Abstract
There are many homeostatic mechanisms for coping with stress conditions in cells, including autophagy. In many studies autophagy, as an intracellular pathway which degrades misfolded and damaged protein, and Mallory-Denk Body(MDB) formation have been shown to be protective mechanisms against stress such as alcoholic hepatitis. Alcohol has a significant role in alteration of lipid homeostasis, sterol regulatory element-binding proteins (SREBPs) and peroxidase proliferator-activated receptors through AMP-activated protein kinase (AMPK)-dependent mechanism. AMPK is one of the kinases that regulate autophagy through the dephosphorylation of ATG1. Activation of ATG1 (ULK kinases family) activates ATG6. These two activated proteins relocate to the site of initial autophagosome and activate the other downstream components of autophagocytosis. Many other proteins regulate autophagocytosis at the gene level. CHOP (C/EBP homologous protein) is one of the most important parts of stress-inducible transcription that encodes a ubiquitous transcription factor. In this report we measure the upregulation of the gene that are involved in autophagocytosis in liver biopsies of alcoholic hepatitis and NASH. Electron microscopy was used to document the presence of autophagosomes in the liver cells. Expression of AMPK1, ATG1, ATG6 and CHOP in ASH were significantly (P value <0.05) upregulated in comparison to control. Electron microscopy findings of ASH confirmed the presence of autophagosomes, one of which contained a MDB, heretofore undescribed. Significant upregulations of AMPK-1, ATG-1, ATG-6, and CHOP, and uptrending of ATG-4, ATG-5, ATG-9, ATR, and ATM in ASH compared to normal control livers indicate active autophagocytosis in alcoholic hepatitis.
Keywords: Mallory- Denk Bodies, Autophagocytosis, Protein quality control, Alcoholic hepatitis
Introduction
Autophagy is one of the essential intracellular pathways that maintains cellular functions and prevents cell death via lysosomal degradation as a cytoprotective path. More than 30 ATG genes are identified for controlling autophagocytosis. (Klionsky et al 2003) ATG1-10 are participating as core proteins in autophagosome formation. (Nakatogawa et al 2009) that have four subgroups: a) ATG1/ ULK1 complex (Chan 2012) that induces autophagocytosis and is negatively modulated by mTOR; b) ATG6/Beclin1 complex that regulates the formation of autophagosome; c) Ubiquitin-like complexes (LC3 and ATG12-5) regulate vesicle expansion; d) ATG9 that is required for delivery of membranes which form autolysosome. (Xie et al 2007, Fujita et al 2008, Hosokawa et al 2009, Yang et al 2009, Yang et al 2010) Autophagy plays a very important role in hepatocytes homeostasis by removing misfolded proteins and damaged organells. Accumulation of proteins in prolonged ER stress may lead to intra hepatic protein aggragate formation such as MDBs. (Komatsu 2012, Liu et al 2014, Peng et al 2014) ATP and energy level reduction in the cell, increase AMPK related autophagy by removing the inhibitory brakes on mTORC1 or activating ULK1 directly. (Mihaylova et al 2011) mTOR has some regulatory effects on liposynthesis; its suppression increases lipophagy and decreases lipogenesis via regulation of SREBP. (Peterson et al 2011) Excess fat deposition, compromised lipid metabolism in NASH and chronic alcohol consumption which plays a significant role in alteration of lipid homeostasis, SREBP and peroxidase proliferator-activated receptors in ASH activate cell stress responses through the AMPK-dependent mechanism. (Ceni et al 2014) AMP binds to the activated part of AMPK when cell energy is low; phosphorylation of AMPK inactivates mTORC1 and activates ULK1 family (ATG1) (Kim et al 2011). ATG1/ULK1 complex with the support of other proteins translocates to the autophagosome formation sites (Hara et al 2008), regulates Beclin1/ ATG6 and translocates ATG9 from Golgi as an additional membrane donor for autophagosome formation (Young et al 2006). ATG4 and ATG5 are other ATG proteins interact with the LC3 complex in autophagocytosis (Zhang et al 2016). Peroxisomes are organelles necessary for β-oxidation of fatty acids, which produces lots of reactive oxygen species (ROS), peroxisomes usually degrade via autophagy. ATM serine/threonine kinase (ATM) is the first responder that is activated by peroxisomal ROS. One of the actions of activated ATM is signaling AMPK and subsequently activates ATG1 (Tripathi et al 2016). Any kind of stress and cell insult may lead to DNA damage. The cell response to DNA damage activates the DNA damage checkpoint, which induces cascades of proteins and autophagocytosis, controled by Mec1 kinase (ATR) (Vinay et al 2015). CHOP expression is upregulated during cell and ER stress. CHOP has been shown to be involved in the cell death via apoptosis pathway. However, CHOP also has been determined as a regulator in autophagy (Zinszner et al 1998, Li et al 2014, Zheng et al 2014, Ohoka et al 2005, Su et al 2008) ) CHOP can modify DNA binding activities in the nucleus and regulate transcriptional factor involving to autophagocytosis or apoptosis. (Bruhat et al 1997) Longevity of stress may increase the expression of cell death mediators as autophagy mediators in the long term stress activate apoptosis regulators (Fimia et al 2010). The aim of this study is to quantitate the expression of the proteins participating in autophagocytosis in alcoholic steatohepatitis (ASH) and non-alcoholic steatohepatitis (NASH) in comparison to normal liver.
