancomycin, which is used as a last resort against many infections. This study followed the release of VRE in a major sewage spill and their persistence over time. Such events can act as a means of spreading vancomycin-resistant bacteria in the environment, which can eventually impact human health.
INTRODUCTION
Antibiotic-resistant bacteria (ARB) are a growing public health threat and an economic burden globally. The Centers for Disease Control and Prevention (CDC) in the United States has placed a high priority on addressing antibiotic resistance because of rising rates of ARB infection and associated disease burden and health care costs (1, 2). Most infections caused by ARB are nosocomial transmissions (i.e., originating in a hospital), but the role of environmental reservoirs in spreading ARB outside clinical settings is poorly understood. Studies have emphasized the role of environmental reservoirs in the spread of antibiotic resistance for decades, but more field and laboratory studies are necessary to address the specific mechanisms and conditions under which ARB survive and antibiotic resistance genes (ARGs) persist or can be transferred (3–5). Wastewater treatment plants (WWTPs) are sources of ARB, ARGs, and antimicrobial compounds through both treated effluent and the unplanned release of raw sewage to surface waters (6–9). ARB, ARGs, and antibiotics can be released into aquatic environments through human and agricultural waste, establishing routes of human exposure and threats to ecosystem health.
Vancomycin is a glycopeptide antibiotic that is used to treat infections caused by Gram-positive bacteria. It is considered a drug of last resort because of its historical success with the most recalcitrant infections caused by Gram-positive bacteria (10, 11). When vancomycin is rendered ineffective (i.e., when target bacteria are resistant), therapeutic treatment may fail and infections can be fatal (12, 13). Intrinsic, low-level resistance to vancomycin is characteristic of Enterococcus casseliflavus and Enterococcus gallinarum but is of less clinical concern than acquired, high-level vancomycin resistance (≥32 μg · ml−1) (14). Acquired vancomycin resistance can occur through the transfer of mobilizable genetic elements (15–17). Nine genes that confer vancomycin resistance in enterococci have been described, eight of which can be acquired (18). The most concerning from a public health perspective is the vanA gene, which is linked to most infections with human vancomycin-resistant enterococci (VRE). vanA is usually carried on a plasmid-borne transposon (Tn1546) (19–21) and confers high-level resistance to vancomycin (>64 μg · ml−1) (22).
The use of the glycopeptide avoparcin in animal agriculture in Europe has been linked to clinical vancomycin resistance (23–25). Although glycopeptides have not been used in animals in the United States, clinical incidence of VRE has steadily increased in past decades (2, 18, 26–29). Detection of VRE in the United States has been predominantly in clinical cases and hospital sewage (30, 31). The monitoring of VRE and associated resistance genes outside the hospital setting is necessary to better understand the spread of resistance and the increased risk to public health (6). Previous studies in Europe and Australia have reported community spread of VRE and fecal colonization of nonhospitalized individuals, but this has not been shown in the United States (32–35).
Antibiotic resistance can spread in bacterial habitats in the external environment, where antibiotics, ARB, and ARG enter water and sediments (6). The influx of sewage-associated microbes and other allochthonous bacteria into an aquatic environment can have ecological impacts, affecting community structure, nutrient cycling, and other ecosystem processes (36–38). In addition, the dynamics of gene exchange in microbial communities can be altered, and transfer of resistance genes may occur (39, 40). VRE and vancomycin resistance genes have been detected globally in the feces of agricultural and wild animals (30, 41–44), surface waters (45–47), WWTPs (48), domestic (community) sewage (49), and hospital sewage (30, 46, 50). Clinically relevant strains and vanA genes have rarely been reported in the environment in the United States (51, 52). The prevalence of genes encoding vancomycin resistance in the environment may increase the frequency of transfer to other Gram-positive pathogens (53), including the opportunistic pathogen Staphylococcus aureus (54). The incidence of vancomycin-resistant S. aureus (VRSA) in hospitals is low; however, 13 incidences have been reported in the United States as of 2014 (55), and the emerging threat is a concern for public health.
Relatively little information is available about the prevalence of clinically relevant VRE and vanA genes in aquatic environments, but many studies that have attempted to detect them have failed to find them in relatively pristine environments. Studies around the world have infrequently and inconsistently detected vanA genes and Enterococcus species isolates with vanA phenotypes in WWTP effluent and surface waters (56–59). One study in the United States isolated Enterococcus faecium carrying vanA genes on a recreational marine beach in Washington (52), but no other confirmation has been established outside hospital settings. In this field study, culturable VRE and/or vanA genes were detected in sediment and water samples after a sewage spill released more than 500,000 gallons of untreated sewage in a residential neighborhood. Illumina next-generation sequencing (NGS) of environmental DNA from sediment and water revealed the temporal changes in the microbial community after a major influx of untreated sewage.
MATERIALS AND METHODS
Sample collection.
A sewer line break in Pinellas County, FL, released more than 500,000 gallons of untreated sewage into a neighborhood drainage ditch beginning 27 September 2014. The line break was repaired with a bypass valve on 30 September 2014 after the sewage leakage was diverted. The site was also washed down, vacuumed, and disinfected with lime. A well-point system was also installed at the site to dewater, which resulted in groundwater discharge. Well-point systems are commonly used in engineering and construction and consist of a series of vacuum pumps designed to draw water up out of the ground. The ditch is connected to estuarine waters through wetlands. Photos of the site are included in Fig. S3A and B in the supplemental material. Water and sediment samples were collected at the spill site, along the drainage ditch for a distance of 800 m, and in adjacent receiving waters. Samples were collected seven times over the course of 7 weeks after the spill (1 October 2014 to 21 November 2014), to determine the persistence of sewage-associated microbes and VRE in the environment.
Six sites (NC-01, NC-02, NC-03, NC-04, NC-05, and NC-06) were selected for spatial assessment, but the majority of reported results are limited to one site that was sampled on all dates, NC-03. The additional sites where early sampling occurred are noted in the maps provided in Fig. S1 in the supplemental material. Site NC-01 became inaccessible after the first 2 weeks of sampling because it was filled in by construction crews. We were not able to collect sediment at the boat ramp in any instance because the site was a dock surrounded by mangroves. The boat ramp was included to represent recreational waters that may have been impacted by the spill. Water samples were collected in 500-ml sterile containers. Sediment samples were collected using a 50-ml sterile, screw-cap tube to scoop up the top 1 to 2 cm of sediments. All samples were transported on ice to the laboratory and processed within 6 h. Enterococci were also quantified by the Pinellas County Water and Sewer Department staff at 16 sites (see Fig. S2A and B in the supplemental material) near the point of the line break for 12 days using standard methods (ASTM D6503-99).
