Abstract
The intracellular parasitic nature of viruses and the emergence of antiviral drug resistance necessitate the development of new potent antiviral drugs. Recently, a method for developing potent inhibitory drugs by targeting biological machines with high stoichiometry and a sequential-action mechanism was described. Inspired by this finding, we reviewed the development of antiviral drugs targeting viral DNA-packaging motors. Inhibiting multisubunit targets with sequential actions resembles breaking one bulb in a series of Christmas lights, which turns off the entire string. Indeed, studies on viral DNA packaging might lead to the development of new antiviral drugs. Recent elucidation of the mechanism of the viral double-stranded DNA (dsDNA)-packaging motor with sequential one-way revolving motion will promote the development of potent antiviral drugs with high specificity and efficiency. Traditionally, biomotors have been classified into two categories: linear and rotation motors. Recently discovered was a third type of biomotor, including the viral DNA-packaging motor, beside the bacterial DNA translocases, that uses a revolving mechanism without rotation. By analogy, rotation resembles the Earth's rotation on its own axis, while revolving resembles the Earth's revolving around the Sun (see animations at http://rnanano.osu.edu/movie.html). Herein, we review the structures of viral dsDNA-packaging motors, the stoichiometries of motor components, and the motion mechanisms of the motors. All viral dsDNA-packaging motors, including those of dsDNA/dsRNA bacteriophages, adenoviruses, poxviruses, herpesviruses, mimiviruses, megaviruses, pandoraviruses, and pithoviruses, contain a high-stoichiometry machine composed of multiple components that work cooperatively and sequentially. Thus, it is an ideal target for potent drug development based on the power function of the stoichiometries of target complexes that work sequentially.
INTRODUCTION
Viruses reproduce themselves in host cells. Their intracellular parasitic nature poses a great challenge to antiviral drug development. Nevertheless, significant progress in antiviral drug discovery, such as new treatments for HIV (1), hepatitis B virus (2), herpesvirus (3), and influenza virus (4), has been made. Even though different approaches to new drug development, such as improving drug target binding affinity (5) and finding new targets with novel pharmacological mechanisms (6), have been explored, new infectious pandemic viruses that greatly threaten human health still emerge sporadically. In addition, the emergence of drug resistance necessitates new drug development. Highly potent drugs with elevated specificity are needed for overcoming viral diseases.
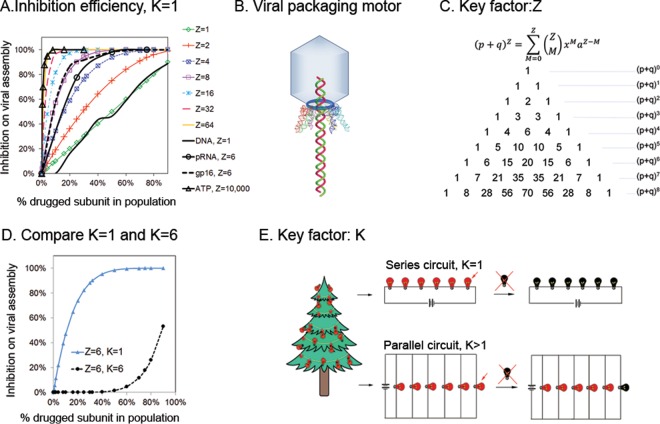
In viral reproduction, the key step of genome packaging is usually accomplished by a biomotor using ATP. Linear double-stranded DNA (dsDNA) or dsRNA viruses package their genomes into preformed procapsids. This group includes dsDNA/dsRNA bacteriophages (7), adenoviruses (8), poxviruses (9, 120), herpesviruses (10, 11), mimiviruses, megaviruses, pandoraviruses, and pithoviruses (52–54). Researchers have been intrigued by the unique viral structure and packaging mechanisms, and extensive studies have been carried out to elucidate the fundamentals in protein-DNA, protein-RNA, and RNA-DNA interactions in the quest for new prototypes of biological machines or new antiviral drugs. Jonathon King's observation several decades ago that studies on viral DNA packaging will lead to the discovery of new antiviral drugs (12) has stimulated scientists to pursue further study on the viral DNA-packaging mechanism with the goal of uncovering better drug targets. Indeed, our study on the phi29 packaging motor revealed that several key motor components composed of multimeric complexes, such as the hexameric pRNA ring, hexameric gp16 ATPase, and ATP with more than 10,000 copies to package one genome (13, 14), may serve as more efficient model drug targets than a monomeric genomic DNA. Comparing the viral packaging inhibition efficiencies of mock drugs targeting each machine with different stoichiometries, we found that inhibition efficiency to the stoichiometry of the targeted bio-complex increased exponentially (Fig. 1A) (14).
FIG 1.
Drug inhibition efficiency is correlated with the stoichiometry of the targeted biocomplex. (A) Targeting 10,000 subunits of ATP showed the strongest inhibition efficiency, compared to those of the 6-subunit pRNA or gp16 and the 1-subunit genomic DNA. (B) Illustration of the viral DNA-packaging motor. (C) One key factor regarding drug potency is the stoichiometry of the homomeric complex serving as a target: the ratio of the functional complex can be calculated from Yang Hui's triangle. (D) Comparison of drug inhibition efficiencies in a complex with a Z equal to 6 and a K equal to 1 and with a Z equal 6 and a K equal to 6. Here, K is the number of blocked subunits required to inhibit the function of the whole complex target. (E) Illustration showing that K is a key factor for drug potency. When K = 1 as in the ATPase sequential motion, the system is similar to a series circuit of Christmas lights; one broken bulb will turn off the whole chain. When K > 1, the chain will not be completely turned off until all bulbs inside the parallel circuit are broken.
Previously, biomotors were classified into two categories: linear motors and rotation motors (15). Recently, a new category of biomotors using a revolving mechanism without rotation was reported and found to be widespread in different biological systems (Fig. 1B) (16, 17). Based on this new mechanism, a method for developing highly efficient inhibitory drugs was described by targeting biological machines with high stoichiometry and exhibiting a sequential-action mechanism (14, 18). Herein, we review the development of antiviral drugs that target viral DNA-packaging motors, a subject that has been investigated for many decades.
NEW METHOD FOR DEVELOPING HIGHLY POTENT DRUGS BY TARGETING HOMOMERIC BIOLOGICAL MACHINES WITH HIGH STOICHIOMETRY TO ACT SEQUENTIALLY
A determining factor for drug inhibition efficiency on its target biological entity is the ratio of the drugged target component (Tinactive) to the undrugged target component (Tactive). Within each virus-infected cell, a high percentage of drugged machines (Tinactive) leads to high inhibitory efficiency. The percentage of drugged machines can be calculated from binomial distribution or Yang Hui's triangle (Fig. 1C), as follows:
| (1) |
Here, p and q represent the fractions of drugged inactive and undrugged active subunits in the population, respectively, Z is the stoichiometry of the target, and M represents drugged subunits in each biocomplex.
