Abstract
Cell growth and proliferation require the coordinated activation of many cellular processes, including cap-dependent mRNA translation. MicroRNAs oppose cap-dependent translation and set thresholds for expression of target proteins. Emerging data suggest that microRNA function is enhanced by cellular activation due in part to induction of the RNA-induced silencing complex (RISC) scaffold protein GW182. In the current study, we demonstrate that increased expression of GW182 in activated or transformed immune cells results from effects of phosphoinositol 3-kinase–Akt–mechanistic target of rapamycin (PI3K-Akt-mTOR) and Jak-Stat-Pim signaling on the translation of GW182 mRNA. Both signaling pathways enhanced polysome occupancy and eukaryotic initiation factor 4E (eIF4E) binding to the 5′ 7mG cap of GW182 mRNA. The effect of Jak-Stat-Pim signaling on polysome occupancy and expression of GW182 protein was greater than that of PI3K-Akt-mTOR signaling, likely resulting from enhanced eIF4A-dependent unwinding of G-quadruplexes in the 5′ untranslated region of GW182 mRNA. Consistent with this, GW182 expression and microRNA function were reduced by inhibition of mTOR or Pim kinases, translation initiation complex assembly, or eIF4A function. Taken together, these data provide a mechanistic link between microRNA function and cap-dependent translation that allows activated immune cells to maintain microRNA-mediated repression of targets despite enhanced rates of protein synthesis.
INTRODUCTION
Cell growth and proliferation are tightly regulated processes that often become dysregulated following oncogenic transformation. Commonly, transforming events activate cellular signaling pathways in a manner that bypasses normal checkpoints used by nontransformed cells to control growth and proliferation. Two intracellular signaling pathways that are constituents of normal immune cell activation and are aberrantly turned on in many malignancies are the phosphoinositol 3-kinase–Akt–mechanistic target of rapamycin (PI3K-Akt-mTOR) pathway and the Jak-Stat-Pim pathway (1, 2). These two pathways converge on the translation initiation complex, termed eukaryotic initiation factor 4F (eIF4F), to facilitate increased protein synthesis required for cells to grow and divide (3).
The translation initiation complex is a multiprotein complex that binds the 7-methyl-guanosine (7mG) cap at the 5′ end of cytoplasmic mRNA through eIF4E, unwinds complex secondary structure within mRNA 5′ untranslated regions (UTRs) through the helicase activity of eIF4A and allows the 43S preinitiation complex to associate with and scan mRNA 5′ UTRs for initiation codons (4). Several lines of evidence indicate that PI3K-Akt-mTOR and Jak-Stat-Pim signaling bolster eIF4F function through phosphorylation of eIF4E-binding proteins (4E-BPs), thus freeing eIF4E from its inhibitory interaction with 4E-BPs (5, 6). A relatively small group of mRNAs containing 5′ terminal oligopyrimidine (TOP) or TOP-like motifs are directly regulated by eIF4E and its essential cofactor eukaryotic initiation factor 4G1 (eIF4G1) (7). TOP motif-containing mRNAs are highly enriched for those coding for ribosomal subunits and other components of translational machinery, resulting in increased cellular capacity for mRNA translation. However, this increased capacity does not enhance translation of all capped transcripts equally. Transcripts associated with cell growth and proliferation often contain complex 5′ UTRs, allowing for an additional level of translational regulation by the RNA helicase eIF4A (8). Helicase activity of eIF4A is regulated by interaction with eIF4B and Pdcd4, both of which are putative targets of the PI3K-Akt-mTOR and Jak-Stat-Pim signaling pathways (9). Therefore, upstream signaling has the potential to regulate two steps of translation initiation, binding of eIF4E to the 7mG cap and unwinding of the 5′ UTR by eIF4A. This mechanism has the potential to enhance translation of specific subsets of mRNAs required for appropriate responses to upstream signals.
In contrast to the translation initiation complex, microRNAs limit the translation and/or stability of specific mRNA transcripts with partially complementary nucleotide sequence in their 3′ UTRs. In recent years, the molecular details of how microRNAs repress translation and cause the degradation of target mRNAs have been extensively characterized (10). Two families of proteins, Argonaute proteins and GW182 family proteins, have emerged as critical mediators of microRNA function (11). Argonaute proteins (Ago1, Ago2, Ago3, and Ago4 in mammals) bind to, stabilize, and allow mature microRNAs to base pair with their targets (11). Recently, we reported that in most adult tissues, Ago2-microRNA complexes are long-lived and not associated with mRNA targets (12, 13). Like Argonaute proteins, GW182 family proteins (GW182, Tnrc6b, and Tnrc6c in mammals) are necessary for microRNA-mediated repression of target mRNA (14, 15). To mediate their effect, GW182 family proteins function as scaffolds to bridge Argonaute-microRNA complexes to proteins involved in mRNA deadenylation and decapping (16). In contrast to Argonaute proteins, GW182 did not exhibit significant expression in most adult tissues but was upregulated upon various forms of cellular activation (12, 13, 17). This suggested that increased GW182 protein might serve to sustain microRNA-mediated repression of target mRNA under conditions where eIF4F activity was stimulated.
In the current study, we set out to determine how increased GW182 expression was coordinated with enhanced eIF4F activity following activation of immune cells. We found that PI3K-Akt-mTOR signaling resulted in enhanced expression of GW182 protein without increased steady-state levels of GW182 mRNA. Maximal GW182 expression also required signaling through the Jak-Stat-Pim pathway. The PI3K-Akt-mTOR and Jak-Stat-Pim pathways converged to elicit translation of GW182 mRNA both by stimulating its association with eIF4E and by facilitating eIF4A-dependent unwinding of G-quadruplexes in its 5′ UTR. Taken together, data from these studies provide a mechanistic link between eIF4F function and cellular microRNA capacity, suggesting that these opposing forces on translation are coregulated to maintain microRNA-mediated repression of targets over a wide range of cap-dependent translation.
MATERIALS AND METHODS
Antibodies and reagents.
Antibodies used for Western blots were obtained from commercial sources as follows: GW182 from Bethyl; Ago2, phospho-4E-BP1(T37/40), 4E-BP1, phospho-Akt(S473), Akt, eIF4G1, eIF4A, Pdcd4, Raptor, Rictor, Jak2, phospho-eIF4B(S422), phospho-mTOR(S2448), phospho-S6K(T389), phospho-rpS6(235/236), phospho-Stat3(Y705), phospho-Stat5(Y649), phospho-Erk(T202/Y204), Erk, and eIF4H from Cell Signaling; eIF4E from BD Biosciences; actin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) from Sigma; panAgo from EMD Millipore; Pim1 and Pim2 from Santa Cruz. Chemicals were obtained from the following sources: Torin1 from Tocris; rapamycin, SGI-1776, CX-6258, 4EGI-1, Jak inhibitor I, and PD98059 from Calbiochem; LY294002 from Cell Signaling; doxycycline and cycloheximide (CHX) from Sigma; MG-132 from Selleckchem. Silvestrol was kindly provided by Neil Rosen.
Isolation and stimulation of primary T cells.
T cells were isolated according to protocols approved by the Memorial Sloan Kettering Cancer Center IACUC or Roswell Park Cancer Institute IACUC from 6- to 12-week-old wild-type C57BL/6 mice purchased from Jackson Laboratory or bred in-house, from miR-22 knockout mice (129S-Mir22tm1.1Arod/J) purchased from Jackson Laboratory, and from wild-type 129S1/SvlmJ mice purchased from Jackson Laboratory. T cell isolations were performed using the Dynabeads Untouched mouse T cell kit (Life Technologies) or Pan T Cell Isolation kit II, mouse (Miltenyl Biotec), with similar results. Isolations were performed essentially as described in the manufacturers' instructions, and fluorescence-activated cell sorter (FACS) analysis indicated >90% CD3+ T cell enrichment (data not shown). Isolated T cells were either washed in cold phosphate-buffered saline (PBS) and used immediately for downstream applications or stimulated with Dynabeads Mouse T-Activator CD3/CD28 beads at a 1:1 bead-to-cell ratio in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (FCS; Gemini Bio-Products), 10 U/ml penicillin-streptomycin, 2 mM l-glutamine, 50 μM β-mercaptoethanol, 10 mM HEPES, and 25 U/ml recombinant murine interleukin-2 (rIL-2; BD Biosciences) and cultured in a humidified incubator maintained at 37°C, 5% CO2. Beads were removed from cultures 24 to 72 h later, and T cells were maintained at a concentration of approximately 1 × 106 cells/ml for the duration of the experiments.
Cell lines and culture conditions.
