Abstract
Throughout biology, function is intimately linked with form. Across scales ranging from subcellular to multiorganismal, the identity and organization of a biological structure’s subunits dictate its properties. The field of molecular morphogenesis has traditionally been concerned with describing these links, decoding the molecular mechanisms that give rise to the shape and structure of cells, tissues, organs, and organisms. Recent advances in synthetic biology promise unprecedented control over these molecular mechanisms; this opens the path to not just probing morphogenesis but directing it. This review explores several frontiers in the nascent field of synthetic morphogenesis, including programmable tissues and organs, synthetic biomaterials and programmable matter, and engineering complex morphogenic systems de novo. We will discuss each frontier’s objectives, current approaches, constraints and challenges, and future potential.
Molecular mechanisms that determine the higher-level structures of biological systems are being elucidated. These mechanisms may be harnessed to engineer systems with complex structures (e.g., synthetic tissues and organs).
What is the underlying basis of biological structure? Speculations were based on macroscopic observation until the 17th century, when Antonie van Leeuwenhoek and Robert Hooke discovered that organisms were composed of microscopic cells (Harris 1999), and that the size and shape of the cells affected the properties of the structures they formed. This hierarchical principle of organization remains a central tenet of the modern study of molecular morphogenesis: higher-level system properties emerge from, and are dependent on, lower-level properties of system components (Davies 2013). Many of these phenomena can be grouped into a small number of recurring motifs or morphogenic “modules,” such as spatial information conveyed by a morphogen gradient or the role of programmed cell death in sculpting tissues and organs (Cachat et al. 2014). As the mechanistic underpinnings of these processes are elucidated, opportunities arise to use this knowledge to direct morphogenesis toward novel, useful ends. We call this emerging field of endeavor “synthetic morphogenesis.” Inspired by and based on natural morphogenic systems, synthetic morphogenesis seeks to engineer, program, grow, and maintain biological systems with complex structures.
The field of synthetic biology is uniquely positioned to enable these efforts. The key to this synergy is the conception of synthetic biology as an engineering discipline (Arpino et al. 2013), combining traditional engineering concepts, such as reusable parts, modularity, and abstraction, with novel rules specifically suited to engineering biology (Slusarczyk et al. 2012). These concepts have served to propel the field’s rapid development. In the last decade, synthetic biology has progressed from simple systems in model prokaryotes (Elowitz and Leibler 2000; Gardner et al. 2000) to sophisticated synthetic gene networks in almost every branch of life, including plants (Schaumberg et al. 2015) and mammalian systems both in vitro (Xie et al. 2011) and in vivo (Ye et al. 2011). As the discipline’s tools become more powerful, they are beginning to enable the construction of systems that have dynamic behavior and nontrivial emergent properties similar to those of natural morphogenetic processes. Synthetic gene circuits can detect a cell’s type (Miki et al. 2015), metabolic state (Callura et al. 2012), (bio-) chemical signals (Weber et al. 2007), and light (Müller et al. 2014; Schmidl et al. 2014); they can use these inputs, combinatorially or sequentially, to alter the cell’s shape (Yeh et al. 2007), motility (Park et al. 2014), differentiation program (Wamaitha et al. 2015; Guye et al. 2016), or even kill the cell outright (Xie et al. 2011). Synthetic intercellular signaling allows cell populations to make decisions and coordinate behaviors both locally (Sprinzak et al. 2010; Matsuda et al. 2012) and globally (Tabor et al. 2009; Prindle et al. 2011; Chen et al. 2015b). These diverse sensors, actuators, and communication channels could implement complex morphogenic systems through a combination of top-down approaches in which cells are patterned by external signals, and bottom-up programs in which collective properties emerge through cells’ local decision-making.
Although synthetic biology supplies tools to implement synthetic morphogenic systems, the design language is inspired by natural molecular morphogenesis. Metazoan development is a marvel of precise, flexible, autocorrecting self-assembly; as synthetic biology strives to improve synthetic gene circuits’ robustness (Brophy and Voigt 2014) and complexity (Moon et al. 2012), natural complex systems inspire and inform best practices. Additionally, the forward design of synthetic morphogenetic systems requires reasoning across levels of scale and complexity. For example, tuning a synthetic tissue’s mechanical properties might involve specifying the molecular components (signaling molecules, receptors, and cell–cell adhesion molecules) in individual cells, as well as the organization and interactions of the cells that compose such a tissue. The field of molecular morphogenesis has developed methods specifically for scaling this hierarchical organization, and many of the theoretical and computational tools used to describe natural systems will find ready use in designing synthetic ones.
Because the field of synthetic morphogenesis is in its infancy, we have structured this article as equal parts review and prospectus, exploring three frontiers that we believe will be particularly fruitful. The first section examines programmable tissues and organs for regenerative medicine, based on precise spatiotemporal control over cell-fate decisions enabled by synthetic gene networks. The second section explores the nascent field of “smart” programmable biomaterials and the impact they will have on the fabrication of the objects in our man-made world. The final section looks further afield, envisioning de novo minimal synthetic morphogenic systems built by rewiring natural morphogenic modules in ways outside the reach of evolution. Arranged in order from concrete to speculative, these three frontiers in synthetic morphogenesis promise an unprecedented opportunity to rationally engineer ourselves and the world around us.
FRONTIER ONE: PROGRAMMABLE TISSUES AND ORGANS
The field of regenerative medicine is based on the pursuit of therapies to repair or replace tissues and organs damaged by disease. Advances in the directed differentiation of stem cells, their development ex vivo into tissue buds or organoids, and advanced materials for tissue engineering all hold promise for future therapies, but each lacks the precise spatiotemporal control over cell fate required to engineer mature, functional tissues and organs for transplant. This section explores how synthetic gene networks synergize with these three emerging technologies, potentially resulting in more structured and mature tissues that bridge the gap between therapeutic promise and medical practice.
A key enabler of proposed regenerative therapies is pluripotent stem cells. Initially derived from human embryos (Thomson et al. 1998), stem cells retain the potential to differentiate into any cell type in the human body. This makes them a promising platform for regenerative medicine, especially because methods for returning terminally differentiated cells to a pluripotent state (Okita et al. 2007) have obviated both the immunogenic and ethical concerns surrounding human embryonic stem cells. Pure cultures of mature differentiated cells offer a promising therapeutic approach for diseases that affect a single cell type, such as Parkinson’s disease (dopaminergic neurons) and heart disease (cardiomyocytes). In such cases, a cellular therapy could be derived from a patient’s own stem cells, expanded and differentiated ex vivo, and then transplanted back into the patient. Although such approaches have met with some success (Kriks et al. 2011; Shiba et al. 2012), challenges surrounding the purity of differentiated cultures (Breitbach et al. 2007) and engraftment of mature cells into existing tissues make the approach difficult to generalize. Diseases causing widespread damages to tissues, such as liver cirrhosis or gross injuries to the skin, are likely best repaired with functional, structured, mature tissue that simple culture systems cannot recreate.