Isolation of and confirmation of VRE.
Water and sediment samples were processed using membrane filtration according to U.S. Environmental Protection Agency (EPA) Method 1600 for culturable enterococci (60), with modifications for the detection of VRE. Water samples were processed in multiple volumes (1 to 300 ml) on each sampling date over the course of the sampling period to account for variability in enterococcal concentrations. Vancomycin stock solution was prepared as an aqueous solution from sodium salt (Acros Organics/Thermo Fisher Scientific, NJ, USA) and sterile nuclease-free water to a final concentration of 10 mg · ml−1 and filter sterilized. To detect culturable VRE, Enterococcus indoxyl-β-d-glucoside (mEI) agar (Becton Dickinson, Sparks, MD) was prepared according to the manufacturer's recommendations. After the medium cooled to 55°C, the vancomycin solution was added to a final concentration of 32 μg · ml−1, the breakpoint for full resistance (14, 61). Sediment samples (30 g wet weight) were diluted 1:10 in phosphate-buffered saline (PBS) and hand shaken for 2 min to detach bacteria from particles (62). Sediment samples of the diluted buffered solution were processed in volumes from 0.1 to 100 ml depending on the sampling date and on previous concentrations of enterococci. Multiple dilutions for water and sediment were processed on each date to obtain viable colony counts.
To confirm culturable VRE as enterococci harboring the vanA gene, colonies with blue halos that grew on vancomycin-amended mEI were transferred to enterococcosel broth (EB) using sterile pipet tips or sterile toothpicks and were grown for 24 h. Wells that turned black were streaked for isolation onto tryptic soy agar (TSA) (Becton Dickinson, Sparks, MD) and then isolated again onto vancomycin-amended mEI (32 μg · ml−1). Isolated colonies were grown overnight in 5 ml Luria-Bertani (LB) broth (Thermo Fisher Scientific, Waltham, MA) that was amended with 32 μg · ml−1 vancomycin. DNA was extracted from overnight cultures using a GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO). Nucleic acid concentration was measured using a NanoDrop spectrophotometer to confirm successful extraction, and DNA was stored at −20°C in aliquots. Quantitative PCR (qPCR) was carried out with an Applied Biosystems 7500 real-time PCR system to confirm isolates as Enterococcus spp. (63) carrying the vanA gene (64). Isolates were identified to the species level by DNA sequencing of the 16S rRNA gene using universal bacterial primers (8F, 1492R) to amplify the 16S rRNA (65, 66); the PCR product was then purified using a GeneJet PCR purification kit (Thermo Fisher Scientific, Waltham, MA), sequenced by Eurofins Genomics (Huntsville, AL), and identified to the genus and species levels by using BLAST to reference the GenBank database (NCBI).
Sequencing and molecular analysis of environmental DNA.
Water (500 ml) was also filtered to obtain environmental DNA, and filters were stored at −80°C for DNA extraction. Sediment samples were also stored for DNA extraction. DNA from environmental water and samples was extracted and purified using the Mo Bio PowerWater kit from 0.45-μm filters. DNA from environmental sediment samples was extracted using Mo Bio PowerSoil kits directly from 0.3-g samples of sediment (Mo Bio Laboratories, Carlsbad, CA). Bacterial communities in those samples were characterized by sequencing the V4 region of the 16S rRNA gene. PCR was carried out to amplify the V4 region with the 515F and 806R primer pair, which included sequencer adapter sequences for Illumina sequencing (67, 68). The forward primer also contained a 12-bp barcode sequence unique to each sample. Each 25-μl PCR mixture contained 12 μl of PCR Water (Mo Bio Laboratories, Carlsbad, CA, USA), 10 μl of 2.5× 5 Prime HotMasterMix (Gaithersburg, MD), 1 μl of each of the primers (5 μM), and 1 μl of template DNA. The conditions for PCR were as follows: 94°C for 3 min, 35 cycles at 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s, and a final extension of 10 min at 72°C. Amplicons were quantified using PicoGreen (Invitrogen) and a plate reader (Infinite 200 Pro; Tecan) and were then pooled in equimolar ratios. This pool was cleaned using the UltraClean PCR clean-up kit (Mo Bio) and sequenced in an Illumina MiSeq run (2 × 150 bp) at Argonne National Laboratory. Sequencing reads were processed using QIIME (69) and USEARCH (70). The forward and reverse reads were merged, and then the merged reads were demultiplexed and filtered with a minimum Phred quality score of 20. Filtering resulted in about 388,000 high-quality reads, averaging about 28,000 reads per sample. Those reads were then clustered into 1,685 operational taxonomical units (OTUs) with a 97% similarity threshold. Chimeric sequences were identified with UCHIME and removed from OTUs (71). The taxonomy of the OTUs was assigned an RDP classifier against the SILVA databases (72, 73). For all downstream analyses, 10,000 reads were randomly selected per sample to correct for differences in sequencing depth.
