Abstract
The ability to site-specifically incorporate noncanonical amino acids (ncAAs) with novel structures into proteins in living cells affords a powerful tool to investigate and manipulate protein structure and function. More than 200 ncAAs with diverse biological, chemical, and physical properties have been genetically encoded in response to nonsense or frameshift codons in both prokaryotic and eukaryotic organisms with high fidelity and efficiency. In this review, recent advances in the technology and its application to problems in protein biochemistry, cellular biology, and medicine are highlighted.
Methods are being developed to site-specifically incorporate noncanonical amino acids with unique features (e.g., novel functional groups or posttranslational modifications) into the proteins of living organisms.
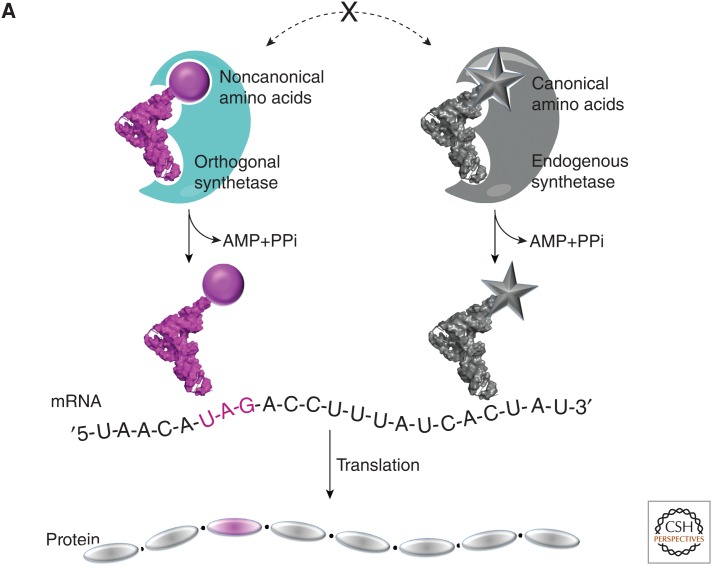
With the rare exceptions of pyrrolysine and selenocysteine, the genetic codes of all known organisms consist of the same 20 canonical amino acids. However, additional functional groups, including organic and inorganic cofactors and posttranslational modifications (PTMs), are required for many of the functions performed by proteins. This observation suggests that an expanded genetic code might allow one to either rationally design or evolve proteins with new or enhanced physical, chemical, and biological properties. To this end, methodology has been developed to exploit the endogenous protein biosynthetic machinery to site-specifically incorporate noncanonical amino acids (ncAAs) into proteins in living organisms with high translational fidelity and efficiency (up to 5 g/L on a commercial scale). The desired ncAA is encoded by a nonsense or frameshift codon using an orthogonal aminoacyl-tRNA synthetase (aaRS)/transfer RNA (tRNA) pair that is specific for the ncAA but does not cross-react with endogenous host aaRSs, tRNAs, or amino acids (Fig. 1A) (Wang et al. 2006b; Liu and Schultz 2010). Using this system, over 200 structurally distinct ncAAs have been genetically encoded in both prokaryotic and eukaryotic organisms. These ncAAs include spectroscopic probes, metal ion chelators, photo-affinity probes and photocaged amino acids, posttranslational modifications, and amino acids with orthogonal chemical activity for the site-specific modification of proteins (Wang et al. 2006b; Liu and Schultz 2010; Wan et al. 2014). They have been used as probes of protein structure and function, both in vitro and in vivo, and in the rational design of proteins with new or enhanced biological or pharmacological activities. Most recently, in vitro evolution experiments have begun to show that additional amino acid building blocks can lead to novel protein structures and activities. Herein, we review a number of recent advances in the field, both in the development and application of this technology.
Figure 1.
Expanding the genetic code. (A) The site-specific incorporation of noncanonical amino acids (ncAAs) using an orthogonal aminoacyl-tRNA synthetase (aaRS)/transfer RNA (tRNA) pair. (B) ncAAs described in this review. mRNA, Messenger RNA.
GENETICALLY ENCODING ncAAs
Evolved Methanococcus jannaschii tyrosyl aaRS/tRNA pairs and Methanosarcina barkeri and mazei pyrrolysyl aaRS/tRNA pairs have enabled the incorporation of a large number of ncAAs into proteins in Escherichia coli (Fig. 1B) (Wang et al. 2006b; Liu and Schultz 2010; Wan et al. 2014). To further expand the number and nature of genetically encoded ncAAs, aaRS/tRNA pairs from other archeal and eukaryotic organisms have been recently developed, including Pyrococcus horikoshii lysyl aaRS/tRNA, P. horikoshii glutamyl aaRS/tRNA, Saccharomyces cerevisiae tryptophanyl aaRS/tRNA, heterogeneous leucyl Mt-tRNA/Hs-aaRS, and proly Af-tRNA/Ph-aaRS pairs (Anderson and Schultz 2003; Santoro et al. 2003; Anderson et al. 2004; Chatterjee et al. 2012, 2013d; Xiao et al. 2014). The structurally distinct active sites of these aaRSs allow one to encode chemically diverse amino acid side chains; however, the efficiencies with which the ncAAs can be incorporated at a given site in the proteome vary—ranging from milligrams to 5+ g/L of mutant protein, most likely because of the differing degrees to which the aaRS/tRNA pair is optimized (of course, for any given protein, efficiency is also affected by the mutation site). Platform vectors, for example, pEvol and pUltra, have been established that allow high-level protein expression of a desired mutant protein in bacteria, and large-scale fermentation (>10,000 L) has yielded mutant proteins on a 5 g/L scale (Young et al. 2010; Chatterjee et al. 2013b). A current focus is the efficient incorporation of multiple identical or distinct ncAAs into the same protein, or the synthesis of an entirely “unnatural” biopolymer. One hypothesis is that the presence of endogenous release factors leads to low overall incorporation efficiency for multiple ncAAs encoded by nonsense codons. Huang et al. (2010) first showed that up to three ncAAs could be genetically incorporated into the same protein by overexpression of the carboxy-terminal domain of ribosomal protein L11 to reduce release factor 1 (RF1)-mediated termination of protein translation. Using an RF1 knockout strain, Johnson et al. (2011) showed that up to 10 p-acetylphenylalanines (pAcF; Fig. 1B1) could be incorporated into the same protein, albeit in relatively low yield (Fig. 2A). Knockout of RF1 improves ncAA incorporation in response to the amber codon; however, impaired growth rates resulting from RF1 deletion may limit the utility of this strain.
Figure 2.
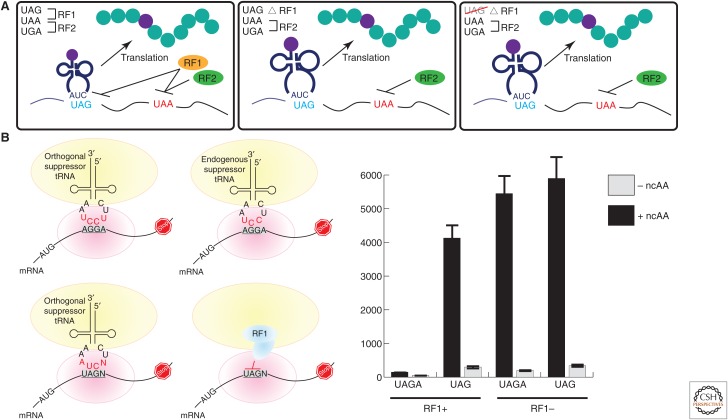
Engineering Escherichia coli strains with a reassigned codon. (A) Escherichia coli genome engineering for increasing noncanonical amino acids (ncAAs) incorporation efficiency. (B) Competing triplet recognition during the translation of quadruplet codons. Deletion of RF1 significantly improves UAGN-suppression levels. mRNA, Messenger RNA; tRNA, transfer RNA.