One development along the path toward mature, stem cell–derived tissues are recent methods for growing organoids. Cultured without pluripotency-maintaining factors, stem cells spontaneously form embryoid bodies (Itskovitz-Eldor et al. 2000), which are heterogeneous, disorganized structures that recapitulate some aspects of early embryogenesis. Subsequent culture with appropriate differentiation factors results in organoids, organized proto-tissues that recapitulate many aspects of their in vivo counterparts (Lancaster and Knoblich 2014). Organoid systems have been developed for many tissues, including cerebrum (Lancaster et al. 2013), liver (Huch et al. 2013), pancreas (Boj et al. 2014), and lung (Dye et al. 2015), and their experimental tractability makes them exciting disease- and drug-screening models (Ranga et al. 2014). For example, hepatic organoids from Alagille syndrome patients recapitulate the in vivo pathology of the disease (Huch et al. 2014), and pancreas organoids have been used to discover new drivers of ductal pancreatic cancer (Boj et al. 2014). Unfortunately, the use of organoids more broadly in regenerative medicine is hampered by their immaturity and heterogeneity. Organoid systems reported thus far only recapitulate early stages of tissue development, they only contain a subset of the cell types present in mature tissue, and many of their fine structures remain disorganized. For example, brain organoids display functional regions similar to those found in naturally developing embryos, but they are not reliably organized relative to one another (Lancaster et al. 2013). This is because organoid structures are developing outside of their natural context: In the developing embryo, cell fate depends on information from neighboring cells, longer-range diffusible signals, and extracellular matrix interactions. Without these signals, organoids develop solely based on local cell–cell interactions, which result in local organization but not large-scale mature tissue.
Tissue engineering approaches offer a promising route toward building larger-scale structures based on recent advances in materials science. Cells are seeded into/onto a functionalized biocompatible scaffold, which promotes their (re)organization into a functional tissue (Dvir et al. 2011). Importantly, the synthetic extracellular matrix is not just a mechanical substrate, but can provide physical and chemical cues to the cells that drive fate decisions and large-scale organization (Lutolf and Hubbell 2005). For example, seeding cardiomyocytes on an anisotropic matrix results in their lengthwise alignment and organization into contractile muscle fibers (Engelmayr et al. 2008), whereas tethering interferon-γ to chitosan hydrogels affects neural progenitor cells’ lineage commitment (Leipzig et al. 2011). Such approaches leverage cells’ innate ability to self-organize and differentiate, but they rely on static environments; in contrast, natural development occurs in a rich milieu of signals that change dynamically in time and space. These signals drive cell-fate decisions in natural development, and their dynamicity is difficult to replicate in engineered environments.
As the field of regenerative medicine matures, synthetic biology is poised to synergize with all three approaches in the development of complex tissues. Synthetic gene networks promise orthogonal control of differentiation and cell-fate decisions by offering programmable control of endogenous gene networks using signals that can be engineered. For example, recent work in our laboratory (Guye et al. 2016) described a simple gene circuit that drives ectopic expression of the master cell-fate regulator Gata6. Because the circuit was transduced with a lentivirus, cells showed a wide range of protein expression levels; a brief pulse of Gata6 led to symmetry breaking and segregation of the isogenic cell line into ectodermal, endodermal, and mesodermal subpopulations. These subpopulations developed further into a structured tissue, resulting in regions of fetal liver-like tissue surrounding populations of neuronal cells. The liver tissue evinces not only hepatocyte-like cells but also stromal cells, tubular structures, and hematopoetic processes, remarkable complexity and organization given the simplicity of the genetic construct used to create it.
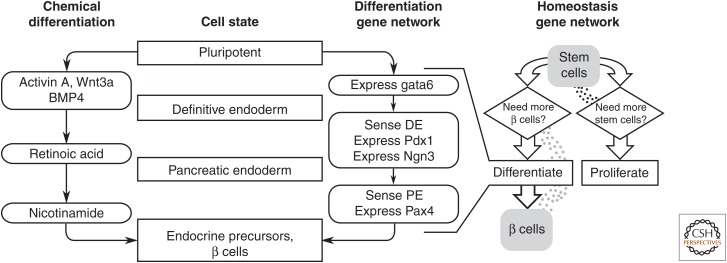
Importantly, synthetic gene circuits can couple these cell-fate actuators to intracellular sensors of cell state. One example application might be to implement an automatic multistep stem cell differentiation protocol without laborious sequential application of chemical inducers (Schiesser et al. 2014). For example, a gene circuit could recapitulate the ontogeny of pancreatic β cells by sequentially expressing Gata6, Pdx1, Ngn3, and Pax4 to drive differentiation of stem cells through definitive endoderm (Wamaitha et al. 2015), pancreatic endoderm (Kubo et al. 2011), and β cells, respectively (Fig. 1, left) (Collombat et al. 2009). Each step of the process could be controlled by the activity of an endogenous transcription factor. For example, Pdx1 and Ngn3 expression could be driven by a transcription factor specific to definitive endoderm such as Fox2A (D’Amour et al. 2005). Such a synthetic multistep “sense-and-differentiate” gene circuit could guide cells down a desired differentiation path more efficiently, and with greater precision, than current methods.
Figure 1.
Synthetic gene networks drive cell-fate decisions using cell state sensors and extracellular signals. Pluripotent stem cells are directed to differentiate into β cells by sequential application of growth factors (Schiesser et al. 2014); the same differentiation program might be performed by a gene network that uses sequential expression of master cell-fate regulators such as Gata6 and Pdx1. Progression through the program could be controlled by endogenous sensors of cell state. This multistep sense-and-differentiate program could be part of a larger gene network, such as one that maintains tissue homeostasis (Miller et al. 2012). A gene network could sense the number of programmed stem cells, and terminally differentiated cells using diffusible signals then proliferate or differentiate to maintain a continuous supply of both.
A synthetic gene circuit for producing pure populations of terminally differentiated cells is useful in some contexts, as discussed above, but it becomes particularly powerful when integrated into a larger synthetic gene network. For example, one could imagine a sense-and-differentiate gene circuit as part of a larger network devoted to maintaining tissue homeostasis in the face of autoimmune assault, as is frequently the case with type 1 diabetes (Zhang and Eisenbarth 2011). The loss of β cells in such patients is frequently a months-to-years-long process (Eisenbarth 2010), and transplanting β cells is ineffective because of the continuing autoimmune attack. A synthetic gene network might maintain β-cell levels by controlling the proliferation of engineered stem cells and their differentiation into β cells (Fig. 1, right). Such a network would sense the population levels of both stem cells and β cells based on the concentrations of diffusible intercellular signaling molecules, inducing proliferation of stem cells and differentiation into β cells when either population needed to be replenished (Miller et al. 2012).
