ABSTRACT
Crown gall disease of grapevine is caused by virulent Agrobacterium strains and establishes a suitable habitat for agrobacteria and, potentially, other bacteria. The microbial community associated with grapevine plants has not been investigated with respect to this disease, which frequently results in monetary losses. This study compares the endophytic microbiota of organs from grapevine plants with or without crown gall disease and the surrounding vineyard soil over the growing seasons of 1 year. Amplicon-based community profiling revealed that the dominating factor causing differences between the grapevine microbiota is the sample site, not the crown gall disease. The soil showed the highest microbial diversity, which decreased with the distance from the soil over the root and the graft union of the trunk to the cane. Only the graft union microbiota was significantly affected by crown gall disease. The bacterial community of graft unions without a crown gall hosted transient microbiota, with the three most abundant bacterial species changing from season to season. In contrast, graft unions with a crown gall had a higher species richness, which in every season was dominated by the same three bacteria (Pseudomonas sp., Enterobacteriaceae sp., and Agrobacterium vitis). For in vitro-cultivated grapevine plantlets, A. vitis infection alone was sufficient to cause crown gall disease. Our data show that microbiota in crown galls is more stable over time than microbiota in healthy graft unions and that the microbial community is not essential for crown gall disease outbreak.
IMPORTANCE The characterization of bacterial populations in animal and human diseases using high-throughput deep-sequencing technologies, such as 16S amplicon sequencing, will ideally result in the identification of disease-specific microbiota. We analyzed the microbiota of the crown gall disease of grapevine, which is caused by infection with the bacterial pathogen Agrobacterium vitis. All other Agrobacterium species were found to be avirulent, even though they lived together with A. vitis in the same crown gall tumor. As has been reported for human cancer, the crown gall tumor also hosted opportunistic bacteria that are adapted to the tumor microenvironment. Characterization of the microbiota in various diseases using amplicon sequencing may help in early diagnosis, to serve as a preventative measure of disease in the future.
INTRODUCTION
Agrobacterium vitis infects domesticated as well as wild grapevines (1) and is the most common cause of crown gall disease in grapevine (2, 3). In addition to A. vitis, other virulent Agrobacterium species are known to induce grapevine crown gall development (4). A. vitis is known to persist in debris from infested grapevine material in soil (5) and can enter grapevines via the root and move through the xylem (6) to wounded parts of the plant, where it transforms the cells (7–9). The pathogen has been detected in the xylem sap of canes, so propagation material of grapevine nurseries serves as an additional risk for distributing A. vitis (10, 11). Virulent A. vitis strains harbor a tumor-inducing (Ti) plasmid, which enables the transfer of transfer DNA (T-DNA) into the plant genome, a process supported by virulence (Vir) genes of the Ti plasmid (12, 13). The T-DNA-located genes express enzymes for opine and plant hormone production (3, 14, 15). Opines are a nutrient source for virulent A. vitis and for other bacterial species that express opine-metabolizing enzymes (16–20). The altered auxin and cytokinin levels at the transformation site induce uncontrolled cell division and, finally, crown gall development. Crown gall development gives rise to an altered tissue morphology and physiology (21).
In nature, both abiotic and biotic factors influence the A. vitis-mediated infection process and, consequently, crown gall disease outbreak. Crown gall disease on grapevine occurs preferentially in regions with cold winters, indicating that cold temperatures are beneficial for disease outbreak (3). In addition, treatments that cause wounding, such as farming devices and the grafting procedure, can also promote outbreak of the disease. Biotic factors that influence crown gall disease in grapevine are both pathogenic and nonpathogenic bacteria. Antagonistic bacteria, which are known as biocontrol agents (e.g., the A. vitis strain F2/5), prevent the transformation of grapevine cells by virulent A. vitis strains (22–24). Among the bacteria isolated from grapevine xylem sap, Pseudomonas sp., for example, showed inhibitory effects on crown gall growth (25).
In recent years, several research groups have studied the bacterial community and structure of grapevine-associated microbiota, focusing on different aspects of viticulture and the taste of the resulting wine. Techniques such as isolation of cultivable bacteria (26), in combination with analysis of fluorescently labeled terminal-restriction fragment analysis (27) and taxon- or genus-specific real-time PCR (28), have been used. The 16S rRNA gene amplicon high-throughput sequencing technique provides a detailed overview of the microbiota and has been employed to resolve bacterial communities to the species level (29). This technique has been used to describe the above- and belowground microbiota of grapevines (30–34). Grapevines from vineyards in Europe (Italy [30, 31] and Portugal [32]) and the United States (New York [33] and California [34]) were sampled to analyze differences in the microbiota resulting from pest management (fungicide versus biocontrol [30] and integrated versus organic [31]), vegetative cycle (32), climate (34), and edaphic factors (33). The leaf and grape microbiota correlate with the vegetative cycle of the plant and the temperature, respectively (32, 34). Moreover, the study on grapevines from Long Island (Suffolk County, NY, USA) observed that the vineyard soil serves as a source for grapevine- and grape must-associated microbiota (33). A better understanding of the microbiota-plant interaction would help improve applications that promote plant growth and protection against pathogens (35–37).
In the present study, we investigated the microbiota of grapevines with and without crown gall disease because the microbiota of diseased and nondiseased grapevines, to our knowledge, have not yet been studied. Employing high-throughput sequencing of 16S rRNA gene amplicons, we analyzed the microbiota of the soil, root, graft union of the trunk, and 1-year-old canes of grapevine plants with and without crown gall over the growing seasons of a year. We also established an infection assay using in vitro-cultivated grapevine plantlets. This assay allowed us to investigate the capability of environmental Agrobacterium isolates to induce crown gall growth and to analyze the role of the microbiota associated with crown gall disease.
MATERIALS AND METHODS
Sample collection.
Grapevine material was collected from four individual plants (Fig. 1A) growing in one row in a stretch of 22 m located at a vineyard at Himmelstadt, Franconia, Germany (49°55′234.78N, 9°49′05.22E). The grapevines of the variety Cabernet Dorsa had been grafted on the rootstock SO4 and planted in 2008 in loamy sand. Four different sampling sites of each grapevine plant were analyzed (Fig. 1B): (i) 1-year-old cane (c), (ii) graft union of the trunk (g), (iii) root (r), and (iv) soil (s) from the root environment. The samples were collected before noon on 30 October 2013 (autumn), 4 April 2014 (spring), and 23 July 2014 (summer). At each time point, the weather was sunny and the soil dry. Three replicates were harvested per sample site and season, resulting in a total of 144 samples (Fig. 1C). Roots were washed with tap water, and the periderm of all plant material was discarded. Half of the wooden grapevine material and the soil was stored at −80°C for DNA extraction and the other half at 4°C for the isolation of bacteria.
FIG 1.
