Abstract
Thyroid hormone (T3) is essential for proper neurological development. The hormone, bound to its receptors, regulates gene transcription in part by modulating posttranslational modifications of histones. Methylation of DNA, which is established by the de novo DNA methyltransferase (DNMT)3a and DNMT3b, and maintained by DNMT1 is another epigenetic modification influencing gene transcription. The expression of Dnmt3a, but not other Dnmt genes, increases in mouse brain in parallel with the postnatal rise in plasma [T3]. We found that treatment of the mouse neuroblastoma cell line Neuro2a[TRβ1] with T3 caused rapid induction of Dnmt3a mRNA, which was resistant to protein synthesis inhibition, supporting that it is a direct T3-response gene. Injection of T3 into postnatal day 6 mice increased Dnmt3a mRNA in the brain by 1 hour. Analysis of two chromatin immunoprecipitation-sequencing datasets, and targeted analyses using chromatin immunoprecipitation, transfection-reporter assays, and in vitro DNA binding identified 2 functional T3-response elements (TREs) at the mouse Dnmt3a locus located +30.3 and +49.3 kb from the transcription start site. Thyroid hormone receptors associated with both of these regions in mouse brain chromatin, but with only 1 (+30.3 kb) in Neuro2a[TRβ1] cells. Deletion of the +30.3-kb TRE using CRISPR/Cas9 genome editing eliminated or strongly reduced the Dnmt3a mRNA response to T3. Bioinformatics analysis showed that both TREs are highly conserved among eutherian mammals. Thyroid regulation of Dnmt3a may be an evolutionarily conserved mechanism for modulating global changes in DNA methylation during postnatal neurological development.
Methylation of cytosine residues in vertebrate genomes (DNA methylation) occurs predominantly in the context of cytosine-guanine (CG) dinucleotides. Approximately 70%–80% of CGs found throughout the genome are methylated, located in intergenic regions, within genes and transposable elements (1–3). The remainder, located near gene promoters (“CpG islands”), are unmethylated or differentially methylated, and methylation of CpG islands can lead to gene repression. The global pattern of DNA methylation in vertebrates is established during embryogenesis by the de novo DNA methyltransferase (DNMT)3a and DNMT3b, and is preserved through rounds of cell division by DNMT1 (4, 5). DNA methylation may impose long-term, stable transcriptional silencing through physical blockade of transcription factor binding, and recruitment of methyl-CpG-binding proteins, which recruit histone modifying enzymes to generate a transcriptionally silent state (4, 6).
The role of DNA methylation in neurological development is an area of intense interest (7, 8). Recently, Lister et al (9) found that non-CG methylation (where G is replaced by A, T, or C) occurs in developing and adult mammalian neurons, but not in other differentiated cell types (10–13), in roughly equal amount to CG methylation. The abundance of methylated cytosines increases dramatically in the developing frontal cortices of human and mouse, mostly in the CA context, coincident with the period of active synaptogenesis and synaptic pruning, suggesting that the acquisition of non-CG methylation is associated with neuronal maturation (9). The increase in non-CG methylation was coordinate with a parallel increase in the expression of Dnmt3a (but not other Dnmt genes) (9, 14) (Supplemental Figure 1), suggesting a role for DNMT3a in establishing appropriate DNA methylation profiles across neural cell genomes during brain development.
Thyroid hormone is essential for normal development in vertebrates. In mammals, T3 is well known to be required for neurological development, and thyroid deficiency during early human development can lead to a range of disorders from mild neurobehavioral deficits to severe mental and growth retardation (cretinism) (15, 16). Plasma T3 titer increases in mouse during the early postnatal period (Supplemental Figure 1), which influences neural cell maturation (17). The actions of T3 are mediated by T3 receptors (TRs), which regulate gene transcription, typically as heterodimers with retinoid X receptor (RXR). The TR-RXR complex binds to T3-response elements (TREs) in the genome, comprised of 2 hexanucleotide half sites (most commonly a direct repeat plus 4-base spacer [DR+4]). The TRs orchestrate modifications to local chromatin structure by recruiting histone-modifying enzymes (18). For genes that are activated by T3, unliganded TR represses transcription through interaction with corepressors, whereas liganded TR activates transcription through recruitment of coactivators (18).
The roles of liganded TR in controlling posttranslational modifications to histones have been extensively studied, but it is not known whether T3 can influence DNA methylation. Here, we investigated T3 regulation of Dnmt3a in the developing mouse brain and in a mouse neuroblastoma cell line (Neuro2a[TRβ1]). Dnmt3a is expressed in parallel with the developmental rise in plasma [T3], and we found that exogenous T3 can induce Dnmt3a mRNA with rapid kinetics in the brain and in Neuro2a[TRβ1] cells. We identified and characterized 2 functional TREs within the mouse Dnmt3a gene. Our findings support that T3 directly regulates transcription of the mouse Dnmt3a gene, and therefore may influence global DNA methylation in the developing brain.
Materials and Methods
Animal care and hormone treatment
We purchased wild-type C57BL/6J mice from The Jackson Laboratory and maintained them at a 12-hour light, 12-hour dark cycle with food and water ad libitum. For the hormone supplementation experiment, we administered ip injections of vehicle (corn oil + ethanol) or T3 (25-μg/kg body weight [BW]) (19). The T3 was first dissolved in 100% ethanol and then added dropwise to corn oil with vigorous mixing (final concentration 0.05% ethanol). Mice were killed by rapid decapitation 1 hour after injection. We dissected the region of the hippocampus, snap froze the tissue in liquid nitrogen, and stored it at −80°C until RNA extraction. We chose postnatal day (PND)6 mice for this experiment because endogenous plasma [T3] is relatively low at this age (20), and we hypothesized that this would enhance our ability to see an early (by 1 h) transcriptional response to T3.
For chromatin immunoprecipitation (ChIP) assays we administered ip injections of T3 (25-μg/kg BW) to PND14 wild-type mice of mixed sex (n = 5). Mice were killed by rapid decapitation 4 hours after injection. The frontal cortex, hippocampus and cerebellum were dissected, and chromatin extracted immediately after dissection, snap frozen in liquid nitrogen and stored at −80°C until ChIP assay (see below). We chose PND14 for this experiment because TR expression (and plasma T3 concentration) is relatively high at this age (17, 20), T3 treatment for 4 hours caused detectable changes in acetylated histone 3 (acH3) in mouse brain (19, 21), and we reasoned that this age/treatment would enhance our ability to detect TR by ChIP.
All procedures involving animals were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Michigan.