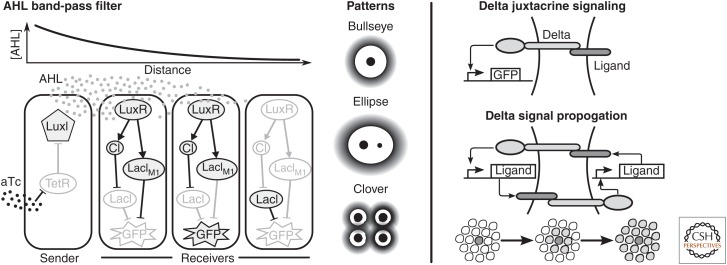
The artificial tissue homeostasis example in Figure 1 relies on engineered intercellular signaling to coordinate cellular responses across a population. Engineered communication channels orthogonal to endogenous signaling will be particularly important in building gene networks for synthetic tissues because they allow engineered networks to recapitulate the larger-scale organization of natural development that many current synthetic tissues lack. For example, engineered diffusible signals can drive spatial patterning of gene expression in prokaryotes and eukaryotes (Basu et al. 2005; Chen and Weiss 2005; Bacchus et al. 2012), whereas synthetic juxtacrine signaling based on the Notch receptor can generate propagating, mutually exclusive states in neighboring cells (Fig. 2) (Sprinzak et al. 2010; Matsuda et al. 2012). The spatial patterns shown in these studies have many similarities to those created by organizing centers in metazoan development, such as the ones that specify the imaginal discs that later form the limbs in Drosophila (Brook et al. 1996). Similar patterns could be used to drive large-scale patterns in cell-fate decisions in developing synthetic tissues.
Figure 2.
Engineered intercellular communication can coordinate spatial changes in gene expression. Communication channels are frequently based on diffusible signals such as acyl-homoserine lactone (AHL) (left) (Basu et al. 2005). The receiver gene network responds to intermediate concentrations of AHL, mimicking the “French flag” model of morphogen patterning (Wolpert 1969). Arranging senders in various starting configurations produces patterns similar to those found in natural development. Communication can also be based on juxtacrine signaling (right), in which a cell presenting a ligand on its surface activates signal transduction in adjacent cells (Sprinzak et al. 2010). A feedback loop between signal activation and ligand production leads to signal propagation (Matsuda et al. 2012). GFP, Green fluorescent protein.
The opportunities to build tissues with synthetic biology present some exceptional challenges as well. Synthetic gene networks promise the ability to precisely direct cell fate by interacting with endogenous cell-fate pathways, but these pathways are frequently complex and sensitive to context. What is worse, the effect of genetic, epigenetic, and environmental context on these endogenous networks is not fully known. Several design strategies will likely prove useful. For example, using orthogonal inter- and intracellular signaling components in synthetic gene networks will help insulate them from the rest of the cellular context (Brophy and Voigt 2014). Additionally, intracellular sensors can allow synthetic gene networks to account for cell state (Xie et al. 2011), whereas extracellular controls via small molecules (Weber et al. 2004), light (Müller et al. 2014), and even electric fields (Levin 2014) can be used to tune gene circuit behavior. These genetic controls nicely complement the macroscale layout imposed by scaffolds of biocompatible materials (Ma 2008) and micropatterned substrates that present extracellular cues at the length scale of single cells (Kinney and McDevitt 2013). Taken together, these strategies will allow synthetic gene networks to operate robustly in the face of uncertain or unknown cellular context.
The ready availability of patient-derived induced pluripotent stem cells has provided us access to the building blocks of our own development. These cells already contain the gene networks necessary to develop into tissues and organs, and the study of molecular development and morphogenesis continues to elucidate their composition and dynamic function. Current developments in synthetic biology promise the ability to precisely control the spatiotemporal activity of these gene networks, harnessing the function of these natural networks toward diagnostic and therapeutic ends. The in vitro, ex vivo, and (eventually) in vivo construction of complex human tissues promises to revolutionize both research into, and treatment of, many common human diseases.
FRONTIER TWO: SYNTHETIC BIOMATERIALS AND PROGRAMMABLE MATTER
Synthetic control of biological morphogenesis has broad applicability outside of biomedicine as well. Biological materials have traits that have proven impossible to duplicate with synthetic materials: the specific strength of wood, the toughness of dragline spider silk, or the selective permeability of endothelium. Efforts in material science to recapitulate these properties synthetically are often “biologically inspired,” but they lack the sophistication and precision of the biological systems that produce natural biomaterials. Because these biological systems are genetically encoded, synthetic biology offers the possibility to interface with and supplement these natural gene networks with synthetic ones to modulate these materials’ properties. This section explores the promises and challenges on the path to “programmable” biological materials.
Biological materials are composed of some combination of cells, biopolymers (e.g., cellulose, collagen, and keratin), and minerals (hydroxyapatite, calcium carbonate, and silica). Living systems assemble these components using a process of guided, feedback-controlled self-assembly (Davies 2013) at scales ranging from molecular to cellular to macroscale tissue arrangement. These multiple scales of feedback mean that biomaterial structure is universally hierarchical (Fratzl and Weinkamer 2007). For example, wood is anisotropic at multiple scales, from the arrangement of cellulose microfibrils in the cell wall, to the cells’ parallel orientation along the shoot, to radial changes in cell morphology that give rise to a tree’s annual rings. This anisotropy is controlled by genetic and molecular mechanisms. For example, cellulose microfibrils are deposited in a continuous, transverse spiral around the cells’ long axis (McFarlane et al. 2014). Their (extracellular) orientation is controlled by the orientation of the (intracellular) microtubule network, which responds, in turn, to growth hormones and changes in turgor pressure. The transverse orientation gives rise to preferential expansion along the longitudinal axis of the stem. Plant hormones also act systemically to promote seasonal changes in the morphology of proliferating cells, leading to nested rings of early- and latewood (Ursache et al. 2013). This hierarchical structure, with its anisotropy at multiple scales, gives wood remarkable specific strength and fracture toughness (Committee on Synthetic Hierarchical Structures 1994); similar hierarchical architectures underlie many other common biomaterials.
