Abstract
The evidence-based review (EBR) process has been widely used to develop standards for medical decision-making and to explore complex clinical questions. This approach can be applied to genetic tests, such as chromosomal microarrays, in order to assist in the clinical interpretation of certain copy number variants (CNVs), particularly those that are rare, and guide array design for optimal clinical utility. To address these issues, the International Standards for Cytogenomic Arrays Consortium has established an EBR Work Group charged with building a framework to systematically assess the potential clinical relevance of CNVs throughout the genome. This group has developed a rating system enumerating the evidence supporting or refuting dosage sensitivity for individual genes and regions that considers the following criteria: number of causative mutations reported; patterns of inheritance; consistency of phenotype; evidence from large-scale case-control studies; mutational mechanisms; data from public genome variation databases; and expert consensus opinion. The system is designed to be dynamic in nature, with regions being reevaluated periodically to incorporate emerging evidence. The evidence collected will be displayed within a publically available database, and can be used in part to inform clinical laboratory CNV interpretations as well as to guide array design.
Keywords: Cytogenetics, DNA copy number variation, evidence-based practice, gene dosage, oligonucleotide array sequence analysis
The complexity of genomic information has dramatically increased since the completion of the international Human Genome Project (1). Advanced technologies such as chromosomal microarray (CMA) and next generation sequencing now allow for comprehensive genome-wide copy number and sequence analyses in the clinical setting. As expected, genome-wide evaluation yields genome-wide variation, and the clinical interpretation of this type of testing has proven in some cases to be challenging. Clinical interpretations of structural and sequence-level variations are often based in part on the perceived clinical significance of the particular gene(s) involved, as supported by the available literature or other supporting evidence. How this evidence is assessed, however, has historically been a subjective process, potentially leading to discrepancies among interpretations. As genome-wide evaluations become more prevalent in the clinical setting, an immediate need for a more objective method of assessing the clinical significance of genes throughout the genome is becoming apparent. It is vital that available information supporting or refuting clinical significance (e.g. case reports, frequencies, penetrance information, etc.) is meaningfully translated in a systematic way for use in the clinical setting.
Evidence-based review process
The evidence-based review (EBR) process is a systematic method for integrating research evidence with clinical expertise in the context of individual patient care (2). It is the preferred method for formulating practice and treatment guidelines across many disciplines because of its unbiased and patient-focused approach (2). In a typical EBR process, a group of experts evaluates data according to its strength, credibility, and reproducibility and translates this information into recommendations for appropriate application in clinical practice. This data may relate to new technology, concepts, or procedures.
The most common use of the EBR process has historically been to evaluate specific treatment and management guidelines. Therefore, existing methods for EBR focus heavily on evidence from clinical trials, cohort, and case-control studies (3). Data from large, prospective randomized clinical trials are typically considered the strongest level of evidence, while information based on ‘expert clinical opinion’, case reports, and case series are considered weaker (3). Applying the EBR process to genomic medicine has been challenging because of the rarity of genetic disorders, the complexity of testing methodologies, and the pace at which advances in the field occur (4). Levels of evidence that may qualify as ‘gold standards’ in EBR processes in other disciplines may not always be achievable in genetic medicine. Despite these potential constraints, the EBR process has been utilized successfully in the genetics community. Several groups have used EBR to generate guidelines for use of specific genetic tests and for the treatment and management of certain disorders. For example, the Evaluation of Genomic Applications in Practice and Prevention Working Group, established by the Centers for Disease Control, uses an EBR process to evaluate genetic tests for public health applications (5). In one instance, this multi-disciplinary group evaluated different testing strategies and the benefit of testing newly diagnosed colon cancer patients for Lynch Syndrome (6). Their recommendations are now routinely followed by clinical laboratories, leading to more consistent testing strategies for colon cancer patients, and likely to improved patient care.
Application of evidence-based review to guide clinical interpretation of copy number variation
With the goal of improved patient care in mind, the results of the EBR process can be applied not only to clinical management but also to decisions related to the optimal application and use of new genetic test assays that support clinical diagnosis. In this era of the genotype-first approach to diagnosing genetic disorders, the EBR process has an important role to play, particularly in regards to the development and implementation of genome-wide assays. To date, CMA has been the most commonly used method for whole genome assessment. It is a prime example of a technology that has been rapidly integrated into clinical use before thorough evidence-based standards could be developed.
CMAs, which detect copy number variants (CNVs) for chromosomal regions throughout the genome, were developed in the research setting in the 1990s (7–9), and clinical laboratories quickly appreciated the potential for this technology in diagnostic applications. It was soon discovered that CNVs occur in all normal individuals (10, 11), but a subset of submicroscopic CNVs were also identified as major causes of birth defects, intellectual disability, and autism spectrum disorders (12–14). Associations between CNVs and an increasing number of diseases continue to be reported. Although CNVs have been identified as a major contributor to genomic variation, there are substantial gaps in our knowledge regarding the biological and medical impact of CNVs in normal human variation and disease.
