Abstract
Targeting of transmembrane proteins to the endoplasmic reticulum (ER) proceeds via either the signal recognition particle (SRP) or the guided entry of tail-anchored proteins (GET) pathway, consisting of Get1 to -5 and Sgt2. While SRP cotranslationally targets membrane proteins containing one or multiple transmembrane domains, the GET pathway posttranslationally targets proteins containing a single C-terminal transmembrane domain termed the tail anchor. Here, we dissect the roles of the SRP and GET pathways in the sorting of homologous, two-membrane-spanning K+ channel proteins termed Kcv, Kesv, and Kesv-VV. We show that Kcv is targeted to the ER cotranslationally via its N-terminal transmembrane domain, while Kesv-VV is targeted posttranslationally via its C-terminal transmembrane domain, which recruits Get4-5/Sgt2 and Get3. Unexpectedly, nascent Kcv recruited not only SRP but also the Get4-5 module of the GET pathway to ribosomes. Ribosome binding of Get4-5 was independent of Sgt2 and was strongly outcompeted by SRP. The combined data indicate a previously unrecognized cotranslational interplay between the SRP and GET pathways.
INTRODUCTION
Protein targeting to the endoplasmic reticulum (ER) can proceed either co- or posttranslationally (1–3). The cotranslational route depends on the signal recognition particle (SRP), which recognizes signal sequences or signal-anchor (SA) sequences within nascent chains and then targets ribosome-bound nascent chain complexes (RNCs) to the ER membrane (1–4). Substrates of SRP include luminal ER proteins, secreted proteins, and membrane proteins, among which the last most significantly benefit from cotranslational targeting, which efficiently circumvents aggregation of transmembrane (TM) domains in the cytosol (1–5). Tail-anchored (TA) proteins possess a single TM domain close to their C termini, which also provides the information for targeting to the appropriate cellular membrane (6). Those TA proteins destined for the ER do not enter the cotranslational pathway, because the TA is not accessible during translation. Instead, ER-targeted TA proteins posttranslationally enter the guided entry of tail-anchored proteins (GET) pathway, which comprises Get1 to -5 and Sgt2 (1–3, 7, 8). The central cytosolic component of the GET pathway is the ATPase Get3, which utilizes nucleotide-linked conformational changes to bind and target TA proteins to the Get1/Get2 receptor complex in the ER membrane (1–3, 7–13). Current models suggest that TA proteins are initially captured by Sgt2 and then transferred to Get3 (1–3, 7, 8, 13, 14). Cargo transfer from Sgt2 to Get3 is facilitated by a stable complex consisting of Get4 and Get5 (Get4-5), which can bind to Sgt2 via Get5 and to Get3 via Get4 (1–3, 7, 11, 13–16).
The currently most poorly understood step of the GET pathway is the initial capture of TA proteins. The process is expected to be highly coordinated, so that exposure of TA domains to the cytosol is minimized. A high-throughput study that identified Get5 among the proteins that are ribosome associated (17) hinted at a transfer of TA proteins to the GET pathway directly upon translation termination. However, it is not known what fraction of Get5 is ribosome associated, if other components of the GET pathway also interact with the ribosome, and how ribosome association of Get5 is coordinated with the functions of other ribosome-bound protein biogenesis factors (18).
The viral K+ channels Kcv and Kesv possess all the functional hallmarks of eukaryotic inwardly rectifying K+ channels, such as the typical membrane-pore-membrane structure (Fig. 1A) and the highly conserved TXXTXG(Y/F)G motif (19, 20). Kcv consists of only 94 residues (Fig. 1B); due to an N-terminal extension, Kesv is slightly larger (Fig. 1B). Because of their minimal size, Kcv and its homologs have proved excellent models for the elucidation of structural and functional properties of K+ channels, including gating and pharmacology (19, 20). When expressed in Saccharomyces cerevisiae or mammalian cells, Kcv localizes to the plasma membrane, while Kesv localizes to the mitochondria (21). Sorting of Kcv requires transmembrane domain 1 (Kcv-TM1), which acts as an SA sequence in vitro and in vivo (22). The sorting of Kesv remained enigmatic, because on one hand the N-terminal domain of Kesv (Fig. 1B) resembles a mitochondrial targeting peptide (mTP), which can direct green fluorescent protein (GFP) to the mitochondria (21), while on the other hand, deletion of the mTP yields a protein (ΔN-Kesv) (Fig. 1B) that is still sorted to the mitochondria (21). Instead, sorting of Kesv to the mitochondria strongly depends on its C-terminal transmembrane domain 2 (TM2) (21, 23). When TM2 of Kesv (Kesv-TM2) was extended by only two valine residues (Kesv-VV) (Fig. 1B), the localization of Kesv-VV was changed from mitochondrial to plasma membrane (21). Here, we show that Kcv is a substrate of the cotranslational SRP-dependent pathway, while Kesv-VV, even though it is not a TA protein, enters the posttranslational GET pathway. The results indicate a previously unrecognized intersection between the SRP and GET targeting pathways and reveal that SRP and Get4-5 compete for the same RNCs.
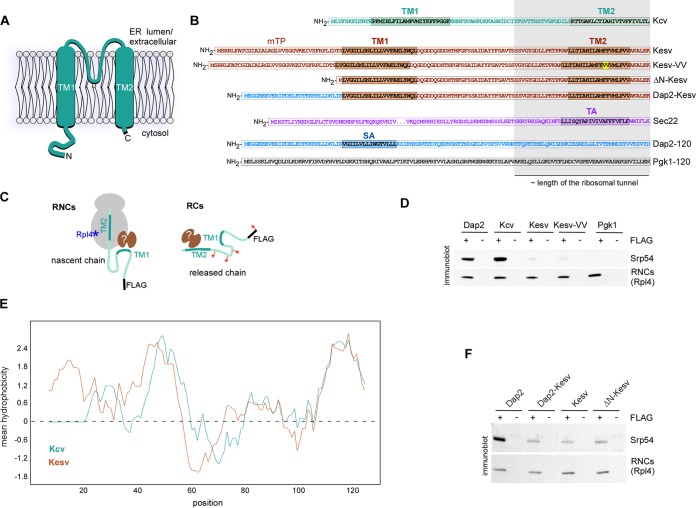
FIG 1.
