Abstract
A novel method for lipid analysis called CTS (collisional activation and traveling wave mass spectrometry) involving tandem mass spectrometry of all precursor ions with ion mobility determinations of all product ions was applied to a sample of human serum. The resulting four dimensional data set (precursor ion, product ion, ion mobility values, and intensity) was found to be useful for characterization of lipids as classes as well as identification of specific species. Utilization of ion mobility measurements of the product ions is a novel approach for lipid analysis. The trends and patterns of product mobility values when visually displayed yield information on lipid classes and specific species independent of mass determination. The collection of a comprehensive set of data that incorporates all precursor-product relationships combined with ion mobility measurements of all products enables data analysis where different molecular properties can be juxtaposed and analyzed to assist with class and species identification. Overall, CTS is powerful, specific, and comprehensive method for lipid analysis.
Introduction
Lipids constitute an important class of biomolecules present in all kingdoms of life. Unlike proteins and nucleic acids which are derived from common molecular building blocks and have diversity in structure based upon sequence of these building blocks, lipids are a diverse family of molecules that can be divided into eight different classes of amphipathic and hydrophobic molecules.1 For most of these lipid classes, biosynthesis results in the production of a large number of closely related structures that differ in the occurrence of fatty acyl moieties of alternative chain length, degree of unsaturation, and double bond positions, but differ in polar structures. Electrospray ionization has provided the means by which all of the known lipid molecules can be analyzed by mass spectrometry2 and hence, the emergence of a strategy to measure numerous lipids present within a cell or tissue system. Such studies of lipids found in biological systems can be both qualitative and quantitative in nature and have been termed “lipidomic” investigations. The challenge in such studies is that the enormous number of lipid species that are found in cells and tissues can number to multiple thousands in aggregate. To address these analytical challenges two major techniques have emerged which are of enormous value. One is LC-MS where chromatographic separation of lipid species takes place prior to mass spectrometric analysis. A second approach, which has been generally termed shotgun analysis,3,4 directly ionizes a lipid extract and uses characteristics of each individual lipid class to generate abundant ions to identify and quantify specific lipids. Such alterations of the direct electrospray analysis could include the polarity of ions detected or modification of the mobile phase to enhance protonation, deprotonation, and adduct ion formation.4 As these different techniques have emerged and been applied, an understanding of the value of general lipidomic analysis is becoming recognized and interest in improving sample throughput for such studies has now become an important consideration.
One recent development in mass spectrometry, namely ion mobility measurements during mass spectrometric analysis, has matured into a powerful technique for separation of ions prior to mass analysis. Ion mobility measurements have been applied to studies of lipids, including detailed analysis of specific lipid classes such as phosphatidylcholine,5 separation of lipids prior to mass spectrometric analysis,6 and the use of ion mobility strategies to separate lipids in a process termed FAIMS.7 A useful advantage of ion mobility is that it offers an additional separation step that can be implemented prior to mass analysis, but ion mobility could offer a way to improve sample throughput in shotgun studies. One of the advances of ion mobility has been in the development of traveling wave ion separations to improve the resolution of ion separation.8,9
An additional strategy, first suggested by Philip Sanders (Eli Lilly Company)10 and implemented commercially by mass spectrometer manufacturers, is the analysis of all electrospray generated ions by tandem mass spectrometry.11,12 In such studies ions are sequentially separated by an initial mass analyzer followed by collisional activation with measurement of all product ions for that mass. This process is repeated for the entire mass range of interest. This is a powerful technique, but does require some time, even with modern fast scanning quadrupole mass spectrometers. Nonetheless, the data obtained is a complete collection of all the collision spectra from every ion generated by the electrospray ionization process. From this collection of data important information such as precursor-product relationships, product ions, as well as neutral losses common to a specific class of lipids, can be discerned with database manipulations.
We have combined tandem mass spectrometry of all ESI-generated ions in conjunction with shotgun analysis of lipids, but have added the additional parameter of ion mobility of all product ions in a collisional activation and traveling wave mass spectrometry (CTS) experiment. We have applied this strategy to the analysis of the neutral lipids present in human serum for which considerable data exist as to the numbers and quantity of molecular species that normally occur.13
Experimental Methods
Materials
Human serum (SRM 909c) was purchased from the National Institute of Standards and Technology (Gaithersburg, MD), divided into 50 μL aliquots and stored at −70°C until used. Solvents and chemicals used for extraction and mass spectrometry were HPLC grade, purchased from Fisher Scientific Company (Pittsburgh, PA), and were used without further purification. Polyalanine was obtained from Sigma Chemical (St. Louis, MO).
Serum extraction protocol
An aliquot of serum was thawed, and extracted for neutral lipids using previously published protocols.13 Briefly, 50 μL of serum was diluted with 1.0 mL Hanks’ balanced salt solution to which was added 0.5 mL hexanes and 0.5 mL methyl t-butyl ether (MTBE). The mixture was vortexed and centrifuged after which the top layer was removed to a separate test tube. This extraction was repeated a second time, and the combined organic layers were dried under a low flow of nitrogen gas and subsequently resuspended in one mL of infusion solvent comprised of a 30:40:7 mixture of hexane: isopropyl alcohol: 10 mM aqueous ammonium formate.