The first intrinsic factor of the target for potent drug development is Z, which is the stoichiometry of the homomeric complex serving as a drug target. The ratio of functional complexes changes dramatically with Z. In equation 1, assuming that the ratio of drugged inactive subunits (p) and undrugged active subunits (q) in the population is fixed, when the target machine contains only one subunit, the ratio equals q, as derived from a Z equal to 1 in binomial distribution (equation 2). Here, p plus q equals 100%.
| (2) |
However, when the homomeric target complex contains multiple subunits, a binomial distribution formula with a higher order (equation 3) is applied. When Z equals 4, then
| (3) |
In this case, the probability of a target machine complex possessing four copies of the inactive subunit is p4. Three copies of the inactive and one copy of the active subunit is 4p3q, two copies of the inactive and two copies of the uninhibited subunits is 6p2q2, three copies of the active subunits is 4pq3, and four copies of the active subunit is q4. Assuming that 70% (p) of subunits are inactivated by drugs, the percentage of active machines containing four copies of active subunits is q4, which is (0.3)4 or 0.8%. However, when Z equals 1 and P equals 70%, the ratio of the uninhibited portion equals 30%. Thus, the stoichiometry (Z) of the targeted machine contributes significantly to the ratio of survival rate, which directly correlates with drug inhibition efficiency upon viral replication.
The second intrinsic factor of the target for potent drug development is K, which is the number of drugged subunits required to block the function of the complex. Stoichiometry has a multiplicative effect on inhibition efficiency only when K equals 1. Reinterpreting this statement with an example of a homo-hexamer machine (Z = 6) as a drug target, where 70% (p) of subunits are inactivated by drugs, if K equals 1, the ratio of active target complexes will be 0.36 or 0.07%, whereas if K equals 6, the ratio will be 0.76 or 88.2%. A K equal to 1 showed much stronger inhibition than a K equal to 6, when the active complex ratio was 88.2% at the same blocking subunit ratio (Fig. 1D). A K equal to 1 is a key factor for multisubunit complexes as potent drug targets.
The third intrinsic factor is that the homomeric complex acts through a sequential or coordination mechanism. Sequential actions or coordination action means that each subunit of the complex works in turn to complete the function of the complex (Fig. 1E). Blocking any step of the sequential actions results in deactivation of the complex. That meets the definition of K equals 1 in a homomeric complex. Analogously to a string of Christmas lights, where one broken light bulb will turn off the entire chain, one inhibited subunit will deactivate the entire complex and consequently its biological activity.
THE PACKAGING MOTORS OF dsDNA VIRUSES MEET THE FIRST INTRINSIC FACTOR OF THE TARGET FOR POTENT DRUG DEVELOPMENT AS HOMOMERIC MULTIMERS WITH HIGH STOICHIOMETRY
Viral DNA packaging motors can generally be divided into two closely related categories (19): (i) terminase and portal channel-mediated DNA translocation, as in herpesviruses (20), adenoviruses (8), and many bacteriophages, including phi29 (21), T4 (22), T3 (23), T5 (24), T7 (25), λ (26), SPP1 (27), and HK97 (16, 28), and (ii) HerA/FtsK-type translocase, as in poxvirus and other nucleocytoplasmic large DNA viruses (NCLDV) (29, 30). Both categories use very similar revolving mechanisms (13, 15, 21, 28). The first category of viral DNA-packaging motors uses a pair of DNA-packaging enzymes: a head and tail connector with high stoichiometry for DNA-packaging activity. For example, the bacterial virus λ motor contains a tetramer terminase composed of two subunits of the large ATPase gpA and two subunits of gpNul1 (31). The phi29 motor contains a hexameric packaging RNA ring (32–34) and a hexameric ATPase gp16 (35). The connector of this group of viruses is a homomeric dodecameric complex. Both the terminase and the connector meet the first requirement: a homomeric complex with high stoichiometry.
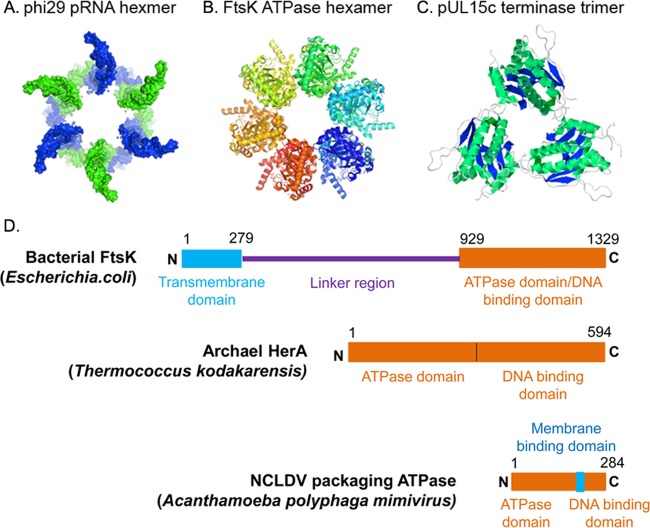
The phi29 motor is the most well-studied model system for viral DNA packaging, the components of which were used as mock drug targets in recent studies to compare the levels of influence of target stoichiometry on drug potency (14). In 1998, the packaging RNA (pRNA) ring in the phi29 motor was first determined to be a hexamer (32, 33) (featured by Cell [34]) (Fig. 2A). Hexameric pRNA formation was verified by cryo-electron microscopy (cryo-EM) (36), biochemical analysis (32–34), single-molecule photobleaching assay (37), gold labeling imaging by EM (38, 39), and RNA crystal structural studies (40). Recent experimental evidence based on binomial distribution analysis, quantitative DNA binding assays, capillary electrophoresis, and electrophoretic mobility shift assays (35, 41, 42) revealed that the ATPase gp16 of the phi29 motor, which belongs to the ASCE superfamily, is an asymmetrical hexamer in its working oligomeric state (35), like the oligomeric ATPase ring of other DNA translocases (43–45) and many other asymmetrical ATPase hexamers (121). Similarly, in the FtsK motor (Fig. 2B), the α and β domains assembled into a hexameric ring-shaped multimer with a central channel through which the dsDNA substrate is translocated (46). All these high-stoichiometric multimeric complexes in dsDNA-packaging motors fit the first requirement on target for potent drug development.
FIG 2.
Structural similarity of multimeric complexes that are widely spread in viral motors. (A) Hexameric packaging RNA in bacteriophage phi29; (B) hexameric FtsK ATPase (Protein Data Bank [PDB] accession number 2IUU); (C) trimeric pUL15C terminase from herpesvirus (PDB accession number 4IOX); (D) domain organization of genome-packaging motors (55). (Adapted from reference 55.)
Adenoviruses (AdV) are a group of well-studied dsDNA viruses that infect eukaryotic cells in vertebrates, including humans. AdV packages the genome using IVa2 and L1 52/55K as two packaging proteins into capsids that contain hexon, penton, and fiber (47). It has been reported that higher-order IVa2-containing complexes formed on adjacent packaging repeats are required for packaging activity (48). Quantitative mass spectrometry, metabolic labeling, and Western blotting revealed that there are approximately six to eight IVa2 molecules in each particle (47). The high-order IVa2 motor protein complex meets the first requirement of a high-stoichiometry target for potent drug development.
Herpes simplex viruses (HSV) package their dsDNA genome into preformed protein shells using terminases (49), which contain a large subunit, pUL15, and a small subunit, pUL28 (50). pUL15 cleaves concatemeric viral DNA during packaging initiation and completion cycles and functions as an ATPase, providing energy to the packaging process. The X-ray structure of the C-terminal domain of pUL15 showed a homo-trimer structure (Fig. 2C) (51), thus fitting the characteristics of a high-stoichiometric homomeric complex for highly potent drugs.