RNA extraction and real-time quantitative PCR (RT-qPCR)
We extracted total RNA from tissues or cells using TRIzol reagent (Invitrogen Life Technologies) and generated cDNA using the High Capacity cDNA Synthesis kit (Applied Biosystems, Inc). For RT-qPCR, we used ABsolute Blue qPCR SYBR Green Low ROX Mix (ABgene Thermo Scientific) and StepOne Real Time PCR Systems (Life Technologies). Primers were designed to span exon/exon boundaries for each gene (Supplemental Table 1). We used a relative quantification method (22, 23) to compare gene expression levels among treatments by generating standard curves using a pool of cDNAs. The level of Dnmt3a (NM_007872) or Krüppel-like factor 9 (Klf9) (NM_010638.4) mRNAs were normalized to GAPDH mRNA (NM_008084), which did not change after hormone treatment (data not shown).
ChIP assay
We conducted ChIP assays as described previously (21). For Neuro2a[TRβ1] cells, we extracted chromatin from cells treated with vehicle (0.1% dimethyl sulfoxide [DMSO]) or T3 (30nM) for 24 hours (n = 4/treatment), and from the frontal cortex of brains of PND14 mice that had received ip injection of T3 (25-μg/kg BW) 4 hours before killing (n = 5). We reduced chromatin to approximately 500 bp using a sonic dismembrator 100 (Fisher) for a total of 200 seconds per sample (20 cycles of 10-s sonication with 1 min between each cycle) at an output rating of 5–6 W. All chromatin samples were snap frozen in liquid nitrogen and stored at −80°C until analysis.
We used the following primary antibodies for ChIP assays: rabbit polyclonal antiserum raised against full-length Xenopus laevis TRβ (PB antiserum recognizes both TRα and TRβ and has been used successfully for studies of mouse TRs; 5 μL) (19, 21), anti-H3 (5 μL, 07–670; Millipore), or anti-acH3 (5 μL, 06–599; Millipore). Negative controls for the assays included the replacement of primary antibody with normal rabbit serum (NRS) (for the PB antiserum) or IgG purified from NRS (24). We analyzed ChIP DNA by real-time, qPCR using SYBR Green assays (primers in Supplemental Table 1).
Plasmid constructs
We generated luciferase reporter constructs with PCR-generated DNA fragments corresponding to genomic sequences containing predicted TREs for analysis in transient transfection assays. We analyzed data from TR ChIP sequencing (ChIP-seq) experiments on mouse liver (25) or the mouse cerebellar cell line C17.2 (26) (described in Results) and identified 2 candidate genomic regions, one at +30.3 kb and another at +49.3 kb relative to the transcription start site (TSS) of mouse Dnmt3a (these fall within the second and third introns, respectively). We subcloned 500-bp DNA fragments corresponding to these genomic regions into pCpGL (27) at the BglII and HindIII sites (+30.3 kb fragment), or into pGL4.23 at the KpnI and SacI sites (+49.3 kb fragment; primer sequences in Supplemental Table 2).
We generated point mutations in the predicted mouse Dnmt3a TREs by site-directed mutagenesis (primer sequences in Supplemental Table 2; QuikChange II Site-Directed Mutagenesis kit; Agilent Technologies).
Electrophoretic mobility shift assay
We conducted EMSA as described by Hoopfer et al (28) with TRβ and RXRα synthesized in vitro from pSp64a-TRβ and pSp64a-RXRα plasmid templates (X. laevis origin; gift of Yun-Bo Shi) using the TnT SP6 Quick-Coupled Translation System (Promega). For the probe we used a DR+4 TRE located within the Xenopus tropicalis dnmt3a gene that we have characterized (Y. Kyono and R. J. Denver, unpublished data; oligonucleotide sequences given in Supplemental Table 2). We used radioinert duplex oligonucleotides (1μM) as competitors (Supplemental Tables 2 and 3).
Cell culture and transient transfection assays
Neuro2a is a neuroblastoma cell line derived from a spontaneous brain tumor of a strain A albino mouse. We used Neuro2a cells engineered to express the TRβ1 isoform (Neuro2a[TRβ1]) (29) cultured in DMEM/F12 supplemented with hygromycin B (500 μg/mL; Invitrogen) and 10% fetal bovine serum that had been stripped of thyroid hormone (21, 30). We plated cells at a density of 0.5 × 106 per well in 6-well plates for RNA extraction, 1.25 × 106 per well in 6-well plates for ChIP assays, and 0.15 × 106 per well in 24-well plates for transient transfection-reporter assays. For RNA extraction or ChIP assay we incubated cells overnight before treatment with vehicle (0.1% DMSO) or T3 (30nM) for different times before harvest.
For reporter assays we transfected cells at 60%–70% confluency with luciferase reporter constructs (200 ng per well plus 10-ng pRenilla-tk) using FuGENE 6 transfection reagent (Promega). Twenty-four hours after transfection, we treated cells with vehicle (0.1% DMSO) or T3 (30nM) for 5 hours before harvest for Dual Luciferase Assay (Promega). Firefly luciferase activity was quantified using a luminometer (Femtometer FB 12; Zylux Corp) and normalized to Renilla luciferase activity. Each transfection experiment was conducted at least 3 times with 4–5 wells per treatment.
To determine whether T3 induction of Dnmt3a mRNA requires ongoing protein synthesis we pretreated cells with 10-μg/mL cycloheximide (CHX) (Sigma-Aldrich; this concentration suppresses ongoing protein synthesis by >95%) (19, 24) for 1 hour, then continued CHX treatment during the 6-hour treatment with T3.
Genome editing in Neuro2a[TRβ1] cells using CRISPR/Cas9
We designed 2 guide RNAs (gRNAs) to introduce mutations at the Dnmt3a +30.3-kb DR+4 TRE in Neuro2a[TRβ1] cells (gRNA-A and gRNA-B; gRNA sequences are given in Supplemental Table 2) and had 2 plasmids synthesized by OriGene (pCas-Guide-EF1a-EGFP) that express one of the gRNAs, Cas9, and enhanced green fluorescent protein (EGFP). We plated 1.5 × 106 cells in 10 cm tissue culture plates (Falcon), and 24 hours later, we transfected them with 10 μg of 1 or both of the gRNA/Cas9 expression plasmids using FuGENE 6 (Promega). Sixteen hours after transfection we sorted EGFP-positive cells (1–2 × 105 cells recovered/sample) by fluorescence-assisted cell sorting at the University of Michigan Flow Cytometry core. We cultured approximately 300 EGFP-positive cells per 10-cm dish for 1 week and isolated clonal colonies using cloning cylinders (Corning). We then transferred these clonal colonies to 6-well plates and expanded them until the cells were approximately 70% confluent. We analyzed mutations at the Dnmt3a +30.3-kb TRE region by isolating genomic DNA using the DNeasy Blood and Tissue kit (QIAGEN), amplifying the region by PCR (oligonucleotide primers given in Supplemental Table 2), subcloning the PCR-amplified fragments into the pGEM T-easy vector (Promega) and conducting direct DNA sequencing. We sequenced plasmid DNA isolated from ten bacterial colonies for each clonal Neuro2a[TRβ1] cell line.