Mass spectrometry
Mass spectrometry and ion mobility measurements involving the CTS analysis were carried out using a Synapt G2-S mass spectrometer (Waters, Manchester, UK) in positive ion mode. Samples were infused directly into the electrospray source of the mass spectrometer using a syringe pump, and a 0.5 mL syringe at a flow rate of 2.0 μL/min. Mass spectrometer parameters for the CTS experiments included: Synapt G2-S set to Resolution Mode, ESI voltage 2500 V, sampling cone 40 V, source offset 80 V, source temperature 120°C, desolvation temperature 150°C, cone gas 40 L/h, desolvation gas 300 L/h, nebulizer 6.0 bar, ion mobility wave height 40 V, ion mobility wave velocity 480 m/s, helium gas in the ion mobility chamber set to 2 mL/min, and N2 collision gas to 180 mL/min. Collisional activation was carried out in the first (trap) of the three traveling wave sections of the Synapt G2-S with collision energy 20 V or as noted. Ion mobility separation was optimized using a mixture of CE(18:2) and TG (52:3) infused as above.
CTS/WREnS
Waters Research Enabled Software (WREnS; Waters Corp) operates alongside MassLynx via a script, written in computer language C#. It allows the user to set voltages and parameters that can be applied over time or on a scan by scan basis for flexible control of the instrument. For the CTS analysis, the script was used to control the precursor mass selection and the rate in a step-wise progression of precursor masses (1.001 amu per step to account for the mass defect accumulation for lipid-like molecules) in an assemblage of CID experiments. The mass range applied to precursor mass selection was m/z 200 – 1200 with acquisition of product ion spectra in the range m/z 50–1200. Quadrupole settings were held for 0.985 s with an interscan delay of 0.015 s before stepping up quadrupole voltage to select the next nominal mass-to-charge-ratio. The resulting run time was 16.67 min/CTS experiment to execute the entire step function of precursor mass selection in the first quadrupole.
Data analysis
The data were analyzed using MassLynx and Driftscope software packages provided by Waters Corporation. Separate software written in MATLAB (Mathworks, Natick, MA) was used to generate extracted ion mobilogram graphs, and selected precursor and product spectra at specific ion mobilities. The full set of data acquired in the CTS analysis (precursor mass, product mass, ion mobility, and intensity) were accessed using Driftscope, and transformed to a tabulated spreadsheet of all recorded values that could be inspected and analyzed in Excel. Precursor m/z value was not recorded in the data output but was calculated using a linear regression line relating elapsed time after initiating the WREnS experiment for several prominent precursor ions (m/z 369.3, 577.5, 671.6. and 879.7) in the serum mixture. In a more general case for other samples, any known ion, that appears both as a precursor and product ion can be used to establish the exact time of the starting mass selected (WREnS time scale) from the exact mass obtained in the time-of-flight measurement. All precursor masses are reported here at nominal mass values since these were actually set by the quadrupole mass settings that increment at a constant 1.00 s for each subsequent mass-to-charge-ratio. This precursor mass assignment based on elapsed time after initiating the WREnS experiment was examined for the four calibration masses listed above on a routine basis to assure reproducibility as a check of reproducibility. Recalibration of precursor mass assignment was carried out for each group of experiments or if instrument ion mobility parameters were altered. Ion mobility values for a specific ion could be found in the collected ion mobility database, or from extracted ion mobilograms of the data after exporting the Driftscope data into MassLynx. Product ion masses from the CTS experiments are reported accurate to 10 mmu and separate accurate mass determinations using the lockspray function with leucine enkephalin as the exact mass calibrant, are reported to 1 mmu.
Collision Cross Section Calculations
Collision cross section (CCS) values for the lipids described were determined using the protocols of Michaelevski et al.14 and Paglia et al.15 After optimizing ion mobility parameters for a mixture of lipid standards to achieve optimal mobility separation for the range of mass values and lipid structures considered in our samples, a calibration curve was established using a solution of polyalanine (0.1 ng/μL) under these optimized conditions. Ion mobility values for the polyalanine oligomers were recorded and transformed as previously described. These values were used in a calibration curve relating the polyalanine oligomer ion mobility and CCS values measured on a non-travelling wave ion mobility instrument. This calibration curve was then used to determine all other collision cross section values for the lipids described in this work.
Results and Discussion
The CTS experiment was carried out by initial selection of electrospray generated ions followed by collisional activation, then analysis of all resulting product ions by traveling wave ion mobility followed by a final time-of-flight high resolution analysis before ion detection. The resultant 4-dimensional dataset (precursor ion, product ion mobility, mass product ion determination, and ion abundance) was obtained for a mass range from m/z 200 – 1,200 in 16.67 min using the first quadrupole set to increment 1 Da/s. This mass range encompassed many of the important lipid substances, including glycerolipids, phospholipids, and steroid esters.