The murine IL-3-dependent lymphoid progenitor cell lines FL5.12, FL5.12.xL, and FL5.12.mAkt were previously described (18–20). To create FL5.12.Jak2 cells, parental FL5.12 cells were transduced with the retroviral vector MSCV-TELJAK2-IRES-GFP (where MSCV is murine stem cell virus, IRES is internal ribosome entry site, and GFP is green fluorescent protein; kindly provided by Ross Levine) and then cultured in the absence of IL-3 until cells began to proliferate in an IL-3-independent manner. Expression of constitutively active TELJAK2 was confirmed by Western blotting (see Fig. S4A in the supplemental material). All FL5.12 variants were propagated in flasks containing RPMI 1640 supplemented with 10% FCS (Gemini Bio-Products), 10 U/ml penicillin-streptomycin, 2 mM l-glutamine, 50 μM β-mercaptoethanol, 10 mM HEPES, and 0.35 ng/ml recombinant murine IL-3 (Gemini Bio-Products) housed in a humidified incubator maintained at 37°C, 5% CO2. Cultures were maintained at densities between 0.01 × 106 and 1 × 106 cells/ml. For IL-3 withdrawal experiments, FL5.12 cell variants were harvested by centrifugation (∼300 × g for 5 min), washed with a medium similar to the one described above except lacking IL-3, and then plated at 0.1 × 106 to 0.2 × 106 cells/ml in the medium described above with or without addition of IL-3 and cultured for 18 to 24 h. The human hematopoietic cell lines Raji and Jurkat were cultured in flasks containing RPMI 1640 supplemented with 10% FCS (Gemini Bio-Products), 10 U/ml penicillin-streptomycin, 2 mM l-glutamine, 50 μM β-mercaptoethanol, and 10 mM HEPES housed in a humidified incubator maintained at 37°C, 5% CO2. Raji cells were engineered to express a doxycycline-inducible variant of 4E-BP1 in which all four mTOR-dependent phosphorylation sites (T37, T46, S65, T70) were mutated to alanines (4E-BP1-4A) (7). This was accomplished by transduction of the lentiviral vector pCW57.1-4EBP1_4xAla (Addgene plasmid number 38240) followed by selection in 2.5 μg/ml puromycin.
Inducible shRNA constructs.
Short hairpin RNAs (shRNAs) targeting GW182 were designed according to the DSIR algorithm, followed by “sensor rules” filtering (21). Each matched control contains the same hairpin sequence except for bases 9 to 11, which were changed to their complements (22). All custom oligonucleotides coding for shRNAs were synthesized by Life Technologies and PCR amplified to generate miR-E shRNAs according to procedures developed by Fellman et al. (21). PCR products were gel purified and cloned into the retroviral backbone RT3GEPIR by standard cloning techniques.
Western blotting.
Western blots were performed on whole-cell lysates made by incubation of cell pellets in 1× radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific) supplemented with Halt protease inhibitors and Halt phosphatase inhibitors (Thermo Scientific) for 20 min on ice followed by centrifugation at 20,000 × g for 20 min at 4°C. Lysates were quantified using the bicinchoninic acid (BCA) method (Thermo Scientific), and 10 to 100 μg of protein per lane was loaded onto NuPAGE gradient gels. Use of NuPAGE 3 to 8% Tris-acetate gels and running buffer (Life Technologies) was necessary for detection of GW182, whereas other proteins could be detected using NuPAGE Novex 4 to 12% bis-Tris gels and either MES (morpholineethanesulfonic acid) or MOPS (morpholinepropanesulfonic acid) running buffer (Life Technologies). Gels were run according to the manufacturer's recommendations and proteins transferred to nitrocellulose membranes for blotting. Membranes were blocked with 3% milk, incubated overnight at 4°C with primary antibodies diluted in 3% bovine serum albumin (BSA), and then washed three times with Tris-buffered saline with Tween 20 (TBST) and incubated with IR-Dye-labeled secondary antibodies (LI-COR) for 2 h at room temperature. Membranes were washed 3 more times with TBST, and bands were detected using a LI-COR Odyssey imaging system.
Polysome fractionation.
Discrimination between monosome- and polysome-associated mRNAs was performed by density centrifugation of cell lysates. Lysates were prepared from 2 × 107 to 10 × 107 cells following 5 min of incubation with 100 μg/ml cycloheximide. Cells were harvested by centrifugation, washed with PBS containing 100 μg/ml CHX, and frozen in liquid nitrogen. Frozen cell pellets were resuspended in freshly prepared, ice-cold polysome buffer (20 mM Tris-Cl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 20 U/ml SUPERase-In [Ambion], and 100 μg/ml CHX) containing 0.02% Triton X-100, incubated for 10 min on ice, passed through a 27-gauge needle, and centrifuged for 10 min at 18,000 × g and 4°C. Resulting lysates were centrifuged through a 10 to 50% sucrose gradient for 2 h at 39,000 rpm in an SW-41 rotor at 4°C. Gradients were then fractionated using a Teledyne ISCO density gradient fractionation system and collected into TRIzol LS. RNA was extracted from resulting fractions with the miRNEasy kit from Qiagen, combined into polysome or monosome pools based on UV spectra (Fig. 1C; see also Fig. S3D and S4C in the supplemental material), and reverse transcribed with SuperScript IV using oligo(dT) primers, and the resulting cDNA was used for quantitative real-time PCR (qRT-PCR) as described below.
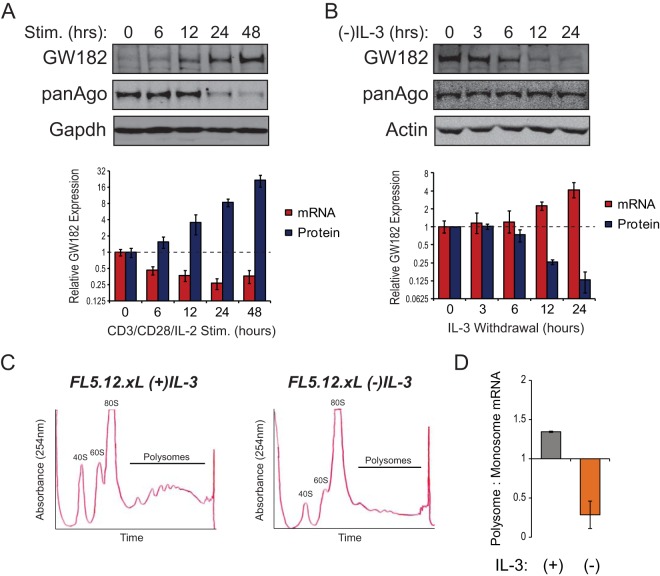
FIG 1.
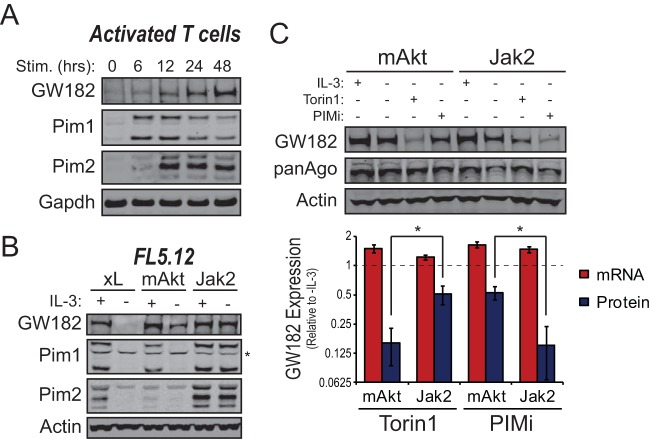
Posttranscriptional regulation of GW182. (A) T cells isolated from spleens of C57BL/6 mice were stimulated for 48 h with microbeads coated with anti-CD3 and anti-CD28 antibodies in the presence of 25 U/ml recombinant IL-2 (rIL-2). Cells were collected at the indicated time points, and protein and RNA were isolated. (Top) Representative Western blots for GW182 and Argonaute protein expression. GAPDH served as an endogenous control. (Bottom) Red bars represent mean relative expression values of the mRNA coding for GW182 determined by quantitative real-time RT-PCR (qRT-PCR) using the ΔΔCT method with data from three independent experiments performed in triplicate or quadruplicate ± 95% confidence intervals of the means. Tbp was used as the endogenous control. Blue bars represent mean relative GW182 protein expression values determined by quantification of data from four Western blots from independent experiments ± standard deviations. GAPDH was used to normalize protein loading between lanes. (B) The IL-3-dependent cell line FL5.12.xL was incubated in medium lacking growth factor (IL-3) for 24 h. Cells were collected at the indicated time points, and protein and RNA were isolated. (Top) Representative Western blots for GW182 and Argonaute protein expression. Actin served as an endogenous control. (Bottom) Red bars represent mean relative expression values of the mRNA coding for GW182 determined by qRT-PCR using the ΔΔCT method with data from two independent experiments performed in quadruplicate ± 95% confidence intervals of the means. 18S rRNA gene was used as the endogenous control. Blue bars represent mean relative GW182 protein expression values determined by quantification of data from three Western blots from independent experiments ± standard deviations. Actin was used to normalize protein loading between lanes. (C) Representative UV absorbance traces recorded during collection of fractions of lysates from FL5.12.xL cells cultured overnight in the presence or absence of IL-3 centrifuged through 10 to 50% sucrose gradients. Fractions collected for qRT-PCR comparison of polysome- and monosome (40S, 60S, and 80S)-associated mRNAs are noted. (D) Enrichment of GW182 mRNA in polysome fractions from FL5.12.xL cells cultured in the presence or absence of IL-3 for 24 h determined by qRT-PCR. Bars represent the ratios of GW182 mRNA detected in polysome fractions to GW182 mRNA detected in monosome fractions. Data were normalized to actin, an endogenous control whose polysome occupancy is unchanged by growth factor (IL-3) withdrawal (see Fig. S1C in the supplemental material).