Recently, multiplex automated genome engineering (MAGE) and conjugative assembly genome engineering (CAGE) were used to replace all of the TAG stop codons in the E. coli MG1655 genome with the TAA ochre nonsense codon (Fig. 2A) (Lajoie et al. 2013); subsequently, RF1 was knocked out to generate E. coli strain C321.ΔA. In the resulting genomically recoded organism (C321.ΔA), UAG was reassigned from a nonsense codon to a sense codon specific for an ncAA in the presence of the cognate orthogonal tRNA/aaRS pair. As a result of genome recoding, RF1 knockout is no longer deleterious and robust ncAA incorporation was achieved in response to the amber codon (Lajoie et al. 2013). In addition, the suppression efficiency of the four-base codon UAGA, which starts with UAG, was significantly increased in the C321.ΔA strain (Fig. 2B) (Chatterjee et al. 2014). This genomically recoded strain was also used to generate a library of chromosomally integrated aaRSs for the selection of more efficient aaRS variants. The evolved aaRSs showed increased ncAA-containing protein production, enabling genetic incorporation of 30 ncAAs into a single protein (Amiram et al. 2015). An alternative approach to minimize the effects of RF1-mediated translational termination is the generation of ribosome-mRNA pairs that function in the presence of, but are orthogonal to, the endogenous translational machinery. The use of orthogonal ribosome–mRNA pairs in E. coli significantly increases the suppression efficiency of ncAA incorporation, and facilitates the use of frameshift codons and incorporation of multiple ncAAs into proteins (Wang et al. 2007; Neumann et al. 2010a,b).
In mammalian cells, the pyrrolysyl pair from archaea, and the tyrosyl and leucyl pairs from E. coli have been widely used to genetically encode novel amino acids, including amino acids with bio-orthogonal chemical reactivities (e.g., azido, alkynyl, and tetrazinyl), fluorescent probes, PTMs, and photocaged amino acids (Fig. 1B) (Wang et al. 2006b; Liu and Schultz 2010; Wan et al. 2014). These orthogonal tRNA/aaRS pairs are either evolved in yeast (for E. coli–derived pairs) or bacteria (for archaea-derived pairs) and then transferred to vectors with appropriate promoters for transient expression in the host cell, typically, HEK293 or CHO cells. Yields of transiently expressed proteins on the order of 5–20 mg/L are common, and yields up to 1 g/L have been reported for stable cell lines expressing mutant full-length antibodies (Tian et al. 2014). However, the application of this technology in mammalian cells is limited by the inherent drawbacks of transient transfection, including the relatively low transfection efficiency of certain cell types (e.g., neurons and embryonic stem cells) and the toxicity of transfection reagents. To overcome these challenges, an enhanced suppression system delivered by a hybrid baculovirus has been reported (Chatterjee et al. 2013c). This baculovirus-based system significantly increases the incorporation efficiency of ncAA into proteins in mammalian cells and allows ncAAs to be encoded in embryonic stem cells, rat cardiac fibroblasts, and neurons. This enhanced suppression system has been used to incorporate up to three identical ncAAs or two distinct ncAAs into a single protein in good yield (Xiao et al. 2013). For example, in a full-length antibody, pAcF (Fig. 1B1) was incorporated in response to a TAG codon in the heavy chain and an azidolysine analog (AzK; Fig. 1B34) was incorporated in response to a TAA codon in the light chain, allowing the mutant antibody to be selectively conjugated to a drug and a fluorophore at these two distinct sites (Xiao et al. 2013). Recently, a Methanosarcina mazei pyrrolysyl aaRS/tRNA pair was stably integrated into the mammalian genome using the PiggyBac transposon system, which enables efficient incorporation of ncAAs into target proteins in diverse mammalian cells. The utility of this system was shown by genetically incorporating six ɛ-N-acetyllysines (AcK; Fig. 1B59) into histone H3 (Elsasser et al. 2016). Methodological advances have also allowed the genetic incorporation of ncAAs into proteins in various multicellular organisms, including Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, and Streptomyces venezuelae (Greiss and Chin 2011; Bianco et al. 2012; Li et al. 2013; Chin 2014; He et al. 2015). Most recently, we have been able to genetically encode ncAAs in Bacillus cereus, allowing the synthesis of ribosomally derived natural products from unnatural building blocks (Luo et al. 2016).
PROBES OF PROTEIN STRUCTURE
The site-specific incorporation of ncAAs with unique spectroscopic properties provides a powerful tool to explore protein structure and dynamics. 13C/15N-labeled p-methoxyphenylalanine (Fig. 1B2) and p-tifluoromethoxyphenylalanine (Fig. 1B13) have been genetically incorporated into human fatty acid synthetase to probe conformational changes that occur on ligand binding by nuclear magnetic resonance (NMR) (Cellitti et al. 2008). Similarly, 19F-labeled p-trifluoroethyl-phenylalanine (Fig. 1B14) was incorporated into nitroreductase and histidinol dehydrogenase to probe structural changes that occur on substrate binding (Jackson et al. 2007; Li et al. 2010). Furthermore, the site-specific incorporation of the metal-binding ncAA, 2-amino-3-(8-hydroxy-quinolin-3-yl)propanoic acid (Fig. 1B21), into membrane proteins (CXCR1 and viroporin p7) allowed long-range paramagnetic relaxation measurements of intra- and intermolecular distance (Park et al. 2014). In related work, to facilitate electron paramagnetic resonance (EPR) studies of proteins, a ketoxime-linked spin label side chain has been site-specifically conjugated to proteins through a reactive chemical handle (Fleissner et al. 2009). However, direct incorporation of these probes is advantageous in many cases and, thus, a spin-labeled ncAA (Fig. 1B51) was recently incorporated into green fluorescent protein (GFP) and thioredoxin in E. coli (Schmidt et al. 2014).
Infrared (IR) spectroscopy is a valuable technique that can be used to characterize protein structure, dynamics, and microenvironment on a range of timescales. To this end, ncAAs with unique IR signatures, including p-cyano-phenylalanine (Fig. 1B15), p-azido-phenylalanine (pAzF, Fig. 1B4), p-nitrophenylalanine (pNO2F; Fig. 1B16), and 3-nitro-tyrosine (3-NO2Y, Fig. 1B19) have been efficiently incorporated into proteins (Schultz et al. 2006; Taskent-Sezgin et al. 2009; Ye et al. 2009; Smith et al. 2011). To study protein dynamics and microenvironments, photocaged 2D-labeled o-nitrobenzyl-tyrosine (oNBTyr, Fig. 1B46) enabled the site-specific deuteration of Tyr100 within dihydrofolate reductase and revealed that Tyr100 stabilizes the positive charge of the nicotinamide moiety in the transition state (TS) via electrostatic interactions (Groff et al. 2009). Photocaged lysine, cysteine, and serine (Fig. 1B46–50) have all been genetically incorporated into proteins, and can be used to site-specifically deuterium-label these amino acids as well (Liu and Schultz 2010; Davis and Chin 2012).
