Abstract
Protein synthesis occurs on macromolecular machines, called ribosomes. Bacterial ribosomes and the translational machinery represent one of the major targets for antibiotics in the cell. Therefore, structural and biochemical investigations into ribosome-targeting antibiotics provide not only insight into the mechanism of action and resistance of antibiotics, but also insight into the fundamental process of protein synthesis. This review summarizes the recent advances in our understanding of protein synthesis, particularly with respect to X-ray and cryoelectron microscopy (cryo-EM) structures of ribosome complexes, and highlights the different steps of translation that are targeted by the diverse array of known antibiotics. Such findings will be important for the ongoing development of novel and improved antimicrobial agents to combat the rapid emergence of multidrug resistant pathogenic bacteria.
Protein synthesis in bacteria can be divided into four main steps: initiation, elongation, termination, and ribosome recycling. Two of these steps—initiation and elongation—are targeted by numerous antibiotics.
THE BACTERIAL RIBOSOME AND PROTEIN SYNTHESIS
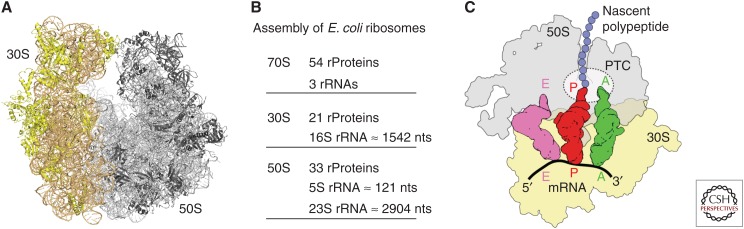
The ribosome is the macromolecular machine that converts the genetic information encoded in the messenger RNA (mRNA) into the polypeptide sequence that comprises the proteins and enzymes of the cell (Schmeing and Ramakrishnan 2009; Voorhees and Ramakrishnan 2013). Bacterial 70S ribosomes are comprised of two subunits, a small 30S subunit and a large 50S subunit (Fig. 1A), both of which are ribonucleoprotein particles. In the bacterium Escherichia coli, the small subunit (SSU) is assembled from 21 ribosomal proteins and a single 16S ribosomal RNA (rRNA) of 1541 nucleotides, whereas the large subunit (LSU) is assembled from 33 ribosomal proteins and two rRNAs, a 5S rRNA of 115 nucleotides, and a 23S rRNA of 2904 nucleotides (Fig. 1B). The ribosome provides the platform for the binding of transfer RNAs (tRNAs), which are adaptor molecules that contain at one end the anticodon to recognize a specific codon of the mRNA, and at the other end the CCA-end that is covalently linked to the amino acid specific for the mRNA codon. There are three tRNA binding sites in the 70S ribosome, termed the A-site, P-site, and E-site (Fig. 1C). The P-site is the peptidyl-tRNA binding site, which is occupied by the tRNA carrying the polypeptide chain during elongation (or the initiator tRNA during initiation). The A-site binds the incoming aminoacylated or charged tRNA, whereas the E-site is the exit site and binds only outgoing deacylated or uncharged tRNA. Thus, during translation elongation, the tRNAs pass consecutively through the A-, P-, and then E-sites before dissociating from the ribosome. There are two main functional centers on the ribosome, namely, the decoding center (DC) and peptidyltransferase center (PTC). The DC is located on the SSU in the A-site and monitors the correctness of the interaction between the codon of the mRNA and the anticodon of the tRNA and thereby ensures that the correct amino acid is delivered, that is, the amino acid corresponding to the codon of the mRNA. The PTC is located on the LSU and catalyzes the process of peptide bond formation. Protein synthesis can be divided into four main steps: initiation, elongation, termination, and ribosome recycling, each of which is targeted by a plethora of different antibiotics (Fig. 2) (Wilson 2009, 2014). Therefore, studies into antibiotic action during translation provide not only insight into the inhibitory action of the antibiotics, but also insight into the fundamental process of protein synthesis. This review provides an overview of the individual steps of protein synthesis and provides a brief overview of how these steps are inhibited by antibiotics.
Figure 1.
The prokaryotic ribosome. (A) Overview of the Escherichia coli 70S ribosome (Dunkle et al. 2011) with 30S subunit colored in yellow and 50S subunit in gray. (B) Table of assembly components of the 70S ribosome as well as the 30S and 50S subunits. (C) Schematic representation of the prokaryotic ribosome bound with three transfer RNAs (tRNAs) showing the 30S subunit (yellow), 50S subunit (gray), A-tRNA (green), P-tRNA (red), E-tRNA (pink), nascent polypeptide chain (violet), and mRNA (black). The peptidyltransferase center (PTC) on the 50S subunit is depicted as a dashed sphere.
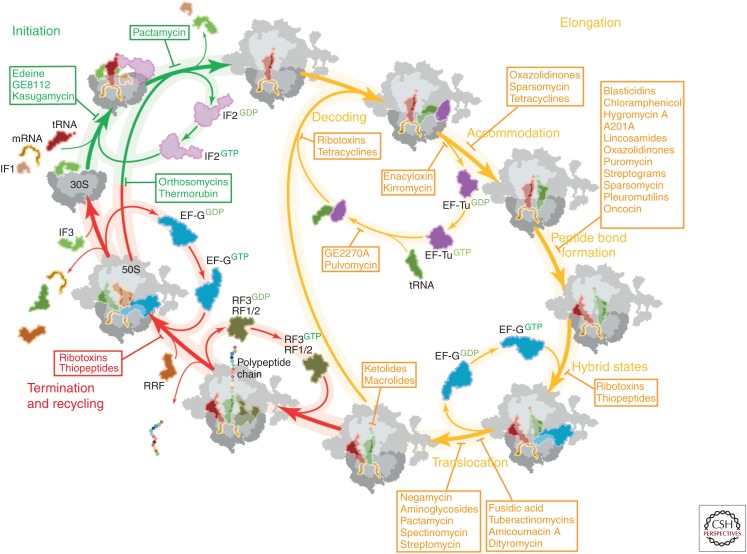
Figure 2.
