ABSTRACT
Human noroviruses (HuNoVs) are the main cause of shellfish-borne gastroenteritis outbreaks. In the absence of routine technical approaches allowing infectious particles to be detected, this viral pathogen is currently targeted by genome research, leading to difficult interpretations. In this study, we investigated the potential of F-specific RNA bacteriophages (FRNAPH) as fecal and viral contamination indicators in shellfish and water from a local harvesting area. FRNAPH were also used as microbial source tracking tools. Constraints imposed by detection limits are illustrated here by the detection of infectious FRNAPH in several samples in the absence of FRNAPH genomes. The opposite situation was also observed, likely explained by the persistence of the genomes being greater than infectivity. Similar considerations may be applied to HuNoVs, suggesting that HuNoV genome targeting is of limited relevance in assessing infectious risks. While FRNAPH did not provide any benefits compared to Escherichia coli as fecal pollution indicators in water, novel observations were made in shellfish: contrary to E. coli, a seasonal trend of infectious FRNAPH concentrations was observed. These concentrations were higher than those found in water, confirming bioaccumulation in shellfish. This study also underlines a relationship between the presence of HuNoV genomes and those of human-specific FRNAPH subgroup II (FRNAPH-II) in shellfish collected throughout Europe. Further research should be undertaken to evaluate FRNAPH potential as an indicator of the presence of infectious HuNoVs. To this end, shellfish involved in HuNoV-caused gastroenteritis outbreaks should be analyzed for the presence of infectious FRNAPH-II.
IMPORTANCE This work provides new data about the use of F-specific RNA phages (FRNAPH) as a tool for evaluating fecal or viral contamination, especially in shellfish. In our case study, FRNAPH did not provide any benefits compared to E. coli as fecal pollution indicators in water but were found to be very useful in shellfish. Their concentrations in shellfish were higher than those found in the surrounding water, confirming bioaccumulation. This study also underlines a relationship between the presence of human norovirus genomes (HuNoVs) and those of FRNAPH subgroup II (FRNAPH-II). Considering that the two virus types have similar behaviors and since FRNAPH infectivity can be investigated, the specific detection of infectious FRNAPH-II could be regarded as an indication of the presence of infectious HuNoVs. The contribution of infectious human FRNAPH targeting for assessing the viral risk associated with HuNoVs in shellfish should thus be investigated.
INTRODUCTION
Human-pathogenic enteric viruses (e.g., human noroviruses [HuNoVs] and hepatitis A virus [HAV]) may affect coastal waters, mainly through human fecal pollution resulting from the disruption of urban wastewater treatment plants or the leaching of agricultural soils amended with urban sewage sludge. A minor part of these viruses may also come from animal fecal pollution (e.g., hepatitis E virus). Oysters, which are able to concentrate such viruses, are implicated in a large number of outbreaks, mainly due to HuNoVs (1–4), despite European regulations on the matter.
In Europe, the microbiological quality of shellfish-harvesting areas is subject to EC regulation 854/2004 (5). Areas are classified according to their levels of fecal contamination estimated by Escherichia coli detection in shellfish flesh and intravalvular liquid (FIL). Areas with less than 230 most probable number (MPN) of E. coli per 100 g of FIL can be harvested for direct human consumption. For higher concentrations, depuration or relaying is needed, and when the limit of 46,000 MPN of E. coli per 100 g of FIL is exceeded, shellfish are considered unfit for human consumption. There is no doubt that for high concentrations of E. coli, the risk of viral infection is significant. Nevertheless, the absence of E. coli, with or without depuration, is no guarantee of the absence of infectious viruses, and shellfish compliant with European Union (EU) legislation may be involved in outbreaks (3, 6, 7). The reasons are well known today: E. coli is (i) less resistant than viruses in the environment (8, 9), (ii) more affected by depuration procedures (10–12), and (iii) also potentially less accumulated than enteric viruses in shellfish because of the presence of specific receptors, especially for HuNoVs (13–15).
Over the past few decades, molecular methods have been greatly improved, and an ISO standard for the detection of HuNoVs and HAV in bivalve molluscan shellfish is now available (16). Such an approach appears to be ideal because of its high sensitivity, rapidity, and specificity. However, molecular methods are not without significant drawbacks. These techniques may underestimate the viral load because of the low sample mass used for detection (17), low extraction yields, the presence of reverse transcription-PCR (RT-PCR) inhibitors (4), and the possible presence of other nontargeted viruses. Consequently, laboratories sometimes fail to detect the presence of HuNoV genomes in oysters involved in global HuNoV gastroenteritis outbreaks (17). On the other hand, a very high overestimation of the infectious risk may also occur (17–19) because of the persistence of viral genomes being greater than viral infectivity (20–23), especially after disinfection treatment. It has been suggested that as shellfish are not able to bioaccumulate NoV RNA fragments present in seawater (24), the genomes detected in shellfish mainly result from infectious virus accumulation. However, genomes in a capsid are not systematically synonymous with infectivity, and recent data have shown that some inactivating treatments, especially UV radiation, cause damage leading to inactivated viral particles but do not affect the binding function (25). In addition to natural UV radiation, this type of radiation is commonly used to treat water used during the shellfish depuration process but also for wastewater treatment, which may impact the shellfish-harvesting areas. It significantly reduces viral infectivity by causing genome damage and, to a lesser extent, by altering capsid proteins. Thereby, the resulting inactivated viral particles should still be able to accumulate in shellfish, and short RNA fragments targeted by molecular techniques remain detectable (26–28), which can explain the high prevalence of HuNoV genomes detected in shellfish. In this context, the 5.0% to 76.2% positive results for the detection of HuNoV genomes in shellfish intended for commercial use (17, 29–37) are difficult to interpret in terms of infectious risk. Although the dose-response concept is often observed (3, 18), HuNoV genome levels may be sometimes low in shellfish commodities involved in HuNoV gastroenteritis outbreaks (19) and may be high in nonoutbreak-related samples (18).
In this context, other indicators have been proposed to evaluate the level of viral pollution and the associated risks. F-specific RNA bacteriophages (FRNAPH) have been proposed as suitable viral indicators, especially in shellfish (38–42), due to their common presence in wastewater and their structural similarity to many enteric viruses (nonenveloped single-stranded RNA viruses, 20 to 30 nm in diameter). They are divided into 4 subgroups, present either in animal (FRNAPH subgroup I [FRNAPH-I] and FRNAPH-IV) or human waste (FRNAPH-II and FRNAPH-III), with various degrees of certainty (43–46). They can be detected by specific RT-PCR methods, allowing the genome of each subgroup to be quantified (47, 48), or by rapid culture that provides information about their infectivity. As with HuNoVs, FRNAPH show greater persistence than E. coli in the environment (49, 50), and several studies underline a relationship between HuNoVs and FRNAPH in shellfish (34, 35, 39, 41, 51) and seasonal variation in FRNAPH concentration, displaying the trend of shellfish-associated gastroenteritis (39, 40, 52), thus attesting to their potential.
The aim of this study was first to evaluate the value of FRNAPH as fecal or viral pollution indicators in shellfish and environmental waters from a designated harvesting area and as specific human fecal pollution markers. To evaluate their value as fecal markers, the level of infectious FRNAPH was compared to those of E. coli and enterococci in waters and mollusks. Their value as viral pollution indicators was evaluated by comparing the level of FRNAPH genomes to those of NoV genogroup I (NoV GI) and NoV GII, taking into account that FRNAPH infectivity can easily be assessed, unlike that of NoV. Because NoV GI and NoV GII are human-specific-pathogenic viruses, the ability of FRNAPH-II and -III to track a human pollution event could also be evaluated. Finally, the investigation of the relationship between the levels of FRNAPH and HuNoV genomes was expanded to include shellfish collected from different harvesting areas throughout Europe.