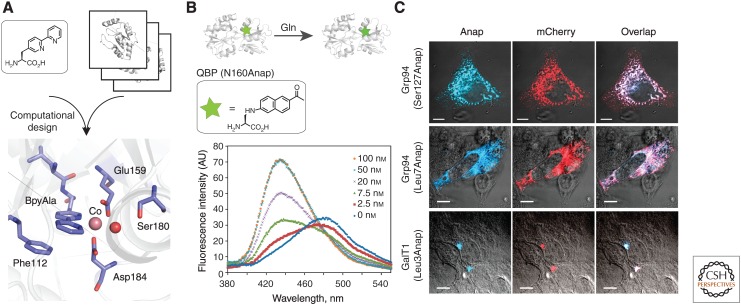
Fluorescent proteins (FPs) have been widely used to study protein expression, localization, and conformation in living cells. By substituting core residues in the chromophores of various FPs with ncAAs, FP variants with distinct spectral properties have been generated, including FP sensors for monitoring pH, reducing and oxidizing environments, and heavy metals (Wang et al. 2003; Ai 2012; Chatterjee et al. 2013d; Niu and Guo 2013; Xiao et al. 2014). For example, a fluorogenic biosensor sensitive to oxidation was generated by substitution of Tyr66 in GFP with p-borono-phenylalanine (Fig. 1B11) (Wang et al. 2012; Chen et al. 2013). In the presence of oxidants, the mutant chromophore is converted to the wild-type chromophore with a change in fluorescence quantum yield. However, fusion of an FP can significantly perturb the structure and function of a target protein, and is largely limited to the amino and carboxyl termini (Zheng et al. 2014). The ability to site-specifically incorporate relatively small fluorescent amino acids at defined sites within proteins would further increase the utility of fluorescence spectroscopy to study protein structure in vitro and in living cells. Several fluorescent ncAAs have been genetically incorporated into proteins in E. coli, including a hydroxycoumarin-derived amino acid (7HC, Fig. 1B42) and acridon-2-ylalanine (Fig. 1B45) (Wang et al. 2006a; Speight et al. 2013). 7HC was inserted into bacterial tubulin FtsZ and used to track FtsZ localization during cell division (Charbon et al. 2011). This fluorophore was also substituted for Trp 564 in activator of transcription 3 to monitor the phosphorylation state of Tyr705 (Lacey et al. 2011). To determine protein localization in yeast and mammalian cells, 2-amino-3-(5-(dimethylamino)naphthalene-1-sulfonamido) propanoic acid (Fig. 1B43) and 3-(6-acetylnaphthalen-2-ylamino)-2-aminopropanoic acid (Anap; Fig. 1B44) have been used by a number of groups (Summerer et al. 2006; Lee et al. 2009; Chatterjee et al. 2013a). Anap was successfully used to detect conformational changes on ligand binding to glutamine-binding protein (QBP) and dynamic internal pore opening in voltage-gated potassium channels (KV) in oocytes (Fig. 3B,C) (Lee et al. 2009; Kalstrup and Blunck 2013). These latter experiments underscore the ability to probe local changes in structure or environment by the site-specific insertion of smaller fluorophores. An important advance in this regard will be the genetic encoding of probes with improved photostability and longer wavelengths.
Figure 3.
Some applications of noncanonical amino acids (ncAAs). (A) X-ray crystal structure of a BipyAla-containing metalloprotein generated by computational design (protein data bank [PDB] code 4IWW). (B) Fluorescence intensities of glutamine-binding protein (QBP) (Asn160Anap) with different concentrations of Gln. (C) Subcellular localization of endoplasmic and Golgi-residue proteins in HEK293 cells visualized using Anap. Scale bars, 10 µm.
PROBING PROTEIN FUNCTION WITH ncAAs
Genetically encoded ncAAs can also be used as mechanistic probes. For example, ncAAs have been used in mechanistic studies of class I ribonucleotide reductases (Cotruvo and Stubbe 2011). The site-specific incorporation of a redox active probe, 3-aminotyrosine (Fig. 1B18), in place of transiently oxidized Tyr356, Tyr730, and Tyr731 residues was used to probe the role of these amino acids in long-range proton coupled electron transfer (PCET) (Seyedsayamdost et al. 2007; Minnihan et al. 2011a). To measure perturbations to the pKas of redox-active tyrosines within the PCET pathway, 3-NO2Y (Fig. 1B19), was substituted for Tyr122, Tyr730, and Tyr731 (Yokoyama et al. 2010). This radical transfer mechanism was also explored using a number of fluorotyrosines (Fig. 1B20) with distinct EPR properties (Minnihan et al. 2011b). Recently, ncAAs have even been used as direct structural probes of the transition states (TSs) of chemical reactions. Specifically, the X-ray crystal structure of the planar TS configuration for rotation about the central C-C bond of biphenyl was directly observed by genetically introducing p-biphenylalanine (BiPhe; Fig. 1B8) into a site in Pyrococcus abyssi threonyl-tRNA synthetase that was designed to bind BiPhe in a planar TS configuration (Pearson et al. 2015). After several rounds of optimization, the crystal structure of the designed protein at 2.05 Å resolution revealed a planar biphenyl TS configuration stabilized by van der Waals interactions with side chains in the pocket.
Photocaged ncAAs can be used to control the temporal and spatial activity of proteins in living cells. To this end, photocaged tyrosine, lysine, cysteine, and serine have been genetically encoded in E. coli, yeast, and mammalian cells (Fig. 1B46–50) (Liu and Schultz 2010; Davis and Chin 2012). In an early example, 4,5-dimethoxy-2-nitrobenzyl-serine (Fig. 1B50) was substituted for Ser114 and Ser128 in the yeast transcription factor Pho4 (Lemke et al. 2007). Uncaging and subsequent phosphorylation of these two sites, which are involved in nuclear export of Pho4, was triggered by a laser pulse and monitored in real time. Similarly, a photocaged lysine, ɛ-N-[(1-(6-nitrobenzo[d][1,3]dioxol-5-yl)ethoxy)-carbonyl]lysine (Fig. 1B47), was used to control the nuclear import of p53 (Gautier et al. 2010). This same lysine analog was also used to control the activation of mitogen-associated protein kinase (MAPK) signaling in mammalian cells by photocaging the MAPK kinase MEK1 (Gautier et al. 2011); and a photocaged tyrosine, oNBTyr (Fig. 1B46), has been used to control phosphorylation of Tyr 701 in human signal transducer and activator of transcription 1 (STAT1) and thereby activate the Janus kinase/STAT signal transduction pathway (Arbely et al. 2012). Recently, a photocaged cysteine (Fig. 1B48) was genetically incorporated into the pore of the inwardly rectifying potassium channel Kir2.1 to generate a light-activated ion channel in the embryonic mouse neocortex (Kang et al. 2013). Photocaged ncAAs have been also used in other studies, including development of light-responsive FPs, light-induced transcription using a light-activated RNA polymerase, photocleavage of the polypeptide backbone, and photoactivatable intein-mediated protein splicing (Peters et al. 2009; Groff et al. 2010; Hemphill et al. 2013; Ren et al. 2015).
To map protein interactions, several ncAAs containing UV-induced cross-linking moieties, including benzophenone, azides, and diazirines (Fig. 1B52, 4, and 53), have been site-specifically incorporated into proteins in both prokaryotic and eukaryotic cells (Liu and Schultz 2010; Davis and Chin 2012). The formation of covalent cross-links by UV-irradiation is especially important for studying relatively weak protein interactions that are not stable to immunoprecipitation or cell lysis, and also allows one to probe intra- or interprotein contacts between specific residues. Numerous studies have been performed using these photo-cross-linking ncAAs, including probing interactions of chaperones with substrates, conformational changes in RNA polymerases, interactions among components within signal transduction pathways, and membrane protein interactions (Schlieker et al. 2004; Mori and Ito 2006; Chen et al. 2007; Mohibullah and Hahn 2008; Ieva and Bernstein 2009; Okuda and Tokuda 2009; Tagami et al. 2010). The use of ncAAs to selectively form covalent bonds not only provides valuable information regarding molecular interactions, but also allows the design of covalent protein-based agonists, antagonists, or inhibitors. For example, p-vinylsulfonamido-phenylalanine (Fig. 1B54), which contains an aza-Michael acceptor that can alkylate nearby cysteine or lysine residues, was site-specifically introduced into Herceptin; the mutant antibody selectively formed a covalent complex with a specific surface lysine of the oncogenic receptor tyrosine kinase ErbB2 on live cells (Furman et al. 2014). In addition, a number of ncAAs containing electrophilic reactive handles (Fig. 1B55 and 56) have been reported by Lei and his coworkers and used to form specific inter- or intramolecular cross-links in proteins (Chen et al. 2014; Xiang et al. 2014).
