Abstract
Thyrotrope hyperplasia and hypertrophy are common responses to primary hypothyroidism. To understand the genetic regulation of these processes, we studied gene expression changes in the pituitaries of Cga−/− mice, which are deficient in the common α-subunit of TSH, LH, and FSH. These mice have thyrotrope hypertrophy and hyperplasia and develop thyrotrope adenoma. We report that cell proliferation is increased, but the expression of most stem cell markers is unchanged. The α-subunit is required for secretion of the glycoprotein hormone β-subunits, and mutants exhibit elevated expression of many genes involved in the unfolded protein response, consistent with dilation and stress of the endoplasmic reticulum. Mutants have elevated expression of transcription factors that are important in thyrotrope function, such as Gata2 and Islet 1, and those that stimulate proliferation, including Nupr1, E2f1, and Etv5. We characterized the expression and function of a novel, overexpressed gene, transcription elongation factor A (SII)-like 5 (Tceal5). Stable expression of Tceal5 in a pituitary progenitor cell line is sufficient to increase cell proliferation. Thus, Tceal5 may act as a proto-oncogene. This study provides a rich resource for comparing pituitary transcriptomes and an analysis of gene expression networks.
Thyrotropes make up to 5%–10% of all cells in the adult pituitary gland. These cells are essential for thyroid development and function, growth, and homeostasis, but our knowledge of how they arise from undifferentiated progenitors is incomplete. Pou1f1 is expressed at embryonic day (E)13.5 in the mouse pituitary gland and is required for development of somatotropes, lactotropes, and thyrotropes (1–4). The signature markers of a thyrotrope are chorionic gonadotropin-α (Cga) and TSH-β (Tshb). Cga is expressed first in the rostral tip at E11.5 and later in the caudo-medial anterior lobe cells, whereas Tshb expression is detected in both areas at E14.5 (5). Tshb expression in the caudo-medial area is Pou1f1 dependent, but POU1F1-negative, TSHβ-positive cells exist in neonatal mice (6, 7). The gonadotropes express Cga and LH-β (Lhb) or FSH-β (Fshb) from E15.5 and E16.5, respectively, and the mature gonadotropes may emerge from TSH, FSH double-positive cells (5, 8).
The factors that drive Pou1f1 progenitors towards a thyrotrope fate are not known. Gata2 is expressed in gonadotropes and thyrotropes, and it acts synergistically with POU1F1 to stimulate Tshb expression (9, 10). However, Gata2 is not essential for thyrotrope or gonadotrope differentiation (11). Mice with a pituitary-specific knockout of Gata2 have fewer gonadotropes and thyrotropes at birth, and the function of these cells is modestly impaired. Several other factors have been implicated in Tshb expression, including LHX3, PITX1/2, Nuclear receptor subfamily 4, group A, member 1, Mediator complex subunit 1, Nuclear receptor co-repressor 1, EYA transcriptional co-activator and phosphotase 3, Sine oculis-related homeobox 1, Thyrotroph embryonic factor, and Hepatic leukemia factor, but none have been shown to be exclusively necessary for the thyrotrope fate (10, 12–15). The Lin11/Isl-1/Mec-3 (LIM)-type homeodomain transcription factor, Islet 1, is expressed in gonadotropes and thyrotropes and is necessary for early pituitary development and maximal thyrotrope response to hypothyroidism (7, 16, 17). However, it is dispensable for thyrotrope and gonadotrope fate (7).
Cga transcription is regulated differently in thyrotropes and gonadotropes. In these 2 cell types, overlapping areas of the promoter region have been implicated for cell-specific expression. In thyrotropes, Cga expression is regulated by GATA2, PITX1, LHX2/3, MSH homeobox, and E26 transformation-specific transcription factor or Trans-acting transcription factor 1 (14, 18–23), but none of these factors are exclusively necessary for thyrotrope fate. In gonadotropes, SF1 (NR5A1), GATA2, and PITX1 are involved in Cga expression (reviewed in Ref. 22). In summary, studies of the regulation of Cga expression have not uncovered thyrotrope critical factors.
Multiple genetic defects can cause congenital central hypothyroidism, and several pituitary cell lineages can be affected, especially somatotropes and lactotropes together with thyrotropes (24). The somatotropes and lactotropes appear to require thyroid hormone (TH) for complete differentiation and/or population expansion. Consistent with this idea, several hypothyroid mouse models exhibit reductions in somatotropes and lactotropes, including the Tshrhyt/hyt, Tpst2grt/grt, and Cga−/− mice (25–29). Cga−/− mice have severe hypothyroidism, hypogonadism, infertility, and develop thyrotrope adenomas by 9 months to 1 year with high penetrance (29). Serum TH is undetectably low, and there is a profound thyrotrope hypertrophy and hyperplasia by 8 weeks. The population sizes of cells in the Pou1f1 lineage are shifted dramatically. Normally the adult pituitary is composed of approximately 40% somatotropes, 30%–40% lactotropes, 10% corticotropes, 7%–10% gonadotropes, and 5% thyrotropes (30). Cga−/− pituitaries have approximately 45%–50% thyrotropes, 12.5% somatotropes, only 3% lactotropes, 10% corticotropes, and 7% gonadotropes (29, 31, 32). This and other features are reversible by TH replacement (32). The profound thyrotrope hypertrophy and hyperplasia in Cga mutants make them a great tool to study thyrotrope cell specification, proliferation, and response to hypothyroidism.
Materials and Methods
Experimental animals, sample collection, RNA, and cDNA preparation
The animal care and use protocol was approved by the University Committee on Use and Care of Animals at the University of Michigan. Cgatm1Sac mice were from our stock (29). For gene expression studies pituitaries were collected from 8-week-old mice of each sex and genotype (see specific numbers at each experiment). For absolute quantification studies, pituitaries were collected from 6 wild-type and 5–6 null mice at birth, and 4 weeks. RNA extraction and cDNA preparation was described previously (33).
Gene expression microarray
RNA was prepared from 24 pituitary samples: 6 males and 6 females per genotype (33). The Illumina TotalPrep RNA Amplification kit was used to prepare biotin-labeled cRNA from 500-ng RNA; 1500-ng cRNA was hybridized to Illumina MouseWG-6 v2.0 Expression BeadChip for 18 hours at 58°C (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6887). BeadChips were scanned, and signal intensity was recorded with an Illumina iScan. Image data were analyzed and quantile-normalized with Illumina Genome Studio (v2011.1, Data Analysis Software package with Gene Expression Module v1.9.0 and manifest MouseWG-6_V2_0_R2_11278593_A). Probes with a detection P ≤ .01 were filtered and genes with a concordance of one were included in the analysis. Our data is available in NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79451). Genes expressed with a fold change of more than or equal to 1.5 or more than or equal to −1.5 in the wild type vs Cga−/− were analyzed with Ingenuity Pathway Analysis software (QIAGEN).
Quantitative PCR
We confirmed gene expression changes with quantitative PCR using either TaqMan or SYBR Green I assays. SYBR Green primers were designed with Primer BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/; National Center for Biotechnology Information) according to the MIQE guidelines (34). Products were confirmed with bidirectional Sanger sequencing. Reactions were performed in triplicate either in the ABI Real-Time PCR 7900HT or 7500 on a 384- or 96-well platform with all default run parameters and standard reagents. Data was processed using MS Excel 2010, GraphPad Prism 6.01 and REST (35). Results were listed as fold change ± SEM in the Cga−/−. For relative quantification, we used pituitaries from 3 male and 3 female 8-week-old mice for each genotype. Reactions were performed with 50-ng cDNA. For absolute quantification of transcription elongation factor A (SII)-like 5 (Tceal5) transcripts, pituitaries of 6 wild-type and 5–6 null mice from the ages of birth, 4, and 8 weeks were collected. A fragment of Tceal5 cDNA was PCR amplified and gel purified (QIAGEN), quantified (Nanodrop) and strand number was defined using http://molbiol.edu.ru/eng/scripts/01_07.html. A calibration curve with 10-fold increments was constructed with Tceal5 strand numbers of 1E9 to 1E1. Primers and TaqMan assays are provided in the Supplemental Materials and Methods.
Cloning of Tceal5 in situ hybridization (ISH) probe, Tceal5-EGFP transgene
We amplified a 336-bp piece of the Tceal5 cDNA (ENSMUST00000066819; primers, 5-TCTCTTCCAGGTACCAGCTACCAGC-3 and 5-TCTAGCTTGCCCTGGCGTGC-3) from a 8-week-old Cga−/− pituitary cDNA and TA-cloned it into the pGEM-T-Easy vector (Promega). The 779-bp Tceal5 cDNA was cloned together with an in-frame 3′ EGFP into the pcDNA3.1(−) vector (Invitrogen). Briefly, the first 779 bp of the Tceal5 cDNA before the stop and the cDNA encoding EGFP (pEGFP; Clontech) were PCR amplified with primers containing extra restriction endonuclease sites. In the final construct, the pieces were ligated through a KasI site and inserted between the XbaI and AflII sites of pcDNA3.1(−) to produce TCEAL5-EGFP. Using a similar strategy, the EGFP sequence alone was inserted between the XbaI/AflII sites in pcDNA3.1(−) to produce EGFP. All plasmids were confirmed by Sanger sequencing.
Tissue processing, ISH, and immunohistochemistry (IHC)
Tissues were fixed for 30 minutes in 4% paraformaldehyde and processed as described previously (7). For generating Tceal5 probes, plasmid template was cleaved using SpeI for sense and SacII for antisense, and the antisense template was blunted. Dioxigenin-labeled probes were synthesized as published and purified with Chroma Spin DEPC-H2O 100 columns (Clontech) (17, 36). ISH was performed at 54°C, and standard alkaline phosphatase detection (17, 36) was modified based on (37): alkaline phosphate was neutralized after 48 hours, slides were counterstained with methyl green (Vector Labs), dehydrated with series of graded ethanols (25%–50%–70%–100%, 1 min each), and mounted using Vectamount (Vector Labs). For IHC, we used antibodies listed in Table 1, and further details are described in the Supplemental Materials and Methods. For sequential ISH followed by IHC, the IHC staining was developed using the Vectastain Elite ABC kit (Vector Biolabs) with 3,3′-diaminobenzidine substrate.
Table 1.
