Abstract
Yeast and fungal prions are infectious proteins, most being self-propagating amyloids of normally soluble proteins. Their effects range from a very mild detriment to lethal, with specific effects dependent on the prion protein and the specific prion variant (“prion strain”). The prion amyloids of Sup35p, Ure2p, and Rnq1p are in-register, parallel, folded β-sheets, an architecture that naturally suggests a mechanism by which a protein can template its conformation, just as DNA or RNA templates its sequence. Prion propagation is critically affected by an array of chaperone systems, most notably the Hsp104/Hsp70/Hsp40 combination, which is responsible for generating new prion seeds from old filaments. The Btn2/Cur1 antiprion system cures most [URE3] prions that develop, and the Ssb antiprion system blocks [PSI+] generation.
Yeast prions are mostly self-propagating amyloids (filamentous polymers) of normally soluble proteins. Chaperone systems can affect their propagation, and antiprion systems (e.g., Btn2/Cur1) can cure them.
In 1989, I was writing a review of nonchromosomal genetic elements of yeast (Wickner 1991). At that time, the infectious protein concept had been proposed to explain the transmissible spongiform encephalopathies (TSEs) (Alper et al. 1967; Griffith 1967; Dickinson et al. 1968; Bolton et al. 1982; Prusiner 1982; Chesebro et al. 1985; Oesch et al. 1985), but there remained considerable debate. In reviewing the nonchromosomal genetic element [URE3], I noted that Aigle and Lacroute (1975) had reported that ure2 strains were unable to propagate the [URE3] element, but that the phenotypes of ure2 and URE2+ [URE3] strains were essentially identical—namely, de-repression of nitrogen catabolism genes. As I had been studying the mak mutants unable to propagate the killer virus (Wickner 1978), I recognized that the relation of ure2 and [URE3] was not what was expected of a chromosomal gene important for propagation of a nucleic acid replicon, but that this was the relation expected if [URE3] was an inactive form of Ure2p able to inactivate the normal form and, thus, act as a prion. This was the beginning of our work showing that [URE3] was a prion of Ure2p, and our inference that [PSI+] was a prion of Sup35p based on the re-interpretation of published experiments (Wickner 1994).
DISCOVERY OF YEAST PRIONS
The genetic criteria that identified yeast prions (infectious proteins) (Wickner 1994) distinguish them from nucleic acid–based infectious elements such as viruses, plasmids, and mitochondria. Assuming that the prion change would inactivate the normal function of the protein, the phenotype of the prion should resemble that of a recessive mutation in the gene for the prion protein, and elimination of the prion protein gene should result in failure of prion propagation (Wickner 1994). Overproduction of the prion protein should result in increased de novo generation of the prion. Finally, if the prion can be cured, it should be possible for it to arise again in the cured strain (at low frequency) because the normal form of the protein is still being produced (Wickner 1994). These genetic criteria are not true for any known mammalian prion, in part because the TSEs are not a consequence of PrP deficiency and in part because some of the required experiments are not feasible in mammals.
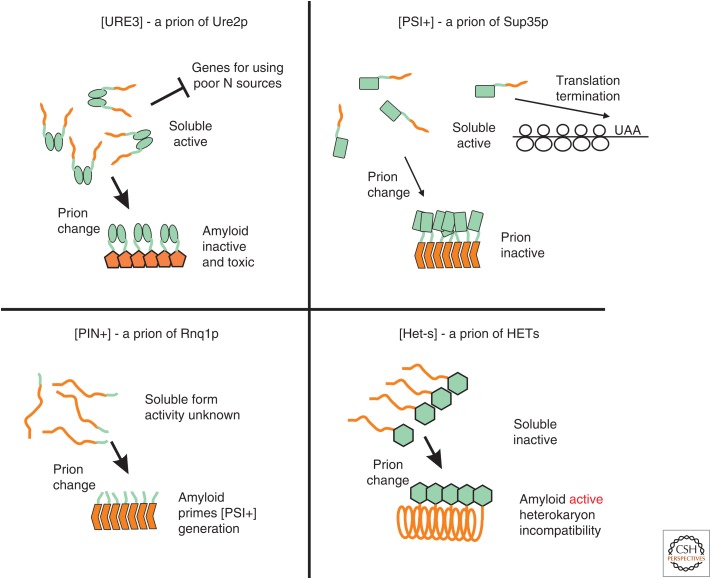
[URE3] is a nonchromosomal genetic element that produces failure of nitrogen catabolite repression (Lacroute 1971), as does recessive chromosomal ure2 mutation (Aigle and Lacroute 1975). Most importantly, ure2 mutants cannot propagate [URE3] (Aigle and Lacroute 1975). [URE3] can be cured by growth in low concentrations of guanidine (Wickner 1994) (much later shown to act by inhibiting Hsp104 [Ferreira et al. 2001; Jung and Masison 2001; Jung et al. 2002]), but it can rarely arise again in the cured strain (Wickner 1994). Finally, transient overproduction of Ure2p increases the frequency with which the [URE3] nonchromosomal genetic element arises. Thus, [URE3] has all of the properties expected of a prion of Ure2p (Fig. 1) (Wickner 1994). DAL5, encoding the allantoate transporter, is the most strongly nitrogen-regulated gene (Turoscy and Cooper 1987), and [URE3] is typically assayed by measuring activity of the DAL5 promoter, either directly as uptake of ureidosuccinate in the presence of ammonia, or fused to the ADE2 gene as adenine prototrophy/auxotrophy (Lacroute 1971; Schlumpberger et al. 2001).
Figure 1.
The yeast and fungal prions [URE3], [PSI+], [PIN+], and [Het-s]. Formation of amyloid by Ure2p, Sup35p, Rnq1p, and HET-s results in either the loss of their normal functions (for Ure2p and Sup35p) or the appearance of novel activity (in Rnq1p and HET-s), all of which are heritable because the amyloid catalyzes the conversion of the nonamyloid to the amyloid form. Most variants of [URE3] are also toxic to the cell, and most [PSI+] prions are lethal or severely impair growth because the essential function of Sup35p is impaired or lost (McGlinchey et al. 2011). The prion domains of Ure2p and Sup35p that form the amyloid cores also have normal nonprion functions (see text).