RIP assay.
RNA bound to eIF4E was purified by using the RNA immunoprecipitation (RIP) assay kit from MBL International essentially as described by the manufacturer. For each RIP, 1 mg of protein lysate was precleared with 25 μl Dynabeads protein G beads (Life Technologies) and then incubated with 15 μg eIF4E antibody (MBL International) or 15 μg normal rabbit IgG (MBL International) previously conjugated to 50 μl Dynabeads protein G beads. Equal volumes of input RNA and RNA isolated from each RIP were used for cDNA synthesis and qRT-PCR as described below.
qRT-PCR.
RNA isolated using TRIzol reagent (Life Technologies) or miRNeasy kit (Qiagen) was quantified on a NanoDrop spectrophotometer (Thermo Scientific) or Qubit fluorimeter (Invitrogen), and 0.2 to 1 μg of total RNA was used for first-strand cDNA synthesis using SuperScript III or SuperScript IV (Life Technologies). Resulting cDNA was PCR amplified using TaqMan primer/probe sets for specified gene products on either an Applied Biosystems 7900HT Fast real-time PCR system or an Applied Biosystems QuantStudio 6 Flex real-time PCR system or using standard primers (see Table S2 in the supplemental material) and the SsoAdvanced Universal SYBR green Supermix (Bio-Rad) master mix to amplify cDNA on a Bio-Rad CFX Connect real-time PCR detection system. Expression of mRNAs was quantified using the ΔΔCT method (where CT is threshold cycle) with the indicated endogenous controls. For RIP and polysome fractionation samples, mRNA enrichment was determined by comparing CT values to standard curves generated from input samples.
DLR assays.
Dual-luciferase reporter (DLR) assays were used to measure microRNA-mediated target repression and the effects of the 5′ untranslated region of GW182 mRNA on protein expression. To measure microRNA-mediated target repression, the CXCR4 microRNA reporter system developed by Doench et al. (23) was transfected into cells (100 ng of reporter vector plus 100 ng of control vector per 106 cells) using an Amaxa 4D Nucleofector. Control or targeting oligonucleotide duplexes were cotransfected at the indicated concentrations. Cells were collected 4 h following transfection and lysed, and the resulting lysates were used to measure activities of renilla and firefly luciferase using the Dual-Luciferase assay system (Promega) and a GlowMax-96 microplate luminometer. Data were normalized for transfection efficiency using firefly luciferase relative light units (RLU) and relative repression values calculated as previously described (12). To study the effects of the 5′ UTR of GW182 mRNA, 5′UTR sequences from human GW182 (wild type or with G-to-A mutations in putative G-quadruplexes) or β-Actin as a control were synthesized as gBlocks by IDT and cloned into the NheI site at the 5′ end of the renilla luciferase gene in pRL-TK (Promega). Resulting vectors were cotransfected with pGL3 into indicated cell types using an Amaxa 4D Nucleofector, and cells were cultured overnight in the presence or absence of 10 nM silvestrol. Data were normalized for transfection efficiency using firefly luciferase RLU and then compared to RLU values given by transfection of the unmodified pRL-TK vector.
RESULTS
Posttranscriptional regulation of GW182 expression.
Mounting evidence suggests that modulation of GW182 protein level allows immune cells to coordinate microRNA function with activation status (12, 13, 17). To gain insight into the mechanism(s) underlying activation-induced changes in GW182 expression, time course experiments were performed using conditions under which GW182 was induced (T cell activation [Fig. 1A]) or depleted (IL-3 withdrawal from the IL-3-dependent hematopoietic progenitor cell line FL5.12.xL [Fig. 1B]) by manipulation of mitogenic signaling. T cell activation led to a time-dependent increase in GW182 protein to its maximal level (∼21-fold higher than basal level) 48 h following ex vivo stimulation (Fig. 1A). Subsequently, GW182 protein expression decreased and returned to the basal level by day 5 following activation (see Fig. S1A in the supplemental material). Surprisingly, the mRNA coding for GW182 decreased over the first 48 h of T cell activation (Fig. 1A, bottom), suggesting that increased transcription was not responsible for the large increase in GW182 protein expression. A similar phenomenon was observed over the first 24 h of IL-3 withdrawal from the IL-3-dependent hematopoietic progenitor cell line FL5.12.xL (19); GW182 protein expression decreased 82% ± 5%, while the mRNA coding for GW182 increased approximately 4-fold (Fig. 1B). A recent study revealed that GW182 expression could be regulated by TRIM65-dependent ubiquitination and degradation by proteasomes (24). This mechanism did not account for changes in GW182 expression upon immune cell stimulation, as GW182 protein expression was unaffected by proteasome inhibition (see Fig. S1B in the supplemental material). Instead, growth factor stimulation led to enrichment of the mRNA coding for GW182 in polysome fractions of sucrose gradients (Fig. 1C and D), indicating that its translation was increased. In contrast, mRNAs coding for other GW182 family proteins Tnrc6b and Tnrc6c were not highly enriched in polysome fractions of stimulated immune cells (see Fig. S1C in the supplemental material).
Changes in GW182 expression regulate microRNA capacity in immune cells.
We previously demonstrated that of the three Tnrc6 family proteins, GW182 was primarily responsible for enhanced microRNA-mediated repression in activated T cells (13). Reanalysis of small RNA sequencing data revealed that association of microRNAs with target mRNA was inversely correlated with microRNA expression in resting T cells (Fig. 2A) (Spearman's r = −0.2201; P = 0.02). This suggested that under conditions where GW182 protein expression was low, a large fraction of highly expressed microRNAs were not in complex with target mRNA. Following T cell activation, a condition under which GW182 protein expression was greatly increased, this correlation was lost and nearly all microRNAs exhibited increased association with target mRNAs (Fig. 2B) (Spearman's r = 0.0853; P = 0.38).
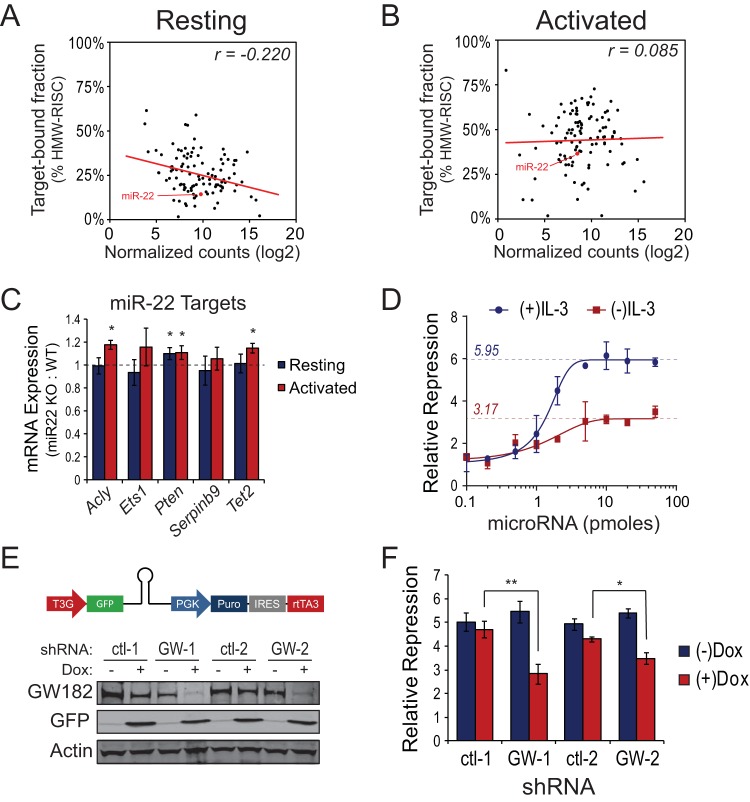
FIG 2.