Overview of antibiotics inhibiting the prokaryotic translation cycle. Overview of antibiotics, inhibiting translation initiation (green), translation elongation (yellow), and translation termination/recycling (red) of the prokaryotic translation cycle (modified from Sohmen et al. 2009). tRNA, Transfer RNA; mRNA, messenger RNA; IF, initiation factor; EF-Tu, elongation factor Tu; GDP, guanosine diphosphate; EF-G, elongation factor G; GTP, guanosine triphosphate; RRF, ribosome recycling factor.
INITIATION OF TRANSLATION
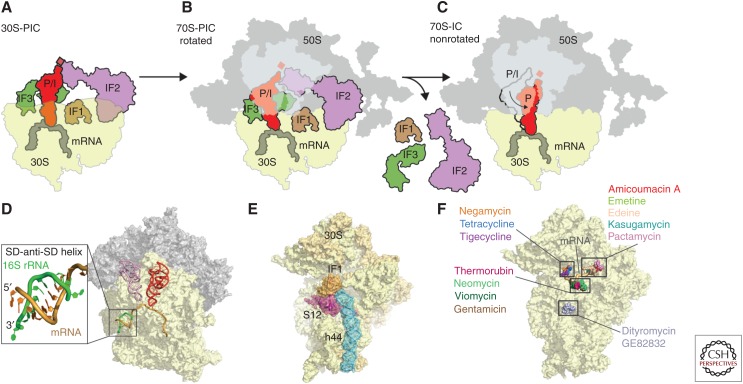
Initiation of translation is the rate-limiting step during translation of mRNA molecules into proteins (Laursen et al. 2005; Simonetti et al. 2009). Prokaryotic translation initiation requires formation of the 30S preinitiation complex (PIC), which in bacteria involves the three initiation factors (IFs), IF1, IF2, and IF3, as well as mRNA and the formylated initiator fMet-tRNAfMet (Fig. 3A). The methionyl-tRNA transformylase-mediated formylation of the initiator tRNA distinguishes initiator fMet-tRNAfMet from elongation Met-tRNAMet. The main goal of 30S-PIC formation is to position the initiator fMet-tRNAfMet in a peptidyl/initiation (P/I) state, such that it is bound to the start codon of the mRNA in the P-site of the SSU (Fig. 3A). Subsequently, the LSU joins to form the 70S preinitiation complex (70S-PIC) (Fig. 3B). 70S-IC formation is accompanied by the dissociation of initiation factors IF1–IF3 from the ribosome, leaving the fMet-tRNAfMet positioned in the P/P state, thereby priming the ribosome for initiation of translation elongation (Fig. 3C).
Figure 3.
Initiation of translation. Schematic assembly of the (A) 30S preinitiation complex (PIC), (B) 70S-PIC, and (C) 70S-IC during translation initiation with 30S subunit (yellow), 50S subunit (gray), messenger RNA (mRNA) (dark gray), initiator transfer RNA (tRNA) (red), initiation factors (IFs): IF1 (brown), IF2 (purple), and IF3 (green). (D) Crystal structure of the prokaryotic ribosome with zoom onto the interaction of the Shine–Dalgarno (SD) sequence of canonical mRNAs (orange) with the anti-SD at the 3′ end of the 16S ribosomal RNA (rRNA) (green), including P-site tRNA (red) and E-site tRNA (pink) (Yusupova et al. 2006). (E) Crystal structure of IF1 (brown) bound to the 30S subunit (yellow) with highlighted h44 (blue) of the 16S rRNA and ribosomal protein S12 (red) (Carter et al. 2001). (F) Binding sites of dityromycin (PDB 4NVU) (Bulkley et al. 2014), gentamicin (PDB 4V53) (Borovinskaya et al. 2007a), thermorubin (PDB 3UXT) (Bulkley et al. 2012), viomycin (PDB 3KNH) (Stanley et al. 2010), neomycin (PDB 2QAL) (Borovinskaya et al. 2007a), negamycin (PDB 4RBH) (Polikanov et al. 2014c), tetracycline (PDB 4G5K) (Jenner et al. 2013), tigecycline (PDB 4G5T) (Jenner et al. 2013), amicoumacin A (PDB 4RB5) (Polikanov et al. 2014a), edeine (PDB 1I95) (Pioletti et al. 2001), kasugamycin (PDB 1VS5) (Schuwirth et al. 2006), pactamycin (PDB 4RBB) (Polikanov et al. 2014c), and emetine (PDB 3J7A) (Wong et al. 2014), on the small subunit (SSU) (yellow). P/I, Peptidyl/initiation.
During initiation, IF1 and IF3 ensure fidelity of the process, whereas IF2 recruits fMet-tRNAfMet. IF3 binds to the E-site of the 30S subunit to form the IF3–30S complex (Dallas and Noller 2001) and, thereby, prevents premature 50S subunit joining before association with IF1, IF2, mRNA, and initiator tRNA (Karimi et al. 1999). The interaction of the Shine–Dalgarno (SD) sequence of canonical mRNAs with the anti-SD at the 3′ end of the 16S rRNA (Fig. 3D) places the start codon in the P-site and allows for subsequent association of fMet-tRNAfMet, IF1, and IF2. Discrimination of the initiator tRNA is performed by IF3 through monitoring of three unique G:C base pairs in fMet-tRNAfMet. Furthermore, the presence of IF3 is required to ensure the fidelity of the codon–anticodon interaction in the P-site of the SSU. IF1 binds at the A-site of the SSU (Fig. 3E) (Carter et al. 2001) where it stabilizes IF2 binding (Julian et al. 2011; Simonetti et al. 2013) and accelerates IF2-dependent fMet-tRNAfMet recruitment (Laursen et al. 2005). Binding of IFs to the SSU stabilizes a swiveled conformation of the head with respect to the body (Julian et al. 2011). The LSU joins the 30S-PIC in ratcheted conformation to form the 70S-IC (Allen et al. 2005). Formation of the 70S-IC activates guanosine triphosphate (GTP) hydrolysis by IF2, which leads to unratcheting of the ribosome, allowing the conformational transition of the fMet-tRNAfMet from the P/I state to the accommodated P/P-state (Fig. 3C). At the same time, the IFs dissociate from the complex, thus readying the ribosome to enter into the elongation cycle.