PTMs can alter protein function, folding, and stability, and thereby regulate numerous biological processes in the cell. The ability to site-specifically incorporate these PTMs into target proteins provides a valuable tool to study their function. Several tyrosine analogs, including p-sulfo-tyrosine (pSY; Fig. 1B58) and p-carboxymethylphenylalanine (pCMF; Fig. 1B57), have been genetically encoded to mimic tyrosine PTMs (Liu and Schultz 2006; Xie et al. 2007). Site-specific incorporation of pSY allowed for the preparation of high-affinity sulfo-hirudin and various sulfated antibodies to the human immunodeficiency virus (HIV) protein GP120 (Liu et al. 2007). Recently, pCMF was used to mimic phosphoTyr291 in protein arginine methyltransferase 1, which plays a role in substrate recognition (Rust et al. 2014). A similar strategy has been used to study the function of lysine PTMs within histones. For example, ɛ-N-acetyllysine (AcK; Fig. 1B59), ɛ-N-methyllysine (MeK; Fig. 1B60), ɛ-N,N-dimethyllysine (Fig. 1B61), ɛ-N-crotonyllysine (Fig. 1B62), and ɛ-N-2-hydroxyisobutyryllysine (Fig. 1B63) have all been site-specifically incorporated into histone proteins (Neumann et al. 2008; Nguyen et al. 2009, 2010; Kim et al. 2012b; Xiao et al. 2015b). Site-specific substitution of AcK for H3K56 revealed that H3K56 has no direct effect on the compaction of chromatin, but leads to an increase in DNA breathing (Neumann et al. 2009); and the substitution of MeK for H3K9 allowed determination of its effects on the binding specificity of heterochromatin protein 1 (Nguyen et al. 2009).
THE DESIGN OF PROTEINS WITH NOVEL PROPERTIES USING ncAAs
There is considerable interest in the site-specific modification of proteins with biological probes, polyethylene glycols (PEGs), drugs, and other polypeptides/proteins for a range of applications, including the generation of antibody–drug conjugates (ADCs) and bispecific antibodies. Unfortunately, it is difficult to do chemistry on proteins with the same degree of control over structure that chemists have with small molecules. Typically, the modification of proteins involves the electrophilic modification of cysteine or lysine residues, which in the case of large, disulfide cross-linked proteins like antibodies, results in chemically heterogeneous products with varying stoichiometries and sites of modification. This heterogeneity makes it difficult to optimize the properties of the resulting conjugates. The site-specific incorporation of ncAAs with bioorthogonal chemical reactivity enables the preparation of homogenous, chemically defined macromolecules. For example, homogenous therapeutic proteins with defined PEG modifications have been prepared by the site-specific incorporation of pAcF (Fig. 1B1), followed by efficient conjugation to alkoxyamine-derivatized PEG by oxime ligation (Cho et al. 2011). Using this strategy, PEG-conjugated human growth hormone, human fibroblast growth factor 21, and bovine granulocyte colony–stimulating factor (GCSF) were prepared and shown in clinical studies to have significantly enhanced serum half-lives while preserving a high degree of biological activity, with little or no observed immunogenicity (Cho et al. 2011; Mu et al. 2012).
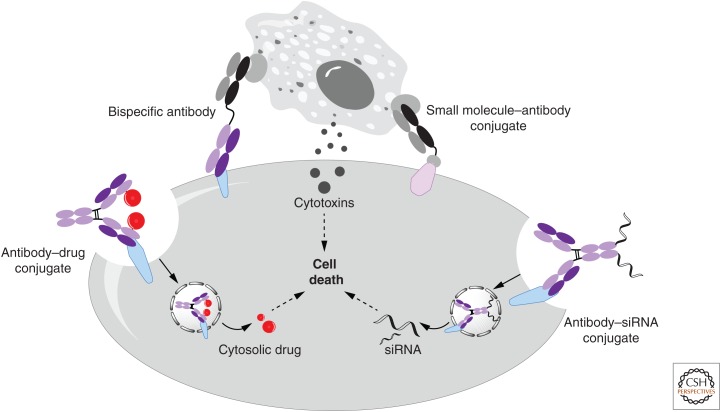
ADCs have emerged as a promising new class of drugs, as evidenced by the clinical efficacy of trastuzumab emtansine for the treatment of Her2-positive breast cancer (Sun et al. 2014; Wals and Ovaa 2014). ADCs allow highly cytotoxic drugs to be selectively delivered to the target tumor, thus, increasing therapeutic efficacy and reducing adverse systemic side effects (Fig. 4). Most ADCs are based on the reactivity of cysteine and lysine residues, which often result in heterogeneous products. Recently, we prepared homogenous ADCs by the site-specific incorporation of pAcF (Fig. 1B1) into Herceptin and conjugation of the mutant antibody to an alkoxy-amine auristatin derivative in good yield (Axup et al. 2012) (a similar ADC developed by Ambrx (San Diego, CA) has entered clinical trials; Tian et al. 2014). Complete regression of Her2-positive tumors in mouse xenograft models was observed without overt toxicity. Importantly, the site and stoichiometry of conjugation were found to affect the pharmacokinetics and efficacy of the ADC. Similarly, site-specific ADCs have been generated that target CXCR4-positive metastatic cancer cells, and this technology is now being applied outside of oncology in the preparation of ADCs for the treatment of inflammatory, autoimmune, and metabolic disease (Kularatne et al. 2014). Other constructs, including antibodies conjugated to imaging probes or two different drugs with synergistic effects, are being explored. Furthermore, the utility of site-specific antibody conjugation was also shown in antibody small-interfering RNA (siRNA) conjugation, which significantly increases the selectivity and efficiency of siRNA delivery (Fig. 4) (Lu et al. 2013).
Figure 4.
Overview of noncanonical amino acid (ncAA)-containing antibody conjugates. siRNA, Small interfering RNA.
Another example of use of ncAAs in the preparation of therapeutic proteins is the generation of bispecific antibodies that selectively deliver cytotoxic T cells to tumor cells (Fig. 4) (Byrne et al. 2013). Bispecific antibodies are typically prepared as genetic fusions or heterodimers, which limits the possible relative spatial orientations of the two antibody variable domains and may affect stability, efficacy, and selectivity (Bluemel et al. 2010; Moore et al. 2011). Again, the use of genetically encoded ncAAs with bio-orthogonal reactivity allows far more control over the relative orientation and distance of the two antibody fragments (Kim et al. 2012a). To illustrate this notion, pAcF-containing anti-Her2 and anti-CD3 Fabs were expressed in E. coli and each was conjugated to short polyethylene linkers terminated with either an azide or cyclooctyne group (Cao et al. 2015). These two Fabs were then conjugated via a copper-free click reaction to afford a bispecific antibody that binds the CD3 protein on T cells and Her2 on tumor cells to form an immunological synapse. The resulting anti-HER2/anti-CD3 bispecific antibody showed good selectivity and high potency with Her2-positive tumor cells in vitro and showed excellent efficacy against both high and low Her2-positive tumors in rodent xenografts in the presence of purified peripheral blood mononuclear cells. Similarly, bispecifics have been made that target CD33 and CLL1 for the treatment of acute myelogenous leukemia (Lu et al. 2014). Tumor-specific small molecules can also be used to recruit cytotoxic T cells to tumor cells (Murelli et al. 2009; Kularatne et al. 2010). For example, 2-[3-(1,3-dicarboxy propyl)-ureido] pentanedioic acid, a small molecule that selectively binds PSMA, which is overexpressed in prostate cancer, was site-specifically conjugated to anti-CD3 to generate a PSMA-targeting small molecule–antibody conjugate (SMAC) (Fig. 4) (Kim et al. 2013). The optimized SMAC showed excellent selectivity and cytotoxicity (EC50 ∼ 100 pm) against patient-derived PSMA-positive prostate cancer cells both in vitro and in vivo and is being advanced to clinical development. A similar strategy was used to recruit T cells to ovarian cancer cells using a folate-anti-CD3 bispecific construct (Kularatne et al. 2013).