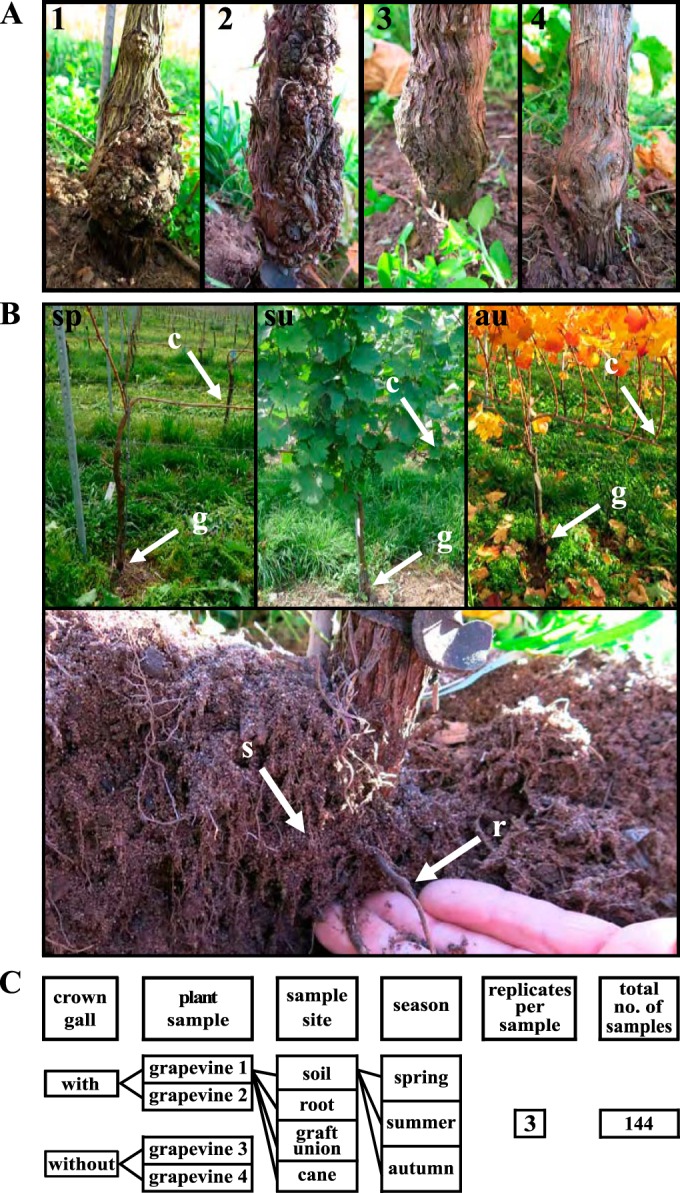
Grapevine plants and sampling procedure used for comparison of grapevine microbiota. (A) Graft unions of the trunk of the two grapevine plants with (1 and 2) and two without (3 and 4) a crown gall. (B) Illustration of the sampling sides; sp, spring; su, summer; au, autumn; c, 1-year-old cane, g, graft union, r, root, and s, soil. (C) Scheme of the experimental setup.
DNA extraction and amplicon sequencing.
The frozen plant and soil samples (−80°C) were shredded in an MM2000 ball mill (Retsch, Hannover, Germany), and DNA was extracted using the FastDNA SPIN kit for soil (MP Biomedicals, Santa Ana, CA, USA). DNA extractions with the kit components and no added sample material served as negative controls. For PCR of the 16S rRNA gene, the primers 515F and 806R, including 2 × 8-bp multiplexing indices and Illumina adapters attached to their 5′ end, were used to amplify the variable region V4 of the 16S rRNA gene (38). The sequence of the forward primer was 5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACX XXX XXX XTA TGG TAA TTG TGT GCC AGC MGC CGC GGT AA-3′, and the reverse primer was 5′-CAA GCA GAA GAC GGC ATA CGA GAT XXX XXX XXA GTC AGT CAG CCG GAC TAC HVG GGT WTC TAA T-3′. XXX XXX XX indicates the index sequences.
Each sample was processed in three technical replicates to reduce random PCR effects (39). PCR was performed in 10-μl reaction mixtures, each containing 5 μl of 2× Phusion high-fidelity PCR master mix (New England BioLabs, Ipswich, MA, USA), 0.33 μl of the 10 μM forward and reverse primers (Eurofins MWG Operon, Huntsville, AL, USA), 3.34 μl of PCR-grade water, and 1 μl of template DNA. The PCR conditions comprised an initial denaturation step at 95°C for 4 min, 35 cycles of denaturation at 95°C for 40 s, annealing at 55°C for 30 s, and elongation at 72°C for 1 min, followed by a final extension step at 72°C for 5 min. We combined the three technical PCR replicates into a 30-μl PCR pool. Successful amplification was verified with agarose gel electrophoresis using 5 μl of the PCR pool. The remaining 25 μl was further processed using the SequalPrep normalization plate kit (Invitrogen, Carlsbad, CA, USA) to remove excess primers and nucleotides, as well as for normalizing the PCR product to quantities of 25 ng. Five microliters of normalized DNA was used for pooling with the samples of other projects for parallel sequencing (38). This final DNA pool was verified for DNA fragment size of the library with a high-sensitivity DNA Chip (Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA) and quantified with the double-stranded DNA (dsDNA) high-sensitivity assay (Life Technologies GmbH, Darmstadt, Germany). The final DNA pool was diluted to 2 nM, and 3 μl of each of the custom sequencing and indexed primers was added to the cartridge of a 2 × 250-bp version 2 paired-end MiSeq sequencing kit (Illumina, San Diego, CA, USA). 16S rRNA amplicons were sequenced according to the manufacturer's protocol for the Illumina MiSeq instrument using a 2 × 250-bp version 2 paired-end sequencing run.
Amplicon sequencing data analysis.
The quality of the sequences was analyzed using FastQC version 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The program fastq-join version 1.8.0 (https://expressionanalysis.github.io/ea-utils/) was used to join forward and reverse reads. The reads were filtered with USEARCH version 8 (40), which included quality filtering according to the Phred score (>Q20) and the sequence length (>250 bp). Clusters of operational taxonomic units (OTUs) were built, and chimeras were removed and taxonomically classified using the UCLUST (40) and UCHIME (41) algorithms, as implemented in USEARCH version 7.0.1090 (42). Using the Ribosomal Database Project (RDP) Classifier version 2.2 (43), we assigned the taxonomy for each OTU. A phylogenetic tree was calculated using FastTree version 2.1.3 (44). Plastids and mitochondrial 16S rRNA gene sequences were removed from the OTU table before continuing the analyses.
The R script of the following downstream analyses, using the packages phyloseq (45) and vegan (https://cran.r-project.org/web/packages/vegan/index.html), are provided in the supplemental material. Using the OTU table without any normalization (46), bacterial community dissimilarities between the individual samples were estimated using the Bray-Curtis distance, and the resulting beta diversity was visualized through nonmetric multidimensional scaling (NMDS). Four outliers were excluded according to the NMDS. The various influential factors (sample site, season, and crown gall disease) were fitted onto the ordination axes, representing the differences in the microbiota, so as to identify significant correlations. A general linear model with the coefficients, soil, root, graft union, cane, and the scores was generated, and the relevance of this model for our data was tested using an analysis of variance (ANOVA) statistical test (NMDS axis one). Fold changes of the sample types were calculated using the R package EdgeR (46, 47). Significant fold changes had a false-discovery rate (FDR) of <0.001. Only OTUs with a mean abundance of ≥20 sequences per sample in at least one group were considered for analysis.
We determined the bacterial species richness as raw counts of the OTUs and calculated the alpha diversity using the Shannon index (48) based on the OTUs. Significant differences in the alpha diversity and bacterial species richness between sample types were calculated using the Wilcoxon test (49). For taxonomic analysis of the microbiota, all samples from one site and all OTUs of the same taxonomic rank were merged. To calculate the relative abundance of a taxonomic rank, the sequences of a taxonomic rank were divided by all sequences of one sample site. We merged the taxonomic ranks that were less abundant than 0.5% to one group called “other.” The relative abundance of each OTU within one sample was used for classification by Random Forest (50), a supervised learning analysis, with 1,500 decision trees. The relative sample counts of the OTUs were used as predictors with season, sample site, and crown gall disease as class labels. The percentage of calculated and actual sample class labels resulted in the out-of-bag error (OOB). A small OOB indicates distinctive microbiota according to the class labels. The VennDiagram package (51) of the R software was used to calculate shared OTUs between the sample sites of galled and nongalled grapevines. Within each season, we randomly paired a galled and a nongalled grapevine plant for a paired Wilcoxon test. This allowed us to calculate any significant differences between the amounts of shared OTUs between, for example, canes and graft unions of plants with and without a crown gall. This calculation was repeated for soils and roots.