[PSI+] is a nonchromosomal genetic element that produces elevated readthrough of nonsense codons (Cox 1965), like chromosomal sup35 mutations, and is typically monitored by readthrough of a nonsense mutation in ADE1 or ADE2. Overexpression of Sup35p increases the frequency of [PSI+] arising de novo (Chernoff et al. 1993), and the curing of [PSI+] by high osmotic strength (Singh et al. 1979) is reversible (Lund and Cox 1981). Finally, [PSI+] propagation requires the N-terminal part of the Sup35 protein (Ter-Avanesyan et al. 1994). Thus, [PSI+] is a prion of Sup35p (Fig. 1) (Wickner 1994).
These genetic criteria continue to be useful in more recent searches for new prions (Derkatch et al. 2001; Du et al. 2008; Alberti et al. 2009; Patel et al. 2009; Rogoza et al. 2010; Suzuki et al. 2012), and prion induction by prion protein overproduction is a standard method for making new prions for study. In some cases, the prion has a novel phenotype distinct from that due to absence of the normal form of the protein. The [PIN+] prion of Rnq1p was detected by its ability to rarely seed formation of the [PSI+] prion (Derkatch et al. 1997), but rnq1Δ has neither this effect nor any other (Fig. 1) (Sondheimer and Lindquist 2000). This Pin+ phenotype was also used in isolation of other prions (Du et al. 2008; Patel et al. 2009; Suzuki et al. 2012).
The presence of protease-resistant Ure2p in extracts of cells carrying the [URE3] prion (Masison and Wickner 1995), the aggregation of Sup35p specifically in [PSI+] strains (Patino et al. 1996; Paushkin et al. 1996) and Ure2p in [URE3] strains (Edskes et al. 1999), the ability of extracts from [PSI+] cells to seed aggregate formation by Sup35p (Glover et al. 1997; Paushkin et al. 1997), and the formation of amyloid in vitro by Sup35p (Glover et al. 1997; King et al. 1997) and by Ure2p (Taylor et al. 1999) suggest that amyloid of these proteins constitutes their prion forms. Transmission of [PSI+] (King and Diaz-Avalos 2004; Tanaka et al. 2004) and later of [URE3] (Brachmann et al. 2005) by amyloids formed in vitro from recombinant Sup35p and Ure2p, respectively, confirmed this conclusion.
PRION DOMAINS
The prion properties of Ure2p and Sup35p are determined by Q/N rich N-terminal domains of these proteins, and these regions have proven to comprise the core of the amyloid structures that are the basis of the [URE3] and [PSI+] prions (Ter-Avanesyan et al. 1994; Masison and Wickner 1995; King et al. 1997; Taylor et al. 1999). Prion domains can be linked to other proteins, with the other protein serving as a reporter of prion formation (Li and Lindquist 2000). This approach has also been useful in searching for new prions (Alberti et al. 2009). Of course, it must be confirmed that a segment acting as a prion domain in a fusion can also do so in its native context.
Surprisingly, the sequence of the prion domains of Ure2p and Sup35p are of minimal importance in their ability to form prions, as randomly shuffling these segments, fusing the shuffled segments to the remainder of the gene, and integrating these constructs in place of the normal gene uniformly (each of five shuffles for each protein), results in proteins that can still form prions (Ross et al. 2004, 2005a). This work made it clear that it is the amino acid composition, and not the sequence, that determines prion-forming ability; and within the Q/N-rich context, this method showed that hydrophobic, aromatic, and hydrophilic residues favor prion formation, whereas charged residues or prolines are unfavorable (Toombs et al. 2010). In fact, artificial prion domains, designed based on these results, actually worked as a prion (Toombs et al. 2012). The oligopeptide repeats in the Sup35 prion domain, reminiscent of the PrP octapeptide repeats, were proposed to play a key role in the [PSI+] prion, but shuffling the amino acid residues of this region did not affect prion propagation or generation, showing that it was the composition of this domain, not the repeats, that made it a prion (Toombs et al. 2011).
THERE ARE MANY YEAST PRIONS
Swi1p, a component of a chromatin-remodeling complex, can form the [SWI+] prion, resulting in poor growth on raffinose, galactose, or glycerol, like an swi1 mutation (Du et al. 2008). Cyc8p, a subunit of a transcription repressor complex, can form the [OCT+] prion, with phenotypes like those of cyc8 mutants (Patel et al. 2009). A large screen of proteins with domains similar in composition to known prion domains revealed several protein domains capable of acting as a prion domain, and the transcription factor Mot3p was shown to actually form a prion called [MOT3+] (Alberti et al. 2009). Like mot3 mutants, [MOT3+] cells show pseudohyphal growth and biofilm formation (Holmes et al. 2013). Sfp1p, a transcription factor regulating expression of many translation-related proteins, can form a prion, [ISP+], named for its phenotype, which is the reverse of [PSI+] (Rogoza et al. 2010). [ISP+] is a largely nuclear-limited prion and therefore shows limited infectivity in cytoplasmic mixing experiments (Rogoza et al. 2010). Mod5p, a tRNA-isopentenyltransferase, can form the [MOD+] prion with a fluconazole-resistant phenotype similar to mot5 mutants (Suzuki et al. 2012). In contrast to the other known yeast prions, the prion domain of Mod5p is not Q/N rich (Suzuki et al. 2012). [Het-s] is a prion of the filamentous fungus Podospora anserina (Fig. 1) and is of interest as a clearly functional prion (for a review, see Saupe 2011).
The definition of the word “prion” as an infectious protein (Prusiner 1982) does not require amyloid formation as its basis. Indeed, the vacuolar protease B, Prb1p (Jones 1991), can form a prion called [β] (Roberts and Wickner 2003), which is simply the active form of the protease. Prb1p is made as an inactive precursor, which is activated by cleavage by protease A. In the absence of protease A, active Prb1p itself can cleave and activate the precursor. This process is self-propagating, and [β] has all of the properties of a prion (Roberts and Wickner 2003). In the absence of protease A, this prion is important for survival in stationary phase and required for meiotic sporulation (Roberts and Wickner 2003). The [GAR] nonchromosomal genetic element conferring glucosamine-resistance (Ball et al. 1976; Kunz and Ball 1977) is also proposed to be a prion based on a self-propagating physiological state (Brown and Lindquist 2009), but the nature of the state and the mechanism of resistance to glucosamine are still unclear.