Quantitative PCR (qPCR) was carried out with an Applied Biosystems 7500 real-time PCR system based on a previously published protocol for the vanA gene (64). Targets in environmental DNA were amplified using the following master mix composition per 25-μl reaction mixture: 12.5 μl TaqMan environmental master mix 2.0 (Thermo Fisher Scientific, Waltham, MA), 3 μl primer/probe mix (composed of 74.5 μl of each primer at 100 μM and 6 μl of target probe at 100 μM), 2.5 μl bovine serum albumin (BSA) (2 mg · ml−1), 2 μl sterile nuclease-free water, and 5 μl template DNA. Temperature cycling consisted of 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C. The lower limit of quantification (LLOQ) for the qPCR assay was 2.5 gene copies per reaction based on successful amplification in 50% of replicates of the lowest concentration on the standard curve (74). Sample LLOQ was 1.67 × 104 gene copies per 100 g for sediment samples and 10 gene copies per 100 ml for water samples. Blanks containing sterile Nanopure water in place of a sample were processed as negative controls (no-template controls [NTCs]). No blank amplified in any vanA qPCR assay. When the quantification cycle (Cq) values for the two replicates were greater than the Cq values for the LLOQ, results were reported as detected but not quantified (DNQ). Samples where neither replicate amplified and samples that did not successfully amplify in the two replicate qPCRs (amplified in 1 of 2) were reported as not detected (ND). The standard curve for vanA was constructed using a synthetic plasmid (IDT, Coralville, IA), containing the target sequence of the pIP816 vanA plasmid as previously published (NCBI accession number X56895) (64). Inhibition of amplification in environmental samples was tested using a qPCR SYBR green assay for the vvhA gene of Vibrio vulnificus (75). V. vulnificus is an autochthonous marine bacterium that does not grow in freshwater environments. Reaction mixtures contained 4 μl of DNA sample and 1 μl of V. vulnificus DNA (20,000 copies) and were compared to a control reaction mixture containing 4 μl of nuclease-free water and 1 μl of V. vulnificus DNA (20,000 copies) using previously published cycling conditions and primers (76).
Accession number(s).
Sequences were deposited in the NCBI BioProject database under BioProject accession number PRJNA322710.
RESULTS
Concentrations of culturable enterococci in water were high at the site of the spill (NC-03) immediately after the event (4.2 × 103 CFU per 100 ml), exceeding the U.S. EPA sample threshold value (STV) standards for recreational waters of 1.3 × 102 CFU per 100 ml (77) (Fig. 1). Levels decreased over time but did not fall below 1.3 × 102 CFU per 100 ml at NC-03 until 30 October 2014, more than 1 month after the event (Fig. 1). Enterococcal levels at the boat ramp in receiving marine waters approximately 3 km from the spill were within regulatory limits at each sampling date, ranging from 5 to 22 CFU per 100 ml. Enterococcal levels were 2 to 3 orders of magnitude higher in sediment than in water at NC-03 and also decreased over time (Fig. 1).
FIG 1.
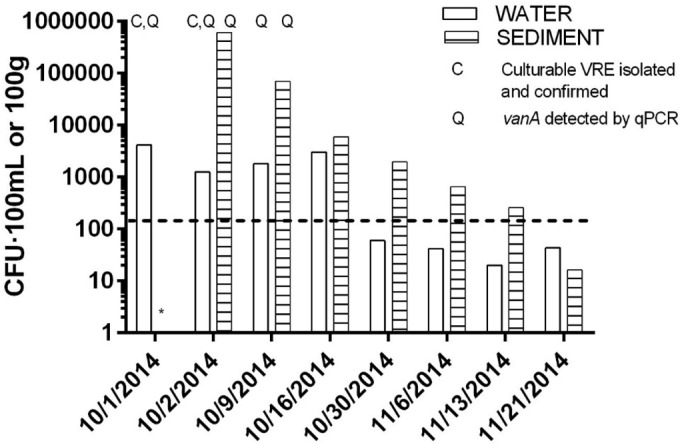
Culturable enterococci in water and sediment at site NC-03, near the site of the sewage spill. The dashed line represents the EPA standard for a single-sample maximum of enterococci in recreational water (130 CFU per 100 ml). Letters indicate where VRE were cultured (C) and where vanA was detected within 800 m of the spill (NC-01, NC-02, and NC-03). Note that on 1 October 2014, no sediment was collected or processed (indicated by *).
Enterococci were also monitored by Pinellas County at eight surface water sites ranging from 1 to 9 km away from the spill (B, C, F, G, H, I, J, and Q) for 12 consecutive days following the spill (see Fig. S2A and B in the supplemental material). Four sites within 4.5 km (from near to far, B, C, J, and Q) displayed enterococcal levels that exceeded recreational water quality standards (130 CFU per 100 ml) (77) for some duration after the spill. Exceedances were recorded at site B for 8 days, site C for 1 day, site J for 2 days, and site Q for 1 day. Maximum enterococcal levels were recorded 6 days after the spill at site B (2,100 CFU per 100 ml) and 1 day after the spill at site C (210 CFU per 100 ml), site J (160 CFU per 100 ml), and site Q (170 CFU per 100 ml). These sites were in the receiving waters directly adjacent to the site of the spill, Long Bayou and Cross Bayou, with the exception of site Q, which was in Boca Ciega Bay. Recreational water quality standards were not exceeded at the other four sites where enterococci were measured (F, G, H, and I) in Boca Ciega Bay, a body of water that mixes with the Gulf of Mexico and that is more than 5 km away from the spill.
VRE were detected by culture and confirmed as Enterococcus faecium in water collected 2 and 3 days after the spill ceased at NC-01, NC-02, and NC-03 (1 October 2014 and 2 October 2014) but could not be confirmed in water or sediment on subsequent dates (Fig. 1). A subset of putative VRE isolates from water sampled on 1 October 2014 and 2 October 2014 (11 of 15) was identified as E. faecium by 16S rRNA gene sequencing. The qPCR assay for vanA also confirmed that all 11 isolates identified as E. faecium carried the vanA gene. The other four putative VRE isolates were identified as Pediococcus spp. by16S rRNA sequencing. Colonies that grew on mEI amended with 32 μg · ml−1 vancomycin but could not be isolated and confirmed with molecular analyses were detected in water until 30 October 2014 and in sediment until 16 October 2014.
The vanA gene was detected in environmental DNA samples extracted from water and sediment up to 12 days after the spill (9 October 2014) at the sites within 800 m of the spill (NC-01, NC-02, and NC-03) (Fig. 1; Table 1) but not at later dates. Concentrations of vanA gene copies were approximately two orders of magnitude higher in sediment than in water (Table 1) but were reported per 100 g (wet weight) versus per 100 ml. In water, the maximum for vanA gene copies was 2.2 log10 gene copies per 100 ml (at site NC-01 on 9 October 2014), and the average was 1.9 log10 gene copies per 100 ml. In sediment, the maximum for vanA gene copies was 5.0 log10 gene copies per 100 g (at site NC-03 on 2 October 2014), and the average was 3.9 log10 gene copies per 100 g.