Early efforts to “program” biomaterials were based on genetically tractable biopolymers, particularly recombinant expression of proteins that could self-assemble into polymers. For example, elastin is an extracellular matrix protein that provides elasticity to connective tissue (Muiznieks et al. 2010). It readily self-assembles, and synthetic hydrogels made from recombinant elastin proteins undergo a temperature-sensitive phase change that makes them useful for drug delivery (Wright et al. 2002). Spider silk, on the other hand, is an extraordinarily tough material (Lazaris 2002); spider silk proteins expressed in Escherichia coli and extruded through an artificial spinneret have mechanical properties similar to the natural fiber (Xia et al. 2010). Their recombinant production allows for these protein-based materials to be readily manipulated, tuned, and functionalized beyond the capabilities of natural systems (Schacht and Scheibel 2014), for example, imbuing synthetic spider silk with antimicrobial properties by fusing recombinant silk proteins with antimicrobial peptides (Gomes et al. 2011). However, the materials form exclusively via entropic molecular self-assembly, which allows for none of the advantages of hierarchical, adaptive assembly seen in natural systems.
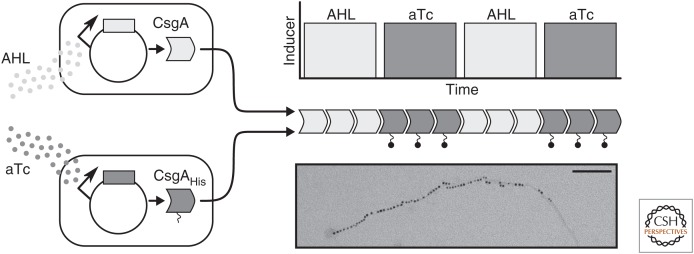
An alternative approach to smart biomaterials is to genetically modify the organism that produces a biomaterial to tune or functionalize it in situ. Diatoms, for example, have a glassy cell wall (“frustule”) made of amorphous silica associated with a matrix of proteins (Gordon et al. 2009); fusing enzymes to these proteins immobilizes them on the diatom’s cell wall, functionalizing this promising biomaterial (Sheppard et al. 2012). Bacterial biofilms have also been functionalized in this way. Recent work has shown functionalization of CsgA, the protein that forms E. coli biofilms, with peptide tags conferring activities such as substrate-specific adhesion and electrical conductivity (Fig. 3) (Chen et al. 2014; Nguyen et al. 2014).
Figure 3.
Chen et al. (2014) created a tunable biomaterial using curli fibers, a constituent of Escherichia coli biofilms. Curli fibers are made up of CsgA subunits, which self-assemble into higher-order structures. The investigators modified CsgA with a His tag that confers metal-binding capability; when they alternated expression of modified CsgA with unmodified CsgA (based on two different small-molecule inducers), the resulting curli fibers had regions that bound gold nanoparticles alternating with regions that did not, as shown with scanning electron microscopy. Scale bar, 100 nm. AHL, Acyl-homoserine lactone. (Image courtesy of Allen Chen and Tim Lu, both at the Massachusetts Institute of Technology.)
Neither approach modifies the actual morphology of a natural biomaterial, however. Why is it so hard to change the shape of a biological structure in a tuned, specific way? For example, diatom cell walls have an array of astonishingly uniform pores, with promising nanotechnology applications ranging from molecular sieves to photonics (Gordon et al. 2009). Many of the molecular details about diatom cell wall biogenesis are known, including the protein scaffolds involved in silicate condensation and large-scale patterning (Kröger and Poulsen 2008; Scheffel et al. 2011), and yet we are still unable to alter pore size and arrangement in any directed fashion. In fact, none of the reported genetic modifications to the cell wall machinery resulted in any morphological changes at all!
The key insight is that the assembly of biomaterials, like most morphogenesis, is not entropic but, instead, guided by interlocking molecular feedback loops at multiple length scales (Davies 2013). A diatom’s frustule, for example, is assembled via a multistep, multiscale process that involves silaic acid import, protein-catalyzed condensation into nanometer-scale particles of amorphous silica, nucleation of the new cell wall inside the silica deposition vesicle, and patterned silica deposition guided by both templating proteins and the diatom’s cytoskeleton (Gordon et al. 2009; Hildebrand and Lerch 2015). In fact, for many biomaterials, we understand the nature of these feedback loops at the molecular level (Canty and Kadler 2002; McFarlane et al. 2014); but the relationships between physical and biochemical processes, molecular mechanisms, genetic control, and environmental factors are so complex as to defy intuition. In particular, specifying a desired “effect” (e.g., smaller pores in a diatom frustule) and then looking for a single molecular change that will “cause” it is challenging because of the difficulty in causal reasoning about feedback-based behavior (Åström and Murray 2008).
One way to program specific morphologies might be to modulate these natural feedback loops with external control instead of allowing structures to emerge from the detailed configuration of biophysical, molecular biological, and genetic processes. Natural biological systems are capable of sensing environmental conditions and responding to them on multiple scales, from single cells that move along chemotactic gradients to trees that grow around boulders. One way to take advantage of these natural morphogenic systems is by directing them using external factors, such as a silken canopy “grown” by silkworms crawling along a scaffolding (Oxman et al. 2014) or a functional large intestine created by seeding a collagen scaffold with intestinal progenitor cells (Shaffiey et al. 2016). External control has been used to control synthetic morphogenic systems as well. A priori arrangement of “sender” and “receiver” cells can create patterns caused by spatial variation in the concentration of a small molecule intercellular signal (Basu et al. 2005; Chen and Weiss 2005), as can spatial activation of synthetic gene networks using light (Gautier et al. 2014). More recent reports have used gene circuits to perform complex spatial computations such as Boolean logic (Tamsir et al. 2011) and edge detection (Tabor et al. 2009). Interfacing these engineered control mechanisms with endogenous gene networks will allow for top-down control of the molecular mechanisms that drive biomaterial morphogenesis.
Ultimately, the ability to synthetically modulate a biomaterial’s structure at the microscale will depend on a detailed systems-level understanding of the natural system’s biogenesis. Because synthetic gene networks act at cellular and subcellular levels, careful modeling and experiments will be needed to elucidate the connection between molecular behavior and material properties. Eventually, these approaches will enable the hybrid design of programmable materials in which microscale control of a material’s properties is provided by a synthetic gene network whose spatiotemporal activity is controlled by external signals. The programmable matter enabled by these synthetic gene networks promises to marry the remarkable functionality of natural biomaterials with the engineerability required by advanced manufacturing and fabrication.
FRONTIER THREE: SYNTHETIC MORPHOGENESIS AND SYNTHETIC LIFE
Synthetic tissues and tunable biomaterials are special cases of a more general goal: the ability to predictably engineer novel biological shape- and pattern-forming systems. This echoes the goal of synthetic biology more broadly, to predictably engineer cells’ behaviors by programming them with synthetic gene networks. Such predictable engineering is difficult in the complex, uncertain environs of naturally evolved cells, spurring recent efforts to engineer a “minimal” life form (Forster and Church 2006) by either minimizing an existing organism’s genome (Glass et al. 2006; Pósfai et al. 2006; Gibson et al. 2010; Hutchison et al. 2016) or by chemically synthesizing autonomously replicating cells de novo (Blain and Szostak 2014). Both approaches aim to better understand the requirements for life in a “simple” minimal system (de Lorenzo and Danchin 2008), and to develop a predictable chassis on which to base further engineering efforts (Keasling 2008; Vickers et al. 2010).