Table of Antibodies
| Peptide/Protein Target | Antigen Sequence (if Known) | Name of Antibody | Manufacturer, Catalog Number, and/or Name of Individual Providing the Antibody | Species Raised in; Monoclonal or Polyclonal | Dilution Used |
|---|---|---|---|---|---|
| TSH | β-Subunit | Anti-r TSH-β | A. F. Parlow, Hormone Distribution Program, AFP967793 | Guinea pig; polyclonal | 1:20 000 |
| Guinea pig IgG | Biotinylated antiguinea pig IgG (H + L) | Jackson ImmunoResearch, 706-065-148 | Donkey; polyclonal | 1:200 | |
| TSH | β-Subunit | Anti-m TSH-β | A. F. Parlow, Hormone Distribution Program | Rabbit; polyclonal | 1:5000 |
| Rabbit IgG | 15-nm gold-conjugated goat antirabbit (H + L) | British Biocell, EM.GAR15 | Goat; polyclonal | 1:60 | |
| POU class 1 homeobox 1 | Rat POU1F1 | Anti-r POU1F1 | S. Rhodes, Indiana University Purdue University-Indianapolis, 1603 | Rabbit; polyclonal | 1:300 |
| Rabbit IgG | Biotinylated antirabbit IgG (H + L) | Jackson ImmunoResearch, 711-066-152 | Donkey; polyclonal | 1:200 |
Cell culture, generation of heterologous cell lines with a stable transgene
The Pit1-triple cell line was a kind gift from S. Liebhaber, Y. Ho and N. Cooke (University of Pennsylvania), and cells were maintained under their published conditions (38). 2E6 Pit1-triple cells were transfected after 24 hours of plating with 4.2 μg of EGFP-pcDNA3.1 or TCEAL5-EGFP-pcDNA3.1 with FuGene6 (3:1 for FuGene6 to DNA ratio; Promega). After 72 hours of transfection, cells were selected with 1250-μg/mL G418 for 7 days (Roche). EGFP-positive cells were collected with a Sony Biotechnology Synergy iCyte flow cytometer and cultured.
Proliferation and programmed cell death assessment with cultured cells
2E5 TCEAL5-EGFP or EGFP cells were plated in 100-mm dishes. Cells were counted in 0.4% trypan blue from 3 dishes per transgene type after 4 and 8 days using a Luna automated cell counter (Firmware 2.5.2; default declustering; Logos Biosystems). Three aliquots were measured from each plate. Nine data points per group were compared with Mann-Whitney U test, with significance level set to 0.05.
1E5 TCEAL5-EGFP or EGFP cells per well were plated in 12-well cluster plates. Twenty-four hours later, apoptosis was detected using the TMR red In Situ Cell Death Detection kit (Roche) in 9 wells along with additional wells for positive and negative controls. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged on the TMR red and DAPI channels with a Leica DMIRB inverted microscope. DAPI+ nuclei were counted automatically and terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL+, TMR red+) cells were manually counted with ImageJ. A third of the DAPI images was manually counted as well. The percentage of TUNEL+/DAPI+ cells was compared with SPSS v22 (IBM). These values were tested for normal distribution and homogeneity with Shapiro-Wilk and Levene test, and the means were compared with one-way ANOVA and Scheffe post hoc test.
Live cell imaging
Cells in culture were imaged using the Olympus FluoView 500 inverted confocal microscope with argon laser (488 nm) and overlaid with bright field images. Dishes were kept in a Pathology Devices LiveCell stage top incubator controlled for 5% CO2 and 37°C. Individual cells at high resolution were imaged using a chambered cover glass (Nunc Lab-Tek). For videos, the images for the z-stack were compiled using 0.5-μm numerical aperture, ×100 oil immersion objective, ×2 digital zoom, at 1024 × 1024-pixel resolution.
Proliferation assessment with 5-ethynyl-2′-deoxyuridine incorporation
Eight-week-old mice (3 wild types and 3 nulls) were administered 50-μg/g body weight 5-ethynyl-2′-deoxyuridine (EdU) dissolved in dimethyl sulfoxide through ip injection. Mice were sacrificed after 2 hours. EdU staining was done on coronal sections from the midrange of the slide series that contained all pituitary lobes. The Click-iT EdU Imaging kit with Alexa Fluor 488 dye (Thermo Fisher Life Technologies) was used as described in Supplemental Materials and Methods. When EdU imaging was combined with ISH the ISH was performed first. Slides were imaged as described before (17). ImageJ was used for manually counting DAPI and EdU-positive cells. EdU to DAPI ratios were compared with SPSS as described above.
Electron microscopy (EM) studies
For EM analysis, pituitary glands were processed and analyzed as described previously (39). Briefly, the tissue was contrasted with uranyl acetate (2% [wt/vol] in distilled water) at room temperature, dehydrated in methanol, and embedded in LR Gold resin at −20°C. Ultrathin sections (50–80 nm) were prepared using a Reichart-Jung ultracut microtome and mounted on nickel grids (Agar Scientific). Sections were immunolabeled for TSH for 2 hours at room temperature with rabbit antimouse TSH primary antibody followed by a 1-hour incubation with a 15-nm gold-conjugated goat antirabbit secondary antibody (British Biocell). Antibodies are further described in Table 1. All antibodies were diluted in 0.1M PBS (containing 0.1% egg albumin). Specificity of antibody labeling was confirmed by the absence of labeling in negative control sections in which the primary antibody was replaced with nonimmune serum. Finally, sections were counterstained with lead citrate and uranyl acetate and examined on a JOEL 1010 transmission EM (JOEL).
Results
Thyrotropes in Cga−/− mice have dilated endoplasmic reticulum (ER) filled with TSH-β-subunit
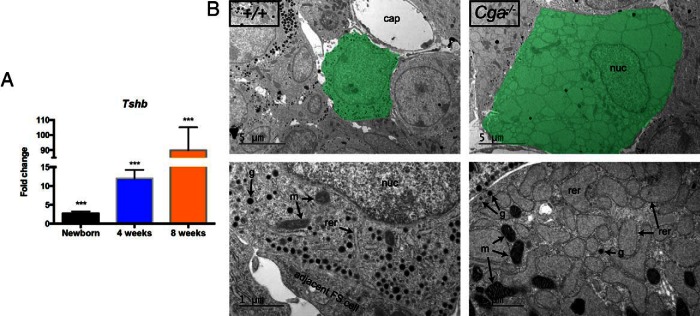
Thyrotrope hypertrophy is evident in 3- and 8-week-old Cga−/− mice, but not at birth, and increased Tshb mRNA has been documented in 8-week-old mutants by ISH and Northern blotting (29, 32). To quantify the progressive increase in Tshb expression during development, we carried out qPCR analysis of pituitaries from mutants and wild types at birth, 4, and 8 weeks. Fold changes in Tshb transcripts were 2.7 ± 0.5 at birth, 12.0 ± 2.3 at 4 weeks, and 89.9 ± 15.2 at 8 weeks (P = .001 for all) (Figure 1A). Thyrotrope hypertrophy in Cga mutants is similar to that observed in radio-thyroidectomized mice, but the thyrotrope hyperplasia is more extensive (29). To compare the cellular effects of hypertrophy in Cga mutants with that reported for other hypothyroid models, we examined the ultrastructure of thyrotropes in 8-week-old Cga−/− by transmission EM. Thyrotropes have about 2 times the diameter of the wild-type ones (Figure 1B, top). The cisternae of the ER are normally thin and elongated and comprise only a small portion of the cytoplasm. The mutant ER appears quite different. Virtually the entire cytoplasm of the cell is filled with dilated ER cisternae (Figure 1B, bottom). Dilated ER is consistently found in most mutant thyrotropes, but about 1 in 10 thyrotropes had a mix of smaller and larger cisternae. There are very few secretory granules in the mutant thyrotropes, probably because heterodimerization of CGA and TSHB is required for TSH secretion (40). The mutant mitochondria are more electron dense than in controls, and are in close contact with the ER. The mutant cristae are markedly electron dense and separated by electro lucent sheets. This mitochondrial phenotype is similar to that reported after exposure to oxidative stress (41, 42). These findings suggest disrupted thyrotrope function in the cellular compartments responsible for energy production and protein synthesis, folding, assembly, and secretion.
Figure 1.
Cga−/− mice show a progressive increase in Tshb mRNA expression from birth to adulthood and present with extremely dilated ER cisternae. A, Quantitative PCR with relative quantification showing the increase of Tshb transcript expression during postnatal life in the Cga−/− vs wild type. B, Transmission EM showing thyrotropes identified by TSHB antibody staining from a normal and Cga−/− mouse pituitary at 8 weeks. Top panels show a low-magnification view of the overall difference in the size of a typical thyrotrope shaded with green. Bottom panels depict an area within a typical thyrotrope with the characteristic thin-layered ER structure in wild types and greatly dilated ER in mutants. Also note the decreased number of secretory granules in the mutants, as well as the more electron dense mitochondria. cap, capillary; nuc, nucleus; g, secretory granule; m, mitochondrion; rer, rough ER.
Increased proliferation in Cga−/− pituitaries
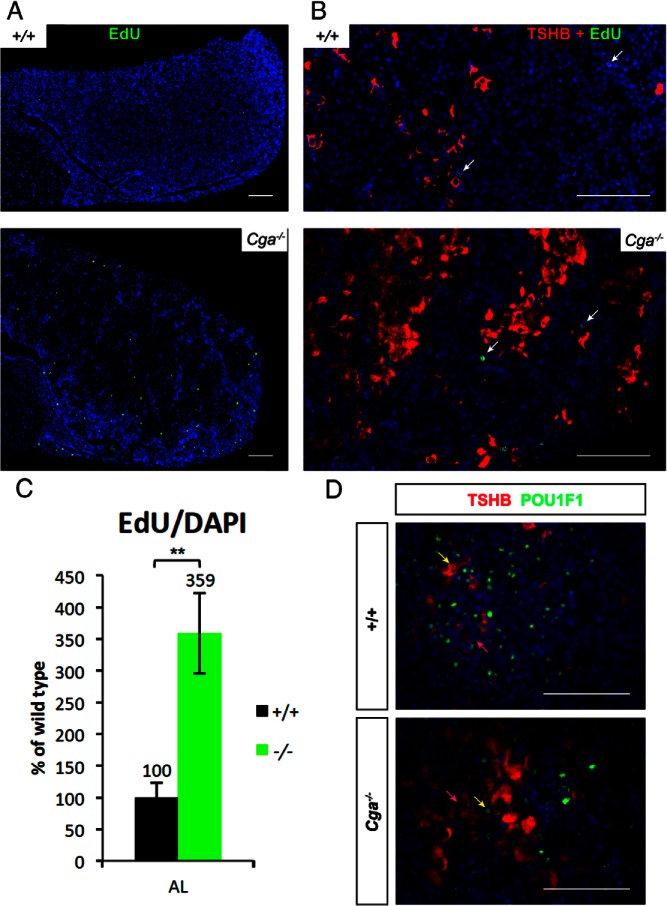
Cga mutants exhibit progressive pituitary hyperplasia, and one year old mutants have pituitaries that are approximately 5–50× normal size with transplantable adenomatous tumors that retain TH responsiveness (43). To assess the proliferation rate at 8 weeks, we injected mice with EdU to mark cells in DNA synthesis or S phase. A small fraction of the anterior pituitary cells were labeled in both genotypes (Figure 2A), but it was increased 3.6-fold in Cga−/− (0.23 ± .05% vs 0.82 ± .14% EdU/DAPI, P = .06) (Figure 2C). No colabeling with TSH antibodies and EdU was observed in either genotype (Figure 2B). The thyrotrope population is comprised of POU1F1 positive and negative cells (Figure 2D). This suggests that the thyrotropes are quiescent at this age and that there may be multiple routes to develop new thyrotropes.