Mimivirus, Megavirus, Pandoravirus, and Pithovirus (52–54) all belong to the NCLDV superfamily and infect a wide range of eukaryotes (55, 56). Nine genes that are shared by all NCLDV families have been identified to encode DNA polymerase: the genes for a capsid protein, 3 helicases, a virion-packaging ATPase, a thiol oxidoreductase, a protein kinase, and a transcription factor (Fig. 2D) (57). Mimiviruses package their 1.2-Mbp dsDNA genome into preformed procapsids through a nonvertex portal (58) driven by the vaccinia virus A32-type virion-packaging ATPase (59). It has been shown that the structure and function of their DNA packaging motors are homologous to those of the FtsK DNA translocase (55).
Poxviruses are large, brick-shaped dsDNA viruses that replicate in the cytoplasm of infected cells. The two DNA strands of the genome are connected at the ends through a hairpin terminus (60). Poxvirus ATPase is coded by the A32 gene (119). Comparative sequence analysis revealed highly conserved N-terminal regions with five motifs among all poxviruses, including ATPase feature Walker A and Walker B motifs, A32L-specific motifs III and IV, and a novel motif V. The secondary-structure predictions of the N terminus of the A32 ATPase protein are homologous to those of the FtsK DNA translocase (19).
THE PACKAGING MOTORS OF dsDNA VIRUSES MEET THE SECOND INTRINSIC FACTOR OF THE TARGET FOR POTENT DRUG DEVELOPMENT, WITH K EQUAL TO 1, AS ONE INACTIVE SUBUNIT BLOCKS THE ENTIRE MACHINE
The K value is a key factor in estimating the probability of inactive nanomachines or biocomplexes by combination and permutation analysis. When K is 1, the uninhibited biocomplexes will equal qZ. Thus, stoichiometry has a multiplicative effect on inhibition. Most multisubunit homomeric biological motors act through sequential coordination mechanisms, where blocking one subunit of the complex inhibits the function of the whole nanomachine. Nucleic acid translocation and duplex unwinding by helicases are coupled to the NTP binding and hydrolysis cycle. This represents an extremely high level of coordination between proteins with their DNA substrates. ATPase undergoes conformational entropy changes upon ATP binding or ATP hydrolysis, leading to a high or low binding affinity toward DNA, respectively (16, 35, 61).
Sequential actions of the phi29 DNA-packaging motor were reported in 1997 (61, 62) by the Hill constant determination and inhibition assay using a binomial distribution (16, 17). ATPase activity has been analyzed by studying the effects of introducing mutant gp16 subunits (41, 42). The cooperative profile of subunits inside the ATPase hexamer mostly overlapped that of a model where a single inactive subunit is able to inactivate the whole oligomer. The asymmetrical ATPase structure has been widely observed, where oligomeric ATPase works through sequential binding and hydrolyzing of ATP to push dsDNA translocation; the subunit of ATPase in the ATP bound state is shown as asymmetric in EM images (121). Thus, the hexameric ATPases in the DNA-packaging motor fit the mathematical model of K equals 1.
In AdV, both L4-22K (63) and L4-33K (64) proteins play important roles in virus assembly (65–67). Removal of the 47 C-terminal amino acids of L4-33K aborted viral assembly function (66). L4-33K also plays a role in bovine AdV assembly (68). Similarly, removal of the 87 amino acids of bovine AdV L4-33K completely blocked capsid assembly (68). It has been reported that L4-33K is a virus-encoded alternative RNA splicing factor and is also involved in viral DNA packaging (64). The presence of IVa2 protein as a hexamer-octamer complex at the unique vertex has been demonstrated and reported to be equivalent to packaging ATPase (47). This assembly process is analogous to common features of portal-translocase systems. The ATPase property of IVa2 protein and oligomeric structures found in the virion vertex indicate that they work cooperatively as ATPases for packaging machinery, which leads to a K equal to 1. The major protein involved in poxvirus DNA packaging is the gene product of A32, which is also an ATPase. This ATPase has sequence similarity to the products of the IVa2 gene of AdV, which is also an ATPase involved in DNA packaging (69).
HSV uses more than seven viral proteins (encoded by the UL6, UL15, UL17, UL25, UL28, UL32, and UL33 genes) for the cleavage and packaging of its genome into procapsids (50). The terminase is comprised of the UL15, UL28, and UL33 proteins, which act in DNA packaging (50, 70, 71). The structure of the pUL15 nuclease domain strongly suggests an evolutionary path from simple phages, such as siphoviruses, to complex phages and, eventually, eukaryotic herpesviruses (51). The structure of C-terminal trimeric pUL15 is easily superimposable upon those of phage terminase large-subunit nuclease domains and also superimposes well onto HCMV pUL89C (51, 72). Also, the full-length protein might be a larger multimer with the motor function of the protein. This evidence suggests that trimeric pUL15 functions cooperatively for its ATPase activity, and one blocked subunit may deactivate the whole multimer.
Mimivirus and other DNA-packaging ATPase motors of NCLDV and prokaryotic viruses with an inner lipid membrane belong to the FtsK/SpoIIIE/HerA superfamily (ATPases of bacteria and archaea) (15, 28, 73). An automated homology model of packaging ATPase has been predicted using bioinformatic tools in vaccinia virus (19). These packaging ATPases possess a conserved domain in addition to the Walker A and Walker B domains. Blocking one component of the genome-packaging machinery might lead to disruption of all genome packaging in these viruses. The blocking agents/drugs targeting genome components in NCLDVS can also be employed in prokaryotes, as they have been predicted to use a similar revolving mechanism for genome packaging (15, 28, 55, 74). Inhibitors targeting the genome-packaging components of NCLDV might even act as potential antibacterial drugs. This research becomes even more important as new antibiotic-resistant strains emerge.
THE DNA-PACKAGING MOTOR OF dsDNA VIRUSES MEETS THE THIRD INTRINSIC FACTOR OF THE TARGET FOR POTENT DRUGS: SEQUENTIAL OR COORDINATED ACTIONS OF HOMOMERIC BIOMACHINES
One key aspect of targeting homomeric biomolecules of viral packaging motors for drug discovery is that the multisubunit biological complexes follow a sequential coordination or cooperative mechanism (16, 62, 75–81), which is widely seen in biological systems (75, 81, 82). Inhibiting any subunit of the motor with sequential actions leads to deactivation of the entire system.