Despite the impressive clinical efficacy of chimeric antigen receptor (CAR)-T-cell therapy, the lack of control in the activation and expansion and in the termination of conventional CAR-T cells has greatly limited its application. To overcome this challenge, a CAR was engineered that binds the bio-orthogonal ligand fluorescein isothiocyanate (FITC). The resulting transduced CAR-T cells are inactive until a switch molecule, consisting of an FITC-conjugated site specifically to a tumor-targeting antibody, are dosed. It was shown that the ability to control the site and stoichiometry of FITC conjugation using ncAAs dramatically impacts effective immunological synapse formation and, as a consequence, in vitro and in vivo activity. These switch molecules allowed dose titration of the CAR-T-cell response and resulted in complete clearance of tumors in rodent xenografts with significantly reduced cytokine levels; the CAR-T-cell response could also be terminated by simply removing the switch. Finally, this single universal CAR can be used for heterogeneous tumors with distinct antigens (or reoccurring tumors with altered antigen expression) by simply using multiple switch molecules. Thus, this platform (which also includes a recombinant version using peptide neoepitopes) should decrease the acute and chronic toxicity associated with this promising new cell therapy, simplify its application, and allow it to be used with both liquid and solid tumors (Ma et al. 2016).
ncAA mutagenesis has also been used for the generation of therapeutic vaccines to break immunological tolerance against a native protein (Sun et al. 2014; Wals and Ovaa 2014). Replacement of a single tyrosine residue (Tyr86) of murine tumor necrosis factor α (mTNF-α) with pNO2F (Fig. 1B16) induced a high-titer, long-lived antibody response to both the mutant and native mTNF-α in vaccinated mice (Gruenewald et al. 2008). Mechanistic studies revealed that the ncAA mutation generated a T-cell neoepitope, which led to a polyclonal antibody response to the autologous antigen (Gauba et al. 2011). Similar results were obtained with other mutant antigens containing ncAAs, including complement component 5α and proprotein convertase subtilisin/kexin type 9. It was also shown that the naturally posttranslationally modified amino acids nitrotyrosine and sulfotyrosine also break immunological tolerance, supporting the hypothesis that such PTMs, induced by viruses or inflammation, could be an underlying cause of autoimmune disease. Recently, an HIV-1 virus that requires ncAA translational machinery for replication has been reported by Wang et al. (2014). Multiple amber codons were introduced at permissive sites of the Gag gene; in the presence of an amber suppressor tRNA/aaRS pair and specific ncAA, full-length Gag protein is generated leading to the assembly of infective virus. However, none of these ncAA translational components are naturally present in humans, thus making the virus unable to reproduce in human cells. This strategy is also being used to create other vaccines—several pyrrolysine analogs have been incorporated into the surface protein of human hepatitis D virus, and MjTyrRS/tRNA has been used to encode a number of tyrosine analogs in Mycobacterium smegmatis and tuberculosis (Wang et al. 2010; Lin et al. 2013). Using a similar strategy, a live-attenuated tuberculosis vaccine is being explored in our laboratory that is dependent on the presence of an ncAA for growth ex vivo, but dies in the absence of the ncAA. Surprisingly, it has not yet been possible to isolate a revertant for some sites of suppression. Alternatively, one could exploit a strategy developed by Church and coworkers, who used an ncAA-based synthetic system to control unintended proliferation of genetically modified organisms (Mandell et al. 2015). A synthetic essential gene that requires the ncAA BiPhe (Fig. 1B8) for translation, folding, and function was generated by computational design. Substitution of the wild-type gene with this synthetic gene in the E. coli genome provides a novel biocontainment mechanism against mutational escape, environmental supplementation, and horizontal gene transfer.
ncAAs have also been used in the preparation of biomaterials with improved or novel properties. For example, a reversible biomechano-responsive surface was generated via covalently coating pAzF (Fig. 1B4)-containing GFPs onto polydimethylsiloxane with PEG brushes (Longo et al. 2015). A reversible decrease in fluorescence intensity was observed on applying mechanical stress to the surface. To prepare homogenous protein–polymer conjugates, an ncAA with a polymer initiator side chain was genetically incorporated into proteins in E. coli. The designed amino acid, 4-(2′-bromoisobutyramido)-phenylalanine (Fig. 1B24) can serve as an initiator in atom-transfer radical polymerization and provides a stable link between target proteins and polymers (Peeler et al. 2010). Metal ion-binding amino acids may allow the generation of enzymes with novel functions, including oxygen transport, electron transfer, and catalytic activity. One such example is the computational design of a metal ion-binding site in the protein using (2,2′-bipyridin-5yl)-alanine (BpyAla; Fig. 1B22) (Fig. 3A) (Mills et al. 2013). Using the software Rosetta, Mills et al. (2013) generated a BpyAla-containing metalloprotein with high affinity (Kd for Zn2+ is ∼ 37 pm) against a number of divalent cations. The three-dimensional structure determined by X-ray crystallography was very close to that of the design model (Fig. 3A).
PROTEIN EVOLUTION WITH ncAAs
Advances in methods for incorporation ncAAs now make it possible to ask whether additional amino acids can confer an evolutionary advantage to a host organism in response to selective pressure. For example, an in vitro antibody phage display system was developed in which ncAAs were available for directed evolution. A germline antibody library containing six randomized residues in CDR3H was subjected to affinity-based phage panning for binding to gp120, an HIV protein that naturally binds the sulfated cytokine receptor CCR5 (Liu et al. 2008). When sulfotyrosine (pSY; Fig. 1B58) was genetically encoded in the host strain for phage production, the highest affinity antibodies identified in the panning experiment all contained sulfotyrosine in CDRH3. A similar phage display system was used to select a Zn(II)-finger-like domain in which two histidine residues involved in Zn binding were substituted with the ncAA, bipyridylalanine (BpyAla; Fig. 1B22). To alter the metal ion-binding site, five residues in the amino terminus of Zif268 zinc-finger protein, including the two histidines, were randomized. Panning this library against the DNA recognition site revealed a mutant in which the wild-type Zn(II) site was replaced with a unique 4-bipyridylalanine.Fe(II) site with no impact on DNA affinity and selectivity (Kang et al. 2014). More recently, a bacterial selection scheme was used to identify cyclic peptides that inhibit HIV protease. Using a split intein system, a library of ncAA-containing ribosomal cyclic peptides was generated in E. coli. This library was further subjected to a selection based on survival of E. coli in response to proteolytic stress. A p-benzoylphenylalanine)-containing (Fig. 1B52) peptide that forms a covalent bond with the ɛ-amino group of Lys14 on the HIV protease and destabilizes the soluble cytoplasmic protein was isolated as the most potent hit (Young et al. 2011). And recently, Hammerling et al. (2014) generated bacteriophage T7 with an expanded genetic code by passaging phage on an E. coli host in which the amber codon was reassigned to 3-iodotyrosine (Fig. 1B17). After serial transfers of lytic phage, it was found that substitution of 3-iodotyrosine in the type II holing protein of bacteriophage T7 leads to increased phage fitness (Hammerling et al. 2014).
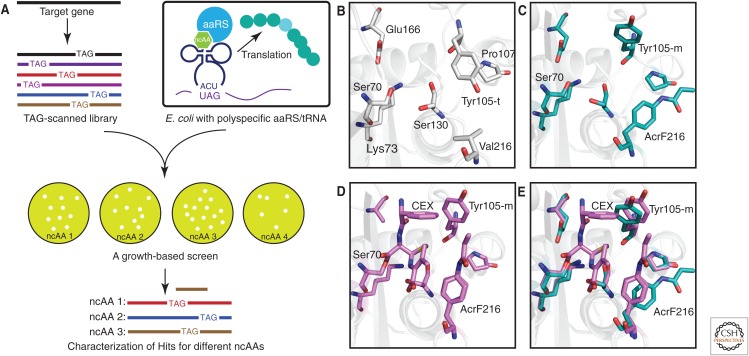
However, a more general rapid in vitro directed evolution for novel functional proteins with ncAAs has remained elusive. To this end, we recently developed a novel in vitro system with ncAAs in which a number of different ncAAs were randomly substituted at each of 144 of the 286 residues in TEM-1 β-lactamase based on the X-ray crystal structure (Fig. 5A) (Xiao et al. 2015a). To simplify construction of a β-lactamase library containing a variety of structurally distinct ncAAs, a polyspecific aminoacyl-tRNA synthetase was used that is orthogonal to the endogenous translational machinery of E. coli, and selectively incorporates 10 distinct ncAAs in response to the amber nonsense codon (Fig. 5A). A screen for growth on the β-lactam antibiotic cephalexin afforded a unique p-acrylamidophenylalanine mutation at Val216 that leads to an increase in catalytic efficiency by increasing kcat without significantly affecting KM. This increase in activity is not recapitulated by substitution with any of the canonical amino acids at residue 216. A comparison of the X-ray crystal structures of the cephalexin complexes of deacylation-defective wild-type and Val216p-AcrPhe mutant β-lactamases suggests that this mutation leads to changes in active site conformation with increased complementarity to the putative reaction TS (Fig. 5B–E). More recently, the impact of ncAA substitution on Phe66, Leu162, and Thr189 of TEM-1 β-lactamase have been systematically studied by Tack et al. (2016), and it has been shown that enzyme activity can be made strictly dependent on the presence of a ncAA. Using a temperature-dependent growth selection system, ncAAs containing long-side-chain thiols that can pair with cysteines to afford extended disulfide bonds have been used to identify enzymes showing enhanced thermostability. A mutant enzyme was identified that is cross-linked by one such extended disulfide bond and is stabilized by 9°C. In addition, a protein has been isolated that appears to form a covalent adduct between a Cys- and keto-containing ncAA and is stabilized by >25°C. These experiments provide more evidence that additional amino acids can confer an evolutionary advantage to a host organism in response to selective pressure.