To determine whether the mutations affected gene expression we conducted T3 dose-response experiments. We plated wild-type or mutant Neuro2a[TRβ1] cells overnight at a density of 0.6 × 105 per well in 6-well plates and then treated them for 6 hours with 0nM, 1nM, 3nM, or 10nM T3 (n = 5 wells per dose) before harvest for RNA isolation and gene expression analysis by RT-qPCR. We repeated each dose-response experiment 2 times.
Data analysis and statistics
We analyzed data using SPSS statistical software (version 19; SPSS, Inc). We conducted one-way ANOVA followed by Fisher's least significant difference (Fisher's least significant difference) post hoc test and Student's independent sample t test (α = 0.05). Data were Log10-transformed before statistical analysis when variances were found to be heterogeneous.
We searched for putative DR+4 TREs using the sequence analysis program Vector NTI (version 10; Invitrogen) with “TGWCCYnnnnTGWCCY” as a search term with 3-bp mismatches allowed. We also used Transfac 6.0 (BIOBASE Biological Databases) based on 0.80 or higher match to the vertebrate transcription factor matrix to search for putative TREs in TR ChIP-enriched regions, and potentially orthologous regions in other vertebrate genomes. The conservation of the identified transcription factor-binding sites and flanking sequences in other vertebrates were assessed using BLAT (http://genome.ucsc.edu/), and extracted sequences were aligned using ClustalW.
Results
Thyroid hormone induces Dnmt3a mRNA in a mouse neuroblastoma cell line and in neonatal mouse brain in vivo
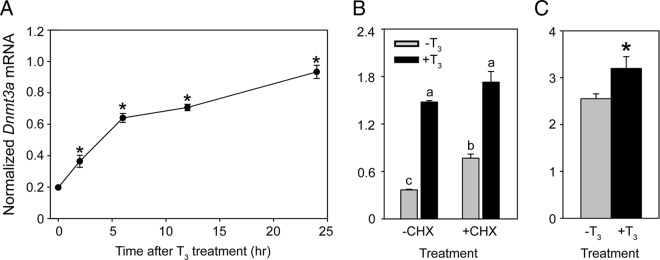
There is a parallel increase in circulating plasma [T3] and [T4] with Dnmt3a mRNA and protein in mouse brain from PND4 to PND14 (31, 32) (Supplemental Figure 1). A recent metaanalysis of published microarray data listed Dnmt3a as one of 734 candidate T3-regulated genes in rodent brain (33). Based on these findings we hypothesized that the mouse Dnmt3a gene is regulated by T3, and so we first treated the mouse neuroblastoma cell line Neuro2a[TRβ1] (29) with T3 (30nM) and analyzed Dnmt3a mRNA by RT-qPCR. Treatment with T3 caused a time-dependent increase in Dnmt3a mRNA, which was statistically significant by 2 hours (Figure 1A). Induction of Dnmt3a mRNA did not require ongoing protein synthesis (Figure 1B), supporting that mouse Dnmt3a is a direct T3-response gene. Intraperitoneal injection of T3 increased Dnmt3a mRNA in PND6 mouse hippocampal region at 1 hour, supporting that T3 regulates the mouse Dnmt3a gene in vivo (Figure 1C).
Figure 1.
Dnmt3a mRNA is induced by T3 in Neuro2a[TRβ1] cells and in PND6 mouse brain in vivo. A, Treatment with T3 induces Dnmt3a mRNA with rapid kinetics in Neuro2a[TRβ1] cells. Cells were treated with vehicle (0.1% DMSO) or T3 (30nM) for 2, 6, 12, or 24 hours before harvest for RNA isolation and analysis of Dnmt3a mRNA by RT-qPCR. Each data point represents the mean ± SEM, and asterisks above the means indicate statistically significant differences compared with the 0-hour time point (n = 4/treatment per time point; F8,25 = 103.017, P < .001; ANOVA). Dnmt3a mRNA levels in vehicle-treated cells were similar at all time points measured (data not shown). B, T3 induction of Dnmt3a mRNA in Neuro2a[TRβ1] cells is resistant to inhibition of protein synthesis. We pretreated cells with the protein synthesis inhibitor CHX (10 μg/mL) for 1 hour and then exposed cells to vehicle (0.1% DMSO) or T3 (30nM) for an additional 6 hours in the continued presence or absence of CHX. Bars represent the mean ± SEM, and letters above the means indicate statistically significant differences among treatments; means with the same letter are not significantly different (n = 4/treatment; F3,12 = 186.490, P < .001; ANOVA). C, Treatment with T3 induces Dnmt3a mRNA in neonatal mouse brain. We measured Dnmt3a mRNA in the hippocampal region of PND6 mice that received ip injections of vehicle (corn oil + 0.05% ethanol) or T3 (25-μg/kg BW) 1 hour before killing. The asterisk indicates a statistically significant difference between vehicle and T3-injected animals (n = 5/treatment; P = .039, Student's independent sample t test). For all experiments, we measured Dnmt3a mRNA using real-time RT-qPCR and normalized to GAPDH mRNA, which did not change after hormone treatment (data not shown).
Identification of TREs at the mouse Dnmt3a locus
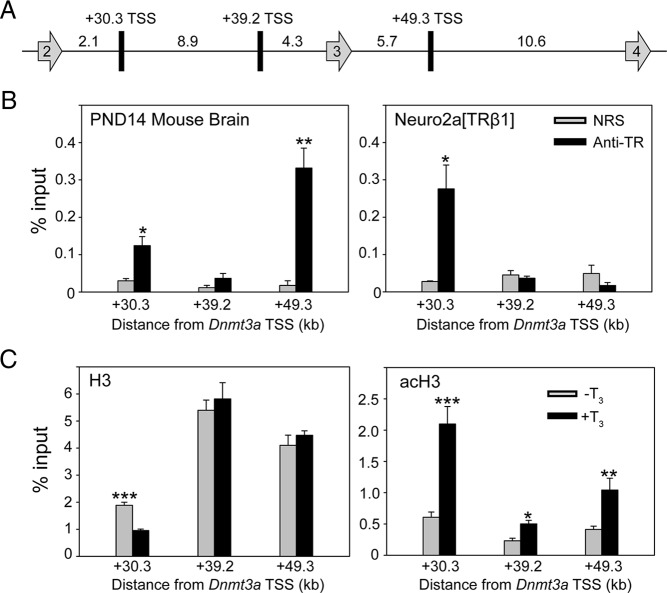
We identified 13 putative TR-binding sites associated with the mouse Dnmt3a locus by analyzing data from TR ChIP-seq experiments conducted in mouse liver (25) and the mouse cerebellar cell line C17.2 (see Supplemental Table 4 for genomic coordinates of putative TR-binding sites) (26). Using this information, we conducted targeted ChIP assays for TR association at these genomic regions using chromatin isolated from mouse frontal cortex; Dnmt3a is expressed throughout the mouse brain with highest mRNA levels in the frontal cortex at PND14 (9, 31) (Supplemental Figure 1). We investigated all 13 predicted TR-binding sites (data not shown) but found that TR associates with only 2, one within the second intron at +30.3 kb and another within the third intron at +49.3 kb from the TSS (Figure 2, A and B, left panel). There was no signal within the second intron at +39.2 kb from the TSS where no TRE was predicted (negative control region). We also conducted ChIP assays for TR on chromatin isolated from Neuro2a[TRβ1] cells that had been treated with vehicle or T3 for 24 hours (there was no difference between treatments, so Figure 2B only shows data from T3-treated cells). We found TR ChIP signal at +30.3 kb, but not at +49.3 kb, in Neuro2a[TRβ1] cells (Figure 2B, right panel).