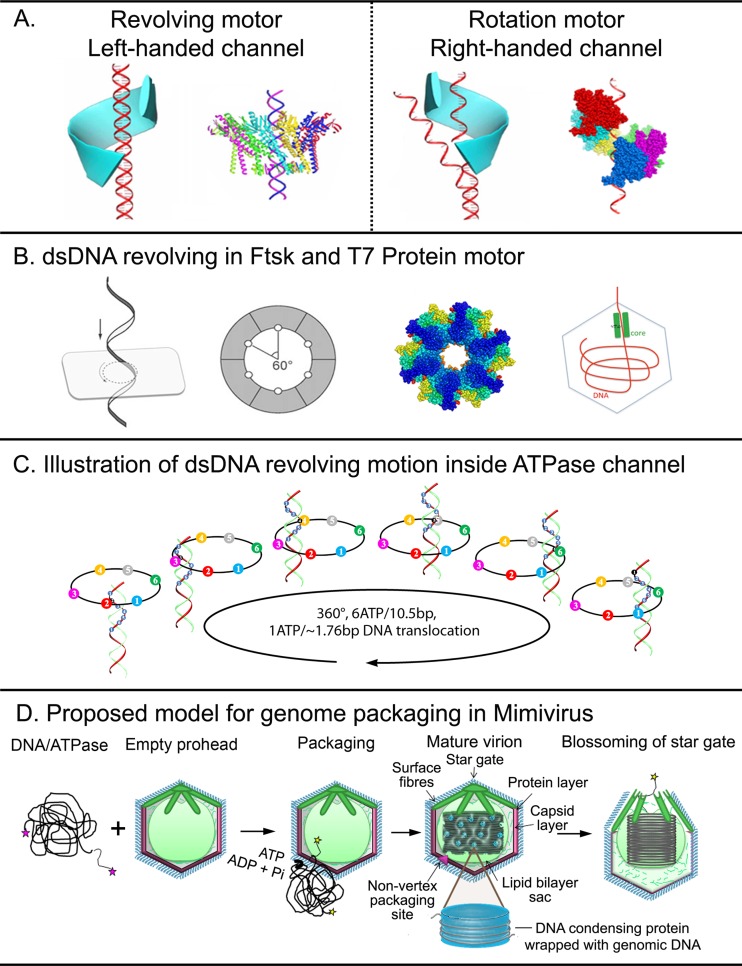
Motors of dsDNA viruses share a revolving mechanism without rotation (Fig. 3A and B) (17). Subunits of these motors coordinate with each other, forming a special structure to drive dsDNA translation through a one-way traffic mechanism with high efficiency (83). Revolving motors can be distinguished from rotation motors by channel size, as all revolving motors have a smaller channel size to allow the bolt and nut to contact, but the channel sizes of revolving motors are bigger, to allow the dsDNA to revolve. The 30° left-handed twist of the channel wall produces an antiparallel arrangement with the right-handed helix of the dsDNA (16, 83). The antichiral property between the left-handed twist of channel protein with the right-handed dsDNA can be a characteristic for revolving motors, as rotation motors have right-handed channels for DNA translocation (Fig. 3A) (16, 17). The electropositive lysine layers in the motor channels interact with a single strand of the electronegative dsDNA phosphate backbone, resulting in a relaying contact that facilitates one-way motion and generation of transitional pauses during dsDNA translocation (83, 84).
FIG 3.
dsDNA translocation motors with revolving mechanisms. (A) Revolving motors and rotation motors can be distinguished by motor chirality. (B) dsDNA translocase of FtsK (PDB accession number 2IUU) and T7 using a revolving mechanism. (C) Phi29 packages dsDNA through a revolving mechanism powered by ATP. (D) Hypothetical model for genome packaging in mimivirus. The packaging ATPase binds at specific sites of the catenated genome and translocates the decatenated genome powered by ATP hydrolysis into the preassembled capsid through a transient nonvertex opening situated on the opposite face of the star gate-like structure. DNA-condensing proteins might be involved in genome condensation. The genome is delivered through the star gate during viral maturation. (Adapted from references 17, 28, 74, 122, and 123 with permission of the publishers.)
Protein molecular motors act as switches that undergo conformational changes, depending on the binding of nucleotides. For example, in the hexameric ring of gp16 ATPase subunits of the phi29 DNA-packaging motor, the ATP-bound subunit has high affinity to dsDNA. Hydrolysis of ATP to ADP generates a conformational change in the subunit, resulting in low affinity to dsDNA. This reduced affinity allows the adjacent ATP-bound subunit to bind the dsDNA, thus translocating the dsDNA forward (Fig. 3C). Sequential firing of subunits around the hexameric ring has been further reported to be regulated by the arginine finger, leading to a highly processive movement of the viral dsDNA into the phage capsid (121).
The main motor protein in AdV, IVa2, is multifunctional. It assists in the assembly of the capsid and activates late transcription. Comparing the IVa2 protein sequences with those of ATPases from different species revealed conserved Walker A and B motifs associated with the binding and hydrolysis of ATP (85). Like the ATPases in bacteriophage-packaging motors, the multimeric ATPase IVa2 motor protein complex also works through sequential actions to provide energy for packaging DNA into their capsids. IVa2 also interacts with a viral L4-22K protein, which is involved in genome encapsidation (86–88). The stoichiometries of IVa2 have been found to be equal for different forms of assembly intermediates, such as the empty capsids, light intermediate particles, and nonmature and mature virions (89). IVa2 mutants were defective in DNA packaging and resulted in an accumulation of empty capsids similar to the procapsid of dsDNA bacteriophages (47).
Herpesvirus terminase contains a large subunit, pUL15, which cleaves concatemeric viral DNA during packaging initiation and completion. The structure of the C-terminal domain of pUL15 resembles those of bacteriophage terminases, RNase H, integrases, DNA polymerases, and topoisomerases, with an active site clustered with acidic residues. The DNA-binding surface is surrounded by flexible loops, indicating that they adopt conformational changes upon DNA binding (51). These conformational changes are similar to the sequential actions of ATPase gp16 observed in phi29 DNA-packaging motors, which provides energy to support the one-way traffic of genomes into procapsids.
Recent microscopic studies provided insight into genome packaging in mimivirus (55, 58). DNA is packaged into preformed procapsids through a nonvertex portal site which is distal to the DNA delivery site. As described earlier, the viral packaging pathway in mimivirus is similar to chromosome segregation in bacteria and requires interaction of the packaging motor with recombinases and topoisomerases (55). It has been proposed that the DNA entry portal is closed once genome packaging is complete, as with the ATPase SpoIIIE, which promotes membrane fusion after bacterial DNA segregation (90). Interestingly, NCLDV members, such as Marseillevirus and Lausannevirus, encode histone-like proteins (91, 92). Genome condensation becomes essential with large genome size (91), and these condensing proteins (histone-like proteins) may be required in mimivirus to package its large genome (Fig. 3D). These proteins are present in most prokaryotes and eukaryotes for genome condensation (93–95).
PERSPECTIVES
In the virology community, significant efforts have been made in the quest for antiviral drugs, focusing on identification of targets for drugs with high specificity. However, little attention has been paid to finding new methods for antiviral drug development. Above, we reviewed a new methodology for antiviral drug development and demonstrated that the inhibition efficiency of a given antiviral drug is also dependent on the stoichiometry of the viral machine with sequential mechanisms. We pointed out that viral DNA-packaging motors are ideal targets for high-efficiency antiviral drugs, since these motors are machines with high stoichiometry and their subunits work sequentially. Here the “stoichiometry” differs from a conventional concept used in drug development. Conventionally, stoichiometry refers to the number of drug molecules bound to each virion or target related to viral replication. In this case, stoichiometry refers to the number of identical subunits in the viral machine that can serve as a target for drug development.
Traditionally, it has been almost impossible to prove this concept by comparing efficacies of two drugs acting on two targets with different stoichiometries. It is very challenging to demonstrate whether the difference in drug efficiency is contributed by the essentiality of the two different targets in viral function, the affinity of drug molecules in target binding, or the stoichiometry of the targeted machine. The phi29 DNA-packaging system has offered an excellent model to prove this concept. It has the sensitivity of 109 PFU for in vitro assays using purified components (96), which made it possible to use inactive mutant components to represent the drugged substrate. Therefore, the ratio of drugged and undrugged subunits can be explicitly defined by a binomial distribution calculation.
The concept described here for highly efficient viral inhibition may have an impact in antiviral therapy by introducing dominant negative proteins (97) or inactive mutant proteins into the cell, either by intracellular expression using viral vectors for gene delivery or direct introduction of proteins into the cell (14, 41, 98, 99). This involves the incorporation of mutant protein subunits into a multimeric complex identified as a drug target. If a multimeric complex is identified with high stoichiometry and a K equal to 1 due to the sequential actions of the homomeric subunits, then incorporating one mutant subunit into the complex would inactivate the complex completely. Since the viral machine is composed of Z subunits, one drugged subunit per complex would work only when the intracellular drug concentration is high. But if the strategy is to apply a dominant negative protein, such as the dominant negative phospholamban in cardiac gene therapy (97), a more augmented effect of the mutant protein subunits will be expected as the value of Z increases.