CTS analysis of human serum neutral lipid extract generated a wealth of precursor/product information, including ion mobility measurements of product ions that could be investigated based on the behavior of the cholesteryl esters (CE) and triacylglycerols (TG) standards. A complex precursor ion/product ion data map (Figure 1A) revealed an abundant population of lines corresponding to common product ions, precursor ions as well as neutral loss behavior for many of the components. Precursor ions (Figure 1B) generated in this human serum sample revealed, as expected, abundant neutral lipids corresponding to a family of TGs, the most abundant of which corresponds to TG(52:2) as the ammoniated ion at m/z 876 and cholesterol esters with the most abundant precursor ion corresponding to CE(18:2), [M+NH4]+ at m/z 666 and corresponding sodiated adduct at m/z 671. Since this study was not set up for quantitation, a rather high concentration of lipid extract was electrosprayed which is known to lead to inaccuracies in ion abundances largely due to cluster ion formation16. Nevertheless, analysis of the ten-fold diluted NIST serum extract (Supplemental Figure S1) was qualitatively identical to the data presented here.
Figure 1.
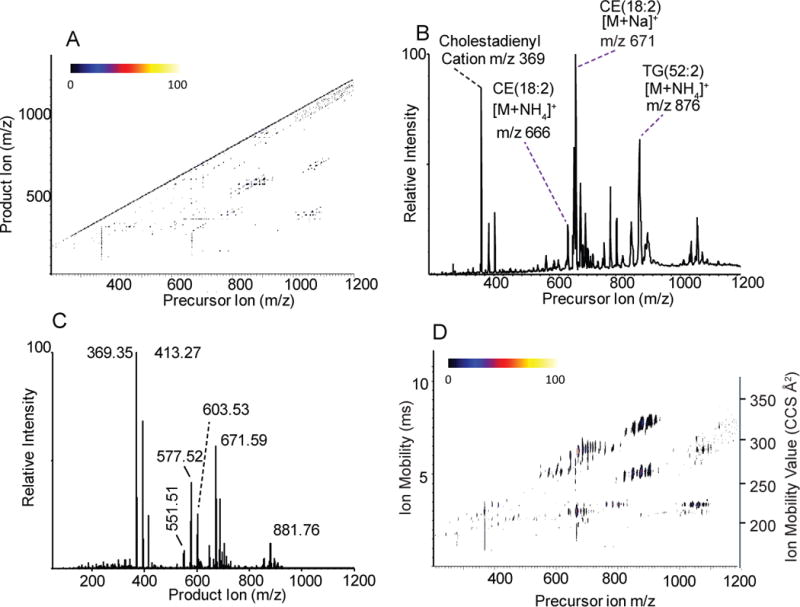
Different displays of CTS analysis of human serum: A) three-dimensional map showing precursor vs. product ion relationships of the full CTS data set. Intensities are indicated by color scale shown. B) two-dimensional plot showing all precursor ions in sample vs. intensity. C) two-dimensional plot showing all product ions in sample vs. intensity. D) three-dimensional map showing all ion mobility values acquired from CID analysis of quadrupole selected precursor ions.
The common product ions (Figure 1C) generated in this neutral lipid extract from human serum revealed a population of high mass ions corresponding to the sodiated adduct of the TGs, abundant diglyceride-like product ions at m/z 551.51, 577.52, and 603.53. A very abundant cholestadiene cation at m/z 369.35 in both Figure 1B and 1C are a result of the instability of the ammonium adduct of cholesteryl esters which undergo partial in-source decomposition during electrospray ionization and whose [M+NH4]+ readily decompose at 20V collision energy. There were also abundant ions at m/z 393 and 413 that gave product ions at m/z 393.30 and m/z 413.27, respectively, due to the presence of contaminants in the serum sample (see below). The absence of a signal at m/z 666 as a product ion was due to the instability of the ammonium adduct ions from CEs to CID to yield m/z 369.3517. The observed collision induced decomposition of CEs as well as TGs was identical to that observed for reference lipids obtained commercially (data not shown).
The ion mobility map of the precursor ions (Figure 1D) revealed a cluster of ions with common ion mobility for cholesterol esters, their associated cholestadiene cation, and ion mobilities of both sodiated and ammoniated TGs with their associated diglyceride-like fragments. An expansion of the precursor ion region, which encompassed the cholesterol esters (Figure 2), revealed a family of cholesterol species that were present as both ammoniated and sodiated adducts, but clearly identified by the abundance of the cholestadiene product ion at 210 Å2 (2.98 ms). The cholesterol esters identified in this CTS experiment corresponded to the known abundant cholesterol esters described in human blood such as 16:0, 18:1, 20:4, and 22:6 fatty acyl components.17,18 In addition to this, there was clearly present a series of oxidized CE species, which were derived from oxidation of a polyunsaturated fatty acyl moiety of the cholesterol esters with addition of 1–4 oxygen atoms. Their identity was confirmed by accurate mass measurements as well as from CID behavior of both the sodiated and ammoniated ions, where the ammoniated ions yielded abundant cholestadiene cations. Since it was unexpected that these oxidized CEs were present, normal phase LC-MS/MS was carried, out as previously described18, to confirm these oxidized neutral lipids (Supplemental Figure S2). The details of preparation and storage of this NIST standard serum sample were not readily available, but it is well known that oxidation of polyunsaturated fatty acyl groups takes place even when serum is store at very low temperatures and a long period of time19.