There are a number of antibiotics that are commonly referred to as translation initiation inhibitors, namely, kasugamycin, pactamycin, edeine, GE81112, which interact with the SSU (Fig. 3F), the orthosomycins evernimicin and avilamycin, which interact with the LSU, as well as thermorubin, the binding site of which comprises components of both the SSU and LSU (Brandi et al. 2008; Wilson 2009; Bulkley et al. 2012). Kasugamycin binds within the E-site of the SSU in a position that overlaps with the mRNA (Fig. 3F) (Schluenzen et al. 2006; Schuwirth et al. 2006). By disturbing the path of the mRNA, kasugamycin prevent the initiator fMet-tRNAfMet binding to the 30S-PIC (Schluenzen et al. 2006; Schuwirth et al. 2006). Similarly, pactamycin has also been reported to prevent 30S-PIC formation by perturbing the path of the mRNA through the E-site (Fig. 3F) (Brodersen et al. 2000); however, subsequent studies suggested that inhibition of translocation, rather than initiation, was the mechanism of action (Dinos et al. 2004). In contrast, edeine and GE81112 are suggested to inhibit 30S-PIC formation by directly blocking the binding of fMet-tRNAfMet (Pioletti et al. 2001; Dinos et al. 2004; Brandi et al. 2006). The binding site of thermorubin is located within a pocket formed by h44 of the SSU and H69 of the LSU (Bulkley et al. 2012) and, therefore, does not directly overlap with the mRNA or initiator-tRNA, but rather inhibits 30S-PIC formation by inducing conformational changes that perturb IF binding (Bulkley et al. 2012). Evernimicin and avilamycin are suggested to block the association of the 30S-PIC with the LSU by preventing the accommodation of IF2 on the LSU during subunit joining (Belova et al. 2001).
ELONGATION PHASE
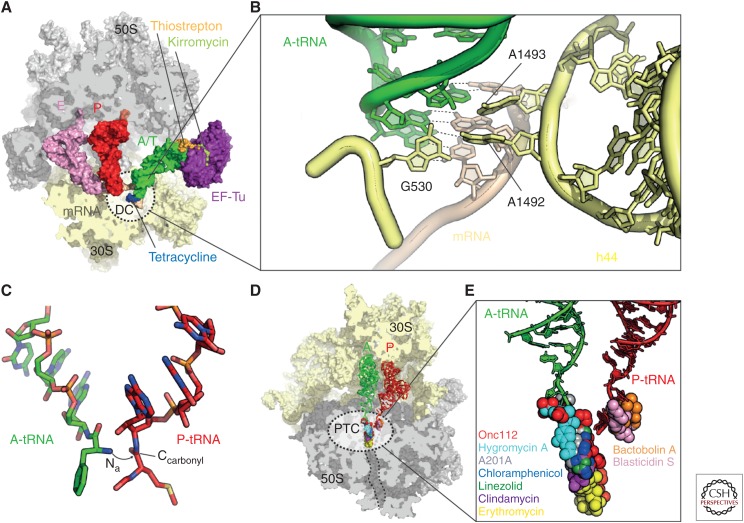
After initiation, the ribosomal P-site is occupied by the initiator fMet-tRNAfMet, whereas the A-site remains empty. The next aminoacyl-tRNA (aa-tRNA) is delivered to the empty A-site as a ternary complex with elongation factor Tu (EF-Tu) and GTP (Schmeing and Ramakrishnan 2009; Voorhees and Ramakrishnan 2013). The ternary complex initially binds with a bent or kinked conformation of the tRNA that allows the anticodon stem loop (ASL) to interact with the codon of the mRNA in the DC, whereas the aminoacylated 3′-acceptor stem remains bounds to EF-Tu and is, hence, termed the A/T state (Fig. 4A) (Stark et al. 1997; Valle et al. 2002; Blanchard et al. 2004a; Schmeing et al. 2009; Schuette et al. 2009; Fischer et al. 2015). To ensure translational fidelity, the ribosome discriminates between cognate and noncognate tRNA binding by monitoring the interaction between the A-site codon of the mRNA and the anticodon of the tRNA (Ogle and Ramakrishnan 2005). During codon recognition, nucleotides A1492 and A1493 (E. coli numbering) adopt a conformation flipped-out of helix 44 (h44) of the 16S rRNA and together with G530 monitor the correct Watson–Crick geometry of the first two base pairs of the codon–anticodon interaction in the form of A-minor motifs (Fig. 4B) (Ogle and Ramakrishnan 2005). At the third nucleotide position of the codon a wobble pair (e.g., G·U) is tolerated. This allows a single tRNA to decode multiple codons that only differ in the third anticodon position, thus providing an explanation for the degeneracy of the genetic code. Recent crystal structures of complete cognate or near-cognate tRNAs bound to the full 70S ribosome have challenged the hypothesis of how the ribosome distinguishes cognate tRNAs from near-cognate or noncognate tRNAs (Demeshkina et al. 2012). Surprisingly, in these structures, even the near-cognate codon–anticodon interactions are observed to form Watson–Crick-like interactions, suggesting that A1492, A1493, and G530 do not discriminate cognate and near-cognate tRNAs (Demeshkina et al. 2012). Rather, the ribosome stabilizes an energetically unfavorable tautomer of the nucleotide to achieve Watson–Crick geometry, leading the investigators to suggest that this energetic penalty is then used by the ribosome to discriminate cognate from near-cognate tRNAs (Demeshkina et al. 2013; Rozov et al. 2015).
Figure 4.