Figure 2.
Thyroid hormone receptors associate in chromatin at the mouse Dnmt3a locus. The schematic diagram (A) shows putative TR-binding sites (rectangles) between exons 2 and 4 (arrows) of the mouse Dnmt3a gene identified by TR ChIP-seq experiments conducted on mouse liver (26) and the mouse cerebellar cell line C17.2 (27) (distances between genomic features are given in kilobases). B, We conducted targeted ChIP assays for TR at +30.3, +39.2, and +49.3 kb from the transcription start site (TSS) of mouse Dnmt3a using chromatin isolated from the brain (frontal cortex) of PND14 mice (left panel) that had received ip injections of T3 (25-μg/kg BW) 4 hours before killing (n = 5/treatment). We conducted similar assays on chromatin isolated from Neuro2a[TRβ1] cells treated with T3 (30nM) for 24 hours before harvest (right panel; similar results were obtained with chromatin isolated from vehicle-treated cells - data not shown). Bars represent the means ± SEM of the ChIP signal expressed as a percentage of input (anti-TR, antiserum to TR; NRS). The asterisks indicate statistically significant differences between anti-TR and NRS (*P < .01, **P < .001; Student's independent sample t test). C, Treatment of Neuro2a[TRβ1] cells with T3 induces nucleosome repositioning and H3 acetylation at the region of the mouse Dnmt3a +30.3-kb TRE. We conducted targeted ChIP assays for H3 and acH3 at +30.3, +39.2, and +49.3 kb from the TSS of the mouse Dnmt3a gene using chromatin isolated from Neuro2a[TRβ1] cells treated with vehicle (0.1% DMSO) or T3 (30nM) for 24 hours before harvest. Protein A-purified IgG (negative control) yielded < 0.03% of the input DNA signal (data not shown). Bars represent the mean ± SEM expressed as a percentage of input DNA. Asterisks above the means indicate statistically significant differences between treatments (*P < .05, **P < .01, ***P < .001; Student's independent sample t test; n = 5/treatment). Treatment with T3 decreased the H3 signal at position +30.3, indicative of nucleosome eviction/repositioning, and caused an approximately 4-fold increase in acH3 signal at this site. The T3 treatment did not alter the H3 signal at +39.2 or +49.3 kb (TR does not associate with these regions in Neuro2a[TRβ1] cells; see B, right panel) but caused small increases in acH3 at these sites, possibly due to spreading of histone acetylation across the locus initiated at the TRE located at +30.3 kb.
ChIP assays for H3 and acH3 showed that T3 reduced H3, indicative of nucleosome repositioning, and increased acH3 at +30.3 kb (Figure 2C, left panel). Treatment with T3 did not alter H3 but increased acH3 at +39.2 and +49.3 kb (Figure 2C, right panel), which may result from spreading of histone acetylation across the locus initiated by T3 action at the +30.3-kb TRE.
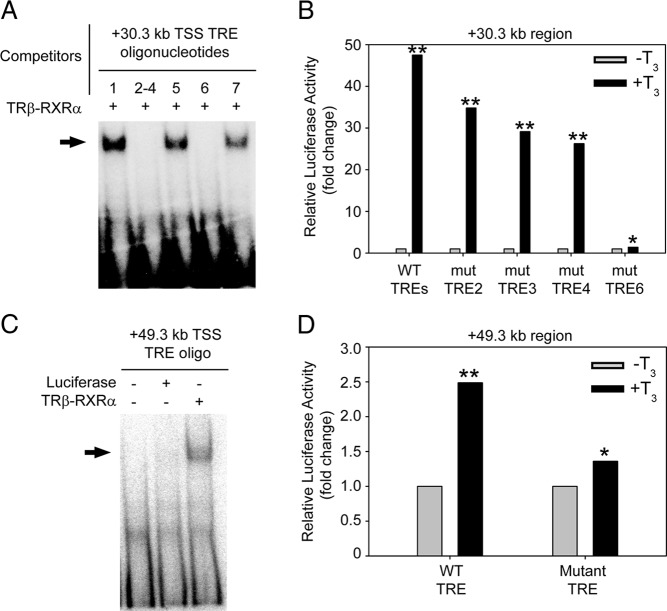
Bioinformatics analysis identified 7 putative DR+4 TREs within a 500-bp region at +30.3 kb from the TSS of mouse Dnmt3a (see Supplemental Figure 2 for location of predicted TREs). Competitive EMSA showed that radioinert duplex oligonucleotides with sequences of the predicted TREs 2, 3, 4 (within a single oligonucleotide because these TREs overlap), and 6 displaced TRβ-RXRα heterodimers from the radiolabeled probe; predicted TREs 1, 5, and 7 did not compete for binding and therefore were not studied further (Figure 3A). We next tested whether this genomic region centered at +30.3 kb (500 bp) could support T3-dependent transactivity. Treatment with T3 for 5 hours induced an approximately 50-fold increase in luciferase activity in Neuro2a[TRβ1] cells transfected with the construct containing the native DNA sequence (Figure 3B). Single base mutations introduced into each of the 2 half sites of the predicted TREs 2, 3, and 4 (2 and 3, and 3 and 4 overlap; see Supplemental Table 3) reduced T3-dependent transactivity by 20%–40%, whereas mutation of the predicted TRE 6 almost completely abolished transactivity (Figure 3B). Competitive EMSA with a radioinert DNA fragment corresponding to the genomic region containing these predicted TREs displaced TRβ-RXRα heterodimers from the radiolabeled probe; however, mutation of the 2 half sites of the predicted TRE 6 destroyed the ability to displace probe (Supplemental Figure 3A). Taken together, these data support a dominant role for TRE 6 within the +30.3-kb region of the mouse Dnmt3a gene. This TRE spans 30 306–30 321 bp downstream of the Dnmt3a TSS (chr12:3,837,286–3,837,301; coordinates based on the Mm9 build of the mouse genome).