It has historically been difficult to assess the clinical significance of certain CNVs, particularly those that are rare. While the clinical utility of microarray-based copy number analysis was immediately appreciated, clinical guidelines related to result interpretation were initially not well defined. The American College of Medical Genetics (ACMG) has recently published guidelines relating to the interpretation and reporting of postnatal constitutional CNVs (15). These guidelines and others state the importance of assessing genomic content when considering the clinical interpretation of CNVs (15, 16), but the process by which this task might be accomplished is ultimately left to the individual medical professional; this can potentially lead to significant interlaboratory variability in CNV interpretation (17). Further ACMG recommendations for minimal performance and design standards for arrays used for copy number detection (18) encourage enhanced array coverage in regions of established clinical relevance, although criteria to define clinical relevance have not been explicitly defined. The International Standards for Cytogenomic Arrays (ISCA) Consortium was organized in part to promote informed and uniform CNV interpretation and, in turn, deliver better patient care. To address these needs, the ISCA Consortium has established an EBR Work Group comprised of individuals with expertise in medical genetics, diagnostic testing, research, and bioinformatics. Although the need to develop standards for clinical CMA interpretation has been well-documented (17, 19), the goal of this group is to actually define and implement an evidence-based process to gather scientific data that links genes and genomic regions with clinical phenotypes caused by haploinsufficiency and/or triplosensitivity, and to make this information available to the genetics community. The tools developed by this group can be used immediately by clinical laboratories to assist in the clinical interpretation of CNVs and to improve interpretation consistency among laboratories, as well as in the future to improve array design.
The ultimate goal of the ISCA EBR Work Group is to evaluate each gene in the human genome with participation from the wider medical genetics community. A readily identifiable starting point for this substantial task is the list of genes currently targeted on the ISCA Consortium array design. As the first arrays transitioned into clinical use, common convention was to target as many genes with suspected clinical relevance as possible. The ISCA Consortium array design was initially developed in 2007 by merging the array designs of several groups, attempting to target all genomic regions that were targeted by various arrays available at the time; though the design has evolved over time, many similar elements have been carried forth (14). The intent was to provide one universal, comprehensive array design that would be available to all interested laboratories, with the goal of standardizing the level of testing that patients were receiving. Over time, it has become clear that many targeted regions lack sufficient evidence for clinical relevance and are therefore challenging to interpret. The approximately 500 genomic regions currently targeted on the ISCA Consortium array design are being evaluated in the systematic manner described below in an effort to provide the genetics community with a tool by which dosage sensitivity for each particular region can be readily assessed. Although the primary goal is to inform the clinical interpretation of CNVs, this information can also be used when considering which genomic regions warrant enriched probe coverage on future ISCA Consortium array designs.
The following is a description of the conceptual framework developed by the ISCA EBR Work Group to evaluate the clinical significance of a gene or region in an evidence-based manner (Fig. 1). Although these guidelines have been formulated with CMA as the primary focus, and clinical significance is being assessed as it relates to dosage sensitivity, the underlying framework can be used as a model to guide the development of any genomic technology. As with any guidelines, there will always be exceptions, and clinical judgment should always be exercised. In addition, since genomics research is progressing at such a rapid pace, new information is produced continually that often has direct bearing on the diagnosis of genetic disorders. As such, any EBR-based strategy for the development of guidelines must be continually revisited and updated.
Fig. 1.
Framework for an evidence-based process by which to evaluate dosage sensitivity.
Definition of ‘evidence’ as it relates to copy number variation
Peer-reviewed literature is considered the gold standard for the primary evidence needed to effectively assess a particular genomic region. When evaluating literature, the quality of the publication should be taken into account; standards have been proposed for manuscripts reporting structural variation data (19). One should consider factors such as whether the study was technically sound, what control populations were used, and whether appropriate confirmatory studies were performed when deciding whether to use a study as evidence for dosage sensitivity of a gene/region. Other factors to consider when comparing multiple literature reports include: are the CNVs focal (i.e. one CNV is not significantly larger or smaller in size than others it is being compared with) and/or associated with a specific phenotype? Are the reported inheritance for the CNV and the severity of the phenotype consistent with other case reports? Does the proband have other CNVs or even other issues that might contribute to his/her phenotype?
Large-scale case-control series are of particular value in assessing clinical relevance. These types of studies can be used to objectively determine frequencies of particular CNVs and compare them between case and control populations. If adequate phenotype information has been collected, it can also be used to inform assertions about the features associated with particular CNVs. When evaluating these types of studies, one must consider things such as how the studied population relates to the phenotype(s) in question (e.g. considering whether a CNV associated with an adult-onset condition is being evaluated in a pediatric population), technical differences between the case analyses and the control analyses (e.g. issues of platform compatibility, coverage, detection issues), etc. Two large case-control series describing CNVs found in clinical populations have recently been published (20, 21). Reports such as these are invaluable for the objective information that they provide, and additional studies replicating such data will continue to strengthen the evidence needed to draw accurate conclusions of CNV frequencies in normal and clinical populations.