Isolation and PCR screening of agrobacteria.
We isolated bacteria from the graft union material used in this study for amplicon sequencing. The wooden parts of the grapevine material were shredded using a ball mill (Retsch, Hannover, Germany). Purified water (RotisolV high-performance liquid chromatography [HPLC] gradient grade; Roth) was added to 300 mg of the processed grapevine material or soil. After 2 h at 28°C, the supernatant was used for 10-fold serial dilutions, and 100 μl was incubated for 5 days on agar plates supplemented with 213 μM cycloheximide (CHX; Sigma-Aldrich, St. Louis, MO, USA) to prevent the growth of fungi. Either yeast extract broth (YEB)-CHX agar plates (0.5% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] sucrose, 1.23% [wt/vol] MgSO4 [AppliChem, Darmstadt, Germany], 1.5% [wt/vol] Agar-Agar Kobe I [Carl-Roth, Karlsruhe, Germany]) or lysogeny broth (LB)-CHX agar plates (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl [AppliChem], 1.5% [wt/vol] Agar-Agar Kobe I [Carl-Roth]) were used for bacterial growth. Single colonies with an Agrobacterium-like morphology were subcultured on YEB-CHX or LB-CHX agar plates. PCR-based screening for Agrobacterium colonies was performed as follows: (i) two different fragments of the 16 rRNA gene were amplified to identify Agrobacterium, (ii) a RecA gene fragment (52) served for differentiation between Agrobacterium vitis and other Agrobacterium species, and (iii) a fragment of the VirD2 was PCR amplified to screen for the presence of the Ti plasmid (11). The following primer sequences were used: (i) 16S rRNA gene primers 27F (5′-AGR GTT YGA TYM TGG CTG AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′), or 515F (5′-GTG YCA GCM GCC GCG GTA A-3′) and 806R (5′-GGA CTA CNV GGG TWT CTA AT-3′), (ii) Agrobacterium species-specific RecA primers F8360 (5′-AGC TCG GTT CCA ATG AAA-3′) and F8361 (5′-GCT TGC GCA GCG CCT GGC T-3′), A. vitis-specific RecA primers G0004F (5′-GAT ATC GCG CTC GGC ATT GGT-3′) and G0005R (5′-CCT TCG ATT TCA GCT TTC G-3′) (52), and (iii) virD2 primers virD2F (5′-TTG GAA TAT CTG TCC CGG AAG-3′) and virD2R (5′-CTT GTA CCA GCA GGG AAG CTT A-3′) (11). A 50-μl PCR mixture contained 1× HF buffer (New England BioLabs, Ipswich, MA, USA), an experimentally determined amount of custom-made Phusion polymerase (53), 0.2 μM each primer, 400 μM dinucleoside triphosphates (dNTPs) (Fermentas, Waltham, MA, USA), and 2 μl of a bacterial colony resuspended and boiled in 100 μl of HPLC-grade water (RotisolV HPLC gradient grade; Roth) for 10 min. The 16S rRNA gene sequences of the PCR products were analyzed by Sanger sequencing (GATC, Constance, Germany), followed by a nucleotide search using the BLAST algorithm (54) from the NCBI database (55).
Infection assay of in vitro-cultivated grapevine.
Four to 8-week-old in vitro-cultivated grapevine plantlets (varieties Mueller Thurgau and 5BB) provided by the vine nursery Steinmann in Sommerhausen, Germany, were used in a virulence assay (see Fig. S1 in the supplemental material). In vitro plantlets originated from 1-year-old cane pieces, with one node of cuttings from environmental grapevine plants from the year 2000. The cane pieces were surface sterilized (96% ethanol, 6% sodium hypochloride), and after root and shoot induction, the plantlets were subcultivated every 8 to 12 weeks in plastic boxes filled with 3 cm of a grapevine-specific agar growth medium. Plantlets were incubated in a growth chamber with a 14-h photoperiod (light, 23°C; dark, 21°C) and a light intensity of 180 μmol s−1 m−2 using universal white lamps (L 36W/25; Osram, Munich, Germany). Agrobacterium isolates were inoculated to induce crown gall development at the second or third internode of the grapevine stems using a sterile needle dipped into a colony. The known virulent Agrobacterium vitis strain S4 (13) and the nonvirulent disarmed Agrobacterium tumefaciens strain GV3101 (56) served as positive and negative controls, respectively. At least eight plantlets were inoculated with the same Agrobacterium strain/isolate. We visually screened the plantlets for the appearance of crown galls on a weekly basis for up to 8 weeks. Four-week-old infection sites were used for 16S rRNA gene amplicon sequencing, as described in “DNA extraction and amplicon sequencing” and “Amplicon sequencing data analysis” above.
Accession number(s).
Raw 16S rRNA gene sequencing data are deposited at the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession number PRJEB12040.
RESULTS
Each sampling site harbors a distinct microbiota.
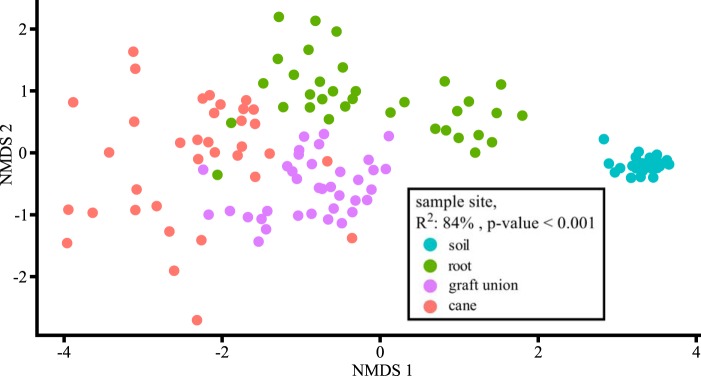
Material of two grapevines with and two without a crown gall (Fig. 1A) from four sampling sites each was collected at three different time points over a year (Fig. 1B), resulting in 144 samples (Fig. 1C). A total of 4,572,415 16S rRNA gene sequences were analyzed. After removing the sequences belonging to chloroplasts and mitochondria, 1,201,593 sequences remained. These were grouped into 8,674 different OTUs. The nonmetric multidimensional scaling (NMDS) ordination shows that the structural differences in the microbial community composition were determined first and foremost by the sample site (Fig. 2, environmental fit, r2 = 84%, P ≤ 0.001). The calculation of a general linear model of the values of NMDS1 resulted in a significant influence of each sampling site on the microbiota (soil, t-value = 32, P < 0.001; root, t-value = 12, P < 0.001; graft union, t-value = 8, P < 0.001; cane, t-value = −18, P value < 0.001). The ANOVA for the general linear model (F = 357, P value < 0.001; residuals degrees of freedom, 136) and Random Forest analysis (see Table S1 in the supplemental material) illustrate that the sample site accounts for the main difference in microbial community composition.
FIG 2.
Distribution of the 144 grapevine-associated microbiota within a nonmetric multidimensional scaling (NMDS) ordination. Analysis is based on the Bray-Curtis distance. The factor sample site explains 84% (R2) of the variation among the microbiota. Significance (P value) was calculated using a permutation test. Colors indicate sample sites.
Microbial structure of the sampling sites.
In terms of richness, the soil microbiota harbored a greater diversity of bacterial taxa than the other sampling sites (Fig. 3A; mean ± standard deviation [SD] richness, 2,712 ± 673). The richness in bacterial taxa decreased with the distance from the soil over the root (richness, 253 ± 170), to the graft union (richness, 166 ± 50), and the cane (richness, 76 ± 41). Similarity analyses of the microbiota from the different sites showed that 410 OTUs (5%) were identical in the soil, root, graft union, and the cane. Each sample site shared most of the OTUs with the soil (root, 88%; graft union, 82%; and cane, 79%). The alpha biodiversity (Shannon index) also changed with the distance from the soil (Fig. 3B) in that the microbiota of the soil was most diverse (mean ± SD Shannon index, 6.8 ± 0.2), followed by the root (Shannon index, 4.0 ± 0.7), graft union (Shannon index, 3.2 ± 0.9), and the cane (Shannon index, 3.0 ± 1).