Kcv-TM1 is a cotranslational substrate of SRP, while Kesv-TM1 is not. (A) Schematic representation of the K+ channel Kcv in the cytoplasmic membrane (20). (B) Reporter proteins. The TM domains are colored. The double valine insertion in TM2 of Kesv-VV is in yellow. The 40 most C-terminal residues of the reporter proteins, corresponding to approximately the portion shielded by the ribosomal exit tunnel, are highlighted in gray. The N-terminal 120 residues of the SRP substrate, Dap2, and of the cytosolic protein Pgk1 served as controls. The middle region of the 214-residue protein Sec22 is not shown. (C) Experimental setup. RNCs carrying full-length Kcv, Kesv, or Kesv-VV were generated via translation of mRNA lacking a stop codon. RCs of full-length Kcv, Kesv, or Kesv-VV were generated via translation of mRNA, including a stop codon. Subsequently, the RNCs or RCs were affinity purified via the FLAG tag (FLAG) fused to the N terminus of the respective reporter protein. The RNCs were detected via immunoblotting of a ribosomal marker protein (e.g., Rpl4 [blue]). In order to detect RCs, [35S]methionine was incorporated into the reporter proteins (red). Factors (brown) associated with FLAG-affinity-purified RNCs or RCs were analyzed via immunoblotting. For details, see Materials and Methods. (D) Affinity-purified RNCs carrying FLAG-tagged (+FLAG) nascent chains as indicated were analyzed for association with SRP via immunoblotting with antibodies directed against the Srp54 subunit. Rpl4 was employed to detect RNCs isolated via the FLAG-tagged nascent chain; untagged (−FLAG) nascent chains served as controls. (E) Kcv and Kesv possess similar hydrophobicity profiles. Shown is a Kyte and Doolittle scale mean hydrophobicity profile of Kcv and Kesv (46). The scan window size was 13. (F) Affinity-purified RNCs carrying nascent chains as indicated were analyzed as described for panel D.
MATERIALS AND METHODS
Plasmids and yeast strains.
Plasmids encoding Dap2 (pSPUTK-Dap2) and Pgk1 (pSPUTK-Pgk1) and the FLAG-tagged versions, pSPUTK-FLAG-Dap2 and pSPUTK-FLAG-Pgk1, which contain an N-terminal DYKDDDDK FLAG peptide, were described previously (24). Genes encoding Kcv, Kesv, and Kesv-VV were amplified from plasmids pYES2-Kcv, pYES2-Kesv, and pYES2-Kesv-113VV (21) and were cloned with or without the FLAG tag into pSP65 (Promega). Dap2-Kesv was constructed by replacing the N-terminal 37 residues of Kesv with the N-terminal 29 residues of Dap2. ΔN-Kesv contains a deletion of residues 2 to 37 of Kesv. Dap2-Kesv, ΔN-Kesv, and full-length Sec22 with or without the FLAG tag were cloned into pSP65. MH272-3f a/α (ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 ade2/ade2) was the parental wild-type strain (25), the Δsrp54 strain was described previously (24). In the Δget4 Δget5 strain (get4::kanMX get5::kanMX), the complete coding sequences of GET4 and GET5 were replaced by the kanMX module amplified by PCR from strain Y02420 or Y16261, respectively (Euroscarf). The Δsgt2 strain carries a replacement of the EcoRI/MfeI fragment within the coding region of SGT2 with the URA3 marker gene. The Δget3 strain carries a replacement of the ClaI/BamH1 fragment within the coding region of GET3 with the HIS3 marker gene. The Δget2 strain (Y10223) and the BY4742 strain (Y10000) employed for the generation of ER microsomes were from Euroscarf.
Generation of RNCs and RCs.
Transcription was performed as described previously using SP6 polymerase (26). DNA templates encoding the N-terminal 120 residues of Dap2 and Pgk1 were generated by PCR using pSPUTK-based plasmids encoding untagged or FLAG-tagged Dap2 or Pgk1, respectively. The pSP65-based plasmids of Kcv, Kesv, Kesv-VV, Dap2-Kesv, ΔN-Kesv, and Sec22 were employed to generate full-length nascent chains or full-length released chains with or without an N-terminal FLAG tag (24, 27). Transcripts lacking a stop codon were employed to generate RNCs, while transcripts containing a stop codon were employed to generate released polypeptide chains (RCs). In the case of Dap2, residue 118 was exchanged for methionine to allow better detection of radiolabeled, released Dap2. Yeast translation extract was prepared as previously described (26) from the wild-type (MH272-3fα), Δsrp54, Δsgt2, or Δget3 strain. Translation reactions were performed for 50 min at 20°C. When released chains were generated, [35S]methionine (Hartmann Analytic GmbH) was added to the translation reaction mixture (24, 27). RNCs were separated from the cytosol by ultracentrifugation at 400,000 × g for 25 min at 4°C. The resulting ribosomal pellets were resuspended in 100 μl pulldown buffer (20 mM HEPES-KOH, pH 7.4, 120 mM potassium acetate [KAc], 2 mM magnesium acetate [MgAc2], 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor mixture [1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, 5 μg/ml pepstatin A]). RCs were separated from ribosomes by two successive ultracentrifugation steps at 400,000 × g for 15 min each at 4°C. Ribosome-free cytosol, which was added to RNCs as indicated, was prepared by ultracentrifugation of glass bead extract (24) at 400,000 × g for 25 min at 4°C. Incubation of RNCs with ribosome-free cytosol was performed at 20°C for 10 min. Subsequently, the RNCs were isolated via ultracentrifugation at 400,000 × g for 15 min at 4°C, and the ribosomal pellets were resuspended in pulldown buffer and used for FLAG pulldown reactions. Low-salt (120 mM KAc)- and high-salt (800 mM KAc)-treated RNCs were isolated via centrifugation through sucrose cushions as previously described (24). Pellets containing ribosomes and RNCs were subsequently resuspended in pulldown buffer and applied to FLAG pulldown reactions.
FLAG pulldown reactions.
FLAG pulldown reactions were performed as described previously (24). In a typical experiment, RNCs or [35S]methionine-labeled RCs were generated in a 120-μl translation reaction mixture as described above. Resuspended RNC pellets or ribosome-free supernatant containing RCs was then added to 40 μl of prewashed anti-FLAG M2 affinity beads (FLAG beads; Sigma) suspended in 500 μl pulldown buffer. Nontagged and FLAG-tagged RNCs/RCs were analyzed in parallel in each experiment to determine background binding. Material bound to FLAG beads was analyzed on 10% Tris-Tricine gels (28), transferred to nitrocellulose membranes, and analyzed via immunoblotting. If 35S-labeled RCs were employed, the lower part of the immunoblot was analyzed via autoradiography. Similar amounts of RNCs or RCs (adjusted with respect to the Rpl4/Rps9 signal or with respect to the intensity of the 35S-labeled chain) were loaded to allow direct comparison of bound SRP (Srp54), Get4-5 (Get4 or Get5), Sgt2, and Get3. The sizes of Srp54 (59.6 kDa), Get5 (23.7 kDa), and Rpl4 (39.1 kDa) allowed the parallel analysis of SRP, Get4-5, and RNCs on a single immunoblot. If RNCs were analyzed for the binding of Sgt2 (37.2 kDa) or Get4 (36.2 kDa), Rps9a (22.7 kDa) was employed as a marker for FLAG affinity purification of RNCs.
ER-targeting assay.