Many clinically relevant but rare CNVs will not occur frequently enough to reach statistical significance in large-scale case-control series; conversely, CNVs that are frequently observed among clinical populations may not yet be associated with distinct clinical phenotypes. Furthermore, such large studies can be limited by lack of a phenotypically homogenous clinical population or a population consisting only of individuals within a certain age range. Because of this, the results of large-scale case-control series cannot be the sole predictors of clinical relevance. Smaller-scale case reports or case series are valuable when assessing the clinical relevance of a genomic region, especially in the absence of large case-control studies. Case reports or case series may provide the additional detailed genotype and phenotype information necessary to suggest a genomic region’s clinical significance. For example, deletions of the PMP22 gene, associated with hereditary neuropathy with liability to pressure palsies (HNPP) (OMIM #162500), did not reach statistical significance in a recently published case-control series (20). This is most likely because the clinical symptoms of HNPP may not be a common indication for CMA testing, and, since age of onset is typically in adulthood, individuals with this deletion would not be expected to be overrepresented in the particular clinical population studied. Numerous reports, however, have been published establishing the link between the deletion of this particular gene and the well-described HNPP phenotype (22–24). Despite the fact that this region was not significantly enriched among case populations in this particular study, smaller case series have showed the clinical relevance of this gene.
Evidence that a particular genomic region is not clinically relevant for the clinical populations in which CMA is typically ordered, or is not subject to dosage sensitivity should also be considered. Examples of this type of evidence include peer-reviewed literature asserting that the region is not associated with a relevant clinical phenotype, or statistically significant representation among controls in case-control series, etc. For example, copy number variation is common at the amylase alpha 1a and alpha 2a loci at 1p13.3 among normal individuals (11). While copy number variation at these loci have been associated with variations in salivary α-amylase levels and the perception of oral starch (25), to date, CNVs in this region have not been associated with neurocognitive disorders or congenital anomalies. Therefore, CNVs in this particular region would not be considered clinically relevant, and, if detected, categorized as ‘benign’ in the context of the currently recommended clinical usage of CMA: the evaluation of individuals with developmental disabilities, multiple congenital anomalies, and/or autism spectrum disorders (26, 27).
Recognizing that many genes/regions will not occur frequently enough for a formal case-control analysis, or have a significant amount of peer-reviewed literature regarding identified mutations, secondary evidence may be used to strengthen or refute the argument for dosage sensitivity. Examples of secondary evidence may include entries in locus-specific databases, the observation that the gene is or is not overly represented in databases of normal variation, and/or models that predict a gene to be haploinsufficient or triplosensitive (28). Cases from databases such as the ISCA Consortium database (http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd37/; www.iscaconsortium.org) or DECIPHER (29) may also be used as supporting evidence.
Development of a rating system for objective review
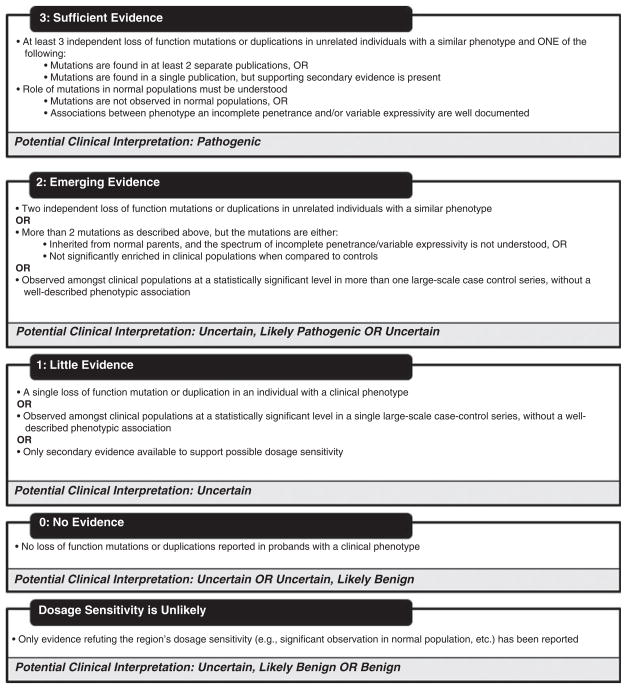
A rating system was developed to quantify the available evidence for standardized decision-making. Since CMA can detect losses and gains of genomic material, each genomic region will have two independent ratings: a haploinsufficiency rating and a triplosensitivity rating. The haploinsufficiency rating will address deletions and loss of function mutations and the ability of subsequent decreased gene dosage to result in a particular phenotype. The triplosensitivity rating will address whole gene duplications and the ability of increased gene dosage (distinct from gain of function, evidence for which is not included in this rating) to result in a particular phenotype. Haploinsufficiency and triplosensitivity ratings range from 0 to 3, with increasing levels of evidence suggesting that dosage-sensitivity results in a particular phenotype (Fig. 2).
Fig. 2.
Levels of evidence suggesting that dosage-sensitivity results in clinical phenotype.
The haploinsufficiency rating incorporates evidence related to phenotypes resulting from loss of function of any particular region. The enrichment of a particular region among cases in large-scale case-control series may be counted as evidence towards that region’s haploinsufficiency rating, although it is not a requirement. In addition, the presence of deletions, nonsense mutations, frameshift mutations, or mutations with demonstrated splicing defects in an affected individual or segregating within an affected family will be considered as evidence toward the haploinsufficiency rating for a particular gene. Each mutation should have evidence suggesting its pathogenicity (e.g. the mutation segregates with disease in cases of familial inheritance, the mutation is de novo in sporadic cases, functional studies have been performed, etc.). Additional mutations such as missense, silent, and intronic changes will only be considered for the haploinsufficiency rating if there are supporting functional studies to show that they truly result in loss of function.