TABLE 1.
Detection and levels of vanA measured by qPCR in water and sediment at three sites near the origin (within 800 m) of the sewage spill over eight sampling datesa
| Sample type | Site | Days postspill |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 9 | 16 | 30 | 36 | 43 | 51 | ||
| Water (log10 gene copies per 100 ml) | NC-01 | 1.96 | 1.79 | 2.21 | — | — | — | — | — |
| NC-02 | NDb | 1.54 | 1.84 | — | — | ND | ND | ND | |
| NC-03 | 1.92 | ND | ND | ND | ND | ND | ND | ND | |
| Sediment (log10 gene copies per 100 g) | NC-01 | —c | ND | DNQd | — | — | — | — | — |
| NC-02 | — | 4.24 | ND | — | — | ND | ND | ND | |
| NC-03 | — | 4.95 | 4.54 | ND | ND | ND | ND | ND | |
Sample limits of detection were 4 gene copies per 100 ml water and 6.7 × 103 gene copies per 100 g sediment. Day 1 postspill is considered to be 1 October 2014. Note that access issues prevented sampling at all sites on all dates; data analysis focuses on site NC-03 where samples were collected on each sampling date.
ND, not detected.
—, not measured.
DNQ, detected but not quantifiable.
Sequencing results from environmental DNA on seven sampling dates where both sediment and water were collected showed distinct bacterial communities in water and sediment samples. In both matrices, dates closest to the spill (2 October 2014 and 9 October 2014 for sediment and water plus 16 October 2014 for water) were distinctly separate from those in the later sampling weeks (Fig. 2). The trend shown by these data suggests that the sediment and water at this site took approximately 2 to 3 weeks to return to a stable structure following the spill. The change in community composition is supported by a similar time frame of noticeable sewage impacts on fecal indicator bacteria and VRE (Fig. 1).
FIG 2.
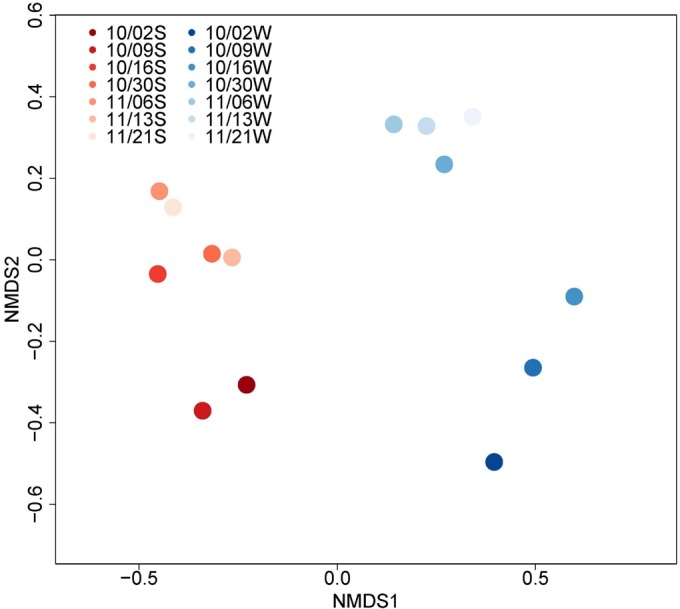
Analysis of DNA sequencing of the 16S rRNA gene in water and sediment on seven dates at site NC-03. The blue gradient represents water samples, and the red gradient represents sediment samples; the color gradient represents different time points in either water (W) or sediment (S) where darker shades are immediately after the spill or earlier in time.
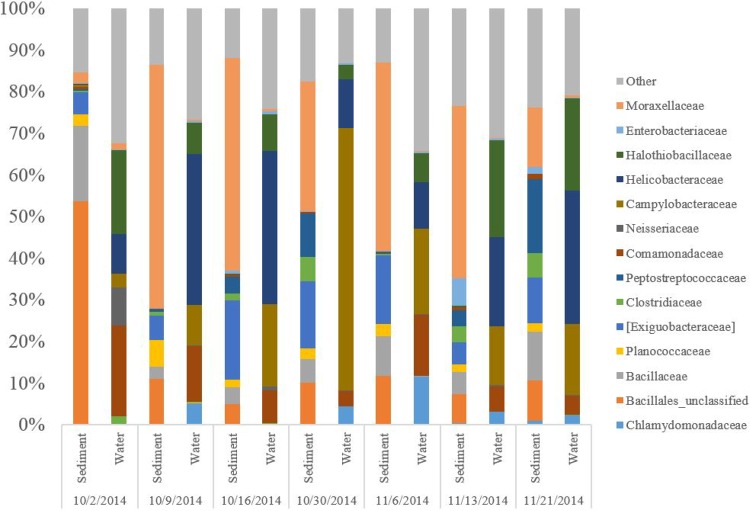
Six bacterial families shown to be highly prevalent in domestic sewage in the United States (Bacteroidaceae, Ruminococcaceae, Lachnospiraceae, Porphyromonadaceae, Veillonellaceae, and Prevotellaceae) (78) decreased in frequency with respect to total 16S rRNA sequences over time at NC-03 in sediment and water (Fig. 3). OTUs identified to the family level represented 85% to 95% of the total OTUs, with the exception of sediment on 2 October 2014, where 43% were identified to the family level. One of the sewage-associated families, Porphyromonadaceae, was found in the dominant taxa (top 10 most abundant) on the first sampling date and not at any later dates. Alpha diversity did not reveal temporal trends during the course of the sampling. The temporal trend of sewage-associated families also aligns with trends demonstrated in enterococci, VRE, and community structure (Fig. 1 to 3). Dominant families in sediment were different from dominant families in water (Fig. 4). Neisseriaceae, a family containing many genera associated with the gut flora of mammals, and Comamonadaceae, a family containing common environmental denitrifiers, had the greatest decline in relative abundance in water from the first to later sampling dates. Similar trends in the distinction between microbial communities in water and sediment were observed in taxonomic diversity based on phyla (see Fig. S4A and B in the supplemental material). Families containing common pathogens (Enterobacteriaceae and Enterococcaceae) were present at low levels in water and sediment throughout the study (see Fig. S5A and B in the supplemental material) and were combined to represent an average of 0.41% and 1.4% of sequence reads over time in water and sediment, respectively.