Figure 2.
Thyrotrope hypertrophy and hyperplasia are associated with proliferation of TSHB-negative cells. A, Pituitaries were stained for EdU, which marks proliferating cells in S phase with green and counterstained with DAPI that makes the nuclei blue. B, Pituitary sections stained for TSHB in red, EdU in green, and nuclei in blue showed no costaining of TSHB and EdU in either genotype. Arrows indicate EdU-positive cells. Clusters of thyrotropes are interspersed with TSHB-negative cells. Scale bar, 100 μm (A, B, and D). Mice were 8 weeks of age. C, The proportion of DAPI positive cells that are EdU positive is increased in the Cga−/− mice. Ratio is compared with wild type. D, Thyrotropes stained with TSHB (red) from representative areas of 8-week-old pituitaries show a mix of POU1F1 positive (green) and negative cells in both genotypes. Nuclei were counterstained with DAPI.
Expected expression changes in cell lineage-specific genes in Cga−/− pituitaries
The increase in thyrotropes at the expense of somatotropes and lactotropes provides a unique opportunity to identify genes that drive thyrotrope fate and play roles in thyrotrope hypertrophy and hyperplasia. We performed a microarray experiment to identify differentially expressed genes in the Cga−/− pituitary. To assess the success of the experimental design, we examined changes in the expression of signature genes for the main differentiated pituitary cell lineages. As expected, the thyrotrope-related genes Trhr, Gata2, and Isl1 were up-regulated, and Cga was down-regulated (Table 2). Transcription factors related to the somatotropes and lactotropes were down-regulated, including Stat5a, Smad3, and Pou1f1. There is no gross change in the gonadotrope cell number (31, 32), and the gonadotrope transcription factor Nr5a1 (44) was unchanged. Gnrhr was down-regulated as expected for hypogonadal mice (45). Tbx19, a marker for corticotropes, was unchanged.
Table 2.
Summary of Gene Expression Changes in the Cga−/− Pituitary
| A |
B |
C |
||||||
|---|---|---|---|---|---|---|---|---|
| Fold Change | Fold Change Cut-Off | Number of Genes | Total Number of Genes | Main Category | Total | Up | Down | |
| Immune | 42 | 36 | 6 | |||||
| Thyrotrope | Protein degradation | 35 | 26 | 9 | ||||
| Cga | −4.55 | ≥+1.5 | 487 | 846 | UPR/ER stress | 41 | 33 | 8 |
| Trhr | 14.58 | ≤−1.5 | 359 | Transcription factor | 60 | 30 | 30 | |
| Gata2 | 2.53 | ≥+2.0 | 157 | 267 | Transport | 70 | 41 | 29 |
| Isl1 | 2.55 | ≤−2.0 | 110 | GPCR | 22 | 11 | 11 | |
| Gonadotrope | Autophagy | 8 | 2 | 6 | ||||
| Gnrhr | −2.56 | Cell-cell interaction | 28 | 17 | 11 | |||
| Nr5a1 | 1.10 | Endocytosis | 23 | 15 | 8 | |||
| Fshb | 1.10 | Exocytosis | 14 | 9 | 5 | |||
| Somato-lactotrope | Metabolism | 144 | 80 | 64 | ||||
| Ghrhr | −2.30 | Signaling | 190 | 108 | 82 | |||
| Stat5a | −2.55 | DNA | 19 | 4 | 15 | |||
| Smad3 | −1.67 | RNA | 42 | 21 | 21 | |||
| Pou1f1 | −3.13 | ECM | 30 | 16 | 14 | |||
| Corticotrope | Cytoskeleton | 49 | 28 | 21 | ||||
| Tbx19 | −1.37 | Vesicle | 22 | 12 | 10 | |||
| Unknown | 118 | 61 | 57 | |||||
Column A shows expression of genes that changed or stayed constant based on expected increased numbers of thyrotropes, decreased numbers of somatotropes and lactotropes, and grossly unchanged numbers of gonadotropes and corticotropes (29, 31). All values represent fold changes in the Cga−/− pituitaries. Column B shows overall numbers of differentially expressed genes with fold change cut-offs of ±1.5 and ±2.0. Column C shows differentially expressed genes grouped into 18 main categories based on their involvements in biological processes and molecular functions. The numbers refer to the total number of genes with altered expression in a category, and the subset that are up- and down-regulated. Some genes can be included in multiple categories. UPR, unfolded protein response; GPCR, G protein-coupled receptor; ECM, extracellular matrix.
Elevated expression of genes involved in immune response, ER stress, unfolded protein response, protein degradation, and transcription
Microarray analysis revealed 846 differentially expressed genes with a fold change of 1.5 or greater (Table 2). IPA and Database for Annotation, Visualization and Integrated Discovery software were limited for pathway analysis because a quarter of the differentially expressed genes were not annotated. To overcome this we performed extensive literature analysis of genes using BioGPS (www.biogps.org), NCBI PubMed Gene RIFs (Reference Into Functions; http://www.ncbi.nlm.nih.gov/gene), and Mouse Genome Informatics (http://www.informatics.jax.org/). The main gene clusters are summarized in Table 2 (see also Supplemental Tables 1 and 2). The adenoma might be responsible for the 42 immune response-related genes (46). Clusters of genes related to ER stress, unfolded protein response, and protein degradation were mostly up-regulated, possibly due to the requirement for breaking down the TSH-β-subunit (Table 3). In addition, there were subclusters of genes in connection with mRNA processing (21 total), amino acid metabolism (13 total), posttranslational modifications (102 total), vesicular transport (22 total), and exocytosis (14 total). We found 17 genes with proteins located in the ER and the Golgi that were associated with posttranslational modification (glycosylation 5 up: Ddost, Stt3b, Colgalt2, Gnptg, and Galnt18 and 6 down: Gyltl1b, B3gnt8, Galnt10/11/16, and Csgalnact1; sialization: St3gal1/6; and sulfatation: Hs6st1/2 and Chst8/10). The growth factor signaling group of genes (30 total) included TGFB (5 total), the Wnt pathway (13 total), the small GTPases (24 total), and the phospholipase C/protein kinase C (13 total) as second messengers. To understand the mechanism of increased cell proliferation, we explored the 60 differentially expressed transcription factor complex genes.
Table 3.
Genes Involved in Unfolded Protein Response/ER Stress and Transcription Factor Complex Are Elevated in the Pituitaries of Cga−/− Mice
| A |
B |
|||||
|---|---|---|---|---|---|---|
| Fold Change | Symbol | Gene Name | qPCR Fold Change | Entrez Gene ID | Symbol | Gene Name |
| 9.74 7.10 4.64 4.18 3.25 3.19 3.07 2.55 2.36 2.31 2.26 2.25 2.24 2.20 2.19 2.13 2.03 1.99 1.94 1.85 1.77 1.67 1.64 1.62 1.61 1.61 1.59 1.58 1.54 1.53 1.52 1.50 1.50 −1.61 −1.68 −1.87 −1.93 −2.09 −2.23 −2.25 −3.61 |
Creld2 Sdf2l1 Pdia4 Pdia6 Hspa5 Hsp90b1 Parm1 Sgtb Fkbp14 Ddit3 Ikbip Fkbp11 Fkbp2 Txndc12 Serp2 Erlec1 Hyou1 C1Galt1c1 Dnajb11 Resp18 Pdia3 Dnajb9 Sil1 Ppib Shisa5 Os9 Ssr4 Sdf2 Shisa2 Creb3l1 Edem2 Serf2 Herpud1 Fkbp4 Ism1 Clgn Irak2 Nnat Homer2 Ssr2 Kiaa1324 |
Cysteine-rich with EGF-like domains 2 Stromal cell-derived factor 2-like 1 PDI family A, member 4 PDI family A, member 6 Heat shock 70-kDa protein5 (Grp78) Heat shock protein 90-kDaβ (Grp94), member 1 Prostate androgen-regulated mucin-like protein 1 Small glutamine-rich tetratricopeptide repeat (TPR)-containing, β FK506-binding protein 14 DNA-damage-inducible transcript 3 Inhibitor of nuclear factor κB kinase-interacting protein FK506-binding FK506-binding protein 2 protein 11 Thioredoxin domain containing 12 (ER) Stress-associated ER protein family member 2 ER lectin 1 Hypoxia up-regulated 1 Core 1 synthase, glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase1 (C1GALT1)-specific chaperone 1 DnaJ (Hsp40) homolog, subfamily B, member 11 Regulated endocrine-specific protein 18 PDI family A, member 3 DnaJ (Hsp40) homolog, subfamily B, member 9 SIL1 nucleotide exchange factor Peptidyl-prolyl isomerase B (cyclophilin B) Shisa family member 5 Osteosarcoma amplified 9, ER lectin Signal sequence receptor, δ Stromal cell-derived factor 2 Shisa family member 2 cAMP-responsive element-binding protein 3-like 1 ER degradation enhancer, mannosidase α-like 2 Small EDRK-rich factor 2 Homocysteine-inducible, ER stress-inducible, ubiquitin-like domain member 1 FK506-binding protein 4 Isthmin 1, angiogenesis inhibitor Calmegin IL-1 receptor-associated kinase 2 Neuronatin Homer scaffolding protein 2 Signal sequence receptor-β KIAA1324 |
46.91 4.05 3.67 3.60 3.12 2.75 2.59 2.56 2.23 1.67 1.55 1.44 1.41 1.17 −1.66 −2.00 −2.08 −2.33 −2.38 −2.38 −2.56 −2.56 −2.63 −2.78 −2.86 −2.94 −2.94 −3.13 −3.23 −3.45 −3.70 −6.67 −12.50 |
56312 331532 26427 13555 104156 14461 16392 241494 13198 20289 72693 270118 14199 406217 231986 16871 17122 192231 67087 20747 15901 18736 11835 56458 68705 17341 60599 20850 13654 68728 16601 15460 66255 |
Nupr1 Tceal5 Creb3l1 E2f1 Etv5 Gata2 Isl1 Znf385b Ddit3 Scx Zcchc12 Maml2 Fhl1 Bex4 Jazf1 Lhx3 Mxd4 Hexim1 Ctnnbip1 Spop Id1 Pou1f1 Ar Foxo1 Gtf2f2 Bhlha15 Tp53inp1 Stat5a Egr2 Tp53inp2 Klf9 Hr Hsbp1l1 |
Nuclear protein, transcriptional regulator, 1 Transcription elongation factor A (SII)-like 5 cAMP-responsive element-binding protein 3-like 1 E2F transcription factor 1 Ets variant 5 GATA-binding protein 2 ISL LIM homeobox 1 Zinc finger protein 385B DNA-damage-inducible transcript 3 Scleraxis basic helix-loop-helix transcription factor Zinc finger, CCHC domain containing 12 Mastermind like 2 Four and a half LIM domains 1 BEX, X-linked 4 JAZF zinc finger 1 LIM homeobox 3 MAX dimerization protein 4 Hexamethylene bis-acetamide inducible 1 Catenin, β-interacting protein 1 Speckle-type POZ protein Inhibitor of DNA binding 1 POU class 1 homeobox 1 Androgen receptor Forkhead box O1 General transcription factor IIF, polypeptide 2 Basic helix-loop-helix family, member a15 Tumor protein p53-inducible nuclear protein 1 Signal transducer and activator of transcription 5A Early growth response 2 Tumor protein p53 inducible nuclear protein 2 Kruppel-like factor 9 Hair growth associated Heat shock factor-binding protein 1-like 1 |
Column A shows unfolded protein response/ER stress genes that are predominantly up-regulated and can explain the largely distended ER cisternae that contributes to thyrotrope hyperplasia in Cga−/− mice. Table includes gene expression changes from the microarray. Column B shows 33 genes from the transcription factor complex category that were confirmed with qPCR in total number of 12 pituitaries (equal number of wild-type and mutant mice with equal number of males and females; P ≤ .004).