PRION VARIANTS/STRAINS AND PRION CLOUDS
A single protein sequence can be the basis for many different prion variants (called strains in mammalian prions), with differing biological and structural properties. Derkatch et al. (1996) first recognized strong (fully Ade+) and weak (leaky Ade+) variants of [PSI+], but there are now variants of [PSI+] and [URE3] differing in their strength and stability (Schlumpberger et al. 2001; Tanaka et al. 2004; Brachmann et al. 2005), the effects of overproduction or deficiency of various chaperones (Kushnirov et al. 2000b; Kryndushkin et al. 2002; Borchsenius et al. 2006), prion–prion interactions (Bradley and Liebman 2003), their ability to be transmitted to other prion protein sequences (interspecies or intraspecies barriers) (King 2001; Edskes et al. 2009; Vishveshwara and Liebman 2009; Chen et al. 2010; Bateman and Wickner 2012), their lethality/toxicity to the host (McGlinchey et al. 2011), and their sensitivity to curing by the Btn2/Cur1 antiprion system (Wickner et al. 2014). Moreover, combinations of these differences multiply the number of possible strains. There are both strong and weak variants with each of the four possible intraspecies transmission variants of [PSI+], all propagated with the same Sup35p sequence (Bateman and Wickner 2012, 2013).
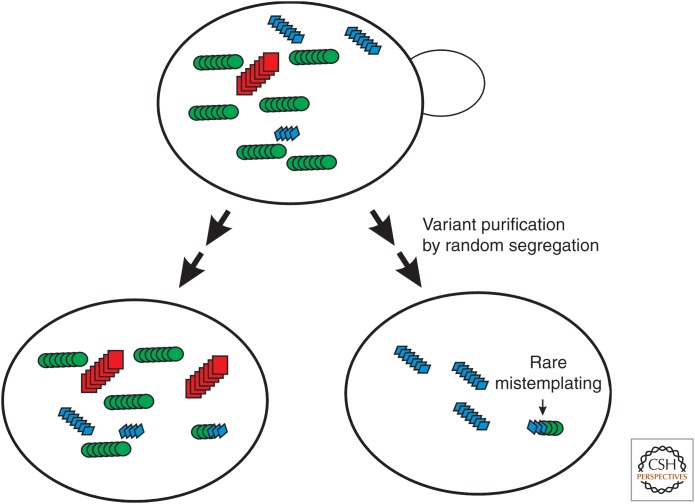
Although prion variants are propagated with relative stability, changes in variant properties can occur under selection pressure in both mammals and yeast, such as in crossing a species barrier or application of a drug (Kimberlin et al. 1987; Vishveshwara and Liebman 2009; Li et al. 2010). However, even without evident selection pressure, variant properties can change, and a mixture of variants can segregate during growth (Bateman and Wickner 2013). Extensive clonal isolation appears to purify single variants, but on further propagation, all of the other variants arise again, showing that variant mutation also occurs, even under nonselective conditions (Fig. 2) (Bateman and Wickner 2013). These results provide strong direct support for the “prion cloud” model proposed by Collinge to explain the selectability of new variants (Collinge and Clarke 2007). Previous data were consistent with this model but could also have been explained by induction of new variants as a result of effects of the selection scheme (new protein sequence or amyloid-binding drug) on the amyloid structure.
Figure 2.
The prion cloud model. A cell with a single predominant prion variant is generally a mixture of variants as a result of occasional mistemplating, and these variants will gradually be purified from each other by random segregation during growth. This model was proven for the yeast prion [PSI+] (Bateman and Wickner 2013) but was first hypothesized to explain the properties of mammalian prions (Collinge and Clarke 2007).
PRION AMYLOID STRUCTURE EXPLAINS INHERITANCE OF PRION VARIANT INFORMATION
Solid-state nuclear magnetic resonance (NMR) and electron microscopic studies of infectious amyloids of the prion domains of Sup35p, Ure2p, and Rnq1p have shown that each has an in-register parallel-folded β-sheet architecture (Fig. 3) (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008; Gorkovskiy et al. 2014; for a review, see Tycko and Wickner 2013). Shuffled versions of the Ure2p and Sup35p prion domains also had this architecture (Shewmaker et al. 2008). Indeed, the fact that shuffled prion domains remained able to form prions was the basis for an early proposal that these domains must have an in-register parallel architecture (Ross et al. 2005b). Mass per unit length measurements show that these filaments have one monomer per ∼4.8 Å, the distance between β strands in a β-sheet (Baxa et al. 2003; Diaz-Avalos et al. 2005; Chen et al. 2009). Electron micrographs of the infectious filaments of the prion domains of Ure2p, Sup35p, and Rnq1p showed filaments that were too narrow to be a single broad β-sheet, implying that the sheet must be folded several times along the long axis of the filaments.
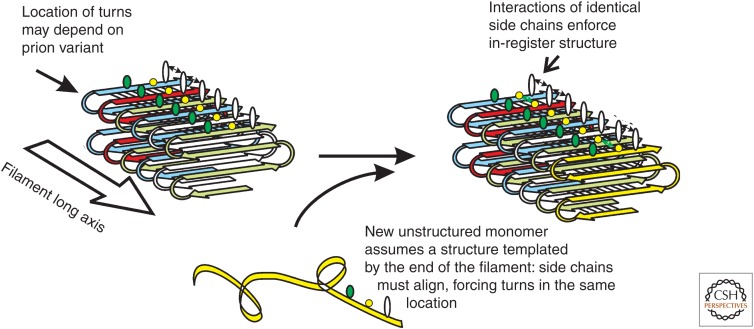
Figure 3.