Mitogen-stimulated GW182 expression regulates cellular microRNA capacity. (A) Log2 normalized small RNA sequencing counts mapped to 110 microRNAs expressed in freshly isolated, resting mouse T cells plotted against their enrichment in target-associated, high-molecular-weight RISC (HMW-RISC), as previously described (13). The line represents the best-fit linear trend line with Spearman's rank correlation coefficient displayed in the upper right corner. (B) Log2 normalized small RNA sequencing counts mapped to 110 microRNAs expressed in mouse T cells following 3 days of ex vivo activation using microbeads coated with anti-CD3 and anti-CD28 antibodies in the presence of 25 U/ml rIL-2 plotted against their enrichment in target-associated, high-molecular-weight RISC (HMW-RISC), as previously described (13). The line represents the best-fit linear trend line with Spearman's rank correlation coefficient displayed in the upper right corner. (C) Change in expression of miR-22 targets in freshly isolated miR-22 knockout T cells (Resting) versus miR-22 knockout T cells activated ex vivo for 2 days as described for panel B. Bars represent mean expression values ± standard deviations of the indicated transcripts in miR-22 knockout T cells relative to wild-type 129S1/SvlmJ T cells as determined by qRT-PCR using the ΔΔCT method with Tbp as the endogenous control. Asterisks indicate a significant (P < 0.05) change in expression determined by the t test comparing 8 resulting data points per transcript. (D) Extensively characterized synthetic microRNA luciferase reporters (23) were transfected into 1 × 106 FL5.12.xL cells cultured in the presence or absence of IL-3 along with increasing concentrations (0.1 to 50 pmol) of targeting microRNA duplexes. Twenty-four hours following initiation of cultures, cells were collected and dual-luciferase (DLR) assays performed. GraphPad software was used to fit four-parameter logistic curves to DLR data from 5 independent experiments. Dashed lines with overlaid values illustrate the repressive capacity for each condition. (E) Western blots showing GW182 protein expression in two inducible GW182 shRNA-expressing FL5.12.xL clones (GW-1 and GW-2) and two inducible control shRNA-expressing FL5.12.xL clones (ctl-1 and ctl-2) cultured in the absence (−) or presence (+) of doxycycline. Above the blots is a schematic of the miR-E-based shRNA construct stably expressed in FL5.12.xL clones. (F) FL5.12.xL cells expressing two independent doxycycline-inducible shRNAs targeting the mRNA coding for GW182 or two independent C9-11 control shRNAs (see Materials and Methods for details) were treated with 1 μg/ml doxycycline overnight and then transfected with microRNA reporters as described for panel C plus a saturating concentration (20 pmol) of targeting or nontargeting microRNA duplexes. Twenty-four hours following addition of doxycycline, cells were collected and DLR assays performed. Bars represent mean microRNA reporter repression values calculated from four independent reporter transfections ± standard deviations of the means. **, P = 0.0005; *, P = 0.003.
To test the functional relevance of increased microRNA-mRNA association following T cell activation, T cells from miR-22 knockout or wild-type mice were activated, and expression of several miR-22 target genes, determined by Ago2 high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) (see Table S1 in the supplemental material and reference 25) and a review of the literature, was measured by qRT-PCR. Target repression by miR-22 was examined because miR-22 was expressed at a high enough level to expect that it would have an effect on target gene expression (∼1% of total microRNA mapped transcriptome sequencing [RNA-seq] reads in both resting and activated T cells) and because its expression did not change following T cell activation, yet its association with target mRNA increased (Fig. 2A and B, red data points). Moreover, a miR-22 knockout mouse model was readily available. Activation of miR-22 knockout T cells resulted in significant increases in expression of miR-22 target genes Acly (26), Pten (27), and Tet2 (28), a trend toward increased expression of putative miR-22 target genes Ets1 and Serpinb9 (Fig. 2C), and no effect on expression of a control gene (Sdha) lacking miR-22 seed sites (see Fig. S2A in the supplemental material) relative to activated wild-type T cells. Importantly, of the five miR-22 target genes tested, only Pten expression was elevated in resting miR-22 knockout T cells compared to resting wild-type T cells (Fig. 2C). These effects were independent of whether miR-22 targets increased or decreased in expression following T cell activation (see Fig. S2B in the supplemental material).
The observations described above are consistent with the possibility that enhanced expression of GW182 protein increased the overall capacity for microRNA-mediated target repression in activated immune cells. To measure the overall capacity for microRNA-mediated repression in resting versus activated immune cells, increasing concentrations of a synthetic microRNA were cotransfected with a constant concentration of an extensively characterized microRNA reporter (23) into FL5.12.xL cells. In the presence of growth factor, FL5.12.xL cells expressed a moderate level of GW182 protein (13) and had a repressive capacity of approximately 6-fold (5.95 ± 0.15). Growth factor withdrawal from the medium of FL5.12.xL cells for 24 h reduced GW182 protein expression (Fig. 1B) and microRNA repressive capacity to (3.17 ± 0.11)-fold (Fig. 2D). However, the ability of the synthetic microRNA used for these assays to repress a reporter with a perfectly complementary target site via the small interfering RNA (siRNA) pathway was similar in growth factor stimulated and withdrawn cells (see Fig. S2C in the supplemental material; see also reference 13), suggesting that Ago2 loading and function were not compromised in growth factor-withdrawn cells.
To test whether growth factor withdrawal-associated reduction in microRNA repressive capacity could be attributed to decreased GW182 expression, doxycycline-inducible shRNAs targeting the mRNA coding for GW182 or control shRNAs were stably expressed in FL5.12.xL cells. After 24 h of doxycycline (1 μg/ml) treatment in the presence of growth factor, GW182 protein expression decreased by 88% ± 8% in cells expressing GW182-targeting shRNA-1, 91% ± 4% in cells expressing GW182-targeting shRNA-2, 3% ± 2% in cells expressing control shRNA-1, and 25% ± 1% in cells expressing control shRNA-2 (Fig. 2E). Concurrently, doxycycline treatment decreased repressive capacity 2.62-fold in cells expressing GW182-targeting shRNA-1, 1.91-fold in cells expressing GW182-targeting shRNA-2, 0.31-fold in cells expressing control shRNA-1, and 0.64-fold in cells expressing control shRNA-2 (Fig. 2F), demonstrating that changes in repressive capacity resulting from growth factor withdrawal correlated closely with changes resulting from GW182 knockdown in the presence of growth factor.
mTOR promotes translation of GW182 mRNA and microRNA function.
Our previous work suggested that GW182 protein expression was linked to PI3K-Akt signaling (13). Consistent with this, introduction of a constitutively active myristoylated Akt (mAkt) rescued ∼60% of GW182 protein expression in FL5.12 cells cultured in the absence of IL-3. A major downstream effector of PI3K-Akt signaling is the serine-threonine kinase mechanistic target of rapamycin (mTOR). Addition of the mTOR inhibitors Torin1 or rapamycin to mAkt-expressing FL5.12 cells (FL.5.12.mAkt [20]) suppressed the ability of mAkt to enhance GW182 protein levels without affecting expression of the mRNA coding for GW182 (Fig. 3A). This effect was conserved between mouse and human cells, as treatment of the human T cell lymphoma line Jurkat with Torin1 or rapamycin decreased expression of GW182 protein (see Fig. S3A in the supplemental material). Moreover, inhibition of mTOR signaling by amino acid withdrawal (see Fig. S3B in the supplemental material) or knockdown of Raptor or Rictor (see Fig. S3C in the supplemental material) decreased expression of GW182 protein. Consistent with the role for GW182 in determining cellular microRNA capacity, treatment of growth factor-starved FL5.12.mAkt cells with Torin1 or rapamycin significantly decreased repression of a reporter targeted by a saturating amount of a transfected synthetic microRNA (Fig. 3B). Taken together, these data suggest that mTOR signaling regulates GW182 protein expression and therefore microRNA repressive capacity via a posttranscriptional mechanism.
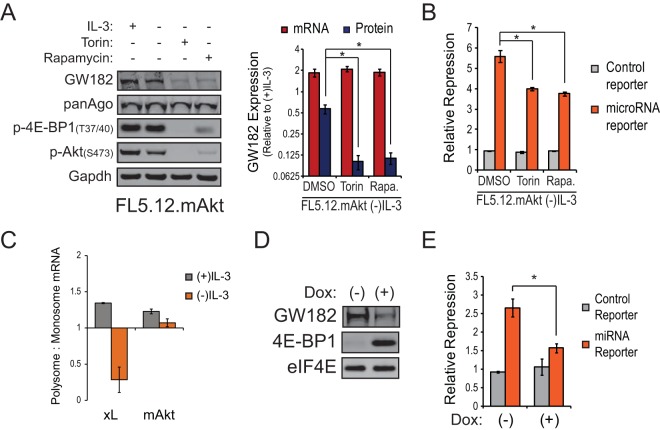
FIG 3.
Regulation of GW182 translation and microRNA capacity by the PI3K-Akt-mTOR signaling pathway. (A) (Left) Representative Western blots for GW182, Argonaute 1-4 (panAgo), and mTOR-dependent phosphorylation events in FL5.12 cells expressing myristoylated Akt (FL5.12.mAkt) cultured in the presence or absence of IL-3 plus Torin1 (500 nM) or rapamycin (100 nM). GAPDH served as an endogenous control. (Right) Red bars represent mean expression values of the mRNA coding for GW182 relative to the (+)IL-3 condition ± 95% confidence intervals of the means from a representative qRT-PCR experiment performed two times in quadruplicate. 18S rRNA gene was used as the endogenous control. Blue bars represent mean GW182 protein expression values relative to the (+)IL-3 condition ± standard deviations determined by quantification of data from Western blots from four independent experiments. GAPDH was used to normalize protein loading between lanes. (B) A renilla luciferase reporter targeted by a synthetic microRNA or a control reporter lacking the microRNA target sequence was cotransfected with pGL3 and 20 pmol of targeting or control microRNA duplex into 1 × 106 FL5.12.mAkt cells cultured in the absence of IL-3 overnight ± 500 nM Torin1 or 100 nM rapamycin. Twenty-four hours following initial incubation, cells were collected and dual-luciferase (DLR) assays performed. Bars represent mean reporter repression values calculated from four independent reporter transfections ± standard deviations. *, P < 0.05. (C) Enrichment of GW182 mRNA in polysome fractions from FL5.12.mAkt cells or FL5.12.xL cells cultured in the presence or absence of IL-3 for 24 h as determined by qRT-PCR. Bars represent the ratios of GW182 mRNA detected in polysome fractions to GW182 mRNA detected in monosome fractions (see Fig. S3D in the supplemental material). Data were normalized to the mRNA coding for actin, an endogenous control whose polysome occupancy is unchanged by growth factor (IL-3) withdrawal (see Fig. S1C in the supplemental material). (D) Representative Western blots for GW182, 4E-BP1, and eIF4E as an endogenous control using proteins extracted from Raji cells expressing doxycycline-inducible 4E-BP1-4A that were cultured in the absence or presence of doxycycline (1 μg/ml) for 48 h. (E) Effects of doxycycline-induced 4E-BP1-4A expression in Raji cells on microRNA reporter repression measured as described for panel B. Bars represent mean reporter repression values from four independent reporter transfections ± standard deviations. *, P < 0.05.