FIG 3.
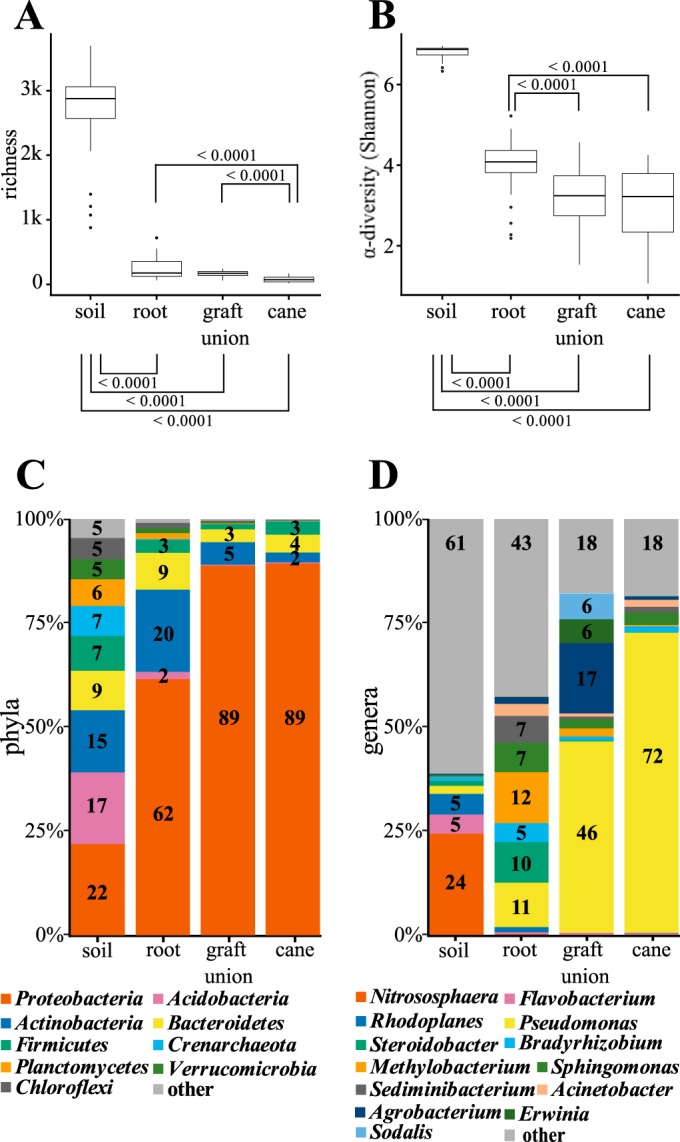
Comparison of the microbial communities from the soil, the grapevine roots, graft unions, and canes. (A and B) Number of bacterial taxa (richness) (A) and Shannon index (α diversity) (B) for each of the sample sites. P values are calculated according to Wilcoxon rank sum test. Significant values are ≤0.01. (C and D) Percentages of phyla (C) and genera (D) in the microbial community for each sample site. Phyla or genera with a relative abundance lower than 0.5% in the microbial community are combined into the group “other.”
A detailed analysis of the bacterial phylum composition revealed that the relative number of Proteobacteria sequences increased with the distance from the soil (Fig. 3C). Proteobacterial sequences comprised 22% of all OTUs in the soil, 62% in the root, 89% in the graft union, and 89% in 1-year-old cane samples. In contrast, Actinobacteria decreased along the plant axis from 20% in the root, 5% in the graft union, to 2% the in cane samples. Sequences of the class Acidobacteria were present only in the soil (17%) and root (2%) microbiota. In addition, Bacteroidetes (soil, 9%; root, 9%), Planctomycetes (soil, 6%; root, 1%), and Verrucomicrobia (soil, 5%; root, 1%) were more represented in soil and root samples than in the graft union and cane microbiota. At the genus level (Fig. 3D), Pseudomonas dominated the aboveground microbiota (graft union, 46%; cane, 72%), while in the soil it was Nitrososphaera (24%), and in the root, it was Methylobacterium (12%). Agrobacterium-related sequences were mainly present in roots (2%) and graft unions (17%, Fig. 3D), while in soil (0.3%) and canes (0.7%), the relative sequence numbers were very low. Taken together, the sample site-specific grapevine-associated microbiota changed with the distance from the soil in diversity, richness, shared operational taxonomic units, composition, and structure.
Impact of the seasons and crown gall disease on the microbiota.
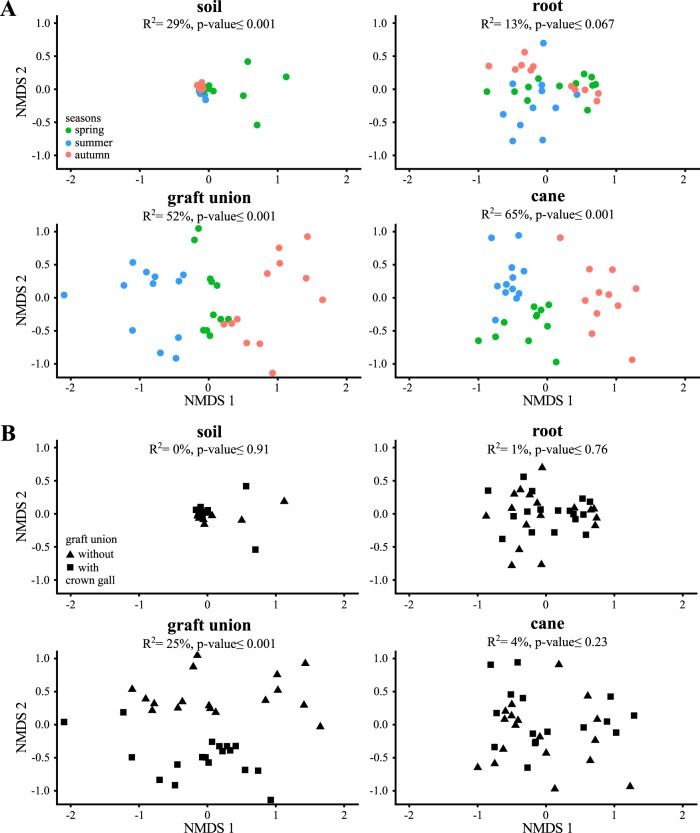
We next analyzed the amplicon data of each sample site with respect to the seasons (Fig. 4A). Separate NMDS ordinations for each sample site demonstrated the effects of the season on the microbiota of soil (environmental fit, r2 = 29%, P < 0.001), graft unions (environmental fit, r2 = 52%, P < 0.001), and canes (environmental fit, r2 = 65%, P < 0.001). The seasons had no significant influence on the root microbiota (root, environmental fit, r2 = 13%, P ≤ 0.067). Computable classification by Random Forest of the samples taking the seasons into account revealed the highest error rate for the root samples (see Table S2 in the supplemental material; out-of-bag estimated error [OOB], 21%), followed by the soil (OOB, 11%), and finally the aboveground samples from the graft union (OOB, 8%) and cane (OOB, 9%). Both the NMDS ordinations and the Random Forest classifications indicated a greater influence of the seasons on aboveground than on belowground microbiota.
FIG 4.
Factors determining the differences between the microbial communities of the sample sites. Nonmetric multidimensional scaling (NMDS) ordinations for the factors season (A) and graft unions (B) without or with a crown gall. The percentage of variation among the microbiota of a sample site was correlated with the factor season or crown gall disease (R2). Significance was calculated using permutation test (P value).
