Abstract
The [Het-s] prion of the fungus Podospora anserina is a well-studied model system to elucidate the action of prions and beyond. The [Het-s] prion works as an activation trigger of a cell death execution protein termed HET-S. Amyloid transconformation of the prion-forming region of HET-S induces activation of its pore-forming cell death execution HeLo domain. The prion motif functions in a signal transduction process by which a nucleotide-binding oligomerization domain (NOD)-like receptor termed NWD2 controls the HET-S cell death effector. This prion motif thus corresponds to a functional amyloid motif, allowing a conformational crosstalk between homologous motif domains in signal transduction processes that appears to be widespread from the fungal to the mammalian animal kingdoms. This review aims to establish a structure–activity relationship of the HET-S/s prion system and sets it in the context of its wider biological significance.
The HET-S/s system of the fungus Podospora anserina underlies heterokaryon incompatibility in this species. The HET-s prion-forming domain may provide the best model for examining the structural basis of amyloid formation.
Amyloids are generally perceived as pathological protein aggregates because they cause devastating neurodegenerative diseases in humans (Jucker and Walker 2013). But amyloids are also involved in various cellular functions such as hormone storage, cell surface structure formation, phenotypic inheritance, and programmed cell death (Fowler et al. 2007; Greenwald and Riek 2010; Li et al. 2012; Syed and Boles 2014). The self-templating ability of amyloids (sometimes referred to as the prion principle) plays a central role in defining the biological properties of both pathologic and functional amyloids. The structural basis of prion formation is probably best understood in the case of the [Het-s] prion model of the fungus Podospora anserina. Upon prion formation, the HET-s prion-forming domain (PFD) undergoes a transition from an unordered flexible monomeric state to a polymeric β-solenoid structure constituted by the almost infinite repetition of an elementary 21-amino acid structural motif (Wasmer et al. 2008a). This fungal prion represents an example of a prion with a function because it is involved in the control of a programmed cell death reaction occurring in the context of non-self recognition (Saupe and Daskalov 2012). It is the aim of this review to summarize the structure–activity relationship of the HET-S/s prion system and discuss it in the context of its broader biological implication.
BASIC BIOLOGY OF [HET-s], A PRION INVOLVED IN A NON-SELF RECOGNITION PROCESS
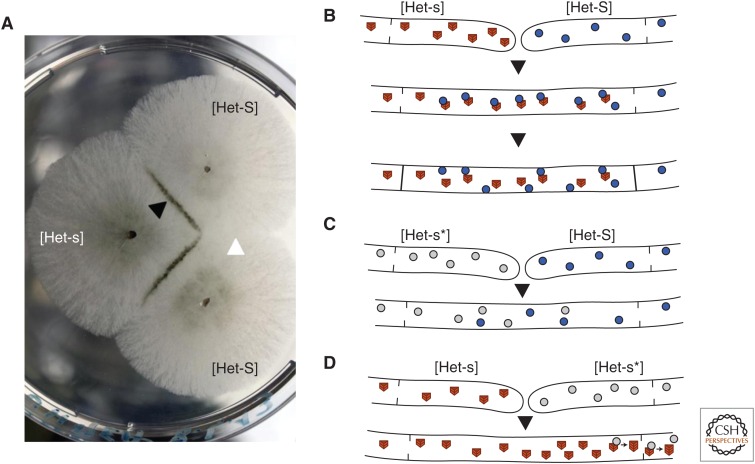
Fungi have the unusual ability to undergo somatic cell fusions. When in close proximity, two individuals belonging to the same species will grow toward each other, fuse, and mix their cytoplasmic content. Yet, in practice in most cases, mixed cells resulting from the fusion will immediately undergo programmed cell death (Glass and Dementhon 2006; Pinan-Lucarre et al. 2007; Aanen et al. 2010). This phenomenon is termed heterokaryon incompatibility. Viable fusion can only occur between strains that are compatible, meaning that they harbor appropriate alleles at particular recognition loci that are termed het loci. The non-self rejection reaction leads to the formation of a demarcation line between the strains, termed barrage (Fig. 1). The [Het-s] prion is involved in this non-self recognition mechanism, which is, in some aspects, analogous to graft rejection occurring in mammals (Saupe 2011). The proposed function of incompatibility is to limit cytoplasmic mixing to genetically similar strains. Uncontrolled cytoplasmic mixing can lead to infection with deleterious genetic elements like virus and senescence plasmids (Pearson et al. 2009) or to conspecific parasitism (parasitic exploitation of an organism by nuclei or cells or of the same species) (Czárán et al. 2014). The P. anserina population can slip into two types of strains, the strain bearing the het-s (lower case s) allele and those bearing the het-S (upper case S) allele (Fig. 1); het-s and het-S are two allelic variants of the same gene, and the corresponding HET-s and HET-S proteins are 95% identical (Turcq et al. 1991). Het-s and het-S strains are incompatible and undergo cell death when they fuse (Fig. 1) (Rizet 1952). But, this cell death reaction occurs if, and only if, the HET-s protein is in a prion form. The HET-s