The triplosensitivity rating incorporates evidence related specifically to whole gene duplications, as they have the potential to cause triplosensitivity (extra dosage of a gene resulting in clinical pathology). Duplications can result in phenotypes distinct from those caused by loss of function mutations in the same gene. Therefore, duplications segregating with a phenotype should be noted separately when evaluating a gene/region. The triplosensitivity rating will be determined by the same criteria as for the loss of function rating described below, i.e., the rating correlates with the available evidence suggesting that triplosensitivity is associated with a clinical phenotype. In general, when assessing evidence for the haploinsufficiency/triplosensitivity rating for a particular region, the phenotype observed among cases considered as evidence supporting dosage sensitivity should be consistent. If a gene/region is associated with multiple, non-overlapping phenotypes, evidence concerning the types of mutations associated with each distinct phenotype should be collected separately. For example, the RET gene has been associated with both Hirschsprung disease and multiple endocrine neoplasia (MEN) type 2A/B (30–33). In this particular example, the evidence (>3 separate probands with loss of function mutations) supports that haploinsufficiency of the RET gene is associated with Hirschsprung disease (34–36), while the MEN2A/B phenotype is associated with gain of function mutations (37).
Genomic regions with a haploinsufficiency or triplosensitivity rating of ‘3’ are those regions with sufficient evidence suggesting that dosage sensitivity is associated with a clinical phenotype. These regions will have loss of function mutations or duplication events reported in at least three unrelated probands with a similar, well-described phenotype. These mutations must come from at least two independent publications; if the three mutations are found in a single compelling publication, some supporting secondary evidence (e.g. unpublished entries in a locus-specific database, cases from databases such as the ISCA database, etc.) must be present. Loss of function mutations or duplications in these regions should not be observed at high frequency within normal populations; or, if there have been associations between the particular clinical phenotype and incomplete penetrance and/or variable expressivity, these relationships should be well understood.
Two different examples of regions with haploinsufficiency ratings of 3 are NSD1 and ZEB2. The 5q35 region encompassing the NSD1 gene would warrant a haploinsufficiency rating of ‘3’, as there is sufficient evidence to suggest that haploinsufficiency of this region is associated with Sotos syndrome (OMIM #117550). This region has been found to be significantly enriched among cases in case-control series (20, 21), and numerous case reports have described associations between deletions/other loss of function mutations of this region and the Sotos phenotype that includes overgrowth, distinct facial features, and learning disability (38, 39). However, regions do not necessarily need to be significantly enriched among cases in large-scale case-control studies to merit a loss of function rating of ‘3’. For example, deletions and other loss of function mutations of the ZEB2 gene associated with Mowat Wilson syndrome (OMIM #235730) have not been described in large-scale case-control studies, but have been frequently reported in association with the distinct phenotype that includes intellectual disability, characteristic facial features, microcephaly, heart defects, and Hirschsprung disease (40–43). These reports describing more than three instances of loss of function mutations in unrelated, phenotypically similar individuals constitute sufficient evidence to show the role of ZEB2 haploinsufficiency in disease.
Genomic regions with a haploinsufficiency or triplosensitivity rating of ‘2’ are those regions with emerging evidence to suggest that dosage sensitivity is associated with a clinical phenotype. These regions may be associated with two different loss of function mutations or duplication events in unrelated probands with a similar phenotype. Alternatively, these regions may be observed among clinical populations at a statistically significant level in more than one large-scale case-control series, but may not have a well-described phenotypic association. Finally, these regions may have more than two loss of function mutations or duplication events with a similar and specific phenotype, but the mutations are inherited, or the regions are not supported as significant in case-control series. For example, four patients with laterality defects or congenital heart defects with two different loss of function mutations in the CFC1 gene have been described (44, 45). However, the mutations were inherited from phenotypically normal parents in at least two of these cases, and the penetrance and expressivity of loss of function mutations in this gene are not yet well understood. In fact, it has been postulated that digenic inheritance or additional modifier mutations may be necessary to illicit the observed phenotypes (44), which underscores the incomplete understanding of the phenotypic consequences of mutations in this gene.
Genomic regions with a haploinsufficiency or triplosensitivity rating of ‘1’ are those regions for which there is little evidence suggesting that dosage sensitivity is associated with a clinical phenotype. This category will encompass regions for which only a single loss of function mutation or duplication has been reported. Regions that have been observed significantly more often in cases compared to controls in large-scale case-control series without a clear phenotypic association may also fall into this category. For example, duplications of the 2q13 region encompassing NPHP1 were observed significantly more often in cases than controls in one large case series (20). To date, duplications of this region have not been reported to be associated with any specific phenotype, and in fact have been reported among patients with discrepant phenotypes, their reportedly normal parents, and in controls (46). Although its significant observation in the case-control series cannot be ignored, additional evidence is required to assess this region’s clinical relevance, warranting its classification as ‘1’.