Figure 2.
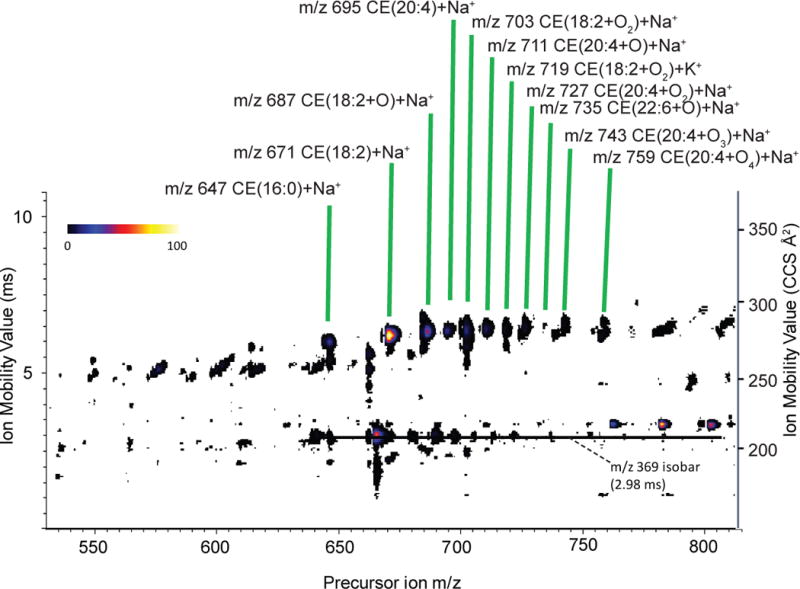
Enlarged region of Figure 1D, the ion mobility map generated from ions produced upon precursor ion selection, showing region corresponding to CEs in greater detail. Data are from analysis of human serum standard.
In a similar manner, detailed investigation of the high mass region of the CTS experiment of human serum between m/z 840 and 920 (Figure 3) revealed a family of TG molecular species as both the sodiated and ammoniated adducts that yielded common diglyceride-like fragment ions with ion mobilities of 248–275 Å2 (4.3–5.4 ms) and the fairly low abundance of fatty acyl related product ions20 between 174–202 Å2 (2.0–2.3 ms).
Figure 3.

Enlarged region of Figure 1D showing region corresponding to TGs in greater detail. Data are from analysis of human serum standard.
A set of four species at precursor mass m/z 848, 850, 853, and 855 represent mixtures of ammoniated (TG(50:2), TG(50:1)) and sodiated (TG(50:2), TG(50:1)), respectively, that were identified by the pattern of ion mobility clusters following CTS analysis and confirmed by accurate mass measurements and CID analysis (Supplemental Figure S3). The most abundant TG species (TG(52:2)) in Figure 3 was a prominent [M+NH4]+ ion at m/z 876 with a sodiated analog 5 Da higher at m/z 881. Like the cholesterol esters, the sodiated species of TGs did not fragment as easily as the ammoniated adduct and present a pattern of mapped entries that supports species confirmation using ion mobility data. This behavior of ammoniated, sodiated, and lithiated adducts following CID has been reviewed2.
Two interesting species at m/z 888 and 890 appeared in Figure 3 corresponding to ammonium adducts of oxidized TGs. These oxidized TG species readily lost ammonia and water from the precursor ions, resulting in large product ion signals 35 Da less than the precursor mass. All product ions could be readily extracted from the CTS data of m/z 890 and 888 shown and reported in Supplemental Figures S4 and S5, respectively. The component reported as TG(16:0_18:1_KODE) refers to an oxidized fatty acid having a keto-group incorporated into 18:2 fatty acyl species (keto-octadecadienoate). Most likely this structural feature arises from an initial hydroperoxy octadecadienoate,21 which rapidly dehydrates to the keto functionality as has previously been reported.22 Thus, the oxidized TGs appear to be identifiable not only from the unique fragmentation in the CID spectra, but also from the subtle patterns of ion mobility, as mapped out relative to precursor ion mass. Approximately 100 TG molecular species and 10 CE molecular species could be identified using the criteria above for this human serum sample (Supplemental Table S1 and S2).
Ion mobilograms
Acquisition of all MS/MS spectra and the corresponding ion mobility values for product ions permitted reconstruction of a novel set of histograms that related the ion mobility behavior to abundance of product ions. The total ion mobilogram of the neutral lipids from human serum extract had surprising abundance features when plotted as the observed ion mobility time. Since mobility time can be converted to collisional cross-section, Figure 4 displays the array of product ions from low to high collisional cross-section.