Decoding and peptide bond formation on the ribosome. (A) Crystal structure of the ribosome in complex with elongation factor Tu (EF-Tu) (purple), A/T-transfer RNA (tRNA) (green), P-tRNA (red), E-tRNA (pink), kirromycin (light green), messenger RNA (mRNA) (brown), 30S subunit (yellow), and 50S subunit (gray) (Voorhees et al. 2010) with superimposed binding positions of thiostrepton (orange) (Harms et al. 2008) and tetracycline (blue) (Jenner et al. 2013). The decoding center (DC) on the 30S subunit is depicted as dashed-lined sphere. (B) DC within the small ribosomal subunit with A-tRNA (green), mRNA (brown), and 16S ribosomal RNA (rRNA) nucleotides G530, A1492, and A1493 (yellow) (Demeshkina et al. 2012). (C) Positions of the A-tRNA (green) and P-tRNA (red) within the peptidyltransferase center (PTC) of the 50S subunit. The nucleophilic attack of the A-tRNA α-amino group (Nα) onto the P-tRNA carbonyl-carbon (Ccarbonyl) is indicated with an arrow. (D) Overview and (E) zoom onto the binding sites of Onc112 (PDB 4ZER) (Seefeldt et al. 2015), hygromycin A (PDB 4Z3R) (Polikanov et al. 2015), A201A (PDB 4Z3S) (Polikanov et al. 2015), chloramphenicol (PDB 3OFC) (Dunkle et al. 2010), linezolid (PDB 3DLL) (Wilson et al. 2008), clindamycin (PDB 3OFZ) (Dunkle et al. 2010), erythromycin (PDB 3OFR) (Dunkle et al. 2010), bactobolin A (PDB 4WWE) (Amunts et al. 2015), and blasticidin S (PDB 4L6J) (Svidritskiy et al. 2013) within the PTC (dashed lines in D) of the 50S subunit (gray) with A-tRNA (green) and P-tRNA (red).
Nevertheless, recognition of a correct codon–anticodon interaction triggers large-scale conformational changes in the ribosome that induce a domain closure of the 30S, involving movement of the SSU shoulder toward EF-Tu (Ogle and Ramakrishnan 2005; Schmeing et al. 2009). These structural rearrangements are propagated to EF-Tu, which ultimately leads to stimulation of GTP hydrolysis (Schmeing and Ramakrishnan 2009; Voorhees and Ramakrishnan 2013). The GTPase activity of EF-Tu is controlled by positioning the catalytic histidine 84 (H84) of EF-Tu in proximity to the phosphate of A2662 of the sarcin–ricin loop (SRL) in H95 of the 23S rRNA. This enables H84 to coordinate a water molecule for nucleophilic attack on the γ-phosphate of GTP, which is then hydrolyzed (Voorhees et al. 2010). GTP hydrolysis and inorganic phosphate (Pi) release cause structural rearrangements in EF-Tu, leading to dissociation of EF-Tu from the ribosome and, thus, allowing the tRNA to transition from the A/T-state into the A/A-state. During accommodation, the acceptor stem of the tRNA moves into the A-site of the PTC in the large ribosomal subunit (Blanchard et al. 2004a; Sanbonmatsu et al. 2005).
There are many antibiotics that inhibit delivery and accommodation of the aminoacyl-tRNA on the ribosome (Wilson 2009). These range from antibiotics that interact with EF-Tu, such as GE2270A and kirromycin, to those that interact directly with the ribosome, such as thiostreptons and tetracyclines (Fig. 4A). In addition, antibiotics of the negamycin and aminoglycoside classes also bind to the ribosome and interfere with the decoding process. Thiopeptide antibiotics, such as GE2270A and related derivatives, such as LFF571, bind to EF-Tu and prevent formation of the complex with the aminoacyl-tRNA, whereas in contrast kirromycin binds to the EF-Tu-aa-tRNA complex and traps it on the ribosome (Fig. 4A) (Wilson 2009). The thiostrepton-like antibiotics interact with the large ribosomal subunit and interfere with the binding of translational GTPases, including EF-Tu and elongation factor G (EF-G) to the ribosome (Fig. 4A) (Wilson 2009). The tetracycline family of antibiotics, including the third-generation glycylcyclines, such as tigecycline, bind to the SSU and sterically block the recognition of the codon of the mRNA by the anticodon of the aa-tRNA (Nguyen et al. 2014). Although negamycin and aminoglycoside antibiotics have distinct binding sites on the SSU, both classes stabilize the binding of aa-tRNAs, including near-cognate aa-tRNAs, thus leading to misreading and stop-codon suppression (Wilson 2009; Olivier et al. 2014; Polikanov et al. 2014c).
PEPTIDE BOND FORMATION AND NASCENT CHAIN PROLONGATION
Peptide bond formation between the incoming amino acid attached to the A-site tRNA and the nascent polypeptide attached to the P-site tRNA represents the main function of the ribosome (Simonovic and Steitz 2009; Rodnina 2013). On accommodation of the A-site tRNA, the peptidyl transfer reaction occurs rapidly, with a rate that is ∼2 × 107-fold faster than the rate of spontaneous peptide bond formation in solution (Sievers et al. 2004). The ribosome acts as an entropy trap by precisely positioning the aminoacylated tRNA CCA-end substrates for trans-esterification and formation of the new peptide bond (Sievers et al. 2004). Positioning of the tRNAs is facilitated by stabilizing interactions between 23S rRNA nucleotides and the tRNA CCA-ends. The P-site CCA end is stabilized by Watson–Crick base pairs of nucleotides C74 and C75 with P-loop nucleotides G2251 and G2252, respectively, whereas the A-tRNA C74 stacks up on U2555, C75 forms a Watson–Crick base pair with G2553 and A76 interacts with G2583 in the form of a class I A-minor motif (Kim and Green 1999; Nissen et al. 2000; Hansen et al. 2002; Voorhees et al. 2009; Polikanov et al. 2014b). Proper tRNA accommodation into the A-site leads to conformational changes within the PTC, namely, 23S rRNA nucleotides G2583-U2585 undergo a shift of 1–2 Å, while U2506 rotates by 90° to provide space for A-tRNA accommodation into the PTC (Schmeing et al. 2005). These conformational changes convert the PTC into its induced state by exposing the peptidyl-tRNA ester for peptide bond formation, which occurs through nucleophilic attack of the α-amino group of the A-tRNA onto the carbonyl-carbon of the aminoacyl ester of the peptidyl-tRNA in the P-site (Fig. 4C). The most recent model for peptide bond formation is based on high-resolution crystal structures of a Thermus thermophilus 70S ribosome in both pre-attack and postcatalysis states (Polikanov et al. 2014b). Three water molecules trapped in the PTC before catalysis allowed the investigators to suggest a proton wire mechanism that couples aa-tRNA accommodation and peptide bond formation. Both tRNAs, 23S rRNA nucleotides A2451, U2584, C2063, and A2602, as well as the amino terminus of ribosomal protein L27 contribute to the coordination of the water molecules. L27and N6 of A2602 activate a water molecule (W1) to initiate the proton wire via the 2′ OH of A2451 to the 2′ OH of the P-site A76, which deprotonates the α-amino group for concerted nucleophilic attack onto the ester carbonyl carbon. The tetrahedral intermediate state is stabilized by a second water molecule (W2), which donates a proton to the negatively charged ester carbonyl carbon. Breakdown of the intermediate state occurs via protonation of the 3′ ester oxygen of the leaving group via a third water (W3) and a partially reversed proton wire via the 2′ OH of P-site A76, the 2′ OH of A2451 back to W1 (Polikanov et al. 2014b).