Genomic regions with a haploinsufficiency or triplosensitivity rating of ‘0’ are those regions for which there is no evidence to assess whether dosage imbalance is associated with a clinical phenotype or is benign in nature. If there is only evidence suggesting that a region is NOT subject to dosage sensitivity, the region will not receive a haploinsufficiency or triplosensitivity rating and will be categorized separately (Fig. 2).
Genes on the sex chromosomes warrant a slightly modified approach. The haploinsufficiency and triplosensitivity ratings for genes on the X chromosome are made in the context of a male genome to account for the effects of hemizygous duplications or nullizygous deletions. This approach is necessary and avoids potential confusion owing to observations that deletions or duplications in X-linked genes may not uniformly result in a phenotype in female individuals. Therefore, for many X-linked conditions the ratings of dosage sensitivity are relevant only for interpreting copy number data in male patients. Examples include congenital adrenal hypoplasia with hypogonadotrophic hypogonadism (NR0B1 deletions) (OMIM #300473), hemophilia A (F8 deletions) (OMIM #306700), or Duchenne muscular dystrophy (DMD deletions) (OMIM #310200). In contrast, disruption of some genes on the X chromosome causes male lethality, and the ratings of dosage sensitivity instead take into account the phenotype in female individuals. Rett syndrome (OMIM #312750) and incontinentia pigmenti (OMIM #308300) are examples of male-lethal disorders. The ratings for haploinsufficiency and triplosensitivity for genes on the Y chromosome are made in the same fashion as for genes linked to autosomal dominant disorders. Duplications of genes on the Y chromosome are typically not associated with overt clinical phenotypes (47). Deletions are pathogenic but only in a very small number of genes, including SRY and the DAZ loci (reviewed in Ref. (48)).
Documentation of evidence and the review process
Both the haploinsufficiency and triplosensitivity ratings, along with all supporting evidence, are being recorded in a web-based database customized for our EBR process. JIRA is a commercially available issue and project tracking system. It is compatible with most enterprise database systems and has a customizable web interface that facilitates project customization. This system has been used by other genome curation groups, such as the Genome Reference Consortium (http://www.ncbi.nlm.nih.gov/pubmed/21750661) and is available from Atlassian (http://www.atlassian.com/software/jira/). We have created custom fields to allow for the tracking of genes/regions and specific pieces of evidence in a structured way. An Application Programming Interface (API) allows for each gene or region in the JIRA system to be populated programmatically (with fields such as genome coordinates, links to outside resources such as Entrez Gene and OMIM, etc.), and a web interface allows for individual curation of each region. The same API allows for genes and regions to be pulled from the system for display on a public web page, which allows broad access to the collected evidence for public review, comment, and use.
To initiate the review process, each gene/region is assigned a primary reviewer, who is responsible for performing the initial literature review and documenting the information in the JIRA system. If the primary reviewer assigns either a haploinsufficiency or triplosensitivity rating of 3, then the gene/region will be brought to full committee review for approval; genes/regions with this level of evidence supporting them will be considered by the group for inclusion on the ISCA Consortium ‘known (or curated) pathogenic’ list. If the highest rating assigned to a gene by its primary reviewer is a 2 or lower, the gene/region is sent to a secondary reviewer. The secondary reviewer is responsible for evaluating the work of the primary reviewer. If the two reviewers agree on the rating of two or lower for the gene or region in question, then this final rating will be documented in the JIRA system. If there is any discrepancy between the two reviewers, the issue will come to the entire committee for review. After discussion, if the entire committee does not come to consensus, a decision will be made via majority vote. The final rating (as decided by group consensus or majority vote), along with points outlining the decision-making process, will be recorded in the JIRA system. As each region is finalized, the ratings, literature citations, and discussion points recorded in the JIRA system will be available on an ongoing basis for public review and comment through an open-access web portal (http://www.ncbi.nlm.nih.gov/projects/dbvar/ISCA).
To incorporate new and emerging evidence and to ensure that the evidence-based recommendations remain up-to-date, each region will be reevaluated on a periodic basis. Regions that have received a haploinsufficiency or triplosensitivity rating of 3 have met the threshold set by this group for significant clinical evidence supporting their dosage sensitivity; as such, these regions will be reviewed less frequently than genes with lower ratings. As mentioned above, these regions will also be brought before the full committee for discussion of inclusion on the ISCA Consortium ‘known pathogenic’ list (http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd45/). The regions currently on the list of ‘known pathogenic’ regions curated by the ISCA Consortium include classic, well-described microdeletion/microduplication syndromes (for example, the 22q11.2 DiGeorge/VCFS region, the Williams syndrome region, the Wolf–Hirschhorn syndrome region, etc.). As a requirement for inclusion on this list is significant, convincing evidence supporting dosage sensitivity, these regions will not be regularly reevaluated unless a specific concern arises (i.e. a seminal manuscript is retracted, new contradictory evidence emerges, etc.). All other regions (i.e. those with ratings of 2 or lower) will be reevaluated on an annual basis.