Figure 3.
Predicted TREs at the mouse Dnmt3a locus bind TR-RXR heterodimers in vitro and support T3-dependent transactivation. A, We conducted competitive electrophoretic mobility shift assay (EMSA) using a [32P]-labeled oligonucleotide probe corresponding to a X. tropicalis dnmt3a TRE (Y. Kyono and R. J. Denver, unpublished data), and radioinert duplex oligonucleotides with sequences of 7 putative TREs found within the region centered at +30.3 kb from the TSS of the mouse Dnmt3a gene (see Supplemental Figure 2 for the locations of the predicted TREs; positions 2, 3, and 4 overlap, so we synthesized a single oligonucleotide that encompasses these 3 predicted TREs; oligonucleotide sequences are given in Supplemental Figure 2). Before gel electrophoresis we incubated the probe with in vitro synthesized Xenopus TRβ plus RXRα with or without radioinert duplex oligonucleotides (1μM). The arrow indicates the location of shifted bands due to TRβ-RXRα binding to the probe. Radioinert oligonucleotides corresponding to TREs 2–4 and 6 completely displaced TRβ-RXRα binding to the probe, whereas those for TREs 1, 5, and 7 had no activity. B, We tested for transcriptional activity of a 500-bp DNA fragment centered at +30.3 kb from the mouse Dnmt3a TSS in transient transfection-reporter assays using Neuro2a[TRβ1] cells. After transfection, we treated cells with vehicle (0.1% DMSO) or T3 (30nM) for 5 hours before conducting dual luciferase assay. This fragment supported T3-dependent transactivation, which was reduced by mutations introduced into TREs 2, 3, and 4, and was almost completely abolished by mutation of TRE 6 (P = .02). Bars represent the mean fold change in firefly luciferase activity (normalized to Renilla luciferase activity) relative to vehicle-treated controls (n = 3/treatment). C, We conducted EMSA using a [32P]-labeled oligonucleotide probe corresponding to the mouse +49.3-kb TRE sequence (Supplemental Table 2) and in vitro synthesized TRβ plus RXRα. In vitro synthesized luciferase was used as a negative control. The arrow indicates the location of the shifted band due to TRβ-RXRα binding to the probe. D, We tested for transcriptional activity of a 500-bp DNA fragment centered at +49.3 kb from the mouse Dnmt3a TSS in transient transfection-reporter assays using Neuro2a[TRβ1] cells. After transfection, we treated cells with vehicle (0.1% DMSO) or T3 (30nM) for 5 hours before conducting dual luciferase assay. This fragment supported T3-dependent transactivation, which was reduced by mutation of the single predicted TRE located in this region. Bars represent the mean fold change in firefly luciferase activity (normalized to Renilla luciferase activity) relative to vehicle-treated controls (n = 4/treatment). Asterisks indicate statistically significant differences between treatments (*, P < .01; **, P < .001; Student's independent sample t test).
Bioinformatics analysis identified a single DR+4 TRE within a 500-bp region at +49.3 kb from the TSS of mouse Dnmt3a. TRβ-RXRα heterodimers bound to this predicted TRE in vitro (Figure 3C), and the 500-bp fragment supported T3-dependent transactivation in transient transfection reporter assay; single base mutations introduced into each of the 2 half sites of the predicted TRE reduced T3-dependent transactivation (Figure 3D). Competitive EMSA showed that radioinert duplex oligonucleotides of the native TRE sequence at +49.3 kb completely displaced TRβ-RXRα heterodimers from the radiolabeled probe, whereas mutation of the 2 half sites reduced the ability of this DNA fragment to displace the probe (Supplemental Figure 3B). This TRE spans 49 266–49 281 bp downstream of the Dnmt3a TSS (chr12:3,856,246–3,856,261; coordinates based on the Mm9 build of the mouse genome).
CRISPR/Cas9-mediated deletion of the +30.3-kb TRE strongly reduced the Dnmt3a mRNA response to T3 in Neuro2a[TRβ1] cells
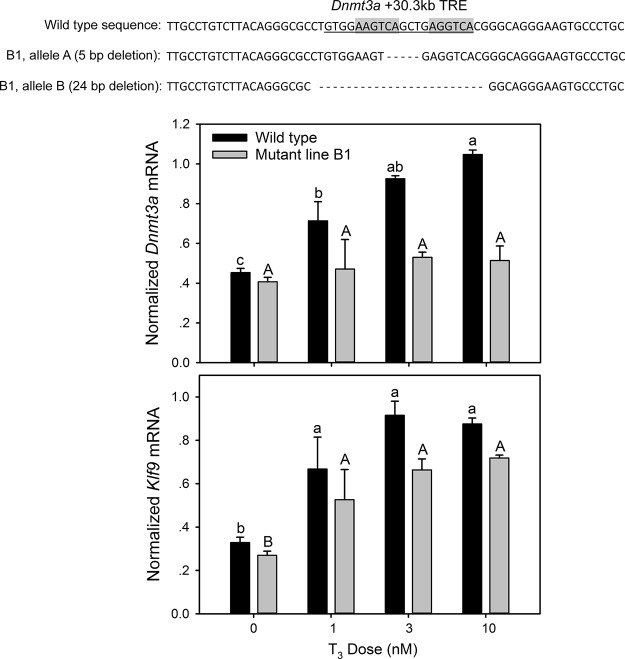
We used 2 gRNAs to generate deletion mutants of the Dnmt3a +30.3-kb TRE region in Neuro2a[TRβ1] cells (the locations of the gRNAs relative to the TRE are shown in Supplemental Figure 4A). The 2 gRNA/Cas9 vectors (gRNA-A and gRNA-B) were transfected alone or together, and clonal lines from each of the 3 transfected pools were isolated for analysis. Shown in Figure 4 are results from mutant line B1, which was generated by transfection with the gRNA-B vector resulting in biallelic deletion of 5 or 24 nucleotides, which disrupted or completely deleted the TRE. Results from 2 other mutant lines are shown in Supplemental Figure 4 (sequences of mutant alleles given in Supplemental Table 5). The AB2 mutant line was generated by cotransfection of gRNA-A and gRNA-B vectors and resulted in identical, biallelic deletion to mutant line B1. The A1 mutant line was generated by transfection of the gRNA-A vector and produced a range of deletions, supporting that this is not a pure clonal line. Nevertheless, all of the deletions in the A1 mutant line disrupt the TRE, and we did not detect any wild-type sequence. We therefore analyzed this line together with the 2 pure clonal lines.
Figure 4.