Materials and Methods
Formalin-fixed paraffin embedded (FFPE) human liver biopsies from patients who were diagnosed with ASH and NASH from Harbor–UCLA hospital archived and a clinical trial funded by NIH/NIAAA grant {“Alcoholic hepatitis pathogenesis as determined from human liver tissue analysis” exempted as determined by the iRIS system} were compared to controls. In this study we used 8-12 ASH, 1-5 NASH and 3 normal liver controls. The slides were double stained for ubiquitin plus AMPK1, ATG1, ATG4, ATG5, ATG6, ATG9, ATM, ATR and CHOP. Texas Red (Millipore, Temecula, CA) was used to detect ubiquitin. The other proteins were detected as green fluorescence by using either donkey-anti mouse or anti rabbit Alex Fluor for the secondary antibody (Jackson Labs, West Grove, PA). The nuclei were stained by DAPI. The staining was done at the same time for all slides together to provide accurate comparisons between groups. We measured the intensity of the fluorescent staining in 3 different areas on each slide with 40x magnifications and 800ms standard exposure time by using a Nikon 400 fluorescent microscope. The Nikon morphometric system was used to a quantitate measurement of the fluorescent intensity morphometrically. The mean, standard error and statistical differences of data achieved from the Nokia were analyzed by Graph pad statistical software. Controls vs. ASH, controls vs NASH and ASH vs. NASH were compared by unpaired t-test with p<0.05. Electron microscopy was used to document the presence of autophagosomes in the liver cells. (See table 1)
Table 1.