Protein templating mechanism of prions. Solid-state nuclear magnetic resonance (NMR) experiments show that the infectious amyloids of the prion domains of Sup35p, Ure2p, and Rnq1p have an in-register parallel β-sheet conformation with the sheets folded along the filament long axis as shown (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008; Gorkovskiy et al. 2014). The energetically favorable interactions between identical hydrophobic or hydrophilic (H-bonding) side chains keep the structure in-register. The same interactions ensure that a new monomer joining the end of the filament will assume the same conformation as molecules already in the filament. This ensures that the location of the folds/turns in the newly joining molecule will be the same as the previous molecules. We have suggested that prion variants differ in the location of the folds/turns in the β-sheet, and by this mechanism (Wickner et al. 2007, 2010, 2013), a protein can template its own conformation, just as DNA templates its own sequence.
The folded, in-register parallel β-sheet architecture of the yeast prion amyloids of Ure2p, Sup35p, and Rnq1p is different than the two-turn β-helix structure of the HET-s prion amyloid (Ritter et al. 2005; Wasmer et al. 2008). These yeast prions have multiple prion variants for a given prion protein sequence, whereas only a single variant of the [Het-s] prion is known. A functional prion such as [Het-s] is expected to have only a single variant, selected for the ability to perform this function. In contrast, the yeast prions appear to be pathological (see discussion below) and are not constrained by the demands of prion function. A knee bends only at one place in one way, but a leg can break at many sites in many ways.
Perhaps the central issue in the prion field is how conformational templating occurs. How can a single protein sequence encode and transmit any of many different amyloid conformations? At present, the only model to explain this phenomenon is our proposal based on the folded, in-register parallel β-sheet architecture of the yeast prion amyloids (Fig. 3) (Wickner et al. 2007, 2010, 2013). We suggest that a primary determinant of prion variants is the locations of the folds/loops in the β-sheet. The parallel in-register architecture is maintained in-register by favorable interactions between identical side chains along the long axis of the filaments. A line of hydrogen bonds between aligned (identical) hydrophilic residues (Q, N, S, T) all along the filament long axis, or hydrophobic interactions between aligned hydrophobic residues (V, L, I, F, Y, W, M), would be possible only if the register was maintained. Only charged amino acid side chains cannot have favorable side chain–side chain interactions with an identical residue, and there are almost no charged residues in these prion domains. The unstructured prion domain of the soluble form of the prion protein (e.g., Pierce et al. 2005) joins the end of the filament in such a way as to maximize these favorable interactions among identical residues, and that requirement results in the new molecule on the end of the filament adopting the same conformation as the molecules already in the filament, with its turns in the same places as the molecules already in the filament (see Fig. 3). Just as DNA templates its sequence, a prion amyloid can template its conformation by this mechanism and so can act as a gene with many different heritable alleles (variants/amyloid conformations). Our recent data identify the locations of some of these folds in the sheets in support of this model (Gorkovskiy et al. 2014).
PRION BIOLOGY: PATHOLOGICAL YEAST PRIONS AND A FUNCTIONAL FUNGAL PRION
The [Het-s] prion of P. anserina is necessary for heterokaryon incompatibility, a process by which the fungus limits fusion of hypae to fusion partners that are genetically identical at a dozen chromosomal loci to limit the spread of detrimental viruses and plasmids (Saupe 2011). This prion plainly serves a function useful for the host and is a case study in what to expect for such “functional prions.” There is only one known variant of [Het-s], presumably because the HETs protein sequence has been selected during evolution to form a prion with the desired characteristics. Accordingly, amyloid of HETs made in vitro has a uniform structure, reflected in sharp peaks in solid-state NMR studies (Wasmer et al. 2008). This functional prion is found in >90% of wild isolates with the het-s allele, as expected for a beneficial prion (Debets et al. 2012).
Lethal or toxic variants of [PSI+] and [URE3] are more common than the mild variants usually studied (McGlinchey et al. 2011). The yeast prions [PSI+], [URE3], and [PIN+] form many variants (see above), and even the mildest variants are rare in wild populations (Chernoff et al. 2000; Resende et al. 2003; Nakayashiki et al. 2005; Halfmann et al. 2012), both implying that they are pathological. Reports of marginal benefits of carrying the mildest [PSI+] prion variants (Eaglestone et al. 1999; True and Lindquist 2000; Halfmann et al. 2012) have not been reproduced (True and Lindquist 2000; Namy et al. 2008; Wickner et al. 2015).
Sup35p from several different species can form [PSI+] prions (Chernoff et al. 2000; Kushnirov et al. 2000a; Santoso et al. 2000; Nakayashiki et al. 2001; Chen et al. 2007; Afanasieva et al. 2011), but many others cannot (Edskes et al. 2014). Ure2p of most species of the genus Saccharomyces can form the [URE3] prion (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003; Edskes et al. 2009), but that of Saccharomyces castellii cannot (Edskes et al. 2009). A careful study showed that the Kluyveromyces lactis Ure2p cannot form [URE3] even in K. lactis itself (Safadi et al. 2011). Ure2p of Candida albicans, although closely related to that of Saccharomyces cerevisiae, cannot form [URE3], whereas that of Candida glabrata, more distant from the S. cerevisiae Ure2p, can form [URE3] with all of its properties (Edskes et al. 2011; Engel et al. 2011; Edskes and Wickner 2013). Thus, [PSI+] and [URE3] prion-forming ability appears to be sporadically distributed. The prion domains of these proteins have nonprion functions, for example, roles in mRNA turnover of the Sup35 prion domain (Hoshino et al. 1999; Hosoda et al. 2003) and microtubule binding (Li et al. 2014) and stabilization of the full-length protein in the case of the Ure2p prion domain (Shewmaker et al. 2007). These functions may explain the persistence of these domains in evolution despite their occasional formation of prions.
PRION ECOLOGY AND EVOLUTION
Yeast and fungal prions are determined by two levels of inheritance. First, the chromosomal gene encoding the prion protein is under positive or negative selection pressure to maintain its normal function and to maintain or lose its ability to form a prion. Second, having formed a prion, the prion itself is under positive or negative selection pressure depending on the phenotype it confers on the cell and its stability. In considering whether prion formation by a particular protein is beneficial or detrimental, one must consider the full range of prion variants that can arise, their relative frequencies, and the frequencies with which yeast will encounter an environmental condition under which there is a prion-specific beneficial or detrimental effect. Likewise, the nonprion functions of the protein may be affected by the prion conversion. This is quite unlike most genes, whose encoded protein usually has one basic heritable state, determined by the protein sequence. The protein activity may be regulated by the environment, but generally not heritably so. The preceding program may be impossible to execute, but a beginning effort has been made to examine some aspects of the evolution and ecology of prion proteins and prions themselves.