Deletion of the Dnmt3a +30.3-kb DR+4 TRE in Neuro2a[TRβ1] cells abolished the response to T3. We used CRISPR/Cas9 genome editing in Neuro2a[TRβ1] cells to generate biallelic mutations at the Dnmt3a DR+4 TRE located +30.3 kb from the TSS. The DNA sequences above the graphs show the deletions that we introduced into both alleles in the mutant line B1 (see Supplemental Figure 4 for results from other mutant lines). Treatment of wild-type cells with T3 caused a dose-dependent increase in Dnmt3a mRNA, which was eliminated in the mutant (top graph). The mRNA response to T3 of the direct T3-response gene Klf9 was unaffected by mutation at the Dnmt3a locus (bottom graph). Bars represent the mean ± SEM, and letters above the means indicate statistically significant differences among treatments; means with the same letter are not significantly different (P < .05; Fisher's LSD test; ANOVA, Dnmt3a mRNA: wild type F3,15 = 40.888, P < .001; B1 mutant F3,14 = 0.977, P = .431; Klf9 mRNA: wild type F3,15 = 20.528, P < .0001; B1 mutant F3,14 = 96.679, P < .0001).
To determine whether disruption of the +30.3-kb TRE altered Dnmt3a transcriptional responses to T3 we conducted hormone dose-response experiments in wild-type and mutant cell lines. Treatment with T3 caused a dose-dependent increase in Dnmt3a mRNA, which was eliminated in the B1 mutant line (Figure 4) and strongly reduced in the AB2 and A1 mutant lines (Supplemental Figure 4, B and C, and Supplemental Table 5). By contrast, T3-dependent induction of the direct T3-response gene Klf9 (19) was unaffected by mutations at the Dnmt3a +30.3-kb TRE.
The 2 mouse Dnmt3a TREs are conserved among mammalian species
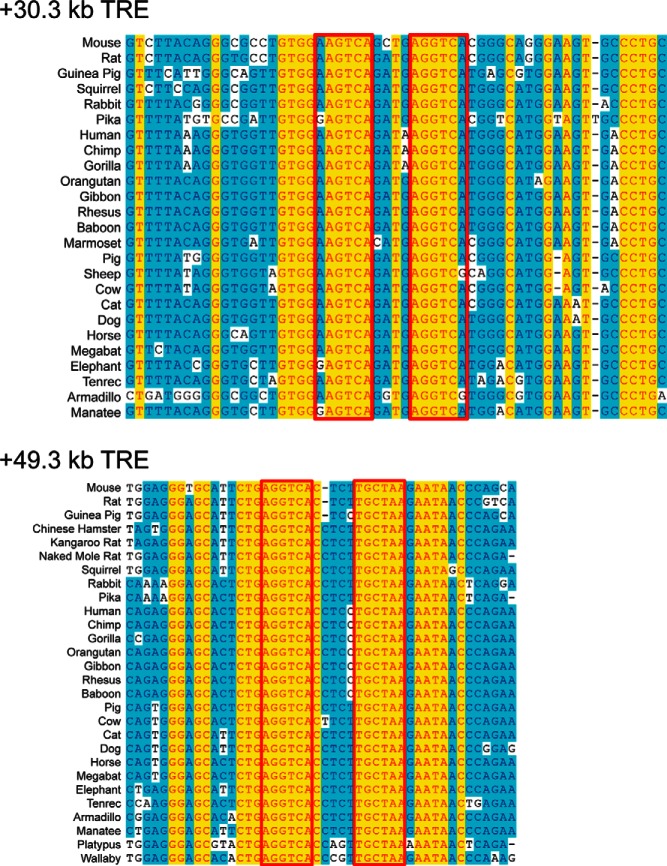
We found strong sequence conservation of both mouse TREs among eutherian mammals (Figure 5). The +49.3-kb, but not the +30.3-kb, TRE is also present in a metatherian, the wallaby, and in a monotreme, the platypus (Figure 5B). The +49.3-kb TRE has a 4-base spacer in mouse, rat, and guinea pig but a 5-base spacer in all other mammalian species analyzed. We found no sequence conservation of these mammalian Dnmt3a TREs in reptiles (subgroups examined: birds, turtles, squamates, and alligators).
Figure 5.
Sequence conservation of Dnmt3a TREs among mammals. Shown are DNA sequence alignments of the genomic regions containing the TREs at +30.3 kb (number 6) (upper panel) and +49.3 kb (lower panel) from the TSS of the mouse Dnmt3a gene. The boxes indicate the location of the predicated TRE half sites based on the mouse sequences. The TRE at +30.3 kb is a DR+4 in all mammalian species analyzed, whereas the TRE at +49.3 kb is DR+4 in mouse, rat, and Chinese hamster but is DR+5 in other mammals.
Discussion
Epigenetic modifications like DNA methylation, which result in heritable changes in gene function that do not involve changes to the DNA sequence, play central roles in human development and disease, from cancer to complex behavioral disorders such as autism and schizophrenia (8, 34–38). DNA methylation plays a critical role in neurological development (7, 8), but very little is known about how DNA methylation is regulated in neural cells. Thyroid hormone has pleiotropic actions in the developing brain, and thyroid deficiency during fetal or neonatal life leads to a condition of severe mental and growth retardation known as cretinism (17). Here, we show that T3, through direct transcriptional regulation of the gene coding for the de novo DNMT3a, has the potential to broadly influence DNA methylation in the developing brain. We found that T3 regulates Dnmt3a mRNA in mouse brain, which may be explained by the existence of at least 2 TREs, at +30.3 and +49.3 kb from the TSS. The gene is also strongly induced by T3 in Neuro2a[TRβ1] cells, but these cells appear to rely predominantly on the +30.3-kb TRE as evidenced by TR ChIP assay and CRISPR/Cas9 genome editing. There is strong conservation of the 2 Dnmt3a TREs among mammalian species. To our knowledge, this is the first demonstration that TRs directly regulate transcription of a gene that influences DNA methylation in the central nervous system.
Hormonal regulation of Dnmt3a in the developing brain and identification of TREs associated with the gene
Because previously published findings showed a parallel increase in circulating plasma [T3] and [T4] with Dnmt3a mRNA and protein in mouse brain (Supplemental Figure 1) (31, 32), we hypothesized that T3 may directly regulate Dnmt3a gene transcription. We first addressed this using the mouse-derived neuroblastoma cell line Neuro2a[TRβ1], and discovered that T3 induced Dnmt3a mRNA with rapid kinetics. This induction was resistant to inhibition of protein synthesis, supporting that mouse Dnmt3a is a direct T3 target gene. We also found that injection of T3 injection induced Dnmt3a mRNA in PND6 mouse brain 1 hour after injection.