Preparation of high-salt-treated microsomal membranes from wild-type or Δget2 strains was performed as described previously (24). Aliquots of microsomal membranes (A280 = 20 ± 2) were stored at −80°C in storage buffer (20 mM HEPES-KOH, pH 7.4, 120 mM KAc, 250 mM sorbitol, 2 mM MgAc2, 1 mM PMSF, 1× protease inhibitor mixture). To generate RCs and RNCs for ER-targeting assays, 80-μl translation reactions were performed in the presence of [35S]methionine in either a wild-type or a Δget3 translation extract (see above). To obtain RCs, ribosome-free supernatants were collected after centrifugation at 400,000 × g at 4°C for 15 min. To obtain RNCs, ribosomal pellets were collected by centrifugation at 400,000 × g at 4°C and subsequently resuspended in 80 μl pulldown buffer (see above). Twenty microliters of ribosome-free RCs or 20 μl resuspended RNCs was then mixed, complemented with a final concentration of 2 mM ATP and 2 mM GTP, incubated for 10 min at 20°C, and subsequently loaded onto a two-step sucrose gradient (5 ml of 1.2 M sucrose/5 ml of 1.5 M sucrose in storage buffer containing 100 mM instead of 250 mM sorbitol). After centrifugation in a swing-out rotor at 100,000 × g, for 1 h at 4°C, microsomes accumulated at the interphase between the 1.2 and 1.5 M sucrose were collected, precipitated by trichloroacetic acid, and subsequently analyzed on 10% Tris-Tricine gels, followed by autoradiography.
Miscellaneous.
Ribosome association assays were performed as described previously (29). Ribosome profiles were performed basically as described previously (30) with the exception that buffers contained 1 mM dithiothreitol (DTT) and 120 mM KAc. Glucose depletion was performed as described previously (31). Immunoblots were developed using enhanced chemiluminescence (ECL) with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce). Autoradiographs were analyzed with a Typhoon 9410 (GE Healthcare Life Sciences). Densitometric analysis of immunoblots was performed using the AIDA image analyzer (Raytest). Polyclonal antibodies directed against Srp54, Sgt2, Get4, Get5, Get3, Rpl4, Rps9, Rpl24, and Sse1 were raised in rabbits (Eurogentec).
RESULTS
Nascent Kcv-TM1 is a substrate of SRP.
When expressed in a Δtrk1 Δtrk2 strain lacking the two yeast plasma membrane K+ transporters, the K+ channel Kcv complements for growth defects connected to K+ uptake deficiency (21). In contrast, the homologous K+ channel Kesv cannot complement for growth defects of a Δtrk1 Δtrk2 strain because Kesv does not localize to the plasma membrane but is imported into mitochondria (21, 23). However, Kesv-VV can rescue growth defects of a Δtrk1 Δtrk2 strain, indicating that the protein is sorted to the cytoplasmic membrane (21). How Kesv is redirected from the mitochondria to the ER due to the insertion of two valine residues into Kesv-TM2 is not understood. We first analyzed the interaction of SRP (employing the Srp54 subunit of the complex) with RNCs carrying Kcv, Kesv, or Kesv-VV (Fig. 1C, RNCs). The N-terminal 120 residues of Dap2, which contain an SA sequence, and of the cytosolic protein Pgk1 served as positive and negative controls, respectively (Fig. 1B) (27). TM2 of the nascent K+ channel proteins is localized inside the ribosomal exit tunnel, while TM1 is exposed on the outside (Fig. 1B). As expected, SRP was recruited to RNCs carrying Dap2 (Dap2 RNCs), while SRP was not recruited to RNCs carrying Pgk1 (Pgk1 RNCs) (24, 27) (Fig. 1D). SRP was also efficiently recruited to RNCs carrying Kcv (Kcv RNCs), indicating that TM1 of Kcv possessed the properties of a bona fide SA sequence (Fig. 1D), as previously suggested (22). In comparison, only slightly more SRP was recruited to RNCs carrying Kesv or Kesv-VV (Kesv RNCs and Kesv-VV RNCs) than to Pgk1 RNCs (Fig. 1D). Importantly, there was no difference between SRP recruitment to Kesv RNCs and Kesv-VV RNCs (Fig. 1D). The data indicate that (i) TM1 of nascent Kesv/Kesv-VV recruited SRP only poorly and (ii) insertion of two valine residues into TM2 of Kesv did not significantly enhance SRP recruitment to RNCs (Fig. 1D).
Consistent with the experimental data, Kcv had a high prediction score for the secretory pathway, while the scores for Kesv, as well as Kesv-VV, predicted similar likelihoods for the mitochondrion or secretory pathway (Table 1). Kesv lacking the mTP, however, possessed a high prediction score to enter the secretory pathway (Table 1). We thus speculated that Kesv-TM1, which displayed a hydrophobicity profile similar to that of Kcv-TM1 (Fig. 1E), might possess the properties of an SA sequence, the function of which was masked by the preceding mTP, as previously described (32). To test this possibility, we generated reporters in which the mTP was either deleted (ΔN-Kesv) or replaced by the N-terminal domain of Dap2, which precedes the SA sequence of Dap2 (Dap2-Kesv) (Fig. 1B). However, recruitment of SRP to RNCs carrying ΔN-Kesv or Dap2-Kesv was not enhanced (Fig. 1F). Thus, SRP possessed a low affinity for Kesv-TM1 even when the mTP was removed or was replaced by a sequence that precedes the SA of the bona fide SRP substrate, Dap2. The combined data did not favor the possibility that Kesv-VV was redirected from the mitochondria to the ER via the SRP pathway.
TABLE 1.
TargetP localization predictions for Kcv and Kesv
| Protein | Length (amino acids) | Scorea |
Predicted localizationb | Reliability classc | ||
|---|---|---|---|---|---|---|
| mTP | SP | Other | ||||
| Kcv | 94 | 0.020 | 0.935 | 0.135 | Secretory pathway | 1 |
| Kesv | 124 | 0.628 | 0.695 | 0.007 | Secretory pathway | 5 |
| Kesv-VV | 126 | 0.629 | 0.690 | 0.007 | Secretory pathway | 5 |
| ΔN-Kesv | 92 | 0.008 | 0.973 | 0.091 | Secretory pathway | 1 |
The location with the highest score is the most likely, according to TargetP, and the relationship between the scores (the reliability class) may be an indication of how certain the prediction is. mTP, mitochondrial targeting peptide; SP, signal peptide; Other, score on which the final prediction was based.
The predicted localization is based on the score; the possible values are mitochondrion, secretory pathway, and other.
The reliability class ranges from 1 to 5, where 1 indicates the strongest prediction. For details compare http://www.cbs.dtu.dk/ and reference 45.
Released Kesv-VV-TM2 is a substrate of the GET pathway.