Besides the viral DNA-packaging motors, viral machines with high stoichiometry that are operated by cooperative sequential actions with a Z greater than 1 and a K equal to 1 are ubiquitous. These viral machines are involved in many aspects of the viral life cycle. These machines include, but are not limited to, viral DNA polymerase, viral RNA polymerase, reverse transcriptase, chaperons, viral genome repair enzymes, viral integrase, membrane pores for viral DNA or RNA trafficking, viral motors for dsRNA packaging, as well as other machines involved in viral entry, motion, and trafficking. The sequential- and cooperative-action mechanism used in these viral machines is similar to the series circuit of Christmas lights, where a single bad bulb will turn off the whole chain. Drugs targeting these viral machines will be highly efficient. Design of potent viral drugs with high specificity is also possible. For example, if a specific high-stoichiometry machine is identified in a virus, specific drugs targeting subunits of this machine will be highly efficient.
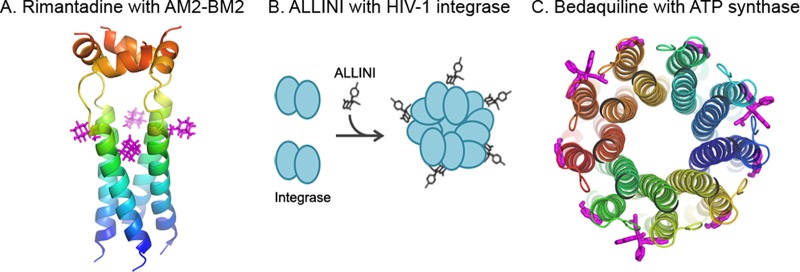
Since viral motors share certain common structures and operation mechanisms, methods of targeting homomeric multisubunit complexes should have general applications in antiviral drug discovery. Homomeric channel proteins, such as the homotetramer M2 proton channel protein, have been shown to be a better target for anti-influenza drugs (100). Amantadine and rimantadine inhibit the influenza virus through this mechanism by entering the barrel of the tetrameric M2 channel and blocking proton translocation function (Fig. 4A) (52). RNA-dependent RNA polymerase NS5B, which plays an important role in hepatitis C virus (HCV) replication, exists as a homomeric oligomer. NS5B-NS5B intermolecular interaction is essential for both initiation and elongation of RNA synthesis (101). Targeting the oligomeric protein NS5B may also be helpful to delineate new and powerful antiviral strategies.
FIG 4.
Drugs that target viral motors with high stoichiometry. (A) Homotetrameric AM2-BM2 channel protein of influenza virus complexed with the inhibitory drug molecule rimantadine (PDB accession number 2RLF) (117); (B) illustration of allosteric HIV-1 integrase inhibitors that act by promoting high-order oligomerization of integrase to block its activity; (C) bedaquiline that targets ATP synthase (PDB accession number 4V1H).
The method for discovering potent dsDNA viral therapeutics can be extended to other viruses. For example, allosteric human immunodeficiency virus type 1 (HIV-1) integrase inhibitors (ALLINIs) that potently impair HIV-1 replication in cell culture are undergoing clinical trials (102, 103). While HIV-1 integrase functions as a tetramer to catalyze covalent insertion of the viral cDNA into human chromosomes, ALLINIs bind at the IN dimer interface and promote cooperative, higher-order oligomerization, resulting in inactive protein (Fig. 4B). Such an innovative mode of action is energetically much more favorable than the competitive mechanism of action of preventing interaction between subunits and may potentially pave a way for developing novel compounds for other viral targets utilizing multiprotein subunits.
The method described here has general applications in other biological systems. In fact, the first drug approved to treat multidrug-resistant tuberculosis, bedaquiline (104), acts on the ATP synthase, which is a multisubunit biomotor (Fig. 4C) (105–116). Although this drug's inventors were not aware of the concept of targeting multisubunit complexes, its success supports the notion of using the multisubunit complex as a potent drug target. Cancer or multisubunit bacterial mutant ATPase can also be used as a target. Drug developers can simply check the published literature and find the multisubunit machine as a drug target. For cancer treatment, the key is to find multisubunit machines with mutations.
ACKNOWLEDGMENTS
Peixuan Guo's Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the C. M. Chen Foundation. P.G. is a consultant of Oxford Nanopore, Nanobio Delivery Pharmaceutical Co., Ltd., and NanoBio RNA Technology Co. Ltd. His inventions at the University of Kentucky have been licensed to Matt Holding and Nanobio Delivery Pharmaceutical Co., Ltd.
Biographies

Fengmei Pi is currently a Ph.D. candidate graduate student at the College of Pharmacy, University of Kentucky. She obtained her B.S. and M.S. from China Pharmaceutical University (2007). She worked as a research scientist at Daewoong Pharmaceuticals Co., South Korea, and as a senior formulation scientist for China GSK Consumer Healthcare before joining the University of Kentucky (2012). She has broad training and working experience in pharmaceutical science. Her recent research focused on RNA nanotechnology for targeted drug delivery and cancer therapy.

Zhengyi Zhao is currently a Ph.D. candidate in the College of Pharmacy, University of Kentucky. She graduated from Shenyang Pharmaceutical University in 2011 in pharmaceutical sciences and then joined Prof. Peixuan Guo's lab in the spring of 2012. She has broad training in molecular biology, nanobiotechnology, biophysics, and pharmaceutics. Her current research focuses on the study of the function, mechanism of action, and application of the bacteriophage phi29 dsDNA-packaging nanomotor.

Venkata Chelikani is a microbiologist with extensive experience in biosensors and nanotechnology. He completed his M.Sc. in microbiology from Andhra University, India, in 2005 and lectured undergraduate microbiology courses for 2 years at Aditya College, India. He then went on to complete an M.Res. in biomedical sciences from the University of Glasgow, United Kingdom. He completed his Ph.D. in biochemistry (developing estrogen-detecting biosensors) from Lincoln University, New Zealand. He worked as a tutor in microbiology both at the University of Canterbury and at Lincoln University for 2 years before carrying out his postdoctoral work on large DNA viruses and their packaging motors at the Indian Institute of Technology (IIT), Mumbai, India. He is currently working as a lab manager in microbiology at Lincoln University, New Zealand. He is interested in working on interdisciplinary projects involving microbiology and nanotechnology.
Kristine Yoder received her B.S. in biology from the Massachusetts Institute of Technology (MIT) in 1993 and her Ph.D. in biology from the University of California, San Diego (UCSD), in 2000. Her graduate work was in the lab of Frederic Bushman at the Salk Institute. Her postdoctoral work was in the Kimmel Cancer Center at Thomas Jefferson University with Richard Fishel and Carlo Croce. She is currently an assistant professor in the Department of Molecular Virology, Immunology, and Medical Genetics at The Ohio State University Wexner Medical Center. Her research uses single-molecule analysis to understand the dynamics of retroviral integration.