Tceal5 is a novel regulator of cell proliferation
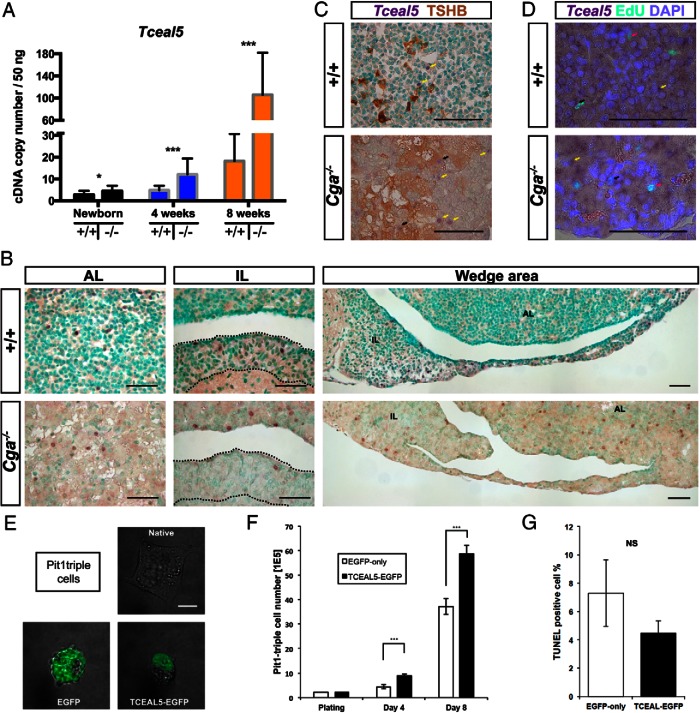
Using qPCR we validated differential expression of 80.5% of the genes with the highest expression levels (33/41) (Table 3). Among these genes were 6 Zn-finger transcription factors, 4 basic helix-loop-helix, 3 basic leucine-zipper, 3 LIM-homeodomain, and 6 in various other transcription factor families. In addition, there were 11 genes that participate in the transcription machinery and elongation. The most up-regulated transcription factor gene was Nupr1 (also known as p8, 46.9 ± 12.4-fold; P = .001), which is not essential for thyrotrope specification but important for cell proliferation and involved in adenoma formation (47–50). Two other highly expressed genes, Etv5 and E2f1 (3.1 ± 0.8- and 3.6 ± 0.9-fold, respectively; P = .001), have known roles in pituitary cell proliferation, (51, 52). We undertook investigation of the second most highly elevated gene, Tceal5 (4.1 ± 1.5-fold; P = .001), because its function is unknown. Although Tceal5 transcripts increase with age in pituitaries of both normal and mutant animals at birth, 4, and 8 weeks, the levels were consistently elevated in the mutants (P ≤ .011 in all pairs) (Figure 3A). We examined the expression of other TCEAL family genes and found that Tceal3 was unchanged (1.0 ± 0.2-fold; P = .323), whereas Tceal6 was slightly decreased (−2.2 ± 0.5-fold; P = .001).
Figure 3.
Characterization of TCEAL5 in the pituitary and in a progenitor cell. A, Tceal5 expression is increased from birth to adulthood in the Cga−/− pituitary. Absolute quantification with qPCR showed an incremental increase in Tceal5 expression from birth through 4 and 8 weeks of age in 50-ng cDNA; P ≤ .011 for all pairs. B, Tceal5 transcripts are localized in the anterior and intermediate lobes of the pituitary. Tceal5 is expressed in few AL and IL cells in the wild-type, and Cga−/− pituitaries show an altered distribution within these lobes. Purple stain marks Tceal5, whereas green is nuclear counterstaining. Scale bar, 50 μm in all panels. C, Cga−/− but not wild-type pituitaries have Tceal5-positive thyrotropes. Combined Tceal5 ISH (purple) with TSHB IHC (brown) and green nuclear counterstaining. In a typical view, yellow arrows mark some Tceal5+/TSHB−, and black arrows mark Tceal5+/TSHB+ cells. Note that single Tceal5+ cells are typically close to thyrotrope clusters. D, A small portion of proliferating cells express Tceal5. Proliferation detected with EdU (green) and Tceal5 mRNA (purple) over nuclear staining with DAPI (blue). Representative views from both genotypes showing cells with arrows as EdU+/Tceal5+ (black), Edu+/Tceal5− (yellow) and EdU−/Tceal5+ (red), respectively. E, Transgenic TCEAL5-EGFP has nuclear expression in the Pit1-triple pituitary progenitor cell line. Individual cells were imaged with confocal microscopy to determine the localization of EGFP in stable, transgenic cell populations. EGFP without TCEAL5 was localized to the cytoplasm, whereas the TCEAL5-GFP fusion localized to the nucleus. Scale bar, 10 μm. F, Stable overexpression of Tceal5 significantly enhances proliferation in a pituitary progenitor cell line. The Pit1-triple line expresses Pou1f1, Gh, Prl, Tshb, and Cga (38). We created heterologous lines stably expressing a Tceal5-Egfp gene fusion or Egfp only and plated the same number of cells on day 0. Six plates with each were monitored and 3 were harvested for cell counting after 4 or 8 days. G, Stable overexpression of Tceal5 does not change the apoptosis rate in a pituitary progenitor line. Nine wells in a cluster plate were assayed for apoptosis with the TUNEL assay heterologous cell lines stably expressing Tceal5-Egfp gene fusion or Egfp only. Nuclei were counterstained with DAPI, and the percentage of TUNEL+ cells was compared. NS, not significant.
Tceal5 is expressed in the anterior and intermediate lobes of the adult pituitary
Robust Tceal5 expression is limited to the adult pituitary, brain, and pancreatic β-cells (NCBI GEO/BioGPS; http://ds.biogps.org/?dataset=GSE10246&gene=331532). We used ISH to assess regional specific expression in the pituitary gland and found scattered distribution of positive cells in the anterior and intermediate lobes but not in the posterior lobe (Figure 3B). The distribution of labeled cells is similar in the intermediate lobes of wild-type and Cga mutant mice. The anterior lobes of wild-type mice had intensely stained cells in the wedge area, and the Cga-nulls had positive cells in the area adjacent to the wedge. These regions are enriched in pituitary progenitors (53). Tceal5 expression did not coincide with TSHβ in the wild types (Figure 3C). In the mutants, a small portion of Tceal5-positive cells was TSHβ positive, and many Tceal5-expressing cells were located near thyrotrope clusters (Figure 3C). About a quarter of the proliferating (EdU+) cells were Tceal5 positive in both genotypes (25% vs 28.6% in the mutant) (Figure 3D).
We examined the expression of stem cell markers because Tceal5 was expressed in a region of the pituitary that is enriched for progenitors. SOX2 immunostaining was indistinguishable in mutant and wild-type pituitaries, and there was no overlap with EdU staining at 8 weeks (data not shown). The microarray did not detect any elevation in expression of stem cell markers. To determine whether there were changes in stem cell marker gene expression that were missed by the microarray, we measured the expression of 16 such genes by qPCR: Ccnd1, c-kit, Egf, Sox2, Sox9, Bmi1, Nestin, Sca1, Hesx1, Pitx2, S100b, CD133, Ki67, Gfra2, Prop1, and Gfap. Most were unchanged, but there were modest changes in Ccnd1 and c-kit (−1.6 ± 0.4; P = .023 and 1.6 ± 0.4; P = .009, respectively). If the stem cell pool is activated to produce new thyrotropes, it is not evident in 8-week-old mice.
Tceal5 increases proliferation of Pou1f1-expressing progenitors
Tceal5 is not expressed in any of the rodent pituitary cell lines that we analyzed, including the rat somato-lactotrope GH3 cells, mouse pregonadotrope αT3–1, mouse corticotrope ATT-20, and Pit1-triple, which expresses POU1F1 and the lineage-specific differentiation markers GH, prolactin, CGA, and TSHβ (data not shown) (38). To study the role of Tceal5 in proliferation, we generated populations of Pit1-triple cells that stably express either a TCEAL5-EGFP fusion protein or EGFP only. Confocal microscopy showed that TCEAL5-EGFP localizes to the nucleus, and EGFP is in the cytoplasm and the secretory vesicles (Figure 3E and Supplemental Videos 1–3). Equal numbers of cells were plated, and 4 and 8 days later, the number of TCEAL5-EGFP cells were significantly increased relative to cells expressing EGFP alone (d 4, 4.4E5 ± 0.8E5 vs 8.7E5 ± 0.9E5; d 8, 37.1E5 ± 6.1E5 vs 58.7E5 ± 3.7E5, respectively; n = 9 at each time point; P < .01) (Figure 3F). To determine whether TCEAL5 decreased the rate of apoptosis we performed TUNEL assays. Twenty-four hours after plating equal numbers of EGFP and TCEAL5-EGFP cells into cluster plates, we found no significant decrease in apoptosis rate (TUNEL+/DAPI+ percentages, 7.30 ± 2.36% vs 4.48 ± 0.88% in the TCEAL5-EGFP; n = 9 each; P = .07) (Figure 3G). Thus, Tceal5 expression is sufficient to enhance proliferation of POU1F1-positive pituitary progenitors.