Applications of an evidence-based dosage-sensitivity map of the human genome
Clinical interpretation
When assessing the clinical consequence of a CNV detected by CMA, a primary concern is the genomic content in the region of gain or loss (15). Evidence that haploinsufficiency or triplosensitivity of a gene is associated with a specific phenotype will aid in the interpretive assessment of CNVs including that particular gene. The public website generated from the work documented in the JIRA system can be used as a primary source for peer-reviewed literature on the clinical consequence of deletion and duplication of the genes within a genomic region and will include links to other relevant databases such as OMIM, GeneReviews, PubMed literature, and mutation specific databases.
In general, the haploinsufficiency rating for an individual gene may be used to guide the clinical interpretation of deletions involving a particular gene. Using the clinical interpretation categories put forth by the ACMG (15), a deletion of a region with a loss of function rating of 3 could be interpreted as ‘pathogenic’ given the level of documentation in the literature regarding the phenotypic consequences of loss of function mutations. A deletion of a region with a haploinsufficiency rating of 2 could be interpreted as either ‘uncertain, probably pathogenic’ or ‘uncertain’. The specific type of evidence available for each gene rating should be considered in the interpretative process, as studies may differ in their potential clinical implications. For example, a deletion in a region with two independent, well-documented loss of function mutations in two unrelated probands with similar, specific phenotypes may warrant a classification of ‘uncertain; probably pathogenic’, while a deleted region significantly overrepresented in cases in two case-control series that lack distinct phenotypic associations may warrant a classification of ‘uncertain’. For the same reasons, deletions of regions with a haploinsufficiency rating of 1 could be interpreted as ‘uncertain’, while deletions of regions with a haploinsufficiency rating of 0 could be interpreted as ‘uncertain’ or ‘uncertain; probably benign’. Regions for which there is only evidence refuting dosage sensitivity could be interpreted as ‘uncertain; probably benign’ or ‘benign’. The same categorization process would apply to duplications of genes based upon their triplosensitivity ratings. CNVs encompassing more than one gene, however, must be evaluated in their totality (e.g. overall size, gain vs loss, presence of other genes, etc). The rating of a single gene within the CNV should not necessarily be the only criteria by which one defines a clinical interpretation. ACMG has published guidelines for the characterization of postnatal CNVs, and these recommendations should be utilized (15). Exceptions to these interpretive correlations will occur, and clinical judgment should always be exercised. Individual interpretations must take into account the phenotype described for the patient as well as issues of penetrance and expressivity of the disorder; the importance of clinical correlation cannot be underestimated.
Development of future microarray designs
The results of the ongoing evidence-based dosage-sensitivity evaluation can be used to direct future microarray designs. If the haploinsufficiency rating is different from the duplication rating, the coverage decisions may be based on the higher of the two ratings. Genomic regions with haploinsufficiency or duplication ratings of 3 may be deemed appropriate for enriched probe coverage on CMA, dependent upon the associated phenotype. Enriched probe coverage may refer to either exon-level targeting or increased probe density as appropriate. Regions with a rating of 2 will be evaluated for enriched probe coverage on a case-by-case basis. In general, regions with ratings of 1 or 0 will be considered inappropriate for enriched probe coverage; however, these regions will be reviewed on an annual basis, and coverage decisions for future array designs may be altered to reflect emerging evidence.
Factors other than the documented dosage sensitivity of genomic regions must be taken into consideration when making decisions regarding CMA design. Technical considerations and other factors related to whether particular genes/regions are appropriate for increased probe coverage on CMA, such as phenotype, age of onset, inheritance patterns, and mutational mechanism, all must be taken into account. In regards to phenotype, one might consider how the phenotype relates to the clinical population for whom arrays are typically ordered. Per ACMG practice guidelines and a consensus statement from the ISCA Consortium, CMA is currently recommended as a first-tier assessment in the postnatal evaluation of individuals with one or more of the following: multiple anomalies not specific to a well-defined genetic syndrome; apparently nonsyndromic developmental delay/intellectual disabilities; or autism spectrum disorders (26, 27). Using these recommendations as a guide, dosage-sensitive regions that lead to phenotypes including at least one of these features will in general be considered appropriate for targeting on CMA. Other phenotypes may still be considered appropriate, particularly those associated with medical intervention strategies (e.g. genes associated with Long QT syndrome). Phenotypes with exclusively adult onset, particularly those in which no effective early intervention exists, will typically be considered inappropriate for purposefully enriched coverage on CMA.
CMA is best suited to diagnose genetic conditions resulting from copy number changes in dosage-sensitive genes, which are generally inherited in a dominant manner. By nature of the design of many currently available CMAs, detection of copy number changes in recessive genes (and therefore, at the very least, carrier status for recessive conditions) is possible. There have been reports of copy number changes in autosomal recessive genes leading to the eventual diagnosis of autosomal recessive conditions in patients (following the confirmation of a mutation in the other allele) (49), as well as reports of autosomal recessive conditions being caused by homozygous copy number changes detected by CMA (50). However, these scenarios are rare. Furthermore, assessing carrier status for recessive conditions is ultimately not the focus of standard CMA testing. It is the opinion of the ISCA Consortium EBR Committee that, in general, standard backbone coverage should not be altered to avoid genes associated with autosomal recessive conditions, but that these genes should not be targeted at the exon-level or with increased probe density throughout the gene without compelling evidence to do so. For example, large homozygous deletions of the NPHP1 gene are frequent causes of nephronophthisis type 1 (51). The frequency of this particular mutation mechanism warrants consideration for enriched probe coverage throughout this genomic region.