With respect to the presence/absence of crown gall disease, the data showed neither a significant effect on the soil microbial community composition (Fig. 4B, environmental fit, r2 = 0%, P ≤ 0.91), on the root (r2 = 1%, P ≤ 0.76), nor on the cane (r2 = 4%, P ≤ 0.23). However, the microbiota differed significantly between the graft unions with a crown gall and those without (Fig. 4B, environmental fit, r2 = 25%, P < 0.001). Computable classification using Random Forest revealed the lowest out-of-bag estimated error rate in the microbiota of the graft unions (see Table S3 in the supplemental material; OOB for graft unions, 8%) compared to the soil (OOB, 40%,), root (OOB, 59%), and cane (OOB, 37%).
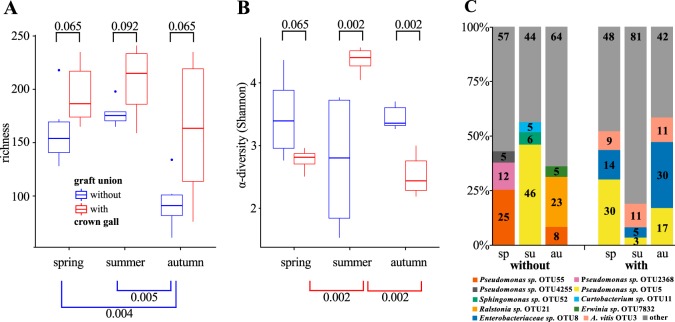
We then compared the microbiota of the two types of graft unions (without and with a crown gall) from each season (spring, summer, and autumn) to each other (Fig. 5). The bacterial richness was higher in the graft unions with a crown gall in spring (P ≤ 0.065), summer (P ≤ 0.092), and autumn (P ≤ 0.065) than in those without (Fig. 5A). Furthermore, the higher richness in the microbiota of the graft unions with a crown gall did not change significantly over the seasons. In contrast, in graft unions without a crown gall, the richness was significantly lower in autumn than in spring and summer (Wilcoxon test, spring-autumn, P ≤ 0.004; summer-autumn, P ≤ 0.005). The richness analysis indicates that the microbial community in graft unions with a crown gall contains additional bacterial taxa and is more stable over the seasons than those without. The alpha diversity (Shannon index) did not change prominently in graft unions without a crown gall over the seasons. In contrast, the alpha diversity differed significantly between the seasons in graft unions with a crown gall (Fig. 5B, Wilcoxon-test, spring-summer P ≤ 0.002; summer-autumn, P ≤ 0.002) and was highest in summer.
FIG 5.
Comparison of the microbial communities of the graft unions without and with crown gall disease. (A) Number of bacterial taxa (richness) and (B) including abundance within each bacterial taxon (α-diversity), calculated according to Shannon for each of the sample sites. P values are calculated according to the Wilcoxon rank sum test. Significant values are ≤0.01. (C) Percentages of the three most abundant operational taxonomic units (OTUs) in the microbial community for each sample site. All OTUs with a relative abundance lower than 0.5% were merged, forming the group “other.”
Bacterial taxa that are affected by the crown gall disease.
We recovered 23 Agrobacterium isolates from the grapevine and soil material used for amplicon sequencing. The screening for agrobacterial virulence resulted in the identification of six virulent Agrobacterium vitis isolates. The remaining 17 non-A. vitis isolates were classified as nonvirulent agrobacteria. A. vitis isolates were found only in crown galls and roots of the diseased grapevine plants, together with nonvirulent Agrobacterium species. According to the RDP Classifier, A. vitis is also one of the three most abundant OTUs in graft unions with a crown gall, the others being Pseudomonas sp. OTU_0005 and Enterobacter OTU_0008. In graft unions with a crown gall, these three most abundant OTUs (A. vitis OTU_0003; Pseudomonas sp. OTU_0005; and Enterobacter OTU_0008) amounted to 53% and 58% of all sequences in spring and autumn, respectively, although in summer, this dropped to 19% of all obtained sequences (Fig. 5C). Nevertheless, these three OTUs still remained the most abundant ones in summer. In contrast, the three most abundant OTUs in graft unions without a crown gall differed in every season (Fig. 5C): in spring, three Pseudomonas species (OTU_0055, OTU_2368, and OTU_4255); in summer, Pseudomonas sp. (OTU_0005), Sphingomonas sp. (OTU_0052), and Curtobacterium sp. (OTU_0011); and in autumn, Pseudomonas sp. (OTU_0055), Ralstonia sp. (OTU_0021), and Erwinia species (OTU_7832).
To record the bacterial taxa that are significantly affected by the crown gall disease, we calculated the fold changes of the sequence numbers for the OTUs detected in graft unions with and without a crown gall separately for each season (EdgeR, FDR < 0.001; see Table S4 in the supplemental material). Of the 28 different OTUs with significant changes in sequence numbers, 24 increased in graft unions with a crown gall compared to those without. Nine OTUs comprised zero sequences in graft unions without a crown gall; hence, these were exclusively present in graft unions with a crown gall. Of the four OTUs of which the sequence numbers decreased in graft unions with a crown gall, three (OTU_0005, OTU_0011, and OTU_0052) were less abundant in the summer. At this time of the year, two other OTUs showed a significant increase: an unknown member of the Proteobacteria phylum (OTU_3436) and A. vitis (OTU_0003). These two were significantly enriched in all seasons and are part of the core microbiota in graft unions with a crown gall. Four additional OTUs contributed to the core microbiota of crown galls: OTU_0005 (Pseudomonas sp.), OTU_0007 (Burkholderiales), OTU_0008 (Enterobacteriales), and OTU_0032 (Agrobacterium sp.). These represented more than 20 sequences per sample in at least 80% of the graft union samples with a crown gall. In graft unions without a crown gall, no OTU met this definition; thus, in graft unions of healthy trunks, the microbiota fluctuated more.
Crown gall induction without core microbiota.
We also analyzed an amplicon sequencing data set of in vitro-cultivated grapevine plantlets to address the question as to whether crown gall development requires a core microbiota and if this in turn profits from the crown gall environment. The in vitro-cultivated plantlets were inoculated either with the virulent A. vitis S7, an isolate from a grapevine crown gall of the same vineyard used for sampling in this study (see Fig. S1A and B in the supplemental material), or with the disarmed Agrobacterium tumefaciens strain GV3101. Uninoculated plantlets served as controls (Fig. S1C). Altogether, amplicon sequencing was performed on 18 samples (Fig. S1D), resulting in a total of 568,855 sequences. After removing the plastid- and mitochondrion-related 16S rRNA gene sequences, 42,700 sequences remained and were grouped into 612 OTUs. In noninoculated in vitro-cultivated grapevines, no OTU was detected with more than 15 amplicon sequences, suggesting an extremely low abundance of bacteria. The stems inoculated with the avirulent A. tumefaciens GV3101 contained an enriched number of sequences of this strain (OTU_0507), another Agrobacterium (OTU_0032), and Curtobacterium (OTU_0011; Table 1, EdgeR, FDR < 0.001). In crown galls of the plantlets inoculated with the virulent A. vitis S7 strain (OTU_0003), no other OTU was significantly increased (Table 1). This experiment indicates that the virulent A. vitis S7 can induce crown gall disease on grapevine without any requirement of a core microbiota.
TABLE 1.
