Abstract
The chromatin environment is modulated by a machinery of chromatin modifiers, required for the specification and maintenance of cell fate. Many mutations in the machinery have been linked to the development and progression of cancer. In this review, we give a brief introduction to Polycomb group (PcG) proteins, their assembly into Polycomb repressive complexes (PRCs) and the normal physiological roles of these complexes with a focus on the PRC2. We review the many findings of mutations in the PRC2 coding genes, both loss-of-function and gain-of-function, associated with human cancers and discuss potential molecular mechanisms involved in the contribution of PRC2 mutations to cancer development and progression. Finally, we discuss some of the recent advances in developing and testing drugs targeting the PRC2 as well as emerging results from clinical trials using these drugs in the treatment of human cancers.
PRC2 is involved in establishing and maintaining gene expression patterns and, in turn, cell fates. Activating and inactivating mutations in genes encoding PRC2 contribute to human cancers, as does PRC2 deregulation.
The genome of eukaryotic cells is organized into chromatin, consisting of DNA wrapped around an octamer of core histones to form nucleosomes. This organization serves both structural and functional purposes; organizing the genome into chromatin enables the linear genome to be packaged into the cell nucleus and protects the DNA strand from physical stresses. In addition, this organization allows specific areas of the genome to be condensed, thereby precluding the transcriptional machinery from gaining access to the underlying genes, while other areas in more open conformations can be actively transcribed, thereby facilitating cell-type specific gene expression patterns. Chromatin-associated proteins modulate the chromatin environment to help establish and maintain gene expression patterns over cell generations (Orkin and Hochedlinger 2011).
Deregulation of the chromatin environment can influence cell fate, and cancer cells often display disrupted chromatin environments with altered levels of various factors of the chromatin machinery. Recently, sequencing studies of human cancers have identified many somatic mutations in genes encoding chromatin-related factors, and intense research is ongoing to further decipher their involvement in cancer development and progression (You and Jones 2012).
The focus of this review is somatic mutations in genes coding for PRC2 subunits as well as recently discovered somatic mutations in the substrate for PRC2. We will discuss the potential roles of these mutations in the development of cancer. Before going into detail, however, we will give a general introduction to Polycomb group proteins, briefly outlining their biochemical and biological functions.
POLYCOMB GROUP PROTEINS
Polycomb group (PcG) proteins were originally identified in Drosophila as important regulators of fly development. The PcG proteins were shown to regulate the spatiotemporal expression pattern of important transcription factors (most notably, the Hox genes) during development, with PcG mutants showing characteristic phenotypes with defects in body segmentation. Orthologs have since been found in species ranging from plants to mammals, where they play key roles in establishing and maintaining correct gene expression patterns (Laugesen and Helin 2014).
POLYCOMB REPRESSIVE COMPLEXES
PcG proteins assemble into large multimeric protein complexes, of which the best characterized ones are Polycomb repressive complexes 1 and 2 (PRC1 and PRC2).
PRC1 complexes all contain either RING1A or RING1B with catalytic activity toward lysine 119 on histone H2A (H2AK119ub1), one of six Polycomb group RING finger (PCGF) subunits as well as additional subunits that define canonical PRC1 (CBX and PHC subunits) and noncanonical PRC1 (RYBP/YAP2) (Gao et al. 2012). The subunit composition affects recruitment to target genes and the catalytic activity of the complex (Blackledge et al. 2015). The mechanism by which PRC1 represses transcription is not entirely clear. Genetic studies have shown that it is only in part dependent on its ubiquitination activity (Illingworth et al. 2015; Pengelly et al. 2015), and, although PRC1 has been shown to promote chromatin compaction (Francis et al. 2004), the specific role of this activity in transcriptional repression remains unclear.
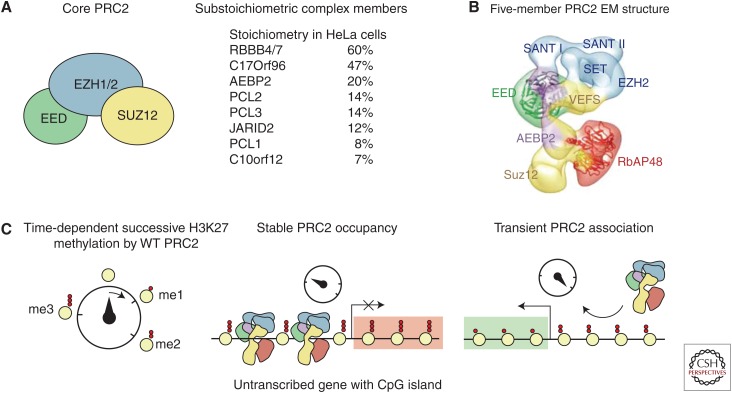
PRC2 is a methyltransferase with activity toward lysine 27 on histone H3 (H3K27). The SET-domain-containing component (EZH1 or EZH2) is closely associated with several other subunits. The core complex necessary for catalytic function consists of EZH1/2, the Zinc-finger protein SUZ12, and the WD40 protein EED, which can be purified from cells with equimolar stoichiometry (Cao et al. 2002; Kuzmichev et al. 2002; Pasini et al. 2004; Smits et al. 2013). The core complex is associated with several additional proteins (RBBP4/7, JARID2, AEBP2, PCL1-3, C17orf96, and C10or12) (Smits et al. 2013) as well as ncRNAs (Blackledge et al. 2015), and the different interaction partners are thought to play roles in regulation of PRC2 activity or recruitment to target genes (Fig. 1) (Di Croce and Helin 2013).
Figure 1.
PRC2 catalyzes H3K27 methylation. (A) The core PRC2 complex and substoichiometric interactors measured in HeLa cells (Smits et al. 2013). (B) Electron micrograph (EM) structure of a PRC2 complex with RBBP4 (RBAP48) and AEBP2 (EMDataBank 2236, deposited image) (Ciferri et al. 2012). (C) Graphic representation of time-dependent successive H3K27 methylation (Sneeringer et al. 2010). A longer residence time at CpG islands at promoters of untranscribed genes allows for establishment of a trimethylated domain. WT, Wild-type.
RECRUITMENT OF POLYCOMB REPRESSIVE COMPLEXES
While the recruitment of PcG proteins in Drosophila to DNA stretches termed Polycomb response elements (PREs) is believed to involve transcription factors (Orsi et al. 2014), the mechanisms governing the recruitment of mammalian PRCs are still unclear. Mammalian cells lack distinct PREs, but mammalian PRCs preferentially bind in CpG-rich contexts and CpG-rich sequences can mediate recruitment of PRCs (Tanay et al. 2007; Ku et al. 2008; Mendenhall et al. 2010; Lynch et al. 2012).