Might a similar approach be applied to engineering multicellular structures? What might such an approach look like? In the same way that minimized cells can help biologists understand the basic functions required for self-sufficient life, minimal morphogenic systems could help us understand the rules underpinning multicellular morphogenesis. In the same way, minimized cells could provide a well-controlled environment for rational engineering; synthetic morphogenic systems could allow us to explore configurations and biological functions not witnessed in evolution (Dichtel-Danjoy and Félix 2004), from prosaic tunable materials for industrial and manufacturing applications to fantastical, functional forms such as trees that grow into tree houses (Ginsberg 2013). These morphogenic systems could be based on engineered or synthetic minimized cells, or they might be built on top of more complex cells with more “built-in” functionality.
Novel biological shapes and patterns have been difficult to engineer due in large part to the complexity of natural morphogenic systems and the way they are organized. As the previous sections discussed, biological structures are hierarchical, emerging from interactions at the molecular, macromolecular, cellular, multicellular, tissue, and organ levels. This hierarchical arrangement is the result of evolutionary processes in which biological complexity evolved stepwise, fixing core cellular processes first and then using them to build higher-order structures (Kirschner and Gerhart 1998). For example, the epithelium is one of the key evolutionary innovations of multicellular organisms (Leys and Riesgo 2012) because it allows organisms to spatially and biochemically isolate an “inside” cavity from the organism’s surroundings. Epithelial cells depend on cadherins and catenins for cell adhesion and on the cytoskeleton to develop cell polarity and directional transport (Fath et al. 1993). Both cadherins (Abedin and King 2008) and catenins (Dickinson et al. 2011) are present in premetazoan organisms, and bacteria have cytoskeleton homologs of both actin and tubulin (Busiek and Margolin 2015), but only their coordinate regulation allows for the formation of higher-order structures. In turn, epithelia serve as the building blocks for larger structures, such as tissues and organs (Jamora et al. 2003), as spatial cues induce their folding, branching, and interactions with other cell types. This kind of hierarchical structure enables rapid evolution and exploration of phenotypic space (Kirschner and Gerhart 1998) by allowing for easy reuse of underlying morphogenic modules. However, it makes engineering lower levels of the hierarchy difficult because changing the behavior of one gene or gene network alters the behavior or structure of every context in which that gene or network is used.
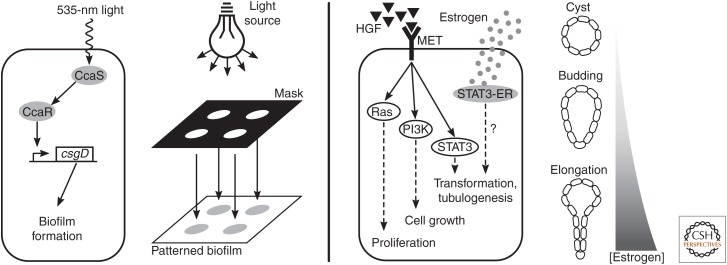
The structure and evolutionary history of morphogenic processes have important implications for building novel morphogenic systems. Instead of attempting to modify or co-opt underlying cellular processes, perhaps it will be easier to follow evolution’s lead and layer synthetic, engineered control mechanisms on top of preexisting morphogenic machinery. One might imagine beginning with a very simple chassis, such as a minimized E. coli or Mycoplasma species, then elaborating multicellular morphogenic modules on top of the existing molecular mechanisms, coordinating multicellular behavior with signals and networks orthogonal to native machinery. For example, the molecular basis of E. coli biofilm formation is relatively well understood (Beloin et al. 2008); placing these mechanisms under the control of both local, short-range intercellular signaling (for bottom-up pattern formation) and global chemical or light-based control could serve to “domesticate” them (Fig. 4, left) (Chen et al. 2015a), giving biological engineers control over both when and where biofilms form and their fine-scale properties.
Figure 4.
Proposed synthetic morphogenic systems. Patterned biofilm formation (left) may be achieved by connecting a synthetic optogenetic sensor, such as CcaS/CcaR (Schmidl et al. 2014), to a master regulator of biofilm production, such as csgD (Brombacher et al. 2003). A model mammalian morphogenic system (right) may offer a richer palette of morphogenic behaviors. For example, hepatocyte growth factor (HGF) induces tubulogenesis in Madin–Darby canine kidney (MDCK) cells grown in three-dimensional culture (Zegers 2014). The HGF receptor, MET, activates a number of downstream signaling pathways; controlling one via synthetic receptor, such as STAT3 fused to an estrogen receptor (ER) (STAT3-ER) (Matsuda et al. 1999), may allow tunable control of some of the morphogenic modules that lead to tubulogenesis.
Layering synthetic control mechanisms on top of preexisting morphogenic modules in a “simple” chassis such as E. coli partially addresses issues of pleiotropy that plague current efforts, because engineers can change the behavior of the morphogenic modules (such as functionalizing the CsgA protein; Nguyen et al. 2014), confident that they understand those changes’ effects in the context of a broader system. However, it may be that the palette of morphogenic behaviors available in a bacterial system is not broad enough. We might be better served by a more complex chassis in which a richer array of multicellular morphogenic modules are present but not yet dynamically regulated. For example, mammalian Madin–Darby canine kidney (MDCK) cells (Gaush et al. 1966) are a common in vitro model for epithelia; in three-dimensional culture, they spontaneously form hollow cysts that mimic in vivo epithelial morphology, and can be then induced to form branching tubes by the addition of hepatocyte growth factor (HGF) (Zegers 2014). Three-dimensional MDCK cultures could be a model system for studying external control of tube formation and extension (Fig. 4, right). Directing tubulogenesis in a system that is already “primed” to do so may be a more attractive engineering prospect than attempting to recapitulate it via the underlying morphogenic building blocks, such as cell–cell adhesion, cell division and apoptosis, and the establishment of apicobasal polarity. This is both because reusing mechanisms that are already present reduces the amount of engineering required to form higher-level structures, and because modulating these processes from the top of the complexity hierarchy is more in line with how these structures evolved naturally. This promises fewer pleiotropic effects, more modularity, greater robustness, and a broader space of phenotypes to explore.
The strategy of overlaying synthetic control on top of high-level morphogenic mechanisms is also poised to take advantage of our increasing control over the differentiation of pluripotent stem cells (Kinney and McDevitt 2013). From this perspective, different cell types have available to them different palettes of morphogenic modules that could be composed synthetically. The kinds of structures we could expect to synthesize from epithelial cells (sheets, domes, tubes) are very different than those we could imagine deriving from fibroblasts (connective and structural tissue components) or myocytes (contractile structures). Not only do the basic morphogenic modules differ between these various cell types, but their coordinate regulation does as well. Unfortunately, for many structures of interest, we do not understand the molecular basis of morphogenesis well enough to co-opt it; but by continuing to build in vitro models of various natural morphogenic systems (Foty and Steinberg 2005; Zegers 2014), we expect to learn about not only the molecular networks that control them but also means by which we can control them synthetically.