OTUs with significant differences in 16S rRNA gene sequence numbers from stems without and with a crown gall of in vitro-cultivated grapevine plantletsa
| Bacterial identity | 16S sequence no. |
logFC | logCPM | Adjusted P value FDRb | |
|---|---|---|---|---|---|
| Without | With | ||||
| Agrobacterium tumefaciens GV3101 (OTU 0507) | 3,591 | 78 | 5.5 | 16.5 | 6.29E−63 |
| Agrobacterium (OTU 0032) | 124 | 1 | 5.8 | 11.8 | 1.03E−64 |
| Agrobacterium vitis isolate S7 (OTU 0003) | 6 | 2,758 | −8.6 | 16.1 | 8.09E−33 |
| Curtobacterium (OTU 0011) | 30 | 1 | 4.1 | 10.1 | 3.45E−07 |
Stems were inoculated with the virulent Agrobacterium vitis isolate S7 (OTU 0003) and the disarmed Agrobacterium tumefaciens strain GV3101 (OTU 0507) 4 weeks before analysis. Displayed are the mean sequence numbers in the samples with (“With”) and without (“Without”) a crown gall, calculated according to the EdgeR package in R. logFC, log2 fold changes; logCPM, log2 counts per million.
P values are adjusted to multiple testing according to Benjamin-Hochberg (FDR < 0.001).
Bacterial taxa shared between crown galls and the other sample sites.
To identify the source of the additional bacterial taxa found in native crown galls, we analyzed the OTUs of the graft unions shared with the other sample sites (soil, root, and cane) separately for diseased and nondiseased native grapevines. We randomly paired a diseased with a nondiseased plant sample from the same season using a paired Wilcoxon test. The microbiota of the graft unions with a crown gall shared significantly more bacteria with the root (P ≤ 0.024) and the soil (P ≤ 0.003) than with the healthy graft unions without a crown gall. In contrast, those without shared more OTUs with the cane (P ≤ 0.009). Thus, the soil and root rather than the cane serve as a source for bacteria in grapevine crown galls.
DISCUSSION
To understand the infection ecology of the crown gall disease, we investigated the endophytic microbial community of grapevines with and without a crown gall. Amplicon-based community profiling revealed a distinct microbial community for each of the sample sites (soil, root, graft union of the trunk, and cane). Distinct microbiota have previously been published for grapevines from vineyards in Long Island (Suffolk county, NY, USA) for soil, root, leaf, flower, and grape berry (33) and from Lussac-Saint-Émilion (Gironde, France) for soil, bark, leaf, and grape berry samples (27). Proteobacteria, Acidobacteria, Actinobacteria, and Bacteroidetes dominated the microbiota in our soil samples and in those from Long Island, while in the samples from Lussac-Saint-Émilion, no Bacteroidetes colonies were isolated. Furthermore, our root samples and those from Long Island (33), as well as our cane samples and those from Trentino, Italy (26), were similar on the phylum level. The microbiota of our sample sites, those from Long Island (33), and those from Lussac-Saint-Émilion (27) all showed a gradient from belowground to aboveground. In all cases, the structure of the soil microbiota was most complex (highest richness and alpha diversity), with that of the aboveground being least complex (lowest richness and alpha diversity). This gradient in the microbial structure and composition is most likely the result of changing environmental factors, such as humidity, distance from the soil, organic substrates, and UV exposure (57–60). The factor of season had an additional impact on the microbial structure, which was stronger on our aboveground than belowground samples. Not only do the seasons have an impact on the microbial composition of the leaf microbiota of grapevine (28, 32), but the time of year also influences endophytic bacteria in woody material, as shown in this study. Thus, we conclude that grapevines have similar phylum compositions in distinct locations and that the crown gall disease does not substantially affect the microbial structure of the soil, root, and cane.
Crown gall disease affected the microbiota only in graft unions. The microbiota of graft unions with a crown gall contained a higher number of different bacterial species in all seasons than graft unions without a crown gall. Agrobacterium vitis, together with eight other bacterial species, caused the difference in the microbial community between graft unions with and without a crown gall. These were exclusively found in graft unions with a crown gall. Likewise, Arabidopsis leaves infected with the fungus Albugo significantly enriched a subset of bacterial endophytes (61). This suggests that both pathogens (A. vitis and Albugo) promote colonization with certain endophytic microbes at the infection site. Compared to healthy graft unions, crown galls share more bacteria with the belowground microbiota and fewer with canes. Therefore, the source for the invasive bacteria in crown galls seems to be the soil and root. This finding supports the idea of the soil as a microbial seed bank for grapevine-associated microbiota, as previously postulated (33).
In graft unions with a crown gall, three OTUs, A. vitis (OTU_0003), Pseudomonas sp. (OTU_0005), and Enterobacteriaceae sp. (OTU_0008), were most abundant in every season. These three, together with three additional OTUs, were present in 80% of graft union samples with a crown gall, indicating that the crown gall microbiota is relatively stable. In summer, the percentage of the three most abundant bacterial species, which included A. vitis, decreased in crown galls, thereby increasing the species evenness in the bacterial communities at this time of year. Other studies have reported that in summer, the CFU of A. vitis are reduced in grapevines (62), and that the isolation of A. vitis from grapevine samples is more difficult (7). Reasons for the reduction in species abundance in summer might be higher temperatures and drought stress. It is known from the model plant Arabidopsis (63, 64) and Ricinus communis (65) that crown gall growth is affected by drought stress. In graft unions without a crown gall, the diversity of the bacterial species was only marginally affected by the season. The three most abundant OTUs encompassed 57% of all sequences in summer, which was only marginally higher than the three in spring (42%) and autumn (36%). However, the three most abundant bacterial species varied between the seasons, indicating that, unlike in graft unions with a crown gall, no core microbiota exists in graft unions without a crown gall. Thus, crown gall disease seems to stabilize the bacterial composition over the seasons, as previously reported for photoplasma-infected grapevine leaves (28). Nonetheless, the striking decrease in the abundance of bacterial species in graft unions with a crown gall in summer seems to be specific for crown gall tissue.
The bacterial species exclusively found or enriched in graft unions with a crown gall may profit from the crown gall environment. Indeed, it is well known from the literature that this habitat provides opines and other accumulating metabolites, as well as additional living space (14, 66). It has been shown that opines serve as common nutrients and cause an increase in the local population of opine-metabolizing bacteria (19). This has also been demonstrated by transgenic opine-producing legume species, which harbored an altered bacterial composition, including an increase in opine-degrading Pseudomonas (67). We also found a Pseudomonas strain (OTU_0005) that was clearly enriched in spring and autumn in crown galls. Pseudomonas is able to cause wounds by producing ice crystals (68, 69). Wounds induce Agrobacterium-mediated processes, such as plant cell transformation, production of opines, and phytohormones (70). For example, indole-3-acetic acid (IAA) is involved in plant and crown gall developmental processes, enriched in crown galls, and can serve as a source of carbon for Pseudomonas putida 1290 (71). Furthermore, Pseudomonas sp. (72, 73), Enterobacteriaceae sp., and many other endophytic grapevine bacteria (26) are able to produce IAA. Interactions of nonpathogenic with pathogenic bacteria are known for tumors of olive trees, induced by Pseudomonas savastanoi (pv. savastanoi), which host a nonpathogenic Erwinia species (74). An Erwinia species (OTU_7832) was enriched in graft unions with a crown gall, and it seems to profit from the crown gall environment.
We used in vitro-cultivated grapevine plantlets to investigate the mechanisms of the infection process and development of crown gall disease. This infection assay demonstrated that A. vitis and no other Agrobacterium species of the environmental isolates, including A. tumefaciens, caused crown gall disease. This indicates that in grapevines, A. vitis retains its virulence machinery. This finding is in accordance with a high-throughput isolation study of agrobacteria from crown galls of herbaceous and woody hosts (75). In this study, only seven out of 5,419 isolates became nonvirulent mutants after being inoculated into host plants to induce crown galls. Furthermore, the induction of crown gall growth on in vitro-cultivated grapevine plantlets proved that A. vitis does not require a microbial community for disease outbreak. This observation suggests that the crown gall-specific bacterial species and those that strongly multiply in crown galls appear to benefit from the crown gall environment provided for them by A. vitis infection.