Several studies show that PRC1 and PRC2 binding patterns overlap, that these complexes maintain the repression of common target genes (Boyer et al. 2006; Bracken et al. 2006), and that PRC1 recruitment is dependent on PRC2 (Rastelli et al. 1993; Cao et al. 2002; Wang et al. 2004). This, along with the fact that PRC1 contains a subunit with affinity for H3K27me3 (the CBX component) (Cao et al. 2002; Fischle et al. 2003; Min et al. 2003), has led to the proposal of a hierarchical recruitment model. According to this model, PRC2 is recruited to chromatin, where it trimethylates H3K27, facilitating PRC1 recruitment and H2AK119 ubiquitination, leading to chromatin compaction and transcriptional repression (Blackledge et al. 2015). In recent years, however, this model has been challenged by several observations. First, in contrast to early studies from Drosophila and mammalian cells, PRC2 and H3K27me3 do not seem to be absolutely required for PRC1 recruitment and its activity (Leeb et al. 2010). This is partly explained by the discovery of “noncanonical” PRC1 lacking a CBX binding partner (Lagarou et al. 2008). Rather, recruitment of these complexes appears to rely on recognition of unmethylated CpG islands through the CXXC-domain of the KDM2B-component (Farcas et al. 2012; Gao et al. 2012; Qin et al. 2012; Tavares et al. 2012; Wu et al. 2013b). Second, recent results have shown that H2AK119ub1 is able to recruit and stimulate the activity of PRC2 (Blackledge et al. 2014; Cooper et al. 2014; Kalb et al. 2014), suggesting that the hierarchy under certain circumstances might be inverted. This suggestion still lacks experimental support, and while H3K27 methylation is essential for normal development and gene repression (Muller et al. 2002; Pengelly et al. 2013), H2AK119ub1 is not required for “global” H3K27 methylation, gene repression and early development in Drosophila and in part in mouse (Illingworth et al. 2015; Pengelly et al. 2015). Thus, while it appears that noncanonical PRC1 can be recruited to chromatin through the binding of KDM2B to CpG-rich stretches and canonical PRC1 can be recruited through binding to H3K27me3, it is currently unclear how PRC2 is recruited to chromatin. Although several of the PRC2 subunits or associated proteins have been shown to possess weak DNA- or RNA-binding activities and other subunits are known to interact with histones, no study has been able to pinpoint one factor being solely responsible for PRC2 recruitment (Blackledge et al. 2015). The recruitment of PRC2 will be discussed further in the next section.
PRC2 AND H3K27 METHYLATION
A low resolution EM structure of a five-member human PRC2 has been solved, which is consistent with a structural composition proposed by biochemical studies (Fig. 1B) (Ketel et al. 2005; Ciferri et al. 2012). Recently, the first crystal structure of an active PRC2 (from the yeast species Chaetomium thermophilum) was solved (Jiao and Liu 2015). A striking feature of the crystal structure is that an amino-terminal region of EZH2 makes extensive contacts with and is wrapped around EED (Fig. 3D). This loop is followed by a region, immediately before the first SANT domain, that is sandwiched by EED and the catalytic SET domain of EZH2. This “pre-SANT” region appears to have important regulatory roles for the enzymatic activity. Previous data have shown that PRC2, more specifically the WD40 domains of EED, have an affinity for H3K27me3, suggesting a potential mechanism for the spreading of H3K27me3 over larger genomic regions (Hansen et al. 2008; Margueron et al. 2009; Xu et al. 2010). Based on the structure, it seems likely that binding of H3K27me3 to EED leads to a conformational change, which, through the pre-SANT region, affects the SET domain of EZH2 and could explain the increased activity of the PRC2 complex observed in the presence of H3K27me3 (Margueron et al. 2009; Xu et al. 2010; Jiao and Liu 2015). The large interaction surfaces observed from the structure also provide an explanation for the stable nature of the PRC2 complex—and why some of the individual components are unstable when not in the complex (Montgomery et al. 2005; Pasini et al. 2007).
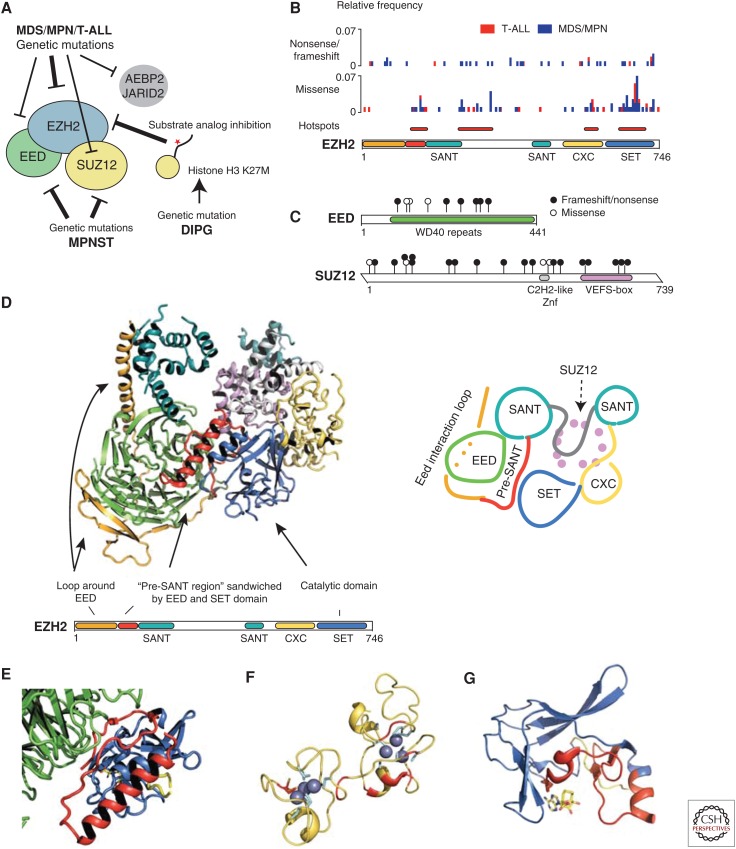
Figure 3.
Loss-of-function PRC2 mutations in cancer. (A) Mutations conferring loss of PRC2 function are recurrent in several cancer types. The mutations are found in the genes coding for PRC2 core subunits or substoichiometric interaction partners, as well as in the genes coding for H3.1 and H3.3. (B) Histogram of EZH2 mutants found in T-cell acute lymphoblastic leukemia (T-ALL) and myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) (Ernst et al. 2010; Nikoloski et al. 2010; Abdel-Wahab et al. 2011; Bejar et al. 2011; Guglielmelli et al. 2011; Jankowska et al. 2011; Score et al. 2012; Muto et al. 2013; Lindsley et al. 2015). The missense mutations cluster in four hotspot regions. (C) Mutations in EED and SUZ12 found in malignant peripheral nerve sheath tumors (MPNSTs) (De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014). (D) Crystal structure of Ezh2 and Suz12 VEFS domain from C. thermophilum (PDB 5CH1) (Jiao and Liu 2015) with Eed replaced by superimposed human EED (PDB3IIW) (Margueron et al. 2009). Right panel shows a schematic representation of the structure. (E) Loss-of-function mutations are frequently found in the part of EZH2 coding for the 50 amino acid pre-SANT region. In the structure D, the pre-SANT region (red) is sandwiched between EED (green) and the catalytic SET domain of Ezh2 (blue), and mutations likely affect SET domain function directly or perturb a putative allosteric communication between EED and Ezh2. (F) Loss-of-function mutations are frequently found in the part of EZH2 coding for the CXC domain. It has nine cysteines (light blue) that coordinate zinc ions in two separate clusters. All missense mutations (red) in this domain map to these clusters. Structure of the human CXC domain is used (PDB 4MI0) (Wu et al. 2013a). (G) The hotspot region for EZH2 mutations in the SET domain maps to the active site and surrounding binding pockets for substrate and cofactor. Structure used is of human SET domain (PDB 4MI0) with substrate and cofactor from EHMT1 superimposed (PDB 2RFI) (Wu et al. 2010). DIPG, Diffuse intrinsic pontine glioma.