Figure 4.
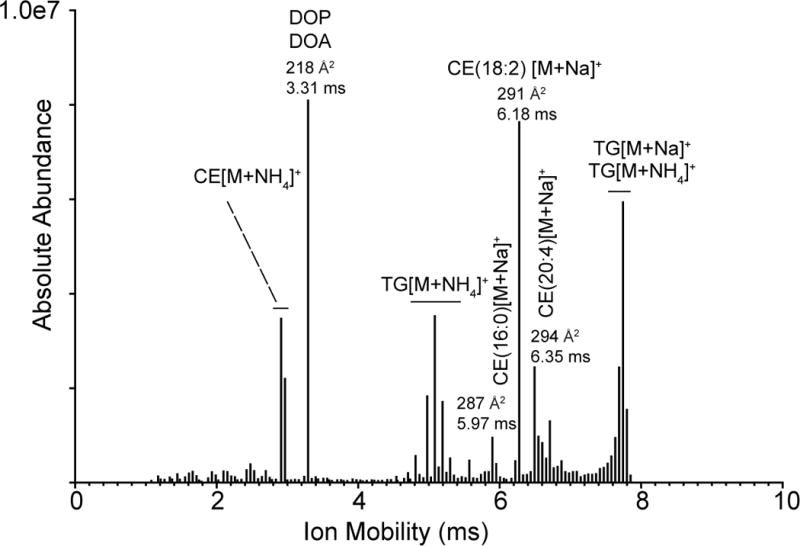
A histogram showing the abundance of all product ions collected from the CTS experiment in the human serum sample as a function of ion mobility. The assignment of the neutral lipids at indicated times corresponds to the precursor ions selected when the product ions were generated with those mobility times.
The identity of the product ions that distribute as to each ion mobility can be ascertained by extraction of the product ions and corresponding precursor ions that have the unique mobility. The abundant ions with the fastest mobility (210–212 Å2, 2.87 to 2.93 ms) were extracted from the CTS database as both the product ions with this mobility (Figure 5A) and the precursor ions that yielded the corresponding product ions (Figure 5B). The product ions observed were essentially of one ion type, namely m/z 369.35, which is the cholestadiene cation discussed above for cholesteryl esters. The most abundant precursor ion was the [M+NH4]+ derived from CE(18:2) at m/z 666. Other cholesterol esters, CE(18:1) and CE(20:4) were present at this same ion mobility since they all yield the identical m/z 369.35 as the major product ion.
Figure 5.
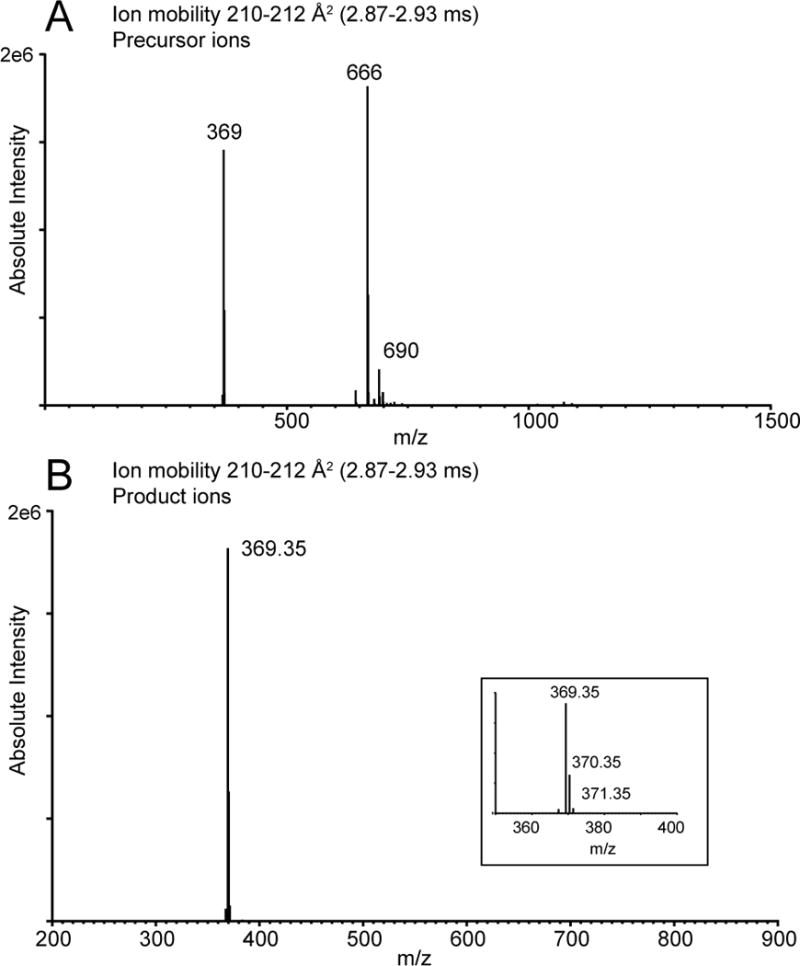
Histograms showing ions extracted from full data set of CTS analysis of human serum related to cholesterol esters. A) Precursor ions related to ion mobility values 2.87–2.93 ms. B) Product ions related to ion mobility values 2.87 – 2.93 ms.