protein is a soluble cytoplasmic protein that can switch to an assembled, self-propagating amyloid prion form (Coustou et al. 1997; Saupe 2011). Strains harboring the protein in its soluble form are designated [Het-s*] (lowercase s, asterisk), whereas strains containing the prion form are designated [Het-s]. [Het-s*] strains switch spontaneously to the [Het-s] prion state at a low but constant frequency during somatic growth. As a consequence, all [Het-s*] strains ultimately acquire the [Het-s] prion phenotype over time. The [Het-s] prion is also systematically transmitted to [Het-s*] strains after cell fusion, thus [Het-s] is systematically transmitted horizontally between individuals and also vertically from maternal parent to progeny with 100% efficiency (Beisson-Schecroun 1962). The prion form propagates in the filamentous cell network at a rate 10 times faster than the radial growth rate. There is no detectable growth alteration associated with the presence of the [Het-s] prion form; [Het-s] has no detrimental effect on fitness in laboratory conditions. Somatically, prion curing never occurs. The [Het-s] prion can, however, be eliminated during a sexual cross with an het-S strain. A large part of the het-s progeny in an [Het-s] × het-S sexual cross is cured of the prion. In this situation, HET-S has a prion-curing effect (Rizet 1952). The prevalence of the [Het-s] prion was analyzed in a natural population of 112 individuals to determine the net result of spontaneous or acquired infection and prion curing on prion prevalence in the wild (Debets et al. 2012). [Het-s] was found to be highly prevalent in wild isolates of the het-s genotype, as 92% of the strains were found to be prion infected. In addition, [Het-s]/HET-S incompatibility was found to limit propagation between strains of a deleterious senescence plasmid leading to premature aging, providing evidence for a beneficial role for [Het-s]/HET-S incompatibility. Typically, alleles of non-self recognition loci, such as het-s and het-S, are expected to be under balancing selection, leading to equilibrated allele frequencies of the two incompatible types. However, the het-s genotype is more frequent than the het-S genotype in an approximate 2:1 ratio. This imbalance is thought to result from an unusual genetic property of the het-s allele. Indeed, in an het-s × het-S sexual cross, more het-s than het-S progeny are recovered. The het-s allele behaves as a meiotic drive element (Dalstra et al. 2003). Meiotic drive elements have been described in plants, fungi, and animals as ultra-selfish genetic elements violating Mendelian segregation laws in their favor to become overrepresented in populations (Hurst and Werren 2001). Het-s meiotic drive only occurs when HET-s is in the prion conformation and results from specific killing of the spore of the het-S genotype.
Figure 1.
[Het-s]/HET-S incompatibility. (A) When confronted on solid medium, [Het-s] and [Het-S] strains fuse and produce a barrage reaction (black arrowhead) indicative of the occurrence on an incompatibility reaction. In contrast, a contact between two [Het-S] strains is normal and shows no demarcation line (white arrowhead). (B) Diagram of the result of an [Het-s]/[Het-S] cell fusion: cytoplasmic mixing occurs, and the prion form of HET-s interacts with HET-S and leads to activation of a pore-forming activity. Upon activation, HET-S is relocated to the plasma membrane, exerts toxicity, and induces cell death. (C) When HET-s is in the soluble (nonprion) state ([Het-s*]), cell fusion with HET-S does not lead to incompatibility. (D) When [Het-s] fuses with [Het-s*], the prion form propagates within the [Het-s*] cells and converts them to the [Het-s] state.
[Het-s] can be described as a highly prevalent prion having no detrimental effect on growth and functioning as part of a non-self rejection system together with its HET-S partner. An het-S allele can be converted to the het-s phenotype by a single amino acid substitution (Deleu et al. 1993). At position 33, one finds a histidine in HET-S and a proline in HET-s; an HET-S H33P point mutant becomes incompatible with HET-S. Thus, the incompatibility between HET-s and HET-S can be stripped down to a single amino acid difference between the two proteins. In vitro, recombinant HET-s spontaneously assembles into amyloid fibrils that display prion infectivity, indicating that the [Het-s] prion conforms to the “protein-only” hypothesis (Dos Reis et al. 2002; Maddelein et al. 2002).