The different methylation states of H3K27 (H3K27me1, H3K27me2, and H3K27me3) are found in different genomic contexts. ChIP-sequencing studies and mass spectrometry of histones isolated from embryonic stem cells show that around 5%–10% of histones carry H3K27me1, and this modification is primarily found within the gene bodies of actively transcribed genes (Ferrari et al. 2014). H3K27me2 is very abundant (about 50%–70% of histones have this modification) and is found in large domains, where it is proposed to exert a protective function against inappropriate transcription or enhancer activity (Jung et al. 2010, 2013; Ferrari et al. 2014). In contrast to H3K27me1 and H3K27me2, the distribution of H3K27me3 overlaps well with PRC2-binding patterns, and 5%–10% of histones carry this modification (Peters et al. 2003; Jung et al. 2010, 2013; Ferrari et al. 2014). These observations bring up several interesting points: The fact that PRC2 catalyzes all the different degrees of H3K27 methylation and H3K27me1 being associated with actively transcribed genes is hard to reconcile with the classic understanding of PRC2 as a transcriptional repressor. Furthermore, the fact that PRC2 binding is only found to overlap with H3K27me3 raises questions about the dynamics and rates of the deposition of each stage of H3K27 methylation, and might help us better understand PRC2 recruitment. With 70% of the genome being methylated on H3K27 by PRC2, it is unlikely that the bulk of PRC2 activity is directed by specific features in DNA sequence or composition. Mono- and dimethylation of H3K27 is reestablished very fast in newly incorporated histones in each cell cycle and show no sequence specificity. In contrast, the reestablishment of H3K27me3 is relatively slow (Zee et al. 2012; Alabert et al. 2015). Genome-wide location analyses have shown that PRC2 is highly enriched at CpG islands, whereas no enrichment of PRC2 is observed at H3K27me1/me2-positive regions. These results are in agreement with in vitro data showing that PRC2 is slower at converting H3K27me2 to H3K27me3 than catalyzing mono- and dimethylation of H3K27, and that it is therefore thought to need longer residence time to catalyze trimethylation (Sneeringer et al. 2010; Yap et al. 2011). Taken together, the current available data suggest that PRC2 interacts with chromatin in a manner independent of the underlying DNA sequence, facilitating mono- and dimethylation of H3K27, whereas conversion to H3K27me3 requires more stable binding of PRC2 (Fig. 1C). This might be achieved by interaction with sequence-specific or CpG island-associated proteins, which in the latter case might even be an intrinsic property of the PRC2 complex. No PRC2 binding is observed at CpG islands by active genes, but appears at many such sites following drug-induced transcriptional blockage. This implies that the transcriptional machinery and associated proteins prevent stable binding of PRC2, which otherwise occurs by default (Riising et al. 2014).
PRC2 AND TRANSCRIPTIONAL REGULATION
While some studies indicate that PRC2 is bound to the stalled promoters of both coding and noncoding transcripts (Enderle et al. 2011; Brookes et al. 2012) with abortive transcripts potentially playing a role in recruitment (Kanhere et al. 2010), one study found the presence of PcG proteins to block transcriptional initiation altogether (Dellino et al. 2004; Kaneko et al. 2014), and it was recently demonstrated that PRC2 is recruited upon transcriptional repression rather than being required for setting up transcriptional repression (Riising et al. 2014). In agreement with the latter results, studies in Drosophila have shown that PRC2 is dispensable for initiating transcriptional repression of homeotic genes, but rather it is required for maintaining the repression of these genes during later stages of development (Struhl and Akam 1985; Jones and Gelbart 1990; Simon et al. 1992).
Whether loss of PRC2 function by itself leads to derepression of its target genes has been debated. Studies of embryonic stem cells lacking functional PRC2 initially showed a certain degree of derepression of PRC2 target genes (Montgomery et al. 2005; Pasini et al. 2007; Chamberlain et al. 2008; Shen et al. 2008). However, subsequent studies from our laboratory in more stable environments (ES cells grown in defined medium) showed that PRC2 loss does not lead to changes in transcription, indicating that the deregulation observed in earlier studies stems from outside stimuli (Riising et al. 2014). Based on these studies, we proposed that, rather than being recruited to target genes to promote their repression, lack of transcription allows a longer residence time of PRC2 at the promoters of repressed genes. Here, PRC2 contributes to maintaining repression of these genes by setting thresholds for their activation, and in this way contributes to preserving cell identity. In accordance with this, loss of PRC2 function should not in itself lead to increased transcription. It would, however, make the chromatin environment more sensitive to stimuli, and abolishing PRC2 function, therefore, greatly influences cells undergoing changes to their transcriptional program, as occurring during cell-fate transitions.
PRC2 IN STEM CELLS AND DEVELOPMENT
As initially observed in Drosophila, PcG mutants display abnormal development in a range of species. Knockout of Ezh2, Eed, or Suz12 in the mouse leads to early embryonic lethality with the embryos failing to undergo gastrulation (Schumacher et al. 1996; O’Carroll et al. 2001; Pasini et al. 2004). Studies of conditional and tissue-specific knockout mice have shown important roles of PRC2 in orchestrating cell-fate transitions in the development of a range of tissues (Laugesen and Helin 2014). Loss of PRC2 integrity impairs the stability of cell identity and makes the differentiation process more labile. With PRC2 being important for balancing differentiation versus proliferation, it is not surprising that PRC2 mutations or misregulation can skew this balance, leading to adverse outcomes including cancer development.
MISREGULATION OF PRC2 IN CANCER
The first observations implicating PRC2 in cancer came from the discoveries that EZH2 is overexpressed and/or its locus amplified in several types of cancer, correlating with poor prognosis, and from knockdown-studies showing that PRC2 is required for proliferation of these cancer cells (Varambally et al. 2002; Bracken et al. 2003; Kleer et al. 2003). Subsequently, a plethora of studies have shown misregulation of PRC2 in human cancers, with PRC2 overexpression often correlating to poor prognosis (Deb et al. 2014; Jiang et al. 2015). Importantly, because of the cell-cycle-coupled expression of PRC2 components (Bracken et al. 2003), and PRC2 expression being high in the stem-cell compartment, increased PRC2 expression in cancer cells may stem from the increased proliferative capacity and/or dedifferentiated phenotype of cancer cells. It is, therefore, difficult to judge whether increased expression levels are a cause or a consequence of oncogenesis. It is clear, however, that many cancers depend on PRC2 expression for proliferation (discussed below). Along with the stronger evidence from EZH2 amplifications (Bracken et al. 2003), the discovery of hyperactivating EZH2 mutants, and the fact that these mutations were found to be an early event through studies of clonality (Bodor et al. 2013), these observations collectively show that increased PRC2 activity is certainly acting as a driver in some cancers. This has recently been supported by numerous sequencing studies that have found the genes encoding the PRC2 members to be mutated in many different cancers. Surprisingly, however, both loss- and gain-of-function mutations have been identified, indicating that PRC2 acts as a tumor suppressor in some cancers and an oncogene in others (Table 1).
Table 1.