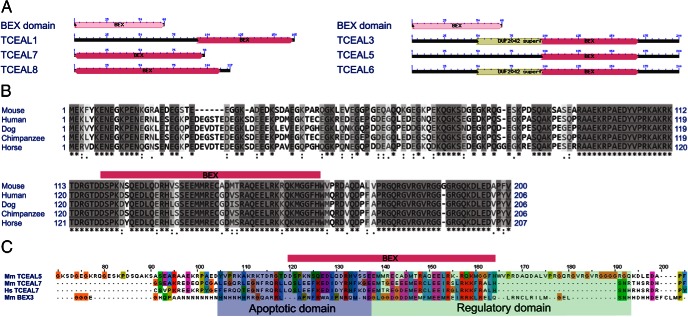
Protein sequence comparisons among TCEAL family members
We compared the amino acid composition of mouse TCEAL proteins and found that they all share the conserved brain expressed (BEX) protein domain (pfam04538) (Figure 4A). TCEAL5 is approximately 80% identical to TCEAL3 and TCEAL6 but only approximately 20%–30% identical to TCEAL1, TCEAL7, and TCEAL8. Mammalian TCEAL5 proteins are highly conserved. Proteins of human, dog, chimpanzee, and horse are 82% (164/200) similar (Figure 4B). The BEX3 proapoptotic and regulatory functions are defined (Figure 4C), and mouse BEX3 and human TCEAL7 tumor suppressors have similar functions (54–57). We compared mouse TCEAL5 with these 2 proteins, and although the BEX domain is conserved, TCEAL5 diverges in adjacent regions. Thus, it is not surprising that the proto oncogenic functions of TCEAL5 are different than TCEAL7.
Figure 4.
Protein sequence comparison suggests similar and divergent functions for the mouse TCEAL5. A, Conserved domain comparison for mouse TCEAL proteins (NCBI Conserved Domain search, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). TCEAL proteins share the BEX domain and TCEAL3/6 include an approximately 70-amino acid-long segment similar to the conserved DUF2042 domain, which has an unknown function. B, The mouse TCEAL5 protein is highly conserved among mammals. Q5H9L2, Q8CCT4, E2RDN6, H2R4M4, and F6UAE4 protein sequences from the UniProt database (http://www.uniprot.org/) for human, mouse, dog, chimpanzee, and horse, respectively. Symbols mark the following: asterisk, identical; colon/semicolon, similar; hyphen, missing amino acids. The BEX domain is marked with magenta box. C, Functional domain comparison suggests different functions in proliferation for TCEAL5 compared with related proteins. Alignment was created with K-Align from the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/msa/kalign/) with default settings and coloring of individual amino acid residues followed the Clustal X color scheme (http://www.jalview.org/help/html/colourSchemes/clustal.html). Magenta, translucent blue, and green boxes are BEX domain, proapoptotic, and regulatory functional domain, respectively.
Discussion
Primary hypothyroidism causes thyrotrope hyperplasia and hypertrophy
Primary hypothyroidism stimulates the pituitary to increase TSH production and secretion because of the loss of negative feedback by TH (58). Fetal thyrotropes become active late in gestation (59), and the size of the population increases in the postnatal period in response to TRH (60). After birth, low serum TH levels affect multiple cell lineages (Supplemental Table 3) (27–29, 32, 61–63). This characteristically includes an increase in overall thyrotrope cell size, first described as the thyroidectomy cell population, and a 1.5- to approximately 10-fold increase in thyrotrope cell number. Interestingly, this is coupled with a substantial decrease in the somatotropes and lactotropes. Because these 3 specialized cell types are Pou1f1 dependent, the expansion of thyrotropes may occur at the expense of the other 2 cell types (1). In support of this idea, hypothyroid mice and rats exhibit similar changes in cell populations whether the hypothyroidism is generated by ablation of the thyroid gland genetically (64), chemically (62, 63), surgically (61), or via a genetic defect in the TSH synthesis (27, 29, 31, 32) and/or action (28, 32) (Supplemental Table 3). However, we cannot rule out the involvement of different mechanisms. To our knowledge, no comprehensive pituitary gene expression analysis has been done previously using any of these models of hypothyroidism. Cga−/− mice have robust thyrotrope hyperplasia, making them a useful model to study gene expression changes associated with expansion of the thyrotrope population. Gene expression changes will also reflect the shift in proportions of other cell types, the reduced or undetectable levels of gonadal steroids and TH, respectively, and cellular changes associated with the inability to secrete heterodimeric glycoprotein hormones.
ER distension and subunit coupling defect in Cga−/− thyrotropes
Hypothyroidism causes thyrotrope hypertrophy, which is partially reversible by TH administration (27, 28, 31, 32, 61–64). Ultrastructure analysis has revealed increased cisternae in the ER and an increase in secretory granules in various hypothyroid mice (28, 61–63, 65). We observed similar effects on the ER in Cga−/− mice, but we observed very few secretory granules. The paucity of secretory granules is likely due to their inability to secrete TSHβ in the absence of the α-subunit (40, 66–69).
The heterodimerization of CGA with TSHB, FSHB, LHB, and chorionic gonadotropin beta subunit occurs in the ER, and β-subunits are required to dimerize with CGA for specific activation of the corresponding receptors (40, 66–70). CGA and LHβ can be secreted without heterodimerization, but they are differentially glycosylated (40, 71). Thus, the mutant gonadotropes are normal sized while being capable of secreting the LHβ monomer and undergoing hypertrophy in response to hypogonadotropic hypogonadism if TH is replaced (31, 32). The Cga−/− mouse thyrotrope is stimulated to overproduce the TSH-β-subunit but cannot secrete it. Thus, TSH-β needs to be eliminated through alternative mechanisms.
Excess, uncoupled TSHβ triggers multiple cellular responses
Protein quality control (QC) starts during the synthesis of the nascent protein at the ER bound ribosome (72). Protein complexes of multiple subunits are common. Many familiar examples come from the immune system, such as the T- and B cell receptors and the main histocompatibility complex (MHC) class I and II molecules (73–75). The inhibin and activin subunit complexes are examples from the endocrine system (76). The machinery used for protein QC includes ER resident heat shock proteins Hspa5, Hsp90b1, Dnajc1, Sec63, and Dnajb9–Dnajb11) and folding enzymes (peptidyl-prolyl isomerases, FK506 [rapamycin[-binding proteins), carbohydrate-binding proteins (calnexins, calreticulins), and protein disulfide isomerases (PDIs) (eg, ERp57) (72, 73). Cga−/− pituitaries have 41 differentially expressed genes in the unfolded protein response/ER stress cluster, including Pdia4/6/3, Hspa5, Hsp90b1, Dnajb11/9, Ppib, and Fkbp14/11/2 (Table 3). Among these, Hspa5 is considered to be the master regulator of the downstream ER stress response (72, 73, 77). We found 17 up-regulated genes related to immune functions, including MHC II antigen presentation, and 17 differentially expressed genes associated with glycosylation, sialization, and sulfation in the ER and Golgi. Transfer of various glycosides, sulfate and sialo groups to CGA and TSHβ are indispensable for heterodimerization (71, 78–84) and receptor activation (66, 80, 85, 86). We hypothesize that some of the same pathways involved in processing TSH subunits are involved in regulating dimerization of other complex proteins. Alternatively, increased expression of immune system genes could be related to responsiveness to tumor development or eliminating stressed pituitary cells. Pituitary folliculostellate cells have phagocytosis functions that remove degenerating cells (87–89), and they present consumed antigens via the MHC II pathway (90).
ER stress response involves proteasomal and lysosomal degradation
There are at least 3 known unfolded protein response regulatory pathways distinguished by the ER sensor protein, such as inositol-requiring transmembrane kinase/endonuclease A, protein kinase R-like ER kinase (PERK), and activating transcription factor 6 (ATF6), which mediate signaling from the stressed ER to the nucleus (91). The response involves transcriptional regulation of genes in ER protein QC, ubiquitin-mediated proteasomal degradation complex (ER-associated degradation), antioxidant response, amino acid metabolism, lipid and membrane lipid biogenesis, and the secretory machinery (92). In Cga−/− pituitaries, the most active pathways involve PERK and ATF6. Downstream target genes for PERK include Ddit3, which we confirmed to be differentially expressed (2.2 ± 0.5-fold; P = .001). Creb3l1 is highly similar to Atf6, and our data confirmed a significant increase in its expression (3.7 ± 1.0-fold; P = .001) (93). We found many up-regulated genes for proteasomal degradation, vesicular transport, and an array of amino acid and lipid metabolism genes. Many genes involved in vesicular transport have roles in ER to Golgi transportation (ie, Copg2, Tmed2/3/9, Sec11). The quantity of genes associated with lysosomal protein degradation clearly implicates this as an operative mechanism. These results are consistent with an active ER stress response along with up-regulated downstream targets, protein degradation, and amplified vesicular transport function.
Chronic ER stress can trigger apoptosis via the mitochondrial pathway (BCL2 protein family members) and by long-term DNA damage inducible transcript 3 expression (91, 94). Cga−/− mice have decreased expression of proapoptotic proteins such as Bag2/3, Bik, and Gadd45a/g. We found very little evidence of apoptosis with activated caspase-3 immunostaining at 8 weeks of age (data not shown). This is consistent with down-regulated Bag2/3 and Bik expression and increased BCL2 expression in the thyrotropes of Cga−/− mice treated with TH (31). Gadd45a is a downstream target of P53, and Gadd45g expression is lost in nonfunctioning pituitary adenomas, and gonadotrope adenomas (95, 96). The chronic ER stress together with low rates of apoptosis suggests that the adult Cga−/− mice manage ER stress with these gene expression changes.
Differentially expressed transcription factors associated with thyrotrope hyperplasia and hypertrophy
We detected increased proliferation in TSHβ-negative anterior lobe cells in the marginal zone, between the anterior lobe and intermediate lobe, where pituitary stem cells reside (53). We did not detect any substantial differences in the proliferation of SOX2 expressing cells or expression of stem cell markers. This argues against an expansion of pituitary stem cells in adult Cga−/− mice, although we cannot rule out an expansion of stem cells in younger animals.
Up-regulated transcriptional regulators may drive cell proliferation and thyrotrope fate (“cell number management”), and down-regulated genes may reflect a loss of TH stimulation (“hypothyroidism genes”) and fewer somatotropes and lactotropes (“lineage-specific genes”). Klf9 and Hr are classic TH-regulated genes (97, 98). Id1 overexpression silences Cga expression in pituitary pregonadotrope (αT3-1) and thyrotrope (αTSH) cell lines; therefore, the lack of Cga transcripts may lead to decreased Id1 expression (99). Foxo1 is expressed in somatotropes (100). Signaling via the GH receptor and PRL receptor activates Stat5a, and these receptors are expressed in endocrine target tissues and somatotropes and lactotropes (101–103). The decrease in Pou1f1 is likely due to the decrease in the number of somatotropes and lactotropes, because the increase in thyrotropes is less than the net decrease in the other 2 cell types. Spop is a positive regulator in gastric, prostate, colorectal, and kidney cancers. The elevated expression of Trp53inp1/2 genes induces cell cycle arrest and inhibits autophagy (104–107). We focused our follow up studies on up-regulated transcription factor genes.