STRUCTURE–FUNCTION OF THE HET-S PRION-FORMING DOMAIN
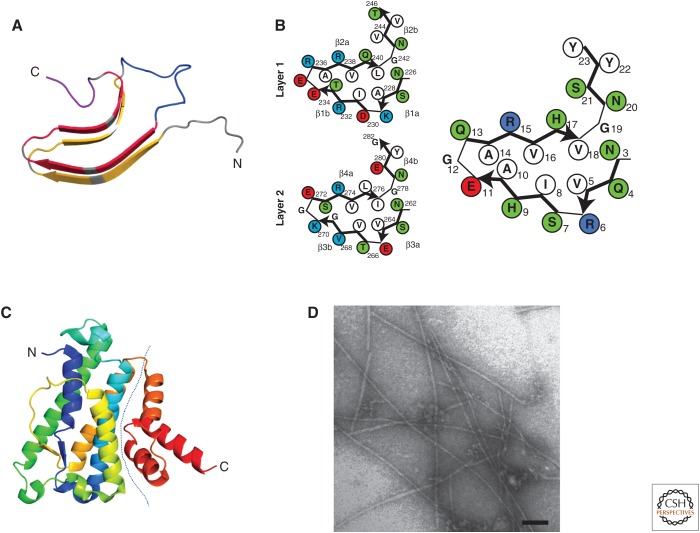
HET-s is a 289-amino acid, two-domain protein. An N-terminal α-helical domain, termed HeLo, spans amino acids 1 to ∼227 and, as detailed below, is responsible for cell death execution in incompatibility (Seuring et al. 2012). A C-terminal PFD occupies position 218 to 289 (Balguerie et al. 2003; Greenwald et al. 2010). The HET-s(218–289) PFD is both necessary and sufficient for amyloid formation and prion propagation and was originally identified as the proteinase K–resistant core of full-length HET-s fibrils (Dos Reis et al. 2002; Balguerie et al. 2003). The HET-s PFD contains two 21-residue imperfect repeats (Ritter et al. 2005). Each of these repeats forms one 4.7-Å layer of a stacked β-sheet structure (Ritter et al. 2005; Sen et al. 2007). In contrast to many amyloids, the HET-s PFD is characterized by the absence of structural polymorphism, as determined by solid-state nuclear magnetic resonance (NMR) as well as being ordered and unique. The two pseudo-repeats adopt a rigid β-sheet structure and are connected by a 15-amino-acid-long flexible loop (Ritter et al. 2005). Each repeat forms one rung of a two-layer β-solenoid structure and comprises four β-strands; the first three β-strands delimit a triangular hydrophobic core, whereas the fourth protrudes from the core (Fig. 2A,B) (Wasmer et al. 2008a; Van Melckebeke et al. 2010). At the C-terminal end of the PFD resides an aromatic loop that folds back into a groove delimited by the third and fourth β-strands to form a semi-hydrophobic pocket (Fig. 2A) (Van Melckebeke et al. 2010). A cryo-electron microscopy (cryo-EM) model of HET-s(218–289) was also obtained and largely agrees with the solid-state NMR structure (Mizuno et al. 2011). The β-solenoid fold displays a number of key structural elements. Two asparagine ladders occur at the beginning of the first and fourth β-strands (N226/N262 and N243/279), and three salt bridges (K229/E265, E234/K270, and R236/E272) stabilize the stacking of the two rungs of β-strands (Fig. 2B). The inside core is composed exclusively of hydrophobic residues with the exception of two hydroxyl residues occupying different layers that can form a hydrogen bond inside the core (T233/S273). Glycine residues occupy the β-arch between the third and the fourth β-strand in both layers. A short C-terminal loop contains two aromatic residues (F286 and W287) and folds back onto the fourth β-strand to form a semi-hydrophobic pocket. Interestingly, all of these key sequence features are conserved (at least to some extent) in the different HET-S/s homologs that could be identified in fungal genomes, which strongly argues for an evolutionary conservation of the β-solenoid fold (Daskalov et al. 2014a). A homolog of HET-S from the phytopathogen Fusarium graminearum was functionally and structurally characterized. HET-s and FgHET-s show 38% identity in the PFD region. The FgHET-s PFD region displays amyloid-forming and prion-forming ability and a structure closely related to the HET-s PFD (Wasmer et al. 2010; Benkemoun et al. 2011). Reciprocal cross-seeding and prion infection occur between the homologous PFDs, but when co-expressed in the same cell, homotypic polymerization is favored over formation of heterotypic aggregates.
Figure 2.
HET-S/s prion-forming domain (PFD) and HeLo domain structure. (A) The β-solenoid structure of HET-s(218–289). The structure of one monomer in the fibrillar form is shown (after PDB 2KJ3). Each rung of β-strands is shown in yellow and the second is in red. The connecting central loop is shown in blue, and the C-terminal loop forming the semi-hydrophobic pocket is in magenta. (B) Individual β-solenoid motifs of HET-s and NWD2. A schematic representation of the β-solenoid form of repeats 1 and 2 of HET-s and NWD2(3–23) are given on the left and right, respectively. The NWD2(3–23) model is based on homology modeling and solid-state nuclear magnetic resonance (NMR) (see text for details). (C) The structure of the HeLo domain of HET-S (after PDB 2WVO). The dotted line delimits the N-terminal and C-terminal bundles forming subdomains. (D) Electron micrograph of NWD2(3–23) fibrils. Scale bar, 50 nm.
The structure–function relationship in the HET-s PFD was analyzed in some detail by site-directed mutagenesis approaches (Ritter et al. 2005; Daskalov et al. 2014a; Wan and Stubbs 2014b). Globally, these studies reveal that the β-solenoid structure is robust and withstands various sequence alterations without the global fold or the prion-forming ability being grossly compromised. Marked exceptions are the glycine residues of the β-arches between the third and fourth strand of each rung, the mutation of which affects the overall fold and abolishes prion function (Daskalov et al. 2014a). Yet the different structural elements mentioned above (N-ladders, salt bridges, hydrophobic core, buried polar residues) do contribute to various extents to the prion function, β-solenoid fold, fibril stability, and fibril formation rate. Interestingly, the C-terminal aromatic loop strongly modulates the prion-forming ability, although this region is not part of the rigid cross-β core of the fold (Daskalov et al. 2014a).