MATERIALS AND METHODS
Water sample preparation.
Thirty-two water samples were collected from a coastal region located in western France between January 2014 and January 2015. Samples came from 4 areas located near a local shellfish-harvesting area (Fig. 1): stream 1 (n = 8 samples of fresh water) flowing into estuary 2 (n = 8 samples of brackish water), and stream 3 (n = 8 samples of fresh water) flowing more slowly into estuary 4 (n = 8 samples of brackish water). For each sampling, 450 ml of water was collected in sterile bottles and kept at −20°C until analyzed.
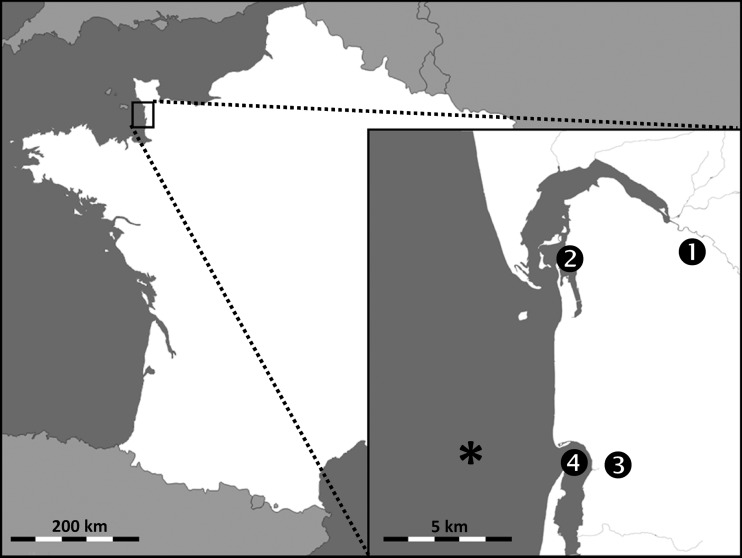
FIG 1.
Geographic locations of the 4 water sampling areas and of the shellfish-harvesting area (*). The figure was generated via Snazzy Maps software, using images from Google Maps.
Analyses were performed either directly from water samples or after concentration by ultracentrifugation. Concentration was performed from 2 × 200 ml of water in 2 polyallomer centrifuge bottles (Thermo Scientific, Waltham, MA, USA) in an A-621 fixed-angle rotor (Thermo Scientific) at 21,000 rpm (67,000 × g), for 18 h at 4°C. The first bottle was used for the enumeration of infectious FRNAPH and the second for the detection of FRNAPH and HuNoV genomes. In the second bottle, 100 μl of a feline calicivirus suspension (FCV) adjusted to 106 genome copies (gc)/ml was added before ultracentrifugation as an internal standard. As described below, for each experiment, the same volume of the FCV suspension was extracted in parallel, and a standard curve was generated from a 10-fold dilution series in order to estimate the yields of the concentration and extraction methods for each sample.
Shellfish sample preparation.
Eight batches of oysters (Crassostrea gigas) and 8 batches of mussels (Mytilus edulis) were collected during the study period from a harvesting area essentially impacted by stream 3 and estuary 4 (Fig. 1). The area is classified as B, according to EC regulation 854/2004 (5). Twenty-eight batches of oysters intended for marketing were additionally collected between January and March 2016 from several harvesting areas throughout Europe (France, Ireland, and Portugal).
After collection, shellfish were kept at 4°C and were shucked and dissected within 12 h. Hepatopancreas (HP) fragments were divided from several specimens, according to ISO/TS standard 15216-1 (16). They were then kept at −20°C until analyzed. A weight estimate based on 5 specimens allowed a determination that the HP fraction represented 5.0% ± 0.7% (wt/wt) of the total FIL mass (data not shown). After thawing, 4 g of digestive tissues was mixed with 4 ml of phosphate-buffered saline (PBS) for 3 min in a DT-20 tube with Ultra-Turrax tube drive (IKA-Werke GmbH & Co. KG, Staufen, Germany). One-quarter of the suspension volume (corresponding to 1 g of HP) was removed for RNA extraction, as described below, and supplemented with 100 μl of the FCV suspension as an internal standard.
For the culture step, a 3-ml volume of PBS–0.3% peptone was added to the remaining suspension to obtain a final concentration of 0.1% peptone, as described in ISO standard 6887-3 (53). The suspension was then mixed for 1 min and kept in ice for 4 h. After centrifugation (2,000 × g for 5 min), 3 ml of supernatant (corresponding to 1 g of HP) was collected, supplemented with 7 ml of PBS, and used for culture.
Infectious FRNAPH enumeration and phage plaque genotyping.
Infectious FRNAPH were detected according to ISO standard 10705-1 (54) using Salmonella enterica serovar Typhimurium WG49 (NCTC 12484) as the host strain (55). Culture was performed in 150-mm-diameter petri dishes, allowing for the analysis of 5-ml samples. Kanamycin and nalidixic acid were added (100 μg/ml) to limit the growth of other bacteria. Viral concentration was expressed in PFU per milliliter of water or per gram of HP after an 18-h incubation period.
Concerning water, detection was performed directly from 20 ml of samples (4 × 5 ml) or after concentration by ultracentrifugation, as described above. In the case of ultracentrifugation, the supernatant was gently eliminated, and the pellet was suspended by two washing and scraping steps with 2 ml of PBS and transferred in a sterile 15-ml tube. The final volume was adjusted to 5 ml and used for titration.
Concerning shellfish, culture was performed from 2 × 5 ml of samples (corresponding to 1 g of HP), prepared as described above.
For infectious FRNAPH genotyping, phage plaques were randomly isolated from positive water and shellfish samples (up to a limit of 12 per sample), collected with tips, and suspended in 1 ml of PBS–15% glycerol. After brief vortexing, the suspension was filtered through sterile Acrodisc syringe filters (pore size, 0.22 μm; Pall Life Sciences, Ann Arbor, MI, USA). RNA was then extracted using NucliSENS easyMAG (bioMérieux, Marcy l'Etoile, France) from 50 μl of phage suspension, eluted in 100 μl of buffer. Genotyping was then performed from RNA by a one-step multiplex quantitative RT-PCR, as used in a previous study (21).
Genome extraction.
Extraction was achieved from waters after ultracentrifugation or from mixed shellfish. Regarding waters, lysis was performed beforehand from the concentrated pellet by adding 1 ml of PBS and 3 ml of Isol-RNA lysis reagent (5 Prime GmbH, Hilden, Germany) in the centrifuge bottle. The pellet was suspended by gentle agitation for 10 min on a tube roller at 70 rpm and, after scraping, the suspension was collected for genome extraction.
Regarding shellfish, a 2-ml volume of the mixture, prepared as described above, was incubated with 220 μl of proteinase K (30 U/ml) at 37°C for 1 h, with stirring. After centrifugation (3,000 × g for 5 min), 1.11 ml of the supernatant was collected (corresponding to 0.5 g of HP) and supplemented with 3 ml of Isol-RNA lysis reagent (5 Prime GmbH, Hilden, Germany).
The 4-ml lysed samples (suspended pellet or supernatant from digested shellfish) were finally collected in 15-ml Phase Lock Gel heavy tubes (5 Prime GmbH, Hilden, Germany) and, after adding 2 ml of chloroform, the mixture was stirred vigorously for 15 s and centrifuged (2,000 × g for 2 min). Extraction was then performed from the whole supernatant using NucliSENS magnetic extraction reagents (bioMérieux, Marcy l'Etoile, France) on a NucliSENS miniMAG in 100 μl of elution buffer, according to the manufacturer's recommendations.
FRNAPH genome quantification.