Mutations in genes coding for PRC2 subunits in cancer
| Gene | Cancer | Notes | References |
|---|---|---|---|
| Hyperactivating mutations | |||
| EZH2 | DLBCL | Y646N/F/S/H/C A682G A692V |
Morin et al. 2010, 2011; Lohr et al. 2012; McCabe et al. 2012a; Okosun et al. 2014 |
| FL | Y646N/F/S/H/C A682G A692V |
Morin et al. 2010, 2011; Bodor et al. 2011, 2013; McCabe et al. 2012a; Okosun et al. 2014 | |
| NHL (“other”) | Y646F/S A692V |
Morin et al. 2011 | |
| B-ALL (patient-derived cell lines) | Y646N A692V |
Ott et al. 2014 | |
| Sporadic parathyroid adenomas | Y646N | Cromer et al. 2012 | |
| Melanoma | Y646N/F/S | Hodis et al. 2012; Krauthammer et al. 2012; Alexandrov et al. 2013; Zingg et al. 2015 | |
| Loss-of-function mutations | |||
| EZH2 | Myeloid malignancies, including MDS, MPN, MF, CMML, and AML | Correlated with poor survival | Ernst et al. 2010; Nikoloski et al. 2010; Abdel-Wahab et al. 2011; Bejar et al. 2011; Guglielmelli et al. 2011; Jankowska et al. 2011; Score et al. 2012; Muto et al. 2013; Lindsley et al. 2015; |
| T-ALL | Ntziachristos et al. 2012; Neumann et al. 2015 | ||
| EED | MDS/MPN | Score et al. 2012 | |
| MPNST | De Raedt et al. 2014; Lee et al. 2014 | ||
| GBM | De Raedt et al. 2014 | ||
| Melanoma | De Raedt et al. 2014 | ||
| SUZ12 | MDS/MPN | Score et al. 2012 | |
| T-ALL | Ntziachristos et al. 2012; Simon et al. 2012; Neumann et al. 2015 | ||
| MPNST | De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014 | ||
| GBM | De Raedt et al. 2014 | ||
| Melanoma | De Raedt et al. 2014 | ||
| JARID2 | MDS/MPN | Puda et al. 2012; Score et al. 2012 | |
| T-ALL | Simon et al. 2012 | ||
| AEBP2 | MDS/MPN | Puda et al. 2012 | |
AML, Acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CMML, chronic myelomonocytic leukemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; GBM, glioblastoma multiforme; MDS, myelodysplastic syndrome; MF, myelofibrosis; MPN, myeloproliferative neoplasm; MPNST, malignant peripheral nerve sheath tumor; NHL, non-Hodgkin lymphoma; T-ALL, T-cell acute lymphoblastic leukemia.
HYPERACTIVATING CATALYTIC MUTANTS OF EZH2
Several studies have identified recurring somatic point mutations in the sequence coding for the SET domain of EZH2 in follicular lymphoma (FL) and the germinal center B-cell (GCB) subset of diffuse large B-cell lymphoma (DLBCL) (Fig. 2). The most frequent of the hyperactivating mutations leads to a substitution of a tyrosine in the SET domain (Y646F/N/S/H/C, initially referred to as Y641), occurring in up to 25% of DLBCL and FLs (Morin et al. 2010, 2011; Bodor et al. 2013; Okosun et al. 2014). Although this mutant was originally reported to inactivate EZH2 activity as assessed by in vitro methylation assays (Morin et al. 2010), subsequent studies have demonstrated that it is a gain-of-function mutation with hyperactive conversion rate of H3K27 di- to trimethylation, and the mutant does indeed confer greatly increased levels of H3K27me3 (Sneeringer et al. 2010; Bodor et al. 2011, 2013; Yap et al. 2011).
Figure 2.
Hyperactive EZH2 mutants in DLBCL and FL. (A) Histogram of EZH2 mutation frequencies in FL and DLBCL. These map to three residues in the catalytic SET domain. (B) The mutated residues (light blue) are found in the active catalytic site of EZH2 near substrate and cofactor (yellow). Y646 coordinates the substrate lysine’s side-chain amine and impedes rotation to allow transfer of the third methyl group, while A682 stabilizes the position of Y646. The crystal structure of the human EZH2 SET domain (PDB 4MI0) (Wu et al. 2013a) is used with substrate and cofactor from EHMT1 superimposed (PDB 2RFI) (Wu et al. 2010). (C) Combined activities of wild-type (WT) and Y646 mutant EZH2 achieves faster H3K27 trimethylation (Swalm et al. 2014).
The initial misperception concerning the biochemical properties of the EZH2-Y646 mutant stems from the fact that the mutant enzyme is very inefficient at the conversions of unmethylated H3K27 to H3K27me1 and of H3K27me1 to H3K27me2. If a wild-type EZH2 is present, however, allowing the mutant enzyme to use H3K27me2 as its substrate, they together very efficiently catalyze the conversion to H3K27me3 (Fig. 2C) (Swalm et al. 2014). Accordingly, the somatic mutation leading to the change of Y646 is always monoallelic and a wild-type allele of EZH2 is always present in these cancers (Sneeringer et al. 2010). Interestingly, this does not seem to be the case for another characterized hyperactivating mutant (A682G), found in 1%–2% of FL, which catalyzes all the steps at a high rate and, consequently, does not seem to require wild-type EZH2 in vitro (Morin et al. 2011; McCabe et al. 2012a). A third recurrent point mutant in the SET domain of EZH2 (A692V) associated with lymphoma also leads to increased H3K27me3, but as it appears to prefer H3K27me1 as its substrate with little-to-no enhanced catalytic activity toward H3K27me2, it does not lead to a global depletion of H3K27me2 as seen in cells expressing Y646 or A682 mutants (Morin et al. 2011; Majer et al. 2012; Bodor et al. 2013; Ott et al. 2014). Interestingly, this mutant is also detected in a patient-derived B-cell acute lymphoblastic leukemia (B-ALL) cell line, and treatment with an EZH2 inhibitor (GSK126) leads to growth arrest and apoptosis (Ott et al. 2014). The same study found another B-cell ALL cell line to contain the Y646 substitution (Ott et al. 2014), and this mutant has also been found in sporadic parathyroid adenomas (Cromer et al. 2012) and in melanomas (Hodis et al. 2012), demonstrating that hyperactive EZH2 mutants are not exclusive to lymphoma. Crystal structures of the human EZH2 SET domain (Antonysamy et al. 2013; Wu et al. 2013a) show that Y646 is placed so that the tyrosine hydroxyl group can make hydrogen bond coordination to the substrate’s terminal amine nitrogen. This could impede free rotation needed to orient the nitrogen properly for transfer of the third methyl group from S-adenosylmethionine (SAM) (Fig. 2B). A682 likely holds Y646 in place, and mutation to glycine may loosen the pocket to decrease suppression of the third methylation step. A692 is also located in the active site, although it is not as clear what exact structural changes occur when it is mutated to valine.
In accordance with Ezh2 being specifically expressed at this stage of B-cell development (Velichutina et al. 2010), Ezh2 is required for the formation of germinal centers. Expression of hyperactive Ezh2 (Ezh2-Y646N) in GC-B cells induces GC hyperplasia, and this hyperplasia is reverted by treatment with an EZH2 inhibitor. Knockin of an Ezh2 allele expressing Ezh2-Y646N alone does not give rise to lymphomas, but collaboration with Bcl2 and sustained activation of GC formation leads to GC-derived lymphomas (Beguelin et al. 2013). Similarly, expressing Ezh2-Y646F from a lymphocytic promoter does not in itself lead to tumor formation. However, crossing these mice with the Eμ-Myc transgenic mice markedly increases the rate of lymphoma formation in the offspring, and the resulting cancers can give rise to tumors in secondary recipients in transplantation experiments (Berg et al. 2014). Collectively, these studies show that Ezh2-Y646N/F can collaborate with known oncogenes to accelerate tumorigenesis.
Both DLBCL and FL arise from GC B cells. As mentioned above, Ezh2 is required for GC formation with Ezh2 levels increasing as B cells enter the GC reaction and then decreasing upon exiting, enabling the expression of genes mediating terminal differentiation (Velichutina et al. 2010; Beguelin et al. 2013). Hyperactive PRC2 in the GC appears to inhibit terminal differentiation, keeping cells in a more stem cell–like state, and allowing continuous rapid proliferation. Thus, the normal physiological role of PRC2 in GC B cells is related to keeping these cells from terminally differentiating—an observation that is mimicked in several other tissues, where PRC2 is often active in the stem-cell compartment and downregulated upon differentiation, seemingly guarding the balance between self-renewal and differentiation (Laugesen and Helin 2014).