Many lines of evidence—such as the evolutionary conservation of β-solenoid-forming sequence, the highly ordered and nonpolymorphic nature of the fold, and the biological function associated with the prion form and its frequent occurrence in the wild—suggest that the β-solenoid fold of HET-s corresponds to the “native” folded state of this domain rather than a misfolded state as observed for disease-causing amyloids. It is of note that, in conditions in which this “native” amyloid fold cannot be attained, HET-s(218–289) can adopt an alternate nonprion amyloid state (Sabate et al. 2007; Wasmer et al. 2008b), which is conceptually analogous to the generic amyloid fold adopted by many proteins in non-native environments (Fandrich et al. 2001).
Since the structure of HET-s(218–289) was solved (Wasmer et al. 2008a), this 72-amino acid PFD has turned into a quite popular amyloid model system and was studied using (in addition to solid-state NMR) a variety of techniques, including hydrogen exchange, fiber diffraction, cryo-EM, and molecular dynamics (Nazabal et al. 2003; Ritter et al. 2005; Wasmer et al. 2008a; Lange et al. 2009; Mizuno et al. 2011; Wan and Stubbs 2014a). The HET-s PFD has in particular been used to structurally identify the binding interface of Congo red, a dye that is commonly used to characterize amyloid aggregates (Schutz et al. 2011).
The prion formation and propagation by the HET-s PFD can also occur in a heterologous host like yeast, indicating that no species-specific cofactors are required for prion propagation (Taneja et al. 2007). The Hsp104 chaperone participates in [Het-s] prion propagation (Malato et al. 2007). This chaperone with amyloid fibril shearing activity makes [Het-s] formation and propagation more efficient but is not strictly required as in the case of the yeast [URE3] and [PSI+] prions. HET-s(218–289) prion aggregates with the β-solenoid fold are also formed during heterologous expression as inclusion bodies in Escherichia coli, a property that HET-s shares with several yeast prions (Wasmer et al. 2009; Espargaro et al. 2012).
HeLo PORE-FORMING DOMAIN IS RESPONSIBLE FOR CELL DEATH EXECUTION
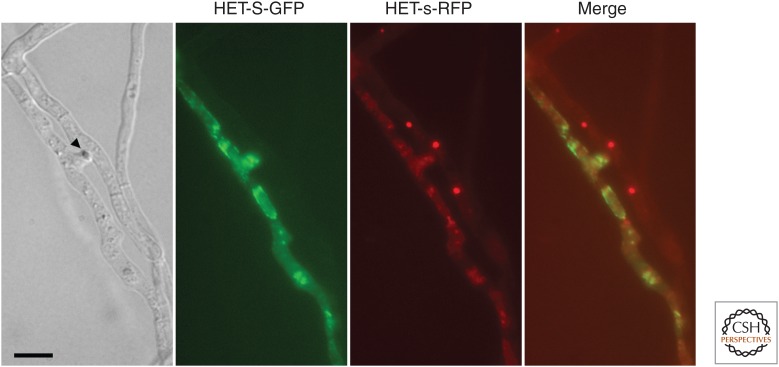
The prion form of HET-s is not toxic to cells; cell death by incompatibility is not caused by amyloid toxicity. Instead, cell death is induced specifically when the prion form of HET-s interacts with the HET-S partner protein. Functionally, the prion-forming regions of HET-S and HET-s are equivalent. The functional difference between HET-S and HET-s lies in the HeLo domain (Deleu et al. 1993; Balguerie et al. 2003). As previously mentioned, a single amino acid substitution in that domain (H33P) is sufficient to turn an HET-S into a protein of het-s specificity. Both the PFD and the HeLo domain of HET-S are required for induction of cell death, whereas the HeLo domain of HET-s is dispensable (Balguerie et al. 2003). The structures of the HeLo domains of HET-s and HET-S have been solved and found to be very similar (Greenwald et al. 2010). Two subregions can be defined in the structure: a first bundle of helices spans residue 1 to ∼170 and a second bundle of shorter helices (from ∼170 to ∼227) leads into the prion-forming region (Fig. 2C) (Greenwald et al. 2010). How does the HET-S/[Het-s] interaction cause cell death? In vivo, the [Het-s]/HET-S interaction causes HET-S to relocate from the cytoplasm to the cell membrane (Fig. 3). This relocation and associated induction of cell death occur both in Podospora and yeast, suggestive of direct toxicity (Mathur et al. 2012). The interaction between HET-S and HET-s occurs via the C-terminal prion-forming region. The molecular interface between HET-S and HET-s PFD regions was characterized by solid-state NMR (Seuring et al. 2012). Mutations that affect the β-solenoid fold abolish HET-S activity, indicating that β-folding of this region is required for induction of toxicity (Greenwald et al. 2010). In vitro, using purified protein and a liposome leakage assay, it was found that, in the presence of HET-s prion seeds, HET-S causes liposome leakage (Seuring et al. 2012). The N-terminal end of the HET-S protein displays a predicted transmembrane region, and mutations that affect this transmembrane region affect both cell death–inducing activity in vivo and liposome leakage in vitro. For instance, the aforementioned HET-S H33P mutant is altered in the predicted transmembrane helix and lacks cell death–inducing activity and liposome-permeabilization activity. Together, these observations lead to a mechanistic model for [Het-s]-induced cell death. Cell death is triggered when the [Het-s] prion templates conversion of the HET-S PFD region into the β-solenoid fold. β-solenoid folding of the HET-S PFD region leads, in turn, to a refolding of the HET-S HeLo domain that acquires pore-forming activity by exposing an N-terminal transmembrane helix, relocating it to the cell membrane, and inducing cell death. In this system, the amyloid fold is thus used as a triggering device that induces refolding and activation of the HeLo pore-forming domain. The conformational switch relies on the partial overlap between the β-solenoid and globular region (Greenwald et al. 2010). Tight coupling between the two domains is critical for this refolding activity, as an artificial construct in which the PFD and the HeLo domain are uncoupled in sequence lacks cell death–inducing activity.