A major function of mTOR is regulation of cap-dependent mRNA translation through phosphorylation of downstream targets such as eIF4E binding proteins (4E-BPs) and ribosomal protein S6 kinase (S6K) (29). Since GW182 was found to be regulated posttranscriptionally in an mTOR-dependent manner, we hypothesized that translation of GW182 mRNA was enhanced by PI3K-Akt-mTOR signaling. To test this hypothesis, polysome profiling experiments were performed on lysates from FL5.12.mAkt cells cultured overnight in the absence of growth factor. In agreement with our hypothesis, sustained activation of the PI3K-Akt-mTOR signaling in the absence of exogenous growth factor led to enhanced occupancy of GW182 mRNA in polysome fractions (Fig. 3C). To further test our hypothesis, we generated Raji cells expressing doxycycline-inducible 4E-BP1 with all four mTOR phosphorylation sites mutated to alanines (4E-BP1-4A), thus rendering it insensitive to inhibition by mTOR (7). Doxycycline treatment of Raji cells expressing inducible 4E-BP1-4A led to marked induction of 4E-BP1-4A and decreased expression of GW182 (Fig. 3D), accompanied by a significant loss of microRNA capacity (Fig. 3E).
Jak-Stat signaling enhances translational upregulation of GW182.
Since signaling through the PI3K-Akt-mTOR pathway only partially accounted for mitogen-stimulated GW182 expression (Fig. 3A) (13), we examined the kinetics of phosphorylation events within additional signaling pathways relative to GW182 induction following ex vivo T cell activation. Activating phosphorylation events in the PI3K-Akt-mTOR, Jak-Stat, and Mek-Erk signaling pathways coincided with GW182 induction (Fig. 4A). To explore the contribution of each pathway to observed increases in GW182 expression following T cell activation, chemical inhibitors were tested for their ability to block GW182 induction. Inhibitors of PI3K (LY294002) and Jak (CAS 457081-03-7) signaling markedly reduced GW182 induction, whereas an inhibitor of the mitogen-activated protein kinase (MAPK) pathway (PD98059) had no effect on GW182 induction in ex vivo-activated T cells (Fig. 4B) or in growth factor-stimulated FL5.12.xL cells (Fig. 4C). Consistent with this, culture of αCD3-stimulated primary T cells with IL-2, which primarily signals via the Jak-Stat pathway (30), induced GW182 protein expression at least as well as costimulation with anti-CD3 and anti-CD28 (Fig. 4D). Neither the combination of anti-CD3 and IL-2 nor that of anti-CD3 and anti-CD28 enhanced steady-state GW182 mRNA levels (Fig. 4D, bottom), suggesting that both stimuli had a posttranscriptional effect on GW182 protein expression.
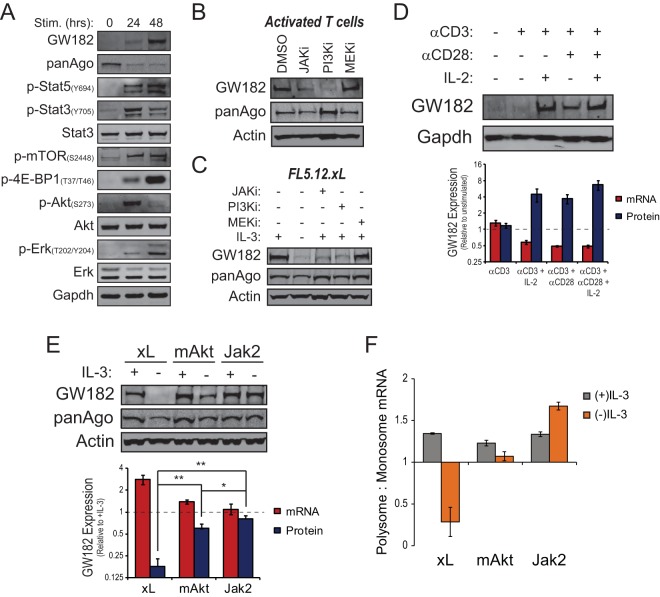
FIG 4.
Regulation of GW182 translation by the Jak-Stat-Pim signaling pathway. (A) Western blots for GW182, Argonaute 1-4 (panAgo), and signaling events associated with ex vivo activation of C57BL/6 splenic T cells with microbeads coated with anti-CD3 and anti-CD28 antibodies in the presence of 25 U/ml recombinant murine interleukin-2 (rIL-2). GAPDH served as an endogenous control. (B) Splenic T cells from C57BL/6 mice were activated as described for panel A in the presence of vehicle control, dimethyl sulfoxide (DMSO), 1 μM Jak inhibitor 1 (JAKi, CAS 457081-03-7), 10 μM LY294002 (PI3Ki), or 20 μM PD98059 (MEKi). Cells were collected 48 h after activation, and lysates were prepared for Western blots using antibodies against GW182, Argonaute1-4 (panAgo), and actin as an endogenous control. (C) Western blots showing GW182 and Argonaute expression in FL5.12.xL cells cultured for 24 h in the presence or absence of IL-3 or in the presence of IL-3 plus 1 μM Jak inhibitor 1 (JAKi, CAS 457081-03-7), 10 μM LY294002 (PI3Ki), or 20 μM PD98059 (MEKi). Actin served as an endogenous control. (D) Expression of GW182 protein and mRNA in C57BL/6 T cells stimulated ex vivo with microbeads coated with anti-CD3 antibody or anti-CD3 and anti-CD28 antibodies in the presence or absence of rIL-2. (Top) Representative Western blots for GW182 protein expression with GAPDH as an endogenous control. (Bottom) Red bars represent mean expression values of the mRNA coding for GW182 relative to freshly isolated, unstimulated T cells ± 95% confidence intervals of the means from a representative qRT-PCR experiment performed two times in quadruplicate. Sdha was used as the endogenous control. Blue bars represent mean GW182 protein expression values relative to freshly isolated, unstimulated T cells ± standard deviations determined by quantification of data from Western blots from three independent experiments. GAPDH was used to normalize protein loading between lanes. (E) Expression of GW182 protein and mRNA in IL-3-dependent FL5.12 cells expressing either Bcl-xL (xL), myristoylated Akt (mAkt), or the TEL-JAK2 fusion protein (Jak2) following 24 h of culture in the presence or absence of IL-3. (Top) Representative Western blots for GW182 and Argonaute 1-4 (panAgo) with actin as an endogenous control. (Bottom) Red bars represent mean expression values of the mRNA coding for GW182 relative to the (+)IL-3 condition ± 95% confidence intervals of the means from a representative qRT-PCR experiment performed two times in quadruplicate. Gapdh was used as the endogenous control. Blue bars represent mean GW182 protein expression values relative to the (+)IL-3 condition ± standard deviations determined by quantification of data from Western blots from five independent experiments. Actin was used to normalize protein loading between lanes. *, P < 0.01; **, P < 0.001. (F) Enrichment of GW182 mRNA in polysome fractions from FL5.12.Jak2, FL5.12.mAkt, or FL5.12.xL cells cultured in the presence or absence of IL-3 for 24 h as determined by qRT-PCR. Bars represent the ratio of GW182 mRNA detected in polysome fractions to GW182 mRNA detected in monosome fractions (see Fig. S4C in the supplemental material). Data were normalized to the mRNA coding for actin, an endogenous control whose polysome occupancy is unchanged by growth factor (IL-3) withdrawal (see Fig. S1C in the supplemental material).
To explore the independent contribution of Jak-Stat signaling to the posttranscriptional regulation of GW182, FL5.12 cells were engineered to stably express the fusion protein TEL-JAK2 (31), leading to the constitutive activation of the Jak-Stat pathway (see Fig. S4A in the supplemental material). Similar to FL5.12.mAkt cells, FL5.12 cells expressing TEL-JAK2 (FL5.12.Jak2) were able to survive and proliferate in the absence of IL-3 stimulation (see Fig. S4B in the supplemental material). A comparison of FL5.12.xL, FL5.12.mAkt, and FL5.12.Jak2 cells cultured in the absence of exogenous growth factor (IL-3) revealed that GW182 mRNA expression was similar in all three cell types while FL5.12.Jak2 cells maintained a significantly higher level of GW182 protein expression in the absence of growth factor stimulation (∼80% of growth factor stimulated) than did FL5.12.xL or FL5.12.mAkt cells (Fig. 4E). Moreover, constitutive Jak-Stat pathway activation in the absence of growth factor stimulation led to further enrichment of GW182 mRNA in polysome fractions compared to PI3K-Akt-mTOR activation (Fig. 4F).