Antibiotics that directly interfere with peptide bond formation generally do so by preventing the accurate placement of the aminoacylated-CCA-end of the A-tRNA at the PTC (Fig. 4D). Therefore, these antibiotics can also be thought of as inhibiting a final stage in the accommodation of the aminoacyl-tRNA at the A-site. Well-known examples include the phenicols (chloramphenicol), oxazolidinones (linezolid), pleuromutilins (tiamulin), and lincosamides (clindamycin) (Wilson 2009, 2014), but was also shown more recently for hygromycin A, the nucleoside antibiotic A201A (Polikanov et al. 2015), and the antimicrobial peptide oncocin (Fig. 4E) (Roy et al. 2015; Seefeldt et al. 2015). Some of the larger macrolide antibiotics, such as josamycin, tylosin, and spiramycin, also interfere with peptide bond formation by perturbing A-tRNA accommodation at the PTC (Wilson 2009), but generally the binding of macrolide antibiotics within the ribosomal tunnel is thought to interfere with prolongation of the nascent polypeptide chain (Kannan et al. 2014). However, recent studies have revealed that some polypeptides manage to bypass the drug in the tunnel and can even become fully synthesized in the presence of the drug (Kannan and Mankin 2012; Kannan et al. 2012).
TRANSLOCATION AND EF-G
After peptide bond formation, the A-site is bound by the peptidyl-tRNA lengthened by one amino acid and the P-site is bound by deacylated tRNA. To allow the next round of elongation, the tRNAs together with the mRNA need to move with respect to the ribosome, namely, to shift the peptidyl-tRNA from the A-site to the P-site and the deacylated tRNA from the P-site to the E-site (Yamamoto et al. 2014). The mRNA shifts precisely by one codon, placing the next codon in the A-site. Translocation is catalyzed by the GTPase elongation factor G (EF-G) and provides an empty A-site, which in turn allows binding and accommodation of the next cognate aa-tRNA and, thus, the elongation cycle to proceed (Fig. 2). However, the ribosome has an intrinsic capability to translocate tRNAs both forward and backward, and therefore, the function of EF-G is to accelerate and direct the process in forward direction (Yamamoto et al. 2014).
In the pretranslocation state, the 3′-ends of A- and P-site tRNA spontaneously move back and forth between the P- and E-sites on the LSU, respectively, while their ASLs remain anchored within the A- and P-sites on the SSU, which creates A/P and P/E hybrid tRNA binding states (Fig. 5A) (Moazed and Noller, 1989; Blanchard et al. 2004b). The spontaneous formation of tRNA hybrid states is driven by decreased affinity for peptidyl-tRNA in the A-site and deacylated tRNA in the P-site by ∼1000-fold (Semenkov et al. 2000). Moreover, the E-site on the 50S subunit sterically occludes binding of peptidyl-tRNA (Rheinberger et al. 1981; Schmeing et al. 2003) and, thus, ensures that translocation occurs only after peptide bond formation is completed. Formation of tRNA hybrid states is coupled to a large-scale conformational rotation (ratcheting) of the SSU ∼3–10° counterclockwise relative to the LSU (Frank and Agrawal 2001; Valle et al. 2003).
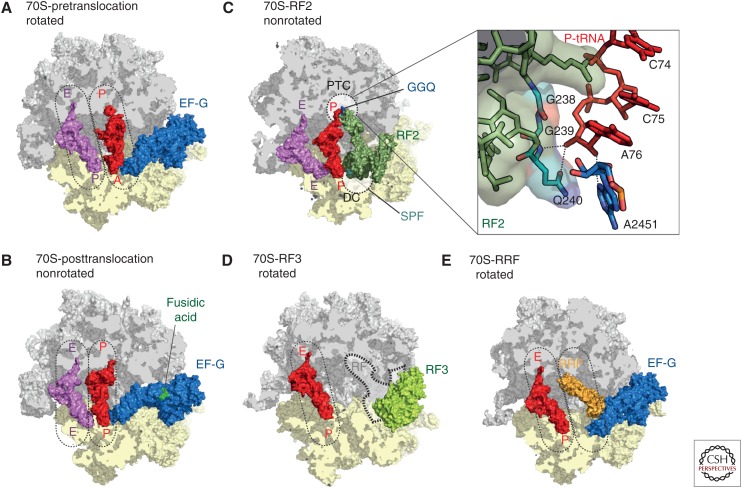
Figure 5.