Figure 3.
[Het-s] triggers HET-S relocalization to the cell membrane. Micrograph of a cell fusion between a strain expressing an HET-S-green fluorescent protein (GFP) fusion and a strain expressing HET-s-red fluorescent protein (RFP). Scale bar, 10 µm.
In contrast to HET-s, HET-S does not form well-ordered fibrils in vitro. In addition, HET-S can inhibit HET-s fibril formation in trans (Greenwald et al. 2010). This observation provides a mechanistic explanation for the prion-curing activity of HET-S occurring in sexual crosses between [Het-S] and [Het-s]. The interaction between HET-s and HET-S can thus lead to toxicity or prion inactivation depending on the cellular context or relative ratio of HET-s and HET-S (Dalstra et al. 2003). In vitro, ordered HET-S fibril formation is probably hindered by the hydrophobic aggregation of the activated HeLo domains. This mechanistic model of HET-S/[Het-s] toxicity also accounts for the functional differences between HET-s/HET-S. In virtue of the amino acid differences in the predicted transmembrane region, HET-s lacks pore-forming activity, and, thus, HET-s prion transconformation of the PFD region does not induce toxicity. This property allows HET-s to be propagated as a nontoxic prion. HET-s can be viewed as a mutant form of HET-S with a dead HeLo domain.
Remarkably, it appears that the HeLo domain is evolutionarily related to the cell death execution domain of a protein termed MLKL responsible for execution of necroptotic cell death in mammals, indicating long-term evolutionary conservation of cell death–inducing mechanisms from fungi to mammals (Daskalov et al. 2016).
NWD2 AND HET-S: NLR SIGNALING BASED ON THE HET-S/s PRION MOTIF
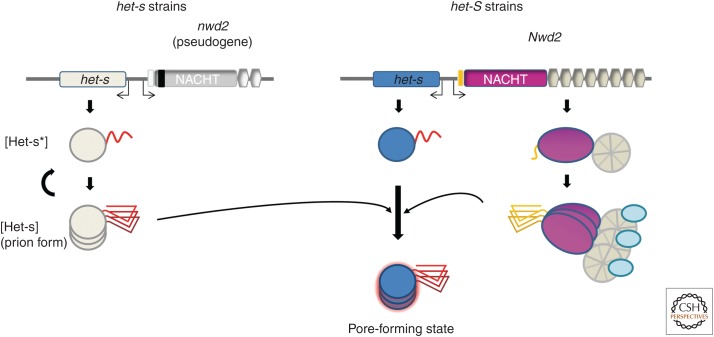
Recently, it was realized that the gene immediately adjacent to het-S in the genome of P. anserina encodes a protein with a short N-terminal region showing homology to the 21-amino acid β-solenoid elementary HET-s motif (Figs. 2B,4) (Daskalov et al. 2012). This protein, termed NWD2, is a fungal nucleotide-binding oligomerization domain (NOD)-like receptor. NOD-like receptors (NLRs) are signal-transducing ATPases (so-called STAND proteins) that control cell fate and host-defense pathways both in animals and plants (Maekawa et al. 2011). Fungi encode vast repertoires of NOD-like receptors, and NWD2 is one of them (Dyrka et al. 2014). STAND proteins, such as NLRs, function by a ligand-induced oligomerization process (Danot et al. 2009). They are typically tripartite proteins with a C-terminal ligand-binding region, a central NOD domain, and an N-terminal effector domain. Binding of the ligand induces oligomerization and downstream signaling via the N-terminal domain. NWD2 displays a central NACHT nucleotide-binding and oligomerization domain and a C-terminal WD-repeat domain. To explain genomic clustering and homology between HET-S and NWD2, it was proposed that NWD2 and HET-S might be functional partners. Ligand-induced oligomerization of NWD2 would lead to the spatial clustering of N-terminal PFD-like regions (Daskalov et al. 2012). This spatial clustering might then allow for cooperative folding of these regions into an HET-s-like β-solenoid fold and induce prion transconformation of the HET-S PFD. Consistent with this hypothesis, in a study on prion-like signaling by immune complexes, Cai et al. (2014) found that the N-terminal HET-S/s-like motif of NWD2 and the HET-s PFD can functionally substitute for the Pyrin-signaling domains in a human NLR and their cognate downstream effectors, respectively, NLRP3 and ASC. Functional evidence of the prion-templating activity of NWD2 was also obtained in vivo in P. anserina. Engineered NWD2 versions responding to known ligands efficiently induce [Het-s] formation in a ligand-dependent manner. The N-terminal region of NWD2 is both necessary and sufficient for prion-inducing activity in vivo. In vitro, a synthetic NWD2(3–23) peptide corresponding to the region homologous to the elementary HET-S/s repeat assembles into amyloid fibrils (Fig. 2D), which display prion infectivity. Critical residues for the formation of the β-solenoid fold are conserved in the NWD2(3–23), and homology modeling predicts a β-solenoid fold for this motif (Fig. 2B) (Daskalov et al. 2012). Solid-state NMR analysis of fibrils of NWD2(3–23) are fully consistent with the proposition that the peptide adopts an HET-S/s-like fold. The NWD2(3–23) peptide is the smallest