Mamuka Kvaratskhelia obtained his M.S. and Ph.D. degrees in the former USSR, received postdoctoral training with Roger Thorneley at the Department of Biological Chemistry, John Innes Center, England (1995 to 1998), and with Malcolm White at the Department of Biochemistry, University of Dundee, Scotland (1998 to 2000). Dr. Kvaratskhelia then worked as a research fellow in Stuart Le Grice's laboratory (2000 to 2003) of the HIV Drug Resistance Program, National Cancer Institute, Frederick, MD, USA, before joining The Ohio State University College of Pharmacy and Center for Retrovirus Research in 2003 as a faculty member. His current research focuses on elucidating the molecular mechanisms for retroviral integration into the host genome, dissecting virus-host interactions, discovering HIV-1 integrase inhibitors with allosteric mechanisms of action, and developing retroviral vectors for human gene therapy. His laboratory employs innovative and complementary biochemical, biophysical, structural biology, molecular biology, and virology approaches.

Peixuan Guo is currently the Sylvan G. Frank Endowed Chair in Pharmaceutics and Drug Delivery at The Ohio State University College of Pharmacy. He was an endowed chair of biomedical engineering at the University of Cincinnati, an endowed chair in nanobiotech, and director of the Nanobiotech Center at the University of Kentucky before moving to OSU in 2016. He received his Ph.D. in microbiology in 1987 from the University of Minnesota and was an NIH postdoctoral fellow in 1990, a Purdue assistant professor in 1990, tenured in 1993, a full professor in 1997, and honored as the Purdue Faculty Scholar in 1998. He was director of the NIH Nanomedicine Development Center from 2006 to 2011. He constructed the first viral DNA-packaging motor, discovered the phi29 motor pRNA, pioneered RNA nanotechnology, incorporated a motor channel into a lipid membrane, and discovered a third class of revolving biomotors without rotation. He received the Pfizer Distinguished Faculty Award in 1995 and a Lions Club Cancer Research Award in 2006 and was recognized as a distinguished alumnus of the University of Minnesota in 2009 and the 100 Years Distinguished Chinese Alumnus of the University of Minnesota in 2014. He is an editorial board member for five nanotech journals, has reported hundreds of times on television (such as ABC, NBC, and BBC), and has been featured many times on the websites of the NIH, NSF, MSNBC, and NCI. He is a member of two prominent national nanotech initiatives sponsored by NIST/NIH/NSF and the National Science and Technology Council. He has been a member of the Foreign Examination Panel of the Chinese Academy of Sciences since 2014.
REFERENCES
- 1.Jurado KA, Engelman A. 2013. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev Mol Med 15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Jie Y, You X, Shi H, Zhang M, Wu Y, Lin G, Li X, Gao Z, Chong Y. 2015. Optimized combination therapies with adefovir dipivoxil (ADV) and lamivudine, telbivudine, or entecavir may be effective for chronic hepatitis B patients with a suboptimal response to ADV monotherapy. Int J Clin Exp Med 8:21062–21070. [PMC free article] [PubMed] [Google Scholar]
- 3.Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM. 2014. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene 33:2928–2937. doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. 2008. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci U S A 105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider A, Corona A, Sporing I, Jordan M, Buchholz B, Maccioni E, Di SR, Bodem J, Tramontano E, Wohrl BM. 2016. Biochemical characterization of a multi-drug resistant HIV-1 subtype AG reverse transcriptase: antagonism of AZT discrimination and excision pathways and sensitivity to RNase H inhibitors. Nucleic Acids Res 44:2310–2322. doi: 10.1093/nar/gkw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Littler E, Oberg B. 2005. Achievements and challenges in antiviral drug discovery. Antivir Chem Chemother 16:155–168. doi: 10.1177/095632020501600302. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Christensen TE, Kamau YN, Tang L. 2013. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc Natl Acad Sci U S A 110:8075–8080. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostapchuk P, Hearing P. 2005. Control of adenovirus packaging. J Cell Biochem 96:25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- 9.Chan KW, Yang CH, Lin JW, Wang HC, Lin FY, Kuo ST, Wong ML, Hsu WL. 2009. Phylogenetic analysis of parapoxviruses and the C-terminal heterogeneity of viral ATPase proteins. Gene 432:44–53. doi: 10.1016/j.gene.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Hwang JS, Bogner E. 2002. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J Biol Chem 277:6943–6948. doi: 10.1074/jbc.M108984200. [DOI] [PubMed] [Google Scholar]
- 11.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJL. 2007. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci 64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King J, Casjens S. 1974. Catalytic head assembly protein in virus morphogenesis. Nature 251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz C, Guo P. 2013. Ultrastable pRNA hexameric ring gearing hexameric phi29 DNA-packaging motor by revolving without rotating and coiling. Curr Opin Biotechnol 24:581–590. doi: 10.1016/j.copbio.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu D, Pi F, Wang C, Zhang P, Guo P. 2015. New approach to develop ultra-high inhibitory drug using the power-function of the stoichiometry of the targeted nanomachine or biocomplex. Nanomedicine (Lond) 10:1881–1897. doi: 10.2217/nnm.15.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo P, Noji H, Yengo CM, Zhao Z, Grainge I. 2016. Biological nanomotors with revolving, linear, or rotation motion mechanism. Microbiol Mol Biol Rev 80:161–186. doi: 10.1128/MMBR.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. 2013. Revolving rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De-Donatis G, Zhao Z, Wang S, Huang PL, Schwartz C, Tsodikov VO, Zhang H, Haque F, Guo P. 2014. Finding of widespread viral and bacterial revolving dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci 4:30. doi: 10.1186/2045-3701-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pi F, Vieweger M, Zhao Z, Wang S, Guo P. 2016. Discovery of a new method for potent drug development using power function of stoichiometry of homomeric biocomplexes or biological nanomotors. Expert Opin Drug Deliv 13:23–36. doi: 10.1517/17425247.2015.1082544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yogisharadhya R, Bhanuprakash V, Venkatesan G, Balamurugan V, Pandey AB, Shivachandra SB. 2012. Comparative sequence analysis of poxvirus A32 gene encoded ATPase protein and carboxyl terminal heterogeneity of Indian orf viruses. Vet Microbiol 156:72–80. doi: 10.1016/j.vetmic.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJ. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc Natl Acad Sci U S A 106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo P, Grainge I, Zhao Z, Vieweger M. 2014. Two classes of nucleic acid translocation motors: rotation and revolution without rotation. Cell Biosci 4:54. doi: 10.1186/2045-3701-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black LW. 2015. Old, new, and widely true: the bacteriophage T4 DNA packaging mechanism. Virology 479-480:650–656. doi: 10.1016/j.virol.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serwer P, Wright ET, Hakala K, Weintraub ST, Su M, Jiang W. 2010. DNA packaging-associated hyper-capsid expansion of bacteriophage T3. J Mol Biol 397:361–374. doi: 10.1016/j.jmb.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett RD. 1981. DNA replication of bacteriophage T5. 3. Studies on the structure of concatemeric T5 DNA. J Gen Virol 52:25–38. [DOI] [PubMed] [Google Scholar]
- 25.Chang CY, Kemp P, Molineux IJ. 2010. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology 398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomka MA, Catalano CE. 1993. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage lambda. J Biol Chem 268:3056–3065. [PubMed] [Google Scholar]