Pim and mTOR kinases transduce parallel signals that boost GW182 protein expression in stimulated immune cells.
Pim kinases (Pim1 to -3) are transcriptionally regulated targets of the Jak-Stat signaling pathway that can phosphorylate many of the same proteins as the AGC kinases, which are integral parts of the PI3K-Akt-mTOR pathway (32). Expression of Pim1 and Pim2 was markedly enhanced following ex vivo activation of T cells and preceded increased GW182 expression (Fig. 5A). Moreover, FL5.12.Jak2 cells cultured in the absence of growth factor maintained expression of GW182, Pim1, and Pim2 (Fig. 5B). Pim 3 could not be detected by Western blotting in T cells or FL5.12 cells.
FIG 5.
Pim kinases mediate Jak2-dependent effects on GW182. (A) Western blots examining kinetic changes in expression of GW182, Pim1, or Pim2 in splenic T cells isolated from C57BL/6 mice and activated ex vivo with microbeads coated with anti-CD3 and anti-CD28 antibodies in the presence of 25 U/ml recombinant murine interleukin-2 (rIL-2). GAPDH served as an endogenous control. (B) Western blots for expression of GW182, Pim1, and Pim2 in IL-3-dependent FL5.12 cells expressing either Bcl-xL (xL), myristoylated Akt (mAkt), or the TEL-JAK2 fusion protein (Jak2) following 24 h of culture in the presence or absence of IL-3. Actin served as an endogenous control, and the asterisk denotes a nonspecific band in the Pim1 blot. (C) Effects of chemical inhibitors of the Pim kinases (PIMi, 2.5 μM SGI-1776) or mTOR kinase (0.5 μM Torin1) on expression of GW182 protein and mRNA in FL5.12.Jak2 or FL5.12.mAkt cells. (Top) Representative Western blots for GW182 and Argonaute 1-4 (panAgo) with actin as an endogenous control. (Bottom) Red bars represent mean expression values of the mRNA coding for GW182 relative to the (−)IL-3 condition ± 95% confidence intervals of the means from two independent qRT-PCR experiments performed in quadruplicate. GAPDH was used as the endogenous control. Blue bars represent mean GW182 protein expression values relative to th (−)IL-3 condition ± standard deviations determined by quantification of data from Western blots from three independent experiments. Actin was used to normalize protein loading between lanes. *, P < 0.05.
Previously published data indicate that Pim2 can enhance cap-dependent mRNA translation in parallel with mTOR (6, 33, 34). Consistent with these observations, Torin1 had only minor effects on GW182 protein expression in FL5.12.Jak2 cells cultured in the absence of IL-3 (Fig. 5C). Conversely, treatment of FL5.12.Jak2 cells with the pan-Pim kinase inhibitor SGI-1776 resulted in a marked reduction in GW182 protein expression without diminished expression of the mRNA coding for GW182 (Fig. 5C). Similar results were obtained using the mTOR inhibitor rapamycin and the Pim kinase inhibitor CX-6258 (see Fig. S5 in the supplemental material). Importantly, effects of Pim and mTOR kinase inhibitors on GW182 expression were opposite in FL5.12.mAkt cells cultured in the absence of growth factor (Fig. 5C), suggesting that Pim and mTOR kinases transduce parallel signals upstream of enhanced GW182 protein expression.
Translation initiation complex assembly on GW182 mRNA is regulated by PI3K-Akt-mTOR and Jak-Stat-Pim signaling and required for enhanced GW182 protein expression and microRNA function in stimulated immune cells.
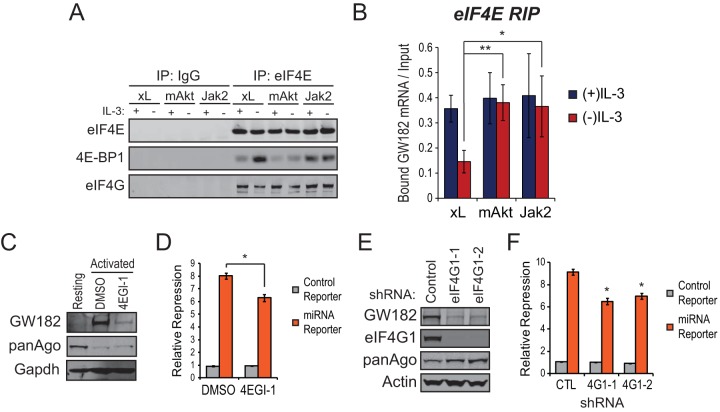
Our data indicate that GW182 protein expression in activated immune cells is enriched primarily through the effects of PI3K-Akt-mTOR and Jak-Stat-Pim signaling on translation of GW182 mRNA. These two signaling pathways act in parallel, suggesting that they share common downstream targets involved in increasing translation of GW182 mRNA. Since both pathways can regulate the rate-limiting step of translation initiation complex assembly (9), eIF4E binding to the 7mG cap of mRNA, we performed RNA-immunoprecipitations (RIPs) to examine association of the GW182 mRNA with eIF4E. Consistent with previous reports, activation of either PI3K-Akt-mTOR or Jak-Stat-Pim signaling resulted in growth factor-independent phosphorylation of 4E-BP1 (see Fig. S6A in the supplemental material) along with decreased 4E-BP1 and increased eIF4G1 association with eIF4E (Fig. 6A). These effects were accompanied by significant enrichment of GW182 mRNA associated with eIF4E in FL5.12.mAkt and FL5.12.Jak2 cells cultured in the absence of growth factor stimulation (Fig. 6B). This enrichment was specific, since of the three mRNAs coding for GW182 family proteins, only association of GW182 mRNA with eIF4E was regulated by growth factor (see Fig. S6B in the supplemental material).
FIG 6.
Convergent PI3K-Akt-mTOR and Jak-Stat-Pim signaling stimulates recruitment of eIF4E to GW182 mRNA. (A) Effects of growth factor stimulation of FL5.12.xL, FL5.12.mAkt, or FL5.12.Jak2 cells on coimmunoprecipitation of 4E-BP1 or eIF4G with eIF4E as determined by Western blotting. Each cell type was cultured overnight in the presence or absence of IL-3 and lysed, and the resulting lysates were immunoprecipitated with antibodies against eIF4E or control rabbit IgG. A representative Western blot is shown, and the experiment was repeated at least three times. (B) eIF4E-associated RNAs were purified by RNA immunoprecipitation (RIP) using antibodies against eIF4E or control rabbit IgG from FL5.12.xL, FL5.12.mAkt, or FL5.12.Jak2 cells that were cultured overnight in the presence or absence of IL-3. Input and RIP-purified RNAs were reverse transcribed, and the resulting cDNA was amplified by qRT-PCR using a TaqMan primer-probe set to detect the mRNA coding for GW182. The fraction of bound GW182 mRNA was determined by comparing CT values from RIPs to a standard curve generated from input RNA. Bars represent mean fractions of bound GW182 mRNA ± 95% confidence intervals of the means from at least two independent experiments performed in quadruplicate. Control IgG RIPs could not be graphed because CT values were not reached for most qRT-PCRs despite running 45 cycles of amplification. (C) Representative Western blots for GW182, Argonaute 1-4 (panAgo), and actin as an endogenous control using proteins extracted from freshly isolated (unstimulated) splenic B57BL/6 T cells or T cells activated for 48 h with microbeads coated with anti-CD3 and anti-CD28 antibodies in the presence of 25 U/ml rIL-2 and 50 μM 4EGI-1 or vehicle control (DMSO). (D) A renilla luciferase reporter targeted by a synthetic microRNA or a control reporter lacking the microRNA target sequence were cotransfected with pGL3 and 20 pmol of targeting or control microRNA duplex into 2 × 106 C57BL/6 T cells activated as described for panel C. Bars represent mean reporter repression values calculated from three independent reporter transfections ± standard deviations. *, P < 0.05. (E) Representative Western blots for GW182, eIF4G1, Argonaute 1-4 (panAgo), and actin as an endogenous control using proteins extracted from FL5.12 cells 5 days following transduction with lentiviral particles harboring two independent shRNAs targeting eIF4G1 mRNA or a control shRNA in pLKO.1. (F) A renilla luciferase reporter targeted by a synthetic microRNA or a control reporter lacking the microRNA target sequence were cotransfected with pGL3 and 20 pmol of targeting or control microRNA duplex into 1 × 106 eIF4G1 or control knockdown FL5.12 cells as described for panel E. Bars represent mean reporter repression values calculated from four independent reporter transfections ± standard deviations. *, P < 0.05.
Upon binding to the 7mG cap, eIF4E recruits the scaffold protein eIF4G, allowing further assembly of the translation initiation complex and loading of the 43S ribosome onto mRNA. To confirm that translation initiation complex assembly enhanced GW182 protein expression in activated immune cells, we disrupted eIF4E interaction with eIF4G using two approaches. Disruption of the eIF4E-eIF4G interaction by treatment of activated T cells with the small-molecule inhibitor 4EGI-1 resulted in a marked reduction in GW182 protein expression (Fig. 6C) and microRNA capacity (Fig. 6D). Similar results were obtained through shRNA-mediated depletion of eIF4G1, the eIF4G isoform primarily responsible for cap-dependent translation (7), from FL5.12 cells (Fig. 6E and F).