molecular entity found to display prion-inducing activity; it thus appears that templating relies strictly on the β-strand regions (and necessitates neither the flexible loop, the C-terminal aromatic loop, nor repetition of the 21-amino acid motif). These results lead to a model in which ligand-induced oligomerization of NWD2 allows for spatial clustering of the HET-S/s-like motif and provides conditions for cooperative nucleation of the β-solenoid fold that can then template HET-S PFD transconformation. NWD2/HET-S is a two-component system in which the signal transduction process from NLR to the effector domain involves propagation of an amyloid prion fold (Daskalov et al. 2012, 2014b; Cai et al. 2014).
Figure 4.
Genomic organization of the het-s and het-S loci. Schematic representations of the domain organization of the het-s and het-S loci are given. The gene adjacent to het-S encodes NWD2, a nucleotide-binding oligomerization domain (NOD)-like receptor with a NACHT domain and WD repeats. Upon binding of a specific ligand, NWD2 acquires β-solenoid templating activity (see text for details). HET-S activation can be achieved either by templating by the prion form of HET-s or by the activated form of NWD2. Note that Nwd2 is in an inactive pseudogene form in all het-s strains because of the insertion of a transposon.
The formation of higher-order signaling machines has been described in the context of innate immunity in mammals (Wu 2013). Among those complexes are the Myddosome (Lin et al. 2010), the MAVS CARD filaments (Hou et al. 2011), the RIP1/RIP3 necrosome (Li et al. 2012), and the ASC-dependent inflammasomes (Lu et al. 2014). These signaling complexes are large filamentous assemblies. The RIP1/RIP3 complex forms via short amyloid motifs, termed RHIM (Li et al. 2012). The polymerization and templating properties of the death domain (DD) fold family, CARD, and PYD domains allow them to function as PFDs in yeast (Cai et al. 2014). The prion-like polymerization process in these signaling machines involves amyloid structures in some instances but not in others, and is conserved from fungi to humans. Higher-order assembly entitles signal transduction cascades with specific properties such as signal amplification, threshold response, and noise reduction (Wu 2013). The strictly cooperative nature of amyloid folds and their templating properties and compactness make amyloid motifs suited for such signal transduction tasks. Several fungal species encode NACHT-WD NOD-like receptors with an N-terminal HeLo domain (Daskalov et al. 2012; Dyrka et al. 2014). At present, it is difficult to determine whether this all-in-one architecture may predate the more complex two-component architecture involving an amyloid-adaptor motif—or whether the converse is true. In addition to the HET-S/s motif, several other functional amyloid motifs located at the N-terminus of NOD-like proteins and at the C-terminus of their putative downstream effector proteins were identified, stressing the evolutionary success of this mode of signal transduction in fungi (Daskalov et al. 2012).
It may now be possible to propose plausible evolutionary scenarios for the emergence of the [Het-s] prion. The HET-S/NWD2 gene pair is largely conserved, whereas apparently the [Het-s]/HET-S incompatibility system is specific to P. anserina (Daskalov et al. 2012; Daskalov et al. 2014a). HET-s can be propagated as a prion because it bears a specific amino acid replacement at position 33 of the HeLo domain, which kills pore-forming activity at the domains (Deleu et al. 1993; Greenwald et al. 2010; Seuring et al. 2012). These observations support the proposal that the [Het-s]/HET-S incompatibility system was derived from the preexisting het-S/nwd2 gene complex by exaptation (Debets et al. 2012; Seuring et al. 2012). An exaptation is an evolutionary mechanism by which a gene or an organ is co-opted to perform a new function that was not originally the target of selection (Gould and Vrba 1982). In that model, a point mutation of the HeLo domain of HET-S has allowed formation of the [Het-s] prion, thus creating the [Het-s]/HET-S incompatibility system (Daskalov et al. 2012; Debets et al. 2012; Seuring et al. 2012). A toxin (HET-S) was turned into a toxin-activation trigger ([Het-s]). This model can explain why nwd2 became useless in this context as evidenced by its pseudogene nature in wild het-s strains (Fig. 4) (Debets et al. 2012).