- 27.Oliveira L, Cuervo A, Tavares P. 2010. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J Biol Chem 285:7366–7373. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo P, Zhao Z, Haak J, Wang S, Wu D, Meng B, Weitao T. 2014. Common mechanisms of DNA translocation motors in bacteria and viruses using one-way revolution mechanism without rotation. Biotechnol Adv 32:853–872. doi: 10.1016/j.biotechadv.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Makarova KS, Koonin EV, Aravind L. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babbar BK, Gold M. 1998. ATP-reactive sites in the bacteriophage lambda packaging protein terminase lie in the N-termini of its subunits, gpA and gpNu1. Virology 247:251–264. doi: 10.1006/viro.1998.9221. [DOI] [PubMed] [Google Scholar]
- 32.Guo P, Zhang C, Chen C, Trottier M, Garver K. 1998. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2:149–155. doi: 10.1016/S1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Lemieux S, Wu X, St-Arnaud S, McMurray CT, Major F, Anderson D. 1998. Function of hexameric RNA in packaging of bacteriophage phi29 DNA in vitro. Mol Cell 2:141–147. doi: 10.1016/S1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 34.Hendrix RW. 1998. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell 94:147–150. doi: 10.1016/S0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz C, De Donatis GM, Fang H, Guo P. 2013. The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA+ superfamily. Virology 443:20–27. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. 2000. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol 298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 37.Shu D, Zhang H, Jin J, Guo P. 2007. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J 26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moll D, Guo P. 2007. Grouping of ferritin and gold nanoparticles conjugated to pRNA of the phage phi29 DNA-packaging motor. J Nanosci Nanotech 7:3257–3267. doi: 10.1166/jnn.2007.914. [DOI] [PubMed] [Google Scholar]
- 39.Xiao F, Zhang H, Guo P. 2008. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res 36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. 2013. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Trottier M, Guo P. 1997. New approaches to stoichiometry determination and mechanism investigation on RNA involved in intermediate reactions. Nucleic Acids Symp Ser 36:190–193. [Google Scholar]
- 42.Trottier M, Guo P. 1997. Approaches to determine stoichiometry of viral assembly components. J Virol 71:487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA plus ATPases. J Struct Biol 146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. 2011. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 45.Willows RD, Hansson A, Birch D, Al-Karadaghi S, Hansson M. 2004. EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: an AAA(+) hexamer. J Struct Biol 146:227–233. doi: 10.1016/j.jsb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell 23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Christensen JB, Byrd SA, Walker AK, Strahler JR, Andrews PC, Imperiale MJ. 2008. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J Virol 82:9086–9093. doi: 10.1128/JVI.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostapchuk P, Yang J, Auffarth E, Hearing P. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J Virol 79:2831–2838. doi: 10.1128/JVI.79.5.2831-2838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Homa FL, Brown JC. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol 7:107–122. doi:. [DOI] [PubMed] [Google Scholar]
- 50.Koslowski KM, Shaver PR, Casey JT, Wilson T, Yamanaka G, Sheaffer AK, Tenney DJ, Pederson NE. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J Virol 73:1704–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvarajan Sigamani S, Zhao H, Kamau YN, Baines JD, Tang L. 2013. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J Virol 87:7140–7148. doi: 10.1128/JVI.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 53.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A 108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie J-M, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 55.Chelikani V, Ranjan T, Zade A, Shukla A, Kondabagil K. 2014. Genome segregation and packaging machinery in Acanthamoeba polyphaga mimivirus is reminiscent of bacterial apparatus. J Virol 88:6069–6075. doi: 10.1128/JVI.03199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghedin E, Fraser CM. 2005. A virus with big ambitions. Trends Microbiol 13:56–57. doi: 10.1016/j.tim.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Raoult D, La Scola B, Birtles R. 2007. The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clin Infect Dis 45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- 58.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. 2008. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol 6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. 2008. Marine mimivirus relatives are probably large algal viruses. Virol J 5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moss B. 1985. Replication of poxviruses, p 685–703. In Fields BN, (ed), Virology. Raven Press, New York, NY. [Google Scholar]
- 61.Schwartz C, Fang H, Huang L, Guo P. 2012. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res 40:2577–2586. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Guo P. 1997. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J Virol 71:3864–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guimet D, Hearing P. 2013. The adenovirus L4-22K protein has distinct functions in the posttranscriptional regulation of gene expression and encapsidation of the viral genome. J Virol 87:7688–7699. doi: 10.1128/JVI.00859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu K, Guimet D, Hearing P. 2013. The adenovirus L4-33K protein regulates both late gene expression patterns and viral DNA packaging. J Virol 87:6739–6747. doi: 10.1128/JVI.00652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fessler SP, Young CS. 1999. The role of the L4 33K gene in adenovirus infection. Virology 263:507–516. doi: 10.1006/viro.1999.9951. [DOI] [PubMed] [Google Scholar]
- 66.Finnen RL, Biddle JF, Flint J. 2001. Truncation of the human adenovirus type 5 L4 33-kDa protein: evidence for an essential role of the carboxy-terminus in the viral infectious cycle. Virology 289:388–399. doi: 10.1006/viro.2001.1130. [DOI] [PubMed] [Google Scholar]
- 67.Gambke C, Deppert W. 1981. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. II. Immunological and protein chemical analysis. J Virol 40:594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulshreshtha V, Babiuk LA, Tikoo SK. 2004. Role of bovine adenovirus-3 33K protein in viral replication. Virology 323:59–69. doi: 10.1016/j.virol.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 69.Koonin EV, Senkevich TG, Chernos VI. 1993. Gene A32 product of vaccinia virus may be an ATPase involved in viral-DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes 7:89–94. doi: 10.1007/BF01702351. [DOI] [PubMed] [Google Scholar]
- 70.White CA, Stow ND, Patel AH, Hughes M, Preston VG. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J Virol 77:6351–6358. doi: 10.1128/JVI.77.11.6351-6358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang K, Baines JD. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J Virol 80:5733–5739. doi: 10.1128/JVI.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheffczik H, Savva CG, Holzenburg A, Kolesnikova L, Bogner E. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res 30:1695–1703. doi: 10.1093/nar/30.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burroughs AM, Iyer LM, Aravind L. 2007. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn 3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 74.Chelikani V, Ranjan T, Kondabagil K. 2014. Revisiting the genome packaging in viruses with lessons from the “Giants.” Virology 466-467:15–26. doi: 10.1016/j.virol.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 75.Stitt BL, Xu Y. 1998. Sequential hydrolysis of ATP molecules bound in interacting catalytic sites of Escherichia coli transcription termination protein Rho. J Biol Chem 273:26477–26486. doi: 10.1074/jbc.273.41.26477. [DOI] [PubMed] [Google Scholar]
- 76.Kammerer RA, Schulthess T, Landwehr R, Lustig A, Fischer D, Engel J. 1998. Tenascin-C hexabrachion assembly is a sequential two-step process initiated by coiled-coil alpha-helices. J Biol Chem 273:10602–10608. doi: 10.1074/jbc.273.17.10602. [DOI] [PubMed] [Google Scholar]