FIG 3.
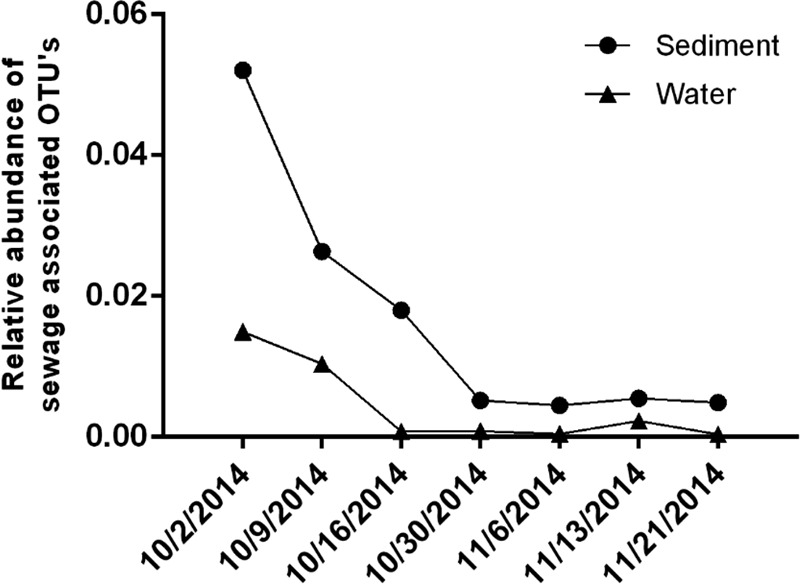
Relative abundance of select sewage-associated OTUs (Bacteroidaceae, Ruminococcaceae, Lachnospiraceae, Porphyromonadaceae, Veillonellaceae, and Prevotellaceae) at site NC-03 in water and sediment samples.
FIG 4.
Dominant families in sediment and water at site NC-03 on all sampling dates.
DISCUSSION
The sewage spill that we studied corresponded with elevated levels of enterococci, VRE, and vanA genes in water and sediment, indicating their release into the environment. All of these levels diminished steadily over the 2 weeks following the spill. No vanA genes were detected in environmental samples after 12 days at the site of the spill. This observation, and the fact that high-level VRE have been infrequently observed in uncontaminated surface waters (30), indicates that their presence in the environment before the spill is unlikely and that these contaminants were sewage associated (i.e., no background levels of vanA or VRE would be expected in the environment). The mitigation measures taken after the spill (vacuum pumping, washing out, lime treatment) probably decreased levels of microorganisms from sewage but left high levels of enterococci that slowly diminished over time in the area directly adjacent to the spill. The plume of the sewage spill was also indicated by the broader sampling effort in the region (as processed by Pinellas County), where enterococcal levels exceeded recreational water quality standards at the site closest to the spill (site B) but decreased after 8 days. Sites downstream from the spill where enterococcal levels were high decreased after 1 to 2 days. Flow rates, temperature, and other environmental conditions may impact the persistence and reach of contamination, but these factors were beyond the scope of this study.
The transfer of resistance through mechanisms such as horizontal gene transfer, demonstrated by the detection of the mobile vanA gene, can impact human health and the spread of resistance in the environment. This study has demonstrated the release of potentially pathogenic VRE and vanA genes into surface waters by sanitary sewer overflow in the United States. High-level VRE and vanA genes have been found in sewage from a hospital in Florida but were not found in other sewage samples that were not directly associated with a hospital (30). The spill in this study was not in close proximity to any hospital; the closest is 2.6 miles from the site of the sewer line break, and sewage from the hospital flows away from the break site. Previous studies have also investigated VRE in aquatic ecosystems, sanitary sewage, and WWTPs (48, 79–81), but community sewage (not associated with a hospital) has not been explicitly linked to vanA genes or highly resistant VRE in the United States. Results confirmed that untreated residential sewage released into aquatic environments can potentially be a route of human exposure to ARB and contribute to environmental reservoirs of ARB and ARGs.
Colonies that resembled VRE were detected in water samples through 30 October 2014 and in sediment samples through 16 October 2014; however, putative VRE colonies observed after 2 October 2014 could not be isolated based on the methods described above for confirmation. In all probability, they were either Enterococcus species or members of other genera that could “struggle” at 32 μg · ml−1 vancomycin on a crowded plate but did not possess vanA and so could not grow when subcultured on vancomycin. This observation reemphasizes the inaccuracy implicit in reporting VRE solely based on culture methods as further evidenced by the identification of Pediococcus spp. in this study. Other studies have demonstrated the isolation of a small percentage of genera other than Enterococcus on mEI (47, 82). The addition of vancomycin in the screening step tends to exacerbate the issue, as selection for intrinsically resistant genera, such as Pediococcus, Weissella, and Leuconostoc, also occurs (30).
DNA sequencing analysis has explored the dominant microbial taxa associated with sewage and human feces (78, 83), but the microbial community in waters impacted by sewage has received less attention. The advantages of this site included limited water input following the initial flushing so that changes in the community could be followed over time without the dilution effect that would occur in a large water body. The influx of sewage at this site produced a bacterial community with a prominent component of sewage- and fecal-associated bacteria that was detectable at the site for at least 2 weeks. The abundance of sewage-associated families declined on a similar time scale to enterococci, but the fate (i.e., death, transport, consumption by predators) of these bacteria and other pathogens was not determined. Some families containing pathogenic members and fecal indicator bacteria (FIB) (Enterococcaceae, Enterobacteriaceae) were represented throughout the sampling period.
Differences in community structure in sediment versus water were evident. The dominant phyla in water were consistent with those found in a study of 10 sites in the Mississippi River (Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and Verrucomicrobia accounting for approximately 94% of sequences) (84). However, Firmicutes (containing pathogen taxa) were more prevalent in water on the days immediately after the sewage spill than at later dates compared to the consistent low levels in the Mississippi River samples. The dominant taxa in sediment were consistent with published research where Proteobacteria and Firmicutes are prevalent phyla (85). It is interesting to note that the communities in water and sediment changed over approximately the same time frame and that they also remained distinct from one another. The relative rate of change in various environmental habitats bears further exploration, particularly given the extensive literature discussion about the potential role of sediments as environmental reservoirs for microbial pathogens and indicators (86–89).