LOSS-OF-FUNCTION MUTATIONS IN GENES ENCODING PRC2 MEMBERS
Loss-of-function mutations in EZH2 in human cancers were first identified in myeloid malignancies (Ernst et al. 2010; Nikoloski et al. 2010). The observed mutations range from chromosomal loss over insertions/deletions to single nucleotide substitutions leading to mis- or nonsense amino acid substitutions, most of them believed to abolish PRC2 activity (Ernst et al. 2010). Subsequently, loss-of-function mutations in genes encoding core PRC2 members (EZH2, SUZ12, EED) and accessory factors (JARID2, AEBP2) have been identified in various myeloid malignancies (Ernst et al. 2010; Nikoloski et al. 2010; Jankowska et al. 2011; Puda et al. 2012; Score et al. 2012; Lindsley et al. 2015) with loss of EZH2 correlating with poor prognosis (Bejar et al. 2011; Guglielmelli et al. 2011), as well as in T-cell lymphoblastic leukemia (T-ALL) (Ntziachristos et al. 2012; Simon et al. 2012; Neumann et al. 2015), malignant peripheral nerve sheath tumor (MPNST) (De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014), and melanoma and glioblastoma multiforme (GBM) (De Raedt et al. 2014).
In myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) and T-ALL, PRC2 is most frequently targeted through mutations of EZH2. This includes mutations leading to homozygous amino-terminal frame shifts, and thus complete loss of EZH2 is tolerated and maybe beneficial for these cancers. Missense mutations are also frequent, and are mostly found in the part of the gene coding for the catalytic domain, but three other regions of the gene appear to have a significant number of mutations clustering as well (Fig. 3B). The first hotspot region lies prior to the first SANT domain. Although sequence homology is poor between the crystallized PRC2 from C. thermophilum and human PRC2, it is plausible that an analogous pre-SANT region sandwiched between EED and the SET domain exists, and mutations may directly affect the SET domain or the allosteric communication from EED (Fig. 3E). The second hotspot region is immediately after the first SANT domain, but the crystal structure currently gives little clues as to how this can affect PRC2 function. Mutations in the CXC domain map to the two zinc-binding clusters and likely disrupt the (unknown) function of the CXC domain (Fig. 3F). In the SET domain mutants, the alterations map to the immediate proximity of the active site where catalysis takes place (Fig. 3G). In MDS/MPN and T-ALL, mutations in EED and SUZ12 are less frequent (1%–2%), but they do occur (Ntziachristos et al. 2012; Score et al. 2012; Simon et al. 2012; Neumann et al. 2015) and are further evidence that PRC2 disruption is the driving event. Missense mutations in SUZ12 map to the VEFS domain, which mediates the binding to EZH2.
In some cancers, both alleles of a gene encoding a PRC2 member carry loss-of-function mutations, leading to complete abrogation of PRC2 activity, while other patients carry one wild-type allele. A recent study of MPNST patients found that 79% with and 34% without NF1 microdeletions are homozygous for loss of either SUZ12 or EED (De Raedt et al. 2014). In an Nf1+/−; Trp53+/− mouse model, heterozygous loss of Suz12 significantly accelerated the development of MPNST and other cancer types, including high-grade glioma (De Raedt et al. 2014). MPNST appears to frequently be derived from a neurofibroma with monoallelic NF1 deletions that include SUZ12 (neighboring gene), and the second SUZ12 mutation drives the event. This observation provides a possible explanation to why SUZ12 is frequently the targeted PRC2 member in MPNST.
It was recently shown that knockout of Suz12 or Eed leads to defects in hematopoietic stem cell (HSC) self-renewal, in part through derepression of the Cdkn2a locus, and that PRC2 function is required for lymphoid development (Xie et al. 2014; Lee et al. 2015). Conversely, partial loss of PRC2 function leads to increased proliferative capacity of HSCs (Lessard et al. 1999; Richie et al. 2002; Majewski et al. 2010; Lee et al. 2015), highlighting the sensitivity of these cells to PRC2 dosage.
Several mouse models have shown tumor suppressive roles of PRC2 in hematological malignancies, with Ezh2 knockout leading to spontaneous development of T-ALL and MDS/MPN (Simon et al. 2012; Mochizuki-Kashio et al. 2015). Importantly, deletion of Ezh1 alone does not cause any hematological malignancies, and concomitant loss of Ezh1/Ezh2 abolishes the myoproliferative effects of Ezh2 loss, indicating that the phenotype of Ezh2 knockouts might rely on Ezh1 compensating to uphold some residual PRC2 activity (Mochizuki-Kashio et al. 2015). In addition, loss of Ezh2 was found to accelerate and exacerbate the development of MDS when deleted in conjunction with Tet2 (Muto et al. 2013), and in a Runx1 mouse model, Ezh2 knockout promotes MDS development but prohibits malignant transformation to AML (Sashida et al. 2014). Interestingly, heterozygous loss of Suz12 was found to accelerate lymphomagenesis in Eμ-myc mice (Lee et al. 2013), a striking observation, since hyperactive Ezh2 was found to induce a similar phenotype (Berg et al. 2014). Furthermore, several studies have shown that deletion or depletion of PRC2 members impedes MLL-AF9-driven AML (Neff et al. 2012; Tanaka et al. 2012; Shi et al. 2013; Danis et al. 2015), and overexpression of Ezh2 leads to myeloproliferative disorders (Herrera-Merchan et al. 2012). Taken together, this underlines the complex nature of PRC2 function in disease progression, and indicates that the consequence of PRC2 loss is highly dependent on the cellular context and exact developmental stage at which the loss occurs, compromising the intricate balance between self-renewal and differentiation. The various observations indicate that HCSs are dependent on a functional PRC2 to prevent activation of the Cdkn2a locus, whereas loss-of-function of one allele coding for a core component of PRC2 predisposes to the development of a hematopoietic malignancy. If these malignancies retain an intact Cdkn2a locus, they could therefore still be dependent on PRC2 activity and might thus respond to treatment with EZH2 inhibitors (see below).
THE SIGNIFICANCE OF H3K27 METHYLATION
Drosophila studies have shown that the catalytic activity of PRC2 is required to prevent derepression of target genes (Muller et al. 2002), and a histone mutant abolishing H3K27 methylation phenocopies of Polycomb mutants, strongly suggesting that H3K27 methylation is the essential physiological function of PRC2 (Pengelly et al. 2013). Additional data from Drosophila show that a mutant with an amino acid substitution (R741K) in the SET domain of E(z) (the Drosophila homolog of EZH2) confers hyper-trimethylation activity of PRC2 with increased levels of H3K27me3 and decreased levels of H3K27me1/me2. Importantly, this increased trimethylation leads to inappropriate repression of PRC2 target genes with phenotypes mimicking those of loss-of-function mutations in trithorax (trx), encoding the Drosophila enzyme orthologous to the MLL H3K4 methyltransferases and key antagonist of Polycomb silencing (Stepanik and Harte 2012). Although the mutation conferring hypertrimethylation of H3K27 is distinct from the mutations observed in mammalian cancers, the data from Drosophila show that increased H3K27me3 can indeed cause inappropriate silencing of PRC2 target genes with consequences for cell-fate decisions.