Based on these findings, we looked for TREs at the mouse Dnmt3a locus, first using ChIP-seq data from mouse liver (25) and the cerebellar cell line C17.2 engineered to express TRs (26). We then conducted targeted ChIP assays on mouse brain chromatin, testing 13 genomic regions with putative TR-binding sites (Supplemental Table 4). This analysis revealed TR association in chromatin at 2 genomic regions in mouse brain, +30.3 and +49.3 kb from the TSS of Dnmt3a. Each region contained DR+4 TREs that were bound by TR-RXR heterodimers in vitro, and that supported T3-dependent transactivation in transient transfection assays. Thus, our findings support the presence of at least 2 functional TREs at the mouse Dnmt3a locus, but it is possible that other TREs exist that were not discovered in this study.
Although Dnmt3a was similarly regulated by T3 in mouse brain and Neuro2a[TRβ1] cells, we only found TR association in chromatin at +30.3 kb in Neuro2a[TRβ1] cells. Also, the failure to detect TR ChIP signal (Figure 2B, right panel), the lack of effect of T3 on nucleosome repositioning at the +49.3 kb site (Figure 2C, left panel), and the elimination or strong abrogation of the Dnmt3a mRNA response to T3 in Neuro2a[TRβ1] cells in which the +30.3 TRE was deleted by CRISPR/Cas9 genome editing provide strong evidence that the +30.3 TRE is the major hormone-response element in these cells. Comparing with the TR ChIP assays conducted in mouse brain chromatin which found TR association at both regions, our findings suggest that different neural cell types may use different TREs to regulate Dnmt3a, or that the neuroblastoma cell line does not recapitulate the in vivo situation. To our knowledge, this is the first demonstration that deletion of a predicted TRE using CRISPR/Cas9 can strongly impact T3 responsiveness of a direct TR target gene.
Sequence conservation of Dnmt3a TREs
Among eutherian mammals, we found strong sequence conservation for both of the mouse Dnmt3a TREs. This conservation extended to a metatherian, the wallaby, and to a monotreme, the platypus for the +49.3-kb, but not the +30.3-kb, TRE. Interestingly, we found that the +49.3-kb TRE is DR+4 in the genomes of mouse, rat, and Chinese hamster, whereas it is DR+5 in other mammals (Figure 5B). We estimate that the transition from DR+5 to DR+4 occurred at least 44 million years ago (divergence between mouse or rat from Chinese hamster), because the TRE sequence is DR+4 in Chinese hamster, whereas it is DR+5 in kangaroo rat, naked mole rat, and several other mammals. Previous work has shown that DR+5 TREs can bind TRβ-RXRα heterodimers in vitro, but whether they can support T3-dependent transactivation may be dependent on the DR+5 sequence context and/or cell type, as one study found support for T3-dependent transactivity, whereas another did not (39, 40). Further study is needed to determine whether the +49.3-kb DR+5 TRE can support T3-dependent transactivation.
Although there is strong conservation of the 2 Dnmt3a TREs that we identified among mammals, we did not find similar sequences in reptiles (including birds). Our failure to find such sequences may be due to divergent, missing or incomplete sequences of genomic regions that could potentially contain TREs in these lineages. Interestingly, we found that the dnmt3a genes of the frogs X. laevis and X. tropicalis are regulated by T3, likely via 2 TREs located in the first and second introns (Y. Kyono and R. J. Denver, unpublished data), but these TREs do not appear to be orthologous to the mammalian TREs that we report here. We do not know whether the orthologous Dnmt3a genes are similarly regulated by T3 in vertebrates other than mouse and Xenopus species. Further study is needed to determine when during the evolution of tetrapods the gene came under thyroid control, whether this evolved once or multiple times using the same regulatory regions, and whether hormone regulation has been lost in some lineages.
Potential roles for T3 regulation of Dnmt3a in the developing brain
Findings in different cell types support that DNMT3a is essential for establishing and/or maintaining cell differentiation and cell cycle exit (41–44). A recent microarray analysis conducted on NSCs derived from the ventricular/subventricular zones of Dnmt3a knockout mice showed impaired neurogenesis in response to a differentiation signal accompanied by dysregulation of cell cycle genes (45). The expression level of Cyclin D1 was inversely correlated with the methylation state of its promoter (46), and several lines of evidence support that Cyclin D1 is directly regulated by DNMT3a-mediated DNA methylation (47, 48). Thyroid hormone, acting via TRβ1 causes cell cycle exit and differentiation, and down regulation of Cyclin D1 (21, 29, 49) (Y. Kyono and R. J. Denver, unpublished data). Considering these findings, we hypothesize that increased DNMT3a (under the direct influence of T3) methylates regulatory regions of cell cycle control genes leading to their repression, and that this is an important pathway for the establishment and/or maintenance of cell differentiation in the developing nervous system.
Acknowledgments
We thank Dr Michael Rehli (University Hospital Regensburg, Germany) for kindly providing the pCpGL plasmids; Dr Yun-Bo Shi (National Institute of Child Health and Development, National Institutes of Health, Bethesda, MD) for the PB antiserum, pSp64a-TRβ, and pSp64a-RXRα plasmids; and Pia Bagamasbad for technical assistance.
Present address for Y.K.: Department of Human Genetics, The University of Michigan, Ann Arbor, MI 48109.
This work was supported by the National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH) Grant R21 NS088062 (to R.J.D.). Y.K. was supported by a Ruth Kirschstein National Research Service Award from National Institute of Neurological Disorders and Stroke. P.R. and A.N.H. received support from NIH Grants R21 DK091941 and R01 DK056123. This research used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center funded by NIH Grant P60 DK20572 from the National Institute of Diabetes and Digestive Kidney Diseases.
Disclosure Summary: The authors have nothing to disclose.
Appendix
Table 1.
Antibody Table
| Peptide/Protein Target | Antigen Sequence (if Known) | Name of Antibody | Manufacturer, Catalog Number, and/or Name of Individual Providing the Antibody | Species Raised in; Monoclonal or Polyclonal | Dilution Used |
|---|---|---|---|---|---|
| X. laevis thyroid hormone receptor-β | Full-length protein | PB | Yun-Bo Shi | Rabbit; polyclonal | 1:100 ChIP |
| Histone H3 (H3) | Anti-H3 | Millipore, 07-670 | Rabbit; polyclonal | 1:100 ChIP | |
| Acetylated histone 3 (acH3) | Anti-acH3 | Millipore, 06-599 | Rabbit; polyclonal | 1:100 ChIP |
Footnotes
- acH3
- acetylated histone 3
- BW
- body weight
- CG
- cytosine-guanine
- ChIP
- chromatin immunoprecipitation
- ChIP-seq
- ChIP sequencing
- CHX
- cycloheximide
- DMSO
- dimethyl sulfoxide
- DNMT
- DNA methyltransferase
- DR+4
- direct repeat plus 4-base spacer
- EGFP
- enhanced green fluorescent protein
- gRNA
- guide RNA
- Klf9
- Krüppel-like factor 9
- NRS
- normal rabbit serum
- PND
- postnatal day
- RT-qPCR
- RT real-time quantitative PCR
- RXR
- retinoid X receptor
- TR
- T3 receptor
- TRE
- T3-response element
- TSS
- transcription start site.