The prokaryotic ribosome. Crystal structures of the ribosome bound with elongation factor G (EF-G) in the (A) pre- (Brilot et al. 2013), and (B) posttranslocation state (Lin et al. 2015), with 30S subunit (yellow), 50S subunit (gray), EF-G (blue), P-transfer RNA (tRNA) (red), E-tRNA (pink), and fusidic acid (green). (C) Crystal structure of RF2 (dark green) bound to the poststate ribosome with P-tRNA (red) and E-tRNA (pink) (Weixlbaumer et al. 2008). The peptidyltransferase center (PTC) on the 50S (gray) and the DC on the 30S subunit (yellow) are indicated as dashed lines. The insert zooms onto the PTC showing the GGQ motif of RF2 interacting with the CCA-end of the P-tRNA. (D) Crystal structure of RF3 (pale green) bound to the rotated 70S ribosome (Zhou et al. 2012b) with superimposed position of the P/E hybrid tRNA (red) (Brilot et al. 2013). The position of class I release factors is indicated with dashed lines. (E) Crystal structure of ribosome recycling factor (RRF, orange) bound to the rotated ribosome (Borovinskaya et al. 2007a) with superimposed positions of P/E hybrid tRNA (red) and prestate EF-G (blue) (Brilot et al. 2013).
It has been shown that EF-G binds to both the ratcheted and nonratcheted states of the ribosome (Chen et al. 2013; Pulk and Cate 2013), but translocation occurs via hybrid state formation for both scenarios. Rotation of the SSU head relative to the body (head swiveling) opens a constriction that allows the passage of the mRNA and ASLs through the SSU (Zhang et al. 2009; Ratje et al. 2010; Yamamoto et al. 2014). Domain IV of EF-G, overlapping the A-site on the SSU, is crucial to facilitate GTP-dependent translocation by disrupting interactions of the codon–anticodon duplex with the DC (Liu et al. 2014). Translocation of tRNAs through the ribosome proceeds via a series of intermediate states including intrasubunit hybrid states on the SSU, where the mRNA and ASLs simultaneously bind to the A- and P-site (ap/P) and to the P- and the E-site (pe/E) (Ratje et al. 2010; Ramrath et al. 2013). Following GTP hydrolysis and Pi release, the 30S head swivel and the ratcheting is reversed, EF-G dissociates and leaves the ribosome in the post-translocation (POST) state with tRNAs bound in the classical P/P and E/E sites (Fig. 5B) (Ratje et al. 2010; Ermolenko and Noller 2011).
There are a plethora of antibiotics that interfere with the process of translocation, including aminoglycosides, tuberactinomycins (viomycin, capreomycin), and spectinomycin (Wilson 2009), as well as more recently characterized translocation inhibitors, such as negamycin, amicoumacin A, and dityromycin (Figs. 2 and 3F) (Bulkley et al. 2014; Olivier et al. 2014; Polikanov et al. 2014a,c). Aminoglycosides, such as kanamycin and gentamycin, bind within h44 of the SSU (Fig. 3F) and stabilize the pretranslocation state (Wilson 2009), whereas aminoglycosides, such as neomycin, have an additional binding site located in H69 of the LSU, and appear to inhibit translocation by trapping intermediate hybrid states (Wang et al. 2012). Similarly, the binding site of viomycin and capreomycin span the ribosomal interface between h44 (Fig. 3F) and H69 (Stanley et al. 2010), and inhibit translocation by stabilizing a distinct intermediate hybrid state (Ermolenko et al. 2007b). In contrast, spectinomycin binds to the neck region of the SSU (Carter et al. 2000), locking a rotated conformation of the SSU head (Borovinskaya et al. 2007b) and, thus, trapping an intermediate state during translocation (Pan et al. 2007). Negamycin inhibits translocation by interacting with the A-tRNA and stabilizing it in the A-site (Olivier et al. 2014; Polikanov et al. 2014c), whereas amicoumacin A interacts with the mRNA in the E-site (Fig. 3F) and, thus, prevents translocation by preventing movement of the mRNA (Polikanov et al. 2014a). In contrast, dityromycin and the related antibiotic GE82832 interact exclusively with ribosomal protein S12 on the SSU (Fig. 3F) and block translocation by preventing EF-G from adopting the final state necessary for translocation of the tRNAs and mRNA on the SSU (Bulkley et al. 2014).
TERMINATION AND RECYCLING
Class I termination release factors 1 (RF1) or 2 (RF2) recognize mRNA stop codons in the A-site of the SSU and trigger translation termination by mediating hydrolysis of the P-site peptidyl-tRNA and subsequent peptide release (Fig. 2) (Klaholz 2011; Rodnina 2013). In a following step, the class I release factors are removed from the ribosome with the help of the GTPase class II release factor RF3 in a GTPase-dependent manner. RF1 and RF2 differ with respect to stop-codon recognition (RF1 recognizes UAG/UAA; RF2 recognizes UGA/UAA), but both mediate peptidyl-tRNA hydrolysis using their universally conserved GGQ-motif (Fig. 5C). Early structures revealed that ribosome binding induces conformational changes in RF1 and RF2 that stabilize an open conformation, which allows the concurrent insertion of the GGQ motif into the PTC on stop-codon recognition (Fig. 5C) (Klaholz 2011; Zhou et al. 2012a). Subsequent X-ray structures of RF1 and RF2 bound to the ribosome provided molecular insight into the structural determinants of specificity of stop-codon recognition (Korostelev et al. 2008; Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010). The exact mechanism by which the GGQ motif coordinates peptidyl-tRNA hydrolysis remains unclear. The amino acid side chain of Q of the GGQ motif is not essential to mediate hydrolysis (Seit-Nebi et al. 2001), but rather the backbone nitrogen of Q240 of RF2 (or Q230 for RF1) that accounts for the catalytic activity by interacting with the 3′ OH of A76 (Fig. 5C) (Laurberg et al. 2008). However, loss of the posttranslational N5-methylation of Q230/Q240 reduces the efficiency of peptide release (Dincbas-Renqvist et al. 2000) and, therefore, favors a model in which the glutamine side chain, together with the backbone amine, the 2′ OH of A76, and A2451, directly coordinate a water molecule for nucleophilic attack (Weixlbaumer et al. 2008; Jin et al. 2010; Klaholz 2011). After hydrolysis of the nascent chain, the class II release factor RF3 binds to the ribosome and promotes dissociation of RF1 and RF2 from the ribosome in a GTP-dependent manner (Freistroffer et al. 1997; Koutmou et al. 2014; Peske et al. 2014). RF3·GTP binding to RF1/RF2·ribosome complexes stabilizes the ratcheted state of the ribosome with tRNAs in hybrid states (Fig. 5D) (Ermolenko et al. 2007a; Gao et al. 2007; Jin et al. 2011; Zhou et al. 2012b). The ratcheted conformation of the ribosome creates a series of steric clashes between RF domains I and IV with the L11 stalk and the 30S head, respectively, which are supposed to contribute to the destabilization of the RFs (Fig. 5D) (Gao et al. 2007).