Genomes of each FRNAPH subgroup were targeted for detection in waters after concentration by ultracentrifugation and in shellfish HP. Detection was performed according to the protocol described by Ogorzaly and Gantzer (47). For each subgroup, quantification was carried out in duplicate using a standard curve range of 101 to 105 gc/reaction. Thereby, quantification limits (LOQ), corresponding to the detection of 101 gc in a PCR well, were 267 gc/100 ml of water after ultracentrifugation and 1,067 gc/g of shellfish HP.
HuNoV genome quantification.
NoV GI and GII genomes were detected in waters after concentration and in shellfish HP, according to ISO/TS standard 15216-1 (16). Quantification was carried out in duplicate for both subgroups using a standard curve range of 101 to 105 gc/reaction. From the volume of RNA suspension used in the reaction, the LOQ were 100 gc/100 ml of water after ultracentrifugation and 400 gc/g of shellfish HP.
Other physicochemical, microbial, and epidemiological parameters.
Other parameters were examined to characterize samples. In waters, dissolved oxygen, temperature, salinity, pH, and turbidity were measured.
E. coli and enterococcal concentrations in waters were also evaluated using a miniaturized MPN method based on ISO standard 9308-3 (56). In oysters and mussels, E. coli was detected by direct impedance measurement in shellfish FIL, according to NF V08-106 (57).
The results were compared to epidemiological data on the incidence rate of gastroenteritis in the region during the study period. In January to February 2014, the gastroenteritis incidence rate was higher than the national epidemic threshold (about 300 cases per 100,000 inhabitants). For the other dates, it was lower (between 50 and 130 cases per 100,000 inhabitants).
Concentration and extraction yields estimated by FCV quantification.
To estimate the yields of the ultracentrifugation and extraction methods, FCV was used as an internal standard. Standard curves were obtained from a 10-fold dilution series of the FCV suspension, corresponding to a yield of 100%, 10%, and 1%. For each sample, the yield was evaluated by an absolute quantification of FCV by quantitative reverse transcription-PCR (RT-qPCR) using the RNA UltraSense one-step quantitative RT-PCR system (Life Technologies, Carlsbad, CA, USA), performed from 5 μl of RNA in a 25-μl reaction volume, according to the manufacturer's recommendations, using primers (500 nM) and a probe (900 nM) designed by Gassilloud et al. (22). The reaction was carried out on StepOne Plus real-time PCR system (Life Technologies) at 55°C for 40 min (reverse transcription), 5 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 65°C. Quantification of FCV was also performed from 10-fold-diluted samples to assess the presence of RT-PCR inhibitors.
PCA.
Principal-component analysis (PCA) is a geometrical and statistical descriptive approach that allows the reduction of multiple quantitative variables into a limited number of factors. The objective is to project variables in a low-dimension space, distorting the facts as little as possible, and to obtain the most relevant summary of initial data. Graphical interpretation can reveal similarities or oppositions between data placed on a 2-dimension plane, defined by two factors (generally F1 and F2) chosen to summarize the maximum amount of information. Only correctly represented variables can be interpreted (i.e., near the correlation circle). Indeed, two points may appear close together in a 2-dimension plane while they are distant in a multiple-dimension space (on both sides of the projection plane). With respect to interpretation, some positions are worth noticing: a positive correlation exists between variables in close proximity (correlation coefficient close to 1), variables moving in opposite directions are negatively correlated (r close to −1), and no correlation is observed between variables at right angles to each other (r close to 0). PCA was performed using an Excel add-in (Revue Modulad 43, 2011, http://www.modulad.fr/excel_macros.htm).
RESULTS
Methodological considerations for the detection of infectious FRNAPH in water samples.
The methods used to detect bacteria and infectious FRNAPH in waters are based on standardized techniques. The theoretical limits of detection (LOD) for E. coli and enterococci (corresponding to the detection of 1 bacterium per final volume analyzed) are 38 MPN/100 ml of water. For infectious FRNAPH, the LOD is 5 PFU/100 ml in the case of direct analysis and 0.5 PFU/100 ml after concentration by ultracentrifugation, if a recovery rate of 100% is considered. Nevertheless, estimation based on the concentration of 28 samples of river water gave a mean recovery rate of only 34.0% (range, 4.7% to 80.0%) for infectious FRNAPH detection. In quantitative terms, the mean concentration detected was 13 PFU/100 ml by direct analysis and 4 PFU/100 ml after ultracentrifugation, and only 3 water samples were positive after concentration, while no infectious FRNAPH were detected in the direct analysis. Conversely, 2 samples were positive by direct detection and negative after concentration. In the present study, the concentration method was of little benefit in the detection of infectious FRNAPH, and therefore, only the results obtained in the direct analysis were considered in a comparison of FRNAPH levels with those of E. coli or enterococci.
Methodological considerations for the detection of genomes in water samples.
Concerning the detection of genomes, the theoretical LOD for FRNAPH (corresponding to 1 gc in a PCR well) is 5.3 × 103 gc/100 ml of water, due to the small sampling volume involved when a direct analysis is performed. Thus, no direct detection can be considered, even if the genome concentration is usually 1- or 2-log10 unit higher than that of infectious particles. Thereby, even if the use of concentration methods is a critical step leading to the loss of viral particles or genomes, they appear to be essential for the analysis of low-concentration samples. After ultracentrifugation, the theoretical LOD values are 26.7 and 10 gc/100 ml of water for FRNAPH and HuNoVs, respectively. Given the structural similarity between FRNAPH and HuNoVs, the same recovery rate may be considered after ultracentrifugation for both virus types, as well as for their various subgroups, with centrifugation parameters being set to pellet FRNAPH that have the lowest sedimentation coefficient. This approach was preferred to the use of adhesion-elution techniques or ultrafiltration through positively charged membranes, which are more dependent on viral and particle surface properties (58). The approach proposed here allows thus a strict comparison between these various parameters in order to evaluate the efficiency of FRNAPH as viral indicators and as a tool to discriminate the source of fecal pollution.
For the detection of viral genomes, nucleic acid extraction yields and the presence of RT-PCR inhibitors are other important parameters to be taken into account. The use of FCV as an internal standard allows the overall efficiency of the method used to be assessed (including ultracentrifugation, RNA extraction, and detection of RT-PCR inhibitors). In water samples, yields were heterogeneous: the average was about 9.5%, but they ranged from 0.01% to 49.6%. Most results were not acceptable according to ISO/TS standard 15216-1 (16), which imposes extraction yields of >1%. For the 10-fold-diluted samples, better yields were observed (ranging from 0.4% to 105%), indicating the presence of RT-PCR inhibitors. No correlation between inhibition and water physicochemical parameters was observed (data not shown). The use of 10-fold-diluted samples is challenging when working with environmental samples, due to genome concentrations close to the LOD, leading to possible overestimation by multiplying the low quantity detected by the dilution factor. For all these reasons, the results for FRNAPH and HuNoV genomes in waters were interpreted from a qualitative standpoint only.
Methodological considerations for the analysis of shellfish samples.
With respect to the shellfish analysis, the LOD was 67 MPN of E. coli/100 g of FIL and 1 PFU/g of HP for infectious FRNAPH. It was 107 gc/g of HP for FRNAPH genomes and 40 gc/g of HP for HuNoV genomes. The extraction yields observed by using FCV were about 14.0%, ranging from 2.0% to 32.6%. All yields were therefore acceptable according to ISO/TS standard 15216-1 (16). Very little variation was observed in the estimated yields between undiluted and 10-fold-diluted samples (range, 2.0% to 52.5%). Inhibition seems therefore to have a minor effect on the quantitative results. Accordingly, infectious bacteria, infectious FRNAPH, as well as viral genome concentrations of FRNAPH and HuNoVs may all be quantitatively compared in shellfish.
Infectious FRNAPH and fecal indicator bacteria in water samples.
The relevance of FRNAPH as fecal and viral indicators, as well as in tracking the source of fecal pollution, was evaluated in waters collected from 4 sampling sites located near the local shellfish-harvesting area.