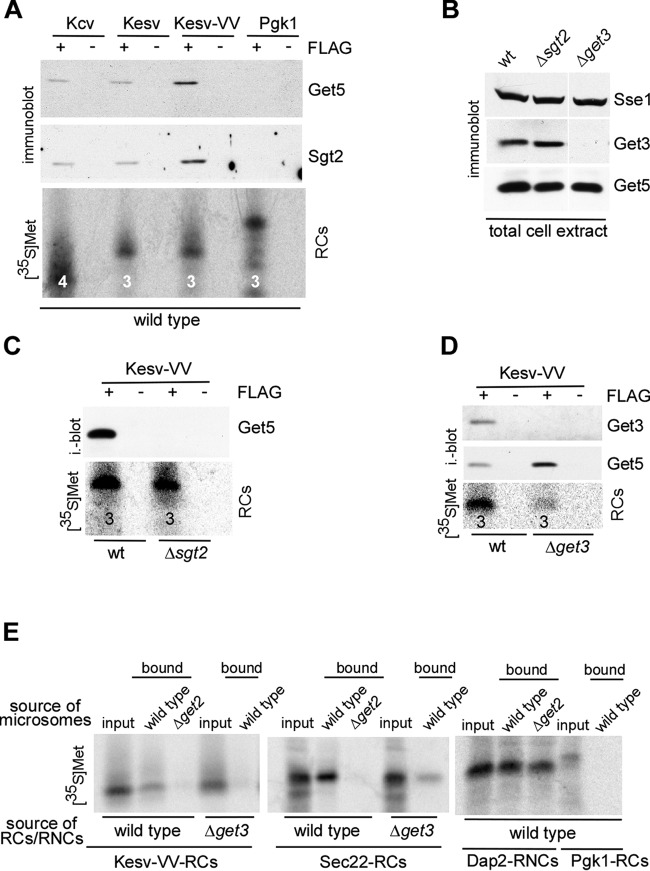
Kesv-TM2 is located very close to the C terminus (Fig. 1B). Thus, we speculated that Kesv, even though it contained two TM domains, may resemble a TA protein that was posttranslationally targeted to the ER via the GET pathway (1–3, 7, 8). To test this possibility, we analyzed the association of Sgt2 and Get4-5 with RCs of Kcv, Kesv, and Kesv-VV (Fig. 1C, RCs). Sgt2 and Get4-5 were most strongly associated with released Kesv-VV and weakly associated with Kcv and Kesv and were not associated with Pgk1 (Fig. 2A). Importantly, direct comparison of Kesv and Kesv-VV revealed that the association of Sgt2 and Get4-5 was significantly enhanced by the insertion of two valine residues into Kesv-TM2 (Fig. 2A). We next tested if binding of Get4-5 to released Kesv-VV was dependent on Sgt2 or Get3, employing translation extracts derived from a Δsgt2 or Δget3 strain. The expression level of Get4-5 was not altered in Δsgt2 or Δget3 cells (Fig. 2B), however, association of Get4-5 with released Kesv-VV was abolished in the absence of Sgt2 (Fig. 2C). In contrast, in the absence of Get3, a larger fraction of released Kesv-VV was associated with Get4-5 (Fig. 2D). The findings suggested that Kesv-VV was targeted to the ER via the GET pathway. To test this directly, we compared the targeting of Kesv-VV RCs to microsomal membranes in a wild-type setup, in the absence of Get3, or in the absence of the GET pathway membrane receptor, Get2 (see the introduction). As a negative control, we employed Pgk1 RCs (Fig. 1B), which, as expected, did not bind to microsomal membranes (Fig. 2E). As a positive control, we employed the tail-anchored natural GET pathway substrate, Sec22 (Fig. 1B) (33), which was efficiently targeted to microsomal membranes (Fig. 2E). Consistent with current models, binding of Sec22 RCs to microsomes was strongly reduced when the microsomes were derived from a Δget2 strain or Sec22 RCs were prepared in a Δget3 translation extract. A side-by-side analysis revealed that Kesv-VV RCs resembled Sec22 RCs and were efficiently targeted to microsomes in a Get2- and Get3-dependent manner (Fig. 2E). In comparison, targeting of the SRP substrate Dap2 RNCs to microsomal membranes was independent of Get2 (Fig. 2E). The combined findings are consistent with current models of the GET pathway (1–3, 7, 8). Released Kesv-VV was bound to Sgt2, which formed a complex with Get4-5 (Fig. 2A and C). From there, a fraction of Kesv-VV was transferred to Get3 (Fig. 2D) and thus remained trapped on Sgt2/Get4-5, when Get3 was absent (Fig. 2D). Subsequent posttranslational targeting of Kesv-VV to the ER membrane resembled the targeting of Sec22 with respect to Get3 and Get2 dependence (Fig. 2E). Based on the data, we suggest that Kesv-VV is a GET pathway substrate and can reach the plasma membrane posttranslationally. This model explains why Kcv and Kesv-VV can both complement growth defects of a yeast Δtrk1 Δtrk2 strain (21), even though Kesv-VV is not a substrate of SRP (Fig. 1D).
FIG 2.
Kesv-VV is targeted to the ER membrane posttranslationally via the GET pathway. (A) Affinity-purified 35S-labeled RCs of the indicated FLAG-tagged reporter proteins (+FLAG) were analyzed for association of Get4-5 (Get5) and Sgt2 via immunoblotting. (B) Total extract derived from wild-type (wt), Δsgt2, and Δget3 strains was analyzed for the expression of Get3 and Get5 via immunoblotting. Sse1 served as a loading control. (C) RCs of FLAG-Kesv-VV (+FLAG) were generated in a wt or Δsgt2 translation extract and subsequently affinity purified and analyzed for association of Get4-5 (Get5). (D) RCs of FLAG-Kesv-VV (+FLAG) were generated in a wt or Δget3 translation extract and subsequently affinity purified and analyzed for association of Get4-5 (Get5) and Get3. Untagged RCs served as controls. The number of methionine residues of each RC is indicated below the corresponding band in the autoradiograph. (E) 35S-labeled RCs or RNCs as indicated were generated in wild-type or Δget3 translation extract and then incubated with ER microsomes derived from wild-type or Δget2 cells. Microsomes and associated RCs/RNCs were subsequently purified via sucrose gradient centrifugation and analyzed via autoradiography.
Get4-5 is ribosome associated while Get3 and Sgt2 are not.