CONCLUDING REMARKS
The HET-S/s prion motif is a functional amyloid motif operating as a conformational switch in a signal transduction process. The achievement of structural characterizations of the HET-s PFD has promoted this prion into a valuable model system for studying various aspects of amyloid formation in a context in which high-resolution structural information is at hand (Wasmer et al. 2008a), a situation still rarely encountered in the amyloid field. In addition, this system, which in the early stages of its characterization may have appeared as not more than an entertaining oddity or convenient structural model, now stands as a clear example of an evolved amyloid motif in which the self-templating activity and cooperativity of the amyloid fold are means directed toward a biological end.
ACKNOWLEDGMENTS
Work on HET-S/s in the Saupe laboratory is supported by a grant from the Agence Nationale de la Recherche (ANR) (STANDPRION), Paris, France.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Aanen DK, Debets AJM, Glass NL, Saupe SJ. 2010. Biology and genetics of vegetative incompatibility in fungi. In Cellular and molecular biology of filamentous fungi (ed. Borkovich KA, Ebbole DJ), pp. 274–288. ASM, Washington, DC. [Google Scholar]
- Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, et al. 2003. Domain organization and structure–function relationship of the HET-s prion protein of Podospora anserina. EMBO J 22: 2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson-Schecroun J. 1962. Incompatibilité cellulaire et interactions nucléocytoplasmique dans les phénomènes de barrage chez le Podospora anserina [Cellular incompatibility and nucleocytoplasmic interactions in the barrage phenomenon in Podospora anserina]. Ann Genet 4: 3–50. [PubMed] [Google Scholar]
- Benkemoun L, Ness F, Sabate R, Ceschin J, Breton A, Clave C, Saupe SJ. 2011. Two structurally similar fungal prions efficiently cross-seed in vivo but form distinct polymers when coexpressed. Mol Microbiol 82: 1392–1405. [DOI] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156: 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci 94: 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czárán T, Hoekstra RF, Aanen DK. 2014. Selection against somatic parasitism can maintain allorecognition in fungi. Fungal Genet Biol 73: 128–137. [DOI] [PubMed] [Google Scholar]
- Dalstra HJ, Swart K, Debets AJ, Saupe SJ, Hoekstra RF. 2003. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci 100: 6616–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. 2009. Wheel of life, wheel of death: A mechanistic insight into signaling by STAND proteins. Structure 17: 172–182. [DOI] [PubMed] [Google Scholar]
- Daskalov A, Paoletti M, Ness F, Saupe SJ. 2012. Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes. Plos ONE 7: e34854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Gantner M, Walti MA, Schmidlin T, Chi CN, Wasmer C, Schutz A, Ceschin J, Clave C, Cescau S, et al. 2014a. Contribution of specific residues of the β-solenoid fold to HET-s prion function, amyloid structure and stability. PLoS Pathog 10: e1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Sabaté R, Saupe SJ. 2014b. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. Plos Biol 10.1371/journal.pbio.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Sabate R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ. 2016. Identification of a novel cell death–inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci 113: 2720–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets AJ, Dalstra HJ, Slakhorst M, Koopmanschap B, Hoekstra RF, Saupe SJ. 2012. High natural prevalence of a fungal prion. Proc Natl Acad Sci 109: 10432–10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleu C, Clave C, Begueret J. 1993. A single amino acid difference is sufficient to elicit vegetative incompatibility in the fungus Podospora anserina. Genetics 135: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Reis S, Coulary-Salin B, Forge V, Lascu I, Begueret J, Saupe SJ. 2002. The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J Biol Chem 277: 5703–5706. [DOI] [PubMed] [Google Scholar]
- Dyrka W, Lamacchia M, Durrens P, Kobe B, Daskalov A, Paoletti M, Sherman DJ, Saupe SJ. 2014. Diversity and variability of NOD-like receptors in fungi. Genome Biol Evol 6: 3137–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espargaro A, Villar-Pique A, Sabate R, Ventura S. 2012. Yeast prions form infectious amyloid inclusion bodies in bacteria. Microb Cell Fact 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandrich M, Fletcher MA, Dobson CM. 2001. Amyloid fibrils from muscle myoglobin. Nature 410: 165–166. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. 2007. Functional amyloid—From bacteria to humans. Trends Biochem Sci 32: 217–224. [DOI] [PubMed] [Google Scholar]
- Glass NL, Dementhon K. 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol 9: 553–558. [DOI] [PubMed] [Google Scholar]
- Gould JS, Vrba ES. 1982. Exaptation—A missing term in the science of form. Paleobiology 8: 4–15. [Google Scholar]
- Greenwald J, Riek R. 2010. Biology of amyloid: Structure, function, and regulation. Structure 18: 1244–1260. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein ML, Ness F, Cescau S, Soragni A, Leitz D, et al. 2010. The mechanism of prion inhibition by HET-S. Mol Cell 38: 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GD, Werren JH. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2: 597–606. [DOI] [PubMed] [Google Scholar]