The discovery of cancers with increased levels of H3K27me3 from hyperactivating mutations in EZH2 point to a definite role of the catalytic activity of PRC2 and particularly H3K27me3 in cancers where PRC2 is playing an oncogenic role. In a case of PRC2 acting as a tumor suppressor, patients with benign neurofibromas retaining some PRC2 function, staining for H3K27me3 is lost upon progression to MPNST (Lee et al. 2014), indicating that complete loss of PRC2 function and H3K27me3 is contributing to malignant transformation in these cancers. In line with this, loss of H3K27me3 was recently shown to be a robust diagnostic marker for distinguishing sporadic and radiation-induced MPNSTs from other cancers in the differential diagnosis for MPNST (Prieto-Granada et al. 2015). In addition, the demethylases responsible for removing H3K27me3 are also deregulated in human cancers with KDM6A/UTX mutations in several cancer types (van Haaften et al. 2009; Gui et al. 2011; Jankowska et al. 2011; Ross et al. 2014; Van der Meulen et al. 2015) and KDM6B/JMJD3 overexpression in Hodgkin’s lymphoma and T-ALL associated with loss of H3K27me3 (Anderton et al. 2011; Simon et al. 2012).
Seemingly direct implications of the histone substrate of PRC2 in cancer development have come from recent findings of somatic mutations, causing a lysine-to-methionine substitution at residue 27 in the H3 tail in genes encoding histone variants H3.1 and H3.3 in pediatric brain tumors (Schwartzentruber et al. 2012; Wu et al. 2012). Global levels of H3K27me3 are diminished in these tumors, putatively because of the inhibitory effect of H3K27M on PRC2 activity (Bender et al. 2013; Chan et al. 2013; Lewis et al. 2013; Venneti et al. 2013), and these mutants might primarily exert their tumorigenic function through their conferred loss-of-function of PRC2. Despite the global reduction in H3K27me3, PRC2 occupancy and H3K27me3 are specifically enriched at certain PRC2 target genes, correlating with lower expression of these genes. Conversely, PRC2 target genes with lower levels of H3K27me3 in cells carrying the H3K27M mutant were found to have reduced levels of DNA methylation and increased expression levels, indicating that the functional consequence of H3K27M mutation includes aberrant transcriptional activation as well as defective repression of PRC2 targets (Bender et al. 2013; Chan et al. 2013).
HOW DOES DEREGULATED PRC2 ACTIVITY CONTRIBUTE TO TUMORIGENESIS?
PRC2 is important for balancing proliferation versus differentiation, and PRC2 has been shown to promote a de-differentiated phenotype of several cancers (Richter et al. 2009; Tanaka et al. 2012; Beguelin et al. 2013). Increased PRC2 activity (be it through overexpression or hyperactive mutants) in stem and progenitor cells might promote self-renewal over differentiation by increasing the thresholds for transcriptional activation of differentiation-associated PRC2 target genes or genes controlling cell proliferation such as CDKN2A. A central role of CDKN2A in relation to PRC2 mutants is supported by findings that knockout of the Cdkn2a locus in combination with Eed knockout partially rescues the growth phenotype of both HSCs (Xie et al. 2014) and leukemic cells in an MLL-AF9 mouse model (Shi et al. 2013; Danis et al. 2015), as well as the co-occurrence of CDKN2A mutations and loss-of-function mutations in EED and SUZ12 in MPNSTs (Lee et al. 2014) and the requirement of PRC2-mediated repression of CDKN2A for proliferation of rhabdoid tumors (Kia et al. 2008).
Another plausible explanation is that deregulation of PRC2 activity in either direction alters thresholds for gene activation, promoting epigenetic instability and giving rise to transcriptional deregulation, thereby increasing the risk of cancer development (Brock et al. 2015). In addition to the direct misregulation of CDKN2A in some cancers with PRC2 alterations, this explanation might provide a more general model of how deregulated PRC2 activity promotes oncogenesis. This would also explain the difficulties with identifying key deregulated genes or conserved gene expression signatures in cancers dependent on PRC2 and with altered PRC2 activity.
TARGETING PRC2 IN CANCER
Knockdown studies in cancer cells with increased PRC2 levels show that many of these cancers depend on PRC2 for proliferation (Albert and Helin 2010). Along with the discovery of mutations in EZH2 leading to hyperactive PRC2, this has prompted the hope of therapeutically targeting PRC2 in cancer. Several companies have developed small molecule inhibitors specific for EZH2, and their efficiency in targeting lymphoma cells harboring hyperactivated EZH2 has been demonstrated both in vitro and in preclinical mouse models (Knutson et al. 2012, 2014a; McCabe et al. 2012b; Qi et al. 2012; Beguelin et al. 2013; Garapaty-Rao et al. 2013; Bradley et al. 2014; Campbell et al. 2015). The inhibitors are all S-adenosylmethionine (SAM, cofactor) competitive inhibitors, and, perhaps because the hyperactivity-conferring mutations map in the substrate-binding pocket (Fig. 2B), these inhibitors are not selective for mutant versus wild-type EZH2.
Three companies (Epizyme, Cambridge, MA; GlaxoSmithKline, Parsippany, NJ; and Constellation Pharmaceuticals, Cambridge, MA) have moved their EZH2 inhibitors forward into phase 1 clinical trials in the last couple of years. Whereas the phase 1 clinical trials are primarily aimed at establishing acceptable safety profiles for the compounds, the three companies have all pointed at B-cell lymphomas with hyperactive mutant EZH2 as the primary indication to target. Recently, the FDA approved that Epizyme can initiate phase 2 clinical trials for the treatment of patients with relapsed DLBCL (Epizyme, press release). Interestingly, the Epizyme EZH2 inhibitor Tazemostat (also known as EPZ-6438) has shown promising antitumor activity in nine out of 15 DLBCL patients of which only one patient expressed hyperactive mutant EZH2 (Epizyme, press release). This intriguing finding is in line with several studies suggesting that targeting EZH2 might be a relevant therapeutic strategy in cancers without hyperactivated EZH2. In addition to EZH2 inhibition effectively targeting lymphomas without hyperactivated EZH2 (Beguelin et al. 2013; Bradley et al. 2014), these studies have shown that EZH2 inhibition shows synthetic lethality with mutations in components of the SWI/SNF remodeling complex (Wilson et al. 2010; Knutson et al. 2013; Bitler et al. 2015; Fillmore et al. 2015; Kim et al. 2015) BAP1 (LaFave et al. 2015), and potentially also with mutations in UTX (Van der Meulen et al. 2015).
With PRC2 mutational status not being sufficient to decide whether a patient will respond to EZH2 inhibition, identification of robust biomarkers that can be used to stratify patients correctly becomes a major challenge. As mentioned above, PRC2 mutations may primarily contribute to oncogenesis through a destabilization of the chromatin environment by altering the thresholds for gene activation and increasing transcriptional noise. Thus, the gene expression changes ultimately promoting cancer development might well be diverse and unique to each patient. This is supported by the identification of very few common gene expression changes following EZH2 inhibition in patient-derived DLBCL cell lines (McCabe et al. 2012b; Beguelin et al. 2013). Analysis of gene expression signatures may thus not be a viable strategy for defining responders to treatment with EZH2 inhibitors.
Collectively, therapeutically targeting PRC2 in cancer through EZH2 inhibition has very promising prospects, and, with the particular requirement of PRC2 in stem and progenitor cells, might present a means of targeting the cancer stem cell compartment. However, the fact that PRC2 acts as a tumor suppressor in some cancer types along with the sensitivity of many cell types to alterations in PRC2 dosage raises some safety concerns and underlines the need to identify potential responders and minimize adverse effects of EZH2 inhibition in the clinic. It has already been demonstrated that secondary mutations can be acquired in tumors carrying either wild-type or mutated EZH2 alleles under prolonged inhibitor treatment, thus making cancer cells refractory to current inhibitors (Baker et al. 2015; Gibaja et al. 2016). Such findings highlight the need to expand the range of EZH2 inhibitors to include a repertoire of inhibitors targeting potential resistance mutants in the clinic. (See also Melnick 2016 on therapy targeting lysine methyltransferases.)