It is not difficult to find detrimental viruses and prions in wild populations. For example, the uniformly fatal chronic wasting disease (CWD) prion is found in ∼10% of wild deer and elk in several areas of the United States. The infectivity of CWD can outweigh the fatality of the disease. For a beneficial virus or prion, infectivity and benefit to the host are working in the same direction, so the infection should spread rapidly in the wild, resulting in a high prevalence of the prion. Mitochondria began as bacterial endosymbionts, infectious elements. Although the mitochondrial genome is rather unstable, it is so beneficial that nearly all wild isolates carry this nonchromosomal DNA. The 2-µm DNA plasmid replicates and, like the yeast prions, is spread by mating. Although three groups have found that the 2-µm DNA plasmid mildly slows growth of yeast about 1%–3% (Futcher and Cox 1983; Mead et al. 1986; Futcher et al. 1988; Kelly et al. 2012), it was found in 38 of 70 wild strains examined (Nakayashiki et al. 2005). This provides a standard of comparison against which prions may be judged. [URE3], [PSI+], and [SWI+] were not found in any of the same 70 wild strains, indicating that they must confer a substantially greater detriment than does 2-µm DNA (Nakayashiki et al. 2005; Bateman and Wickner 2012; Kelly et al. 2012). Note that this is the detriment of the most mild variants of [URE3] or [PSI+] or other prions. The [PIN+] prion was found in ∼15% of these wild strains (Nakayashiki et al. 2005), and [MOT3+] was detected in ∼6% of a different set of 96 wild isolates (Halfmann et al. 2012), indicating that both are also detrimental on the net.
The preceding discussion shows that even the mildest variants of the prions examined are detrimental to their hosts. Nonetheless, they are found in nature at frequencies above their frequency of de novo generation. For [PIN+], two possible scenarios were considered: (1) there may be some part of the ecological niche of S. cerevisiae in which [PIN+], should it arise, would be beneficial to its host, and this has led to expansion of the line in which it arose because of this benefit; and (2) [PIN+] is mildly detrimental in all niches, but it has spread by mating despite this detriment. Detailed examination of the occasional wild [PIN+] strains has shown that the presence of [PIN+] is associated with the recent occurrence of outcross mating, as shown by heterozygosity (Kelly et al. 2014). This result favors the second explanation.
The polymorphs of Sup35 among wild S. cerevisiae that limit the spread of [PSI+] are also a reflection of the ecology of this prion (Bateman and Wickner 2012). It would be of interest to know whether certain ecological niches favor certain prion domain sequences.
ANTIPRION SYSTEMS
Ribosome-Associated Hsp70s (Ssbs) Inhibit [PSI+] Generation
Ssb1 or Ssb2 are members of the Hsp70 family associated with the ribosome and involved in folding of nascent proteins (Fig. 4) (Nelson et al. 1992; Pfund et al. 1998). Hsp104 overproduction cures the [PSI+] prion by an as-yet uncertain mechanism (Chernoff et al. 1995). Overproduction of Ssb1p or Ssb2p facilitates the curing of [PSI+] by overproduced Hsp104 (Chernoff et al. 1999). Moreover, in an ssb1Δ ssb2Δ strain, the frequency of [PSI+] formation is elevated, whether spontaneous or induced by Sup35p overexpression (Chernoff et al. 1999). Restoration of Ssb levels to [PSI+] strains generated in the ssb1Δ ssb2Δ strain did not cure [PSI+], indicating that Ssb1 and Ssb2, at their normal levels, inhibit the generation of [PSI+] rather than its propagation (Chernoff et al. 1999). Since yeast prions are genes and the generation of [PSI+] constitutes a “mutation” from [psi-], it is proposed that Ssbs are analogous to DNA repair systems, or an antimutator system (Chernoff et al. 1999).
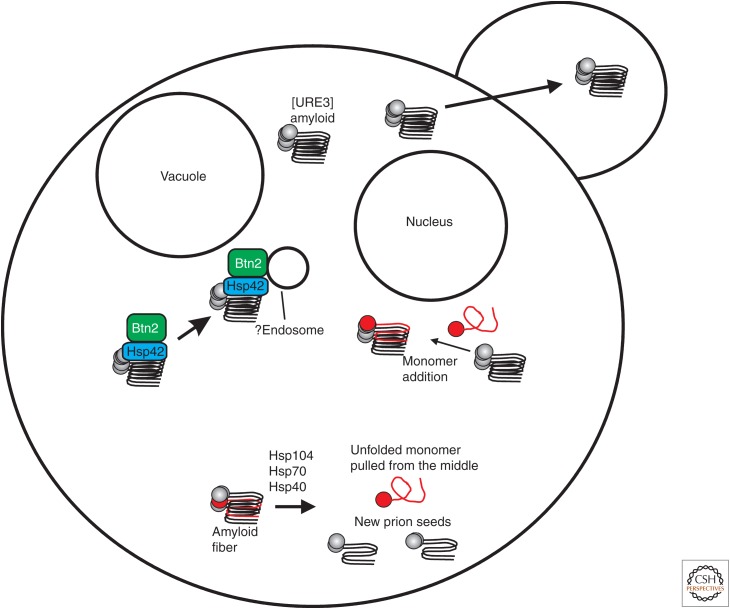
Figure 4.
Prion-handling systems. Prion protein monomers are synthesized and are soon incorporated into amyloid fibers (see Fig. 2). These fibers grow and are split by the ability of the Hsp104–Hsp70–Hsp40 machine to remove a monomer from the middle of the fiber. This ensures sufficient prion seeds to distribute to daughter cells, allowing prion propagation. Some filaments are sequestered by the Btn2p–Hsp42 system. If the seed number is sufficiently low, this results in curing the prion from progeny cells. The Ssb1/2 Hsp70s act to inhibit conversion of the normal form (of Sup35p) to the prion form, an inhibition of prion generation not of propagation (Chernoff et al. 1999).