- Jucker M, Walker LC. 2013. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501: 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Gattin Z, Van Melckebeke H, Wasmer C, Soragni A, van Gunsteren WF, Meier BH. 2009. A combined solid-state NMR and MD characterization of the stability and dynamics of the HET-s(218–289) prion in its amyloid conformation. Chembiochem 10: 1657–1665. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. 2012. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. 2010. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156: 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. 2002. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci 99: 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. 2011. NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol 12: 817–826. [DOI] [PubMed] [Google Scholar]
- Malato L, Dos Reis S, Benkemoun L, Sabate R, Saupe SJ. 2007. Role of Hsp104 in the propagation and inheritance of the [Het-s] prion. Mol Biol Cell 18: 4803–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Seuring C, Riek R, Saupe SJ, Liebman SW. 2012. Localization of HET-S to the cell periphery, not to [Het-s] aggregates, is associated with [Het-s]-HET-S toxicity. Mol Cell Biol 32: 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Baxa U, Steven AC. 2011. Structural dependence of HET-s amyloid fibril infectivity assessed by cryoelectron microscopy. Proc Natl Acad Sci 108: 3252–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazabal A, Dos Reis S, Bonneu M, Saupe SJ, Schmitter JM. 2003. Conformational transition occurring upon amyloid aggregation of the HET-s prion protein of Podospora anserina analyzed by hydrogen/deuterium exchange and mass spectrometry. Biochemistry 42: 8852–8861. [DOI] [PubMed] [Google Scholar]
- Pearson MN, Beever RE, Boine B, Arthur K. 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinan-Lucarre B, Paoletti M, Clave C. 2007. Cell death by incompatibility in the fungus Podospora. Semin Cancer Biol 17: 101–111. [DOI] [PubMed] [Google Scholar]
- Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. 2005. Correlation of structural elements and infectivity of the HET-s prion. Nature 435: 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G. 1952. Les phénomènes de barrage chez Podospora anserina. I: Analyse de barrage entre les souches s et S [The barrage phenomena in Podospora. I: Analysis of the barrage between s and S strains]. Rev Cytol Biol Veg 13: 51–92. [Google Scholar]
- Sabate R, Baxa U, Benkemoun L, Sanchez de Groot N, Coulary-Salin B, Maddelein ML, Malato L, Ventura S, Steven AC, Saupe SJ. 2007. Prion and non-prion amyloids of the HET-s prion forming domain. J Mol Biol 370: 768–783. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. 2011. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol 22: 460–468. [DOI] [PubMed] [Google Scholar]
- Saupe SJ, Daskalov A. 2012. The [Het-s] prion, an amyloid fold as a cell death activation trigger. PLoS Pathog 8: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AK, Soragni A, Hornemann S, Aguzzi A, Ernst M, Bockmann A, Meier BH. 2011. The amyloid–Congo red interface at atomic resolution. Angew Chem Int Ed Engl 50: 5956–5960. [DOI] [PubMed] [Google Scholar]
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ, Steven AC. 2007. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J Biol Chem 282: 5545–5550. [DOI] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. 2012. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10: e1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AK, Boles BR. 2014. Fold modulating function: Bacterial toxins to functional amyloids. Front Microbiol 5: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. 2007. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell 27: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcq B, Deleu C, Denayrolles M, Begueret J. 1991. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet 228: 265–269. [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Bockmann A, Meier BH. 2010. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc 132: 13765–13775. [DOI] [PubMed] [Google Scholar]
- Wan W, Stubbs G. 2014a. Fiber diffraction of the prion-forming domain HET-s(218–289) shows dehydration-induced deformation of a complex amyloid structure. Biochemistry 53: 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Stubbs G. 2014b. Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc Natl Acad Sci 111: 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008a. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science 319: 1523–1526. Erratum in Science320: 50. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Soragni A, Sabate R, Lange A, Riek R, Meier BH. 2008b. Infectious and noninfectious amyloids of the HET-s(218–289) prion have different NMR spectra. Angew Chem Int Ed Engl 47: 5839–5841. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Benkemoun L, Sabate R, Steinmetz MO, Coulary-Salin B, Wang L, Riek R, Saupe SJ, Meier BH. 2009. Solid-state NMR spectroscopy reveals that E. coli inclusion bodies of HET-s(218–289) are amyloids. Angew Chem Int Ed Engl 48: 4858–4860. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Zimmer A, Sabate R, Soragni A, Saupe SJ, Ritter C, Meier BH. 2010. Structural similarity between the prion domain of HET-s and a homologue can explain amyloid cross-seeding in spite of limited sequence identity. J Mol Biol 402: 311–325. [DOI] [PubMed] [Google Scholar]
- Wu H. 2013. Higher-order assemblies in a new paradigm of signal transduction. Cell 153: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]