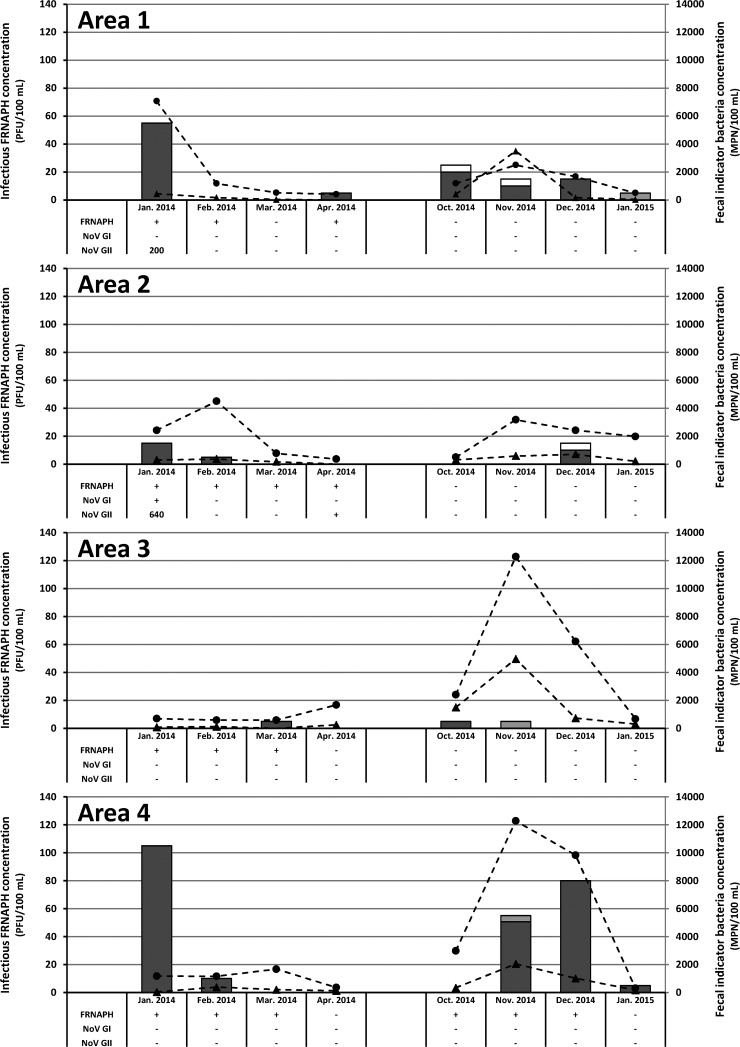
Fecal indicator bacteria (E. coli or enterococci) were detected in all water samples throughout the year (Fig. 2). Infectious FRNAPH were present in only 53% of the samples, with a mean concentration 100-fold lower than that of E. coli. Genotyping of phage plaques showed that infectious FRNAPH were mainly FRNAPH-I (93%). In other samples, infectious FRNAPH did not allow the fecal contamination highlighted by the presence of E. coli to be detected. Overall, all areas were affected by fecal pollution, and the concentrations of indicators (bacteria or FRNAPH) were higher in January, November, and December 2014 than in March, April, and October 2014 and January 2015. Except in rare cases, infectious FRNAPH and E. coli concentrations demonstrated similar trends. By associating the two parameters, 3 peaks of fecal pollution were distinguishable: (i) in area 1, concentrations detected in January 2014 were 7,060 MPN/100 ml and 55 PFU/100 ml for E. coli and FRNAPH, respectively; (ii) in area 4, concentrations of 1,170 MPN/100 ml and 105 PFU/100 ml were detected during the same period; and (iii) in area 4, concentrations were higher than 9,830 MPN/100 ml and 55 to 80 PFU/100 ml in November to December 2014.
FIG 2.
Infectious FRNAPH-I (dark gray bar), FRNAPH-II (light gray bar), and undetermined phage (white) concentrations estimated by relating phage plaque genotyping results to the total FRNAPH, as well as E. coli (black circles) and enterococcal (black triangles) concentrations detected in water samples collected from the 4 study areas. For each sample, FRNAPH, NoV GI, and NoV GII genome detection results are also given. Only qualitative information is given for FRNAPH genomes (presence [+] or absence [−]). HuNoV genome concentrations are expressed in genome copies per 100 ml of water; −, HuNoV genomes undetected; +, HuNoV genome concentrations above the detection limit but unquantifiable (<100 gc/100 ml).
FRNAPH and HuNoV genomes in water samples.
FRNAPH genomes were detected in 50% of the samples (Fig. 2), which confirmed on the whole the results obtained with infectious FRNAPH. The higher concentrations of infectious FRNAPH (i.e., >40 PFU/100 ml) were always associated with the presence of genomes (n = 4), as opposed to the lower ones (n = 13), for which genomes were sometimes not detected, presumably due to the low concentration or extraction recovery rates and the presence of RT-PCR inhibitors. Conversely, in 22% of the samples (n = 7), genomes were detected in the absence of infectious FRNAPH. In 25% of the samples, neither genomes nor infectious FRNAPH were detected.
NoV GI and GII were targeted here as specific human viral pollution markers. Three water samples from area 1 (January 2014) and area 2 (January and April 2014) were positive for the detection of both HuNoV and FRNAPH genomes. NoV GII genome concentrations were above the LOQ in samples collected in January 2014 (200 and 640 gc/100 ml in areas 1 and 2, respectively). In these 2 samples, infectious FRNAPH were also detected. Interestingly, during the sampling period of January 2014, the incidence of gastroenteritis in the population was above the epidemic threshold for the region.
Infectious FRNAPH and E. coli in shellfish samples.
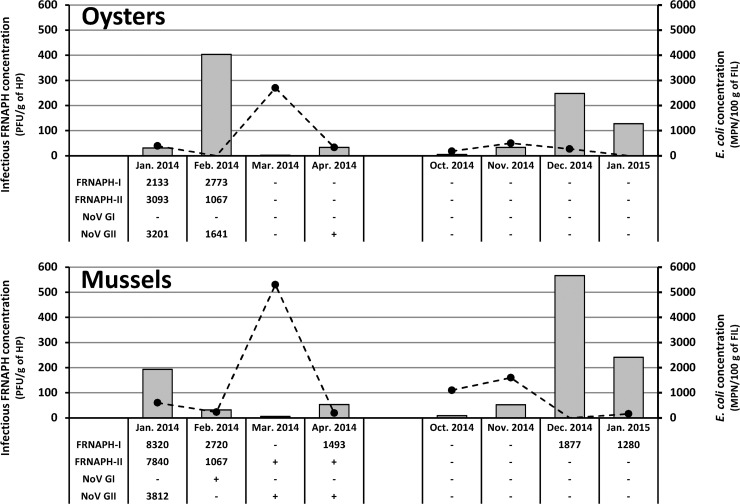
E. coli was targeted in FIL of oysters (8 batches) and mussels (8 batches) collected from the growing area affected by contaminated waters coming from areas 1 to 4 (Fig. 1). As in waters, E. coli was detected throughout the year (Fig. 3) in 81% of the samples (n = 13). For the 2 sample types, the batches collected in March 2014 presented the highest concentrations (2,700 and 5,300 MPN of E. coli/100 g of FIL for oysters and mussels, respectively). According to EC regulation 854/2004 (5), the harvesting area is classified as B. Our results confirm such classification when taking into account E. coli concentration levels detected in oysters but not for mussels, for which classification would be C (>4,600 MPN of E. coli/100 g of FIL detected in March 2014).
FIG 3.