Grapevine organs and the vineyard soil harbor a distinct microbial community, which is not affected by crown gall disease, except at the site of graft union and gall formation. Graft unions with a crown gall stabilize core microbiota and host opportunistic bacteria over the seasons. These, however, are not essential for the induction of crown gall growth. Our in vitro assay showed that the induction of crown gall growth requires no other bacterium than Agrobacterium vitis. This finding suggests that none of the invasive endophytic bacteria, including A. tumefaciens, are obligate for crown gall development. Nonetheless, a supportive role in the performance of crown gall development cannot be excluded and will be addressed in future studies. The invasive bacterial species most likely profit from the crown gall environment in that they have an advantage, nutritional or otherwise, by living within crown gall tissues. Unraveling the role of the opportunistic bacteria in crown gall performance may help support disease management in the future.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks go to Peter Schwappach (Bavarian Regional Office for Viticulture and Horticulture, Veitshoechheim, Germany) for providing grapevine plants from the vineyard. We are very grateful to Gabriele Brendel (Vine Nursery Steinmann, Sommerhausen, Germany) for providing in vitro-cultivated grapevine plantlets, as well as for ongoing discussions and practical advice. Many thanks go also to our colleagues from the University of Wuerzburg (Wiebke Sickel, Gudrun Grimmer, and Lisa Walther [for support in the laboratory] and Hannes Horn [for support in bioinformatics]). We also acknowledge Anne Müller and Lorenz Hoffmann, who performed their Master's thesis (2012) and Diploma thesis (2013) at the University of Würzburg, respectively, on this topic. Finally, we thank Rainer Hedrich (University of Würzburg, Germany) for his support during this study and Tracey A. Cuin (University of Würzburg, Germany) for critical reading of the manuscript.
Funding Statement
This work was funded by the DFG Graduiertenkolleg (GK1342 “Progress in lipid signaling”; TPs A8 [U. Hentschel] and A5 [R. Deeken]) and by a development grant of the Chamber of Industry and Commerce 2012, Schweinfurt-Wuerzburg, Germany, to U. Hentschel and R. Deeken. The funders had no role in the study design, data collection and interpretation, or decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01131-16.
REFERENCES
- 1.Genov I, Atanassov I, Tsvetkov I, Atanassov A. 2006. Isolation and characterization of Agrobacterium strains from grapevines in Bulgarian vineyards and wild grapes, V. vinifera ssp. silvestris. Vitis 45:97–101. [Google Scholar]
- 2.Burr TJ, Katz BH. 1983. Isolation of Agrobacterium tumefaciens biovar 3 from grapevine galls and sap, and from vineyard soil. Phytopathology 73:163–165. doi: 10.1094/Phyto-73-163. [DOI] [Google Scholar]
- 3.Burr TJ, Otten L. 1999. Crown gall of grape: biology and disease management. Annu Rev Phytopathol 37:53–80. doi: 10.1146/annurev.phyto.37.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Genov N, Llop P, Lopez MM, Bobev SG, Alvarez B. 2015. Molecular and phenotypic characterization of Agrobacterium species from vineyards allows identification of typical Agrobacterium vitis and atypical biovar 1 strains. J Appl Microbiol 118:1465–1477. doi: 10.1111/jam.12791. [DOI] [PubMed] [Google Scholar]
- 5.Burr TJ, Reid CL, Yoshimura M, Momol EA, Bazzi C. 1995. Survival and tumorigenicity of Agrobacterium vitis in living and decaying grape roots and canes in soil. Plant Dis 79:677–682. doi: 10.1094/PD-79-0677. [DOI] [Google Scholar]
- 6.Tarbah FA, Goodman RN. 1988. Ultrastructural observations of the process of Agrobacterium tumefaciens biovar 3 infection of grape cv. Chancellor. Physiol Mol Plant Pathol 32:437–453. doi: 10.1016/S0885-5765(88)80036-5. [DOI] [Google Scholar]
- 7.Pu XA, Goodman RN. 1993. Effects of fumigation and biological control on infection of indexed crown gall free grape plants. Am J Enol Vitic 44:241–248. [Google Scholar]
- 8.Bishop AL, Katz BH, Burr TJ. 1988. Infection of grapevines by soilborne Agrobacterium tumefaciens biovar 3 and population dynamics in host and nonhost rhizospheres. Phytopathology 78:945–948. doi: 10.1094/Phyto-78-945. [DOI] [Google Scholar]
- 9.Filo A, Sabbatini P, Sundin GW, Zabadal TJ, Safir GR, Cousins PS. 2013. Grapevine crown gall suppression using biological control and genetic engineering: a review of recent research. Am J Enol Vitic 64:1–14. [Google Scholar]
- 10.Szegedi E, Bottka S. 2002. Detection of Agrobacterium vitis by polymerase chain reaction in grapevine bleeding sap after isolation on a semiselective medium. Vitis 41:37–42. [Google Scholar]
- 11.Johnson KL, Zheng D, Kaewnum S, Reid CL, Burr T. 2013. Development of a magnetic capture hybridization real-time PCR assay for detection of tumorigenic Agrobacterium vitis in grapevines. Phytopathology 103:633–640. doi: 10.1094/PHYTO-10-12-0267-R. [DOI] [PubMed] [Google Scholar]
- 12.Otten L, de Ruffray P, Momol EA, Momol MT, Burr TJ. 1996. Phylogenetic relationships between Agrobacterium vitis isolates and their Ti plasmids. Mol Plant Microbe Interact 9:782–786. doi: 10.1094/MPMI-9-0782. [DOI] [Google Scholar]
- 13.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu H, Wood DW. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191:2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szegedi E. 2003. Opines in naturally infected grapevine crown gall tumors. Vitis 42:39–41. [Google Scholar]
- 15.Huss B, Tinland B, Paulus F, Walter B, Otten L. 1990. Functional analysis of a complex oncogene arrangement in biotype III Agrobacterium tumefaciens strains. Plant Mol Biol 14:173–186. doi: 10.1007/BF00018558. [DOI] [PubMed] [Google Scholar]
- 16.Ride M, Ride S, Petit A, Bollet C, Dessaux Y, Gardan L. 2000. Characterization of plasmid-borne and chromosome-encoded traits of Agrobacterium biovar 1, 2, and 3 strains from France. Appl Environ Microbiol 66:1818–1825. doi: 10.1128/AEM.66.5.1818-1825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell CR, Moore LW, Canfield ML. 1990. Growth of octopine-catabolizing Pseudomonas spp. under octopine limitation in chemostats and their potential to compete with Agrobacterium tumefaciens. Appl Environ Microbiol 56:2834–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dairi T, Asano Y. 1995. Cloning, nucleotide sequencing, and expression of an opine dehydrogenase gene from Arthrobacter sp. strain 1C. Appl Environ Microbiol 61:3169–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt TG, Fuqua C, Bever JD. 2012. Resource and competitive dynamics shape the benefits of public goods cooperation in a plant pathogen. Evolution 66:1953–1965. doi: 10.1111/j.1558-5646.2011.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szegedi E, Czako M, Otten L, Koncz CS. 1988. Opines in crown gall tumours induced by biotype 3 isolates of Agrobacterium tumefaciens. Physiol Mol Plant Pathol 32:237–247. doi: 10.1016/S0885-5765(88)80020-1. [DOI] [Google Scholar]
- 21.Gohlke J, Deeken R. 2014. Plant responses to Agrobacterium tumefaciens and crown gall development. Front Plant Sci 5:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaewnum S, Zheng DS, Reid CL, Johnson KL, Gee JC, Burr TJ. 2013. A host-specific biological control of grape crown gall by Agrobacterium vitis strain F2/5: its regulation and population dynamics. Phytopathology 103:427–435. doi: 10.1094/PHYTO-07-12-0153-R. [DOI] [PubMed] [Google Scholar]
- 23.Zheng D, Burr TJ. 2016. Inhibition of grape crown gall by Agrobacterium vitis F2/5 requires two nonribosomal peptide synthetases and one polyketide synthase. Mol Plant Microbe Interact 29:109–118. doi: 10.1094/MPMI-07-15-0153-R. [DOI] [PubMed] [Google Scholar]