Translation of GW182 mRNA is further regulated by G-quadruplexes in its 5′ UTR.
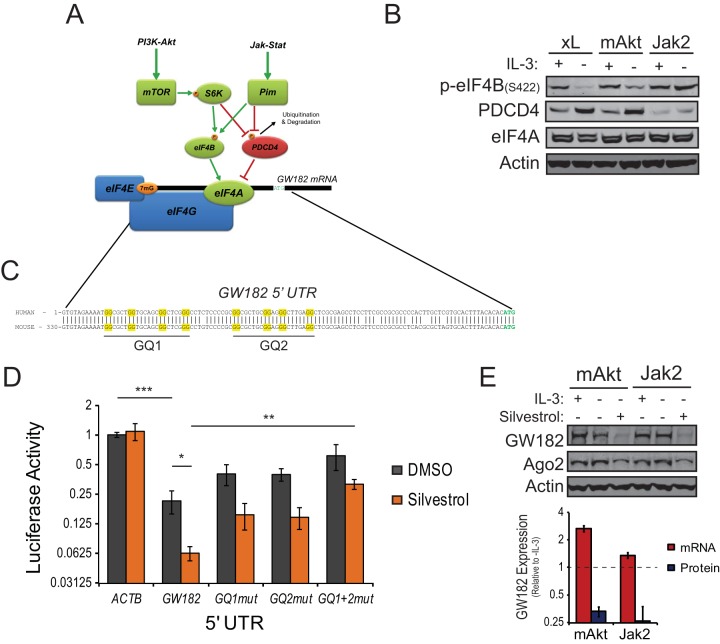
Data from polysome fractionation and RIP experiments suggest that efficient translation of the GW182 mRNA relies on eIF4E association with its 7mG cap and a subsequent regulatory step that was more efficiently enhanced by Jak-Stat-Pim signaling than by PI3K-Akt-mTOR signaling, potentially explaining the difference in GW182 protein expression upon activation of each pathway in the absence of exogenous growth factor. Since eIF4E-GW182 mRNA and eIF4E-eIF4G interactions did not differ in growth factor-withdrawn FL5.12.mAkt and FL5.12.Jak2 cells (Fig. 6A and B) and both signaling pathways could have stimulatory effects on the RNA helicase component of the translation initiation complex, eIF4A (Fig. 7A), we examined the effects of both pathways on eIF4A regulatory proteins. Jak-Stat-Pim signaling had a greater effect on phosphorylation of eIF4B, an upstream eIF4A activator, and suppression of Pdcd4, an upstream repressor of eIF4A activity (Fig. 7B), than did PI3K-Akt-mTOR signaling, suggesting that eIF4A activity may be higher in FL5.12.Jak2 cells cultured in the absence than in the presence of exogenous growth factor, potentially leading to enhanced translation of GW182 mRNA.
FIG 7.
Unwinding of G-quadruplexes in the 5′ UTR of GW182 mRNA by eIF4A enhances GW182 protein expression. (A) Model for regulation of eIF4A activity by convergent signaling through the PI3K-Akt-mTOR and Jak-Stat-Pim signaling pathways. (B) Western blots for phosphorylation of eIF4B or expression of Pdcd4 and eIF4A in IL-3-dependent FL5.12 cells expressing either Bcl-xL (xL), myristoylated Akt (mAkt), or the TEL-JAK2 fusion protein (Jak2) following 24 h of culture in the presence or absence of IL-3. Actin served as an endogenous control. (C) Conserved sequence in the 5′ untranslated region (UTR) of the mRNA coding for GW182 contains two potential G-quadruplexes (GQ1 and GQ2), highlighted in yellow. (D) Effects of the 5′ UTR from GW182 mRNA on translation of pRL-TK-based reporter constructs in the absence (DMSO) or presence of silvestrol. The 5′ UTR from GW182, mutant variants in which two of the four guanines forming putative G-quadruplexes were mutated to alanines, or the 5′ UTR from β-actin were cloned into pRL-TK upstream of the start codon for renilla luciferase. These constructs were transfected into FL5.12 cells along with pGL3, and dual-luciferase (DLR) assays were performed following overnight incubation in complete medium in the absence or presence of silvestrol (10 nM). Bars represent mean normalized luciferase activities ± standard deviations from three independent experiments performed in duplicate. ***, P = 0.00003; **, P = 0.005; *, P = 0.02. (E) Effects of silvestrol (10 nM) on expression of GW182 protein and mRNA in FL5.12.mAkt or FL5.12.Jak2 cells. (Top) Representative Western blots for GW182 and Ago2 with actin as an endogenous control. (Bottom) Red bars represent mean expression values of the mRNA coding for GW182 relative to the (−)IL-3 condition ± 95% confidence intervals of the means from two independent qRT-PCR experiments performed in quadruplicate. GAPDH was used as the endogenous control. Blue bars represent mean GW182 protein expression values relative to the (−)IL-3 condition ± standard deviations determined by quantification of data from Western blots from three independent experiments. Actin was used to normalize loading between lanes.
Many proteins involved in growth and proliferation are coded by mRNAs containing complex 5′ untranslated regions (UTRs) that limit translation in the absence of mitogenic stimulation (8). Emerging evidence suggests that a common feature of many of these transcripts is the presence of G-quadruplex structures in their 5′ UTRs that must be unwound by RNA helicases, such as eIF4A, to promote efficient ribosome binding (35). Examination of the 5′ UTR of GW182 mRNA revealed highly conserved sequence containing two putative G-quadruplexes (Fig. 7C). The effect of these G-quadruplexes on translation was tested using a reporter system in which the 5′ UTR from human GW182 mRNA was cloned at the 5′ end of the renilla luciferase gene in pRL-TK. Transfection of the wild-type human GW182 5′ UTR reporter into FL5.12.xL cells resulted in inhibition of reporter expression by approximately 80% compared to empty vector or a control vector with the 5′ UTR of β-actin mRNA cloned into pRL-TK (Fig. 7D). Inhibition of eIF4A helicase function with silvestrol led to further inhibition of reporter expression, suggesting that eIF4A contributed to unwinding of mRNA structure in the 5′ UTR of GW182 mRNA. Mutation of the G-quadruplexes in the GW182 mRNA 5′ UTR resulted in a significant decrease in the ability of silvestrol to inhibit reporter expression (Fig. 7D), consistent with the proposed role of eIF4A in unwinding G-quadruplexes (35). Similar results were obtained using Jurkat cells (see Fig. S7A in the supplemental material).
Since eIF4A is recruited to the translation initiation complex by eIF4G (36), it follows that eIF4A-dependent unwinding of the G-quadruplexes in the GW182 mRNA 5′ UTR may be the second control point subsequent to eIF4E-eIF4G assembly that regulates the translation of GW182 mRNA. To test this, FL5.12.mAkt and FL5.12.Jak2 cells cultured in the absence of growth factor or Jurkat cells cultured in complete medium were treated with silvestrol to inhibit eIF4A function. Expression of GW182 protein was significantly inhibited by treatment with silvestrol, regardless of species of origin or whether PI3K-Akt-mTOR or Jak-Stat-Pim signaling was constitutively active (Fig. 7E; see also Fig. S7B in the supplemental material). Silvestrol treatment did not decrease expression of GW182 mRNA (Fig. 7E, red bars).
DISCUSSION
GW182 family proteins are necessary and sufficient mediators of microRNA function in mammalian cells (14, 15). It therefore stands to reason that modulation of GW182 family protein expression has the potential to globally alter the ability of microRNAs to repress target mRNAs, regardless of which of the many repressive mechanisms attributed to microRNAs (37) are employed. The current study provides further evidence supporting this notion along with mechanistic detail to previous observations that GW182 protein expression is dramatically enhanced in activated T cells (12, 13, 17). Enhanced GW182 expression was sufficient to increase the activity of microRNAs in the absence of increased expression (Fig. 2) (13) and despite proteasome-mediated degradation of Argonaute proteins in activated T cells (38), suggesting that GW182 is limiting for microRNA activity in resting T cells. Additionally, data demonstrate that stimulation of PI3K-Akt-mTOR and Jak-Stat-Pim signaling networks downstream of activating surface receptors on immune cells enhanced translation of the mRNA coding for GW182 via effects on the translation initiation complex (eIF4F), resulting in augmented GW182 protein expression and increased capacity for microRNAs to repress targets in activated immune cells. We propose that this mechanism allows activated immune cells to maintain microRNA function in the face of activation-induced mRNA translation.