Genes known to cause disease by other mechanisms (such as gain of function) will generally not be appropriate for targeting on CMA, unless there is compelling reason to do so. For example, although mutations in SOS1 have been associated with Noonan syndrome, virtually all reported mutations are thought to result in gain of function (52), and dosage sensitivity has not been found to be a significant mutational mechanism for this condition (53). A deletion or duplication of this particular gene on CMA would not be expected to result in a Noonan syndrome phenotype. The ability to detect small copy number changes within this gene by targeting with exon-level coverage or increased probe density throughout the gene would not add information of clinical relevance for a patient, and is therefore not warranted.
Conclusions
The EBR process is well-suited to address the challenges related to the emergence of new clinical genome-wide testing strategies, although the processes necessary to accumulate and evaluate the evidence needed are constantly evolving. Powerful applications such as CMA should be constructed and interpreted using the best available scientific evidence, which, at this time, includes case reports and case series, large-scale case-control series, and other secondary data such as population databases and predictive models (3). Efforts are underway by the ISCA Consortium to obtain additional large-scale case-control evidence via the publically available ISCA Clinical CNV Database (http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd37/; www.iscaconsortium.org), which has been established to leverage data from the thousands of patients with developmental disabilities, congenital anomalies, and other phenotypes being tested through ISCA member laboratories. The clinically relevant information gleaned from case reports and case series will continue to be important in the evidence-based assessment of particular genes, but participation from all clinical cytogenetics laboratories in the database effort is still necessary to obtain reproducible large-scale, objective frequency data.
Participation by the genetics community at large is also necessary to contribute to the ongoing development of the evidence-based dosage-sensitivity map project. Through the public website (http://www.ncbi.nlm.nih.gov/projects/dbvar/ISCA), users will be encouraged to comment on particular genomic regions, provide evidence supporting or refuting dosage sensitivity, and/or suggest genomic regions for immediate evaluation. The results of this important effort is intended to be a valuable reference tool for both clinicians and laboratorians, compiling relevant evidence in a convenient, easy-to-use format for use in clinical practice.
Acknowledgments
This work was supported, in part, by NIH Grant HD064525 (D. H. L. and C. L. M.) and by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Conflict of interest
S. A. is employed by GeneDx (a subsidiary of Bioreference Laboratories, Inc.). S. S. is the Medical Director at ARUP Laboratories, a not-for-profit organization owned by the University of Utah offering among other tests genomic microarray. She also serves as a consultant to Lineagen, Inc., a for-profit company, which offers genomic microarray testing. Dr S. S. has received honoraria for speaking engagements on behalf of Affymetrix Inc. D. L. is a consultant for Roche Nimblegen. The other authors declare no conflict of interest.
References
- 1.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Toriello HV, Goldenberg P. Evidence-based medicine and practice guidelines: application to genetics. Am J Med Genet C Semin Med Genet. 2009;151C:235–240. doi: 10.1002/ajmg.c.30222. [DOI] [PubMed] [Google Scholar]
- 3.Kruer MC, Steiner RD. The role of evidence-based medicine and clinical trials in rare genetic disorders. Clin Genet. 2008;74:197–207. doi: 10.1111/j.1399-0004.2008.01041.x. [DOI] [PubMed] [Google Scholar]
- 4.Tuckson RV. Challenges and opportunities for evidence-based genetics practice. Genet Med. 2009;11:1–2. doi: 10.1097/GIM.0b013e31819251b2. [DOI] [PubMed] [Google Scholar]
- 5.Teutsch SM, Bradley LA, Palomaki GE, et al. The evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group EW. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor SP, Read JL, Pirrung MC, et al. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 8.Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 9.Solinas-Toldo S, Lampel S, Stilgenbauer S, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 10.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 11.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 12.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin EL, Lee JY, Blake DM, et al. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet Med. 2008;10:415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 15.Kearney HM, Thorland EC, Brown KK, et al. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 16.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39:S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 17.Tsuchiya KD, Shaffer LG, Aradhya S, et al. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet Med. 2009;11:866–873. doi: 10.1097/GIM.0b013e3181c0c3b0. [DOI] [PubMed] [Google Scholar]
- 18.Kearney HM, South ST, Wolff DJ, et al. American College of Medical Genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med. 2011;13:676–679. doi: 10.1097/GIM.0b013e31822272ac. [DOI] [PubMed] [Google Scholar]