This study confirms that potentially pathogenic ARB and associated ARGs can be released into the environment through untreated sewage and can persist for days or weeks after the initial introduction. Although the study area was flushed with water immediately after the spill, the sewage signal, as measured by enterococcal levels, persisted for 2 weeks after the event. This study supports the need for more mechanistic, empirical studies to address the role of environmental variability in the survival of ARB and ARGs, including parameters such as temperature and flow rates. Later sampling events, when no vanA genes were detected and no VRE were detected, support the previous studies suggesting their sewage association and absence of environmental background levels (30, 50, 90). While this study lacks a “before” sampling date for this site, the temporal sampling and the current literature support the idea that the vanA genes and VRE were derived from sewage. Immediately following the spill, E. faecium isolates harboring the vanA gene were identified in water samples at the site. The probability of human exposure outside of the cleanup crew was minimal in this case study, but sewage contamination events that occur at popular beaches and recreational areas may put more people at risk of exposure to antibiotic-resistant pathogens. In this study, antibiotic-resistant, opportunistic pathogens (VRE) associated with sewage entered the environment through a contamination event and persisted, potentially contributing to the spread of antibiotic resistance in the environment. Environmental reservoirs of ARB need further research and should be considered in frameworks designed to assess the spread of antibiotic resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Maccini of the Pinellas County Utilities Department for sharing data collected at additional sites, as well as the maps indicating their locations. We also thank undergraduate students Rochelle Etienne and Jenna Hindsley for their assistance processing samples and members of the Rohr Lab and Harwood Lab who provided useful comments on the manuscript. Anthony Carbo of St. Petersburg, FL, created the map of the sewage spill area, which is included as supplementary information. Sarah Owens of Argonne National Laboratory provided assistance with processing and method description for the Illumina sequencing.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01927-16.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2011. Antimicrobial resistance posing growing health threat. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. U.S. Department of Health and Human Services, Washington, DC. [Google Scholar]
- 3.Walsh F. 2013. Investigating antibiotic resistance in non-clinical environments. Front Microbiol 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy SB, Fitzgerald GB, Macone AB. 1976. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N Engl J Med 295:583–588. doi: 10.1056/NEJM197609092951103. [DOI] [PubMed] [Google Scholar]
- 5.McLain JE, Cyntryn E, Durso LM, Young S. 2016. Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J Environ Qual 45:432–440. doi: 10.2134/jeq2015.06.0317. [DOI] [PubMed] [Google Scholar]
- 6.Baquero F, Martinez JL, Canton R. 2008. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Michael I, Rizzo L, McArdell CS, Manaia CM, Merlin C, Schwartz T, Dagot C, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Res 47:957–995. doi: 10.1016/j.watres.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz T, Kohnen W, Jansen B, Obst U. 2003. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol 43:325–335. doi: 10.1111/j.1574-6941.2003.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 9.Young S, Juhl A, O'Mullan GD. 2013. Antibiotic-resistant bacteria in the Hudson River Estuary linked to wet weather sewage contamination. J Water Health 11:297–310. doi: 10.2166/wh.2013.131. [DOI] [PubMed] [Google Scholar]
- 10.Bassetti M, Ginocchio F, Giacobbe DR. 2011. New approaches for empiric therapy in Gram-positive sepsis. Minerva Anestesiol 77:821–827. [PubMed] [Google Scholar]
- 11.Bode C, Muenster S, Diedrich B, Jahnert S, Weisheit C, Steinhagen F, Boehm O, Hoeft A, Meyer R, Baumgarten G. 2015. Linezolid, vancomycin and daptomycin modulate cytokine production, Toll-like receptors and phagocytosis in a human in vitro model of sepsis. J Antibiot (Tokyo) 68:485–490. doi: 10.1038/ja.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmond MB, Ober JF, Dawson JD, Weinbaum DL, Wenzel RP. 1996. Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin Infect Dis 23:1234–1239. doi: 10.1093/clinids/23.6.1234. [DOI] [PubMed] [Google Scholar]
- 13.Cetinkaya Y, Falk P, Mayhall CG. 2000. Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707. doi: 10.1128/CMR.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI M100-20. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Arthur M, Courvalin P. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother 37:1563–1571. doi: 10.1128/AAC.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa K, Marchaim D, Martin ET, Tiwari N, Yousuf A, Sunkara B, Pulluru H, Kotra H, Hasan A, Bheemreddy S, Sheth P, Lee DW, Kamatam S, Bathina P, Nanjireddy P, Chalana IK, Patel S, Kumar S, Vahia A, Ku K, Yee V, Swan J, Pogue JM, Lephart PR, Rybak MJ, Kaye KS. 2012. Comparison of the clinical characteristics and outcomes associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant E. faecium bacteremia. Antimicrob Agents Chemother 56:2452–2458. doi: 10.1128/AAC.06299-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray BE. 2000. Vancomycin-resistant enterococcal infections. N Engl J Med 342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 18.O'Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 175:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guardabassi L, Dalsgaard A. 2004. Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol 70:984–990. doi: 10.1128/AEM.70.2.984-990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner G, Strommenger B, Witte W. 2008. Acquired vancomycin resistance in clinically relevant pathogens. Future Microbiol 3:547–562. doi: 10.2217/17460913.3.5.547. [DOI] [PubMed] [Google Scholar]
- 22.Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin Infect Dis 42(Suppl):S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 23.Bager F, Aarestrup FM, Madsen M, Wegener HC. 1999. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb Drug Resist 5:53–56. doi: 10.1089/mdr.1999.5.53. [DOI] [PubMed] [Google Scholar]