Btn2/Cur1 at Normal Levels Cure Most [URE3] Variants
Overproduction of Btn2p, or its paralog Cur1p, efficiently cures [URE3-1], the original [URE3] variant isolated by Lacroute (Kryndushkin et al. 2008). Remarkably, in the process of curing, Ure2p aggregates are concentrated at a single cellular site, co-localized with Btn2p, suggesting that overproduced Btn2p collects the prion aggregates, preventing their distribution to both daughter cells (the sequestration model) (Fig. 4) (Kryndushkin et al. 2008). Btn2p also dramatically co-localizes with several nonamyloid aggregates in yeast (Kryndushkin et al. 2012). Direct measures of [URE3] seed-number by the methods developed by Cox et al. (2003) showed that normal levels of Btn2p and Cur1p lower the seed number of [URE3-1] without curing it (Kryndushkin et al. 2008). Nearly all [URE3] variants isolated in a btn2Δ cur1Δ strain were cured by restoring normal levels of either or both of these proteins by mating with a wild-type strain or introducing a single-copy plasmid with the normal gene(s) (Wickner et al. 2014). This shows that these proteins are normally curing most [URE3] variants as they arise, so that usually one only sees those variants whose curing requires overproduction of Btn2/Cur1. What distinguishes those [URE3]s curable by normal levels of Btn2/Cur1 from those only cured by overproduction of these proteins? Again, seed (propagon) number measurements (Cox et al. 2003) showed that [URE3] variants eliminated by just restoring normal levels of Btn2 and Cur1 were several-fold lower than those only cured by overproduction of those proteins (Wickner et al. 2014). This result is also what one would predict from the sequestration model of curing. Normal levels of the proteins would only be able to sequester a limited number of prion seeds/propagons and so should only be able to cure variants with a low seed number. Other variants with higher seed number would require overproduction of the curing proteins, as observed (Wickner et al. 2014).
Btn2p interacts with and co-localizes with Hsp42 (Malinovska et al. 2012), Hsp42 co-localizes with and sequesters certain nonamyloid aggregates (Specht et al. 2011), Hsp42 is necessary for curing of [URE3] by overproduction of Btn2p (Wickner et al. 2014), and overproduction of Hsp42p itself cures [URE3] (Wickner et al. 2014). Thus, Btn2p appears to be part of a group of proteins involved in aggregate sequestration.
Btn2p was first identified as a protein overproduced in mutants lacking Btn1p, the yeast homolog of human CLN3, whose mutation leads to juvenile neuronal ceroid lipofuscinosis, also known as Batten disease (Pearce and Sherman 1997; Pearce et al. 1999). Btn2p is localized to late endosomes, binds to endocytic v-SNAREs, and is necessary for proper retrieval of Kex2p and Yif1p from late endosomes to the Golgi (Kama et al. 2007). Overexpression of Btn3p interferes with both the protein-trafficking role of Btn2 and its ability to cure [URE3] (Kanneganti et al. 2011). Does Btn2p move Ure2p aggregates to the endosomes or Golgi?
Chaperones and Yeast Prions
Chaperones play diverse roles in prion propagation (see Chernova et al. 2016). The Hsp104–Hsp70–Hsp40 machine breaks amyloid filaments by extracting a molecule from within the filament and thereby creates new prion “seeds” or “propagons” (Fig. 4) (Chernoff et al. 1995; Paushkin et al. 1996; Glover et al. 1997; Jung et al. 2000; Higurashi et al. 2008; Haslberger et al. 2010; for a review, see Reidy and Masison 2011). Overproduction of Hsp104 cures [PSI+] (Chernoff et al. 1995), whereas overproduction of Ydj1p or Sse1p cures [URE3] (Moriyama et al. 2000; Kryndushkin and Wickner 2007). Hsp90 and Sti1p are involved in the Hsp104 overproduction curing of [PSI+] (Reidy and Masison 2010). As mentioned above, the small heat-shock protein Hsp42 is involved in the Btn2/Cur1 system of aggregate segregation, and its overproduction cures [URE3] (Wickner et al. 2014).
FUTURE DIRECTIONS
With the growing evidence that the common human amyloidoses involve prion or prion-like mechanisms, study of the yeast prions has taken on added importance. The ease with which genetic manipulations are performed in yeast and the vast array of genomic tools available have made rapid progress in the yeast prion area possible. It is unlikely that the existence of many prions in S. cerevisiae is unique to this organism, and the prion-like features of many human amyloidoses may reflect this fact. Searches for new prions in other organisms will certainly reveal interesting new biology (e.g., the P. anserina [Het-s] prion) and pathology. Structural work on yeast prions has produced for the first time a working model that can explain how prion variants are inherited, but additional structural detail will require extensive further studies. The mechanisms of the Btn2/Cur1 prion curing will be particularly important because this antiprion system normally cures prions as they arise, and enhancement of homologous or analogous human system(s) may be utilized for treatment of amyloidoses. Continued dissection of the mechanisms by which chaperones shape prion formation and propagation is revealing important aspects of these complex systems.