Figure 5.
Directed evolution by noncanonical amino acid (ncAA) mutagenesis. (A) A TAG-scanned library was transformed into cells expressing a polyspecific amber suppressor aminoacyl-tRNA synthetase (aaRS)/transfer RNA (tRNA) pair and a growth selection was performed with various ncAAs. (B) X-ray crystal structure of ligand-free wild-type β-lactamase (protein data bank [PDB] code 1BTL). (C) X-ray crystal structure of ligand-free Val216AcrF mutant enzyme (PDB code 4ZJ1). (D) X-ray crystal structure of cephalexin (CEX) acyl-enzyme intermediate for the Val216AcrF mutant enzyme (PDB code 4ZJ3). (E) Overlay of active site residues of cephalexin-bound and ligand-free Val216AcrF mutant enzyme.
CONCLUSIONS AND PERSPECTIVES
The canonical 20 amino acid genetic code has significantly limited the chemical diversity of proteins throughout evolution. Nature has added additional chemistries to proteins by the use of cofactors and PTMs and, in a small number of cases, pyrrolysine and selenocysteine. Using a relative simple strategy, we and others have been able to augment the natural protein biosynthetic machinery with a large number of ncAAs with novel chemical, physical, and biological properties. These new amino acids have been genetically encoded in both prokaryotic and eukaryotic organisms with excellent fidelity and, in some cases, efficiencies that allow mutant proteins to be produced on a commercial scale. This expanded genetic code has provided powerful new tools to probe protein structure and function both in vitro and in living cells, and is allowing the rational design and evolution of proteins with useful new properties for a variety of applications, including the creation of new medicines. Many opportunities exist to further expand and exploit this technology, including methods for the generation of proteins consisting of a large number (or entirely) of distinct ncAAs, methods to evolve whole proteomes with novel properties rather than individual proteins, the use of computational methods together with ncAAs to generate new function, and the use of this technology to create a new generation of therapeutic proteins whose pharmacology can be optimized with medicinal chemistry like control over chemical structure and homogeneity. Clearly, the synergistic use of tools and principles from both chemistry and biology has made it possible to remove the constraints of nature’s 20 amino acid genetic code.
ACKNOWLEDGMENTS
We acknowledge Kristen Williams for her assistance in manuscript preparation. This work is supported by DOE Grant DE-SC0011787 (P.G.S.). This is manuscript number 29056 of The Scripps Research Institute.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Ai HW. 2012. Biochemical analysis with the expanded genetic lexicon. Anal Bioanal Chem 403: 2089–2102. [DOI] [PubMed] [Google Scholar]
- Amiram M, Haimovich AD, Fan C, Wang YS, Aerni HR, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, et al. 2015. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol 33: 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Schultz PG. 2003. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry 42: 9598–9608. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. 2004. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci 101: 7566–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E, Torres-Kolbus J, Deiters A, Chin JW. 2012. Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J Am Chem Soc 134: 11912–11915. [DOI] [PubMed] [Google Scholar]
- Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, et al. 2012. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci 109: 16101–16106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. 2012. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol 8: 748–750. [DOI] [PubMed] [Google Scholar]
- Bluemel C, Hausmann S, Fluhr P, Sriskandarajah M, Stallcup WB, Baeuerle PA, Kufer P. 2010. Epitope distance to the target cell membrane and antigen size determine the potency of T cell-mediated lysis by BiTE antibodies specific for a large melanoma surface antigen. Cancer Immunol Immunother 59: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne H, Conroy PJ, Whisstock JC, O’Kennedy RJ. 2013. A tale of two specificities: Bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol 31: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Axup JY, Ma JS, Wang RE, Choi S, Tardif V, Lim RK, Pugh HM, Lawson BR, Welzel G, et al. 2015. Multiformat T-cell-engaging bispecific antibodies targeting human breast cancers. Angew Chem Int Ed Engl 54: 7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellitti SE, Jones DH, Lagpacan L, Hao X, Zhang Q, Hu H, Brittain SM, Brinker A, Caldwell J, Bursulaya B, et al. 2008. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc 130: 9268–9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon G, Brustad E, Scott KA, Wang JY, Lobner-Olesen A, Schultz PG, Jacobs-Wagner C, Chapman E. 2011. Subcellular protein localization by using a genetically encoded fluorescent amino acid. Chembiochem 12: 1818–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Schultz PG. 2012. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci 109: 14841–14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Guo J, Lee HS, Schultz PG. 2013a. A genetically encoded fluorescent probe in mammalian cells. J Am Chem Soc 135: 12540–12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. 2013b. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52: 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Bollong M, Ai HW, Schultz PG. 2013c. Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc Natl Acad Sci 110: 11803–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Yang PY, Soundararajan G, Schultz PG. 2013d. A tryptophanyl–tRNA synthetase/tRNA pair for unnatural amino acid mutagenesis in E. coli. Angew Chem Int Ed Engl 52: 5106–5109. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Lajoie MJ, Xiao H, Church GM, Schultz PG. 2014. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. Chembiochem 15: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Chen HT, Warfield L, Hahn S. 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14: 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Ren W, Wright QE, Ai HW. 2013. Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J Am Chem Soc 135: 14940–14943. [DOI] [PubMed] [Google Scholar]
- Chen XH, Xiang Z, Hu YS, Lacey VK, Cang H, Wang L. 2014. Genetically encoding an electrophilic amino acid for protein stapling and covalent binding to native receptors. ACS Chem Biol 9: 1956–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW. 2014. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem 83: 379–408. [DOI] [PubMed] [Google Scholar]
- Cho H, Daniel T, Buechler YJ, Litzinger DC, Maio Z, Putnam AM, Kraynov VS, Sim BC, Bussell S, Javahishvili T, et al. 2011. Optimized clinical performance of growth hormone with an expanded genetic code. Proc Natl Acad Sci 108: 9060–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo JA, Stubbe J. 2011. Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem 80: 733–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Chin JW. 2012. Designer proteins: Applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol 13: 168–182. [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Ernst RJ, Walker OS, Chin JW. 2016. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat Methods 13: 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. 2009. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Natl Acad Sci 106: 21637–21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman JL, Kang M, Choi S, Cao Y, Wold ED, Sun SB, Smider VV, Schultz PG, Kim CH. 2014. A genetically encoded aza-Michael acceptor for covalent cross-linking of protein–receptor complexes. J Am Chem Soc 136: 8411–8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauba V, Grunewald J, Gorney V, Deaton LM, Kang MC, Bursulaya B, Ou WJ, Lerner RA, Schmedt C, Geierstanger BH, et al. 2011. Loss of CD4 T-cell-dependent tolerance to proteins with modified amino acids. Proc Natl Acad Sci 108: 12821–12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Nguyen DP, Lusic H, An WA, Deiters A, Chin JW. 2010. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc 132: 4086–4088. [DOI] [PubMed] [Google Scholar]