- 24.Klare I, Badstubner D, Konstabel C, Bohme G, Claus H, Witte W. 1999. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb Drug Resist 5:45–52. doi: 10.1089/mdr.1999.5.45. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson O. 2012. Vancomycin resistant enterococci in farm animals—occurrence and importance. Infect Ecol Epidemiol 2:16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martone WJ. 1998. Spread of vancomycin-resistant enterococci: why did it happen in the United States? Infect Control Hosp Epidemiol 19:539–545. doi: 10.2307/30141777. [DOI] [PubMed] [Google Scholar]
- 27.Rubinstein E, Keynan Y. 2013. Vancomycin-resistant enterococci. Crit Care Clin 29:841–852. doi: 10.1016/j.ccc.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Kirst HA, Thompson DG, Nicas TI. 1998. Historical yearly usage of vancomycin. Antimicrob Agents Chemother 42:1303–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 58:163–170. doi: 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 30.Harwood VJ, Brownell M, Perusek W, Whitlock JE. 2001. Vancomycin-resistant Enterococcus spp. isolated from wastewater and chicken feces in the United States. Appl Environ Microbiol 67:4930–4933. doi: 10.1128/AEM.67.10.4930-4933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK, National Healthcare Safety Network Team, Participating National Healthcare Safety Network Facilities. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson KB, Searle K, Stoddard GJ, Samore M. 2005. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci in rural communities, western United States. Emerg Infect Dis 11:895–903. doi: 10.3201/eid1106.050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padiglione AA, Grabsch EA, Olden D, Hellard M, Sinclair MI, Fairley CK, Grayson ML. 2000. Fecal colonization with vancomycin-resistant enterococci in Australia. Emerg Infect Dis 6:534–536. doi: 10.3201/eid0605.000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endtz HP, van den Braak N, van Belkum A, Kluytmans JA, Koeleman JG, Spanjaard L, Voss A, Weersink AJ, Vandenbroucke-Grauls CM, Buiting AG, van Duin A, Verbrugh HA. 1997. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol 35:3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendt C, Krause C, Xander LU, Loffler D, Floss H. 1999. Prevalence of colonization with vancomycin-resistant enterococci in various population groups in Berlin, Germany. J Hosp Infect 42:193–200. doi: 10.1053/jhin.1999.0597. [DOI] [PubMed] [Google Scholar]
- 36.Costanzo SD, Murby J, Bates J. 2005. Ecosystem response to antibiotics entering the aquatic environment. Mar Pollut Bull 51:218–223. doi: 10.1016/j.marpolbul.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 37.Mallin MA, Cahoon LB, Toothman BR, Parsons DC, McIver MR, Ortwine ML, Harrington RN. 2007. Impacts of a raw sewage spill on water and sediment quality in an urbanized estuary. Mar Pollut Bull 54:81–88. doi: 10.1016/j.marpolbul.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Barcina I, Lebaron P, Vives-Rego J. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol Ecol 23:1–9. doi: 10.1111/j.1574-6941.1997.tb00385.x. [DOI] [Google Scholar]
- 39.Martinez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 40.Nazaret S, Aminov R. 2014. Role and prevalence of antibiosis and the related resistance genes in the environment. Front Microbiol 5:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klibi N, Ben Amor I, Rahmouni M, Dziri R, Douja G, Ben Said L, Lozano C, Boudabous A, Ben Slama K, Mansouri R, Torres C. 2015. Diversity of species and antibiotic resistance among fecal enterococci from wild birds in Tunisia. Detection of vanA-containing Enterococcus faecium isolates. Eur J Wildl Res 61:319–323. [Google Scholar]
- 42.Lozano C, Gonzalez-Barrio D, Garcia JT, Ceballos S, Olea PP, Ruiz-Fons F, Torres C. 2015. Detection of vancomycin-resistant Enterococcus faecalis ST6-vanB2 and E. faecium ST915-vanA in faecal samples of wild Rattus rattus in Spain. Vet Microbiol 177:168–174. doi: 10.1016/j.vetmic.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 43.Oravcova V, Zurek L, Townsend A, Clark AB, Ellis JC, Cizek A, Literak I. 2014. American crows as carriers of vancomycin-resistant enterococci with vanA gene. Environ Microbiol 16:939–949. doi: 10.1111/1462-2920.12213. [DOI] [PubMed] [Google Scholar]
- 44.Goncalves A, Igrejas G, Radhouani H, Correia S, Pacheco R, Santos T, Monteiro R, Guerra A, Petrucci-Fonseca F, Brito F, Torres C, Poeta P. 2013. Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Lett Appl Microbiol 56:268–274. doi: 10.1111/lam.12044. [DOI] [PubMed] [Google Scholar]
- 45.Ben Said L, Klibi N, Lozano C, Dziri R, Ben Slama K, Boudabous A, Torres C. 2015. Diversity of enterococcal species and characterization of high-level aminoglycoside resistant enterococci of samples of wastewater and surface water in Tunisia. Sci Total Environ 530–531:11–17. [DOI] [PubMed] [Google Scholar]
- 46.Novais C, Coque TM, Ferreira H, Sousa JC, Peixe L. 2005. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl Environ Microbiol 71:3364–3368. doi: 10.1128/AEM.71.6.3364-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishiyama M, Iguchi A, Suzuki Y. 2015. Identification of Enterococcus faecium and Enterococcus faecalis as vanC-type vancomycin-resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J Environ Sci Health A Tox Hazard Subst Environ Eng 50:16–25. doi: 10.1080/10934529.2015.964599. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg Goldstein ER, Micallef SA, Gibbs SG, George A, Claye E, Sapkota A, Joseph SW, Sapkota AR. 2014. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci Total Environ 466–467:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanthier M, Scott A, Lapen DR, Zhang Y, Topp E. 2010. Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can J Microbiol 56:715–729. doi: 10.1139/W10-046. [DOI] [PubMed] [Google Scholar]
- 50.Iversen A, Kuhn I, Franklin A, Mollby R. 2002. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl Environ Microbiol 68:2838–2842. doi: 10.1128/AEM.68.6.2838-2842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts MC, No DB, Marzluff JM, Delap JH, Turner R. 3 February 2016. Vancomycin resistant Enterococcus spp. from crows and their environment in metropolitan Washington State, U S A: is there a correlation between VRE positive crows and the environment? Vet Microbiol doi: 10.1016/j.vetmic.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 52.Roberts MC, Soge OO, Giardino MA, Mazengia E, Ma G, Meschke JS. 2009. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. J Appl Microbiol 107:300–307. doi: 10.1111/j.1365-2672.2009.04207.x. [DOI] [PubMed] [Google Scholar]