ACKNOWLEDGMENTS
This work is supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Afanasieva EG, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. 2011. Molecular basis for transmission barrier and interference between closely related prion proteins in yeast. J Biol Chem 286: 15773–15780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigle M, Lacroute F. 1975. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol Gen Genet 136: 327–335. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper T, Cramp WA, Haig DA, Clarke MC. 1967. Does the agent of scrapie replicate without nucleic acid? Nature 214: 764–766. [DOI] [PubMed] [Google Scholar]
- Ball AJ, Wong DK, Elliott JJ. 1976. Glucosamine resistance in yeast. I: A preliminary genetic analysis. Genetics 84: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman DA, Wickner RB. 2012. [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: Intraspecies “species barriers.” Genetics 190: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman D, Wickner RB. 2013. The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet 9: e1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. 2003. Conservation of the prion properties of Ure2p through evolution. Mol Biol Cell 14: 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven A. 2003. Architecture of Ure2p prion filaments: The N-terminal domain forms a central core fiber. J Biol Chem 278: 43717–43727. [DOI] [PubMed] [Google Scholar]
- Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau WM, Tycko R. 2007. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46: 13149–13162. [DOI] [PubMed] [Google Scholar]
- Bolton DC, McKinley MP, Prusiner SB. 1982. Identification of a protein that purifies with the scrapie prion. Science 218: 1309–1311. [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. 2006. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet 49: 21–29. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. 2005. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J 24: 3082–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. 2003. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics 165: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Lindquist S. 2009. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev 23: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Newnam GP, Chernoff YO. 2007. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci 104: 2791–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. 2009. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci 106: 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bruce KL, Newnam GP, Gyoneva S, Romanyuk AV, Chernoff YO. 2010. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol Microbiol 76: 1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. 1993. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 24: 268–270. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono BI, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. 1999. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI] prion. Mol Cell Biol 19: 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. 2000. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 35: 865–876. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Wilkinson KD, Chernoff YO. 2016. Prions, chaperones, and proteostasis in yeast. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith JM, et al. 1985. Identification of scrapie prion protein-specific mRNA in scrapie-infected brain. Nature 315: 331–333. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. 2007. A general model of prion strains and their pathogenicity. Science 318: 930–936. [DOI] [PubMed] [Google Scholar]
- Cox BS. 1965. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20: 505–521. [Google Scholar]
- Cox BS, Ness F, Tuite MF. 2003. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics 165: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets AJ, Dalstra HJ, Slakhorst M, Koopmanschap B, Hoekstra RF, Saupe SJ. 2012. High natural prevalence of a fungal prion. Proc Natl Acad Sci 109: 10432–10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144: 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. 2001. Prions affect the appearance of other prions: The story of [PIN]. Cell 106: 171–182. [DOI] [PubMed] [Google Scholar]
- Diaz-Avalos R, King CY, Wall JS, Simon M, Caspar DLD. 2005. Strain-specific morphologies of yeast prion amyloids. Proc Natl Acad Sci 102: 10165–10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AG, Meikle VMH, Fraser H. 1968. Identification of a gene which controls the incubation period of some strains of scrapie in mice. J Comp Path 78: 293–299. [DOI] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J 18: 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Wickner RB. 2002. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full-length protein. Proc Natl Acad Sci 99: 16384–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Wickner RB. 2013. The [URE3] prion in Candida. Eukaryot Cell 12: 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Gray VT, Wickner RB. 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci 96: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, McCann LM, Hebert AM, Wickner RB. 2009. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 181: 1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Engel A, McCann LM, Brachmann A, Tsai HF, Wickner RB. 2011. Prion-forming ability of Ure2 of yeasts is not evolutionarily conserved. Genetics 188: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Khamar HJ, Winchester CL, Greenler AJ, Zhou A, McGlinchey RP, Gorkovskiy A, Wickner RB. 2014. Sporadic distribution of prion-forming ability of Sup35p from yeasts and fungi. Genetics 198: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Shewmaker F, Edskes HK, Dyda F, Wickner RB. 2011. Amyloid of the Candida albicans Ure2p prion domain is infectious and has a parallel in-register β-sheet structure. Biochemistry 50: 5971–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Futcher AB, Cox BS. 1983. Maintenance of the 2 μm circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol 154: 612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Reid E, Hickey DA. 1988. Maintenance of the 2 μm circle plasmid of Saccharomyces cerevisiae by sexual transmission: An example of selfish DNA. Genetics 118: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Shirmer EC, Patino MM, Liu JJ, Lindquist S. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819. [DOI] [PubMed] [Google Scholar]
- Gorkovskiy A, Thurber KR, Tycko R, Wickner RB. 2014. Locating the folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc Natl Acad Sci 111: E4615–E4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JS. 1967. Self-replication and scrapie. Nature 215: 1043–1044. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslberger T, Bukau B, Mogk A. 2010. Towards a unifying mechanism for the ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol 88: 63–75. [DOI] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci 105: 16596–16601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. 2013. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 153: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kobayashii T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 278: 38287–38291. [DOI] [PubMed] [Google Scholar]
- Jones EW. 1991. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem 266: 7963–7966. [PubMed] [Google Scholar]
- Jung G, Masison DC. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr Microbiol 43: 7–10. [DOI] [PubMed] [Google Scholar]
- Jung G, Jones G, Wegrzyn RD, Masison DC. 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Jones G, Masison DC. 2002. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci 99: 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R, Robinson M, Gerst JE. 2007. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol Cell Biol 27: 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V, Kama R, Gerst JE. 2011. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell 22: 1648–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Shewmaker FP, Kryndushkin D, Wickner RB. 2012. Sex, prions and plasmids in yeast. Proc Natl Acad Sci 109: E2683–E2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Busby B, Wickner RB. 2014. Effect of domestication on the spread of the [PIN+] prion in Saccharomyces cerevisiae. Genetics 197: 1007–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin RH, Cole S, Walker CA. 1987. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol 68: 1875–1881. [DOI] [PubMed] [Google Scholar]
- King CY. 2001. Supporting the structural basis of prion strains: Induction and identification of [PSI] variants. J Mol Biol 307: 1247–1260. [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. 2004. Protein-only transmission of three yeast prion strains. Nature 428: 319–323. [DOI] [PubMed] [Google Scholar]