Antibodies used in the study with animal source and company/Vendor
| Antibody | Company/Vendor |
|---|---|
| AMPK1 | SIGMAALDRICH, ST. LUOIS, MO 63103 |
| ATG1 | THERMO-FISHER ROCKFORD, IL 61105 |
| ATG4 | MYBIOSOURCE, SAN DIEGO, CA 92195 |
| ATG5 | NOVUSBIO, LITTLETON, CO 80120 |
| ATG6 | SIGMAALDRICH, ST. LUOIS, MO 63103 |
| ATG9 | ABCAM, CAMBRIGE, MA, 02139 |
| ATR | LSBIO, SEATTLE, WA 98121 |
| ATM | ABCAM, CAMBRIDGE, MA 02139 |
| CHOP | THERMO-FISHER, ROCKFORD, IL 61105 |
Results
ASH
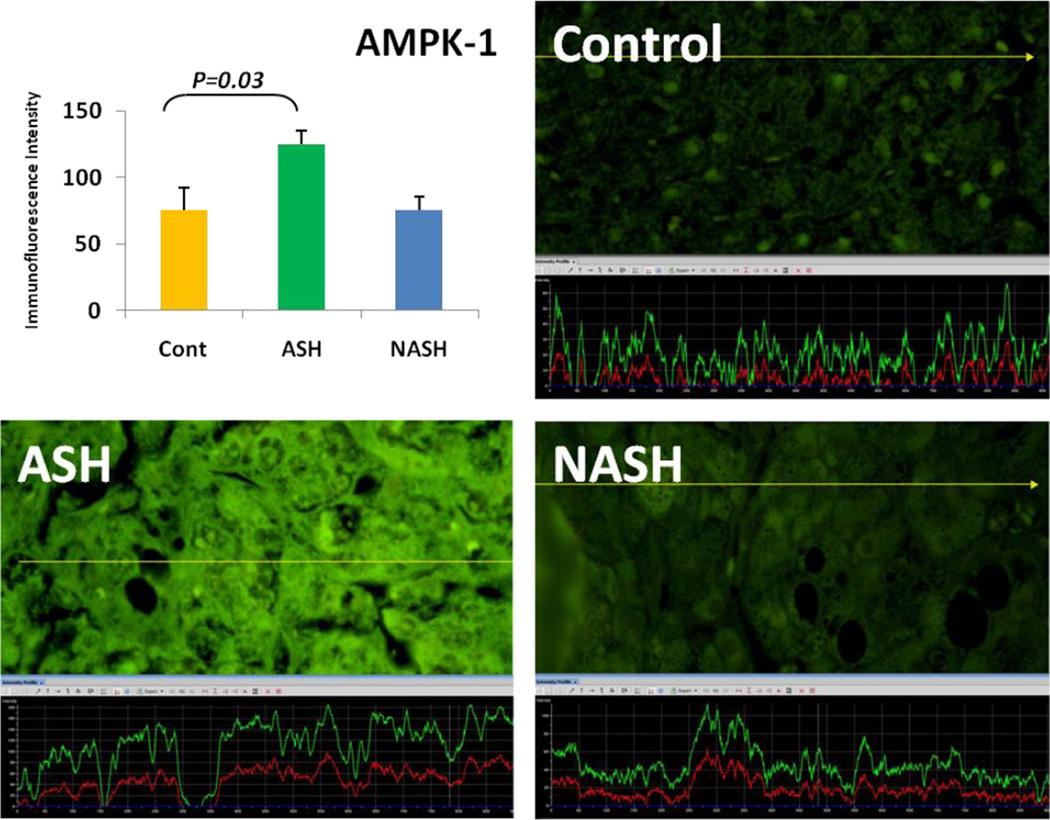
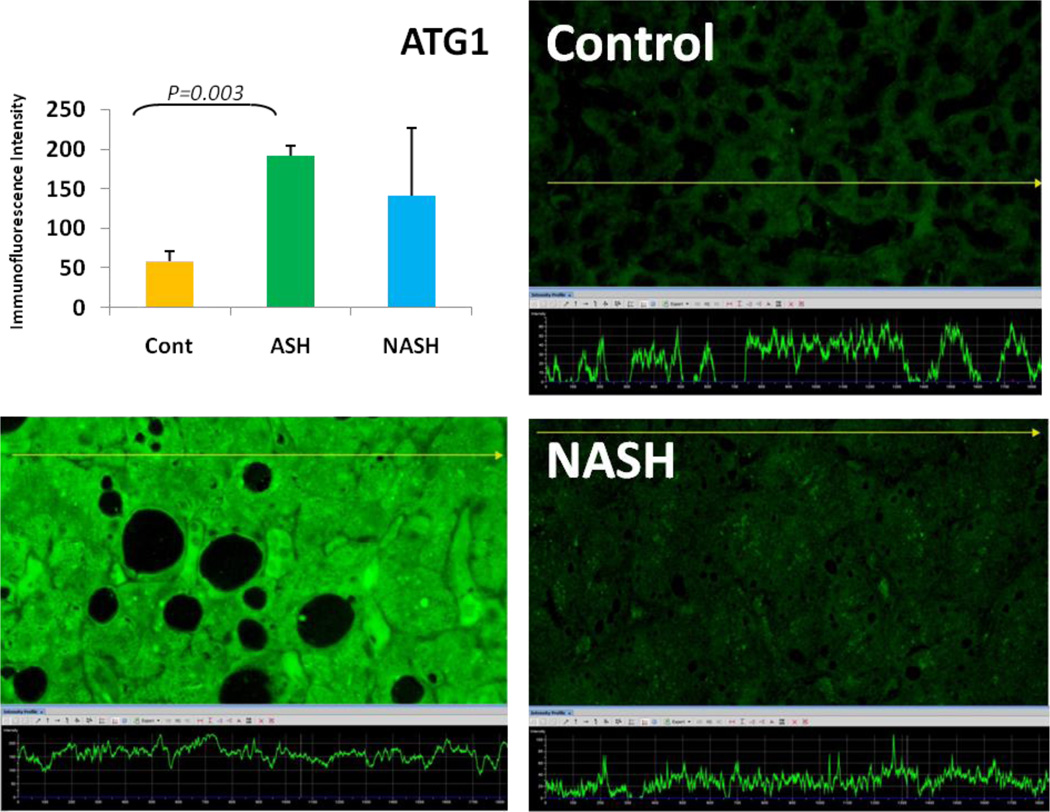
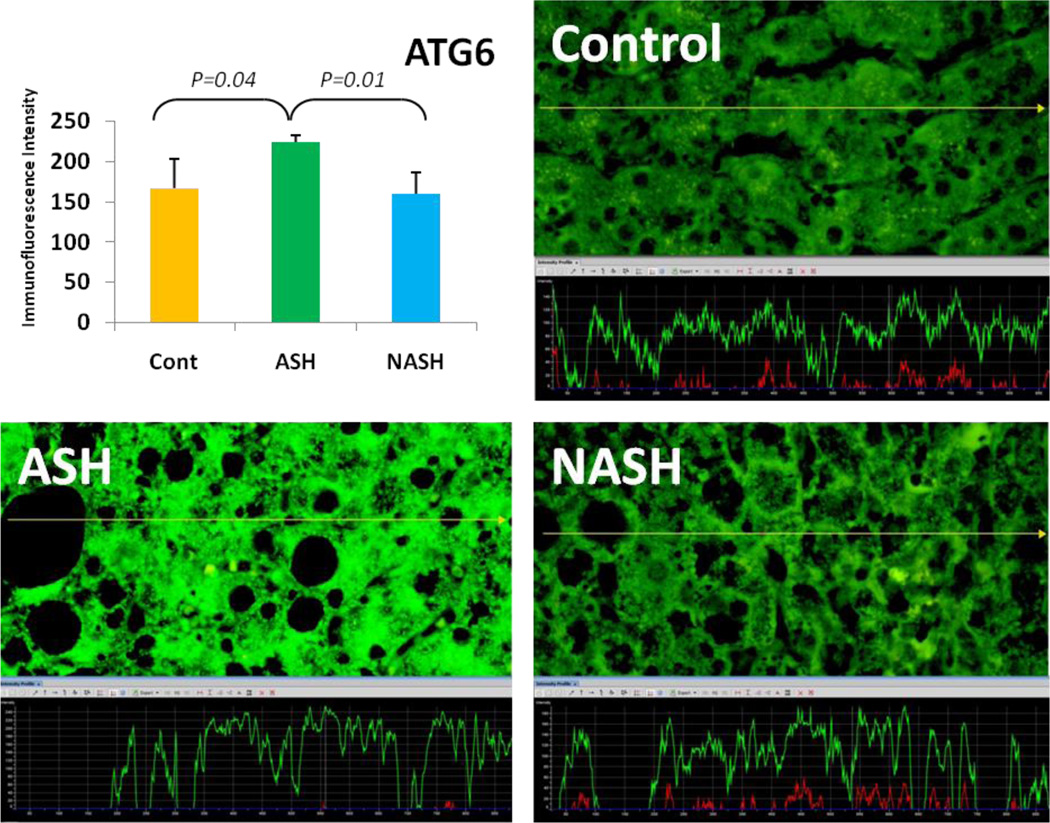
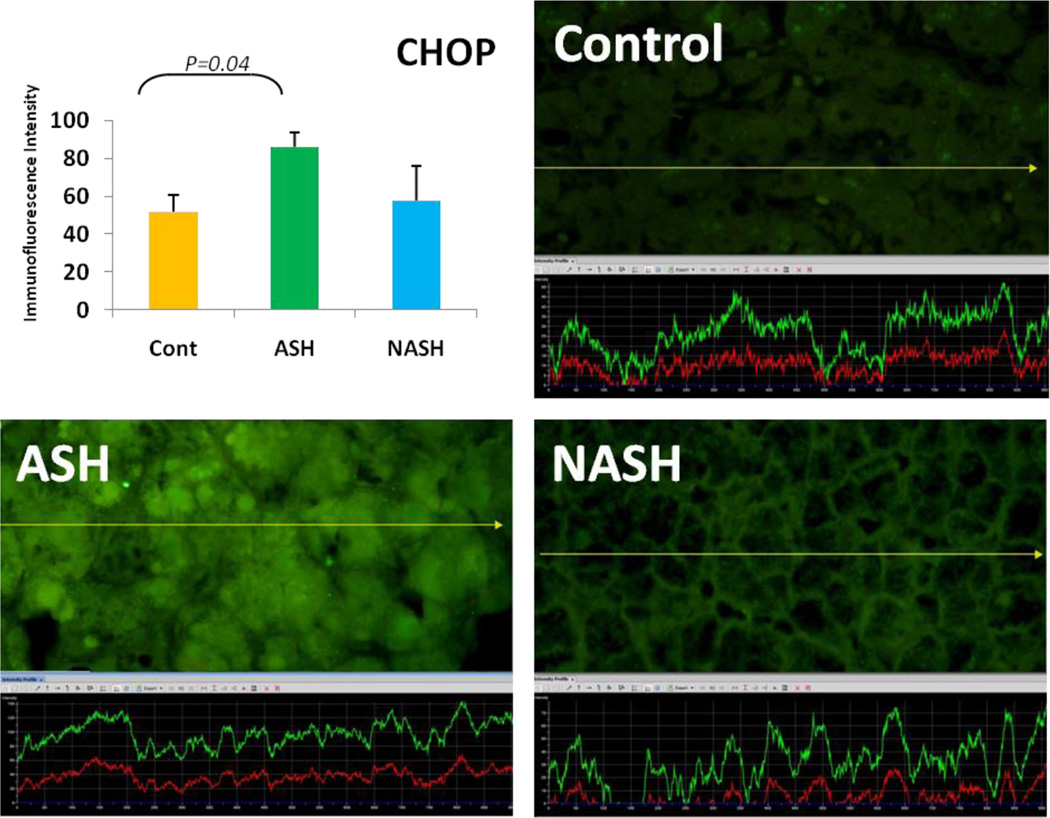
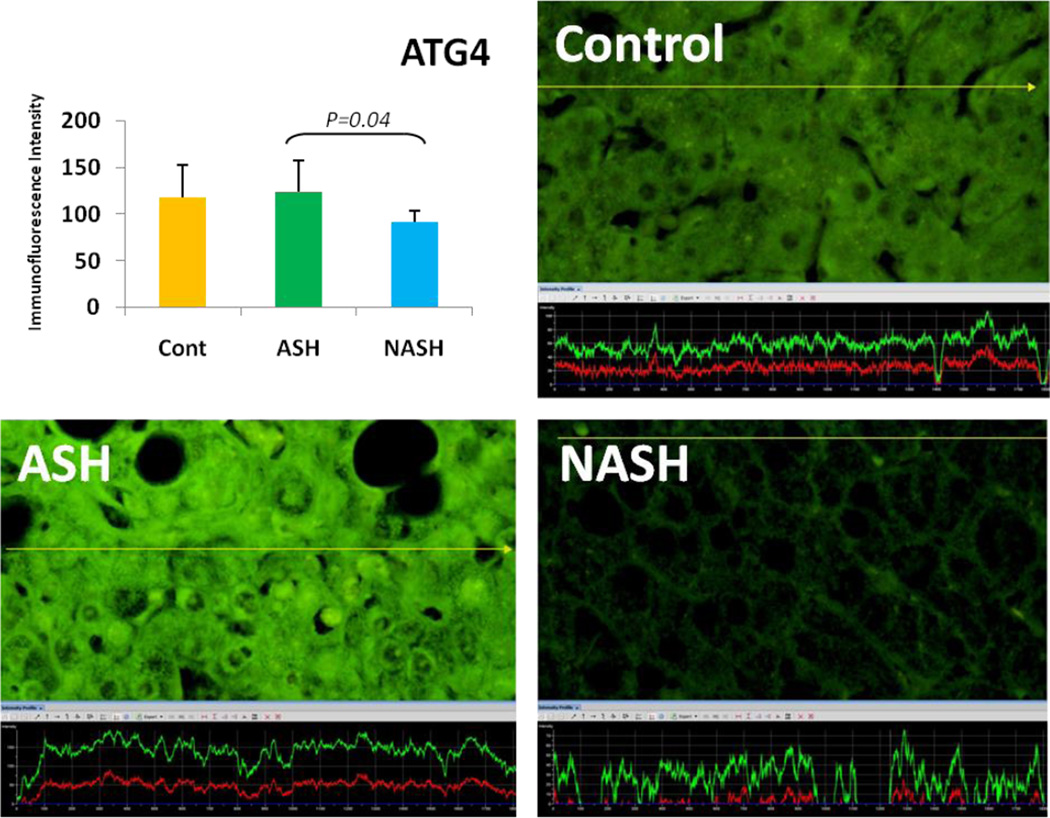
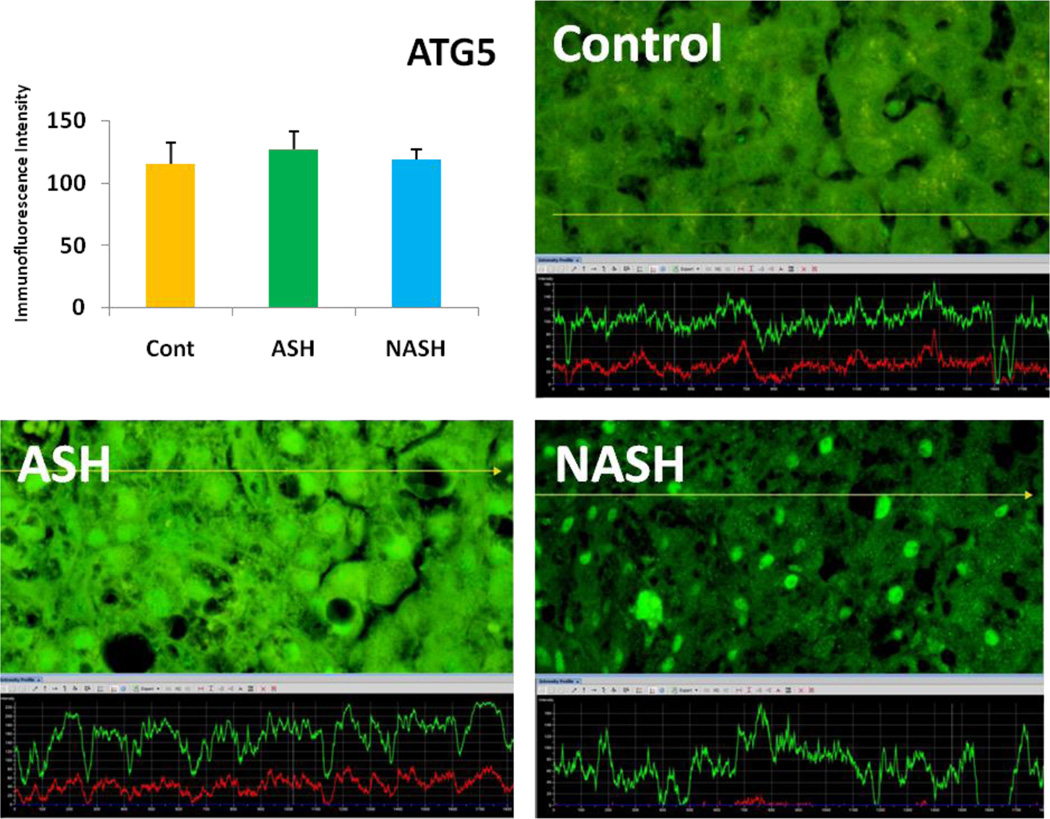
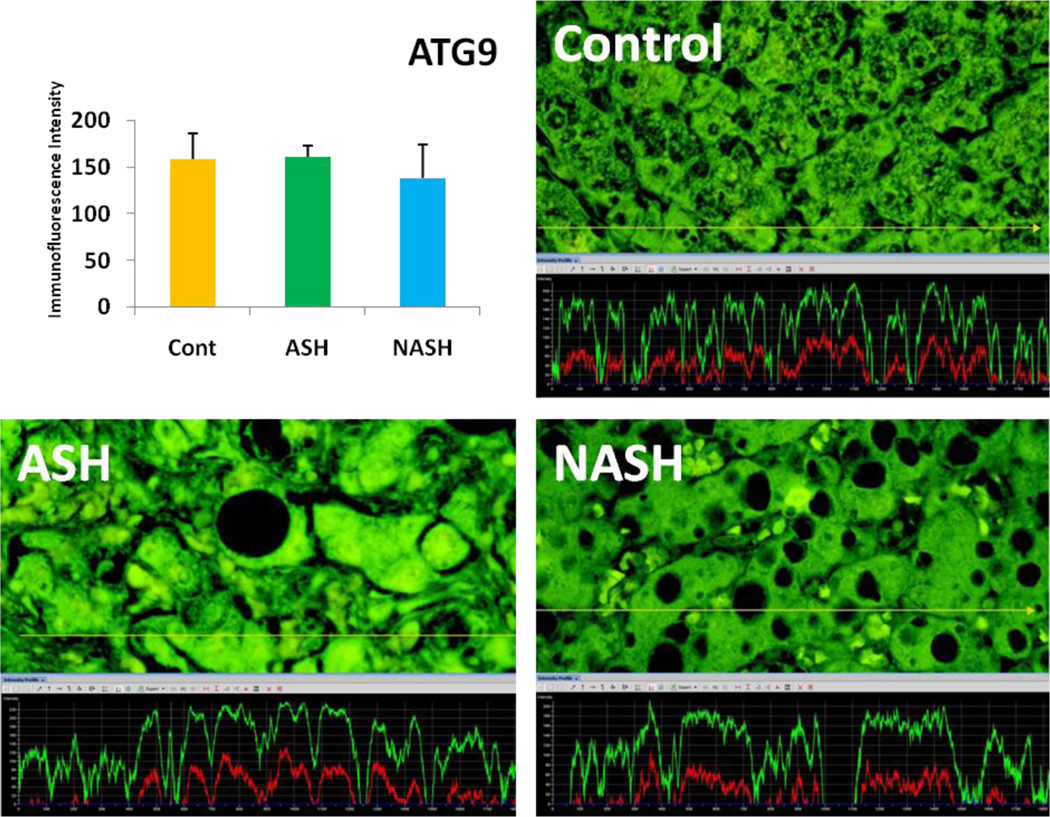
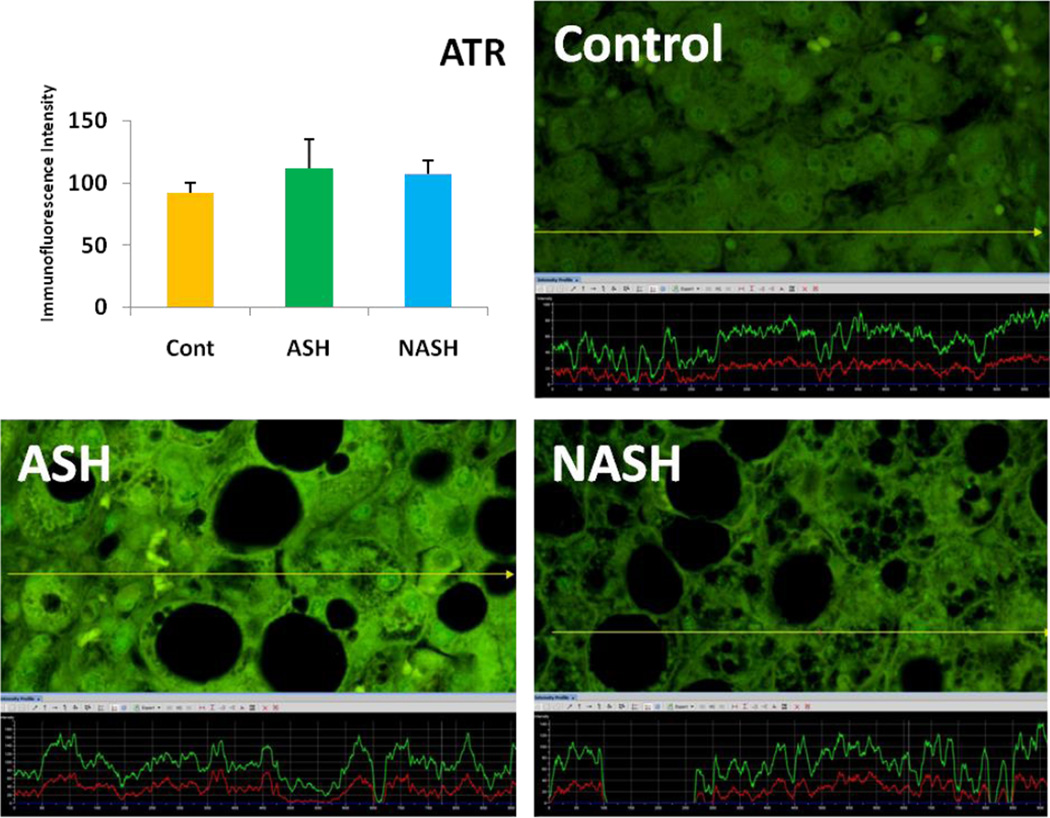