- 19.Scherer SW, Lee C, Birney E, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chance PF, Alderson MK, Leppig KA, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzetti D, Pareyson D, Sghirlanzoni A, et al. A 1.5-Mb deletion in 17p11. 2-p12 is frequently observed in Italian families with hereditary neuropathy with liability to pressure palsies. Am J Hum Genet. 1995;56:91–98. [PMC free article] [PubMed] [Google Scholar]
- 24.LeGuern E, Gouider R, Lopes J, et al. Constant rearrangement of the CMT1A-REP sequences in HNPP patients with a deletion in chromosome 17p11.2: a study of 30 unrelated cases. The French CMT Collaborative Research Group. Hum Mol Genet. 1995;4:1673–1674. doi: 10.1093/hmg/4.9.1673. [DOI] [PubMed] [Google Scholar]
- 25.Mandel AL, Peyrot des Gachons C, Plank KL, et al. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One. 2010;5:e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang N, Lee I, Marcotte EM, et al. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulligan LM, Kwok JB, Healey CS, et al. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature. 1993;363:458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 31.Donis-Keller H, Dou S, Chi D, et al. Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet. 1993;2:851–856. doi: 10.1093/hmg/2.7.851. [DOI] [PubMed] [Google Scholar]
- 32.Edery P, Lyonnet S, Mulligan LM, et al. Mutations of the RET proto-oncogene in Hirschsprung’s disease. Nature. 1994;367:378–380. doi: 10.1038/367378a0. [DOI] [PubMed] [Google Scholar]
- 33.Romeo G, Ronchetto P, Luo Y, et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature. 1994;367:377–378. doi: 10.1038/367377a0. [DOI] [PubMed] [Google Scholar]
- 34.Fewtrell MS, Tam PK, Thomson AH, et al. Hirschsprung’s disease associated with a deletion of chromosome 10 (q11.2q21.2): a further link with the neurocristopathies? J Med Genet. 1994;31:325–327. doi: 10.1136/jmg.31.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attie T, Pelet A, Edery P, et al. Diversity of RET proto-oncogene mutations in familial and sporadic Hirschsprung disease. Hum Mol Genet. 1995;4:1381–1386. doi: 10.1093/hmg/4.8.1381. [DOI] [PubMed] [Google Scholar]
- 36.Yin L, Barone V, Seri M, et al. Heterogeneity and low detection rate of RET mutations in Hirschsprung disease. Eur J Hum Genet. 1994;2:272–280. doi: 10.1159/000472371. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi M, Asai N, Iwashita T, et al. Molecular mechanisms of development of multiple endocrine neoplasia 2 by RET mutations. J Intern Med. 1998;243:509–513. [PubMed] [Google Scholar]
- 38.Kurotaki N, Harada N, Shimokawa O, et al. Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum Mutat. 2003;22:378–387. doi: 10.1002/humu.10270. [DOI] [PubMed] [Google Scholar]
- 39.Tatton-Brown K, Douglas J, Coleman K, et al. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zweier C, Temple IK, Beemer F, et al. Characterisation of deletions of the ZFHX1B region and genotype-phenotype analysis in Mowat-Wilson syndrome. J Med Genet. 2003;40:601–605. doi: 10.1136/jmg.40.8.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amiel J, Espinosa-Parrilla Y, Steffann J, et al. Large-scale deletions and SMADIP1 truncating mutations in syndromic Hirschsprung disease with involvement of midline structures. Am J Hum Genet. 2001;69:1370–1377. doi: 10.1086/324342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishihara N, Yamada K, Yamada Y, et al. Clinical and molecular analysis of Mowat-Wilson syndrome associated with ZFHX1B mutations and deletions at 2q22-q24. 1. J Med Genet. 2004;41:387–393. doi: 10.1136/jmg.2003.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dastot-Le Moal F, Wilson M, Mowat D, et al. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- 44.Bamford RN, Roessler E, Burdine RD, et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 45.Goldmuntz E, Bamford R, Karkera JD, et al. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet. 2002;70:776–780. doi: 10.1086/339079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baris H, Bejjani BA, Tan WH, et al. Identification of a novel polymorphism–the duplication of the NPHP1 (nephronophthisis 1) gene. Am J Med Genet A. 2006;140A:1876–1879. doi: 10.1002/ajmg.a.31390. [DOI] [PubMed] [Google Scholar]
- 47.Jobling MA. Copy number variation on the human Y chromosome. Cytogenet Genome Res. 2008;123:253–262. doi: 10.1159/000184715. [DOI] [PubMed] [Google Scholar]
- 48.Carvalho CM, Zhang F, Lupski JR. Structural variation of the human genome: mechanisms, assays, and role in male infertility. Syst Biol Reprod Med. 2011;57:3–16. doi: 10.3109/19396368.2010.527427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivera-Brugues N, Albrecht B, Wieczorek D, et al. Cohen syndrome diagnosis using whole genome arrays. J Med Genet. 2011;48:136–140. doi: 10.1136/jmg.2010.082206. [DOI] [PubMed] [Google Scholar]
- 50.Moreno-De-Luca A, Helmers SL, Mao H, et al. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J Med Genet. 2011;48:141–144. doi: 10.1136/jmg.2010.082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konrad M, Saunier S, Heidet L, et al. Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet. 1996;5:367–371. doi: 10.1093/hmg/5.3.367. [DOI] [PubMed] [Google Scholar]
- 52.Lepri F, De Luca A, Stella L, et al. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum Mutat. 2011;32:760–772. doi: 10.1002/humu.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nystrom AM, Ekvall S, Thuresson AC, et al. Investigation of gene dosage imbalances in patients with Noonan syndrome using multiplex ligation-dependent probe amplification analysis. Eur J Med Genet. 2010;53:117–121. doi: 10.1016/j.ejmg.2010.03.001. [DOI] [PubMed] [Google Scholar]