The ions with abundant, but unique mobilities corresponding to the sodiated adducts of these cholesterol esters were observed between 287 to 304 Å2 (5.97 and 6.67 ms). As discussed above, the sodiated adduct of cholesterol esters are more stable than the ammoniated adducts. These intact CE adducts did not yield abundant product ions and thus the ion mobilities were in fact the precursor ion mobility (data not shown). Each sodiated CE adduct had a rather unique ion mobility seen for CE(16:0)+Na+ at 287 Å2 (5.97 ms), CE(18:2)+Na+ at 291 Å2 (6.18 ms), and CE(20:4)+Na+ at mobility 294 Å2 (6.35 ms) (Table S2). The exact ion mobility measured for product ions was found to be quite reproducible. For example the same sample was analyzed, using the same instrument parameters for ion mobility, and the ion mobility values did not change for four target product ions after a 5-month interval between measurements (Table S3).
Abundant ion mobility was observed at 293 Å2 (6.29 ms) (Figures 2 and 4) and the ion generating this mobility was observed to be m/z 687.56 (C45H76O3Na) as a product ion, but an ammoniated ion (as a product ion) at m/z 682.61 was not observed, likely due to facile collision induced dissociation. These data were consistent with a cholesteryl ester that contained one additional oxygen compared to the expected CEs and suggested the presence of a hydroxyl group in the fatty acyl portion of the molecule. The corresponding precursor ammoniated ion (m/z 682) readily yielded m/z 369.35 as the cholesterol cation after collisional activation, revealing no additional oxygen atom on the cholesteryl ring structure. This feature added to the likelihood of oxidation of the fatty acyl linoleate, suggesting this component in the human serum was CE(HODE) [M+Na]+. It was not possible to determine the position of hydroxyl group, but most likely oxidation of linoleate preceded by autooxidation at both carbons-9 and -13.21 which was confirmed by LC-MS/MS analysis of this serum sample (Supplemental Figure S2).
The abundant ions of longer ion mobility 323–327 Å2 (7.65–7.81 ms) (Figure 6) were due to a complex mixture of sodiated and ammoniated adducts of triacylglycerol species that included TG(52:3) m/z 879 and TG(52:2) m/z 881. Ammonium adduct precursor ions from the TG molecular species were observed because they are somewhat more stable to collisional activation than ammoniated adducts of CEs. There was also evidence for oxidized triacylglycerols TG(52:3-O) and TG(52:2-O) at m/z 895 and 897 as the sodiated adducts and ammoniated adducts at m/z 890 (discussed previously) and at m/z 892. There was an additional ion observed at m/z 893 likely as the sodium adduct of a further oxidized species that had a predicted precursor mass for the ammoniated ion at m/z 888 (discussed previously), respectively.
Figure 6.
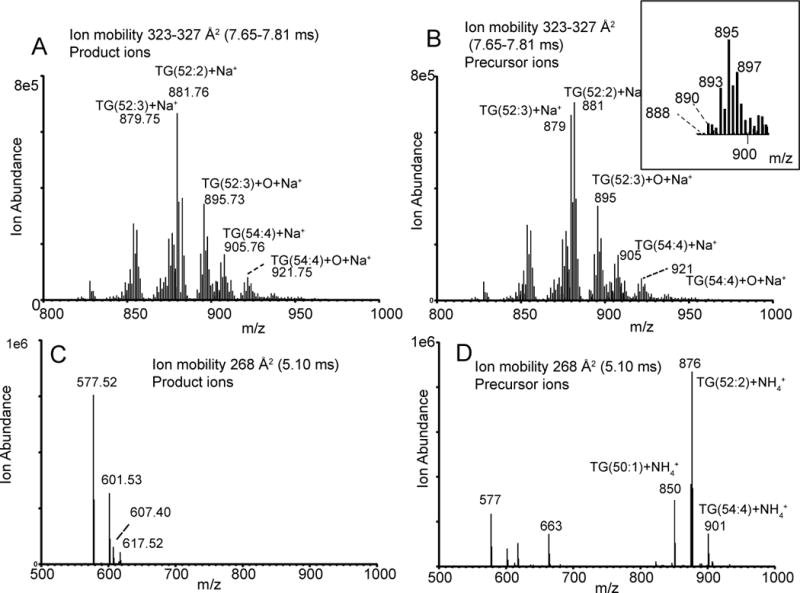
Histograms showing ions extracted from the full data set of the CTS analysis of human serum related to TGs. A) Product ions related to ion mobility values 7.60–7.76 ms. B) Precursor ions related to ion mobility values 7.60–7.76 ms. C) Product ions related to ion mobility values 4.94–5.15 ms. D) Precursor ions related to ion mobility values 4.94–5.15 ms
The product ions with ion mobilities of 265, 268 and 271 Å2 (4.99, 5.10 and 5.21 ms) were due to the diglyceride-like product ions derived from collisional activation of the ammoniated TG adducts. Collecting the ion mobilities of such diglyceride ions along with the high resolution mass measurements to determine elemental composition, the fatty acyl make up in terms of the total fatty acyl carbon atoms and double bonds could be determined (Supplemental Table S1). For example, the diglyceride-like product ions appearing with ion mobility of 268 Å2 (5.10 ms) are m/z 577.63 and m/z 601.53 (Figure 6C). The precursor ions for these diglyceride-like ions are from TG(50:1) [M+NH4]+, TG(52:2) [M+NH4]+, and TG(54:4) [M+NH4]+ (Figure 6D). When the ion mobility of all the diglyceride-like ions collided in the CTS experiment was plotted but grouped as to number of double bonds, a linear relationship was found for each group (Figure 7). Thus, at one ion mobility value, one could observe several diglyceride-like ions where the higher masses would contain a larger total number of double bonds than those of lower masses.