Alcoholic steatohepatitis (ASH) is severe liver injury due to excessive alcohol consumption. Chronic alcohol intake activates cytoprotective pathways in liver cells. Autophagocytosis as a cytoprotective pathway balance the liver cell function and viability. The pathology of ASH, the presence of Mallory- Denk bodies (MDBs) , excess fat and misfolded proteins, ballooning degeneration and degrees of fibrosis indicates severe cell responses. In this study the expression of AMPK1, the initiator of autophagocytosis, ATG1, induces autophagocytosis, ATG6, helps autophagosome formation and CHOP, which controls autophagocytosis in the gene level by increasing transcriptional factors, were significantly (P value <0.05) upregulated in ASH compared to controls. (Fig. 1, 4, 7, 10) Expression of ATG4, ATG5, both interact with Ubiquitin-like complex LC3 in vesicle expansion, ATG9, helps in autolysosome formation and ATR, activates autophagocytosis in DNA damage, was increased in ASH in comparison to controls. (Fig. 5, 6, 8, 9). As evidence of the function of AMPK1, ATG1, ATG6, and CHOP in protein quality control, the following proteins are found to be colocalized within the Mallory Bodies (Fig. 2, 3) as a consequence of the failure of the proteasome protein quality control. As a result, we found that Mallory Bodies are digested by the autophagosome protein quality control system to compensate for proteasome inhibition.
Fig. 1.
AMPK-1 was significantly upregulated compared to controls (p<0.05). The yellow tracer line converted the fluorescence intensity in to the green line in a graph. (x690)
Fig. 4.