- Gautier A, Deiters A, Chin JW. 2011. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc 133: 2124–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S, Chin JW. 2011. Expanding the genetic code of an animal. J Am Chem Soc 133: 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff D, Thielges MC, Cellitti S, Schultz PG, Romesberg FE. 2009. Efforts toward the direct experimental characterization of enzyme microenvironments: Tyrosine100 in dihydrofolate reductase. Angew Chem Int Ed Engl 48: 3478–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff D, Wang F, Jockusch S, Turro NJ, Schultz PG. 2010. A new strategy to photoactivate green fluorescent protein. Angew Chem Int Ed Engl 49: 7677–7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenewald J, Tsao ML, Perera R, Dong LQ, Niessen F, Wen BG, Kubitz DM, Smider VV, Ruf W, Nasoff M, et al. 2008. Immunochemical termination of self-tolerance. Proc Natl Acad Sci 105: 11276–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerling MJ, Ellefson JW, Boutz DR, Marcotte EM, Ellington AD, Barrick JE. 2014. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol 10: 178–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Van Treeck B, Nguyen HB, Melancon CE III. 2015. Development of an unnatural amino acid incorporation system in the actinobacterial natural product producer Streptomyces venezuelae ATCC 15439. ACS Synth Biol 5: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J, Chou CJ, Chin JW, Deiters A. 2013. Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J Am Chem Soc 135: 13433–13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Russell WK, Wan W, Pai PJ, Russell DH, Liu W. 2010. A convenient method for genetic incorporation of multiple noncanonical amino acids into one protein in Escherichia coli. Mol Biosyst 6: 683–686. [DOI] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD. 2009. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci 106: 19120–19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JC, Hammill JT, Mehl RA. 2007. Site-specific incorporation of a 19F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J Am Chem Soc 129: 1160–1166. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. 2011. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol 7: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalstrup T, Blunck R. 2013. Dynamics of internal pore opening in KV channels probed by a fluorescent unnatural amino acid. Proc Natl Acad Sci 110: 8272–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Kawaguchi D, Coin I, Xiang Z, O’Leary DDM, Slesinger PA, Wang L. 2013. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron 80: 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Light K, Ai HW, Shen W, Kim CH, Chen PR, Lee HS, Solomon EI, Schultz PG. 2014. Evolution of iron(II)-finger peptides by using a bipyridyl amino acid. Chembiochem 15: 822–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Axup JY, Dubrovska A, Kazane SA, Hutchins BA, Wold ED, Smider VV, Schultz PG. 2012a. Synthesis of bispecific antibodies using genetically encoded unnatural amino acids. J Am Chem Soc 134: 9918–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Kang M, Kim HJ, Chatterjee A, Schultz PG. 2012b. Site-specific incorporation of ɛ-N-crotonyllysine into histones. Angew Chem Int Ed Engl 51: 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Axup JY, Lawson BR, Yun H, Tardif V, Choi SH, Zhou Q, Dubrovska A, Biroc SL, Marsden R, et al. 2013. Bispecific small molecule-antibody conjugate targeting prostate cancer. Proc Natl Acad Sci 110: 17796–17801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kularatne SA, Venkatesh C, Santhapuram HK, Wang K, Vaitilingam B, Henne WA, Low PS. 2010. Synthesis and biological analysis of prostate-specific membrane antigen-targeted anticancer prodrugs. J Med Chem 53: 7767–7777. [DOI] [PubMed] [Google Scholar]
- Kularatne SA, Deshmukh V, Gymnopoulos M, Biroc SL, Xia J, Srinagesh S, Sun Y, Zou N, Shimazu M, Pinkstaff J, et al. 2013. Recruiting cytotoxic T cells to folate-receptor-positive cancer cells. Angew Chem Int Ed Engl 52: 12101–12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kularatne SA, Deshmukh V, Ma J, Tardif V, Lim RK, Pugh HM, Sun Y, Manibusan A, Sellers AJ, Barnett RS, et al. 2014. A CXCR4-targeted site-specific antibody-drug conjugate. Angew Chem Int Ed Engl 53: 11863–11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey VK, Parrish AR, Han SL, Shen ZX, Briggs SP, Ma YG, Wang L. 2011. A fluorescent reporter of the phosphorylation status of the substrate protein STAT3. Angew Chem Int Ed Engl 50: 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, et al. 2013. Genomically recoded organisms expand biological functions. Science 342: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Guo J, Lemke EA, Dimla RD, Schultz PG. 2009. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J Am Chem Soc 131: 12921–12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. 2007. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol 3: 769–772. [DOI] [PubMed] [Google Scholar]
- Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RA, Mehl RA, Pielak GJ. 2010. Protein 19F NMR in Escherichia coli. J Am Chem Soc 132: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang H, Sun Y, Pan Y, Zhou J, Wang J. 2013. Expanding the genetic code for photoclick chemistry in E. coli, mammalian cells, and A. thaliana. Angew Chem Int Ed Engl 52: 9700–9704. [DOI] [PubMed] [Google Scholar]
- Lin SX, Yan H, Li L, Yang MY, Peng B, Chen S, Li WH, Chen PR. 2013. Site-specific engineering of chemical functionalities on the surface of live hepatitis D virus. Angew Chem Int Ed Engl 52: 13970–13974. [DOI] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. 2006. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol 24: 1436–1440. [DOI] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. 2010. Adding new chemistries to the genetic code. Annu Rev Biochem 79: 413–444. [DOI] [PubMed] [Google Scholar]
- Liu CC, Brustad E, Liu W, Schultz PG. 2007. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J Am Chem Soc 129: 10648–10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Mack AV, Tsao ML, Mills JH, Lee HS, Choe H, Farzan M, Schultz PG, Smider VV. 2008. Protein evolution with an expanded genetic code. Proc Natl Acad Sci 105: 17688–17693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo J, Yao C, Rios C, Chau NT, Boulmedais F, Hemmerle J, Lavalle P, Schiller SM, Schaaf P, Jierry L. 2015. Reversible biomechano-responsive surface based on green fluorescent protein genetically modified with unnatural amino acids. Chem Commun (Camb) 51: 232–235. [DOI] [PubMed] [Google Scholar]
- Lu H, Wang D, Kazane S, Javahishvili T, Tian F, Song F, Sellers A, Barnett B, Schultz PG. 2013. Site-specific antibody-polymer conjugates for siRNA delivery. J Am Chem Soc 135: 13885–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhou Q, Deshmukh V, Phull H, Ma J, Tardif V, Naik RR, Bouvard C, Zhang Y, Choi S, et al. 2014. Targeting human C-type lectin-like molecule-1 (CLL1) with a bispecific antibody for immunotherapy of acute myeloid leukemia. Angew Chem Int Ed Engl 53: 9841–9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XZ, Zambaldo C, Liu T, Zhang YH, Xuan WM, Wang C, Reed SA, Yang PY, Wang RSE, Javahishvili T, et al. 2016. Recombinant thiopeptides containing noncanonical amino acids. Proc Natl Acad Sc 113: 3615–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JS, Kim JY, Kazane SA, Choi SH, Yun HY, Kim MS, Rodgers DT, Pugh HM, Singer O, Sun SB, et al. 2016. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc Natl Acad Sci 113: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM. 2015. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JH, Khare SD, Bolduc JM, Forouhar F, Mulligan VK, Lew S, Seetharaman J, Tong L, Stoddard BL, Baker D. 2013. Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy. J Am Chem Soc 135: 13393–13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan EC, Seyedsayamdost MR, Uhlin U, Stubbe J. 2011a. Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases. J Am Chem Soc 133: 9430–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan EC, Young DD, Schultz PG, Stubbe J. 2011b. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J Am Chem Soc 133: 15942–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22: 2994–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PA, Zhang WJ, Rainey GJ, Burke S, Li H, Huang L, Gorlatov S, Veri MC, Aggarwal S, Yang YH, et al. 2011. Application of dual affinity retargeting molecules to achieve optimal redirected T-cell killing of B-cell lymphoma. Blood 117: 4542–4551. [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K. 2006. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci 103: 16159–16164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Pinkstaff J, Li Z, Skidmore L, Li N, Myler H, Dallas-Yang Q, Putnam AM, Yao J, Bussell S, et al. 2012. FGF21 analogs of sustained action enabled by orthogonal biosynthesis demonstrate enhanced antidiabetic pharmacology in rodents. Diabetes 61: 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murelli RP, Zhang AX, Michel J, Jorgensen WL, Spiegel DA. 2009. Chemical control over immune recognition: A class of antibody-recruiting small molecules that target prostate cancer. J Am Chem Soc 131: 17090–17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. 2008. Genetically encoding Nɛ-acetyllysine in recombinant proteins. Nat Chem Biol 4: 232–234. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. 2009. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Slusarczyk AL, Chin JW. 2010a. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J Am Chem Soc 132: 2142–2144. [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. 2010b. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464: 441–444. [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. 2009. Genetically encoding Nɛ-methyl-l-lysine in recombinant histones. J Am Chem Soc 131: 14194–14195. [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Garcia Alai MM, Virdee S, Chin JW. 2010. Genetically directing ɛ-N,N-dimethyl-l-lysine in recombinant histones. Chem Biol 17: 1072–1076. [DOI] [PubMed] [Google Scholar]