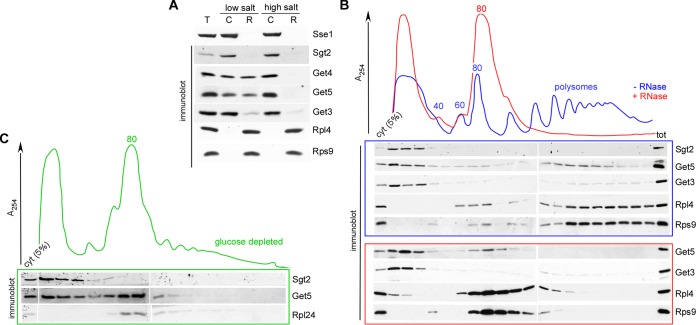
A possible cotranslational role of the GET pathway is suggested by the finding that the mammalian homologs of Get4 and Get5, termed TRC35 and Ubl4, together with an additional subunit termed Bag6, interact with ribosomes (2, 3, 7, 34). We thus tested ribosome association of the GET pathway components in a total yeast extract. In this experiment, ribosomes were separated from the cytosol under low- or high-salt conditions. Cytosolic proteins are recovered in the supernatant, while ribosomal proteins are recovered in the pellet independent of the salt concentration. Ribosome-associated proteins, however, are released to the supernatant under high-salt conditions. Sgt2 behaved like a typical cytosolic protein (Fig. 3A), while Get4-5 was equally distributed between ribosomes and cytosol under low-salt conditions and was fully released under high-salt conditions (Fig. 3A). In the case of Get3, a minor fraction was detected in the pellet under low- as well as high-salt conditions. Thus, Sgt2 was cytosolic and Get4-5 behaved like a ribosome-associated factor, while in the case of Get3, the outcome was not fully conclusive. Ribosome profiles confirmed the cytosolic localization of Sgt2 and the distribution of Get4-5 between a cytosolic and a ribosome-associated pool (Fig. 3B). The major fraction of Get3 was detected in the cytosolic fractions of the ribosome profile; however, a small fraction of Get3 smeared through the high-molecular-mass fractions of the sucrose gradient (Fig. 3B). To determine if this minor pool of Get3 was bound to translating ribosomes, cell extracts were treated with RNase A to destroy polysomes and accumulate 80S ribosomes prior to the analysis (30). Under these conditions, ribosome-associated proteins accumulate in the 80S peak. This was indeed the case for Get4-5; however, the minor fraction of Get3 was still distributed throughout the high-molecular-mass fractions of the gradient (Fig. 3B). The data indicate that Sgt2 and Get3 are localized to the cytosol, while a substantial fraction of Get4-5 was ribosome associated at steady state.
FIG 3.
Get4-5 is distributed between a cytosolic and a ribosome-bound pool. (A) Ribosome association of GET pathway components under low- and high-salt conditions. Total (T) extract prepared from wild-type cells was separated into a cytosolic supernatant (C) and a ribosomal pellet (R) via a sucrose cushion under low- or high-salt conditions. Aliquots were subsequently analyzed via immunoblotting using the antibodies indicated. The ribosomal proteins Rpl4 and Rps9 served as ribosomal markers, and Sse1 served as a cytosolic marker. (B) Ribosome profiles. Cytosolic extract prepared from glucose-grown wild-type cells was separated into polysomes, 80S ribosomes, and ribosomal subunits (60S and 40S) via sucrose density gradient centrifugation. Ribosome profiles were performed with untreated (blue) or RNase-treated (red) cell extract. Due to RNase treatment, polysomes are destroyed and accumulate in an 80S ribosomal peak. Fractions were analyzed via immunoblotting using the antibodies indicated. cyt, 5% of the cytosolic extract loaded onto the gradient; tot, total cell extract of the wild-type strain, which was loaded to confirm proper immunodetection of proteins in the polysome fractions. (C) Ribosome profile after glucose depletion. The experiment was performed as described for panel B, with the exception that cytosolic extracts were prepared after 10 min of glucose depletion. Under these conditions, the 80S peak represents nontranslating ribosomes.
The ribosome-bound fraction of Get4-5 was distributed between the 80S and polysome fractions when cell extracts were derived from glucose-grown cells (Fig. 3B, − RNase). Under these conditions, the 80S peak represents a mixture of translating and nontranslating ribosomes. When cells are briefly depleted of glucose, translation initiation is blocked and the 80S peak consists mainly of nontranslating ribosomes (35). To test if Get4-5 was associated with nontranslating ribosomes, we analyzed ribosome profiles from glucose-depleted cells (Fig. 3C). A significant fraction of Get4-5 but not of Sgt2 comigrated with nontranslating 80S ribosomes, indicating that Get4-5 was able to interact with ribosomes in the absence of nascent substrate proteins. If one takes into account that a yeast cell contains about 300,000 ribosomes but only about 6,000 Get4-5 heterodimers (18, 36), the findings indicate that a small subpopulation of ribosomes (about every hundredth one) was associated with Get4-5 at steady state. The data suggest that Get4-5 may be bound to ribosomes dynamically in order to scan the ribosomes for possible GET pathway substrates.
The TA of Sec22, but not Kesv-VV-TM2, enhances Get4-5 recruitment to RNCs.
In the mammalian system, the TRC35/Ubl4/Bag6 complex is recruited to ribosomes synthesizing membrane proteins when the TM domain is still inside the ribosomal tunnel (34). Because after ribosome release Kesv-VV-TM2 was a substrate of Sgt2/Get4-5 (Fig. 2A), we speculated that Kesv-VV RNCs might recruit Get4-5 to ribosomes above the basal binding level (see above) (Fig. 3). While Get4-5 was indeed associated at a low basal level with any of the RNCs tested, the occupation of Kesv-VV RNCs was not enhanced compared to Kesv RNCs or Pgk1 RNCs (Fig. 4A, panel 1). The finding suggested that the heterologous viral protein Kesv-VV was not fully functional with respect to Get4-5 recruitment to RNCs. To test this possibility, we employed the GET pathway substrate Sec22 (Fig. 1B). Indeed, recruitment of Get4-5 to Sec22 RNCs was enhanced about 2-fold compared to Pgk1 RNCs (Fig. 4B, panel 1). Moreover, Sec22 RNCs recruited not only Get4-5 but also SRP (Fig. 4B, panel 1). These observations resemble previous findings in the mammalian system (34, 37) and indicate that a bona fide TA segment inside the yeast ribosomal tunnel can enhance Get4-5 binding to ribosomes above the basal level. Kesv-VV-TM2 was seemingly too weak to trigger recruitment of Get4-5 from within the ribosomal tunnel (Fig. 4A, panel 1). Note, however, that after release from the ribosome, Kesv-VV-TM2 recruited Get4-5 efficiently (Fig. 2A and C).
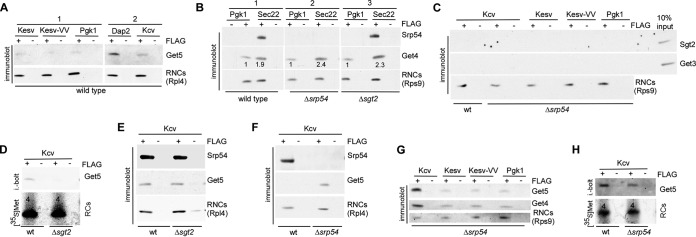
FIG 4.