The expression of ATG1 in ASH was significantly elevated in comparison to controls. (p<0.05) Its expression in NASH tend to be decreased compared to ASH and tend to be increased in comparison to controls. (x690)
Fig. 7.
The expression of ATG6 in ASH was significantly upregulated compared with NASH and controls. (p<0.05) Its expression in NASH was slightly decreased in comparison to controls. (x690)
Fig. 10.
The expression of CHOP in ASH was significantly increased in ASH compared with controls (p<0.05) and tend to be increased compared to NASH. Its expression in NASH tend to be increased in comparison to controls. (x690)
Fig. 5.
The expression of ATG4 in ASH was elevated in comparison to NASH and controls. Its expression in NASH tend to be decreased compared to controls. (x690)
Fig. 6.
ATG5 expression in ASH was slightly elevated compared to NASH and controls. (x690)
Fig. 8.
ATG9 expression in ASH was slightly elevated in comparison to NASH. Its expression in NASH tend to be decreased compared to controls. (x690)
Fig. 9.
The expression of ATR tend to be elevated in ASH compared to NASH and controls. In NASH ATR tend to show more expression compared to controls. (x690)
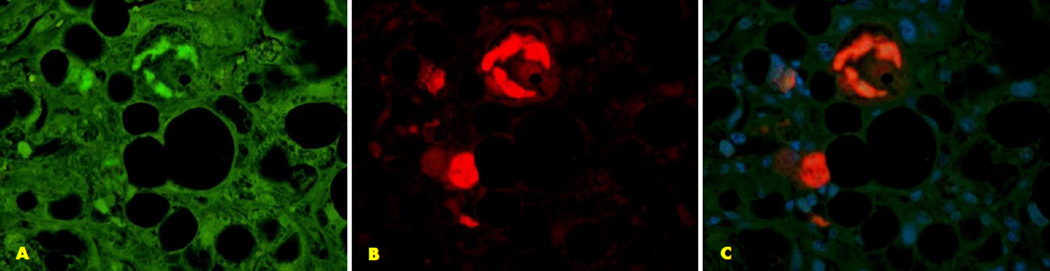
Fig. 2.
The double stain for AMPK1 and UB is shown (A,B). Colocalization of AMPK1 and UB is seen within the Mallory body. Also shown is an increase in the fluorescent intensity in the adjacent liver cells. (x520)
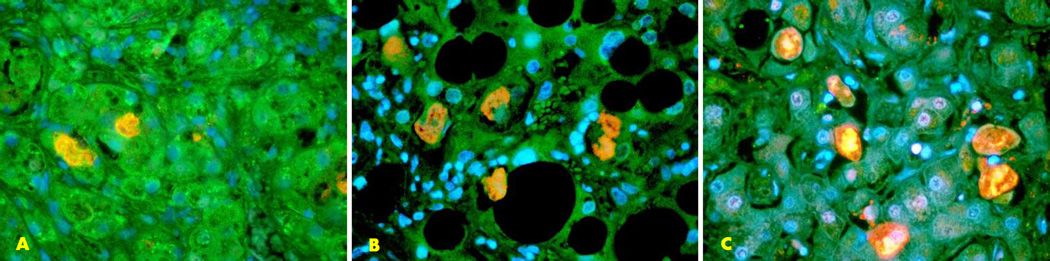
Fig. 3.
The double stain for ATG1, ATG6, and CHOP with UB is shown. Colocalization of ATG1 and UB (A), ATG6 and UB (B), and CHOP and UB (C) are seen within the Mallory body. Also shown is an increase in the fluorescent intensity in the adjacent liver cells. (x520)
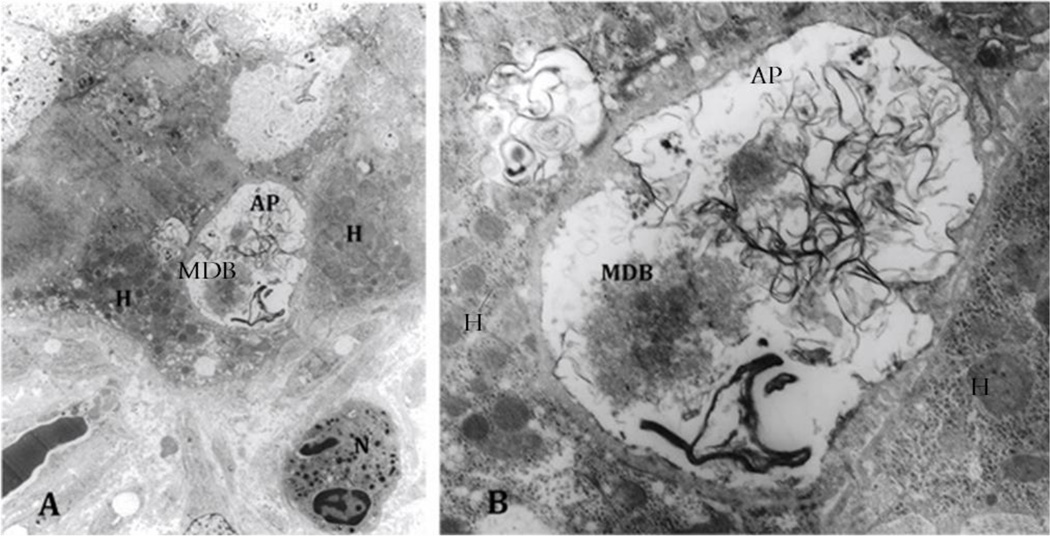
Electron microscopy findings of ASH confirmed the presence of autophagosomes, one of which contained a MDB, heretofore undescribed. (Fig. 11)
Fig. 11.