- 24.Burr TJ, Reid CL. 1994. Biological control of grape crown gall with nontumorigenic Agrobacterium vitis strain F2/5. Am J Enol Vitic 45:213–219. [Google Scholar]
- 25.Bell CR, Dickie GA, Chan JWYF. 1995. Variable response of bacteria isolated from grapevine xylem to control grape crown gall disease in planta. Am J Enol Vitic 46:499–508. [Google Scholar]
- 26.Campisano A, Pancher M, Puopolo G, Puddu A, Lopez-Fernandez S, Biagini B, Yousaf S, Pertot I. 2015. Diversity in endophyte populations reveals functional and taxonomic diversity between wild and domestic grapevines. Am J Enol Vitic 66:12–21. doi: 10.5344/ajev.2014.14046. [DOI] [Google Scholar]
- 27.Martins G, Lauga B, Miot-Sertier C, Mercier A, Lonvaud A, Soulas ML, Soulas G, Masneuf-Pomarede I. 2013. Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS One 8:e73013. doi: 10.1371/journal.pone.0073013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulgari D, Casati P, Quaglino F, Bianco PA. 2014. Endophytic bacterial community of grapevine leaves influenced by sampling date and phytoplasma infection process. BMC Microbiol 14:198. doi: 10.1186/1471-2180-14-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perazzolli M, Antonielli L, Storari M, Puopolo G, Pancher M, Giovannini O, Pindo M, Pertot I. 2014. Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl Environ Microbiol 80:3585–3596. doi: 10.1128/AEM.00415-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campisano A, Antonielli L, Pancher M, Yousaf S, Pindo M, Pertot I. 2014. Bacterial endophytic communities in the grapevine depend on pest management. PLoS One 9:e112763. doi: 10.1371/journal.pone.0112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto C, Pinho D, Sousa S, Pinheiro M, Egas C, Gomes AC. 2014. Unravelling the diversity of grapevine microbiome. PLoS One 9:e85622. doi: 10.1371/journal.pone.0085622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA. 2015. The soil microbiome influences grapevine-associated microbiota. mBio 6(2):e02527-14. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 36.Lebeis SL. 2015. Greater than the sum of their parts: characterizing plant microbiomes at the community-level. Curr Opin Plant Biol 24:82–86. doi: 10.1016/j.pbi.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Guttman DS, McHardy AC, Schulze-Lefert P. 2014. Microbial genome-enabled insights into plant-microorganism interactions. Nat Rev Genet 15:797–813. doi: 10.1038/nrg3748. [DOI] [PubMed] [Google Scholar]
- 38.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fierer N, Hamady M, Lauber CL, Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, Smyth GK. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 48.Spellerberg IF, Fedor PJ. 2003. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon-Wiener’ index. Global Ecol Biogeogr 12:177–179. doi: 10.1046/j.1466-822X.2003.00015.x. [DOI] [Google Scholar]
- 49.Bauer DF. 1972. Constructing confidence sets using rank statistics. J Am Stat Assoc 67:687–690. doi: 10.1080/01621459.1972.10481279. [DOI] [Google Scholar]
- 50.Breiman L. 2001. Random Forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 51.Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shams M, Vial L, Chapulliot D, Nesme X, Lavire C. 2013. Rapid and accurate species and genomic species identification and exhaustive population diversity assessment of Agrobacterium spp. using recA-based PCR. Syst Appl Microbiol 36:351–358. [DOI] [PubMed] [Google Scholar]
- 53.Nørholm MH. 2010. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol 10:21. doi: 10.1186/1472-6750-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2015. GenBank. Nucleic Acids Res 43:D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koncz C, Schell J. 1986. The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 57.Joux F, Jeffrey WH, Lebaron P, Mitchell DL. 1999. Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol 65:3820–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diab S, Bashan Y, Okon Y, Henis Y. 1982. Effects of relative humidity on bacterial scab caused by Xanthomonas campestris pv. vesicatoria on pepper. Phytopathology 72:1257–1260. doi: 10.1094/Phyto-72-1257. [DOI] [Google Scholar]
- 59.Leben C. 1988. Relative humidity and the survival of epiphytic bacteria with buds and leaves of cucumber plants. Phytopathology 78:179–185. doi: 10.1094/Phyto-78-179. [DOI] [Google Scholar]
- 60.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 61.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM. 2016. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bauer C, Schulz TF, Lorenz D, Eichhorn KW, Plapp R. 1994. Population dynamics of Agrobacterium vitis in 2 grapevine varieties during the vegetation period. Vitis 33:25–29. [Google Scholar]
- 63.Efetova M, Zeier J, Riederer M, Lee CW, Stingl N, Mueller M, Hartung W, Hedrich R, Deeken R. 2007. A central role of abscisic acid in drought stress protection of Agrobacterium-induced tumors on Arabidopsis. Plant Physiol 145:853–862. doi: 10.1104/pp.107.104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klinkenberg J, Faist H, Saupe S, Lambertz S, Krischke M, Stingl N, Fekete A, Mueller MJ, Feussner I, Hedrich R, Deeken R. 2014. Two fatty acid desaturases, stearoyl-acyl carrier protein Δ9-desaturase6 and fatty acid desaturase3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol 164:570–583. doi: 10.1104/pp.113.230326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schurr U, Schuberth B, Aloni R, Pradel KS, Schmundt D, Jahne B, Ullrich CI. 1996. Structural and functional evidence in xylem-mediated water transport and high transpiration in Agrobacterium tumefaciens-induced tumors of Ricinus communis. Botanica Acta 109:405–411. doi: 10.1111/j.1438-8677.1996.tb00590.x. [DOI] [Google Scholar]
- 66.Deeken R, Engelmann JC, Efetova M, Czirjak T, Muller T, Kaiser WM, Tietz O, Krischke M, Mueller MJ, Palme K, Dandekar T, Hedrich R. 2006. An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18:3617–3634. doi: 10.1105/tpc.106.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oger P, Petit A, Dessaux Y. 1997. Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol 15:369–372. doi: 10.1038/nbt0497-369. [DOI] [PubMed] [Google Scholar]
- 68.Süle S, Seemüller E. 1987. The role of ice formation in the infection of sour cherry leaves by Pseudomonas syringae pv. syringae. Phytopathology 77:173–177. doi: 10.1094/Phyto-77-173. [DOI] [Google Scholar]
- 69.Lindow SE. 1983. The role of bacterial ICE nucleation in frost injury to plants. Annu Rev Phytopathol 21:363–384. doi: 10.1146/annurev.py.21.090183.002051. [DOI] [Google Scholar]
- 70.Pitzschke A, Hirt H. 2010. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leveau JH, Lindow SE. 2005. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl Environ Microbiol 71:2365–2371. doi: 10.1128/AEM.71.5.2365-2371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. 1998. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 11:156–162. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 73.Gardan L, David C, Morel M, Glickmann E, Abu-Ghorrah M, Petit A, Dessaux Y. 1992. Evidence for a correlation between auxin production and host plant species among strains of Pseudomonas syringae subsp. savastanoi. Appl Environ Microbiol 58:1780–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buonaurio R, Moretti C, da Silva DP, Cortese C, Ramos C, Venturi V. 2015. The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front Plant Sci 6:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Llop P, Murillo J, Lastra B, Lopez MM. 2009. Recovery of nonpathogenic mutant bacteria from tumors caused by several Agrobacterium tumefaciens strains: a frequent event? Appl Environ Microbiol 75:6504–6514. doi: 10.1128/AEM.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.