Binding of Get4-5 is enhanced if RNCs contain a TA segment inside the ribosomal tunnel or expose an SA outside the ribosomal tunnel. (A) Get4-5 shows basal binding to a variety of RNCs and enhanced binding to Dap2 RNCs. The Rpl4 blot is identical to the Rpl4 blot shown in Fig. 1D, because Srp54 and Get5 were analyzed in the same experiment. The immunoblots shown were derived from a single exposure of the same blot. (B) Get4-5 is recruited to Sec22 RNCs. The immunoblots shown were derived from a single exposure of the same blot. The numbers below the Get4 bands indicate the relative amounts of Get4 bound to Sec22 RNCs compared to Pgk1 RNCs. To allow a direct comparison, each Get4 band was normalized to the corresponding Rps9 band. (C) Sgt2 and Get3 do not interact with RNCs. Ten percent of the translation reaction mixture (10% input) was loaded to control for proper detection of Sgt2 and Get3. The Δsrp54 strain was employed, because Get4-5 binding in the absence of SRP was enhanced. (D) Sgt2 is required for the posttranslational binding of Get4-5 to Kcv RCs. (E) Sgt2 is not required for the cotranslational binding of Get4-5 to Kcv RNCs. (F) Get4-5 binding to Kcv RNCs is enhanced in the absence of SRP. (G) Get4-5 binding to Kesv RNCs, Kesv-VV RNCs, or Pgk1 RNCs is not affected by the absence of SRP. (H) Posttranslational binding of Get4-5 to Kcv RCs is not affected in the absence of SRP. (A to H) RNCs of FLAG-tagged (+FLAG) or untagged (−FLAG) reporter proteins as indicated were generated in translation extracts derived from a wt, Δsrp54, or Δsgt2 strain; isolated via FLAG affinity purification; separated on 10% Tris-Tricine gels; and subsequently analyzed for the association of Get4-5 (Get4 or Get5), SRP (Srp54), Get3, or Sgt2 via immunoblotting. Rpl4 or Rps9 was employed to detect RNCs isolated via the FLAG-tagged nascent chain. The number of methionine residues within 35S-labeled RCs is indicated above the corresponding band in the autoradiograph.
Consistent with the cytosolic localization of Sgt2 and Get3 in ribosome binding experiments (Fig. 3), neither Sgt2 nor Get3 was associated with RNCs (Fig. 4C). Because Sgt2 was required for the association of Get4-5 with released Kesv-VV (Fig. 2C) and also released Kcv (Fig. 4D), we tested if Sgt2 affected the recruitment of Get4-5 to RNCs without becoming stably associated with the RNCs. However, this was not the case. Get4-5 binding to Sec22 RNCs (Fig. 4B, panel 3), as well as to Kcv RNCs (Fig. 4E), was not reduced in the absence of Sgt2. Thus, while Sgt2 was strictly required for the posttranslational association of Get4-5 with RCs (Fig. 2C and 4D), Sgt2 itself was not associated with RNCs (Fig. 4C) and also was not required for the association of Get4-5 with RNCs (Fig. 4B, panel 3, and E).
SRP and Get4-5 compete for RNCs exposing nascent Kcv-TM1.
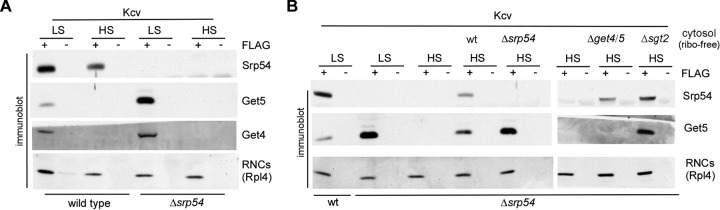
Rather unexpectedly, Dap2 RNCs, and to a lesser extent also Kcv RNCs, recruited Get4-5 above the basal level (Fig. 4A, panel 2). Because exactly these RNCs expose an SA segment outside the ribosomal tunnel (Fig. 1B) and recruit SRP (Fig. 1D), the finding suggested possible binding competition between SRP and Get4-5. Indeed, binding of Get4-5 to Kcv RNCs was significantly enhanced when the RNCs were generated in a Δspr54 translation extract (Fig. 4F). In contrast, Get4-5 binding to Kesv RNCs, Kesv-VV RNCs, Pgk1 RNCs (Fig. 4G), and Sec22 RNCs (Fig. 4B, panel 2), and also to released Kcv RCs (Fig. 4H), was not affected by SRP. The data are compatible with a model in which Get4-5 and SRP compete specifically for the RNCs that expose a nascent SA segment outside the ribosomal tunnel. To further characterize the competition between SRP and Get4-5, we generated RNCs free of SRP and Get4-5. Because SRP was bound to Kcv RNCs in a salt-resistant manner (Fig. 5A), translation was performed in a Δsrp54 translation extract, and Get4-5 was subsequently stripped off RNCs via high-salt treatment (Fig. 5). The SRP/Get4-5-free Kcv RNCs were then complemented with ribosome-free cytosol derived from wild-type, Δsrp54, Δget4 Δget5, or Δsgt2 cells (Fig. 5B). Wild-type ribosome-free cytosol allowed the rebinding of SRP and Get4-5 (Fig. 5B). When Δsrp54 ribosome-free cytosol was added, significantly more Get4-5 was able to rebind to Kcv RNCs, while addition of a Δget4 Δget5 ribosome-free cytosol did not enhance rebinding of SRP (Fig. 5B). Addition of Δsgt2 ribosome-free cytosol allowed the efficient rebinding of Get4-5 to RNCs (Fig. 5B), confirming that Get4-5 binding to RNCs occurred independently of Sgt2. The combined data indicate that the binding of SRP and Get4-5 to Kcv RNCs was competitive. In this competition, SRP binding was strongly favored, and thus, Get4-5 was efficiently recruited only when SRP was absent.
FIG 5.
SRP and Get4-5 compete for binding to Kcv RNCs. (A) Binding of SRP to Kcv is salt resistant, while binding of Get4-5 in the absence of SRP is salt sensitive. RNCs carrying FLAG-tagged (+FLAG) or untagged (−FLAG) Kcv were generated in a wild-type or Δsrp54 translation extract. Ribosomes and RNCs were then isolated via a low-salt (LS) or a high-salt (HS) sucrose cushion. Subsequently, the RNCs were affinity purified. Binding of Get4-5 (Get4 and Get5) and SRP (Srp54) to RNCs was analyzed via immunoblotting. (B) RNCs carrying FLAG-tagged (+FLAG) Kcv were generated in a wt or Δsrp54 translation extract. The RNCs were then isolated via an LS or an HS sucrose cushion and then applied to FLAG affinity purification reactions. If indicated, ribosome-free cytosol (cyt) prepared from wt, Δsrp54, Δsgt2, or Δget4 Δget5 cells was added to the ribosomes/RNCs prior to affinity purification. Binding of Get4-5 (Get5) and SRP (Srp54) to RNCs was analyzed via immunoblotting. Rpl4 was employed to detect RNCs isolated via the FLAG-tagged nascent chain.
DISCUSSION
TA proteins contain an N-terminal functional domain, which is exposed to the cytosol; a single TM domain, which can be sorted to a variety of cellular compartments; and a short C-terminal domain of no more than 30 residues (6). Because the C-terminal TM domain contains the targeting information, TA proteins cannot be sorted to the ER via a cotranslational route (1, 2, 6–8). Instead, TA proteins are delivered to the ER via the posttranslational GET pathway, which involves specific protein machinery identified only recently (1–3, 7, 8). In contrast, sorting of TA proteins to the mitochondrial outer membrane (MOM) seemingly does not require specific machinery. For example, targeting of the mitochondrial TA protein Fis1 depends on the lipid composition and does not require protein components (6, 38). Whether a TA protein is sorted to the mitochondria or to the ER critically depends on the properties of its TM domain (6, 39). Moderate hydrophobicity favors targeting to the MOM; the TM length and C-terminal charge also contribute to targeting (6, 39).