To date, the major focus of research on GW182 family proteins has involved examination of the molecular details of how they interact with other RNA-induced silencing complex (RISC) components to repress mRNA translation and stability (39–51). Only recently has attention been paid to the regulation of expression of individual GW182 family proteins. In activated immune cells, GW182 is the predominant family member that contributes to microRNA-mediated repression, and its expression can be upregulated by PI3K signaling (13). Here we demonstrate that PI3K signaling functions to enhance GW182 expression through mTOR-dependent effects on translation initiation. Furthermore, we demonstrate that Jak-Stat-Pim signaling is an independent regulator of GW182 translation. While GW182 has been reported to be regulated by ubiquitin-dependent proteasome degradation in some cell types (24), GW182 expression does not appear to be regulated by the ubiquitin-proteasome system in the cell types and under the conditions used for these studies, as indicated in Fig. S1B in the supplemental material (also, data not shown). Instead, our data continue to support a model in which microRNAs are able to rapidly respond to cellular signals in the absence of de novo biogenesis in order to modulate gene expression programs. Since GW182 mRNA is expressed in resting cells in the absence of high levels of GW182 protein, it appears that this microRNA response system is poised to respond to changes in protein translation stimulated by upstream signaling.
It has long been appreciated that the expression of many proteins involved in cell growth and proliferation is translationally regulated (52). Groundbreaking work from the Sonenberg lab in the early 1990s defined eIF4E as a proto-oncogene (53), linking protein synthesis and cellular transformation. In retrospect, it seems obvious that enhanced protein synthesis is required for cell growth and proliferation that accompanies transformation, especially since many more links between transformation and regulators of protein synthesis have been discovered (8). Here we define a novel link, mediated by translational regulation of GW182, between two cancer-associated signaling pathways and the activity of microRNAs that negatively regulate translation of specific mRNAs. Constitutive activation of either the PI3K-Akt or the Jak-Stat signaling pathway stimulates GW182 translation through activity of the downstream kinase mTOR or Pim, respectively. Inhibition of each kinase blocks GW182 expression only in cells where the associated upstream pathway is active (Fig. 4D), suggesting that mTOR and Pim kinases are functionally redundant for their ability to regulate GW182 translation. Whether or not shared downstream effectors of mTOR and Pim kinases stimulate GW182 translation and whether translation of other mRNAs is regulated similarly are areas of active investigation.
A confounding aspect of this work is that GW182 is one of three functionally redundant GW182 family proteins. Therefore, expression of the other two family members, Tnrc6b and Tnrc6c, has the potential to influence microRNA function independent of GW182 expression. Commercial antibodies against Tnrc6b and Trnc6c have been unreliable in determining their expression by Western blotting. However, enrichment of Tnrc6b or Tnrc6c mRNAs in polysome fractions (see Fig. S1C in the supplemental material) or eIF4E RIPs (see Fig. S6B in the supplemental material) showed little change in activated immune cells, suggesting that the mechanism by which GW182 is induced is unique among GW182 family proteins. Since expression of mRNAs coding for Tnrc6b or Tnrc6c does not increase in activated immune cells (data not shown), it is unlikely that expression of either protein is significantly enhanced. Additional evidence for this comes from functional studies showing that GW182 is primarily responsible for determining microRNA capacity in activated immune cells, as knockdown of either Tnrc6b or Tnrc6c had little effect on the repression of microRNA reporters (reference 13 and data not shown). However, different cell types may rely more on Tnrc6b or Tnrc6c expression to determine their microRNA capacity than on GW182. Therefore, continued study of this critical family of microRNA regulators is necessary to better understand how microRNA function is regulated.
Data suggest that microRNA function is bolstered to compensate for high translational output, thus maintaining a balance between protein synthesis and the ability of microRNAs to regulate their targets. Importantly, individual microRNAs can act as oncogenes or tumor suppressors (54), making the outcome of translational upregulation of GW182, and thus microRNA function, dependent on which microRNAs and which targets are expressed in a given cell. Therefore, use of translation inhibitors to treat cancers with microRNA expression patterns that favor tumor suppression could actually promote oncogenic gene expression. It is therefore critical that we continue to explore the oncogenic and tumor suppressor capabilities of individual microRNAs and to determine how microRNAs interact with one another to influence cell fate.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge members of the Thompson lab for critical review of data presented in the manuscript, John Panepinto, Dithi Banerjee, and Amanda Bloom for assistance with polysome fractionation, Paige Yellen for insights into translation regulation, Sanjay Mehta for technical support, and Ross Levine, Paul Coffer, Neil Rosen, and Sandra Gollnick for sharing reagents. Quantitative real-time PCR for miR-22 targets was performed by the Genomics Shared Resource supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) grant P30CA016056.
This work was supported by the following grants from the NCI: K99CA175189 (to S.H.O.), R01CA105463 (to C.B.T.), and Memorial Sloan-Kettering Cancer Center Support Grant P30CA008748.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01011-15.
REFERENCES
- 1.Nawijn MC, Alendar A, Berns A. 2011. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 2.Thorpe LM, Yuzugullu H, Zhao JJ. 2015. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, Porco JA Jr, Teruya-Feldstein J, Rosen N, Zelenetz AD, Pelletier J, Wendel HG. 2011. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. 2014. Translational control of immune responses: from transcripts to translatomes. Nat Immunol 15:503–511. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- 5.von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 1996. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci U S A 93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. 2003. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev 17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat Rev Drug Discov 14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary C, Mann M. 2010. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 10.Wilson RC, Doudna JA. 2013. Molecular mechanisms of RNA interference. Annu Rev Biophys 42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaff J, Meister G. 2013. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem Soc Trans 41:855–860. doi: 10.1042/BST20130047. [DOI] [PubMed] [Google Scholar]
- 12.Olejniczak SH, La Rocca G, Gruber JJ, Thompson CB. 2013. Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proc Natl Acad Sci U S A 110:157–162. doi: 10.1073/pnas.1219958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Rocca G, Olejniczak SH, Gonzalez AJ, Briskin D, Vidigal JA, Spraggon L, DeMatteo RG, Radler MR, Lindsten T, Ventura A, Tuschl T, Leslie CS, Thompson CB. 2015. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci U S A 112:767–772. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Rivas FV, Wohlschlegel J, Yates JR III, Parker R, Hannon GJ. 2005. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazzaretti D, Tournier I, Izaurralde E. 2009. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun JE, Huntzinger E, Izaurralde E. 2013. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol 768:147–163. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 18.McKearn JP, McCubrey J, Fagg B. 1985. Enrichment of hematopoietic precursor cells and cloning of multipotential B-lymphocyte precursors. Proc Natl Acad Sci U S A 82:7414–7418. doi: 10.1073/pnas.82.21.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 20.Plas DR, Talapatra S, Edinger AL, Rathmell JC, Thompson CB. 2001. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J Biol Chem 276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- 21.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, Sinha N, Zuber J. 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 22.Buehler E, Chen YC, Martin S. 2012. C911: a bench-level control for sequence specific siRNA off-target effects. PLoS One 7:e51942. doi: 10.1371/journal.pone.0051942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench JG, Petersen CP, Sharp PA. 2003. siRNAs can function as miRNAs. Genes Dev 17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Wang L, Fu B, Berman MA, Diallo A, Dorf ME. 2014. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc Natl Acad Sci U S A 111:6970–6975. doi: 10.1073/pnas.1322545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koufaris C, Valbuena GN, Pomyen Y, Tredwell GD, Nevedomskaya E, Lau CH, Yang T, Benito A, Ellis JK, Keun HC. 2016. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene 35:2766–2776. doi: 10.1038/onc.2015.333. [DOI] [PubMed] [Google Scholar]
- 27.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, Rameh L, Loda M, Pandolfi PP. 2010. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, Delwel R, Pandolfi PP. 2013. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma XM, Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 30.Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL. 1995. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity 2:321–329. doi: 10.1016/1074-7613(95)90140-X. [DOI] [PubMed] [Google Scholar]
- 31.Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. 1997. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 32.Su B, Jacinto E. 2011. Mammalian TOR signaling to the AGC kinases. Crit Rev Biochem Mol Biol 46:527–547. doi: 10.3109/10409238.2011.618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox CJ, Hammerman PS, Thompson CB. 2005. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med 201:259–266. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB. 2005. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood 105:4477–4483. doi: 10.1182/blood-2004-09-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, Schatz JH, Rodrigo CM, Zhao C, Rondou P, de Stanchina E, Teruya-Feldstein J, Kelliher MA, Speleman F, Porco JA Jr, Pelletier J, Ratsch G, Wendel HG. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imataka H, Sonenberg N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 17:6940–6947. doi: 10.1128/MCB.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 38.Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, Rao A, Ansel KM. 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. 2014. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 42.Christie M, Boland A, Huntzinger E, Weichenrieder O, Izaurralde E. 2013. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol Cell 51:360–373. doi: 10.1016/j.molcel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Eulalio A, Huntzinger E, Izaurralde E. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 44.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 45.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huntzinger E, Kuzuoglu-Ozturk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. 2013. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 49.Yao B, Li S, Lian SL, Fritzler MJ, Chan EK. 2011. Mapping of Ago2-GW182 functional interactions. Methods Mol Biol 725:45–62. doi: 10.1007/978-1-61779-046-1_4. [DOI] [PubMed] [Google Scholar]
- 50.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. 2013. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flynn A, Proud CG. 1996. The role of eIF4 in cell proliferation. Cancer Surv 27:293–310. [PubMed] [Google Scholar]
- 53.Lazaris-Karatzas A, Montine KS, Sonenberg N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 54.Lujambio A, Lowe SW. 2012. The microcosmos of cancer. Nature 482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.