The phrase “de novo synthetic morphogenesis” conjures synthetic cells created in the laboratory, living systems entirely orthogonal to natural ones, designed rationally, and freed from evolution’s series of “bad hacks.” Although promising advances are being made in the field of artificial life, it seems wasteful and presumptuous to throw away four billion years of evolution and the dynamic, robust, flexible set of core molecular mechanisms, gene networks, and morphogenic modules that have resulted. By choosing carefully the level of the complexity hierarchy with which our synthetic gene circuits interact, we can minimize the issues of pleiotropy and context sensitivity that hamper current efforts.
CONCLUSION
Complex synthetic multicellular structures could lead to advances in medicine, manufacturing, and many other human endeavors. To deliver on these promises, however, the field needs a set of design rules akin to the ones the broader field of synthetic biology is developing (Brophy and Voigt 2014). We close with a look at some of the challenges that are common not only to the three frontiers discussed in this review but indeed the entire field of synthetic morphogenesis. We also consider lessons learned from the study of natural morphogenic systems, as well as subjects further afield.
First, why are natural morphogenic systems difficult to alter rationally? One reason lies in their structure. Such systems are modular and hierarchical, just like many complex engineered systems (Csete and Doyle 2002). This structure is the result of biological systems’ evolutionary history, which evolved complexity by fixing core molecular processes and then overlaying dynamic regulation on top of them (Kirschner and Gerhart 1998). This resulted in the evolution of multicellularity, followed by intercellular signaling, the fixation of phylotypic body plans, and then elaborations on those body plans: appendages, the neural crest, and reproductive specializations (Kuratani 2009). Each level of complexity was selected for robustness and stability, which supported phenotypic exploration of additional complexity. This modular hierarchy is a key evolutionary driver of morphogenesis because rewiring modules’ connectivity (via the signals that regulate them) requires fewer genetic changes and is less likely maladaptive than altering the molecular species that compose them (Pigliucci 2008).
Unfortunately, this evolutionary history also makes rational changes to natural morphogenesis difficult because these morphogenic modules are not isolable from each other. Morphogenic motifs are reused in many different contexts, as are the signals that activate them; this reuse leads to pleiotropy and muddies the boundaries between morphogenic modules. Instead, these modules assume much of their identity from the system in which they are embedded (Csete and Doyle 2002). The formation of a structure is not controlled by a single gene but the time-varying interactions of many molecular species. A prospective biological engineer is forced to contend with complexity and feedback control between modules and across levels of hierarchy; the global behavior of the system emerges from the interactions of its constituent parts.
Why are morphogenic systems so complex? One might intuit that such complexity would result in systems that are fragile, but of course natural morphogenesis is marvelously robust. The study of biology through the lens of complex systems seems to indicate several reasons for this reliability. First, morphogenic and developmental systems make wide use of feedback (Davies 2013). Negative feedback makes systems robust to perturbations, and positive feedback can amplify stimuli to move systems from one state to another. Second, natural biological systems are composed of overlapping and redundant parts, and there are frequently several mechanisms that give rise to the same phenotypic output (Kitano 2004). These increases in complexity actually serve to make morphogenic systems more robust, not less.
In fact, it may be that the tradeoff between robustness, performance, and complexity is an underlying law not only of biological systems but of complex systems more generally (Csete and Doyle 2002). This may be the reason, for example, that development is slow; complex programs, executed slowly, lead to a more robust outcome than simple processes proceeding quickly. Conversely, simulations of minimal metabolic networks suggest that they are more fragile than naturally evolved ones (Gabaldón et al. 2007). As engineered systems begin to achieve complexity comparable to biological ones, explorations of the tradeoff between complexity and robustness in natural and engineered biological systems may have lessons to teach engineers of complex mechanical and electronic systems too.
Another constellation of open questions concerns modularity and context sensitivity. This review has used the term “module” without giving it a precise definition; comparisons of biological organisms at both the gene (Stuart et al. 2003) and protein network (Wuchty et al. 2003) levels show evolutionarily conserved networks of interacting genes and, of course, cells coordinately regulate suites of genes in response to both intracellular and extracellular conditions. In spite of this, we do not understand the organization of biological systems well enough to harness the abstraction that is implied by a modular organization. How can molecular and morphogenic modules be rearranged, rewired, or replaced to modify existing systems or build new ones? How can one best account for pleiotropy and context sensitivity, or isolate engineered systems from it? What is the right balance between leveraging natural systems and building synthetic, orthogonal systems alongside or on top of them?
Biological systems manipulate matter, perform chemistry, and process information precisely, robustly, and efficiently; the promise of synthetic biology, writ broad, is to harness this sophistication to engineer biological systems reliably and predictably. Great strides have been made in engineering useful, predictable functions into single cells, but life in nature is never unicellular. Life is structured, and cells are organized to maximize productivity, protect against environmental assault, and thus improve the transmission of genetic information. Synthetic biologists are just beginning to explore the design and control of biological structures, leading to a new field of synthetic morphogenesis. The field is still embryonic, but we look forward excitedly to its development.