Infectious FRNAPH (gray bars) and E. coli (black circles) concentrations detected in shellfish collected from the local harvesting area. For each sample, FRNAPH-I, FRNAPH-II, NoV GI, and NoV GII genome detection results are also given, expressed in genome copies per gram of hepatopancreas (HP). −, genomes undetected; +, concentration above the detection limit but unquantifiable (<1,067 and <400 gc/g of HP for FRNAPH and HuNoVs, respectively).
Concerning infectious FRNAPH, they were detected in all shellfish samples. Concentrations ranged from 2 to 566 PFU/g of HP (average concentration, 144 PFU/g), with FRNAPH-I being in the majority (99% of the 12 phage plaques analyzed per sample). Higher concentrations were observed in samples collected in January to February 2014 and December 2014 to January 2015 (average, 230 PFU/g of HP). Considering that FRNAPH are mainly found in shellfish HP and that the HP fraction corresponds approximately to 5% of the total FIL mass, the mean ratio between E. coli and infectious FRNAPH in total FIL is 3.6 (geometric mean).
According to these indicators, shellfish were constantly affected by fecal pollution in this area, and the large amounts of FRNAPH in shellfish compared to concentrations detected in nearby environmental waters indicated that shellfish are able to accumulate FRNAPH in digestive tissues.
FRNAPH and HuNoV genomes in shellfish samples.
Concerning genomes, only FRNAPH-I and -II were detected in shellfish. An FRNAPH-II genome was detected in 75% of the samples (Fig. 3; n = 6/8) during the first study period (January 2014 to April 2014), but it was not detected during the second study period (October 2014 to January 2015).
HuNoV genomes were targeted in shellfish HP, and they were detected in 44% of the samples collected from the studied area (3/8 oysters and 4/8 mussels). Six batches were positive for NoV GII and only 1 for NoV GI (mussels from February 2014). Quantifiable NoV GII genome concentrations (>400 gc/g of HP) were detected in only 3 positive samples (oysters from January and February 2014 and mussels from January 2014).
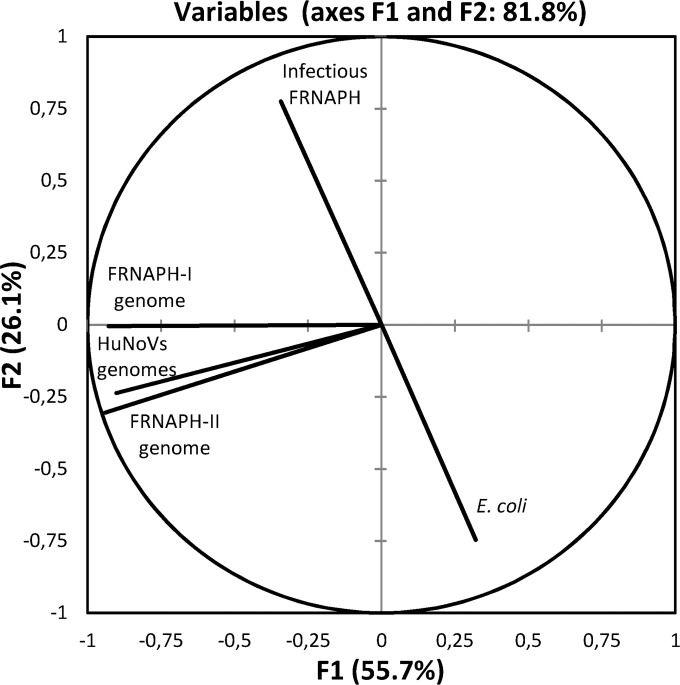
Principal-component analysis (PCA) was performed to describe associations between the different parameters targeted in shellfish collected from the local harvesting area. A representation along axes F1 and F2 allows a description of 81.8% of the total information (Fig. 4). Several of the parameters considered in this study are close to the correlation circle, allowing the interpretations of no relationship being observed between (i) E. coli and HuNoV genome levels, (ii) infectious FRNAPH and HuNoV genome concentrations, and (iii) infectious FRNAPH and their genomes; however, (iv) a relationship is underlined between HuNoV and FRNAPH-II genomes.
FIG 4.
Principal-component analysis of parameters targeted in shellfish collected from the local harvesting area.
These findings have led to the further investigation of the association between FRNAPH and HuNoV genomes in shellfish collected from other European harvesting areas. Among the 28 batches collected, 43% were positive for the presence of NoV GII (n = 12), while NoV GI was never detected. Concerning the detection of FRNAPH genomes, 11 batches were positive, and it was FRNAPH-II in each case. In line with ISO/TS standard 15216-1 (16) for extraction and genome detection, HuNoV genome concentrations ranged between the LOD and LOQ values for 92% of the positive samples, and FRNAPH-II genome concentrations were also above the LOQ, while extraction yields were in agreement with the ISO standard. Therefore, the association between the two parameters was investigated in qualitative terms (Table 1). Considering the 44 specimens monitored in this study, a highly significant association is observed between HuNoV and FRNAPH-II genomes (Pearson's χ2 test with Yates' continuity correction, P < 0.01).
TABLE 1.
Two-by-two contingency table for the detection of FRNAPH-II and HuNoV genomes in shellfish batches
| FRNAPH-II genome detection | HuNoV genome detection |
|
|---|---|---|
| No. of positive samples | No. of negative samples | |
| No. of positive samples | 14 | 3 |
| No. of negative samples | 5 | 22 |
DISCUSSION
Fecal indicator bacteria are commonly investigated in waters or shellfish, and their detection has been standardized and managed for a long time (51, 56, 57). Techniques used for the detection of HuNoV and FRNAPH genomes and for those of infectious FRNAPH have also been standardized or published for several years (16, 47, 48, 54). However, concentration methods, which are usually essential for the detection of viruses in environmental samples, are far from easy, and the use of an internal standard is required to estimate the extraction yield of the methods and the inhibition rates. In waters collected from the studied area, infectious FRNAPH levels were sufficient to be detected by direct analysis. As for genomes (FRNAPH or HuNoV), a concentration step was needed, and they were detected after ultracentrifugation. The average concentration yield is low (9.5%), especially in freshwater (2%), and several rates are below the 1% threshold set by ISO/TS standard 15216-1 (16), thereby allowing a qualitative interpretation only. For shellfish, extraction yields are >10%, allowing a quantitative analysis.
The first step to ranking viral hazards is to evaluate the extent of fecal pollution using appropriate indicators. Indeed, high levels of fecal pollution are often associated with the presence of pathogenic viruses, in the case of circulating strains in the population (epidemic period). It is also suggested that high levels of fecal pollution may be considered in the absence of pathogenic viruses, without compromising the potential of the indicators used. Conversely, these indicators must always be present when pathogens are detected. Because indicator bacteria (E. coli and enterococci) are commonly used (59, 60), the potential of FRNAPH (genomes and infectious phages) to evaluate the level of fecal pollution must be compared to them.
In this study, fecal pollution was observed in waters no matter the indicator used, and E. coli levels led to classify the studied site as being of poor quality, according to EC directive 2006/7/EC (61). E. coli was detected in all samples, while FRNAPH were absent from almost half of them (15/32). Nevertheless, some relationships appeared between the levels of E. coli, enterococci, and infectious FRNAPH. Area 4 seemed to be the most affected by fecal pollution, according to the levels of E. coli (3,720 MPN/100 ml) and FRNAPH (32 PFU/100 ml). According to the levels of enterococci, high fecal pollution was observed in areas 1 and 3. From these data, infectious FRNAPH did not provide novel information about the fecal pollution of waters of the studied site: as with E. coli, concentration peaks were observed in winter (January 2014 for areas 1 and 2, November and December 2014 for area 4), and although correlated on the whole with E. coli, their concentrations were about 100 times lower, thus explaining their nondetection in several samples. A ratio of about 10 to 100 is classically observed between these 2 indicators in raw wastewater or in environmental waters (46, 62, 63), restricting their potential as fecal pollution indicators. Given that E. coli is less resistant than FRNAPH in the environment, high concentrations of this indicator may be indicative of a recent fecal pollution event. Finally, no benefit was observed either regarding FRNAPH genome detection.