Figure 7.
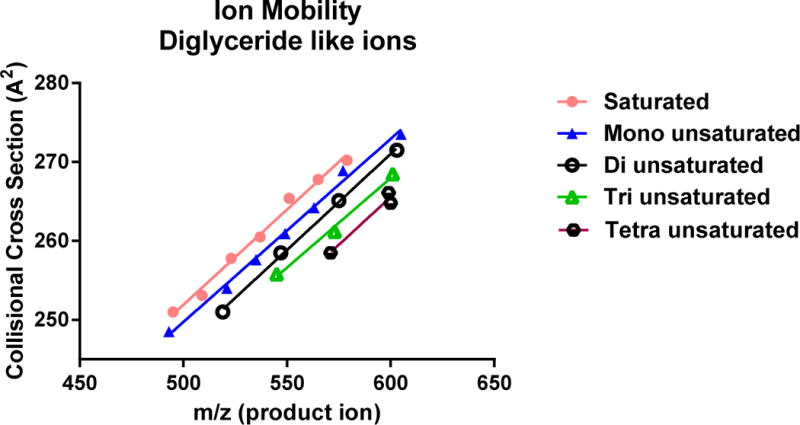
Triacylglycerol molecular species identified from CTS analysis of human serum sample plotted to show relationship between levels of total fatty acyl group unsaturation (n=0 to 4) and collisional cross section ion mobility.
The ions of highest ion mobility abundance occurred at 218 Å2 (3.31 ms) (Figure 4). Two product ions had this ion mobility and they were m/z 393.30 and m/z 413.27, which differed by 20 Da (Supplemental Figure S6A). From their exact masses, the difference could be assigned to a gain of two carbon atoms, but loss of four hydrogen atoms from the high mass product ion relative to low mass product ion. This high mass species had been previously reported in neutral lipid extracts as due to a dioctyl phthalate (DOP) ester,23 most likely ethyl hexyl phthalate, a very common vinyl plasticizer found in clinical apparatus such as blood bags. The lower mass product ion was consistent with another closely related plasticizer, dioctyl adipate (DOA) as a sodium adduct.
Of more interest were the precursor ions observed (Supplemental Figure S6B) that in addition to the lower mass ions observed as product ions, had a triplet set of ions at m/z 763, 783, and 803 that all differed by the 20 Da seen for the lower mass product ions. These ions were consistent with the sodium adduct of a dimer of DOA, a heterodimer of the sodium adduct of DOA and DOP, and a sodium ion adduct of the dimer of DOP, respectively. At even higher mass was a complex multiplet with major ions at m/z 1042 and 1062 that was consistent with the sodium adducts of the heterodimers of DOP and DOA with a major cholesterol ester, CE(18:2). Thus, these very abundant neutral lipids were forming dimer sodiated adducts with each other during the electrospray process. Of interest was that the extracted ion mobility plot (Figure 4) readily revealed these unique product ions derived from the plasticizer from that of CEs and TAGs, present in the serum sample.
Conclusion
The CTS experiment is readily interfaced with shotgun-type lipid analysis. Acquisition of MS/MS data for all ions generated by electrospray ionization adds considerable more detail to lipid studies than just data-dependent MS/MS data approaches. While the 16.67 min acquisition of this dataset may seem long, the advances made in nanoelectrospray engineering makes this relatively easy to implement in the shotgun strategy for multiple samples. The ion mobility portion of this CTS dataset also adds additional information that is quite useful for lipid analysis. In large part this was due to the strategy of obtaining ion mobility measurements on product ions rather than the precursor ions. While this has not been the usual application of ion mobility for lipid analysis,6 it should be recognized that many facile protocols that have been developed over the past 40 years for selecting the specific lipid classes for analysis. This includes normal phase chromatographic techniques such as TLC, specific solvent extractions (as used in this study), as well as stability of specific lipids to saponification followed by extraction. Thus, to a large extent, the use of product ion mobility provides unique information somewhat unique in that product ions would not have the identical mobility compared to the starting lipid, but ion mobilities of product ions may be common to a lipid class. For example, the compact nature of the cholestadiene cation generated from all cholesteryl ester molecular species can be compared to a lipid fragment ion containing the same number of carbon atoms, but having an open and rather flexible structure.