ACKNOWLEDGMENTS
We thank Samira Kiani for stimulating discussions as this manuscript was being developed. This work is supported by National Science Foundation grants CBET-0939511, EEC-0540879, CNS-1446607, and EFMA-1137269.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Abedin M, King N. 2008. The premetazoan ancestry of cadherins. Science 319: 946–948. [DOI] [PubMed] [Google Scholar]
- Arpino JA, Hancock EJ, Anderson J, Barahona M, Stan GB, Papachristodoulou A, Polizzi K. 2013. Tuning the dials of Synthetic Biology. Microbiology 159: 1236–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström KJ, Murray RM. 2008. Feedback systems: An introduction for scientists and engineers. Princeton University Press, Princeton, NJ. [Google Scholar]
- Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. 2012. Synthetic two-way communication between mammalian cells. Nat Biotechnol 30: 991–996. [DOI] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. 2005. A synthetic multicellular system for programmed pattern formation. Nature 434: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr Top Microbiol Immunol 322: 249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain JC, Szostak JW. 2014. Progress toward synthetic cells. Annu Rev Biochem 83: 615–640. [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. 2014. Organoid models of human and mouse ductal pancreatic cancer. Cell 160: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbach M, Bostani T, Roell W, Xia Y, Dewald O, Nygren JM, Fries JWU, Tiemann K, Bohlen H, Hescheler J, et al. 2007. Potential risks of bone marrow cell transplantation into infarcted hearts. Blood 110: 1362–1369. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Dorel C, Zehnder AJB, Landini P. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149: 2847–2857. [DOI] [PubMed] [Google Scholar]
- Brook WJ, Diaz-Benjumea FJ, Cohen SM. 1996. Organizing spatial pattern in limb development. Annu Rev Cell Dev Biol 12: 161–180. [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA. 2014. Principles of genetic circuit design. Nat Methods 11: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Margolin W. 2015. Bacterial actin and tubulin homologs in cell growth and division. Curr Biol 25: R243–R254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E, Liu W, Hohenstein P, Davies JA. 2014. A library of mammalian effector modules for synthetic morphology. J Biol Eng 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. 2012. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci 109: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty E, Kadler K. 2002. Collagen fibril biosynthesis in tendon: A review and recent insights. Comp Biochem Physiol A Mol Integr Physiol 133: 979–985. [DOI] [PubMed] [Google Scholar]
- Chen M-T, Weiss R. 2005. Artificial cell–cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol 23: 1551–1555. [DOI] [PubMed] [Google Scholar]
- Chen AY, Deng Z, Billings AN, Seker UOS, Lu MY, Citorik RJ, Zakeri B, Lu TK. 2014. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater 13: 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Zhong C, Lu TK. 2015a. Engineering living functional materials. ACS Synth Biol 4: 8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR. 2015b. Emergent genetic oscillations in a synthetic microbial consortium. Science 349: 986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. 2009. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into α and subsequently β cells. Cell 138: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Synthetic Hierarchical Structures. 1994. Hierarchical structures in biology as a guide for new materials technology, National Academy of Sciences, Washington, DC. [Google Scholar]
- Csete ME, Doyle JC. 2002. Reverse engineering of biological complexity. Science 295: 1664–1669. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541. [DOI] [PubMed] [Google Scholar]
- Davies JA. 2013. Mechanisms of morphogenesis, 2nd ed Academic, Cambridge, MA. [Google Scholar]
- de Lorenzo V, Danchin A. 2008. Synthetic biology: Discovering new worlds and new words. EMBO Rep 9: 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy M-L, Félix M-A. 2004. Phenotypic neighborhood and micro-evolvability. Trends Genet 20: 268–276. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI. 2011. A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331: 1336–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir T, Timko BP, Kohane DS, Langer R. 2011. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol 6: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MAH, Tsai Y-H, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, et al. 2015. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4: e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth GS. 2010. Banting Lecture 2009: An unfinished journey: Molecular pathogenesis to prevention of type 1A diabetes. Diabetes 59: 759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338. [DOI] [PubMed] [Google Scholar]
- Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. 2008. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat Mater 7: 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Mamajiwalla SN, Burgess DR. 1993. The cytoskeleton in development of epithelial cell polarity. J Cell Sci Suppl 17: 65–73. [DOI] [PubMed] [Google Scholar]
- Forster AC, Church GM. 2006. Towards synthesis of a minimal cell. Mol Syst Biol 2: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS. 2005. The differential adhesion hypothesis: A direct evaluation. Dev Biol 278: 255–263. [DOI] [PubMed] [Google Scholar]
- Fratzl P, Weinkamer R. 2007. Nature’s hierarchical materials. Prog Mater Sci 52: 1263–1334. [Google Scholar]
- Gabaldón T, Peretó J, Montero F, Gil R, Latorre A, Moya A. 2007. Structural analyses of a hypothetical minimal metabolism. Philos Trans R Soc Lond B Biol Sci 362: 1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342. [DOI] [PubMed] [Google Scholar]
- Gaush CR, Hard WL, Smith TF. 1966. Characterization of an established line of canine kidney cells (MDCK). Proc Soc Exp Biol Med 122: 931–935. [DOI] [PubMed] [Google Scholar]
- Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S. 2014. How to control proteins with light in living systems. Nat Chem Biol 10: 533–541. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329: 52–56. [DOI] [PubMed] [Google Scholar]
- Ginsberg AD. 2013. The pre future of synthetic biology, www.nextnature.net/2013/05/the-prefuture-of-synthetic-biology.
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci 103: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SC, Leonor IB, Mano JF, Reis RL, Kaplan DL. 2011. Antimicrobial functionalized genetically engineered spider silk. Biomaterials 32: 4255–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R, Losic D, Tiffany MA, Nagy SS, Sterrenburg FAS. 2009. The glass menagerie: Diatoms for novel applications in nanotechnology. Trends Biotechnol 27: 116–127. [DOI] [PubMed] [Google Scholar]
- Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, Weiss R. 2016. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat Commun 7: 10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. 1999. The birth of the cell. Yale University Press, New Haven, CT. [Google Scholar]
- Hildebrand M, Lerch SJL. 2015. Diatom silica biomineralization: Parallel development of approaches and understanding. Semin Cell Dev Biol 46: 27–35. [DOI] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. 2013. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MMA, Ellis E, van Wenum M, Fuchs SA, de Ligt J, et al. 2014. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. 2016. Design and synthesis of a minimal bacterial genome. Science 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. 2000. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med 6: 88–95. [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. 422: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling JD. 2008. Synthetic biology for synthetic chemistry. ACS Chem Biol 3: 64–76. [DOI] [PubMed] [Google Scholar]
- Kinney MA, McDevitt TC. 2013. Emerging strategies for spatiotemporal control of stem cell fate and morphogenesis. Trends Biotechnol 31: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. 1998. Evolvability. Proc Natl Acad Sci 95: 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. 2004. Biological robustness. Nat Rev Genet 5: 826–837. [DOI] [PubMed] [Google Scholar]