- King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wüthrich K. 1997. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci 94: 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB. 2007. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell 18: 2149–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. 2002. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem 277: 23702–23708. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D, Shewmaker F, Wickner RB. 2008. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J 27: 2725–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Ihrke G, Piermartiri TC, Shewmaker F. 2012. A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol Microbiol 86: 1531–1547. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Ball AJ. 1977. Glucosamine resistance in yeast. II: Cytoplasmic determinants conferring resistance. Mol Gen Genet 153: 169–177. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Kochneva-Pervukhova NV, Cechenova MB, Frolova NS, Ter-Avanesyan MD. 2000a. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J 19: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin D, Boguta M, Smirnov VN, Ter-Avanesyan MD. 2000b. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 10: 1443–1446. [DOI] [PubMed] [Google Scholar]
- Lacroute F. 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol 106: 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S. 2000. Creating a protein-based element of inheritance. Science 287: 661–664. [DOI] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. 2010. Darwinian evolution of prions in cell culture. Science 327: 869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rayman JB, Kandel ER, Derkatch IL. 2014. Functional role of Tia1/Pub1 and Sup35 prion domains: Directing protein synthesis machinery to the tubulin cytoskeleton. Mol Cell 55: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PM, Cox BS. 1981. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet Res 37: 173–182. [DOI] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spaciotemporal distribution of protein aggregates. Mol Biol Cell 23: 3041–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95. [DOI] [PubMed] [Google Scholar]
- McGlinchey R, Kryndushkin D, Wickner RB. 2011. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci 108: 5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DJ, Gardner DCJ, Oliver SG. 1986. The yeast 2 μm plasmid: Strategies for the survival of a selfish DNA. Mol Gen Genet 205: 417–421. [DOI] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. 2000. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol 20: 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. 2001. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell 7: 1121–1130. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci 102: 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset C. 2008. Epigenetic control of polyamines by the prion [PSI+]. Nat Cell Biol 10: 1069–1075. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegilhoffer T, Nicolet C, Werner-Washburne M, Craig EA. 1992. The translation machinery and 70 kDal heat shock protein cooperate in protein synthesis. Cell 71: 97–105. [DOI] [PubMed] [Google Scholar]
- Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Templow DB, Hood LE, et al. 1985. A cellular gene encodes scrapie PrP 27–30 protein. Cell 40: 735–746. [DOI] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 11: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273: 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15: 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. 1997. In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277: 381–383. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Sherman F. 1997. BTN1, a yeast gene corresponding to the human gene responsible for Batten’s disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast 13: 691–697. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Ferea T, Nosel SA, Das B, Sherman F. 1999. Action of BTN1, the yeast ortholog of the gene mutated in Batten disease. Nat Genet 22: 55–58. [DOI] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J 17: 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. 2005. Is the prion domain of soluble Ure2p unstructured? Biochemistry 44: 321–328. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. [DOI] [PubMed] [Google Scholar]
- Reidy M, Masison DC. 2010. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol Cell Biol 30: 3542–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Masison DC. 2011. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion 5: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. 2003. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol 49: 1005–1017. [DOI] [PubMed] [Google Scholar]
- Ritter C, Maddelein ML, Siemer AB, Lührs T, Ernst M, Meier BH, Saupe SJ, Riek R. 2005. Correlation of structural elements and infectivity of the HET-s prion. Nature 435: 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Wickner RB. 2003. A class of prions that propagate via covalent auto-activation. Genes Dev 17: 2083–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. 2010. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci 107: 10573–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Baxa U, Wickner RB. 2004. Scrambled prion domains form prions and amyloid. Mol Cell Biol 24: 7206–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. 2005a. Primary sequence independence for prion formation. Proc Natl Acad Sci 102: 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton AP, Wickner RB. 2005b. Prion domains: Sequences, structures and interactions. Nat Cell Biol 7: 1039–1044. [DOI] [PubMed] [Google Scholar]
- Safadi RA, Talarek N, Jacques N, Aigle M. 2011. Yeast prions: Could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res 11: 151–153. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. 2000. Molecular basis of a yeast prion species barrier. Cell 100: 277–288. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. 2011. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Sem Cell Dev Biol 22: 460–468. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M, Prusiner SB, Herskowitz I. 2001. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol 21: 7035–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. 2006. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci 103: 19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Ross ED, Tycko R, Wickner RB. 2008. Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry 47: 4000–4007. [DOI] [PubMed] [Google Scholar]
- Singh AC, Helms C, Sherman F. 1979. Mutation of the non-Mendelian suppressor ψ in yeast by hypertonic media. Proc Natl Acad Sci 76: 1952–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. 2000. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell 5: 163–172. [DOI] [PubMed] [Google Scholar]
- Specht S, Miller SBM, Mogk A, Bukau B. 2011. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336: 355–359. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283: 1339–1343. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan A, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137: 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED. 2010. Compositional determinants of prion formation in yeast. Mol Cell Biol 30: 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, Liss NM, Cobble KR, Ben-Musa Z, Ross ED. 2011. [PSI+] maintenance is dependent on the composition, not the primary sequence, of the oligopeptide repeat domain. PLoS ONE 6: e21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, Petri M, Paul KR, Kan GY, Ross ED. 2012. De novo design of synthetic prion domains. Proc Natl Acad Sci 109: 6519–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- Turoscy V, Cooper TG. 1987. Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J Bacteriol 169: 2598–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Wickner RB. 2013. Molecular structures of amyloid and prion fibrils: Consensus versus controversy. Acc Chem Res 46: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Liebman SW. 2009. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s(218–279) prion form a β solenoid with a triangular hydrophobic core. Science 319: 1523–1526. [DOI] [PubMed] [Google Scholar]
- Wickner RB. 1978. Twenty-six chromosomal genes needed to maintain the killer double-stranded RNA plasmid of Saccharomyces cerevisiae. Genetics 88: 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. 1991. Methods in classical genetics. In Saccharomyces (ed. Tuite MF and Oliver SG), pp. 101–147. Plenum, New York. [Google Scholar]
- Wickner RB. 1994. [URE3] as an altered URE2 protein: Evidence for a prion analog in S. cerevisiae. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. 2007. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol 5: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Dyda F, Tycko R. 2008. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci 105: 2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemecek J, McGlinchey R, Bateman D, Winchester CL. 2010. Prion amyloid structure explains templating: How proteins can be genes. FEMS Yeast Res 10: 980–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman DA, Kelly AC, Gorkovskiy A, Dayani Y, Zhou A. 2013. Amyloids and yeast prion biology. Biochemistry 52: 1514–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Beszonov E, Bateman DA. 2014. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc Natl Acad Sci 111: E2711–E2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Shewmaker FP, Bateman DA, Edskes HE, Gorkovskiy A, Dayani Y, Beszonov EE. 2015. Yeast prions: Structure, biology and prion-handling systems. Microbiol Mol Biol Rev 79: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]