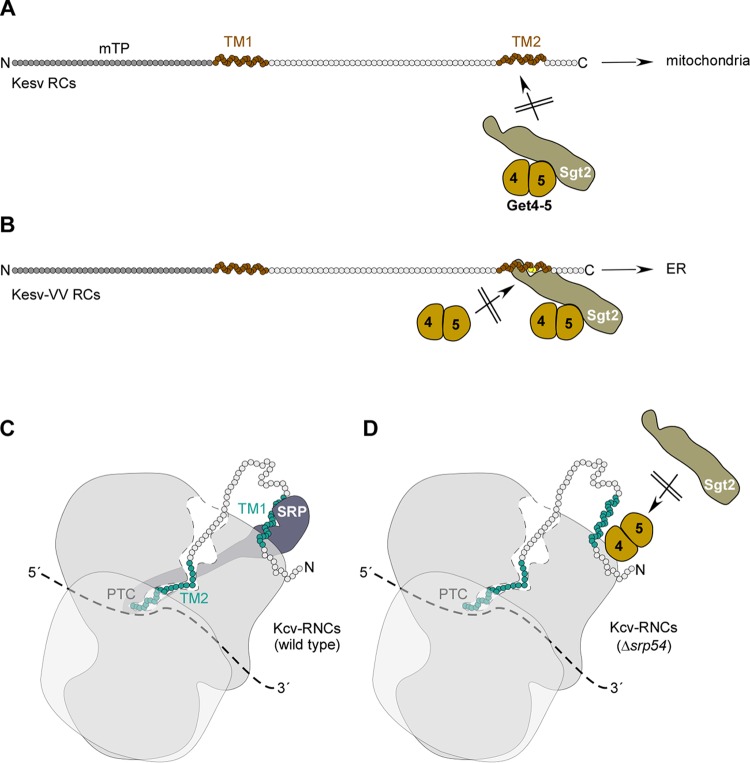
In this work, we asked how the insertion of only two valine residues into the C-terminal TM2 of Kesv can redirect the protein from the mitochondria to the ER (21). Kesv contains two TM domains and thus does not resemble a classical TA protein. Moreover, the C terminus of Kesv-VV TM2 is localized in the cytosol, which is opposite to the orientation of TA domains. We therefore initially did not favor the idea that Kesv-VV is a GET pathway substrate. Rather, we speculated that SRP was responsible for redirecting Kesv-VV from the mitochondria to the ER, because recruitment of SRP to RNCs was possibly affected by the nascent chain inside the ribosomal tunnel, as previously shown for other TM domains (27, 34, 37, 40). However, SRP was not recruited to Kesv-VV RNCs. Instead, after ribosome release, Kesv-VV, but not Kesv, was a substrate of the soluble GET pathway components Sgt2, Get4-5, and Get3. Moreover, ER targeting of Kesv-VV was dependent on Get3 and the membrane receptor Get1-2. The sorting of Kesv versus Kesv-VV resembles the sorting of Fis1 versus the mutant Fis1-4L, in which 4 residues within the TA are replaced by leucine residues, resulting in redirection from the mitochondria to the ER (41). Consistent with our observations, Sgt2 binds to Fis1-4L but does not bind to Fis1 (14).
Our findings also provide an explanation for the puzzling observation that the mTP of Kesv can direct a reporter to the mitochondria but is not essential for mitochondrial targeting of Kesv itself (21). Seemingly, Kesv contains two independent mitochondrial targeting signals, namely, the mTP and TM2 (Fig. 6A and B). This finding suggests that sorting of Kesv to the mitochondria is crucial for the propagation of ectocarpus siliculosus virus 1 (EsV-1), which encodes Kesv (20, 21). Indeed, it was suggested that Kesv may be part of an early antiapoptotic system relevant for viral persistence during the lysogenic phase (42). This would also explain why paramecium bursaria chlorella virus 1 (PBCV-1), which encodes Kcv, does not require a mitochondrially localized K+ channel (42). Rather, Kcv is located in the internal viral membrane, which presumably originates from the host ER membrane and fuses with the host plasma membrane upon infection. In this case, the K+ channel is required for the lytic life style of the virus, because it leads to depolarization, K+ loss, and a decrease of internal turgor, facilitating viral-DNA ejection (20, 42, 43).
FIG 6.
Model of Sgt2/Get4-5 association with RCs versus RNCs. (A) Kesv does not interact with Sgt2/Get4-5 posttranslationally and is targeted to the mitochondria via the N-terminal mTP and possibly also via TM2. (B) Kesv-VV interacts posttranslationally with Sgt2/Get4-5 via TM2 and enters the GET pathway to the ER. Get4-5 does not bind to Kesv-VV directly but only via Sgt2. Thus, in the absence of Sgt2, Get4-5 does not associate with Kesv-VV. (C) Kcv RNCs recruit SRP cotranslationally via TM1, which acts as a signal-anchor sequence and targets Kcv to the ER. (D) In the absence of SRP, Kcv RNCs recruit Get4-5. Cotranslational recruitment of Get4-5 to Kcv RNCs is Sgt2 independent and competitive with SRP. For details, see Discussion.
Current models of the GET pathway suggest that Sgt2 might functionally replace mammalian Bag6, which does not possess a yeast homolog, in the ribosome-associated TRC35/Ubl4/Bag6 complex (1, 3). If this model was correct, we expected to find ribosome-associated Get4-5 in a complex with Sgt2. However, this was not the case; while a significant fraction of Get4-5 was associated with ribosomes, Sgt2 was not. One possible explanation for this observation is that the interaction of Sgt2 with Get4-5 was too transient to be detected. However, because Sgt2 was not even required for the recruitment of Get4-5 to RNCs, we favor a model in which the cotranslational role of Get4-5 is Sgt2 independent (Fig. 6C and D) and in which Sgt2 is not a functional counterpart of mammalian Bag6. This model is consistent with the presence of a human Sgt2 homolog termed hSgt2, which itself functionally interacts with Bag6 (7, 44). It should be emphasized, however, that, consistent with numerous previous observations (1–3, 7, 8), the posttranslational interaction of Get4-5 with the GET pathway substrate Kesv-VV was strictly dependent on Sgt2.
It was previously reported that Get5 (also called Tma24) copurifies with ribosomal complexes (17). However, it remained unclear if other components of the GET pathway also interact with ribosomes. Moreover, it was not known what fraction of Get5 is ribosome bound and if the association depends on nascent substrate proteins. Here, we followed up on these questions and showed that a significant fraction of Get4-5 but not of Sgt2 or Get3 was associated with vacant as well as with translating ribosomes. We found that this basal occupation of ribosomes and RNCs by Get4-5 was enhanced in two different situations: first, if a TA segment of a GET pathway substrate was inside the ribosomal tunnel and, second, if an SA segment of an SRP pathway substrate was exposed outside the ribosomal tunnel. In the latter case, the binding of Get4-5 was competitive with the binding of SRP (Fig. 6C and D). The ability of Get4-5 to recognize and interact cotranslationally with SRP substrate RNCs suggests that the GET pathway may serve as a backup system if SRP function is compromised or limiting. Alternatively, recruitment of Get4-5 to the ribosome might be a mechanism to transfer membrane proteins that do not belong to the TA protein family and at the same time are only poorly recognized by SRP to the GET pathway (5). Interestingly, not only was the Get4-5 module of the GET pathway recruited to RNCs carrying an SRP substrate, but SRP was also efficiently recruited to RNCs carrying a bona fide GET pathway substrate. The interplay between the GET and SRP targeting pathways appears to be more complex than previously anticipated.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 746, DFG RO 1028/5-1, and DFG TH 558/18-2) and by the Excellence Initiative of the German federal and state governments (BIOSS-2).