Concerning shellfish collected from the studied area, massive fecal pollution was observed according to E. coli and infectious FRNAPH levels (detected in 81% and 100% of the samples, respectively). However, in 3 samples, fecal pollution was revealed by the presence of infectious FRNAPH, but no E. coli was detected. More than 99% of infectious FRNAPH isolated were FRNAPH-I. Genotyping was, however, performed on a maximum of 12 phage plaques per specimen, representing sometimes less than 2% of the listed phage plaques. Interestingly, and contrary to the observations made in waters, infectious FRNAPH concentrations were higher than the concentration of E. coli in shellfish. The fact that HP was used for the detection of FRNAPH while E. coli was targeted in whole shellfish (i.e., FIL) was not sufficient to justify such a change compared to the concentrations detected in water (the levels of E. coli were up to 100 times higher than those of infectious FRNAPH in water samples, while a geometric mean of only 3.6 was calculated in shellfish). These results indicated better survival and/or better accumulation of FRNAPH in shellfish than those of bacteria, as previously suggested (11, 12, 52). This observation is an important argument for the benefit of using FRNAPH to estimate fecal contamination in shellfish compared to E. coli. Besides that, no relationships were found between the concentrations of these 2 indicators. High concentrations were observed during the winter months for infectious FRNAPH (January to February 2014 and December 2014 to January 2015), while E. coli concentration peaks were observed in spring and fall (March and October to November 2014). Other studies also reported better accumulation of FRNAPH in shellfish in winter and no seasonal variation for E. coli (40, 64).
To sum up the findings from the present study on the use of FRNAPH as fecal pollution indicators conducted in a specific study area, no benefits were observed in water compared to E. coli: infectious FRNAPH provided no relevant input compared to the usual indicator, and low concentrations and poor yields prevented any conclusions with regard to FRNAPH genomes. However, FRNAPH provided novel information in shellfish compared to E. coli.
In this study, FRNAPH were also evaluated as indicators of viral pollution and as microbial source tracking tools. In this context, we compared the presence of FRNAPH and HuNoV genomes in water and shellfish samples, taking into account the aforementioned methodological limitations. Surrogates must behave as pathogenic viruses, and FRNAPH genome detection appears interesting due to their persistence being greater than infectivity (20–23, 65), allowing the source of fecal pollution to be tracked for a longer period of time, despite the presence of inactivating factors in the environment. Unfortunately, low genome levels in water require a concentration step, and poor yields and inhibition cause them to be around their detection limits. For this reason, a qualitative analysis appeared more appropriate here. Microbial source tracking was undertaken based on the approach described by Hartard et al. (21). With the RT-PCR method that was used, FRNAPH-II and -III seemed to be specific indicators of human pollution, while FRNAPH-I was associated with both animal and urban pollution. FRNAPH genomes were occasionally detected in water collected from the 4 areas. The genomes detected in almost half of the samples collected during the first winter (7/16) were those of FRNAPH-II, thus suggesting that the pollution could be of urban origin. Such an assumption was strengthened by the detection of HuNoV genome levels above the LOQ in areas 1 and 2 during the first winter, a time that is also characterized by the gastroenteritis epidemic threshold being surpassed for the studied region. During the second winter, neither of these two viruses was detected, and the gastroenteritis epidemic threshold was not crossed.
The association between FRNAPH-II and HuNoV genomes observed in water was even more marked in shellfish, whether in specimens from the studied area (n = 16) or in others collected in several European harvesting areas (n = 28). In the studied area, HuNoV genomes were qualitatively and quantitatively more present in shellfish than in waters during the first winter, confirming a storage capacity in shellfish, as previously described (29, 66, 67). No HuNoV or FRNAPH-II genomes were detected during the following winter, suggesting, as in water, a limited impact of human pollution, while the seasonal peak of infectious FRNAPH-I was also observed. Concerning shellfish collected from other European harvesting areas, which were probably less impacted by fecal pollution, only a qualitative analysis was conducted for the presence of FRNAPH and HuNoV genomes, due to concentrations close to the LOQ. Finally, concerning E. coli detection in shellfish, the absence of a peak during the winter months, especially when HuNoVs were detected, confirmed that this conventional indicator is not suitable to monitor viral pollution in shellfish. The contribution of FRNAPH detection in shellfish and the lack of relevance regarding the monitoring of E. coli had already been observed by Flannery et al. (34). By grouping shellfish specimens according to their infectious FRNAPH content, a correlation was observed with the level of NoV GII genome, unrelated to that of E. coli. In another study, Flannery et al. (41) reported comparable kinetics of accumulation between infectious FRNAPH-II and the genomes of probably infectious NoV GII in oysters following combined sewer overflow (CSO) discharge events. Finally, FRNAPH-II, generally present in low concentrations in treated water effluents (21, 68), has been found to be the most appropriate indicator of viral persistence during water treatment processes (69) and may thus be proposed as a specific indicator of urban pollution in the environment (21).
To conclude, assessing viral hazard risks using molecular tools raises the question of whether the genomes detected in water and/or food commodities correspond to infectious viruses able to infect humans. In this study, infectious FRNAPH were detected in shellfish samples in the absence of any FRNAPH genomes, which clearly illustrated technical limitations (i.e., high limit of detection, low extraction yields, and RT-PCR inhibitors), even when using standardized methods. These limitations are well known, and in this respect, infectious HuNoVs may easily be present in a specimen even when no genome is detected. The reverse situation is also possible, in view of genome persistence being greater than infectivity. For these 2 reasons, the detection of HuNoV genomes provides limited relevance for assessing the associated infectious risk. The findings of this study show a relationship between the presence of HuNoV and FRNAPH-II genomes, especially in shellfish. Taking into account this observation, further experiments are now needed to investigate the relationship between the presence of infectious FRNAPH and that of infectious HuNoVs in shellfish, and to conclude whether the specific detection of human infectious FRNAPH-II could be used as an indicator of infectious HuNoV contamination in shellfish. The relative scarcity of FRNAPH-II compared to nonhuman-specific FRNAPH-I requires, however, the development of more-specific detection methods. Finally, with HuNoVs being noncultivable, the analysis of shellfish involved in outbreaks of HuNoV gastroenteritis will be the only guarantee of the presence of infectious particles.
ACKNOWLEDGMENTS
This study was funded by the Seine-Normandy Water Agency and conducted within the framework of the Joint Technological Unit VIROcontrol.
We thank RiskManche (funded by the European Regional Development Fund Interreg IVA Programme) for their supply of samples, as well as the Zone Atelier du Bassin de la Moselle (ZAbM) and the Institut Carnot Energie et Environnement en Lorraine (ICEEL) for their financial and technical support.
Funding Statement
This work was funded by the Seine Normandy Water Agency and by Institut Carnot Energie et Environnement en Lorraine.