- Kriks S, Shim J-W, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. 2011. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger N, Poulsen N. 2008. Diatoms—From cell wall biogenesis to nanotechnology. Annu Rev Genet 42: 83–107. [DOI] [PubMed] [Google Scholar]
- Kubo A, Stull R, Takeuchi M, Bonham K, Gouon-Evans V, Sho M, Iwano M, Saito Y, Keller G, Snodgrass R. 2011. Pdx1 and Ngn3 overexpression enhances pancreatic differentiation of mouse ES cell-derived endoderm population. PLoS ONE 6: e24058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani S. 2009. Modularity, comparative embryology and evo–devo: Developmental dissection of evolving body plans. Dev Biol 332: 61–69. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345: 1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris A. 2002. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science 295: 472–476. [DOI] [PubMed] [Google Scholar]
- Leipzig ND, Wylie RG, Kim H, Shoichet MS. 2011. Differentiation of neural stem cells in three-dimensional growth factor–immobilized chitosan hydrogel scaffolds. Biomaterials 32: 57–64. [DOI] [PubMed] [Google Scholar]
- Levin M. 2014. Molecular bioelectricity: How endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo. Mol Biol Cell 25: 3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Riesgo A. 2012. Epithelia, an evolutionary novelty of metazoans. J Exp Zool B Mol Dev Evol 318: 438–447. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Hubbell JA. 2005. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol 23: 47–55. [DOI] [PubMed] [Google Scholar]
- Ma PX. 2008. Biomimetic materials for tissue engineering. Adv Drug Deliv Rev 60: 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 18: 4261–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Koga M, Nishida E, Ebisuya M. 2012. Synthetic signal propagation through direct cell–cell interaction. Sci Signal 5: ra31. [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Döring A, Persson S. 2014. The cell biology of cellulose synthesis. Annu Rev Plant Biol 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, et al. 2015. Efficient detection and purification of cell populations using synthetic MicroRNA switches. Cell Stem Cell 16: 699–711. [DOI] [PubMed] [Google Scholar]
- Miller M, Hafner M, Sontag E, Davidsohn N, Subramanian S, Purnick PEM, Lauffenburger D, Weiss R. 2012. Modular design of artificial tissue homeostasis: Robust control through synthetic cellular heterogeneity. PLoS Comput Biol 8: e1002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. 2012. Genetic programs constructed from layered logic gates in single cells. Nature 491: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiznieks LD, Weiss AS, Keeley FW. 2010. Structural disorder and dynamics of elastin. Biochem Cell Biol 88: 239–250. [DOI] [PubMed] [Google Scholar]
- Müller K, Engesser R, Timmer J, Zurbriggen MD, Weber W. 2014. Orthogonal optogenetic triple-gene control in mammalian cells. ACS Synth Biol 3: 796–801. [DOI] [PubMed] [Google Scholar]
- Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. 2014. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317. [DOI] [PubMed] [Google Scholar]
- Oxman N, Laucks J, Kayser M, Nuro-Royo J, Gonzales-Uribe C. 2014. Silk pavilion: A case study in fiber-based digital fabrication. In Proceedings of the Fabricate Conference, pp. 248–255. Zurich, Switzerland. [Google Scholar]
- Park JS, Rhau B, Hermann A, McNally KA, Zhou C, Gong D, Weiner OD, Conklin BR, Onuffer J, Lim WA. 2014. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc Natl Acad Sci 111: 5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. 2008. Is evolvability evolvable? Nat Rev Genet 9: 75–82. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Plunkett G, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312: 1044–1046. [DOI] [PubMed] [Google Scholar]
- Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. 2011. A sensing array of radically coupled genetic “biopixels.” Nature 481: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A, Gjorevski N, Lutolf MP. 2014. Drug discovery through stem cell-based organoid models. Adv Drug Deliv Rev 69–70: 19–28. [DOI] [PubMed] [Google Scholar]
- Schacht K, Scheibel T. 2014. Processing of recombinant spider silk proteins into tailor-made materials for biomaterials applications. Curr Opin Biotechnol 29: 62–69. [DOI] [PubMed] [Google Scholar]
- Schaumberg KA, Antunes MS, Kassaw TK, Xu W, Zalewski CS, Medford JI, Prasad A. 2015. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat Methods 13: 94–100. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Poulsen N, Shian S, Kröger N. 2011. Nanopatterned protein microrings from a diatom that direct silica morphogenesis. Proc Natl Acad Sci 108: 3175–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiesser JV, Micallef SJ, Hawes S, Elefanty AG, Stanley EG. 2014. Derivation of insulin-producing β-cells from human pluripotent stem cells. Rev Diabet Stud 11: 6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl SR, Sheth RU, Wu A, Tabor JJ. 2014. Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth Biol 3: 820–831. [DOI] [PubMed] [Google Scholar]
- Shaffiey SA, Jia H, Keane T, Costello C, Wasserman D, Quidgley M, Dziki J, Badylak S, Sodhi CP, March JC, et al. 2016. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen Med 11: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard VC, Scheffel A, Poulsen N, Kröger N. 2012. Live diatom silica immobilization of multimeric and redox-active enzymes. Appl Environ Microbiol 78: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Fernandes S, Zhu W-Z, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, et al. 2012. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489: 322–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarczyk AL, Lin A, Weiss R. 2012. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13: 406–420. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. 2010. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255. [DOI] [PubMed] [Google Scholar]
- Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. 2009. A synthetic genetic edge detection program. Cell 137: 1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamsir A, Tabor JJ, Voigt CA. 2011. Robust multicellular computing using genetically encoded NOR gates and chemical “wires.” Nature 469: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Shapiro SS, Waknitz MA, Marshall VS. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Ursache R, Nieminen K, Helariutta Y. 2013. Genetic and hormonal regulation of cambial development. Physiol Plant 147: 36–45. [DOI] [PubMed] [Google Scholar]
- Vickers CE, Blank LM, Krömer JO. 2010. Grand challenge commentary: Chassis cells for industrial biochemical production. Nat Chem Biol 6: 875–877. [DOI] [PubMed] [Google Scholar]
- Wamaitha SE, del Valle I, Cho LTY, Wei Y, Fogarty NME, Blakeley P, Sherwood RI, Ji H, Niakan KK. 2015. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev 29: 1239–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. 2004. Gas-inducible transgene expression in mammalian cells and mice. Nat Biotechnol 22: 1440–1444. [DOI] [PubMed] [Google Scholar]
- Weber W, Daoud-El Baba M, Fussenegger M. 2007. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc Natl Acad Sci 104: 10435–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25: 1–47. [DOI] [PubMed] [Google Scholar]
- Wright ER, McMillan RA, Cooper A, Apkarian RP, Conticello VP. 2002. Thermoplastic elastomer hydrogels via self-assembly of an elastin-mimetic triblock polypeptide. Adv Funct Mater 12: 149–154. [Google Scholar]
- Wuchty S, Oltvai ZN, Barabási A-L. 2003. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nat Genet 35: 176–179. [DOI] [PubMed] [Google Scholar]
- Xia X-X, Qian Z-G, Ki CS, Park YH, Kaplan DL, Lee SY. 2010. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc Natl Acad Sci 107: 14059–14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. 2011. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333: 1307–1311. [DOI] [PubMed] [Google Scholar]
- Ye H, Daoud-El Baba M, Peng R-W, Fussenegger M. 2011. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332: 1565–1568. [DOI] [PubMed] [Google Scholar]
- Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. 2007. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature 447: 596–600. [DOI] [PubMed] [Google Scholar]
- Zegers MM. 2014. 3D in vitro cell culture models of tube formation. Semin Cell Dev Biol 31: 132–140. [DOI] [PubMed] [Google Scholar]
- Zhang L, Eisenbarth GS. 2011. Prediction and prevention of Type 1 diabetes mellitus. J Diabetes 3: 48–57. [DOI] [PubMed] [Google Scholar]