References
- 1. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. [DOI] [PubMed] [Google Scholar]
- 2. Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107(19):8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. [DOI] [PubMed] [Google Scholar]
- 4. Rottach A, Leonhardt H, Spada F. DNA methylation-mediated epigenetic control. J Cell Biochem. 2009;108(1):43–51. [DOI] [PubMed] [Google Scholar]
- 5. Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev. 2002;118(1–2):187–190. [DOI] [PubMed] [Google Scholar]
- 6. MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88(3):170–183. [DOI] [PubMed] [Google Scholar]
- 7. Shin J, Ming G-l, Song H. DNA modifications in the mammalian brain. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652):20130512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Isles AR. Neural and behavioral epigenetics; what it is, and what is hype. Genes Brain Behav. 2015;14(1):64–72. [DOI] [PubMed] [Google Scholar]
- 9. Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varley KE, Gertz J, Bowling KM, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie W, Barr CL, Kim A, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148(4):816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ziller MJ, Muller F, Liao J, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79(6):734–746. [DOI] [PubMed] [Google Scholar]
- 15. Bernal J. Action of thyroid hormone in brain. J Endocrinol Invest. 2002;25(3):268–288. [DOI] [PubMed] [Google Scholar]
- 16. Anderson GW, Schoonover CM, Jones SA. Control of thyroid hormone action in the developing rat brain. Thyroid. 2003;13(11):1039–1056. [DOI] [PubMed] [Google Scholar]
- 17. Bernal J. Thyroid hormones and brain development. Vitam Horm. 2005;71:95–122. [DOI] [PubMed] [Google Scholar]
- 18. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31(2):139–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bagamasbad PD, Bonett MR, Sachs L, et al. Deciphering the regulatory logic of an ancient, ultraconserved nuclear receptor enhancer module. Mol Endocrinol. 2015;29(6):856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hadj-Sahraoui N, Seugnet I, Ghorbel MT, Demeneix B. Hypothyroidism prolongs mitotic activity in the post-natal mouse brain. Neurosci Lett. 2000;280(2):79–82. [DOI] [PubMed] [Google Scholar]
- 21. Denver RJ, Williamson KE. Identification of a thyroid hormone response element in the mouse Kruppel-like factor 9 gene to explain its postnatal expression in the brain. Endocrinology. 2009;150(8):3935–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crespi EJ, Denver RJ. Leptin (ob gene) of the South African clawed frog Xenopus laevis. Proc Natl Acad Sci USA. 2006;103(26):10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao M, Stenzel-Poore M, Denver RJ. Structural and functional conservation of vertebrate corticotropin-releasing factor genes: evidence for a critical role for a conserved cyclic AMP response element. Endocrinology. 2007;148:2518–2531. [DOI] [PubMed] [Google Scholar]
- 24. Bagamasbad P, Ziera T, Borden SA, et al. Molecular basis for glucocorticoid induction of the Kruppel-like factor 9 gene in hippocampal neurons. Endocrinology. 2012;153(11):5334–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramadoss P, Abraham BJ, Tsai L, et al. Novel mechanism of positive versus negative regulation by thyroid hormone receptor β 1 (TR β 1) Identified by genome-wide profiling of binding sites in mouse liver. J Biol Chem. 2014;289(3):1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatonnet F, Guyot R, Benoît G, Flamant F. Genome-wide analysis of thyroid hormone receptors shared and specific functions in neural cells. Proc Natl Acad Sci USA. 2013;110(8):E766–E775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1(3):127–130. [DOI] [PubMed] [Google Scholar]
- 28. Hoopfer ED, Huang LY, Denver RJ. Basic transcription element binding protein is a thyroid hormone-regulated transcription factor expressed during metamorphosis in Xenopus laevis. Dev Growth Diff. 2002;44(5):365–381. [DOI] [PubMed] [Google Scholar]
- 29. Lebel JM, Dussault JH, Puymirat J. Overexpression of the β-1 thyroid receptor induces differentiation in Neuro-2a cells. Proc Natl Acad Sci USA. 1994;91(7):2644–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samuels HH, Stanley F, Casanova J. Depletion of L-3,5,3′-triiodothyronine and L-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105(1):80–85. [DOI] [PubMed] [Google Scholar]
- 31. Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31(46):16619–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor α gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;151(2):731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chatonnet F, Flamant F, Morte B. A temporary compendium of thyroid hormone target genes in brain. Biochim Biophys Acta. 2015;1849(2):122–129. [DOI] [PubMed] [Google Scholar]
- 34. Ausió J, Martínez de Paz A, Esteller M. MeCP2: the long trip from a chromatin protein to neurological disorders. Trends Mol Med. 2014;20(9):487–498. [DOI] [PubMed] [Google Scholar]
- 35. Barrow TM, Michels KB. Epigenetic epidemiology of cancer. Biochem Biophys Res Commun. 2014;455(1–2):70–83. [DOI] [PubMed] [Google Scholar]
- 36. Hall L, Kelley E. The contribution of epigenetics to understanding genetic factors in autism. Autism. 2014;18(8):872–881. [DOI] [PubMed] [Google Scholar]
- 37. Falahi F, Sgro A, Blancafort P. Epigenome engineering in cancer: fairytale or a realistic path to the clinic? Front Oncol. 2015;5:22–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emi Y, Ikushiro S, Kato Y. Thyroxine-metabolizing rat uridine diphosphate-glucuronosyltransferase 1A7 is regulated by thyroid hormone receptor. Endocrinology. 2007;148(12):6124–6133. [DOI] [PubMed] [Google Scholar]
- 40. Shulemovich K, Dimaculangan DD, Katz D, Lazar MA. DNA bending by thyroid hormone receptor: influence of half-site spacing and RXR. Nucleic Acids Res. 1995;23(5):811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–U43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):U423–U437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236(6):1663–1676. [DOI] [PubMed] [Google Scholar]
- 44. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. [DOI] [PubMed] [Google Scholar]
- 45. Wu H, Coskun V, Tao J, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329(5990):444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kitazawa S, Kitazawa R, Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J Biol Chem. 1999;274(40):28787–28793. [DOI] [PubMed] [Google Scholar]
- 47. Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4(7):487–499. [DOI] [PubMed] [Google Scholar]
- 48. Brenner C, Deplus R, Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24(2):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perez-Juste G, Aranda A. The cyclin-dependent kinase inhibitor p27(Kip1) is involved in thyroid hormone-mediated neuronal differentiation. J Biol Chem. 1999;274(8):5026–5031. [DOI] [PubMed] [Google Scholar]