Following termination, the ribosome still contains mRNA and deacylated tRNA in the P-site, which needs to be recycled to allow a new round of translation. During recycling, the ribosome is split into subunits by the combined action of ribosome recycling factor (RRF) and EF-G in a GTP-dependent manner (Fig. 2) (Kaji et al. 2001). Recent crystal structures of RRF bound to the ribosome show RRF bound in the ribosomal P-site stabilizing the ratcheted conformation of the ribosome and would, therefore, presumably also stabilize a deacylated tRNA in a P/E hybrid state (Fig. 5E) (Weixlbaumer et al. 2007; Dunkle et al. 2011). Binding of EF-G to the RRF ·mRNA·tRNA·70S complex subsequently splits the ribosome into subunits on GTP hydrolysis (Peske et al. 2005; Zavialov et al. 2005; Barat et al. 2007). Binding of IF3 to the 30S subunit leads to dissociation of mRNA and deacylated tRNA and simultaneously prevents re-association with the 50S subunit (Zavialov et al. 2005) and thereby links the last steps in translation termination to the first steps of translation initiation (Fig. 2).
To date, there are no antibiotics that specifically target the termination and recycling phases of translation. Many antibiotics that act during elongation to inhibit factor binding, for example, thiostrepton, or prevent peptide bond formation, for example, chloramphenicol, also inhibit RF binding or peptidyl-tRNA hydrolysis (Wilson 2009). However, a few antibiotics have been suggested to act preferentially during termination, rather than elongation, namely, blasticidin S (Svidritskiy et al. 2013) and fusidic acid (Savelsbergh et al. 2009). Fusidic acid does not bind to the free form of EF-G, but rather when EF-G–GTP is in complex with the ribosome (Fig. 5B). Fusidic acid allows GTP hydrolysis but prevents the associated changes in EF-G that are necessary for dissociation and, thus, traps EF-G on the ribosome. Inhibition of EF-G’s function during ribosome recycling, rather than during elongation, has been reported to be more sensitive to the inhibitory action of fusidic acid (Savelsbergh et al. 2009). Blasticidin S binds to the P-site of the LSU and overlaps the binding position of C75 of the CCA-end of the P-tRNA (Fig. 4E) (Hansen et al. 2003; Svidritskiy et al. 2013). As expected, the presence of blasticidin S reduces the rate of peptide bond formation, but is even more effective at inhibiting peptidyl-tRNA hydrolysis by RF1 (Svidritskiy et al. 2013).
ACKNOWLEDGMENTS
The Wilson laboratory is supported by grants from the Deutsche Forschungsgemeinschaft (FOR1805, WI3285/4-1, and GRK1721 to D.N.W.).
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Allen G, Zavialov A, Gursky R, Ehrenberg M, Frank J. 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712. [DOI] [PubMed] [Google Scholar]
- Amunts A, Fiedorczuk K, Truong TT, Chandler J, Peter Greenberg E, Ramakrishnan V. 2015. Bactobolin A binds to a site on the 70S ribosome distinct from previously seen antibiotics. J Mol Biol 427: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK. 2007. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell 27: 250–261. [DOI] [PubMed] [Google Scholar]
- Belova L, Tenson T, Xiong LQ, McNicholas PM, Mankin AS. 2001. A novel site of antibiotic action in the ribosome: Interaction of evernimicin with the large ribosomal subunit. Proc Natl Acad Sci 98: 3726–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. 2004a. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S. 2004b. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci 101: 12893–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. 2007a. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14: 727–732. [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. 2007b. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol 2: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L, Fabbretti A, La Teana A, Abbondi M, Losi D, Donadio S, Gualerzi C. 2006. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc Natl Acad Sci 103: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L, Fabbretti A, Pon CL, Dahlberg AE, Gualerzi CO. 2008. Initiation of protein synthesis: A target for antimicrobials. Expert Opin Ther Targets 12: 519–534. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. 2013. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci 110: 20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103: 1143–1154. [DOI] [PubMed] [Google Scholar]
- Bulkley D, Johnson F, Steitz TA. 2012. The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge B2a of the ribosome. J Mol Biol 416: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulkley D, Brandi L, Polikanov YS, Fabbretti A, O’Connor M, Gualerzi CO, Steitz TA. 2014. The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation. Cell Rep 6: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. [DOI] [PubMed] [Google Scholar]
- Chen Y, Feng S, Kumar V, Ero R, Gao YG. 2013. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol 20: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Dallas A, Noller HF. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2013. New structural insights into the decoding mechanism: Translation infidelity via a G·U pair with Watson–Crick geometry. FEBS Lett 587: 1848–1857. [DOI] [PubMed] [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19: 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. 2004. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: The universally conserved residues G693 and C795 regulate P-site tRNA binding. Mol Cell 13: 113–124. [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci 107: 17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Noller HF. 2011. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 18: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. 2007a. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 370: 530–540. [DOI] [PubMed] [Google Scholar]
- Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. 2007b. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol 14: 493–497. [DOI] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. 2015. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520: 567–570. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal R. 2001. Ratchet-like movements between the two ribosomal subunits: Their implications in elongation factor recognition and tRNA translocation. Cold Spring Harb Symp Quant Biol 66: 67–75. [DOI] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. 1997. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J 16: 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. 2007. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129: 929–941. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Schmeing TM, Moore PB, Steitz TA. 2002. Structural insights into peptide bond formation. Proc Natl Acad Sci 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol 330: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. 2008. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell 30: 26–38. [DOI] [PubMed] [Google Scholar]
- Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci 110: 3812–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Ramakrishnan V. 2011. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci 108: 15798–15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M. 2011. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol 9: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji A, Kiel M, Hirokawa G, Muto A, Inokuchi Y, Kaji H. 2001. The fourth step of protein synthesis: Disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb Symp Quant Biol 66: 515–529. [DOI] [PubMed] [Google Scholar]
- Kannan K, Mankin AS. 2012. Macrolide antibiotics in the ribosome exit tunnel: Species-specific binding and action. Ann NY Acad Sci 1241: 33–47. [DOI] [PubMed] [Google Scholar]
- Kannan K, Vazquez-Laslop N, Mankin AS. 2012. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151: 508–520. [DOI] [PubMed] [Google Scholar]
- Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci 111: 15958–15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov M, Buckingham R, Ehrenberg M. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609. [DOI] [PubMed] [Google Scholar]
- Kim D, Green R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4: 859–864. [DOI] [PubMed] [Google Scholar]