CONCLUDING REMARKS
In accordance with their crucial functions in specification and preservation of cell fate, the genes encoding the PRC2 components are often found mutated or deregulated in human cancers. The fact that both activating and inactivating mutations can contribute to the development of cancer is consistent with the general role of PRC2 in maintaining transcriptional programs and thereby guarding cell identity. Deregulation of PRC2 activity does not in itself lead to changes in transcriptional programs, but it alters the thresholds for gene activation. Therefore, the apparent role of PRC2 becomes context-dependent, that is, the phenotypic consequences of deregulated PRC2 activity are determined by the cellular environment and other mutations contributing to tumor development. With this in mind, it might not be surprising that efforts to identify common gene expression patterns in PRC2-dependent tumors have been futile. Apart from a crucial role of PRC2 in maintaining the transcriptional repression of the CDKN2A locus, genes have not been identified that indicate PRC2-dependency in tumors. However, despite this complexity, several companies have developed specific EZH2 inhibitors that have shown promising results in both preclinical and clinical trials. Thus far, the clinical trials have focused on cancers overexpressing EZH2 (e.g., DLBCL and FL), cancers expressing hyperactivating mutants of EZH2 (e.g., DLBCL and FL), and cancers with mutations in genes coding for the SWI/SNF complex (e.g., rhabdoid tumors and synovial sarcomas). If further success is achieved in these clinical trials, they could be extended to other cancers with somatic mutations that show synthetic lethality with PRC2 inhibition (e.g., BAP1 mutated mesotheliomas [LaFave et al. 2015]). While EZH2 inhibitors have until now been used as single agents, several studies have suggested that combination therapies might be a viable strategy (Beguelin et al. 2013; De Raedt et al. 2014; Knutson et al. 2014b; Fillmore et al. 2015), which could be exploited.
ACKNOWLEDGMENTS
We thank members of the Helin laboratory for discussions. The work in the Helin laboratory is supported by The European Research Council (294666_DNAMET), the 7th framework program of the European Union (4DCellFate and INGENIUM), the Danish Cancer Society, the Danish National Research Foundation (DNRF 82), the Danish Council for Strategic Research, the Danish Medical Research Council, the Novo Nordisk Foundation, The Lundbeck Foundation, and through a center grant from the Novo Nordisk Foundation (The Novo Nordisk Foundation Section for Stem Cell Biology in Human Disease).
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abdel-Wahab O, Pardanani A, Patel J, Wadleigh M, Lasho T, Heguy A, Beran M, Gilliland DG, Levine RL, Tefferi A. 2011. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia 25: 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. 2015. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29: 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Helin K. 2010. Histone methyltransferases in cancer. Semin Cell Dev Biol 21: 209–220. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, Helin K, Christensen J, Rowe M, Murray PG, et al. 2011. The H3K27me3 demethylase, KDM6B, is induced by Epstein–Barr virus and over-expressed in Hodgkin’s lymphoma. Oncogene 30: 2037–2043. [DOI] [PubMed] [Google Scholar]
- Antonysamy S, Condon B, Druzina Z, Bonanno JB, Gheyi T, Zhang F, MacEwan I, Zhang A, Ashok S, Rodgers L, et al. 2013. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain. PLoS ONE 8: e84147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T, Nerle S, Pritchard J, Zhao B, Rivera VM, Garner A, Gonzalvez F. 2015. Acquisition of a single EZH2 D1 domain mutation confers acquired resistance to EZH2-targeted inhibitors. Oncotarget 6: 32646–32655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, et al. 2013. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23: 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, et al. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. 2013. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24: 660–672. [DOI] [PubMed] [Google Scholar]
- Berg T, Thoene S, Yap D, Wee T, Schoeler N, Rosten P, Lim E, Bilenky M, Mungall AJ, Oellerich T, et al. 2014. A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood 123: 3914–3924. [DOI] [PubMed] [Google Scholar]
- Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, et al. 2015. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 21: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: Modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor C, O’Riain C, Wrench D, Matthews J, Iyengar S, Tayyib H, Calaminici M, Clear A, Iqbal S, Quentmeier H, et al. 2011. EZH2 Y641 mutations in follicular lymphoma. Leukemia 25: 726–729. [DOI] [PubMed] [Google Scholar]
- Bodor C, Grossmann V, Popov N, Okosun J, O’Riain C, Tan K, Marzec J, Araf S, Wang J, Lee AM, et al. 2013. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 122: 3165–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WD, Arora S, Busby J, Balasubramanian S, Gehling VS, Nasveschuk CG, Vaswani RG, Yuan CC, Hatton C, Zhao F, et al. 2014. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol 21: 1463–1475. [DOI] [PubMed] [Google Scholar]
- Brock A, Krause S, Ingber DE. 2015. Control of cancer formation by intrinsic genetic noise and microenvironmental cues. Nat Rev Cancer 15: 499–509. [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JE, Kuntz KW, Knutson SK, Warholic NM, Keilhack H, Wigle TJ, Raimondi A, Klaus CR, Rioux N, Yokoi A, et al. 2015. EPZ011989, a potent, orally-available EZH2 inhibitor with robust in vivo activity. ACS Med Chem Lett 6: 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. 2012. Molecular architecture of human Polycomb repressive complex 2. eLife 1: e00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. 2014. Targeting Polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7: 1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer MK, Starker LF, Choi M, Udelsman R, Nelson-Williams C, Lifton RP, Carling T. 2012. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J Clin Endocrinol Metab 97: E1774–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E, Yamauchi T, Echanique K, Haladyna J, Kalkur R, Riedel S, Zhu N, Xie H, Bernt KM, Orkin SH, et al. 2015. Inactivation of Eed impedes MLL-AF9-mediated leukemogenesis through Cdkn2a-dependent and Cdkn2a-independent mechanisms in a murine model. Exp Hematol 43: 930–935.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb G, Singh AK, Gupta S. 2014. EZH2: Not EZHY (easy) to deal. Mol Cancer Res 12: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. 2004. Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893. [DOI] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. 2014. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20: 1147–1155. [DOI] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. 2011. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res 21: 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726. [DOI] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. 2012. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 1: e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, Pasini D. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62. [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, et al. 2015. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 520: 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. 2004. Chromatin compaction by a Polycomb group protein complex. Science 306: 1574–1577. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty-Rao S, Nasveschuk C, Gagnon A, Chan EY, Sandy P, Busby J, Balasubramanian S, Campbell R, Zhao F, Bergeron L, et al. 2013. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem Biol 20: 1329–1339. [DOI] [PubMed] [Google Scholar]
- Gibaja V, Shen F, Harari J, Korn J, Ruddy D, Saenz-Vash V, Zhai H, Rejtar T, Paris CG, Yu Z, et al. 2016. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene 35: 558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmelli P, Biamonte F, Score J, Hidalgo-Curtis C, Cervantes F, Maffioli M, Fanelli T, Ernst T, Winkelman N, Jones AV, et al. 2011. EZH2 mutational status predicts poor survival in myelofibrosis. Blood 118: 5227–5234. [DOI] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. 2011. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. 2012. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun 3: 623. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. 2015. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 29: 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, Visconte V, Sugimoto Y, Prince C, O’Keefe C, et al. 2011. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118: 3932–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Wang Y, Zhou F, Gao G, Ren S, Zhou C. 2015. Prognostic value of high EZH2 expression in patients with different types of cancer: A systematic review with meta-analysis. Oncotarget 7: 4584–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Liu X. 2015. Structural basis of histone H3K27 trimethylation by an active Polycomb repressive complex 2. Science 350: aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. 1990. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HR, Pasini D, Helin K, Jensen ON. 2010. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9: 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HR, Sidoli S, Haldbo S, Sprenger RR, Schwammle V, Pasini D, Helin K, Jensen ON. 2013. Precision mapping of coexisting modifications in histone H3 tails from embryonic stem cells by ETD-MS/MS. Anal Chem 85: 8232–8239. [DOI] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. 2014. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28: 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed Polycomb target genes and interact with Polycomb repressive complex-2. Mol Cell 38: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. 2005. Subunit contributions to histone methyltransferase activities of fly and worm Polycomb group complexes. Mol Cell Biol 25: 6857–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. 2008. SWI/SNF mediates Polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 28: 3457–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, Wang W, Haswell JR, Walensky LD, Hahn WC, et al. 2015. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 21: 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci 100: 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. 2012. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8: 890–896. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, et al. 2013. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci 110: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, et al. 2014a. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2 mutant non-Hodgkin lymphoma. Mol Cancer Ther 13: 842–854. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Johnston LD, Klaus CR, Wigle TJ, Iwanowicz D, Littlefield BA, Porter-Scott M, Smith JJ, Moyer MP, et al. 2014b. Synergistic anti-tumor activity of EZH2 inhibitors and glucocorticoid receptor agonists in models of germinal center non-Hodgkin lymphomas. PLoS ONE 9: e111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 44: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, et al. 2015. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 21: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22: 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14: 735–751. [DOI] [PubMed] [Google Scholar]