Furthermore, the ion mobility of specific fragments such as diglycerides-like ions derived from the TAGs, were found to be highly reproducible under specific and standard conditions applied in the traveling wave experiment. Such ion mobility information for diglyceride-like ions could be very powerful in assisting structural assignment of unknown lipid components present in a particular molecular species. It is quite easy to identify common as well as very different molecular species of lipids in mixtures such as this neutral lipid mixture of human serum. There was clearly no problem in assigning the ion at m/z 393 and 413 to contaminant ions, since these ions did not behave as any other constituents of serum by either MS/MS or ion mobility.
In summary, the CTS experiment provides a complete 4-dimensional dataset of tandem mass spectrometry and ion mobility for all product ions obtained from electrospray generated precursor ions in a relatively rapid time frame. This dataset enables identification of not only expected molecules, but also unexpected molecules such as oxidized CEs, oxidized TGs and contaminants present in this serum sample.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of Environmental Health Sciences of the National Institutes of Health (ES022172) and does not necessarily represent the official views of NIH. Jeff Brown and Emmy Hoyes provided WREnS and programming support from Waters Corporation, Wilmslow, U.K.
Footnotes
Author Contributions
Joseph A Hankin, Robert M. Barkley, Karin Zemski-Berry, and Yiming Deng performed experiments and contributed to the data analyses. Joseph A Hankin, Robert M. Barkley, Karin Zemski-Berry, Yiming Deng, and Robert C. Murphy were involved in the experimental design, data analysis, and writing of the manuscript.
Disclosures
There are no financial conflicts of interest related to this work, to disclose by any of the authors.
References
- 1.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van MG, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Murphy RC. Tandem Mass Spectrometry of Lipids: Molecular Analysis of Complex Lipids; Royal Society of Chemistry; London, UK: 2015. [Google Scholar]
- 3.Han X, Yang K, Gross RW. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Wang C, Han RH, Han X. Prog Lipid Res. 2015;61:83–108. doi: 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridenour WB, Kliman M, McLean JA, Caprioli RM. Anal Chem. 2010;82:1881–1889. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Anal Chem. 2014;86:2107–2116. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shvartsburg AA, Isaac G, Leveque N, Smith RD, Metz TO. J Am Soc Mass Spectrom. 2011;22:1146–1155. doi: 10.1007/s13361-011-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 9.Shvartsburg AA, Smith RD. Anal Chem. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YQ, Kuo M-S, Li S, Bui HH, Peake DA, Sanders PE, Thibodeaux SJ, Chu S, Qian Y-W, Zhao Y, Bredt DS, Moller DE, Konrad RJ, Beigneux AP, Young SG, Cao G. J Biol Chem. 2008;283:10048–10057. doi: 10.1074/jbc.M708151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons B, Kauhanen D, Sylvanne T, Tarasov K, Duchoslav E, Ekroos K. Metabolites. 2012;2:195–213. doi: 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokol E, Ulven T, Faergeman NJ, Ejsing CS. Eur J Lipid Sci Technol. 2015;117:751–759. doi: 10.1002/ejlt.201400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaelevski I, Kirshenbaum N, Sharon M. J Vis Exp. 2010;41:e1985. doi: 10.3791/1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, Grant DF, Palsson BO, Langridge J, Geromanos S, Astarita G. Anal Chem. 2015;87:1137–1144. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins PM, Barkley RM, Murphy RC. J Lipid Res. 2008;49:804–813. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchins PM, Moore EE, Murphy RC. J Lipid Res. 2011;52:2070–2083. doi: 10.1194/jlr.M019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne GL, Dai Q, Roberts LJ., 2nd Biochim Biophys Acta. 2015;1851:433–445. doi: 10.1016/j.bbalip.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng C, Gross ML, Pittenauer E. Anal Chem. 1998;70:4417–4426. doi: 10.1021/ac9805192. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Yin H, Akazawa YO, Yoshida Y, Niki E, Porter NA. Chem Res Toxicol. 2010;23:986–995. doi: 10.1021/tx1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacMillan DK, Murphy RC. J Am Soc Mass Spectrom. 1995;6:1190–1201. doi: 10.1016/1044-0305(95)00505-6. [DOI] [PubMed] [Google Scholar]
- 23.Kato K, Silva MJ, Reidy JA, Hurtz D, III, Malek NA, Needham LL, Nakazawa H, Barr DB, Calafat AM. Environ Health Perspect. 2004;112:327–330. doi: 10.1289/ehp.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