- 53.Davison J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 54.de Niederhausern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, Anacarso I. 2011. Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr Microbiol 62:1363–1367. doi: 10.1007/s00284-011-9868-6. [DOI] [PubMed] [Google Scholar]
- 55.Limbago BM, Kallen AJ, Zhu W, Eggers P, McDougal LK, Albrecht VS. 2014. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J Clin Microbiol 52:998–1002. doi: 10.1128/JCM.02187-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander J, Bollmann A, Seitz W, Schwartz T. 2015. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci Total Environ 512–513:316–325. [DOI] [PubMed] [Google Scholar]
- 57.Bessa LJ, Barbosa-Vasconcelos A, Mendes A, Vaz-Pires P, Martins da Costa P. 2014. High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J Water Health 12:426–435. doi: 10.2166/wh.2014.160. [DOI] [PubMed] [Google Scholar]
- 58.Nam S, Kim MJ, Park C, Park JG, Maeng PJ, Lee GC. 2013. Detection and genotyping of vancomycin-resistant Enterococcus spp. by multiplex polymerase chain reaction in Korean aquatic environmental samples. Int J Hyg Environ Health 216:421–427. doi: 10.1016/j.ijheh.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Santiago-Rodriguez TM, Rivera JI, Coradin M, Toranzos GA. 2013. Antibiotic-resistance and virulence genes in Enterococcus isolated from tropical recreational waters. J Water Health 11:387–396. doi: 10.2166/wh.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.U.S. Environmental Protection Agency. 2002. Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-d-glucoside agar (mEI). EPA-821-R-02-022. U.S. Environmental Protection Agency, Cincinnati, OH. [Google Scholar]
- 61.Gold HS. 2001. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis 33:210–219. doi: 10.1086/321815. [DOI] [PubMed] [Google Scholar]
- 62.Boehm AB, Griffith J, McGee C, Edge TA, Solo-Gabriele HM, Whitman R, Cao Y, Getrich M, Jay JA, Ferguson D, Goodwin KD, Lee CM, Madison M, Weisberg SB. 2009. Faecal indicator bacteria enumeration in beach sand: a comparison study of extraction methods in medium to coarse sands. J Appl Microbiol 107:1740–1750. doi: 10.1111/j.1365-2672.2009.04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.U.S. Environmental Protection Agency. 2015. Method 1611.1: enterococci in water by TaqMan quantitative polymerase chain reaction (qPCR). EPA-820-R-15-008. U.S. Environmental Protection Agency, Cincinnati, OH. [Google Scholar]
- 64.Volkmann H, Schwartz T, Bischoff P, Kirchen S, Obst U. 2004. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J Microbiol Methods 56:277–286. doi: 10.1016/j.mimet.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 65.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl):S4516–S4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 71.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Layton BA, Cao Y, Ebentier DL, Hanley K, Balleste E, Brandao J, Byappanahalli M, Converse R, Farnleitner AH, Gentry-Shields J, Gidley ML, Gourmelon M, Lee CS, Lee J, Lozach S, Madi T, Meijer WG, Noble R, Peed L, Reischer GH, Rodrigues R, Rose JB, Schriewer A, Sinigalliano C, Srinivasan S, Stewart J, Van De Werfhorst LC, Wang D, Whitman R, Wuertz S, Jay J, Holden PA, Boehm AB, Shanks O, Griffith JF. 2013. Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res 47:6897–6908. doi: 10.1016/j.watres.2013.05.060. [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto K, Wright AC, Kaper JB, Morris JG Jr. 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae E1 Tor hemolysin gene. Infect Immun 58:2706–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright AC, Garrido V, Debuex G, Farrell-Evans M, Mudbidri AA, Otwell WS. 2007. Evaluation of postharvest-processed oysters by using PCR-based most-probable-number enumeration of Vibrio vulnificus bacteria. Appl Environ Microbiol 73:7477–7481. doi: 10.1128/AEM.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.U.S. Environmental Protection Agency. 2012. Recreational water quality criteria. 820-F-12–058. Office of Water, U.S. Environmental Protection Agency, Cincinnati, OH. [Google Scholar]
- 78.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6:. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furukawa T, Hashimoto R, Mekata T. 2015. Quantification of vancomycin-resistant enterococci and corresponding resistance genes in a sewage treatment plant. J Environ Sci Health A Tox Hazard Subst Environ Eng 50:989–995. doi: 10.1080/10934529.2015.1038150. [DOI] [PubMed] [Google Scholar]
- 80.Morris D, Galvin S, Boyle F, Hickey P, Mulligan M, Cormican M. 2012. Enterococcus faecium of the vanA genotype in rural drinking water, effluent, and the aqueous environment. Appl Environ Microbiol 78:596–598. doi: 10.1128/AEM.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Torres C, Reguera JA, Sanmartin MJ, Perez-Diaz JC, Baquero F. 1994. vanA-mediated vancomycin-resistant Enterococcus spp. in sewage. J Antimicrob Chemother 33:553–561. doi: 10.1093/jac/33.3.553. [DOI] [PubMed] [Google Scholar]
- 82.Nayak BS, Badgley B, Harwood VJ. 2011. Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by BOX-PCR typing and 16S rRNA gene sequencing. Appl Environ Microbiol 77:5050–5055. doi: 10.1128/AEM.00130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. 2010. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ. 2013. Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115:1147–1158. doi: 10.1111/jam.12323. [DOI] [PubMed] [Google Scholar]
- 85.Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Badgley BD, Nayak BS, Harwood VJ. 2010. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res 44:5857–5866. doi: 10.1016/j.watres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 87.Anderson KL, Whitlock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol 71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferguson CM, Coote BG, Ashbolt NJ, Stevenson IM. 1996. Relationships between indicators, pathogens and water quality in an estuarine system. Water Res 30:2045–2054. doi: 10.1016/0043-1354(96)00079-6. [DOI] [Google Scholar]
- 89.Burton GA Jr, Gunnison D, Lanza GR. 1987. Survival of pathogenic bacteria in various fresh-water sediments. Appl Environ Microbiol 53:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luczkiewicz A, Jankowska K, Kurlenda J, Olanczuk-Neyman K. 2010. Identification and antimicrobial resistance of Enterococcus spp. isolated from surface water. Water Sci Technol 62:466–473. doi: 10.2166/wst.2010.909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.