- 77.Lisal J, Tuma R. 2005. Cooperative mechanism of RNA packaging motor. J Biol Chem 280:23157–23164. doi: 10.1074/jbc.M502658200. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Z, Lewis D, Strock C, Inesi G, Nakasako M, Nomura H, Toyoshima C. 2000. Detailed characterization of the cooperative mechanism of Ca(2+) binding and catalytic activation in the Ca(2+) transport (SERCA) ATPase. Biochemistry 39:8758–8767. doi: 10.1021/bi000185m. [DOI] [PubMed] [Google Scholar]
- 79.Sun H, Squier TC. 2000. Ordered and cooperative binding of opposing globular domains of calmodulin to the plasma membrane Ca-ATPase. J Biol Chem 275:1731–1738. doi: 10.1074/jbc.275.3.1731. [DOI] [PubMed] [Google Scholar]
- 80.Hiller R, Carmeli C. 1990. Kinetic analysis of cooperative interactions induced by Mn2+ binding to the chloroplast H(+)-ATPase. Biochemistry 29:6186–6192. doi: 10.1021/bi00478a011. [DOI] [PubMed] [Google Scholar]
- 81.Persechini A, Hartshorne DJ. 1982. Cooperative behavior of smooth muscle myosin. Fed Proc 41:2868–2872. [PubMed] [Google Scholar]
- 82.Lee CS, Guo P. 1995. Sequential interactions of structural proteins in phage phi29 procapsid assembly. J Virol 69:5024–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao Z, Khisamutdinov E, Schwartz C, Guo P. 2013. Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano 7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fang H, Jing P, Haque F, Guo P. 2012. Role of channel lysines and “push through a one-way valve” mechanism of viral DNA packaging motor. Biophys J 102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ostapchuk P, Hearing P. 2008. Adenovirus IVa2 protein binds ATP. J Virol 82:10290–10294. doi: 10.1128/JVI.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ostapchuk P, Anderson ME, Chandrasekhar S, Hearing P. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J Virol 80:6973–6981. doi: 10.1128/JVI.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ewing SG, Byrd SA, Christensen JB, Tyler RE, Imperiale MJ. 2007. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J Virol 81:12450–12457. doi: 10.1128/JVI.01470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tyler RE, Ewing SG, Imperiale MJ. 2007. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J Virol 81:3447–3454. doi: 10.1128/JVI.02097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winter N, D'Halluin JC. 1991. Regulation of the biosynthesis of subgroup C adenovirus protein IVa2. J Virol 65:5250–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu NJ, Dutton RJ, Pogliano K. 2006. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol 59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La SB, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G. 2011. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol 13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 93.Teif VB, Bohinc K. 2011. Condensed DNA: condensing the concepts. Prog Biophys Mol Biol 105:208–222. doi: 10.1016/j.pbiomolbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 94.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. 2010. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci U S A 107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Holde KE. 1989. Chromatin, p 69–180. Springer Verlag, New York, NY. [Google Scholar]
- 96.Lee CS, Guo P. 1994. A highly sensitive system for the assay of in vitro viral assembly of bacteriophage phi29 of Bacillus subtilis. Virology 202:1039–1042. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- 97.Lee JY, Finkelstein IJ, Arciszewska LK, Sherratt DJ, Greene EC. 2014. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Mol Cell 54:832–843. doi: 10.1016/j.molcel.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trottier M, Zhang CL, Guo P. 1996. Complete inhibition of virion assembly in vivo with mutant pRNA essential for phage phi29 DNA packaging. J Virol 70:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fang H, Zhang P, Huang LP, Zhao Z, Pi F, Montemagno C, Guo P. 2014. Binomial distribution for quantification of protein subunits in biological nanoassemblies and functional nanomachines. Nanomedicine 10:1433–1440. doi: 10.1016/j.nano.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang C, Takeuchi K, Pinto LH, Lamb RA. 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol 67:5585–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lopez-Jimenez AJ, Clemente-Casares P, Sabariegos R, Llanos-Valero M, Bellon-Echeverria I, Encinar JA, Kaushik-Basu N, Froeyen M, Mas A. 2014. Hepatitis C virus polymerase-polymerase contact interface: significance for virus replication and antiviral design. Antiviral Res 108:14–24. doi: 10.1016/j.antiviral.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 102.Garmann RF, Gopal A, Athavale SS, Knobler CM, Gelbart WM, Harvey SC. 2015. Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA 21:877–886. doi: 10.1261/rna.047506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 104.Lakshmanan M, Xavier AS. 2013. Bedaquiline—the first ATP synthase inhibitor against multi drug resistant tuberculosis. J Young Pharm 5:112–115. doi: 10.1016/j.jyp.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Watanabe R, Matsukage Y, Yukawa A, Tabata KV, Noji H. 2014. Robustness of the rotary catalysis mechanism of F-1-ATPase. J Biol Chem 289:19331–19340. doi: 10.1074/jbc.M114.569905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yasuda R, Noji H, Kinosita K Jr, Yoshida M. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93:1117–1124. doi: 10.1016/S0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 107.Kinosita K Jr, Yasuda R, Noji H, Ishiwata S, Yoshida M. 1998. F1-ATPase: a rotary motor made of a single molecule. Cell 93:21–24. doi: 10.1016/S0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 108.Stock D, Leslie AG, Walker JE. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 109.Boyer PD. 1999. What makes ATP synthase spin? Nature 402:247–249. doi: 10.1038/46193. [DOI] [PubMed] [Google Scholar]
- 110.Adachi K, Yasuda R, Noji H, Itoh H, Harada Y, Yoshida M, Kinosita K Jr. 2000. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc Natl Acad Sci U S A 97:7243–7247. doi: 10.1073/pnas.120174297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hara KY, Noji H, Bald D, Yasuda R, Kinosita K Jr, Yoshida M. 2000. The role of the DELSEED motif of the beta subunit in rotation of F1-ATPase. J Biol Chem 275:14260–14263. doi: 10.1074/jbc.275.19.14260. [DOI] [PubMed] [Google Scholar]
- 112.Masaike T, Mitome N, Noji H, Muneyuki E, Yasuda R, Kinosita K, Yoshida M. 2000. Rotation of F(1)-ATPase and the hinge residues of the beta subunit. J Exp Biol 203(Part 1):1–8. [DOI] [PubMed] [Google Scholar]
- 113.Wada Y, Sambongi Y, Futai M. 2000. Biological nano motor, ATP synthase F(o)F(1): from catalysis to gammaepsilonc(10-12) subunit assembly rotation. Biochim Biophys Acta 1459:499–505. doi: 10.1016/S0005-2728(00)00189-4. [DOI] [PubMed] [Google Scholar]
- 114.Okazaki K, Hummer G. 2013. Phosphate release coupled to rotary motion of F-1-ATPase. Proc Natl Acad Sci U S A 110:16468–16473. doi: 10.1073/pnas.1305497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ito Y, Yoshidome T, Matubayasi N, Kinoshita M, Ikeguchi M. 2013. Molecular dynamics simulations of yeast F-1-ATPase before and after 16 degrees rotation of the gamma subunit. J Phys Chem B 117:3298–3307. doi: 10.1021/jp312499u. [DOI] [PubMed] [Google Scholar]
- 116.Arai HC, Yukawa A, Iwatate RJ, Kamiya M, Watanabe R, Urano Y, Noji H. 2014. Torque generation mechanism of F-1-ATPase upon NTP binding. Biophys J 107:156–164. doi: 10.1016/j.bpj.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schnell JR, Chou JJ. 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reference deleted. [Google Scholar]
- 119.Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol 72:5769–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grubisha O, Traktman P. 2003. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J Virol 77:10929–10942. doi: 10.1128/JVI.77.20.10929-10942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao Z, De-Donatis GM, Schwartz C, Fang H, Li J, Guo P. Arginine finger regulates sequential action of asymmetrical hexameric ATPase in double-stranded DNA translocation motor. Mol Cel Biol, in press. doi: 10.1128/MCB.00142-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Crozat E, Grainge I. 2010. FtsK DNA translocase: the fast motor that knows where it's going. Chembiochem 11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 123.Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. 2013. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A 110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]