Among the top up-regulated transcription factor genes were Nupr1, Tceal5, Creb3l1, E2f1, Etv5, Gata2, and Isl1. The increase in Gata2 transcripts was expected because of the increase in thyrotropes, and we recently demonstrated the role of Isl1 in thyrotrope differentiation and function (7, 9). The increases in Nupr1, Creb3l1, E2f1, and Etv5 expression were not anticipated, but elevated expression of these genes is consistent with their established roles in cell proliferation. Nupr1 is implicated in hypertrophy, cell proliferation, and cancer in the pituitary, pancreas and muscle, although it is not essential for thyrotrope fate (48–50, 108). Atf4 is the target of the PERK-mediated unfolded protein response, which is highly active in Cga mutants, and ATF4 up-regulates Nupr1 transcription (91, 109). Creb3l1 also ties into the ER stress regulated pathway (ATF6) (77, 91, 93). Nonfunctioning lactotrope and somatotrope adenomas have increased E2f1 expression (52). Increased E2f1 expression reduces Wnt/β-catenin signaling, and we found 12 genes related to Wnt signaling that would be associated with reduced activity (Ctnna1, Wisp1, Wnt10a, and Tle2) and up-regulated Wnt inhibitor (Dkk3 2.8 ± 0.7-fold; P = .001). However, Ctnnbip1, a key node between E2f1 and Wnt signaling, was down-regulated (−2.4 ± 0.6-fold; P = .001) (110), and there was no evidence for alteration in Ctnnb, Axin2, or Lef1 (data not shown). High Etv5 expression in endometrial carcinoma positively regulates Nupr1 transcription, epithelial to mesenchymal transition, and myometrial invasion (111). In spermatogonial stem cells, Etv5 regulates miR-21, which is critical for self-renewal (51). Thus, transcription factor genes involved in thyrotrope fate and cell proliferation were highly expressed in Cga−/− pituitaries.
Tceal5 is the least studied of the most up-regulated transcription factor genes. Tceal genes share a BEX domain. TCEAL1 is widely expressed in normal tissues, and low expression exists in squamous cell esophageal cancer (112, 113). TCEAL4 is ubiquitously expressed, and expression is decreased in anaplastic thyroid cancer samples and cell lines of the ovarium, cervix, lung, and bladder cancer (114). TCEAL7 is the most well studied family member. Its expression is regulated by CpG methylation and it exerts tumor suppressor functions by modulating expression of the transcription factor nuclear factor κ-light-chain-enhancer of activated B cells target genes, C-MYC and CCND1, in ovarian carcinoma (55–57).
Tceal5 orthologs have been described in mammals (http://www.ncbi.nlm.nih.gov/gene/?term=tceal5). Tceal5 is predominantly expressed in the brain and pituitary gland. It is expressed in the wedge area of the pituitary, which is thought to contain progenitors. We demonstrated increasing expression of Tceal5 in the Cga−/− pituitary over time from birth to 8 weeks of age. Stable overexpression of Tceal5 in a Pou1f1 expressing pituitary progenitor cell line was sufficient to increase cell proliferation, consistent with the idea that it might act as a proto-oncogene. Because the related TCEAL7 is thought to act as a tumor suppressor (55), we compared its protein sequences with mouse TCEAL5 and found many notable structural dissimilarities.
There are several other mouse models that are deficient in a unique subunit of a complex protein. Because these subunits may be as critical as the CGA is for the TSH assembly, we postulate that these mice may also present with protein misfolding, which leads to ER stress in the cells of a tissue where the complex protein would be normally expressed. In the endocrine system, the inhibin α-subunit-deficient mice are especially suggestive of such a mechanism, because they exhibit ovarian granulosa cell or Sertoli cell hyperplasia/tumors at the main site of the normal inhibin α-subunit expression, and have a constant drive for more inhibin A/B synthesis due to unrestrained FSH production (76). The same concept of improper complex protein assembly has already been leveraged in many nonendocrine disorders as a disease of protein conformation leading to ER stress and tissue-specific phenotypes (115).
In our current work, the robust thyrotrope hyperplasia and hypertrophy in Cga−/− pituitaries is intriguing. The thyrotropes of hypothyroid mice consistently present dilated ER associated with the increased production of TSH. Cga−/− mice are unable to couple subunits for secretion, which results in ER stress and up-regulation of the unfolded protein response. Not much is known about these pathways in pituitary tumor formation (92). We found up-regulation of thyrotrope-enriched genes (Islet 1, Gata2) and numerous, relatively novel regulators of cell proliferation: Nupr1, Etv5, E2f1, and Tceal5. Tceal5 is sufficient to increase the growth rate of pituitary progenitors, suggesting a role as a proto-oncogene. Further studies in pituitary adenomas are warranted to clarify its role in normal proliferation as well as in human pituitary adenomas. Tceal5 is encoded on the X chromosome, which may make it compelling to find out whether it is randomly or nonrandomly inactivated in females with pituitary hyperplasia or adenomas. Thyrotrope adenomas are extremely rare, but they mostly follow the histological and ultrastructural phenotype of the Cga−/− pituitary (116). Therefore, thyrotrope adenoma patients are the top ranked candidates in which to look for previously undiscovered CGA deficiency and clarify the proliferation features of TCEAL5 in the human.
Acknowledgments
We thank the University of Michigan Core Facilities: DNA Sequencing (Bob Lyons, Susan Dagenais), Microscopy, and Imaging and Flow Cytometry; Michelle Brinkmeier, Shannon Davis, and Alisha John for contributions to the early stages of the work; and Simon J. Rhodes at Indiana University-Purdue University Indianapolis for providing the POU1F1 antibody. P.G. is a participant of the International Endocrine Scholars Program of The Endocrine Society.
This work was funded by the National Institute of Child Health and Human Development Grant R01HD34283 (to S.A.C.) and the National Institutes of Health Grant 2T32DK007245-39 (to A.A.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ATF6
- activating transcription factor 6
- BEX
- brain expressed
- Cga
- choriogonadotropin-α
- DAPI
- 4′,6-diamidino-2-phenylindole
- E
- embryonic day
- EdU
- 5-ethynyl-2′-deoxyuridine
- EGFP
- enhanced green fluorescent protein
- EM
- electron microscopy
- ER
- endoplasmic reticulum
- GATA
- GATA binding protein
- IHC
- immunohistochemistry
- ISH
- in situ hybridization
- LIM
- Lin11/Isl-1/Mec-3
- MHC
- main histocompatibility complex
- PDI
- protein disulfide isomerase
- PERK
- protein kinase R-like ER kinase
- PITX
- paired-like homeodomain
- POU1F1
- POU domain, class 1, transcription factor 1
- QC
- quality control
- qPCR
- quantitative polymerase chain reaction
- Tceal5
- transcription elongation factor A (SII)-like 5
- TH
- thyroid hormone
- TMR
- tetramethyl rhodamine
- TUNEL
- terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
References
- 1. Camper SA, Saunders TL, Katz RW, Reeves RH. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics. 1990;8:586–590. [DOI] [PubMed] [Google Scholar]
- 2. Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. [DOI] [PubMed] [Google Scholar]
- 3. Sornson MW, Wu W, Dasen JS, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. [DOI] [PubMed] [Google Scholar]
- 4. Simmons DM, Voss JW, Ingraham HA, et al. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711. [DOI] [PubMed] [Google Scholar]
- 5. Japón MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. [DOI] [PubMed] [Google Scholar]
- 6. Lin SC, Li S, Drolet DW, Rosenfeld MG. Pituitary ontogeny of the Snell dwarf mouse reveals Pit-1-independent and Pit-1-dependent origins of the thyrotrope. Development. 1994;120:515–522. [DOI] [PubMed] [Google Scholar]
- 7. Castinetti F, Brinkmeier ML, Mortensen AH, et al. ISL1 is necessary for maximal thyrotrope response to hypothyroidism. Mol Endocrinol. 2015;29:1510–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wen S, Ai W, Alim Z, Boehm U. Embryonic gonadotropin-releasing hormone signaling is necessary for maturation of the male reproductive axis. Proc Natl Acad Sci USA. 2010;107:16372–16377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dasen JS, O'Connell SM, Flynn SE, et al. Reciprocal interactions of Pit1 and GATA2 mediate signaling gradient-induced determination of pituitary cell types. Cell. 1999;97:587–598. [DOI] [PubMed] [Google Scholar]
- 10. Gordon DF, Lewis SR, Haugen BR, et al. Pit-1 and GATA-2 interact and functionally cooperate to activate the thyrotropin β-subunit promoter. J Biol Chem. 1997;272:24339–24347. [DOI] [PubMed] [Google Scholar]
- 11. Charles MA, Saunders TL, Wood WM, et al. Pituitary-specific Gata2 knockout: effects on gonadotrope and thyrotrope function. Mol Endocrinol. 2006;20:1366–1377. [DOI] [PubMed] [Google Scholar]
- 12. García M, Fernández A, Moreno JC. Central hypothyroidism in children. Endocr Dev. 2014;26:79–107. [DOI] [PubMed] [Google Scholar]
- 13. Ohba K, Sasaki S, Matsushita A, et al. GATA2 mediates thyrotropin-releasing hormone-induced transcriptional activation of the thyrotropin β gene. PLoS One. 2011;6:e18667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sloop KW, Meier BC, Bridwell JL, Parker GE, Schiller AM, Rhodes SJ. Differential activation of pituitary hormone genes by human Lhx3 isoforms with distinct DNA binding properties. Mol Endocrinol. 1999;13:2212–2225. [DOI] [PubMed] [Google Scholar]
- 15. Masumoto KH, Ukai-Tadenuma M, Kasukawa T, et al. Acute induction of Eya3 by late-night light stimulation triggers TSHβ expression in photoperiodism. Curr Biol. 2010;20:2199–2206. [DOI] [PubMed] [Google Scholar]
- 16. Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. [DOI] [PubMed] [Google Scholar]
- 17. Gergics P, Brinkmeier ML, Camper SA. Lhx4 deficiency: increased cyclin-dependent kinase inhibitor expression and pituitary hypoplasia. Mol Endocrinol. 2015;29:597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto K, Yamada M, Monden T, Satoh T, Wondisford FE, Mori M. Thyrotropin-releasing hormone (TRH) specific interaction between amino terminus of P-Lim and CREB binding protein (CBP). Mol Cell Endocrinol. 2005;229:11–20. [DOI] [PubMed] [Google Scholar]