REFERENCES
- 1.Bellou M, Kokkinos P, Vantarakis A. 2013. Shellfish-borne viral outbreaks: a systematic review. Food Environ Virol 5:13–23. doi: 10.1007/s12560-012-9097-6. [DOI] [PubMed] [Google Scholar]
- 2.Rajko-Nenow P, Waters A, Keaveney S, Flannery J, Tuite G, Coughlan S, O'Flaherty V, Doré W. 2013. Norovirus genotypes present in oysters and in effluent from a wastewater treatment plant during the seasonal peak of infections in Ireland in 2010. Appl Environ Microbiol 79:2578–2587. doi: 10.1128/AEM.03557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doré B, Keaveney S, Flannery J, Rajko-Nenow P. 2010. Management of health risks associated with oysters harvested from a norovirus contaminated area, Ireland, February–March 2010. Euro Surveill 15:pii=19567. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19567. [PubMed] [Google Scholar]
- 4.Koopmans M, Duizer E. 2004. Foodborne viruses: an emerging problem. Int J Food Microbiol 90:23–41. doi: 10.1016/S0168-1605(03)00169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Parliament. 2004. Regulation (EC) no. 854/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption. Off J Eur Union L226:83–127. [Google Scholar]
- 6.Le Guyader FS, Bon F, DeMedici D, Parnaudeau S, Bertone A, Crudeli S, Doyle A, Zidane M, Suffredini E, Kohli E, Maddalo F, Monini M, Gallay A, Pommepuy M, Pothier P, Ruggeri FM. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J Clin Microbiol 44:3878–3882. doi: 10.1128/JCM.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalmers JWT, McMillan JH. 1995. An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiol Infect 115:163–167. doi: 10.1017/S0950268800058222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havelaar AH, van Olphen M, Drost YC. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl Environ Microbiol 59:2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doré WJ, Lees DN. 1995. Behavior of Escherichia coli and male-specific bacteriophage in environmentally contaminated bivalve molluscs before and after depuration. Appl Environ Microbiol 61:2830–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formiga-Cruz M, Tofino-Quesada G, Bofill-Mas S, Lees DN, Henshilwood K, Allard AK, Conden-Hansson AC, Hernroth BE, Vantarakis A, Tsibouxi A, Papapetropoulou M, Furones MD, Girones R. 2002. Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden, and the United Kingdom. Appl Environ Microbiol 68:5990–5998. doi: 10.1128/AEM.68.12.5990-5998.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lees D. 2000. Viruses and bivalve shellfish. Int J Food Microbiol 59:81–116. doi: 10.1016/S0168-1605(00)00248-8. [DOI] [PubMed] [Google Scholar]
- 13.Le Guyader FS, Loisy F, Atmar RL, Hutson AM, Estes MK, Ruvoën-Clouet N, Pommepuy M, Le Pendu J. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg Infect Dis 12:931–936. doi: 10.3201/eid1206.051519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian P, Bates AH, Jensen HM, Mandrell RE. 2006. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett Appl Microbiol 43:645–651. doi: 10.1111/j.1472-765X.2006.02010.x. [DOI] [PubMed] [Google Scholar]
- 15.Tian P, Engelbrektson AL, Jiang X, Zhong W, Mandrell RE. 2007. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J Food Prot 70:2140–2147. [DOI] [PubMed] [Google Scholar]
- 16.International Organization for Standardization. 2013. ISO/TS 15216-1. Microbiology of food and animal feed—horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR—part 1: method for quantification. International Organization for Standardization, Geneva, Switzerland: http://www.iso.org/iso/catalogue_detail.htm?csnumber=55382. [Google Scholar]
- 17.European Food Safety Authority. 2012. Scientific opinion on norovirus (NoV) in oysters: methods, limits and control options. EFSA J 10:2500–2539. doi: 10.2903/j.efsa.2012.2500. [DOI] [Google Scholar]
- 18.Lowther JA, Gustar NE, Hartnell RE, Lees DN. 2012. Comparison of norovirus RNA levels in outbreak-related oysters with background environmental levels. J Food Prot 75:389–393. doi: 10.4315/0362-028X.JFP-11-360. [DOI] [PubMed] [Google Scholar]
- 19.Lowther JA, Avant JM, Gizynski K, Rangdale RE, Lees DN. 2010. Comparison between quantitative real-time reverse transcription PCR results for norovirus in oysters and self-reported gastroenteric illness in restaurant customers. J Food Prot 73:305–311. [DOI] [PubMed] [Google Scholar]
- 20.Ogorzaly L, Bertrand I, Paris M, Maul A, Gantzer C. 2010. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl Environ Microbiol 76:8019–8025. doi: 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartard C, Rivet R, Banas S, Gantzer C. 2015. Occurrence of and sequence variation among F-specific RNA bacteriophage subgroups in feces and wastewater of urban and animal origins. Appl Environ Microbiol 81:6505–6515. doi: 10.1128/AEM.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gassilloud B, Schwartzbrod L, Gantzer C. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl Environ Microbiol 69:3965–3969. doi: 10.1128/AEM.69.7.3965-3969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi S, Jiang SC. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl Environ Microbiol 71:7426–7433. doi: 10.1128/AEM.71.11.7426-7433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dancer D, Rangdale RE, Lowther JA, Lees DN. 2010. Human norovirus RNA persists in seawater under simulated winter conditions but does not bioaccumulate efficiently in Pacific oysters (Crassostrea gigas). J Food Prot 73:2123–2127. [DOI] [PubMed] [Google Scholar]
- 25.Wigginton KR, Pecson BM, Sigstam T, Bosshard F, Kohn T. 2012. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ Sci Technol 46:12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 26.Simonet J, Gantzer C. 2006. Inactivation of poliovirus 1 and F-specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl Environ Microbiol 72:7671–7677. doi: 10.1128/AEM.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos CJA, Lees DN. 2014. Environmental transmission of human noroviruses in shellfish waters. Appl Environ Microbiol 80:3552–3561. doi: 10.1128/AEM.04188-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pecson BM, Ackermann M, Kohn T. 2011. Framework for using quantitative PCR as a nonculture based method to estimate virus infectivity. Environ Sci Technol 45:2257–2263. doi: 10.1021/es103488e. [DOI] [PubMed] [Google Scholar]
- 29.Lowther JA, Gustar NE, Powell AL, Hartnell RE, Lees DN. 2012. Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl Environ Microbiol 78:5812–5817. doi: 10.1128/AEM.01046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polo D, Varela MF, Romalde JL. 2015. Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas. Int J Food Microbiol 193:43–50. doi: 10.1016/j.ijfoodmicro.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Suffredini E, Lanni L, Arcangeli G, Pepe T, Mazzette R, Ciccaglioni G, Croci L. 2014. Qualitative and quantitative assessment of viral contamination in bivalve molluscs harvested in Italy. Int J Food Microbiol 184:21–26. doi: 10.1016/j.ijfoodmicro.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 32.Cheng PKC, Wong DKK, Chung TWH, Lim WWL. 2005. Norovirus contamination found in oysters worldwide. J Med Virol 76:593–597. doi: 10.1002/jmv.20402. [DOI] [PubMed] [Google Scholar]
- 33.Boxman ILA, Tilburg JJHC, Te Loeke NAJM, Vennema H, Jonker K, de Boer E, Koopmans M. 2006. Detection of noroviruses in shellfish in the Netherlands. Int J Food Microbiol 108:391–396. [DOI] [PubMed] [Google Scholar]
- 34.Flannery J, Keaveney S, Doré W. 2009. Use of FRNA bacteriophages to indicate the risk of norovirus contamination in Irish oyster. J Food Prot 72:2358–2362. [DOI] [PubMed] [Google Scholar]
- 35.Lowther JA, Henshilwood K, Lees DN. 2008. Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semiquantitative real-time reverse transcription PCR. J Food Prot 71:1427–1433. [DOI] [PubMed] [Google Scholar]
- 36.Nishida T, Nishio O, Kato M, Chuma T, Kato H, Iwata H, Kimura H. 2007. Genotyping and quantitation of noroviruses in oysters from two distinct sea areas in Japan. Microbiol Immunol 51:177–184. doi: 10.1111/j.1348-0421.2007.tb03899.x. [DOI] [PubMed] [Google Scholar]