- Klaholz BP. 2011. Molecular recognition and catalysis in translation termination complexes. Trends Biochem Sci 36: 282–292. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, McDonald ME, Brunelle JL, Green R. 2014. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA 20: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gagnon MG, Bulkley D, Steitz TA. 2015. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 160: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Song G, Zhang D, Zhang D, Li Z, Lyu Z, Dong J, Achenbach J, Gong W, Zhao XS, et al. 2014. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon–anticodon duplex. Nat Struct Mol Biol 21: 817–824. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148. [DOI] [PubMed] [Google Scholar]
- Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. 2014. Tetracycline antibiotics and resistance mechanisms. Biol Chem 395: 559–575. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177. [DOI] [PubMed] [Google Scholar]
- Olivier NB, Altman RB, Noeske J, Basarab GS, Code E, Ferguson AD, Gao N, Huang J, Juette MF, Livchak S, et al. 2014. Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome. Proc Natl Acad Sci 111: 16274–16279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. 2007. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 25: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina M, Wintermeyer W. 2005. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412. [DOI] [PubMed] [Google Scholar]
- Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. 2014. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res 42: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20: 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Osterman IA, Szal T, Tashlitsky VN, Serebryakova MV, Kusochek P, Bulkley D, Malanicheva IA, Efimenko TA, Efremenkova OV, et al. 2014a. Amicoumacin A inhibits translation by stabilizing mRNA interaction with the ribosome. Mol Cell 56: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Steitz TA, Innis CA. 2014b. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol 21: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Szal T, Jiang F, Gupta P, Matsuda R, Shiozuka M, Steitz TA, Vazquez-Laslop N, Mankin AS. 2014c. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol Cell 56: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Starosta AL, Juette MF, Altman RB, Terry DS, Lu W, Burnett BJ, Dinos G, Reynolds KA, Blanchard SC, et al. 2015. Distinct tRNA Accommodation intermediates observed on the ribosome with the antibiotics Hygromycin A and A201A. Mol Cell 58: 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Cate JH. 2013. Control of ribosomal subunit rotation by elongation factor G. Science 340: 1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. 2013. Visualization of two transfer RNAs trapped in transit during elongation factor G–mediated translocation. Proc Natl Acad Sci 110: 20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. 2010. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468: 713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinberger HJ, Sternbach H, Nierhaus KH. 1981. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci 78: 5310–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV. 2013. The ribosome as a versatile catalyst: Reactions at the peptidyl transferase center. Curr Opin Struct Biol 23: 595–602. [DOI] [PubMed] [Google Scholar]
- Roy RN, Lomakin IB, Gagnon MG, Steitz TA. 2015. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol 22: 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2015. Structural insights into the translational infidelity mechanism. Nat Commun 6: 7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung CS. 2005. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci 102: 15854–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W. 2009. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, Nierhaus KH, et al. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13: 871–878. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA. 2003. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9: 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. 2005. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438: 520–524. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVT, Weir JR, Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. 2009. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JH, Vila-Sanjurjo A. 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 13: 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt AC, Nguyen F, Antunes S, Perebaskine N, Graf M, Arenz S, Inampudi KK, Douat C, Guichard G, Wilson DN, et al. 2015. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol 22: 470–475. [DOI] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L. 2001. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res 29: 3982–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. 2000. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol 7: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Sievers A, Beringer M, Rodnina MV, Wolfenden R. 2004. The ribosome as an entropy trap. Proc Natl Acad Sci 101: 7897–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M. 2009. A structural view of translation initiation in bacteria. Cell Mol Life Sci 66: 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Billas IM, Tsai A, Fabbretti A, Myasnikov AG, Roblin P, Vaiana AC, Hazemann I, Eiler D, et al. 2013. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc Natl Acad Sci 110: 15656–15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA. 2009. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim Biophys Acta 1789: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohmen D, Harms JM, Schlunzen F, Wilson DN. 2009. Enhanced SnapShot: Antibiotic inhibition of protein synthesis II. Cell 139: 211e–212e. [DOI] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. 2010. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol 17: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinkeappel J, Brimacombe R, Wintermeyer W, Vanheel M. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389: 403–406. [DOI] [PubMed] [Google Scholar]
- Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. 2013. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci 110: 12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. 2002. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. 2003. Locking and unlocking of ribosomal motions. Cell 114: 123–134. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. 2013. Structural basis of the translational elongation cycle. Annu Rev Biochem 82: 203–236. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JH, Blanchard SC. 2012. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol 19: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. 2007. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14: 733–737. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees R, Petry S, Kelley A, Ramakrishnan V. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. 2009. The A–Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 44: 393–433. [DOI] [PubMed] [Google Scholar]
- Wilson DN. 2014. Ribosome-targeting antibiotics and bacterial resistance mechanisms. Nat Rev Microbiol 12: 35–48. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci 105: 13339–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. 2014. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife 3: e03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Qin Y, Achenbach J, Li C, Kijek J, Spahn CM, Nierhaus KH. 2014. EF-G and EF4: Translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol 12: 89–100. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. 2006. Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M. 2005. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell 18: 675–686. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH. 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Korostelev A, Lancaster L, Noller HF. 2012a. Crystal structures of 70S ribosomes bound to release factors RF1, RF2 and RF3. Curr Opin Struct Biol 22: 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Trakhanov S, Noller HF. 2012b. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 18: 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]