- Lee SC, Phipson B, Hyland CD, Leong HS, Allan RS, Lun A, Hilton DJ, Nutt SL, Blewitt ME, Smyth GK, et al. 2013. Polycomb repressive complex 2 (PRC2) suppresses Eμ-myc lymphoma. Blood 122: 2654–2663. [DOI] [PubMed] [Google Scholar]
- Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, Zhu S, Cao Z, Liang Y, Sboner A, et al. 2014. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 46: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Miller S, Hyland C, Kauppi M, Lebois M, Di Rago L, Metcalf D, Kinkel SA, Josefsson EC, Blewitt ME, et al. 2015. Polycomb repressive complex 2 component Suz12 is required for hematopoietic stem cell function and lymphopoiesis. Blood 126: 167–175. [DOI] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. 1999. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev 13: 2691–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, Pigneux A, Wetzler M, Stuart RK, Erba HP, et al. 2015. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. 2012. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci 109: 3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. 2012. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, Smith JJ, Moyer MP, Richon VM, Copeland RA, et al. 2012. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett 586: 3448–3451. [DOI] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, et al. 2010. Opposing roles of Polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116: 731–739. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. 2012a. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci 109: 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012b. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. [DOI] [PubMed] [Google Scholar]
- *.Melnick AM. 2016. Lysine methyltransferases. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026682. [DOI] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kashio M, Aoyama K, Sashida G, Oshima M, Tomioka T, Muto T, Wang C, Iwama A. 2015. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 126: 1172–1183. [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. 2005. The murine Polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, et al. 2013. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med 210: 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, Xie H, Orkin SH, Armstrong SA. 2012. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci 109: 5028–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Vosberg S, Schlee C, Heesch S, Schwartz S, Gokbuget N, Hoelzer D, Graf A, Krebs S, Bartram I, et al. 2015. Mutational spectrum of adult T-ALL. Oncotarget 6: 2754–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. 2010. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42: 665–667. [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. 2012. Genetic inactivation of the Polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. 2001. The Polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Kasinathan S, Hughes KT, Saminadin-Peter S, Henikoff S, Ahmad K. 2014. High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res 24: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott HM, Graves AP, Pappalardi MB, Huddleston M, Halsey WS, Hughes AM, Groy A, Dul E, Jiang Y, Bai Y, et al. 2014. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hypertrimethylation. Mol Cancer Ther 13: 3062–3073. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. 2007. The Polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. [DOI] [PubMed] [Google Scholar]
- Pengelly AR, Kalb R, Finkl K, Muller J. 2015. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev 29: 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589. [DOI] [PubMed] [Google Scholar]
- Prieto-Granada CN, Wiesner T, Messina JL, Jungbluth AA, Chi P, Antonescu CR. 2015. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol 40: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puda A, Milosevic JD, Berg T, Klampfl T, Harutyunyan AS, Gisslinger B, Rumi E, Pietra D, Malcovati L, Elena C, et al. 2012. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am J Hematol 87: 245–250. [DOI] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, et al. 2012. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci 109: 21360–21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, et al. 2012. The Polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11: 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, Pirrotta V. 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J 12: 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie ER, Schumacher A, Angel JM, Holloway M, Rinchik EM, Magnuson T. 2002. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene 21: 299–306. [DOI] [PubMed] [Google Scholar]
- Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, et al. 2009. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci 106: 5324–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. 2014. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55: 347–360. [DOI] [PubMed] [Google Scholar]
- Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, He J, Palmer G, Yelensky R, Lipson D, et al. 2014. Advanced urothelial carcinoma: Next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol 27: 271–280. [DOI] [PubMed] [Google Scholar]
- Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, Tanaka S, Mochizuki-Kashio M, Wang C, Saraya A, et al. 2014. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun 5: 4177. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Faust C, Magnuson T. 1996. Positional cloning of a global regulator of anterior–posterior patterning in mice. Nature 384: 648. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Score J, Hidalgo-Curtis C, Jones AV, Winkelmann N, Skinner A, Ward D, Zoi K, Ernst T, Stegelmann F, Dohner K, et al. 2012. Inactivation of Polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 119: 1208–1213. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, Lowe SW, Vakoc CR. 2013. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;NrasG12D acute myeloid leukemia. Oncogene 32: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505. [DOI] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, et al. 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 26: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. 2013. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. 2010. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci 107: 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanik VA, Harte PJ. 2012. A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes. Dev Biol 364: 249–258. [DOI] [PubMed] [Google Scholar]
- Struhl G, Akam M. 1985. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J 4: 3259–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalm BM, Knutson SK, Warholic NM, Jin L, Kuntz KW, Keilhack H, Smith JJ, Pollock RM, Moyer MP, Scott MP, et al. 2014. Reaction coupling between wild-type and disease-associated mutant EZH2. ACS Chem Biol 9: 2459–2464. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, Suzuki Y, Sugano S, Nakaseko C, Yokote K, et al. 2012. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 120: 1107–1117. [DOI] [PubMed] [Google Scholar]
- Tanay A, O’Donnell AH, Damelin M, Bestor TH. 2007. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci 104: 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, et al. 2015. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. 2002. The Polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629. [DOI] [PubMed] [Google Scholar]
- Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. 2010. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116: 5247–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. 2013. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. 2004. Hierarchical recruitment of Polycomb group silencing complexes. Mol Cell 14: 637–646. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. 2010. Epigenetic antagonism between Polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, et al. 2010. Structural biology of human H3K9 methyltransferases. PLoS ONE 5: e8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zeng H, Dong A, Li F, He H, Senisterra G, Seitova A, Duan S, Brown PJ, Vedadi M, et al. 2013a. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS ONE 8: e83737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. 2013b. Fbxl10/Kdm2b recruits Polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49: 1134–1146. [DOI] [PubMed] [Google Scholar]
- Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, Orkin SH. 2014. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. 2010. Binding of different histone marks differentially regulates the activity and specificity of Polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci 107: 19266–19271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Jones PA. 2012. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 22: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. 2012. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 32: 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Jones S, Sausen M, McMahon K, Sharma R, Wang Q, Belzberg AJ, Chaichana K, Gallia GL, et al. 2014. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet 46: 1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M, et al. 2015. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun 6: 6051. [DOI] [PubMed] [Google Scholar]