- 19. Sarapura VD, Strouth HL, Wood WM, Gordon DF, Ridgway EC. Activation of the glycoprotein hormone α-subunit gene promoter in thyrotropes. Mol Cell Endocrinol. 1998;146:77–86. [DOI] [PubMed] [Google Scholar]
- 20. Sarapura VD, Strouth HL, Gordon DF, Wood WM, Ridgway EC. Msx1 is present in thyrotropic cells and binds to a consensus site on the glycoprotein hormone α-subunit promoter. Mol Endocrinol. 1997;11:1782–1794. [DOI] [PubMed] [Google Scholar]
- 21. Wood WM, Dowding JM, Gordon DF, Ridgway EC. An upstream regulator of the glycoprotein hormone α-subunit gene mediates pituitary cell type activation and repression by different mechanisms. J Biol Chem. 1999;274:15526–15532. [DOI] [PubMed] [Google Scholar]
- 22. Brinkmeier ML, Gordon DF, Dowding JM, et al. Cell-specific expression of the mouse glycoprotein hormone α-subunit gene requires multiple interacting DNA elements in transgenic mice and cultured cells. Mol Endocrinol. 1998;12:622–633. [DOI] [PubMed] [Google Scholar]
- 23. Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone α-subunit gene expression. J Biol Chem. 1999;274:36159–36167. [DOI] [PubMed] [Google Scholar]
- 24. Schoenmakers N, Alatzoglou KS, Chatterjee VK, Dattani MT. Recent advances in central congenital hypothyroidism. J Endocrinol. 2015;227:R51–R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Samuels HH, Forman BM, Horowitz ZD, Ye ZS. Regulation of gene expression by thyroid hormone. J Clin Invest. 1988;81:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pernasetti F, Caccavelli L, Van de Weerdt C, Martial JA, Muller M. Thyroid hormone inhibits the human prolactin gene promoter by interfering with activating protein-1 and estrogen stimulations. Mol Endocrinol. 1997;11:986–996. [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi K, Yamamoto K, Kikuyama S, Machida T, Kobayashi T. Impaired development of somatotropes, lactotropes and thyrotropes in growth-retarded (grt) mice. J Toxicol Pathol. 2009;22:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noguchi T, Kudo M, Sugisaki T, Satoh I. An immunocytochemical and electron microscopic study of the hyt mouse anterior pituitary gland. J Endocrinol. 1986;109:163–168. [DOI] [PubMed] [Google Scholar]
- 29. Kendall SK, Samuelson LC, Saunders TL, Wood RI, Camper SA. Targeted disruption of the pituitary glycoprotein hormone α-subunit produces hypogonadal and hypothyroid mice. Genes Dev. 1995;9:2007–2019. [DOI] [PubMed] [Google Scholar]
- 30. Nuñez L, Villalobos C, Senovilla L, García-Sancho J. Multifunctional cells of mouse anterior pituitary reveal a striking sexual dimorphism. J Physiol. 2003;549:835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kulig E, Camper SA, Kuecker S, Jin L, Lloyd RV. Remodeling of hyperplastic pituitaries in hypothyroid α-subunit knockout mice after thyroxine and 17β-estradiol treatment: role of apoptosis. Endocr Pathol. 1998;9:261–274. [DOI] [PubMed] [Google Scholar]
- 32. Stahl JH, Kendall SK, Brinkmeier ML, et al. Thyroid hormone is essential for pituitary somatotropes and lactotropes. Endocrinology. 1999;140:1884–1892. [DOI] [PubMed] [Google Scholar]
- 33. Mortensen AH, MacDonald JW, Ghosh D, Camper SA. Candidate genes for panhypopituitarism identified by gene expression profiling. Physiol Genomics. 2011;43:1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. [DOI] [PubMed] [Google Scholar]
- 35. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polk RC, Gergics P, Steimle JD, et al. The pattern of congenital heart defects arising from reduced Tbx5 expression is altered in a Down syndrome mouse model. BMC Dev Biol. 2015;15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaston-Massuet C, Henderson DJ, Greene ND, Copp AJ. Zic4, a zinc-finger transcription factor, is expressed in the developing mouse nervous system. Dev Dyn. 2005;233:1110–1115. [DOI] [PubMed] [Google Scholar]
- 38. Sizova D, Ho Y, Cooke NE, Liebhaber SA. Research resource: T-antigen transformation of pituitary cells captures three novel cell lines in the Pit-1 lineage. Mol Endocrinol. 2010;24:2232–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abel MH, Charlton HM, Huhtaniemi I, Pakarinen P, Kumar TR, Christian HC. An investigation into pituitary gonadotrophic hormone synthesis, secretion, subunit gene expression and cell structure in normal and mutant male mice. J Neuroendocrinol. 2013;25:863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matzuk MM, Kornmeier CM, Whitfield GK, Kourides IA, Boime I. The glycoprotein α-subunit is critical for secretion and stability of the human thyrotropin β-subunit. Mol Endocrinol. 1988;2:95–100. [DOI] [PubMed] [Google Scholar]
- 41. Cole NB, Daniels MP, Levine RL, Kim G. Oxidative stress causes reversible changes in mitochondrial permeability and structure. Exp Gerontol. 2010;45:596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grönblad M, Akerman KE. Electron-dense endoplasmic reticulum-like profiles closely associated with mitochondria in glomus cells of the carotid body after fixation with oxalate. Exp Cell Res. 1984;152:161–168. [DOI] [PubMed] [Google Scholar]
- 43. Brinkmeier ML, Stahl JH, Gordon DF, et al. Thyroid hormone-responsive pituitary hyperplasia independent of somatostatin receptor 2. Mol Endocrinol. 2001;15:2129–2136. [DOI] [PubMed] [Google Scholar]
- 44. Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. [DOI] [PubMed] [Google Scholar]
- 45. Wu S, Chen Y, Fajobi T, et al. Conditional knockout of the androgen receptor in gonadotropes reveals crucial roles for androgen in gonadotropin synthesis and surge in female mice. Mol Endocrinol. 2014;28:1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woo SR, Corrales L, Gajewski TF. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 2015;36:250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Million Passe CM, White CR, King MW, Quirk PL, Iovanna JL, Quirk CC. Loss of the protein NUPR1 (p8) leads to delayed LHB expression, delayed ovarian maturation, and testicular development of a sertoli-cell-only syndrome-like phenotype in mice. Biol Reprod. 2008;79:598–607. [DOI] [PubMed] [Google Scholar]
- 48. Quirk CC, Seachrist DD, Nilson JH. Embryonic expression of the luteinizing hormone β gene appears to be coupled to the transient appearance of p8, a high mobility group-related transcription factor. J Biol Chem. 2003;278:1680–1685. [DOI] [PubMed] [Google Scholar]
- 49. Mohammad HP, Seachrist DD, Quirk CC, Nilson JH. Reexpression of p8 contributes to tumorigenic properties of pituitary cells and appears in a subset of prolactinomas in transgenic mice that hypersecrete luteinizing hormone. Mol Endocrinol. 2004;18:2583–2593. [DOI] [PubMed] [Google Scholar]
- 50. Chowdhury UR, Samant RS, Fodstad O, Shevde LA. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009;28:225–232. [DOI] [PubMed] [Google Scholar]
- 51. Niu Z, Goodyear SM, Rao S, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci USA. 2011;108:12740–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou C, Wawrowsky K, Bannykh S, Gutman S, Melmed S. E2F1 induces pituitary tumor transforming gene (PTTG1) expression in human pituitary tumors. Mol Endocrinol. 2009;23:2000–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garcia-Lavandeira M, Diaz-Rodriguez E, Bahar D, et al. Pituitary cell turnover: from adult stem cell recruitment through differentiation to death. Neuroendocrinology. 2015;101:175–192. [DOI] [PubMed] [Google Scholar]
- 54. Mukai J, Shoji S, Kimura MT, et al. Structure-function analysis of NADE: identification of regions that mediate nerve growth factor-induced apoptosis. J Biol Chem. 2002;277:13973–13982. [DOI] [PubMed] [Google Scholar]
- 55. Chien J, Staub J, Avula R, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. [DOI] [PubMed] [Google Scholar]
- 56. Chien J, Narita K, Rattan R, et al. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008;27:7223–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rattan R, Narita K, Chien J, et al. TCEAL7, a putative tumor suppressor gene, negatively regulates NF-κB pathway. Oncogene. 2010;29:1362–1373. [DOI] [PubMed] [Google Scholar]
- 58. Roelfsema F, Pereira AM, Adriaanse R, et al. Thyrotropin secretion in mild and severe primary hypothyroidism is distinguished by amplified burst mass and Basal secretion with increased spikiness and approximate entropy. J Clin Endocrinol Metab. 2010;95:928–934. [DOI] [PubMed] [Google Scholar]
- 59. Beamer WG, Cresswell LA. Defective thyroid ontogenesis in fetal hypothyroid (hyt/hyt) mice. Anat Rec. 1982;202:387–393. [DOI] [PubMed] [Google Scholar]
- 60. Shibusawa N, Yamada M, Hirato J, Monden T, Satoh T, Mori M. Requirement of thyrotropin-releasing hormone for the postnatal functions of pituitary thyrotrophs: ontogeny study of congenital tertiary hypothyroidism in mice. Mol Endocrinol. 2000;14:137–146. [DOI] [PubMed] [Google Scholar]
- 61. Surks MI, DeFesi CR. Determination of the cell number of each cell type in the anterior pituitary of euthyroid and hypothyroid rats. Endocrinology. 1977;101:946–958. [DOI] [PubMed] [Google Scholar]
- 62. Stratmann IE, Ezrin C, Sellers EA, Simon GT. The origin of thyroidectomy cells as revealed by high resolution radioautography. Endocrinology. 1972;90:728–734. [DOI] [PubMed] [Google Scholar]
- 63. DeFesi CR, Astier HS, Surks MI. Kinetics of thyrotrophs and somatotrophs during development of hypothyroidism and L-triiodothyronine treatment of hypothyroid rats. Endocrinology. 1979;104:1172–1180. [DOI] [PubMed] [Google Scholar]
- 64. Friedrichsen S, Christ S, Heuer H, et al. Expression of pituitary hormones in the Pax8−/− mouse model of congenital hypothyroidism. Endocrinology. 2004;145:1276–1283. [DOI] [PubMed] [Google Scholar]
- 65. Farquhar MG, Rinehart JF. Cytologic alterations in the anterior pituitary gland following thyroidectomy: an electron microscope study. Endocrinology. 1954;65:857–876. [DOI] [PubMed] [Google Scholar]
- 66. Fares F. The role of O-linked and N-linked oligosaccharides on the structure-function of glycoprotein hormones: development of agonists and antagonists. Biochim Biophys Acta. 2006;1760:560–567. [DOI] [PubMed] [Google Scholar]
- 67. Fares FA, Yamabe S, Ben-Menahem D, Pixley M, Hsueh AJ, Boime I. Conversion of thyrotropin heterodimer to a biologically active single-chain. Endocrinology. 1998;139:2459–2464. [DOI] [PubMed] [Google Scholar]