Funding Statement
This work was funded by grants from the Deutsche Forschungsgemeinschaft (DFG) to Sabine Rospert (BIOSS-2, SFB 746, and RO 1028/5-1) and to Gerhard Thiel (TH 558/8-2).
REFERENCES
- 1.Shao S, Hegde RS. 2011. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol 27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegde RS, Keenan RJ. 2011. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ast T, Schuldiner M. 2013. All roads lead to Rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit Rev Biochem Mol Biol 48:273–288. doi: 10.3109/10409238.2013.782999. [DOI] [PubMed] [Google Scholar]
- 4.Nyathi Y, Wilkinson BM, Pool MR. 2013. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim Biophys Acta 1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Ast T, Cohen G, Schuldiner M. 2013. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Borgese N, Fasana E. 2011. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Chartron JW, Clemons WM Jr, Suloway CJ. 2012. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol 22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denic V, Dotsch V, Sinning I. 2013. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb Perspect Biol 5:a013334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rome ME, Chio US, Rao M, Gristick H, Shan SO. 2014. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc Natl Acad Sci U S A 111:E4929–E4935. doi: 10.1073/pnas.1411284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gristick HB, Rao M, Chartron JW, Rome ME, Shan SO, Clemons WM Jr. 2014. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol 21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Chan C, Weir NR, Denic V. 2014. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 512:441–444. doi: 10.1038/nature13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. 2015. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Brown EC, Mak G, Zhuang J, Denic V. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YW, Chuang YC, Ho YC, Cheng MY, Sun YJ, Hsiao CD, Wang C. 2010. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem 285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM Jr. 2010. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci U S A 107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev 20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raue U, Oellerer S, Rospert S. 2007. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem 282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- 19.Plugge B, Gazzarrini S, Nelson M, Cerana R, Van Etten JL, Derst C, DiFrancesco D, Moroni A, Thiel G. 2000. A potassium channel protein encoded by chlorella virus PBCV-1. Science 287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 20.Thiel G, Baumeister D, Schroeder I, Kast SM, Van Etten JL, Moroni A. 2011. Minimal art: or why small viral K(+) channels are good tools for understanding basic structure and function relations. Biochim Biophys Acta 1808:580–588. doi: 10.1016/j.bbamem.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Balss J, Papatheodorou P, Mehmel M, Baumeister D, Hertel B, Delaroque N, Chatelain FC, Minor DL Jr, Van Etten JL, Rassow J, Moroni A, Thiel G. 2008. Transmembrane domain length of viral K+ channels is a signal for mitochondria targeting. Proc Natl Acad Sci U S A 105:12313–12318. doi: 10.1073/pnas.0805709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson HR, Wunderley L, Andreou T, Warwicker J, High S. 2013. Reorientation of the first signal-anchor sequence during potassium channel biogenesis at the Sec61 complex. Biochem J 456:297–309. doi: 10.1042/BJ20130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Charpuis C, Meckel T, Moroni A, Thiel G. 2015. The sorting of a small potassium channel in mammalian cells can be shifted between mitochondria and plasma membrane. Cell Calcium 58:114–121. doi: 10.1016/j.ceca.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Berndt U, Gölz H, Tais A, Oellerer S, Wölfle T, Fitzke E, Rospert S. 2012. NAC functions as a modulator of SRP during the early steps of protein targeting to the ER. Mol Biol Cell 23:3027–3040. doi: 10.1091/mbc.E12-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heitman J, Movva NR, Hiestand PC, Hall MN. 1991. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 88:1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia PD, Hansen W, Walter P. 1991. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol 194:675–682. doi: 10.1016/0076-6879(91)94049-I. [DOI] [PubMed] [Google Scholar]
- 27.Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. 2009. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci U S A 106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci U S A 98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiabudini M, Conz C, Reckmann F, Rospert S. 2012. RAC/Ssb is required for translational repression induced by polylysine segments within nascent chains. Mol Cell Biol 32:4769–4779. doi: 10.1128/MCB.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Plehwe U, Berndt U, Conz C, Chiabudini M, Fitzke E, Sickmann A, Petersen A, Pfeifer D, Rospert S. 2009. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev 23:2102–2115. doi: 10.1101/gad.529409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyazaki E, Kida Y, Mihara K, Sakaguchi M. 2005. Switching the sorting mode of membrane proteins from cotranslational endoplasmic reticulum targeting to posttranslational mitochondrial import. Mol Biol Cell 16:1788–1799. doi: 10.1091/mbc.E04-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamagata A, Mimura H, Sato Y, Yamashita M, Yoshikawa A, Fukai S. 2010. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells 15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 34.Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. 2010. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashe MP, De Long SK, Sachs AB. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 37.Voorhees RM, Hegde RS. 2015. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 4:e07975. doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D. 2008. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci 121:1990–1998. doi: 10.1242/jcs.024034. [DOI] [PubMed] [Google Scholar]
- 39.Fueller J, Egorov MV, Walther KA, Sabet O, Mallah J, Grabenbauer M, Kinkhabwala A. 2015. Subcellular partitioning of protein tyrosine phosphatase 1B to the endoplasmic reticulum and mitochondria depends sensitively on the composition of its tail anchor. PLoS One 10:e0139429. doi: 10.1371/journal.pone.0139429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Wölfle T, Rospert S. 2013. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J Biol Chem 288:33697–33707. doi: 10.1074/jbc.M113.508283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. 2003. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem 278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- 42.Hamacher K, Greiner T, Ogata H, Van Etten JL, Gebhardt M, Villarreal LP, Cosentino C, Moroni A, Thiel G. 2012. Phycodnavirus potassium ion channel proteins question the virus molecular piracy hypothesis. PLoS One 7:e38826. doi: 10.1371/journal.pone.0038826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neupartl M, Meyer C, Woll I, Frohns F, Kang M, Van Etten JL, Kramer D, Hertel B, Moroni A, Thiel G. 2008. Chlorella viruses evoke a rapid release of K+ from host cells during the early phase of infection. Virology 372:340–348. doi: 10.1016/j.virol.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Winnefeld M, Grewenig A, Schnolzer M, Spring H, Knoch TA, Gan EC, Rommelaere J, Cziepluch C. 2006. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res 312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 46.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98. [Google Scholar]