A. Electron microscopy of an alcoholic steatohepatitis specimen showing autophagosome (AP) adjacent to a neutrophil (N) and hepatocytes (H) (x1458.). B. Closer look of the autophagosome containing Mallory-Denk Bodies (MDB) and tangles of membranes (x6250). Autophagocytosis of a MDB has never been reported or documented in vivo.
NASH
Non-Alcoholic Steatohepatitis is one of the causes of fatty liver, occurring when fat is deposited in the liver due to obesity, metabolic syndromes or insulin resistance. Steatosis is usually combined with ballooning degeneration, MDB formation, inflammation and fibrosis. In NASH the expression of ATG1, ATG5, ATR and CHOP tended to be elevated compared to controls. ATG9, ATG6 and ATG4 expression tended to show some reduction in NASH in comparison to controls.
ASH vs. NASH
The expression of ATG1, ATG4, ATG5, ATG6, ATG9, ATR and CHOP tended to be increased in ASH compared to NASH.
Conclusion
Autophagy is one of the homeostatic mechanisms that is activated during cell stress condition. In many studies autophagy, as an intracellular pathway which degrades unfolded and damaged protein, (Puri et al 2014 Mar) and Mallory-Denk Body(MDB) formation (Liu et al 2014, Peng et al 2014) have been shown as two protective mechanisms caused by stress (Hiura et al 2015, Mendoza et al 2015). Significant upregulations of proteins participating in autophagy pathway such as AMPK-1, ATG-1, ATG-6, and CHOP, and uptrending of ATG-4, ATG-5, ATG-9, ATR, and ATM in ASH compared to normal control livers indicates active autophagocytosis in alcoholic hepatitis. The EM findings in ASH showed a MDB in an autophagosome has never been reported before in human liver biopsies.
Acknowledgments
This study was funded by NIH/AAA grant # UO-21898-04
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement:
The authors declare that there are no conflicts of interest.
References
- Bruhat A, Jousse C, Wang XZ, Ron D, Ferrara M, Fafournoux P. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and posttranscriptional levels. J Biol Chem. 1997;272(28):17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J Gastroenterol. 2014;20(47):17756–17772. doi: 10.3748/wjg.v20.i47.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY. Regulation and Function of Uncoordinated-51 Like Kinase Proteins. Antioxid Redox Signal. 2012;17(5):775–785. doi: 10.1089/ars.2011.4396. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, D'Amelio M, Nardacci R, Romagnoli A, Piacentini M, Cecconi F, Fimia GM. The Dynamic Interaction of Ambra1 with the Dynein Motor Complex Regulates Mammalian Autophagy. J Cell Biol. 2010;191(1):155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V, Waterman D, Bernard A, Ang J, Klionsky D, Haber J. The DNA Damage Response Induces Autophagy via Mec1(ATR) and Tel1(ATM) to Regulate the Initiation of Anaphase. The FASEB Journal. 2015;29(1) Supplement 879.816. [Google Scholar]
- Fimia GM, Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci. 2010;67(10):1581–1588. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura M, Honma Y, Miyagawa K, Oe S, Shimajiri S, Mihara H, Oe M, Sato-Morita M, Katsuki Y, Harada M. Alleviation mechanisms against hepatocyte oxidative stress in patients with chronic hepatic disorders. Hepatol Res. 2015;45(11):1124–1135. doi: 10.1111/hepr.12478. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M. Liver autophagy: physiology and pathology. J Biochem. 2012;152(1):5–15. doi: 10.1093/jb/mvs059. [DOI] [PubMed] [Google Scholar]
- Li Y, Guo Y, Tang J, Jiang J, Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin (Shanghai) 2014;46(8):629–640. doi: 10.1093/abbs/gmu048. [DOI] [PubMed] [Google Scholar]
- Mendoza AS, Dorce J, Peng Y, French BA, Tillman B, Li J, French SW. Levels of metacaspase1 and chaperones related to protein quality control in alcoholic and nonalcoholic steatohepatitis. Exp Mol Pathol. 2015;98(1):65–72. doi: 10.1016/j.yexmp.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24(6):1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P, Chandra A. Autophagy modulation as a potential therapeutic target for liver diseases. J Clin Exp Hepatol. 2014 Mar;4(1):51–59. doi: 10.1016/j.jceh.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. Ulk1 Induces Autophagy by Phosphorylating Beclin-1 and Activating Vps34 Lipid Kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283(50):35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi DN, Zhang J, Jing J, Dere R, Walker CL. A new role for ATM in selective autophagy of peroxisomes (pexophagy) Autophagy. 2016;12(4):711–712. doi: 10.1080/15548627.2015.1123375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119(pt18):3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li J, Ouyang L, Liu B, 2, Cheng Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett. 2016;373(1):19–26. doi: 10.1016/j.canlet.2016.01.022. [DOI] [PubMed] [Google Scholar]
- Zheng YZ, Cao ZG, Hu X, Shao ZM. The endoplasmic reticulum stress markers GRP78 and CHOP predict disease-free survival and responsiveness to chemotherapy in breast cancer. Breast Cancer Res Treat. 2014;145(2):349–358. doi: 10.1007/s10549-014-2967-x. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]