- 68. Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. [DOI] [PubMed] [Google Scholar]
- 69. Grossmann M, Wong R, Szkudlinski MW, Weintraub BD. Human thyroid-stimulating hormone (hTSH) subunit gene fusion produces hTSH with increased stability and serum half-life and compensates for mutagenesis-induced defects in subunit association. J Biol Chem. 1997;272:21312–21316. [DOI] [PubMed] [Google Scholar]
- 70. Szkudlinski MW, Thotakura NR, Weintraub BD. Subunit-specific functions of N-linked oligosaccharides in human thyrotropin: role of terminal residues of α- and β-subunit oligosaccharides in metabolic clearance and bioactivity. Proc Natl Acad Sci USA. 1995;92:9062–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Parsons TF, Bloomfield GA, Pierce JG. Purification of an alternate form of the α subunit of the glycoprotein hormones from bovine pituitaries and identification of its O-linked oligosaccharide. J Biol Chem. 1983;258:240–244. [PubMed] [Google Scholar]
- 72. Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. [DOI] [PubMed] [Google Scholar]
- 73. Feige MJ, Hendershot LM. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol Cell. 2013;51:297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Kasteren SI, Overkleeft H, Ovaa H, Neefjes J. Chemical biology of antigen presentation by MHC molecules. Curr Opin Immunol. 2014;26:21–31. [DOI] [PubMed] [Google Scholar]
- 75. Vettermann C, Herrmann K, Jäck HM. Powered by pairing: the surrogate light chain amplifies immunoglobulin heavy chain signaling and pre-selects the antibody repertoire. Semin Immunol. 2006;18:44–55. [DOI] [PubMed] [Google Scholar]
- 76. Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. α-Inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. [DOI] [PubMed] [Google Scholar]
- 77. Greenwood M, Greenwood MP, Paton JF, Murphy D. Transcription factor CREB3L1 regulates endoplasmic reticulum stress response genes in the osmotically challenged rat hypothalamus. PLoS One. 2015;10:e0124956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Magner JA, Weintraub BD. Thyroid-stimulating hormone subunit processing and combination in microsomal subfractions of mouse pituitary tumor. J Biol Chem. 1982;257:6709–6715. [PubMed] [Google Scholar]
- 79. Magner JA. Thyroid-stimulating hormone: biosynthesis, cell biology, and bioactivity. Endocr Rev. 1990;11:354–385. [DOI] [PubMed] [Google Scholar]
- 80. Grossmann M, Szkudlinski MW, Tropea JE, et al. Expression of human thyrotropin in cell lines with different glycosylation patterns combined with mutagenesis of specific glycosylation sites. Characterization of a novel role for the oligosaccharides in the in vitro and in vivo bioactivity. J Biol Chem. 1995;270:29378–29385. [DOI] [PubMed] [Google Scholar]
- 81. Weintraub BD, Stannard BS, Linnekin D, Marshall M. Relationship of glycosylation to de novo thyroid-stimulating hormone biosynthesis and secretion by mouse pituitary tumor cells. J Biol Chem. 1980;255:5715–5723. [PubMed] [Google Scholar]
- 82. Wopereis S, Lefeber DJ, Morava E, Wevers RA. Mechanisms in protein O-glycan biosynthesis and clinical and molecular aspects of protein O-glycan biosynthesis defects: a review. Clin Chem. 2006;52:574–600. [DOI] [PubMed] [Google Scholar]
- 83. Grossmann M, Szkudlinski MW, Wong R, Dias JA, Ji TH, Weintraub BD. Substitution of the seat-belt region of the thyroid-stimulating hormone (TSH) β-subunit with the corresponding regions of choriogonadotropin or follitropin confers luteotropic but not follitropic activity to chimeric TSH. J Biol Chem. 1997;272:15532–15540. [DOI] [PubMed] [Google Scholar]
- 84. Fairlie WD, Stanton PG, Hearn MT. Contribution of specific disulphide bonds to two epitopes of thyrotropin β-subunit associated with receptor recognition. Eur J Biochem. 1996;240:622–627. [DOI] [PubMed] [Google Scholar]
- 85. Thotakura NR, LiCalzi L, Weintraub BD. The role of carbohydrate in thyrotropin action assessed by a novel approach using enzymatic deglycosylation. J Biol Chem. 1990;265:11527–11534. [PubMed] [Google Scholar]
- 86. Weintraub BD, Stannard BS, Meyers L. Glycosylation of thyroid-stimulating hormone in pituitary tumor cells: influence of high mannose oligosaccharide units on subunit aggregation, combination, and intracellular degradation. Endocrinology. 1983;112:1331–1345. [DOI] [PubMed] [Google Scholar]
- 87. Harrisson F, Van Hoof J, Vakaet L. Processing of cell debris suggestive and phagocytosis in the follicular cavities of the avian adenohypophysis. Cell Biol Int Rep. 1982;6:153–161. [DOI] [PubMed] [Google Scholar]
- 88. Orgnero de Gaisán EM, Maldonado CA, Aoki A. Fate of degenerating lactotrophs in rat pituitary gland after interruption of lactation: a histochemical and immunocytochemical study. Histochem J. 1993;25:150–165. [DOI] [PubMed] [Google Scholar]
- 89. Claudius L, Yoshimi Y, Yoichiro H, Gabriel M, Koichi M. Phagocytotic removal of apoptotic endocrine cells by folliculostellate cells and its functional implications in clusterin accumulation in pituitary colloids in helmeted guinea fowl (Numida meleagris). Acta Histochem. 2006;108:69–80. [DOI] [PubMed] [Google Scholar]
- 90. Allaerts W, Vankelecom H. History and perspectives of pituitary folliculo-stellate cell research. Eur J Endocrinol. 2005;153:1–12. [DOI] [PubMed] [Google Scholar]
- 91. Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. 2011;91:1219–1243. [DOI] [PubMed] [Google Scholar]
- 92. Chaudhari N, Talwar P, Parimisetty A, Lefebvre d'Hellencourt C, Ravanan P. A molecular web: endoplasmic reticulum stress, inflammation, and oxidative stress. Front Cell Neurosci. 2014;8:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Murakami T, Saito A, Hino S, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205–1211. [DOI] [PubMed] [Google Scholar]
- 94. Iurlaro R, Muñoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2015;283:2640–2652. [DOI] [PubMed] [Google Scholar]
- 95. Michaelis KA, Knox AJ, Xu M, et al. Identification of growth arrest and DNA-damage-inducible gene β (GADD45β) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology. 2011;152:3603–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang X, Sun H, Danila DC, et al. Loss of expression of GADD45 γ, a growth inhibitory gene, in human pituitary adenomas: implications for tumorigenesis. J Clin Endocrinol Metab. 2002;87:1262–1267. [DOI] [PubMed] [Google Scholar]
- 97. Dugas JC, Ibrahim A, Barres BA. The T3-induced gene KLF9 regulates oligodendrocyte differentiation and myelin regeneration. Mol Cell Neurosci. 2012;50:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Thompson CC. Thyroid hormone-responsive genes in developing cerebellum include a novel synaptotagmin and a hairless homolog. J Neurosci. 1996;16:7832–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jackson SM, Gutierrez-Hartmann A, Hoeffler JP. Upstream stimulatory factor, a basic-helix-loop-helix-zipper protein, regulates the activity of the α-glycoprotein hormone subunit gene in pituitary cells. Mol Endocrinol. 1995;9:278–291. [DOI] [PubMed] [Google Scholar]
- 100. Majumdar S, Farris CL, Kabat BE, Jung DO, Ellsworth BS. Forkhead Box O1 is present in quiescent pituitary cells during development and is increased in the absence of p27 Kip1. PLoS One. 2012;7:e52136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mertani HC, Pechoux C, Garcia-Caballero T, Waters MJ, Morel G. Cellular localization of the growth hormone receptor/binding protein in the human anterior pituitary gland. J Clin Endocrinol Metab. 1995;80:3361–3367. [DOI] [PubMed] [Google Scholar]
- 102. Morel G, Ouhtit A, Kelly PA. Prolactin receptor immunoreactivity in rat anterior pituitary. Neuroendocrinology. 1994;59:78–84. [DOI] [PubMed] [Google Scholar]
- 103. Teglund S, McKay C, Schuetz E, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. [DOI] [PubMed] [Google Scholar]
- 104. Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121:626–633. [DOI] [PubMed] [Google Scholar]
- 105. Li G, Ci W, Karmakar S, et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tomasini R, Seux M, Nowak J, et al. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene. 2005;24:8093–8104. [DOI] [PubMed] [Google Scholar]
- 107. Sala D, Ivanova S, Plana N, et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J Clin Invest. 2014;124:1914–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kong DK, Georgescu SP, Cano C, et al. Deficiency of the transcriptional regulator p8 results in increased autophagy and apoptosis, and causes impaired heart function. Mol Biol Cell. 2010;21:1335–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Passe CM, Cooper G, Quirk CC. The murine p8 gene promoter is activated by activating transcription factor 4 (ATF4) in the gonadotrope-derived LβT2 cell line. Endocrine. 2006;30:81–91. [DOI] [PubMed] [Google Scholar]
- 110. Wu Z, Zheng S, Li Z, Tan J, Yu Q. E2F1 suppresses Wnt/β-catenin activity through transactivation of β-catenin interacting protein ICAT. Oncogene. 2011;30:3979–3984. [DOI] [PubMed] [Google Scholar]
- 111. Pedrola N, Devis L, Llauradó M, et al. Nidogen 1 and nuclear protein 1: novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin Exp Metastasis. 2015;32:467–478. [DOI] [PubMed] [Google Scholar]
- 112. Pillutla RC, Shimamoto A, Furuichi Y, Shatkin AJ. Genomic structure and chromosomal localization of TCEAL1, a human gene encoding the nuclear phosphoprotein p21/SIIR. Genomics. 1999;56:217–220. [DOI] [PubMed] [Google Scholar]
- 113. Makino H, Tajiri T, Miyashita M, et al. Differential expression of TCEAL1 in esophageal cancers by custom cDNA microarray analysis. Disea Esoph. 2005;18:37–40. [DOI] [PubMed] [Google Scholar]
- 114. Akaishi J, Onda M, Okamoto J, et al. Down-regulation of transcription elogation factor A (SII) like 4 (TCEAL4) in anaplastic thyroid cancer. BMC Cancer. 2006;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Parmar JS, Lomas DA. α-1-antitrypsin deficiency, the serpinopathies and conformational disease. J R Coll Physicians Lond. 2000;34:295–300. [PMC free article] [PubMed] [Google Scholar]
- 116. Alkhani AM, Cusimano M, Kovacs K, Bilbao JM, Horvath E, Singer W. Cytology of pituitary thyrotroph hyperplasia in protracted primary hypothyroidism. Pituitary. 1999;1:291–295. [DOI] [PubMed] [Google Scholar]