- 37.Costantini V, Loisy F, Joens L, Le Guyader FS, Saif LJ. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl Environ Microbiol 72:1800–1809. doi: 10.1128/AEM.72.3.1800-1809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mieszkin S, Caprais MP, Le Mennec C, Le Goff M, Edge TA, Gourmelon M. 2013. Identification of the origin of faecal contamination in estuarine oysters using Bacteroidales and F-specific RNA bacteriophage markers. J Appl Microbiol 115:897–907. doi: 10.1111/jam.12260. [DOI] [PubMed] [Google Scholar]
- 39.Doré WJ, Henshilwood K, Lees DN. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl Environ Microbiol 66:1280–1285. doi: 10.1128/AEM.66.4.1280-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doré WJ, Mackie M, Lees DN. 2003. Levels of male-specific RNA bacteriophage and Escherichia coli in molluscan bivalve shellfish from commercial harvesting areas. Lett Appl Microbiol 36:92–96. doi: 10.1046/j.1472-765X.2003.01268.x. [DOI] [PubMed] [Google Scholar]
- 41.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2013. Norovirus and FRNA bacteriophage determined by RT-qPCR and infectious FRNA bacteriophage in wastewater and oysters. Water Res 47:5222–5231. doi: 10.1016/j.watres.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2012. Concentration of norovirus during wastewater treatment and its impact on oyster contamination. Appl Environ Microbiol 78:3400–3406. doi: 10.1128/AEM.07569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole D, Long SC, Sobsey MD. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol 69:6507–6514. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaper M, Jofre J, Uys M, Grabow WOK. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J Appl Microbiol 92:657–667. doi: 10.1046/j.1365-2672.2002.01600.x. [DOI] [PubMed] [Google Scholar]
- 45.Harwood VJ, Boehm AB, Sassoubre LM, Vijayavel K, Stewart JR, Fong TT, Caprais MP, Converse RR, Diston D, Ebdon J, Fuhrman JA, Gourmelon M, Gentry-Shields J, Griffith JF, Kashian DR, Noble RT, Taylor H, Wicki M. 2013. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res 47:6929–6943. doi: 10.1016/j.watres.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 46.Sundram A, Donnelly L, Ehlers MM, Vrey A, Grabow WOK, Bailey IW. 2002. Evaluation of F-RNA coliphages as indicators of viruses and the source of faecal pollution, p 86–91. In WISA 2002 Biennial Conf Exhib Proc, Durban, South Africa, 19 to 23 May 2002. [Google Scholar]
- 47.Ogorzaly L, Gantzer C. 2006. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J Virol Methods 138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Wolf S, Hewitt J, Greening GE. 2010. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl Environ Microbiol 76:1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinton LW, Hall CH, Lynch PA. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline water. Appl Environ Microbiol 68:1122–1131. doi: 10.1128/AEM.68.3.1122-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noble RT, Lee IM, Schiff KC. 2004. Inactivation of indicator micro-organisms from various sources of faecal contamination in seawater and freshwater. J Appl Microbiol 96:464–472. doi: 10.1111/j.1365-2672.2004.02155.x. [DOI] [PubMed] [Google Scholar]
- 51.Formiga-Cruz M, Allard AK, Conden-Hansson AC, Henshilwood K, Hernroth BE, Jofre J, Lees DN, Lucena F, Papapetropoulou M, Rangdale RE, Tsibouxi A, Vantarakis A, Girones R. 2003. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl Environ Microbiol 69:1556–1563. doi: 10.1128/AEM.69.3.1556-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doré WJ, Henshilwood K, Lees DN. 1998. The development of management strategies for control of virological quality in oysters. Water Sci Technol 38:29–35. doi: 10.1016/S0273-1223(98)00796-3. [DOI] [Google Scholar]
- 53.International Organization for Standardization. 2003. ISO 6887-3. Microbiology of food and animal feeding stuffs—preparation of test samples, initial suspension and decimal dilutions for microbiological examination—part 3: specific rules for the preparation of fish and fishery products. International Organization for Standardization, Geneva, Switzerland: http://www.iso.org/iso/catalogue_detail.htm?csnumber=31589. [Google Scholar]
- 54.International Organization for Standardization. 2001. ISO 10705-1. Water quality—detection and enumeration of bacteriophages—part 1: enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland: http://www.iso.org/iso/catalogue_detail.htm?csnumber=18794. [Google Scholar]
- 55.Havelaar AH, Hogeboom WM. 1984. A method for the enumeration of male-specific bacteriophages in sewage. J Appl Bacteriol 56:439–447. doi: 10.1111/j.1365-2672.1984.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 56.International Organization for Standardization. 1998. ISO 9308-3. Water quality—detection and enumeration of Escherichia coli and coliform bacteria in surface and waste water—part 3: miniaturized method (most probable number) by inoculation in liquid medium. International Organization for Standardization, Geneva, Switzerland: https://www.iso.org/obp/ui/#iso:std:iso:9308:-3:ed-1:v1:en. [Google Scholar]
- 57.AFNOR. 2010. NF V08-106. Enumeration Escherichia coli in live shellfish—indirect technique using direct impedance measurement. Association Française de Normalisation, La Plaine Saint-Denis, France: http://standards.globalspec.com/std/1282477/afnor-nf-v08-106. [Google Scholar]
- 58.Dika C, Ly-Chatain MH, Francius G, Duval JFL, Gantzer C. 2013. Non-DLVO adhesion of F-specific RNA bacteriophages to abiotic surfaces: importance of surface roughness, hydrophobic and electrostatic interactions. Colloids Surf A 435:178–187. doi: 10.1016/j.colsurfa.2013.02.045. [DOI] [Google Scholar]
- 59.World Health Organization, Organisation for Economic Co-operation and Development. 2003. Assessing microbial safety of drinking water: improving approaches and methods. OECD Publishing, London, United Kingdom: http://www.who.int/water_sanitation_health/dwq/9241546301full.pdf. [Google Scholar]
- 60.Tallon P, Magajna B, Lofranco C, Leung KT. 2005. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut 166:139–166. doi: 10.1007/s11270-005-7905-4. [DOI] [Google Scholar]
- 61.European Parliament. 2006. Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC. Off J Eur Union L 64:37–51. [Google Scholar]
- 62.Lucena F, Méndez X, Morón A, Calderón E, Campos C, Guerrero A, Cárdenas M, Gantzer C, Shwartzbrood L, Skraber S, Jofre J. 2003. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J Appl Microbiol 94:808–815. doi: 10.1046/j.1365-2672.2003.01812.x. [DOI] [PubMed] [Google Scholar]
- 63.Contreras-Coll N, Lucena F, Mooijman K, Havelaar A, Pierzo V, Boque M, Gawler A, Höller C, Lambiri M, Mirolo G, Moreno B, Niemi M, Sommer R, Valentin B, Wiedenmann A, Young V, Jofre J. 2002. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Res 36:4963–4974. doi: 10.1016/S0043-1354(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 64.Myrmel M, Berg EMM, Rimstad E, Grinde B. 2004. Detection of enteric viruses in shellfish from the Norwegian coast. Appl Environ Microbiol 70:2678–2684. doi: 10.1128/AEM.70.5.2678-2684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maalouf H, Zakhour M, Le Pendu J, Le Saux JC, Atmar RL, Le Guyader FS. 2010. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl Environ Microbiol 76:5621–5630. doi: 10.1128/AEM.00148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richards GP, McLeod C, Le Guyader FS. 2010. Processing strategies to inactivate enteric viruses in shellfish. Food Environ Virol 2:183–193. doi: 10.1007/s12560-010-9045-2. [DOI] [Google Scholar]
- 68.Haramoto E, Fujino S, Otagiri M. 2015. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci Total Environ 520:32–38. doi: 10.1016/j.scitotenv.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 69.Boudaud N, Machinal C, David F, Fréval-Le Bourdonnec A, Jossent J, Bakanga F, Arnal C, Jaffrezic MP, Oberti S, Gantzer C. 2012. Removal of MS2, Qβ and GA bacteriophages during drinking water treatment at pilot scale. Water Res 46:2651–2664. doi: 10.1016/j.watres.2012.02.020. [DOI] [PubMed] [Google Scholar]