- Niu W, Guo J. 2013. Expanding the chemistry of fluorescent protein biosensors through genetic incorporation of unnatural amino acids. Mol Biosyst 9: 2961–2970. [DOI] [PubMed] [Google Scholar]
- Okuda S, Tokuda H. 2009. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci 106: 5877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Wang VS, Radoicic J, De Angelis AA, Berkamp S, Opella SJ. 2014. Paramagnetic relaxation enhancement of membrane proteins by incorporation of the metal-chelating unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQA). J Biomol NMR 61: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson AD, Mills JH, Song Y, Nasertorabi F, Han GW, Baker D, Stevens RC, Schultz PG. 2015. Transition states. Trapping a transition state in a computationally designed protein bottle. Science 347: 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeler JC, Woodman BF, Averick S, Miyake-Stoner SJ, Stokes AL, Hess KR, Matyjaszewski K, Mehl RA. 2010. Genetically encoded initiator for polymer growth from proteins. J Am Chem Soc 132: 13575–13577. [DOI] [PubMed] [Google Scholar]
- Peters FB, Brock A, Wang JY, Schultz PG. 2009. Photocleavage of the polypeptide backbone by 2-nitrophenylalanine. Chem Biol 16: 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Ji A, Ai HW. 2015. Light activation of protein splicing with a photocaged fast intein. J Am Chem Soc 137: 2155–2158. [DOI] [PubMed] [Google Scholar]
- Rust HL, Subramanian V, West GM, Young DD, Schultz PG, Thompson PR. 2014. Using unnatural amino acid mutagenesis to probe the regulation of PRMT1. ACS Chem Biol 9: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Anderson JC, Lakshman V, Schultz PG. 2003. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res 31: 6700–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. 2004. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Borbas J, Drescher M, Summerer D. 2014. A genetically encoded spin label for electron paramagnetic resonance distance measurements. J Am Chem Soc 136: 1238–1241. [DOI] [PubMed] [Google Scholar]
- Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. 2006. A genetically encoded infrared probe. J Am Chem Soc 128: 13984–13985. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Xie J, Chan CTY, Schultz PG, Stubbe J. 2007. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: Direct evidence for involvement of Y730 and Y731 in radical propagation. J Am Chem Soc 129: 15060–15071. [DOI] [PubMed] [Google Scholar]
- Smith EE, Linderman BY, Luskin AC, Brewer SH. 2011. Probing local environments with the infrared probe: l-4-nitrophenylalanine. J Phys Chem B 115: 2380–2385. [DOI] [PubMed] [Google Scholar]
- Speight LC, Muthusamy AK, Goldberg JM, Warner JB, Wissner RF, Willi TS, Woodman BF, Mehl RA, Petersson EJ. 2013. Efficient synthesis and in vivo incorporation of acridon-2-ylalanine, a fluorescent amino acid for lifetime and Förster resonance energy transfer/luminescence resonance energy transfer studies. J Am Chem Soc 135: 18806–18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. 2006. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci 103: 9785–9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SB, Schultz PG, Kim CH. 2014. Therapeutic applications of an expanded genetic code. Chembiochem 15: 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack DS, Ellefson JW, Thyer R, Wang B, Gollihar J, Forster MT, Ellington AD. 2016. Addicting diverse bacteria to a noncanonical amino acid. Nat Chem Biol 12: 138–140. [DOI] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. 2010. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468: 978–982. [DOI] [PubMed] [Google Scholar]
- Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. 2009. Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side-chain quenchers: A combined fluorescence, IR and molecular dynamics study. Biochemistry 48: 9040–9046. [DOI] [PubMed] [Google Scholar]
- Tian F, Lu Y, Manibusan A, Sellers A, Tran H, Sun Y, Phuong T, Barnett R, Hehli B, Song F, et al. 2014. A general approach to site-specific antibody drug conjugates. Proc Natl Acad Sci 111: 1766–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wals K, Ovaa H. 2014. Unnatural amino acid incorporation in E. coli: Current and future applications in the design of therapeutic proteins. Front Chem 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Tharp JM, Liu WR. 2014. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta 1844: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie J, Deniz AA, Schultz PG. 2003. Unnatural amino acid mutagenesis of green fluorescent protein. J Org Chem 68: 174–176. [DOI] [PubMed] [Google Scholar]
- Wang JY, Xie JM, Schultz PG. 2006a. A genetically encoded fluorescent amino acid. J Am Chem Soc 128: 8738–8739. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. 2006b. Expanding the genetic code. Annu Rev Biophys Biomol Struct 35: 225–249. [DOI] [PubMed] [Google Scholar]
- Wang K, Neumann H, Peak-Chew SY, Chin JW. 2007. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol 25: 770–777. [DOI] [PubMed] [Google Scholar]
- Wang F, Robbins S, Guo JT, Shen WJ, Schultz PG. 2010. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PloS ONE 5: e9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Niu W, Guo J, Schultz PG. 2012. Unnatural amino acid mutagenesis of fluorescent proteins. Angew Chem Int Ed Engl 51: 10132–10135. [DOI] [PubMed] [Google Scholar]
- Wang NX, Li Y, Niu W, Sun M, Cerny R, Li QS, Guo JT. 2014. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew Chem Int Ed Engl 53: 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Ren HY, Hu YS, Coin I, Wei J, Cang H, Wang L. 2014. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods 10: 885–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Chatterjee A, Choi SH, Bajjuri KM, Sinha SC, Schultz PG. 2013. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew Chem Int Ed Eng 52: 14080–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Peters FB, Yang PY, Reed S, Chittuluru JR, Schultz PG. 2014. Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase. ACS Chem Biol 9: 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Nasertorabi F, Choi SH, Han GW, Reed SA, Stevens RC, Schultz PG. 2015a. Exploring the potential impact of an expanded genetic code on protein function. Proc Natl Acad Sci 112: 6961–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Xuan W, Shao S, Liu T, Schultz PG. 2015b. Genetic incorporation of ɛ-N-2-hydroxyisobutyryl-lysine into recombinant histones. ACS Chem Biol 10: 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Supekova L, Schultz PG. 2007. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol 2: 474–478. [DOI] [PubMed] [Google Scholar]
- Ye S, Huber T, Vogel R, Sakmar TP. 2009. FTIR analysis of GPCR activation using azido probes. Nat Chem Biol 5: 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Uhlin U, Stubbe J. 2010. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J Am Chem Soc 132: 8385–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TS, Ahmad I, Yin JA, Schultz PG. 2010. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395: 361–374. [DOI] [PubMed] [Google Scholar]
- Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. 2011. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci 108: 11052–11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Zhang G, Li J, Chen PR. 2014. Monitoring endocytic trafficking of anthrax lethal factor by precise and quantitative protein labeling. Angew Chem Int Ed Engl 53: 6449–6453. [DOI] [PubMed] [Google Scholar]